Abstract
Oxylipins are bioactive mediators that play diverse roles in (patho)physiology. We developed a sensitive and selective ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) method for the simultaneous profiling of 57 targeted oxylipins derived from five major n-6 and n-3 polyunsaturated fatty acids (PUFAs) that serve as oxylipin precursors, including linoleic (LA), arachidonic (AA), alpha-linolenic (ALA), eicosapentaenoic (EPA), and docosahexaenoic (DHA) acids. The targeted oxylipin panel provides broad coverage of lipid mediators and pathway markers generated from cyclooxygenases, lipoxygenases, cytochrome P450 epoxygenases/hydroxylases, and non-enzymatic oxidation pathways. The method is based on combination of protein precipitation and solid-phase extraction (SPE) for sample preparation, followed by UPLC-MS/MS. This is the first methodology to incorporate four hydroxy-epoxy-octadecenoic acids and four keto-epoxy-octadecenoic acids into an oxylipin profiling network. The novel method achieves excellent resolution and allows in-depth analysis of isomeric and isobaric species of oxylipin extracts in biological samples. The method was quantitatively characterized in human plasma with good linearity (R = 0.990–0.999), acceptable reproducibility (relative standard deviation (RSD) < 20% for the majority of analytes), accuracy (67.8 to 129.3%) for all analytes, and recovery (66.8–121.2%) for all analytes except 5,6-EET. Ion enhancement effects for 28% of the analytes in tested concentrations were observed in plasma, but were reproducible with RSD < 17.2%. Basal levels of targeted oxylipins determined in plasma and serum are in agreement with those previously reported in literature. The method has been successfully applied in clinical and preclinical studies.
Keywords: Oxylipins, Ultra-performance liquid chromatography-tandem mass spectrometry, Quantitation characteristics, Lipidomics, Plasma
Introduction
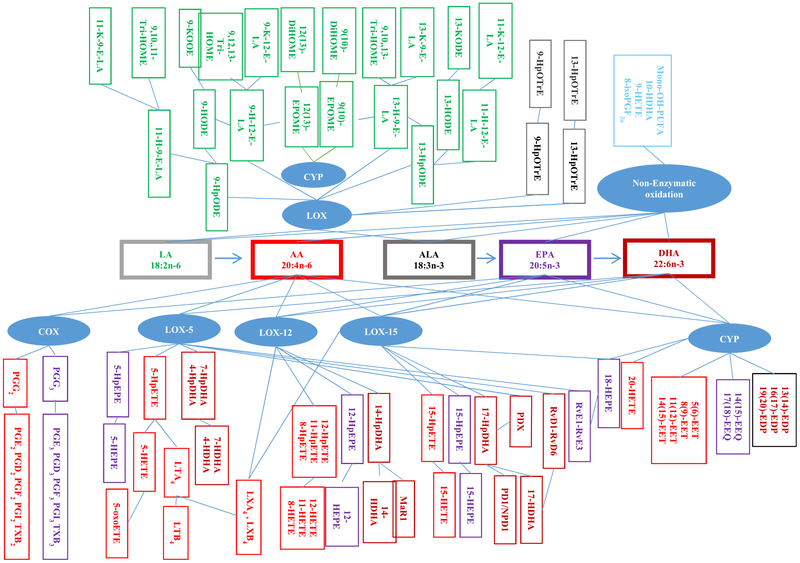
Oxylipins are biosynthesized by enzymatic and non-enzymatic oxidation of polyunsaturated fatty acids (PUFAs) forming eicosanoids from metabolism of 20-carbon arachidonic acid (AA) and eicosapentaenoic acid (EPA), 22-carbon docosanoids from docosahexaenoic acid (DHA), and 18-carbon oxidized metabolites of linoleic acid (LA) and linolenic acid (ALA). The three major enzymatic oxidation pathways comprise cyclooxygenase (COX), lipoxygenase (LOX), and cytochrome P450 (CYP) epoxygenases/hydroxylases. Non-enzymatic generation can occur via free radical-mediated oxidation by reactive oxygen/nitrogen species (ROS) [1, 2]. The metabolic pathways and networks used to generate oxylipins (Fig. 1) are dynamic and interconnected, often utilizing the same metabolic enzymes. Therefore, there may be competition with multiple n-6 and n-3 PUFAs acting as substrates for enzymes in these metabolic pathways. COX enzymes convert 20-carbon AA and EPA into endoperoxide prostaglandin PGG2 and PGG3, respectively, which are further metabolized to a variety of prostaglandins and thromboxanes (class 2 and 3 PGs and TXs are formed from AA and EPA, respectively) [1]. LOX enzymes (5-LOX, 12-LOX, 15-LOX) catalyze regio- and stereospecific oxidation to produce PUFA-hydroperoxides, which are then converted to leukotrienes, protectins (e.g., PD1/NPD1), maresins (e.g., MaR1), or mono-hydroxylated PUFAs (HODEs, HOTrEs, HETEs, HEPEs, HDHAs), which can be further oxidized to corresponding ketones (e.g., oxo-ODEs, oxo-ETEs) [1, 3, 4]. Sequential oxidation steps biotransform PUFAs to generate lipoxins, E and D series resolvins, and PDX (10S,17S-diHDHA) [4, 5]. CYP epoxygenase enzymes