Abstract
Tuberculosis (TB) remains a public health crisis and a leading cause of infection-related death globally. Although in high demand, imaging technologies that enable rapid, specific, and nongenetic labeling of live Mycobacterium tuberculosis (Mtb) remain underdeveloped. We report a dual-targeting strategy to develop a small molecular probe (CDG-DNB3) that can fluorescently label single bacilli within 1 hour. CDG-DNB3 fluoresces upon activation of the β-lactamase BlaC, a hydrolase naturally expressed in Mtb, and the fluorescent product is retained through covalent modification of the Mtb essential enzyme decaprenylphosphoryl-β-D-ribose 2’-epimerase (DprE1). This dual-targeting probe not only discriminates live from dead Bacillus Calmette–Guérin (BCG) but also shows specificity for Mtb over other bacterial species including 43 nontuberculosis mycobacteria (NTM). In addition, CDG-DNB3 can image BCG phagocytosis in real time, as well as Mtb in patients’ sputum. Together with a low-cost, self-driven microfluidic chip, we have achieved rapid labeling and automated quantification of live BCG. This labeling approach should find many potential applications for research toward TB pathogenesis, treatment efficacy assessment, and diagnosis.
Introduction
Tuberculosis (TB), an infectious disease caused by the slow-growing pathogen Mycobacterium tuberculosis (Mtb), kills an estimated 2 million people per year according to World Health Organization (1-3). Emergence of multidrug resistance with synergistic interaction with HIV/AIDS pandemic exacerbated this problem and reactivated a global concerted effort on TB research (4, 5). Microbial cell culture is considered as the gold standard for TB diagnosis, however it is time consuming – usually taking 1 to 2 months to complete because of the extremely slow growth rate of Mtb. Although nucleic acid amplification technology for detecting the DNA material from Mtb has advanced (6-8), imaging technologies that allow rapid and specific labeling of live Mtb, have seen little progress (9). Since the introduction of fluorochrome staining by Hagemann in 1937, auramine O has been widely adopted for Mtb fluorescent microscopic examination. Auramine O interacts with the mycolic acids within the cell wall of acid-fast microorganisms like mycobacteria, but is not specific to Mtb and cannot discriminate viable from dead cells. Auramine O sensitivity is also undesirable and varies because of tedious staining, decolorizing, and counterstaining procedures (10, 11). Replacing this century-old technology has proven not a trivial effort: Molecular probes for live Mtb labeling have been developed to target the capsular components such as esterases (12, 13), the D-Ala-D-Ala motif of peptidoglycan (14, 15), trehalose mycolyltransesterases (16-19), and sulfatases (20), yet none has been shown to be specific for labeling Mtb. Esterases, the D-Ala-D-Ala motif and sulfatases exist not only in mycobacteria but also in many other bacterial strains. Trehalose mycolyltransesterases are expressed in Actinobacteria phylum including mycobacteria; thus, trehalose-based probes do not have Mtb specificity either (19).
Recently, electron-deficient nitroaromatic compounds have been discovered as a new class of potent anti-TB agents (21-23) that target decaprenylphosphoryl-β-D-ribose 2’-epimerase (DprE1), a periplasmic enzyme highly conserved among actinobacteria and required for the synthesis of the cell wall arabinans. Specifically, DprE1 reduces one nitro group of these compounds to a nitroso derivative, and covalently modifies this nitroso to form a stable semi-mercaptal complex using the cysteine residue in the active site. This mechanism presents a possibility to design a fluorescent probe to image Mtb at the single-cell level through DprE1-mediated signal retention.
Mtb is intrinsically resistant to nearly all β-lactam antibiotics, largely because of the production of an ambler class A β-lactamase named BlaC that is highly conserved through clinical isolates (24, 25). By engineering the core structure of a β-lactam cephalosporin, we recently developed a fluorogenic probe that is specific to BlaC (26-28), but because of signal diffusion, it cannot be used for single-cell Mtb labeling. Here, we report a fluorogenic probe that targets both BlaC and DprE1 to achieve specific labeling of single live Mtb in less than 1 hour.
Results
Design of dual-targeting fluorogenic probes
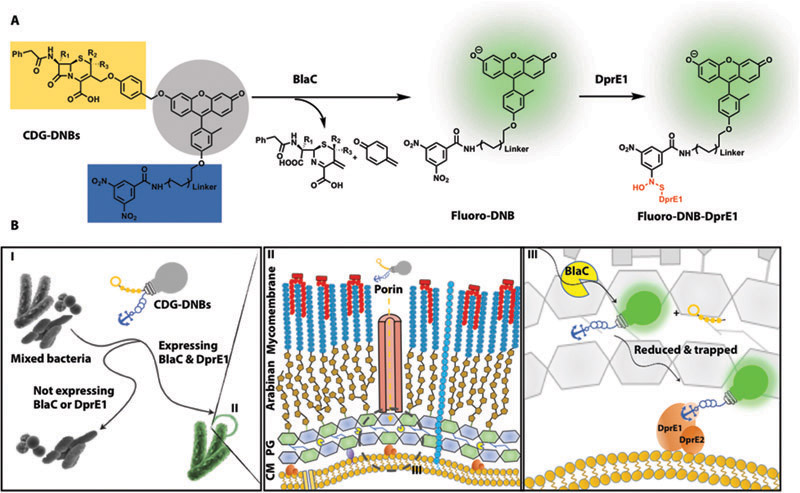
Our dual-targeting fluorogenic probes (CDG-DNBs) contain three functional units: a BlaC-sensing unit, a caged fluorescent reporter, and a DprE1-binding unit for signal trapping (Fig. 1A). We reasoned that CDG-DNBs would pass the Mtb cell wall (mycolate arabinogalactan layer) through porins, because the uptake of cephalosporin antibiotics in mycobacteria is reported to be mediated by porins (Fig. 1B) (29). BlaC and DprE1 enzymes located in the peptidoglycan layer and at the outer membrane would react with the probes: BlaC would hydrolyze the lactam ring to activate the fluorophore, and DprE1 would covalently bind the anchor unit for fluorescence immobilization. The combined actions of BlaC and DprE1 would enable fluorescent labeling of single Mtb. Bacteria without any BlaC and/or DprE1 activity would not fluorescently label due to the lack of fluorescence activation (no BlaC) or signal retention (no DprE1) in cells (Fig. 1B).
Fig. 1. Strategy for specific labeling of Mtb by targeting BlaC and DprE1.
(A) Structure of dual-targeting fluorogenic probes (CDG-DNBs) and their reaction with BlaC and DprE1: blue, DprE1 targeting group; gray, caged fluorophore; orange, BlaC sensing group; red, the covalent bond formed between DprE1 and uncaged fluorophore. (B) Mechanism of Mtb imaging by CDG-DNBs: (I) CDG-DNBs specifically label Mtb complex expressing BlaC and DprE1 but other bacterial species not expressing BlaC or DprE1. Gray bulb with a light string and an anchor, corresponding to CDG-DNBs structure in (A), indicates the lactam (orange) caged fluorophore (gray bulb) with a trapping unit (blue). (II) Cartoon illustration of Mtb cell wall and membrane layer with glycolipids and mycolic acids (Mycomembrane), lipoarabinomannan (LAM), arabinogalactan (Arabinan), peptidoglycan (PG), and cytoplasmic membrane (CM). CDG-DNBs traverse the mycolate arabinogalactan layer through porins and are activated by BlaC and DprE1. (III) Proposed dual processing of CDG-DNBs by BlaC and DprE1 in periplasm (dashed circle in II). Upon BlaC (yellow pacman) activation, the lactam is hydrolyzed to uncage the fluorophore, which restores fluorescing (the string is pulled away to switch the light bulb green). The uncaged flurophore is retained in Mtb through DprE1 modification (the green bulb is anchored on DprE1).
Targeting DprE1 to label DprE1-expressing bacteria
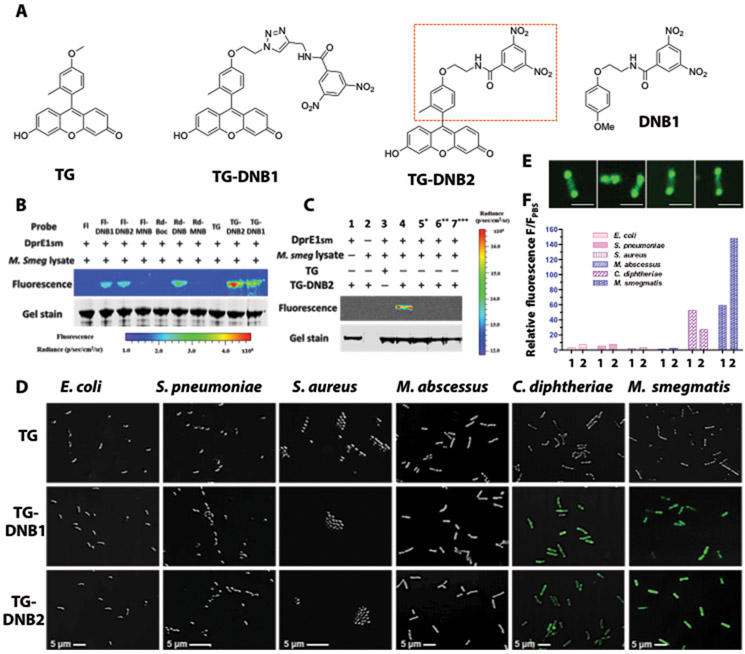
DprE1 has been extensively studied as a target for anti-TB reagents but less explored for bacterial imaging (30). We started by preparing different electron-deficient nitroaromatic compounds conjugated with fluorophores (see synthesis in Supplementary Materials and Methods, figs. S1 to S7 and appendix S1) and assessed their labeling efficiency. Fluorophores including rhodol, fluorescein and Tokyo Green (TG) were tethered with nitrobenzene analogs that have one or two nitro groups (Fig. 2A, fig. S1, S2). The binding affinity upon covalent modification by these analogues was first studied in vitro with purified recombinant Mycobacterium smegmatis DprE1 (DprE1SM) (fig. S8). As shown in Fig. 2B, fluorophore-dinitrobenzene (DNB) analogs labeled DprE1SM with different efficiency: Mono-nitro probes (Rd-MNB and FI-MNB) showed negligible fluorescence compared to di-nitro analogs, and TG-DNB2 exhibited the highest DprE1SM labeling efficiency. This result is consistent with the literature indicating that an electron-withdrawing substituent in the meta position is required for DprE1SM binding (23). TG-DNB2 is favored because it closely resembles the optimal DprE1-binding candidate DNB1 (Fig. 2A) (22). The covalent labeling of DprE1 SM by TG-DNB2 was further validated with varying conditions. The fluorescent complex TG-DNB2-DprE1SM was observed only when DprE1SM, TG-DNB2 and the lysate were present simultaneously (Fig. 2C, lane 4). Conditions missing the lysate [thus the substrate decaprenylphosphoryl-β-D-ribofuranose (DPR) and the cofactor flavin adenine dinucleotide (FAD); lane 1], DprE1SM (lane 2), or the DNB function group (lane 3) failed to produce the fluorescent complex. As expected, functional DprE1SM protein was required: There was no labeling when it was denatured by heat (lane 5) or 7 M urea/ dithiothreitol (DTT) reduction (lane 6). Preincubation with DNB1 also blocked the labeling of DprE1SM by TG-DNB2 (lane 7). Collectively, these data support that labeling of DprE1SM by TG-DNB2 proceeded through the reduction-covalent modification mechanism.
Fig. 2. TG-DNBs as the optimal candidates for labeling DprE1-expressing bacteria.
(A) Structures of TG, DprE1 inhibitor DNB1, and designed fluorophore-DNB analogs (TG-DNB1 and TG-DNB2). Red rectangle in TG-DNB2 indicates the structural similarity to DNB1. (B) Fluorescent labeling of DprE1SM by fluorescein-DNB (FI-DNB), rhodol-DNB (Rd-DNB) and TG-DNB analogs (10 μM). The whole lysate of freshly cultured M. smegmatis provided the natural substrate DPR and the cofactor FAD that are essential for the generation of DNB-DprE1 covalent product. (C) Fluorescence labeling of DprE1SM by TG-DNB2 (10 μM). 5*: DprE1SM was preheated at 90°C for 1 hour to denature the protein. 6**: Sample buffer was adjusted to 7 M urea and 20 mM DTT after incubation. These samples were further incubated for 1 hour at 37°C. 7***: DprE1SM was incubated with 50 μM DNB1 before addition of TG-DNB2. (D) Overlaid confocal images (bright-field and green fluorescence; 63X/oil excitation, Ex-490 nm; emission, 520 nm) of freshly cultured E. coli, S. pneumoniae, S. aureus, M. abscessus, C. diphtheriae, and M. smegmatis stained with 10 μM of TG, TG-DNB1 or TG-DNB2 at room temperature for 1 hour. (E) Confocal images of individual M. smegmatis bacilli treated with TG-DNB2 showing polarized localization of green fluorescence. Scale bars, 2 μm. (F) Histogram of fluorescence-activated flow cytometry data from bacteria in (D). 1: TG-DNB1 treated group; 2: TG-DNB2 treated group. The relative fluorescence (F/FPBS) was calculated by normalizing the mean fluorescence intensity (MFI) against the autofluorescence intensity of PBS-treated bacteria (table S2).
We further tested the fluorescence labeling and retention of TG-DNBs in different bacteria strains including M. smegmatis, Mycobacterium abscessus, Escherichia coli, Streptococcus pneumoniae, Staphylococcus aureus, and Corynebacteria diphtheriae. M. smegmatis is one of the most commonly used fast growing mycobacteria in the laboratory. M. abscessus is a prevalent human-infecting nontuberculosis mycobacterium (NTM) (31). Unlike M. smegmatis containing Cys394 in the DprE1SM (MSMEG_6382) active pocket (Cys387 in Mtb), the active cysteine residue (Mabs_19977) in M. abscessus DprE1 is replaced by an alanine (Ala395; fig. S9); thus M. abscessus is naturally resistant to TG-DNB modification. C. diphtheriae, the pathogenic bacterium also belonging to actinobacteria, carries a similar capsular structure to mycobacterium and the DprE1 homolog DIP1062 (72% identical/82% similar to DprE1SM) that contains a conserved cysteine residue (Cys414) in its active site (fig. S9) (32). S. pneumoniae and S. aureus as human respiratory pathogens were included as control strains to test for specificity (33, 34). As shown in Fig. 2D, TG-DNB1 and TG-DNB2 both exhibited distinct fluorescence labeling in M. smegmatis and C. diphtheriae but not in other strains, whereas none of the phosphate-buffered saline (PBS) or TG treated bacteria showed any fluorescence retention (fig. S10). Confocal fluorescence imaging of M. smegmatis after TG-DNB2 treatment revealed a polarized distribution of DprE1 in M. smegmatis (Fig. 2E). The treated bacteria were further analyzed by flow cytometry. Similar to the results of fluorescence microscopy, TG-DNB2 gave a 150-fold fluorescence enhancement relative to the PBS-treated M. smegmatis and TG-DNB1 showed a 50-fold fluorescence enhancement (Fig. 2F). One-hour incubation with 10 µM TG-DNB2 at room temperature could reach the maximal fluorescence in M. smegmatis without affecting its growth curve (figs. S11 and S12). An abolishment of the fluorescence by preincubating M. smegmatis with DNB1 before TG-DNB2 treatment further validated the specific retention through DprE1 covalent modification (fig. S13). Together, these data established the use of DprE1 as the target for trapping fluorophores through a covalent adduct formation.
Evaluation of dual-targeting probes CDG-DNB1/2 for labeling mycobacteria
Because mycobacteria generally express β-lactamases (BlaC in Mtb, BlaS in M. smegmatis and Blamab in M. abscessus), we sought to convert TG-DNB1 and TG-DNB2 into fluorogenic probes that could fluoresce only upon β-lactamase activation. Such dual-targeting strategy should further enhance selectivity and also lower the background from nonspecific interactions. On the basis of TG-DNB1 and TG-DNB2, we designed, synthesized, and tested two dual-targeting probes: CDG-DNB1 and CDG-DNB2 (Fig. 3A, see synthesis in figs. S5 and S6). Intense fluorescence was observed in M. smegmatis expressing both DprE1 and BlaS after incubating with CDG-DNB1 (fig. S14A) and CDG-DNB2 (Fig. 3B). A control probe (CDG-1) without DprE1 targeting showed no fluorescence retention (fig. S14A). Flow cytometry analysis revealed that CDG-DNB2-treated M. smegmatis could generate an 80-fold enhancement in fluorescent intensity (Fig. 3C), and about 30-fold for CDG -DNB1 owing to lower DprE1SM labeling efficiency by its uncaged product TG-DNB1 (fig. S14, B and D). Control strains expressing β-lactamase but not DprE1 (β-lactamase+/DprE1−) included TEM-1 β-lactamase (TEM-1 Bla) transformed E. coli (figs. S14 and 15A), S. pneumoniae, S. aureus and M. abscessus (fig. S14). Although they could hydrolyze CDG-DNB2 to uncage the fluorophore, no fluorescently labeled cells were observed because of the lack of or mutated DprE1 (Fig. 3B). C. diphtheriae (β-lactamase−/DprE1+), on the other hand, was unable to release the caged fluorophore. An inhibition study with a β-lactamase inhibitor (clavulanic acid) and DprE1 inhibitor (DNB1) decreased the fluorescent signals by 60% in M. smegmatis, offering further evidence supporting this dual-targeting mechanism (Fig. 3D). M. smegmatis expresses a major β-lactamase called BlaS and a minor β-lactamase called BlaE. BlaE is a type-1 β-lactamase and is not as sensitive to the clavulanic acid inhibition (24), which may explain why some β-lactamase activity remained in M. smegmatis in the presence of clavulanic acid.