catalyze epoxidation giving rise to epoxides (e.g., EpOMEs, EETs, EEQs, EDPs) that are hydrolyzed by soluble epoxide hydro-lase (sEH) or in an acidic milieu to form dihydrodiols (e.g., DiHOMEs). CYP hydroxylases catalyze the formation of midchain hydroxides and ω-hydroxylation (e.g., 20-HETE, 18-HEPE) [1, 6]. The two-enzyme systems LOX and CYP can convert PUFAs to epoxyalcohols (e.g., hydroxy-epoxyoctadecenoic acids), which can be subsequently hydrolyzed to trihydroxy PUFAs (e.g., TriHOMEs) or further oxidized to keto-epoxy-octadecenoic acids (e.g., keto-epoxyoctadecenoic acids) [7, 8]. Non-enzymatic ROS-mediated reactions can generate the isoprostanes and neuroprostanes, such as 8-isoPGF2α derived from AA. These compounds and related ROS-generated DHA derivatives, which can be subsequently released by phospholipases, are considered to be reliable biomarkers for in situ oxidative stress [9]. Alternatively, ROS-mediated reactions can generate hydro-peroxides from unesterified and esterified PUFA; hydroperoxides are then rapidly reduced to mono-hydroxy PUFAs as racemic mixtures [10]. Furthermore, both levels of oxylipins and the composition of their precursor PUFAs in membrane phospholipids can be modulated by diet [1, 11–18].
Fig. 1.
Generation of oxylipins derived from n-6 LA and AA, and n-3 ALA, EPA, and DHA via lipoxygenase (LOX), cyclooxygenase (COX), cytochrome P450 (CYP), and non-enzymatic oxidation. Colors depict the precursor pathway metabolite families: LA (green), AA (red), ALA (tan), EPA (purple), DHA (dark red), LA, linoleic acid; AA, arachidonic acid; ALA, α-linolenic acid, EPA, eicosapentaenoic acid; DHA, docosahexaenoic acid; EpODE, epoxy-octadecadienoic acid; HODE, hydroxy-octadecadienoic acid; oxoODE, keto-octadecadienoic acid; EPOME, epoxy-octadecenoic acid; DiHOME, dihydroxy-octadecenoic acid, TriHOME, trihydroxy-octadecenoic acid; H-E-LA, hydroxyepoxy-octadecenoic acid; K-E-LA, keto-epoxy-octadecenoic acid; HPOTrE, hydroperoxy-octadecatrienoic acid, HOTrE, hydroxyoctadecatrienoic acid; PGG, hydroperoxide-endoperoxide prostaglandin; PG, prostaglandin; TX, thromboxane; LT, leukotriene; LX, lipoxin; HpETE, hydroperoxyeicosatetraenoic acids; HETE, hydroxypentaenoic acid; oxoETE, keto-eicosatetraenoic acid; EET, epoxyeicosatrienoic acids; HpEPE, hydroperoxyeicosapentaenoic acids; HEPE, hydroxyeicosapentaenoic acid; EEQ, epoxy eicosatetraenoic acid; HpDHA, hydroperoxy-docosahexaenoic acid, HDHA, hydroxyl-docosahexaenoic acid; EDP, epoxy docosapentaenoic acid; DiHDHA, hydroxydocosahexaenoic acid; PDX, 10S,17S-DiHDHA; PD/NPD, protectin; Rv, resolving
Oxylipins play important roles in physiology and are synthesized in a tightly regulated manner [19]. Alterations in oxylipin concentrations have been described for numerous diseases, including Alzheimer’s disease [20, 21], chronic pain [15, 22], diabetes [23], and cardiovascular disease [24]. Although these diseases are complex and multifactorial, they may share common pathological processes including neuroinflammation/inflammation [25], oxidative stress [2, 26], nociception [27], disrupted glucose metabolism [28, 29], and endothelial cell activation [30]. These processes consist of distinct biochemical cascades, but are closely intertwined with each other [27, 31–35]. Oxylipins are reported to stimulate, oppose, or otherwise modulate these biochemical cascades, and influence these processes in an interactive ways. Therefore, a targeted profiling platform capable of quantifying multiple precursors, bioactive mediators, and inactivation products can provide a more thorough understanding of these complex, interconnected molecular pathways.
Various methods including immunoassays, gas chromatography-mass spectrometry (GC-MS), and high performance liquid chromatography-ultraviolet detector (HPLCUV) have been applied in the analysis of oxylipins. These methods lack either specificity, and/or sensitivity, or require derivatization [36–40], and are capable of analyzing only a limited numbers of oxylipins. Due to the versatility, specificity, sensitivity, large dynamic range, and absence of a need for derivatization, the tandem LC-MS has become the method of choice for oxylipin determination [3, 12, 41–43], and also offers superior options to structural elucidation of metabolites [43]. Historically, there have been numerous LC-MS-based methods to quantify oxylipins, and often these methods are limited to detecting only a small number of eicosanoids generated from AA. These approaches often do not adequately take into account the importance and interconnectedness of metabolic networks. Furthermore, extensive validation or quantitative characterization is often lacking in these publications. Recently, tandem LC-MS lipidomic approaches have been widely used for profiling oxylipins in biological systems and to correlate the changes with biological responses. For instance, Yang et al. [43] performed profiling of 52 eicosanoids and docosanoids with extensive inflammatory and specialized pro-resolving mediators. Yang et al. [44] quantified 39 oxylipins originating from AA and LA including EETs and 20-HETE that regulate vascular function [45, 46]. Wolfer et al. [47] profiled 43 oxylipins generated from LA, EPA, DHA, and AA, including 8-isoPGF2α, a biomarker for oxidative stress. A few methods papers have targeted over 100 oxylipins for quantitative lipidomic investigation [3, 48, 49]. Although not as broad, our method constructed a 57 oxylipin panel that included lipid mediators and major pathway markers derived from COX, LOX, CYP epoxygenases/hydroxylases enzymes from three major PUFA precursors LA, AA, and DHA, 18-HEPE (the precursor of E-series resolving) and two EEQs from EPA, and two HOTrEs from ALA. These targeted oxylipins encompass not only pro-inflammatory eicosanoids, and anti-inflammatory and pro-resolving eicosanoids and docosanoids, but also mediators in regulation of nociceptive sensitivity [16], glucose metabolism [28] and vascular tone [30], as well as biomarkers of oxidative stress derived from AA (8-isoPGF2α) and DHA (10-HDHA). Most importantly, we incorporated two newly discovered LA-derived hydroxyepoxides, keto-epoxides, and triols into the novel methodology. To the best of our knowledge, we are the first to successfully separate and quantify isomeric groups of H-E-LAs and K-E-LAs.
We developed a sensitive and selective UPLC-MS/MS method for the simultaneous profiling of 57 targeted oxylipins derived from five major PUFAs including LA, AA, ALA, EPA, and DHA. In this method, the surrogate matrix, phosphate buffer saline-methanol containing BHT/EDTA cocktail (PMC) was used for calibration and calibrators were prepared by solid-phase extraction (SPE). The oxylipins were extracted from human plasma using protein precipitation, followed by SPE. The separation was performed on a reverse-phase C18 column. Detection was achieved using a QTrap 5500 mass spectrometer, employing the negative ion mode along with scheduled multiple reaction monitoring (sMRM). Additionally, the optimized method was characterized for linearity, limit of detection, lower limit of quantitation, precision, accuracy, extraction recovery, matrix effects, and autosampler stability. The method has been successfully applied to human clinical studies in plasma, serum, and skin matrices.