Fig. 3. Characterization of the dual-targeting probe CDG-DNB2.
A) Structure of CDG-DNB2. B) Overlaid confocal images (bright field and green fluorescence, 63X/oil, Ex-490 nm/Em-520 nm) of freshly cultured E. coli expressing TEM-1-β-lactamase, S. pneumoniae, S. aureus, M. abscessus, C. diphtheriae, and M. smegmatis stained with 10 μM CDG-DNB2 in PBS at room temperature for 1 hour. C) Histogram of normalized fluorescence from fluorescence activated flow cytometry data with bacteria in B). D) Normalized fluorescence from M. smegmatis treated with CDG-DNB2 with and without β-lactamase inhibitor clavulanic acid (10 mM) and DNB1 (50 μM), analyzed by flow cytometry. M. smegmatis treated with CDG-DNB2 exhibited an average 90-fold increase of MFI over PBS, which was arbitrarily set to 1 to normalize the other groups with inhibitors. E) Overlaid confocal images of freshly cultured BCG and H37Rv Mtb stained with CDG-DNB2. F) Real-time confocal imaging of CDG-DNB2 treated BCG aggregates showing a time-dependent enhancement of green fluorescence.
Next, we tested CDG-DNB2 in freshly cultured Mycobacterium bovis Bacillus Calmette–Guérin (BCG) and Mtb H37Rv, both of which expressed BlaC and DprE1. After 1 hour incubation at room temperature both bacilli exhibited sharp green fluorescence, but not in CDG-1 or PBS treated groups (Fig. 3E and fig. S16). Further quantification by flow cytometry showed an 80-fold increase in the fluorescence intensity with CDG-DNB2 treated BCG and 52-fold increase in H37Rv (fig. S16). In a BlaC knockout (KO) H37Rv strain stained with CDG-DNB2, the fluorescence labeling was compromised by 55% but could be restored by BlaC transformation (fig. S17). We note that this BlaC KO strain was reported to have additional β-lactamase activity (24), which may contribute to the remaining signal.
To demonstrate rapid labeling of live BCG, we tested a nonwash CDG-DNB2 staining protocol was tested for real-time imaging. A time-dependent enhancement of green fluorescence was observed in continuous imaging at an interval of 2.5 min for 65 min (movie S1 and S2). Prominent green fluorescence in BCG was observed as early as 15 min. This rapid labeling indicated fast penetration of CDG-DNB2 into BCG (Fig. 3F), which prompted investigation into whether CDG-DNB2 could also label single Mtb in patient sputum. Four sputum specimens (two GeneXpert/smear positive and two negative) were collected, processed and neutralized, and then stored at 4°C for 2 days before incubation in 10 μM CDG-DNB2/PBS solution at room temperature for 1 hour. Sediments by centrifugation were washed three times, then resuspended in acidic buffer for confocal imaging. Single rod-shaped bacilli with emerald green fluorescence were observed in both TB-positive specimens (fig. S18).
Rapid and specific labeling of live Mtb by CDG-DNB3
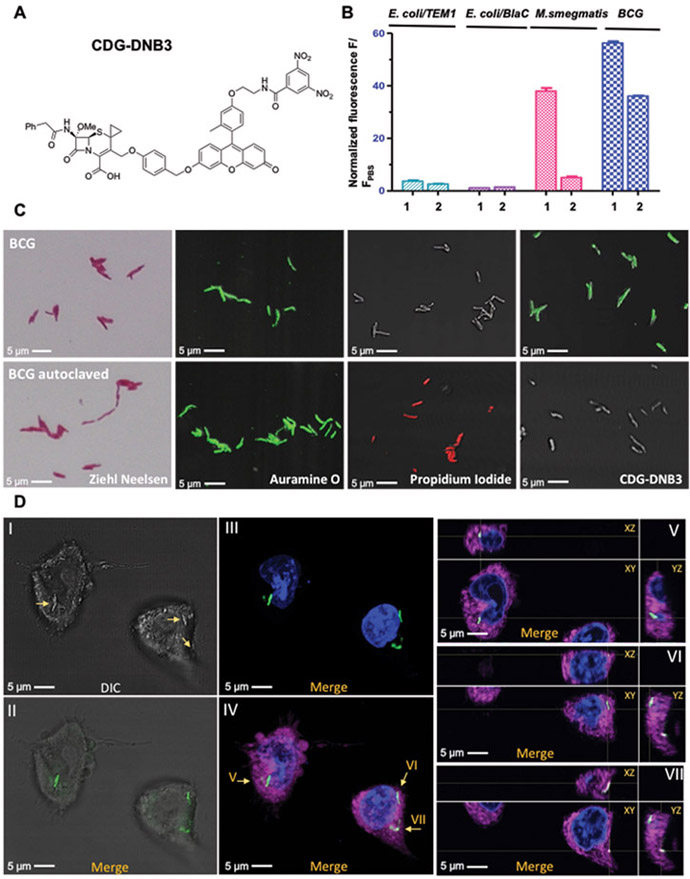
To enhance the specificity of dual-targeting probes for Mtb over other mycobacteria, we introduced a methoxy substitution to the lactam ring at 7- position on CDG-DNB2 (26, 27) to produce CDG-DNB3 (Fig. 4A and fig. S7). The quantum yields of CDG-DNB3 (φ = 0.01) and its fluorogenic product TG-DNB2 (φ = 0.77) were measured in 0.1 M phosphate buffer, using fluorescein as a reference standard (φfl = 0.85; excitation, 490 nm) (35). When incubated with CDG-DNB3, TEM-1 Bla showed negligible activity whereas BlaC activated fluorescence at a 200-fold lower concentration. CDG-DNB2, in contrast, could be activated by both β-lactamases (fig. S19). CDG-DNB3 and its product TG-DNB2, with or without clavulanic acid or DNB1, showed little change in their fluorescence (fig. S20), which confirmed that observed fluorescence activation with CDG-DNB3 was from the BlaC enzymatic activity. No fluorescence increase was observed when CDG-DNB3 was treated with DprE1, indicating that DprE1 most likely plays the role of a signal-trapping unit and alone does not cause the fluorescence increase in CDG-DNB3 (fig. S20). CDG-DNB3 was also characterized in bacteria. The findings were congruent with the enzymatic study: that CDG-DNB3 selectively stained BCG but not M. smegmatis, TEM-1 Bla or BlaC transformed E. coli (Fig. 4B and fig. S21).
Fig. 4. Specific labeling of live Mtb by CDG-DNB3.
A) Structure of CDG-DNB3. B) Histogram of fluorescence activated flow cytometry analysis with freshly cultured E. coli (TOP10) expressing TEM-1 β-lactamase, E. coli (TOP10) expressing BlaC, M. smegmatis and BCG stained with 10 μM CDG-DNB2 or CDG-DNB3 in PBS at room temperature for 1 hour. 1: CDG-DNB2 treated group; 2: CDG-DNB3 treated group. C) Microscopic imaging of freshly cultured (top row) and 121°C autoclaved (bottom row) BCG stained by Ziehl-Neelsen reagents, auramine O (green; excitation, 490 nm; emission, 520 nm), PI (red; excitation, 535 nm; emission, 617 nm), or CDG-DNB3 (10 μM, 1 hour). D) Infection of macrophages by CDG-DNB3-labeled individual BCG bacilli: (I) bright-field image of BCG-infected macrophages (bacilli are indicated by arrows); (II) overlay of bright-field and fluorescence images showing the localization of CDG-DNB3 stained bacilli (green; excitation, 490 nm; emission, 520 nm); (III) overlay of fluorescence images showing the bacilli (green) and 4’,6-diamidino-2-phenylindole (DAPI) stained nucleus (blue; excitation, 358 nm; emission, 461 nm); (IV) overlay of fluorescence image showing the bacilli, nucleus, and the macrophage plasma membrane (magenta; excitation, 649 nm; emission, 666 nm); (V to VII) orthogonal views (XY, XZ, YZ) of the bacilli indicated by arrows in (IV). ImageJ was used for image processing and stack projection.
Current staining probes, including auramine O and the trehalose probe, cannot differentiate Mtb from NTMs. To evaluate the selectivity of CDG-DNB3 for Mtb over NTMs, we tested 45 live NTMs, including both rapidly- and slowly-growing mycobacteria (table S1), 43 of which showed negligible fluorescent staining except Mycobacterium kansasii and Mycobacterium phlei, indicating the unprecedented selectivity of CDG-DNB3 toward Mtb. M. kansasii is a slowly growing mycobacterium and a common cause of pulmonary infections; however, M. phlei is a rapidly growing mycobacterium and does not cause pulmonary infections (36). Both M. kansasii and M. phlei carry DrpE1 with a conserved cysteine residue at the active site (32) and are known for their excessive β-lactamase activity (37). It is likely that their β-lactamase activity may resemble that of BlaC. Illustration of their detailed mechanism may assist future probe optimization to discriminate M. kansasii.
As a unique dual-targeting, activity-based probe, the labeling mechanism of CDG-DNB3 suggests its potential to differentiate live from dead Mtb. We stained freshly cultured or heat-killed (autoclaved) BCG and E. coli with carbol fuchsin (Ziehl-Neelsen stain), auramine O, propidium iodide (PI), and CDG-DNB3 for comparison (Fig. 4C and fig. S22). The red fluorescence observed in PI-stained BCG and E. coli indicated the loss of membrane integrity after autoclave. Carbol fuchsin and auramine O could discriminate between BCG and E. coli, however, they were unable to differentiate live from dead. CDG-DNB3-labeled BCG showed an identical growth curve to unlabeled BCG, unlike auramine O staining with phenol that considerably compromised the viability of stained BCG (fig. S12B). We further examined whether CDG-DNB3 could fluorescently label BCG for macrophage infection. BCG was preincubated with CDG-DNB3 before macrophage infection (two to three bacteria per macrophage). As shown in Fig. 4D, green fluorescence of internalized individual bacilli in macrophages was observed after 4 hours of infection (movie S3). Although Mtb may be genetically labeled with fluorescent proteins for real-time imaging of phagocytosis (38), the CDG-DNB3 method can be simply applied to wild-type (WT) Mtb and clinical isolates without any genetic manipulation that may pose unexpected effects such as changes in virulence.
Last, we applied CDG-DNB3 for staining of sputum. Single rod-shaped and green fluorescent bacilli were observed in all three clinically confirmed positive processed sputum specimens (fig. S23). One of the TB patient’s sputum that labeled with CDG-DNB3 (patient 5) was GeneXpert positive but auramine O negative, implicating higher sensitivity of CDG-DNB3 than auramine O.
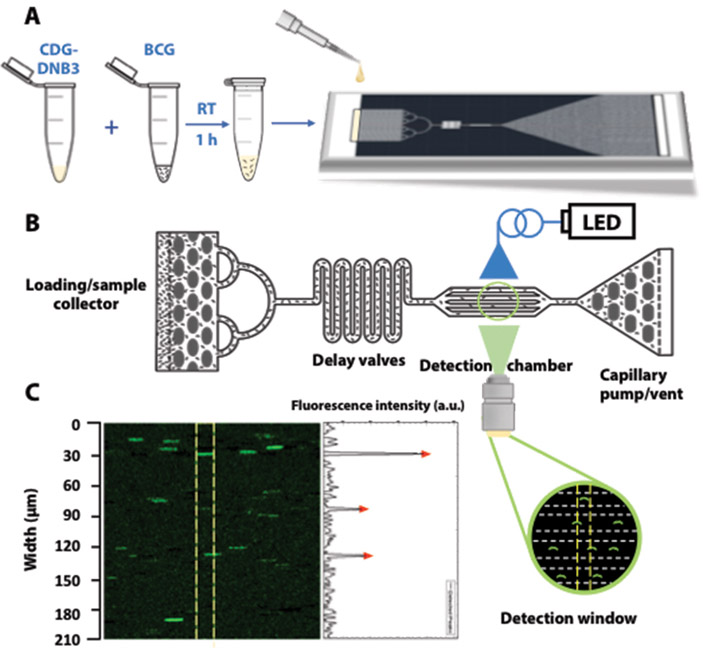
Automated counting of the number of stained BCG with a microfluidic chip
Clinically, after smear staining with auramine O, slides are manually examined under a fluorescence microscope to count the number of stained Mtb, which is a tedious and labor-intensive process. To evaluate whether CDG-DNB3 labeling could provide a solution for rapid counting of Mtb in combination with microfluidic technology, we designed a quantification strategy using a self-driven microfluidic chip (Fig. 5 and fig. S24). When the CDG-DNB3-labeled BCG was loaded on the chip, the fluid was dragged to flow along the channel by the capillary pump with no need for any extra pumping device (“self-driven” feature) (Fig. 5A and B). Four parallel channels across the detection window allowed for rapid detection and automatic counting of fluorescent BCG using a publicly available software (Fig. 5C). In an exemplary test run (movie S4), a total of 306 bacteria were detected in 2.42 s with a flow rate of 2.5 nl/s, corresponding to a concentration of 1.6 X 107 colony-forming units (CFU)/mL in a volume of 6 nl, consistent with calculation by measuring optical density of the BCG solution [optical density at 600 nm (OD600)].
Fig. 5. Rapid and specific quantification of live BCG with a microfluidic chip.
A) Overview of workflow. BCG was stained with CDG-DNB3 in PBS at room temperature (RT) for 1 hour, and then 10 µl of incubated mixture was applied onto the loading/sample collector in the chip. B) Schematic of image capture using a microfluidic device. The capillary pump with a vent drove the flow of the sample from the loading/sample collector, whereas the collector and delay valves filtrated and retarded the mixture to allow single BCG imaging in detection chamber under fluorescence illumination. Cartoon shows a microscopic view of the channels with flowing BCG. LED, light-emitting diode. C) Representative frame of image captured by the camera over the detection chamber (green fluorescence; excitation, 490 nm; emission, 520 nm) with fluorescence intensity peaks quantified in arbitrary units (a.u.) of up to 250 and 50 per scale (right). Yellow lines define the detection window used by the software to analyze the fluorescent signal. Each peak (red triangle) indicates one fluorescently labeled bacterium.
Discussion
Noninvasive fluorescence imaging technologies for bacteria have revealed greater insights into the small organisms (39), from membrane and intracellular structures to specific protein localization and trafficking dynamics. A common approach for fluorescent labeling of live bacteria uses the genetic fusion of a fluorescent protein or epitope tag to a protein of interest. In comparison, small molecule fluorescent probes can enable visualization of many nonprotein targets, but the small size and thick cell envelope of bacteria present big challenges for probe development. In particular, Mtb is known for having a nearly impenetrable cell wall. Here, we report a powerful dual-targeting approach in designing small molecule probes for rapid and specific labeling of live Mtb.
We propose the use of two enzymes in the cell wall and membrane layers of Mtb as imaging markers: BlaC, an ambler class A β-lactamase highly conserved through clinical isolates; and DprE1, a periplasmic enzyme required for the biosynthesis of the cell wall arabinans and highly conserved among actinobacteria. BlaC has been explored for Mtb detection, and preliminary testing with sputum samples showed promises for TB diagnosis (27), but targeting BlaC alone does not allow for single Mtb labeling (fig. S7A). The combination of BlaC and DprE1 in a dual-targeting probe offers considerable advantages. As our results demonstrate, this strategy is specific for Mtb over other clinically prevalent strains, including 43 of 45 closely related NTMs we tested. Fluorophore and lactam conjugates have been reported to target penicillin-binding proteins (PBPs) for bacterial labeling (40, 41). However, it is unknown whether PBPs can serve as the target for specific labeling of Mtb. It is unlikely that our probes are processed by PBPs due to the modifications on the lactam; those bacteria without β-lactamase cannot be fluorescently labeled with our probes even with the expression of DprE1 (for example, C. diphtheriae).
Another important feature provided by the dual-targeting approach is the ability to label single-cell, live Mtb. Live BCG was successfully labeled, without any genetic modification, to image phagocytosis by macrophages. It is likely that our probe could similarly rapidly and specifically label live Mtb in clinical samples to detect and isolate labeled mycobacteria using cell sorting for further analysis, such as culturing or drug sensitivity assays. We also designed a microfluidic device and workflow to automate quantification of individual fluorescently labeled BCG; it took about 20 min for a total volume of 10 µl of stained-bacteria mixture to flow through the detection window. In principle, this method could lead to a detectable concentration of 100 CFU/ml assuming one copy of BCG in the loading mixture. Further optimization, such as increasing the number of channels on the chip to load larger sample volumes, may allow an even more rapid detection and a lower detection limit. The materials and fabrication costs of each off-the-shelf chip are estimated to be $0.65 in an academic research setting but may be reduced by 5 to 10 times in a large-scale manufacturing facility via the use of thermoplastic polymer chips and injection molding. The combination of rapid, specific CDG-DNB3 labeling and the low-cost microfluidic chip provide a simple and cost-effective solution to automated counting of live Mtb in samples.
In summary, this study reports a dual-targeting approach using the expression of BlaC and DprE1 in Mtb to develop CDG-DNB3, a fluorogenic probe that enables rapid, specific fluorescent labeling and quantification of single live Mtb. A series of biochemical assays and microscopic imaging experiments have successfully validated the BlaC/DprE1 dual-targeting mechanism, and demonstrated that CDG-DNB3 can differentiate Mtb from other clinically prevalent strains, distinguish live from dead BCG, and image BCG infection in macrophages. Finally, a low-cost, self-driven microfluidic chip was successfully developed to rapidly count the number of CDG-DNB3 stained BCG. This rapid, noninvasive, specific labeling and counting of live Mtb positions CDG-DNB3 as a potentially powerful probe for pathogenesis study, drug and vaccine screening, drug susceptibility testing, treatment efficacy assessment, and diagnosis of TB.