Method
Materials and chemicals
All HPLC solvents and reagents were purchased from Fisher Scientific Co. (Pittsburg, PA). Acetic acid (> 99%), formic acid (≥ 99%), and 7.5 M ammonium acetate solution (ACS reagent grade), ethylenediaminetetraacetic acid (EDTA, ≥ 99%), butylated hydroxytoluene (≥ 99%), and glycerol (≥ 99.5) were obtained from Sigma (Sigma-Aldrich, St. Louis, MO, USA). Solid-phase extraction cartridges were purchased from Phenomenex (Strata-X, 200 mg/mL; CA, USA). Deionized water (18 mΩcm) from a Millipore Milli-Q water purification system was used to prepare all aqueous solutions. Forty-seven non-deuterated and nine deuterated standards of target oxylipins were ordered from Cayman Chemicals (Ann Arbor, MI, USA), or Larodan (Malmo, Sweden) with purity of ≥ 98% according to the manufacturer. Protectin 1 (PD1), resolvin D3 (RvD3) and resovlin D4 (RvD4) were generous gifts from Dr. Charles Serhan at Brigham and Women’s Hospital, Harvard Medical School, Boston, MA with purity > 95%. Four hydroxy-epoxy-octadecenoic acids [9-hydroxy-12,13-epoxy-octadecenoic acid (9-H-12-E-LA), 11-hydroxy-12,13-epoxy-octadecenoic acid (11-H-12-E-LA), 11-hydroxy-9,10-epoxy-octadecenoic acid (11-H-9-E-LA), 13-hydroxy-9,10-epoxy-octadecenoic acid (13-H-9-E-LA)] and three keto-epoxy-octadecenoic acids [11-keto-12,13-epoxyoctadecenoic acid (11-K-12-E-LA), 11-keto-9,10-epoxyoctadecenoic acid (11-K-9-E-LA), 13-keto-9,10-epoxyoctadecenoic acid (13-K-9-E-LA)] employed in this study were custom-synthesized by Cayman Chemical with purities of > 95%. We conducted impurity interference checks by injection of individual standards and deuterated standards, and monitor other selected MRM channels in the initial stage of method development. Impurity interferences from reference standards at the selected MRM transitions were not found. The occurrences of analyte cross-talk interferences have been minimized and discussed in sections “Resolution of isomers to prevent inter channel cross-talk” and “Resolution of targeted hydroxy-epoxy-octadecenoic acids and keto-epoxyoctadecenoic acids” in Results and Discussion.
Research participants and plasma sample collection
Informed consent was obtained from healthy premenopausal female volunteers participating in an ongoing National Institute on Alcohol Abuse and Alcoholism (NIAAA) and National Institute of Health Clinical Center dietary protocol (NCT01251887). Baseline fasting whole blood was collected into 10 ml ethylenediaminetetraacetic acid (EDTA) tubes. Samples were immediately centrifuged at 2000g for 10 min at 25 °C with centrifuge on slow brake setting. Plasma aliquots were stored in a − 80 °C freezer until analysis.
Liquid chromatography and mass spectrometry
The Shimadzu chromatographic system consisted of two LC-20AD pumps, a SIL-20AC autosampler (4 °C), a CTO-20AC column oven (30 °C), and a CBM-20A controller (Shimadzu Scientific Instruments, Kyoto, Japan). A ZorBAX RRHD Eclipse Plus C18 column (100 mm × 4.6 mm; 1.8 μm, part number: 959964–902) was used. Mobile phase A consisted of 12 mM ammonium acetate solution and acetic acid (100:0.02 v/v). Mobile phase B contained 12 mM ammonium acetate and was composed of acetonitrile/water/acetic acid (90:10:0.02, v/v/v). The flow rate was 0.5 mL/min. The elution gradient conditions were as follows: 25–40% B from 0 to 2.0 min, 40–46% B from 2 to 8 min, 46–57% B from 8 to 9 min, 57–66% B from 9 to 20 min, 66–76% B from 20 to 22 min, 76–100% B from 22 to 27 min, held at 100% B from 27 to 33 min, 100–25% B from 33.1 to 35 min.
The UPLC system was coupled to a Qtrap 5500 mass spectrometer (AB Sciex, USA) via electrospray ionization (ESI) source and was operated in negative ion mode. The various compound-dependent parameters, namely declustering potential (DP), collision energy (CE), and collision cell exit potential (CXP) for MRM transitions of each analyte and internal standards were optimized by infusion of each standard solution. The optimum source parameters were as follows: curtain gas (CUR), 20 psi; nebulizer gas (GS1), 65 psi; turbo-gas (GS2), 70 psi; electrospray voltage, − 4500 V; turbo ion spray source temperature (TEM), 500 °C. Quantitative analysis was performed using scheduled multiple reaction monitoring (sMRM) scans with the retention time window of 90 s (Table 1). The MS/MS spectra were obtained by using enhanced product ion scan mode at a scan speed of 1000 Da/s. Collision-induced dissociation (CID) was performed using the collision energy of 35 V with collision energy spread of 10. Data processing was performed with Analyst Software™ (version 1.6.2).
Table 1.
Mass spectrometry parameters, MRM transitions, diagnostic ions, and retention times of targeted oxylipins and matching internal standards
| Compound | Retention time (min) | MRM transition | Qualitative diagnostic ions | DP | CE | CXP | IS | |
|---|---|---|---|---|---|---|---|---|
| Q1 | Q3 | |||||||
| 9-HODE | 21.30 | 295.2 | 170.9 | 207, 277 | −130 | −24 | −17 | 13-HODE-d4 |
| 13-HODE | 21.30 | 295.2 | 195.0 | 113, 277 | −140 | −30 | −0 | 13-HODE-d4 |
| 9-oxoODE | 23.89 | 293.2 | 185.1 | 125, 149 | −110 | −28 | −15 | 15-HETE-d8 |
| 13-oxoODE | 23.41 | 293.2 | 195.1 | 113, 177 | −120 | −28 | −13 | 15-HETE-d8 |
| 9,10-EpOME | 25.97 | 295.2 | 171.1 | 125, 183 | −100 | −22 | −13 | 15-HETE-d8 |
| 12,13-EPOME | 25.81 | 295.2 | 195.0 | 113, 183 | −65 | −26 | −9 | 15-HETE-d8 |
| 9,10-DiHOME | 14.49 | 313.2 | 201.1 | 125, 171 | −140 | −28 | −17 | LTB4-d4 |
| 12,13-DiHOME | 14.17 | 313.2 | 183.1 | 129, 195 | −115 | −28 | −13 | LTB4-d4 |
| 9,12,13-TriHOME | 6.64 | 329.2 | 211.1 | 119, 229 | −90 | −30 | −11 | LXA4-d5 |
| 9,10,13-TriHOME | 6.86 | 329.2 | 171.0 | 127, 139 | −100 | −30 | −13 | LXA4-d5 |
| 9,10,11-TriHOME | 8.59 | 329.2 | 201.1 | 139, 171 | −50 | −30 | −11 | LXA4-d5 |
| 9-HOTrE | 17.59 | 293.2 | 171.1 | 185, 277 | −105 | −22 | −11 | 13-HODE-d4 |
| 13-HOTrE | 18.24 | 293.2 | 195.0 | 205, 277 | −80 | −24 | −15 | 13-HODE-d4 |
| PGE2 | 7.49 | 351.2 | 271.2 | 189,217 | −15 | −24 | −15 | PGE2-d4 |
| PGF2α | 6.68 | 353.2 | 193.0 | 165, 247 | −50 | −34 | −13 | LXA4-d5 |
| 8-IsoPGF2α | 5.76 | 353.2 | 193.0 | 165, 247 | −50 | −34 | −13 | LXA4-d5 |
| TXB2 | 5.81 | 369.2 | 169.1 | 177, 195 | −100 | −15 | −15 | TXB2-d4 |
| LTB4 | 12.81 | 335.2 | 194.9 | 123, 151 | −60 | −22 | −15 | LTB4-d4 |
| LXA4 | 8.60 | 351.2 | 114.9 | 217, 235 | −70 | −20 | −9 | LXA4-d5 |
| LXB4 | 7.33 | 351.2 | 221.1 | 189,233 | −90 | −22 | −25 | LXA4-d5 |