Materials and Methods
Study Design
The goal of this study was to develop a rapid and specific fluorogenic probe for the detection of Mtb in both laboratorial and clinical settings. The probe CDG-DNB3 carries a caged-fluorophore that fluoresces only upon enzymatic activation by Mtb-specific BlaC. The fluorescence product generated by BlaC cleavage was retained through a covalent modification at the cysteine residue 387 of DprE1, an essential enzyme for Mtb cell wall biosynthesis. To discover the optimal structure to label DprE1, we synthesized a series of fluorophore-conjugated dinitrobenzamide (DNB) analogs and tested them against various bacterial strains including DprE1+ mycobacterial and corynebacterial strains, and other clinically prevalent but DprE1− or cysteine-mutated bacterial and mycobacterial strains. Next, the DprE1 targeting unit was introduced into the BlaC-targeting cephalosporin derivative to generate CDG-DNB3, selective for Mtb over other non-Mtb mycobacterial strains (NTMs). As an enzyme-dependent probe, CDG-DNB3 was evaluated for differentiating NTMs and viability of BCG, and labeling of live BCG for imaging phagocytosis in real time. The utility of the dual-targeting probes was also tested in two small groups of TB patient sputum. In the United States, fresh patient sputum specimens were collected by the Clinical Microbiology Laboratory at Stanford University Medical Center. Experiments in this work used discarded sputum samples and no institutional review board approval was required. In China, ethics approval for the use of the six patient samples was provided by the Ethics Committee of the Beijing Chest Hospital affiliated to the Capital Medical University. The experiments with biohazard materials were designed and performed adhering to the biosafety guidelines at Stanford University and Beijing Chest Hospital. Finally, a portable microfluidic device was designed and a workflow applied for automated counting of CDG-DNB3-labeled BCG.
Expression and purification of M. smegmatis DprE1
The general expression and purification procedures were described previously (42). In brief, a single colony of BL21(DE3) containing the pET-SUMO-DprE1 plasmid was inoculated into 100 ml of lysogeny broth (LB) with kanamycin (50 µg/ml), followed by incubation at 37°C, 250 rpm overnight. The overnight culture was added into 500 ml of fresh LB broth with kanamycin (50 µg/ml) and 1 mM isopropyl β-D-1-thiogalactopyranoside. After incubation for up to 6 hours at 37°C and 250 rpm, bacteria were harvested and pellet was frozen at −80°C before resuspension in 22.5 ml of lysis buffer [25 mM Hepes (pH 7.4), 300 mM KCl, and 10% glycerol]. Lysozyme (2.5 ml of 7.5 mg of lysozyme per milliliter in lysis buffer) was used to lyse the bacteria. Halt proteinase inhibitor cocktail (Thermo Fisher Scientific) was added to the bacterial lysate before two rounds of metal-affinity purification using TALON metal affinity resin. The affinity purified DprE1sm fraction was eluted using lysis buffer containing up to 500 mM imidazole. DprE1sm was then dialyzed into Hepes buffer [25 mM Hepes and 10% glycerol (pH 7.4)] and stored at −80°C. Tag cleavage was achieved by overnight incubation with SUMO protease (Thermo Fisher Scientific), followed by a third-round affinity purification to remove both His6-SUMO tag and SUMO protease.
Sample preparation for SDS-PAGE analysis
Typically, aliquots of the purified protein DprE1sm [50 μM in 50 mM Hepes/10% glycerol, (pH 7.5)] were incubated with 10 μM FL, FL-DNB1/2, FL-MNB, Rd-Boc, Rd-DNB, Rd-MNB, or TG-DNB1/2 [from 1 mM stock in dimethyl sulfoxide (DMSO)] and 100 μg whole lysate of M. smegmatis containing phenylmethylsulfonyl fluoride (1 mM) in a final volume of 30 μl at 37°C for 1 hour. The whole lysate of freshly cultured M. smegmatis provided the natural substrate DPR and the cofactor FAD that are essential for the generation of DNB-DprE1 covalent products. As negative controls, (i) DprE1sm was preheated for 1 hour at 90°C, to denature the protein. (ii) The sample buffer was adjusted to 7 M urea and 20 mM DTT after incubation. These samples were further incubated for 1 hour at 37°C. (iii) DprE1sm was incubated with 50 μM DNB1 for 1 hour before addition of TG-DNB2. Samples were treated with 4x LDS loading buffer (Life Technologies) without heating and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (NuPAGE, Life Technologies). The analysis was performed in duplicate.
Bacteria growth and staining
BCG [American Type Culture Collection (ATCC) 35734], Mtb H37Rv (ATCC 25618), M. smegmatis MC2155 (ATCC 700084), S. pneumoniae (ATCC 49619), S. aureus (ATCC 25923), M. abscessus (ATCC 19977) and C. diphtheriae (ATCC 13812) were purchased from ATCC. All other NTMs were purchased from ATCC or other specified DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen).
In general, M. smegmatis, BCG and H37Rv were cultured in Middlebrook 7H9 broth supplemented with ADC enrichment and 0.1% Tween 80. E. coli (TOP10) was grown in LB medium. S. pneumoniae was grown on tryptic soy agar plates supplemented with defibrinated sheep blood at 5% CO2 and 37°C. S. aureus, M. abscessus and C. diphtheriae were grown on tryptic soy agar plates supplemented with defibrinated sheep blood at 37°C. The unit CFU/ml was determined by measuring the ultraviolet (UV) absorbance at OD600.
For growth curve, M. smegmatis was pre-treated with PBS, 10 μM TG-DNB2, and 10 μM CDG-DNB2 for 1 hour, or 2% NaOH for 20 min at room temperature. Culture of pretreated M. smegmatis was started at OD600=0.1 in 7H9 medium for all groups at 37°C and 220 rpm. OD600 of each sample was measured every 2 hours over 14 hours. Similarly, BCG was pre-treated with PBS and 10 μM CDG-DNB3 at room temperature for 1 hour or auramine O (AlphaTec). Culture of pretreated BCG was started at OD600=0.4 to 0.5 in 7H9 medium for all conditions at 37°C and 80 rpm. OD600 of each sample was measured daily for 10 days. Experiments were repeated three times.
For TG, TG-DNB1/2 and CDG-DNB2/3 analog staining in the laboratory, 500 μM probes in pure DMSO were frozen at −80°C as stock solutions. Working solution (20 μM) was prepared freshly by diluting stock solution in PBS. Staining was done by mixing bacteria in PBS resuspension and 20 μM working solution in a 1:1 volume ratio and left in room temperature for 1 h, then centrifuged at 3000g, and washed three times stringently with PBS. A light safe tube or foil wrap was used to prevent photobleaching during incubation. Bacteria were fixed in 10% formalin for 30 min before centrifugation and resuspension in saline for flow cytometry analysis. For CDG-DNB3 staining to differentiate live and autoclaved BCG or E. coli, a wash step with alcohol/water was performed immediately after staining and two additional PBS washes were performed before formalin fixing.
For Ziehl-Neelsen staining, briefly, freshly cultured or 121°C/20 min autoclaved BCG and E. coli were smeared and fixed by gentle heat on glass slides and then stained with carbol fuchsin for 3 min. The smears were decolorized with acidic alcohol and then rinsed with sterile water, followed by counterstaining with methylene blue for 1 to 2 min, rinsing and drying for immediate microscope imaging.
For auramine O staining, briefly, freshly cultured or 121°C/20 min autoclaved BCG and E. coli were smeared and fixed by methanol on glass slides and then stained with auramine O for 15 min. Smears were rinsed with sterile water and decolorized by acidic alcohol for 3 min. The smears were rinsed again with sterile water, counterstained by potassium permanganate for 4 min, and finally rinsed and dried for immediate microscope imaging.
For PI staining, briefly, freshly cultured or 121°C/20 min autoclaved BCG and E. coli were incubated in 20 μM PI solution at room temperature for 1 hour then washed three times with PBS. Bacteria were fixed in 10% formalin solution for 30 min before inspected under a microscope.
Bacteria analysis using flow cytometry and microscope
Flow cytometry analysis was done with instrumentation and assistance provided by the Stanford Shared FACS Facility. Bacteria were analyzed using a FACScan with a four-laser, 12-color DxP12 Cytek upgrade (Becton Dickinson, Cytek Biosciences) and evaluated according to their granularity, [side Scatter (SSC)] property on a log scale, excited by 488 nm later, and filtered with 560 nm short-pass filter and 525/50 nm band-Pass filter. Flow cytometry data analysis was done using FlowJo v10 software. The MFI was collected and plotted against the natural autofluorescence of bacteria.
All bacteria images were taken either with a Zeiss AxioImager M1 upright widefield fluorescence/DIC microscope or with a Zeiss LSM710 inverted confocal microscope. Images were processed and converted by ImageJ.