| 5-HETE | 23.26 | 319.2 | 115.0 | 161,203 | −75 | −18 | −9 | 5-HETE-d8 |
| 8-HETE | 23.20 | 319.2 | 155.0 | 163, 257 | −80 | −20 | −11 | 15-HETE-d8 |
| 9-HETE | 23.46 | 319.2 | 123.0 | 167, 179 | −85 | −22 | −11 | 15-HETE-d8 |
| 11-HETE | 23.09 | 319.1 | 167.2 | 149, 195 | −65 | −22 | −11 | 15-HETE-d8 |
| 12-HETE | 23.63 | 319.2 | 135.0 | 163, 179 | −85 | −20 | −17 | 15-HETE-d8 |
| 15-HETE | 22.41 | 319.2 | 219.1 | 175, 203 | −75 | −18 | −17 | 15-HETE-d8 |
| 20-HETE | 18.77 | 319.2 | 245.1 | 163, 289 | −115 | −20 | −23 | 15-HETE-d8 |
| 5-oxoETE | 25.19 | 317.2 | 203.2 | 163, 175 | −90 | −24 | −15 | 15-HETE-d8 |
| 5,6-EET | 26.18 | 319.2 | 191.2 | 115, 163 | −25 | −16 | −13 | 5,6-EET-d11 |
| 8.9-EET | 26.66 | 319.2 | 155.0 | 123, 179 | −95 | −16 | −11 | 15-HETE-d8 |
| 11,12-EET | 26.62 | 319.2 | 208.0 | 167, 179 | −105 | −20 | −15 | 15-HETE-d8 |
| 14,15-EET | 25.96 | 319.2 | 219.1 | 113, 175 | −90 | −16 | −15 | 14,15-EET-d11 |
| 18-HEPE | 18.07 | 317.2 | 259.1 | 161,215 | −70 | −16 | −21 | 13-HODE-d4 |
| 14,15-EEQ | 24.12 | 317.2 | 207.0 | 163, 175 | −75 | −18 | −19 | 15-HETE-d8 |
| 17,18-EEQ | 23.14 | 317.1 | 215.2 | 201, 259 | −90 | −14 | −17 | 15-HETE-d8 |
| 4-HDHA | 23.42 | 343.2 | 101.0 | 133, 241 | −95 | −18 | −7 | 15-HETE-d8 |
| 7-HDHA | 22.79 | 343.2 | 141.1 | 113, 201 | −70 | −18 | −11 | 15-HETE-d8 |
| 10-HDHA | 22.71 | 343.2 | 153.1 | 161,261 | −95 | −22 | −17 | 15-HETE-d8 |
| 14-HDHA | 22.96 | 343.2 | 205.1 | 161,233 | −85 | −16 | −17 | 15-HETE-d8 |
| 17-HDHA | 22.45 | 343.2 | 245.1 | 147, 201 | −85 | −20 | −15 | 15-HETE-d8 |
| PDX (10S,17S-DiHDHA) | 12.86 | 359.1 | 153.0 | 137, 261 | −90 | −22 | −13 | LTB4-d4 |
| PD1/NPD1 | 12.47 | 359.2 | 153.0 | 137, 261 | −115 | −20 | −13 | LTB4-d4 |
| MaR1 | 12.58 | 359.2 | 250.1 | 177, 221 | −90 | −20 | −19 | LTB4-d4 |
| RvD1 | 8.95 | 375.2 | 215.1 | 141, 233 | −70 | −26 | −15 | LXA4-d5 |
| RvD2 | 7.75 | 375.2 | 141.0 | 175, 215 | −70 | −26 | −15 | LXA4-d5 |
| RvD3 | 7.28 | 375.3 | 147.1 | 137, 181 | −145 | −26 | −39 | LXA4-d5 |
| RvD4 | 10.06 | 375.3 | 101.0 | 225, 131 | −75 | −28 | −9 | LXA4-d5 |
| 16,17-EDP | 25.96 | 343.2 | 245.2 | 147, 201 | −100 | −15 | −15 | 15-HETE-d8 |
| 19,20-EDP | 25.30 | 343.2 | 241.2 | 133, 187 | −115 | −16 | −15 | 15-HETE-d8 |
| 11-H-9-E-LA 1 (3c) | 14.61 | 311.2 | 201.1 | −90 | −24 | −29 | ||
| 11-H-9-E-LA2 (3d) | 15.41 | 311.2 | 201.1 | 171, 185 | −90 | −24 | −29 | LTB4-d4 |
| 13-H-9-E-LA | 14.21 | 311.2 | 171.0 | 139,211 | −90 | −28 | −15 | LTB4-d4 |
| 9-H-12-E-LA | 14.52 | 311.2 | 193.0 | 171,211 | −110 | −24 | −13 | LTB4-d4 |
| 11-H-12-E-LA 1 (3 g) | 14.81 | 311.2 | 197.1 | −80 | −24 | −15 | ||
| 11-H-12-E-LA 2 (3 h) | 15.46 | 311.2 | 197.1 | 181,211 | −90 | −26 | −15 | LTB4-d4 |
| 11-K-9-E-LA 1 (3n) | 19.85 | 309.1 | 185.1 | −60 | −22 | −9 | ||
| 11-K-9-E-LA2 (3o) | 22.14 | 309.1 | 185.1 | 155, 171 | −140 | −22 | −9 | 13-HODE-d4 |