Inhibition study with M. smegmatis
Freshly cultured M. smegmatis (100 µl) was pretreated with PBS, clavulanic acid (2 mg/ml), 50 µM DNB1 or clavulanic acid + DNB1 at 37°C for 2 hours and then incubated with PBS or 10 µM CDG-DNB2 in a final volume of 200 µl at room temperature for 3 hours. Cells were washed with PBS twice before analysis by flow cytometry. M. smegmatis treated with CDG-DNB2 exhibited averagely 90-fold increase of MFI over PBS, which was arbitrarily set to 1 to normalize the other samples with inhibitors. Inhibition study with TG-DNB2 by DNB1 was done under similar conditions.
Inhibition study with BlaC knockout H37Rv
Two milliliters of freshly cultured WT, BlaC KO, and vector control H37Rv at OD600 = 1 was pelleted and pretreated with 50 µM DNB1 at 37°C in a 200 µl volume for 1 hour. Two hundred microliters of (20 µM) CDG-DNB2 was added into each tube to reach a 10 µM final concentration and then incubated at room temperature for 2 hours. Mycobacteria were washed three times with PBS, fixed in 10% (v/v) formalin for 30 min, and then analyzed by flow cytometry. PBS-treated WT, BlaC KO, BlaC compensated and vector control H37Rv were also analyzed to show the fold increase of fluorescence signal by CDG-DNB2. The fold increase of fluorescence in WT H37Rv by CDG-DNB2 over PBS was arbitrarily set to 1 to normalize the other groups.
Western blot analysis
The procedures for Western blot have been previously described (43). In brief, E. coli TOP10 strains expressing TEM-1 β-lactamase (TEM-1) or BlaC were lysed with a bead beater in PBS containing proteinase inhibitors. Twenty-five micrograms of lysates was loaded on NuPAGE gel for electrophoresis at 200 V for 90 min. Wet transfer was performed using the Bio-Rad transfer kit at 300 mA for 90 min. The transferred nitrocellulose membrane was blocked in PBS containing 5% bovine serum albumin and 0.1% Tween-20 for 1 hour. Primary antibody (His-probe mouse monoclonal IgG, Invitrogen) incubation (1: 2000) was performed in the blocking buffer overnight at 4°C. The membrane was then washed four times with PBS containing 0.1% Tween 20. Secondary antibody incubation (LI-COR donkey anti-mouse IgG IRDye 680 or anti-rabbit IgG IRDye 800CW, 1: 10,000) was performed in the blocking buffer for 2 hours at room temperature. After washing four times with PBS containing 0.1% Tween 20, the membrane was analyzed using a LI-COR Odyssey imaging system.
Visualization of BCG in macrophage by CDG-DNB3
Mouse RAW 264.7 macrophages were grown on sterile coverslips in a six-well plate in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% (v/v) fetal bovine serum at 5% CO2 and 37°C to 70 to 80% confluence. Freshly cultured BCG were treated with 10 μM CDG-DNB3 at room temperature for 1 hour and then resuspended in serum-free DMEM to infect macrophages (two to three bacteria per macrophage). After 4 hours of infection, macrophages were washed to remove free bacteria and then incubated with CellMask Deep Red Cell Plasma staining reagent in 1X dilution at 37°C for 10 min. Cells were washed and fixed in 10% formalin. Cells were permeabilized then incubated with 300 nM DAPI solution for 5 to 10 min and washed immediately before mounting for imaging. Images of infected macrophages were obtained by confocal microscopy (Zeiss LSM710 inverted confocal microscope), using a 63X/oil immersion objective. The bright-field image was from a single image capture. All fluorescence images were gathered sequentially and stacked. Essential sequential Z sections of stained cells were recorded for generation of stacked images through cells. Multichannel three-dimensional projections of fluorescent images were constructed from sequential Z sections of cells assembled in ImageJ. All collected fluorescence images were deconvoluted by ImageJ [W. Rasband, National Institute of Health (NIH)]. Percentage colocalization of material was calculated using the colocalization function in ImageJ.
Staining of processed sputum by CDG-DNB2 and CDG-DNB3
Sputum specimens were processed with NaOH/N-acetyl-L-cysteine, neutralized, and stored in PBS at 4°C for 2 days before incubation in 10 μM CDG-DNB2/PBS solution at room temperature for 1 h. Sediments were obtained by centrifugation at 4000g for 5 min before washing three times with PBS. Stained samples were fixed in 10% formalin for 30 min, washed once by PBS with last resuspension in citrate buffer (pH 3), and imaged with a Zeiss LSM710 inverted confocal microscope.
For CDG-DNB3, patients’ sputum specimens were collected freshly by the National Tuberculosis Clinical Laboratory at Beijing, China. Specimens were processed with NaOH/N-acetyl-L-cysteine, neutralized, and then incubated in 10 μM CDG-DNB3/PBS solution at room temperature for 1 h. Sediments were obtained by centrifugation at 4000g for 5 min then washed three times by PBS before resuspension in citrate buffer (pH3) and imaged with a Nikon fluorescence microscope.
Microfluidic device design and fabrication
The design and fabrication of the capillary pump driven bacteria counting microfluidic device were performed using photoresist molds and standard soft lithography methods. The device layout was designed using a CAD drawing program and the final design was printed on high resolution transparency sheets which were used as photolithography masks during the fabrication of the polydimethylsiloxane (PDMS; Dow Corning Sylgard 184) microfluidic chips with the features shown in fig. S24. The device has a sample loading area (a), a fluid flow delay region (b), an observation chamber with four separate channels (c), and a capillary pump (d) that drives the fluid flow. The channel is 100 µm wide and 100 µm high in the flow-delaying region (b), and each channel is 30 µm wide and 100 µm high in the observation chamber (c). Briefly, a clean and dry silicon wafer (100 mm in diameter and 500 µm in thickness) was treated with hexamethyldisilazane (Sigma-Aldrich) vapor for 2 min in a clean room environment. Subsequently, the wafer was spin-coated with photoresist (Shipley SJR 5740) at 2000 rpm (ramping time, 15 s; spin time, 60 s). The wafer was then soft-baked in an oven at 90°C for 60 min. The transparency mask from above was used for the photolithography. The wafer was then exposed to UV light and developed in 20% Microposit Developer 2401 for 1.5 min to form the final molds for elastomeric chip fabrication. The microfluidic devices were prepared via negative casting on the molds using PDMS. Sylgard 184 A and B components were mixed at a ratio of 20:1 (w/w), degassed in a vacuum chamber, and spin-coated onto the photoresist molds at 5000 rpm. The curing was carried out in an 80°C oven for 1 to 3 hours and at the end of the curing period the fabricated devices were peeled off from the molds gently and bonded to microscope slides immediately by conformal contact pressure followed by an additional baking at 100°C from 2 hours to overnight.
Optical setup, image capturing and bacteria counting by image analysis
The bacteria counting device on a standard glass microscope slide was mounted on an inverted microscope with a 63X objective (Olympus America Inc.) for observation under fluorescence illumination. The chip was then loaded with the BCG/PBS to be analyzed using the capillary pumping action of the microfluidic device. The florescence pictures of the stained bacteria were captured at rates of 33 frames/s by a digital video camera mounted on the microscope (ORCA-Flash4.0, Hamamatsu) and attached to a computer. In total, 80 frames were captured in 2.42 s. It took 11 frames (1/3 second) for a bacterium to pass through the imaging window (with a length of 210 µm). Movie S4 contains a four times slower version of the captured video defined in green.
For counting the fluorescently-stained bacteria, images were collected in grayscale, analyzed, and processed by ImageJ. Every image was subtracted from its next frame to remove the background noise. The generated hyperstack was median filtered with 2-pixel dimensions to further remove the noise before the sum image of the stack was subtracted from each frame to highlight moving objects in the image. Then, the final frames were automatically analyzed by a custom plugin filter [Broadly Applicable Routines (BAR) for ImageJ] to find the peaks in the gray intensity profile plot values. A baseline threshold for each image stack was manually set to obtain the peak counts corresponding to bacterial counts for each frame, and this was tallied to a total count across the entire loaded sample volume processed.
Statistical Analysis
GraphPad Prism 5 was used for plotting and statistical analysis. Significant difference was determined by performing one-way (Fig. 3D and fig. S13C) or two-way (fig. S17) analysis of variance (ANOVA) followed by post hoc Bonferroni’s multiple comparison test to determine the statistical significance with 95% confidence intervals with *P < 0.05, **P < 0.01 and ***P < 0.001. All error bars in figures represent ± SD (n = 3).
Supplementary Material
Fig. S1. Synthesis of FI-DNB1, FI-DNB2 and FI-MNB.
Fig. S2. Synthesis of Rd-DNB and Rd-MNB.
Fig. S3. Synthesis of TG-DNB1.
Fig. S4. Synthesis of TG-DNB2.
Fig. S5. Synthesis of CDG-DNB1.
Fig. S6. Synthesis of CDG-DNB2.
Fig. S7. Synthesis of CDG-DNB3
Fig. S8. SDS-PAGE analysis of purified M. smegmatis DprE1.
Fig. S9. DprE1 DNA sequence alignment.
Fig. S10. Microscopic imaging and flow cytometry analysis of bacteria labeled by TG and TG-DNB analogs.
Fig. S11. Characterization of TG-DNB2 in M. smegmatis.
Fig. S12. Growth curve of M. smegmatis and BCG.
Fig. S13. Inhibition study of TG-DNB2 with DNB1.
Fig. S14. Characterization of CDG-1 and CDG-DNB analogs in bacteria.
Fig. S15. Western blots detecting the expression of TEM-1 Bla and BlaC in transformed E. coli.
Fig. S16. Labeling of BCG and H37Rv by CDG-DNB2.
Fig. S17. Inhibition study with BlaC KO, BlaC compensated and control vector transformed H37Rv and DNB1.
Fig. S18. Labeling of Mtb in processed sputum by CDG-DNB2.
Fig. S19. Evaluation of selectivity of CDG-DNB2/3 for BlaC.
Fig. S20. Fluorescence intensity of CDG-DNB3 and TG-DNB2 with or without clavulanic acid, DNB1 or DprE1.
Fig. S21. Characterization of CDG-DNB3 in M. smegmatis, BCG and TEM-1 Bla or BlaC transformed E. coli.
Fig. S22. Specific labeling of viable BCG by CDG-DNB3.
Fig. S23. Labeling of Mtb in processed sputum by CDG-DNB3.
Fig. S24. Overall layout and the functional regions of the microfluidic bacteria counting device.
Table S1. List of NTMs stained with CDG-DNB3.
Table S2. List of individual subject-level data shown on bar graphs in all figures.
Movie S1. Real-time fluorescence imaging of CDG-DNB2 treated BCG aggregates.
Movie S2. Real-time bright-field imaging of CDG-DNB2 treated BCG aggregates.
Movie S3. Infection of macrophages by CDG-DNB3 labeled individual BCG bacilli.
Movie S4. Automated counting of CDG-DNB3 labeled BCG bacilli with microfluidic chip.
Appendix S1. 1H and 13C nuclear magnetic resonance and mass spectroscopy spectra.
Acknowledgments:
We thank K. Johnsson (École Polytechnique Fédérale de Lausanne, Switzerland) for providing pET-SUMO-DprE1, A. Olson at Stanford Neuroscience Microscopy Service (supported by NIH NS069375) for help on confocal imaging, C. Carswell-Crumpton at Stanford Shared FACS Facility for assistance with flow cytometry, Functional Genomics Facility at Stanford for access to LI-COR Odyssey for Western blots, and T. C. Doyle at Stanford Center for Innovation in In-Vivo Imaging (SCI3) for help on IVIS imaging.
Funding: This work was supported by NIH grant 1R01AI125286-01A1 and the Stanford Center at Peking University Faculty Fellowship. J.X. thanks the Molecular Imaging Program at Stanford for the Molecular Imaging Young Investigator (MIYI) Prize. T.D. thanks the support from the Center for Molecular Analysis and Design (CMAD) at Stanford.
Footnotes
Competing interests: Y.C., J.X., and J.R. are inventors on U.S. patent application US20170044593A1 submitted by Leland Junior Stanford University that covers “Methods for rapid and specific detection of Mycobacterium tuberculosis with a dual targeting fluorogenic probe.” All other authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or Supplementary Materials. Materials are available and will be provided under the material transfer policies of Stanford University. These requests should be directed to the corresponding author. Requests for the option to license the technology can be addressed to the Stanford University Office of Technology Licensing.
References
- 1.Zumla A, Raviglione M, Hafner R, von Reyn CF, Tuberculosis. N. Engl. J. Med. 368, 745–755 (2013). [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization, Global tuberculosis report 2015, (World Health Organization; 2016). [Google Scholar]
- 3.Dye C, Bassili A, Bierrenbach AL, Broekmans JF, Chadha VK, Glaziou P, Gopi PG, Hosseini M, Kim S, Manissero D, Onozaki I, Rieder HL, Scheele S, van Leth F, van der Werf M, Williams BG, Measuring tuberculosis burden, trends, and the impact of control programmes. Lancet Infect. Dis 8, 233–243 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Zumla A, Abubakar I, Raviglione M, Hoelscher M, Ditiu L, McHugh TD, Squire SB, Cox H, Ford N, McNerney R, Marais B, Grobusch M, Lawn SD, Migliori GB, Mwaba P, O’Grady J, Pletschette M, Ramsay A, Chakaya J, Schito M, Swaminathan S, Memish Z, Maeurer M, Atun R, Drug-resistant tuberculosis-current dilemmas, unanswered questions, challenges, and priority needs. J. Infect. Dis. 205, S228–S240 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Gandhi NR, Nunn P, Dheda K, Schaaf HS, Zignol M, van Soolingen D, Jensen P, Bayona J, Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet 375, 1830–1843 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Dheda K, Ruhwald M, Theron G, Peter J, Yam WC, Point-of-care diagnosis of tuberculosis: past, present and future. Respirology 18, 217–232 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Bates M, Zumla A, The development, evaluation and performance of molecular diagnostics for detection of Mycobacterium tuberculosis. Expert Rev. Mol. Diagn. 16, 307–322 (2016). [DOI] [PubMed] [Google Scholar]
- 8.McNerney R, Daley P, Towards a point-of-care test for active tuberculosis: obstacles and opportunities. Nat. Rev. Microbiol. 9, 204–213 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Chan J, Dodani SC, Chang CJ, Reaction-based small-molecule fluorescent probes for chemoselective bioimaging. Nat. Chem. 4, 973–984 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagemann PKH, Fluoreszenzfärbung von tuberkelbakterien mit auramin. Münch. Med. Wochenschr. 85, 1066–1067 (1938). [Google Scholar]
- 11.Richards OW, The staining of acid-fast tubercle bacteria. Science 93, 190–190 (1941). [DOI] [PubMed] [Google Scholar]
- 12.Kvach JT, Veras JR, A fluorescent staining procedure for determining the viability of mycobacterial cells. Int. J. Lepr. Other Mycobact. Dis. 50, 183–192 (1982). [PubMed] [Google Scholar]
- 13.Datta S, Sherman JM, Bravard MA, Valencia T, Gilman RH, Evans CA, Clinical evaluation of tuberculosis viability microscopy for assessing treatment response. Clin. Infect. Dis. 60, 1186–1195 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thanky NR, Young DB, Robertson BD, Unusual features of the cell cycle in mycobacteria: Polar-restricted growth and the snapping-model of cell division. Tuberculosis 87, 231–236 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Yang D, Ding F, Mitachi K, Kurosu M, Lee RE, Kong Y, A fluorescent probe for detecting mycobacterium tuberculosis and identifying genes critical for cell entry. Front. Microbiol. 7, 13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Backus KM, Boshoff HL, Barry CS, Boutureira O, Patel MK, D’Hooge F, Lee SS, Via LE, Tahlan K, Barry CE, Davis BG, Uptake of unnatural trehalose analogs as a reporter for mycobacterium tuberculosis. Nat. Chem. Biol. 7, 228–235 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swarts BM, Holsclaw CM, Jewett JC, Alber M, Fox DM, Siegrist MS, Leary JA, Kalscheuer R, Bertozzi CR, Probing the mycobacterial trehalome with bioorthogonal chemistry. J. Am. Chem. Soc. 134, 16123–16126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foley HN, Stewart JA, Kavunja HW, Rundell SR, Swarts BM, Bioorthogonal chemical reporters for selective in situ probing of mycomembrane components in mycobacteria. Angew. Chem.-Int. Ed. 55, 2053–2057 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Kamariza M, Shieh P, Ealand CS, Peters JS, Chu B, Rodriguez-Rivera FP, Sait MRB, Treuren WV, Martinson N, Kalscheuer R, Kana BD, Bertozzi CR, Rapid detection of Mycobacterium tuberculosis in sputum with a solvatochromic trehalose probe. Sci. Transl. Med. 10, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beatty KE, Williams M, Carlson BL, Swarts BM, Warren RM, van Helden PD, Bertozzi CR, Sulfatase-activated fluorophores for rapid discrimination of mycobacterial species and strains. Proc. Natl. Acad. Sci. U. S. A. 110, 12911–12916 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makarov V, Manina G, Mikusova K, Mollmann U, Ryabova O, Saint-Joanis B, Dhar N, Pasca MR, Buroni S, Lucarelli AP, Milano A, De Rossi E, Belanova M, Bobovska A, Dianiskova P, Kordulakova J, Sala C, Fullam E, Schneider P, McKinney JD, Brodin P, Christophe T, Waddell S, Butcher P, Albrethsen J, Rosenkrands I, Brosch R, Nandi V, Bharath S, Gaonkar S, Shandil RK, Balasubramanian V, Balganesh T, Tyagi S, Grosset J, Riccardi G, Cole ST, Benzothiazinones kill mycobacterium tuberculosis by blocking arabinan synthesis. Science 324, 801–804 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ribeiro A, Degiacomi G, Ewann F, Buroni S, Incandela ML, Chiarelli LR, Mori G, Kim J, Contreras-Dominguez M, Park YS, Han SJ, Brodin P, Valentini G, Rizzi M, Riccardi G, Pasca MR, Analogous mechanisms of resistance to benzothiazinones and dinitrobenzamides in mycobacterium smegmatis. PLoS One 6, 7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trefzer C, Rengifo-Gonzalez M, Hinner MJ, Schneider P, Makarov V, Cole ST, Johnsson K, Benzothiazinones: prodrugs that covalently modify the decaprenylphosphoryl-beta-D-ribose 2’-epimerase dprE1 of mycobacterium tuberculosis. J. Am. Chem. Soc. 132, 13663–13665 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Flores AR, Parsons LM, Pavelka MS, Genetic analysis of the beta-lactamases of Mycobacterium tuberculosis and Mycobacterium smegmatis and susceptibility to beta-lactam antibiotics. Microbiology 151, 521–532 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Kurz SG, Wolff KA, Hujer AM, Nguyen L, Bonomo RA, Sequence analysis of the beta-lactamase gene blac in clinical strains of Mycobacterium tuberculosis reveals a conserved therapeutic target. Am. J. Respir. Crit. Care Med. 187, 2 (2013).23281347 [Google Scholar]
- 26.Xie HX, Mire J, Kong Y, Chang MH, Hassounah HA, Thornton CN, Sacchettini JC, Cirillo JD, Rao J, Rapid point-of-care detection of the tuberculosis pathogen using a BlaC-specific fluorogenic probe. Nat. Chem. 4, 802–809 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng YF, Xie HX, Sule P, Hassounah H, Graviss EA, Kong Y, Cirillo JD, Rao J, Fluorogenic probes with substitutions at the 2 and 7 positions of cephalosporin are highly blaC-specific for rapid mycobacterium tuberculosis detection. Angew. Chem.-Int. Ed. 53, 9360–9364 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, Kong Y, Cheng Y, Janagama H, Hassounah H, Xie H, Rao J, Cirillo J, Real-time imaging of mycobacterium tuberculosis, using a novel near-infrared fluorescent substrate. J. Infect. Dis. 215, 405–414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikaido H, Jarlier V, Permeability of the mycobacterial cell-wall. Res. Microbiol. 142, 437–443 (1991). [DOI] [PubMed] [Google Scholar]
- 30.Mikusova K, Makarov V, Neres J, DprE1-from the discovery to the promising tuberculosis drug target. Curr. Pharm. Design 20, 4379–4403 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Bryant JM, Grogono DM, Rodriguez-Rincon D, Everall I, Brown KP, Moreno P, Verma D, Hill E, Drijkoningen J, Gilligan P, Esther CR, Noone PG, Giddings O, Bell SC, Thomson R, Wainwright CE, Coulter C, Pandey S, Wood ME, Stockwell RE, Ramsay KA, Sherrard LJ, Kidd TJ, Jabbour N, Johnson GR, Knibbs LD, Morawska L, Sly PD, Jones A, Bilton D, Laurenson I, Ruddy M, Bourke S, Bowler I, Chapman SJ, Clayton A, Cullen M, Dempsey O, Denton M, Desai M, Drew RJ, Edenborough F, Evans J, Folb J, Daniels T, Humphrey H, Isalska B, Jensen-Fangel S, Jonsson B, Jones AM, Katzenstein TL, Lillebaek T, MacGregor G, Mayell S, Millar M, Modha D, Nash EF, O’Brien C, O’Brien D, Ohri C, Pao CS, Peckham D, Perrin F, Perry A, Pressler T, Prtak L, Qvist T, Robb A, Rodgers H, Schaffer K, Shafi N, van Ingen J, Walshaw M, Watson D, West N, Whitehouse J, Haworth CS, Harris SR, Ordway D, Parkhill J, Floto RA, Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium. Science 354, 751–757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Incandela ML, Perrin E, Fondi M, Ribeiro A, Mori G, Moiana A, Gramegna M, Fani R, Riccardi G, Pasca MR, DprE1, a new taxonomic marker in mycobacteria. FEMS Microbiol. Lett. 348, 66–73 (2013). [DOI] [PubMed] [Google Scholar]
- 33.van der Poll T, Opal SM, Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet 374, 1543–1556 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Cole AM, Tahk S, Oren A, Yoshioka D, Kim YH, Park A, Ganz T, Determinants of staphylococcus aureus nasal carriage. Clin. Diagn. Lab. Immunol. 8, 1064–1069 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ueno T, Urano Y, Setsukinai K, Takakusa H, Kojima H, Kikuchi K, Ohkubo K, Fukuzumi S, Nagano T, Rational principles for modulating fluorescence properties of fluorescein. J. Am. Chem. Soc. 126, 14079–14085 (2004). [DOI] [PubMed] [Google Scholar]
- 36.García-Agudo L, García-Martos P, Clinical significance and antimicrobial susceptibility of rapidly growing mycobacteria Science against microbial pathogens: communicating current research and technological advances, Mendez-Vilas A, Ed. (Formatex Research Center, 2011) pp. 363–377. [Google Scholar]
- 37.Kwon HH, Tomioka H, Saito H, Distribution and characterization of beta-lactamases of mycobacteria and related organisms. Tubercle Lung Dis. 76, 141–148 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Dhandayuthapani S, Via LE, Thomas CA, Horowitz PM, Deretic D, Deretic V, Green fluorescent protein as a marker for gene expression and cell biology of mycobacterial interactions with macrophages. Mol. Microbiol. 17, 901–912 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Kocaoglu O, Carlson EE, Progress and prospects for small-molecule probes of bacterial imaging. Nat. Chem. Biol. 12, 472–478 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao GS, Meier TI, Kahl SD, Gee KR, Blaszczak LC, BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrobial Agents and Chemotherapy 43, 1124–1128 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kocaoglu O, Calvo RA, Sham LT, Cozy LM, Lanning BR, Francis S, Winkler ME, Kearns DB, Carlson EE, Selective Penicillin-Binding Protein Imaging Probes Reveal Substructure in Bacterial Cell Division. ACS Chemical Biology 7, 1746–1753 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie J, Huang X, Park MS, Pham HM, Chan WK, Differential suppression of the aryl hydrocarbon receptor nuclear translocator-dependent function by an aryl hydrocarbon receptor PAS-A-derived inhibitory molecule. Biochem. Pharm. 88, 253–265 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park MS, Chu F, Xie J, Wang Y, Bhattacharya P, Chan WK, Identification of cyclophilin-40-interacting proteins reveals potential cellular function of cyclophilin-40. Anal. Biochem. 410(2):257–265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Synthesis of FI-DNB1, FI-DNB2 and FI-MNB.
Fig. S2. Synthesis of Rd-DNB and Rd-MNB.
Fig. S3. Synthesis of TG-DNB1.
Fig. S4. Synthesis of TG-DNB2.
Fig. S5. Synthesis of CDG-DNB1.
Fig. S6. Synthesis of CDG-DNB2.
Fig. S7. Synthesis of CDG-DNB3
Fig. S8. SDS-PAGE analysis of purified M. smegmatis DprE1.
Fig. S9. DprE1 DNA sequence alignment.
Fig. S10. Microscopic imaging and flow cytometry analysis of bacteria labeled by TG and TG-DNB analogs.
Fig. S11. Characterization of TG-DNB2 in M. smegmatis.
Fig. S12. Growth curve of M. smegmatis and BCG.
Fig. S13. Inhibition study of TG-DNB2 with DNB1.
Fig. S14. Characterization of CDG-1 and CDG-DNB analogs in bacteria.
Fig. S15. Western blots detecting the expression of TEM-1 Bla and BlaC in transformed E. coli.
Fig. S16. Labeling of BCG and H37Rv by CDG-DNB2.
Fig. S17. Inhibition study with BlaC KO, BlaC compensated and control vector transformed H37Rv and DNB1.
Fig. S18. Labeling of Mtb in processed sputum by CDG-DNB2.
Fig. S19. Evaluation of selectivity of CDG-DNB2/3 for BlaC.
Fig. S20. Fluorescence intensity of CDG-DNB3 and TG-DNB2 with or without clavulanic acid, DNB1 or DprE1.
Fig. S21. Characterization of CDG-DNB3 in M. smegmatis, BCG and TEM-1 Bla or BlaC transformed E. coli.
Fig. S22. Specific labeling of viable BCG by CDG-DNB3.
Fig. S23. Labeling of Mtb in processed sputum by CDG-DNB3.
Fig. S24. Overall layout and the functional regions of the microfluidic bacteria counting device.
Table S1. List of NTMs stained with CDG-DNB3.
Table S2. List of individual subject-level data shown on bar graphs in all figures.
Movie S1. Real-time fluorescence imaging of CDG-DNB2 treated BCG aggregates.
Movie S2. Real-time bright-field imaging of CDG-DNB2 treated BCG aggregates.
Movie S3. Infection of macrophages by CDG-DNB3 labeled individual BCG bacilli.
Movie S4. Automated counting of CDG-DNB3 labeled BCG bacilli with microfluidic chip.
Appendix S1. 1H and 13C nuclear magnetic resonance and mass spectroscopy spectra.