| 11-K-9-E-LA3 (3p) | 22.96 | 309.1 | 185.1 | −150 | −22 | −9 | ||
| 13-K-9-E-LA 1 (3 l) | 19.21 | 309.1 | 171.1 | −150 | −22 | −15 | ||
| 13-K-9-E-LA 2 (3 m) | 20.18 | 309.1 | 171.1 | 137, 155 | −150 | −22 | −15 | 13-HODE-d4 |
| 9-K-12-E-LA | 18.49 | 309.1 | 209.2 | 165, 193 | −70 | −26 | −15 | 13-HODE-d4 |
| 11-K-12-E-LA 1 (3r) | 19.51 | 309.1 | 181.2 | −80 | −28 | −9 | ||
| 11-K-12-E-LA 2 (3 s) | 21.67 | 309.1 | 181.2 | 151,209 | −150 | −21 | −9 | 13-HODE-d4 |
| 11-K-12-E-LA 3 (3 t) | 22.53 | 309.1 | 181.2 | −140 | −22 | −9 | ||
| 13-HODE-d4 | 21.25 | 299.2 | 198.0 | 198 | −120 | −26 | −15 | |
| PGE2-d4 | 7.40 | 355.2 | 275.3 | 275 | −75 | −24 | −23 | |
| TXB2-d4 | 5.71 | 373.2 | 173.0 | 173 | −85 | −22 | −11 | |
| LTB4-d4 | 12.71 | 339.2 | 197.0 | 197 | −140 | −22 | −15 | |
| 5-HETE-d8 | 23.16 | 327.2 | 116.0 | 116 | −70 | −18 | −15 | |
| 15-HETE-d8 | 22.31 | 327.2 | 226.1 | 226 | −30 | −18 | −17 | |
| LXA4-d5 | 8.50 | 356.0 | 115.1 | 115 | −70 | −20 | −9 | |
| 14,15-EET-d11 | 25.90 | 330.3 | 219.2 | 219 | −100 | −15 | −15 | |
| 5,6-EET-d11 | 26.10 | 330.3 | 202.2 | 202 | −100 | −15 | −15 | |
DP, declustering potential; CE, collision energies; CXP, collision cell exit potential; IS, internal standard
Preparation of standard and quality control solutions
Stock solutions of standard compounds were prepared in ethanol at a concentration of 100 ng/μL for all analytes except 1000 ng/uL for 9-HODE and 9-HOTrE. An intermediate stock solution of the 50 analytes was prepared at 1 ng/uL for all analytes except 10 ng/uL for 9-HODE and 9-HOTrE by mixing stock solutions of each analyte in methanol. We used methanol to make standard working solutions since methanol is the common organic solvent used for the preparation of oxylipin calibration standards [44, 49, 50]. Also, it has been reported that the solubilities in methanol are 3.04 g/L at −70 °C for LA [51], 13.8 g/L at − 62 °C for ALA [51], and 50 mg/ml for AA and EPA (Product Information Sheet from Sigma-Aldrich). DHA is soluble in methanol (LipidBank DHA). Oxylipins are oxidative metabolites of PUFA and are known to be more polar than their respective precursor PUFA. The increased polarities cause them to dissolve more readily in methanol based on the principle that “like dissolve like.” Therefore, oxylipin solubilities in methanol at the concentrations used in our work will not be a problem. Calibrator working solutions were prepared by dilution from this intermediate stock solution with methanol to achieve concentrations of 0.2, 0.5, 1, 2, 5, 10, 20, 50, 100, 200, 500, and 1000 ng/mL for all analytes except 2, 5, 10, 20, 50, 100, 200, 500, 1000, 2000, 50,000, and 10,000 ng/mL for 9-HODE and 9-HOTrE. The quality control (QC) working solutions were prepared separately in the same fashion at five concentrations of 2, 5, 10, 100, and 800 ng/mL for all analytes except 20, 50, 100, 1000, and 8000 ng/mL for 9-HODE and 9-HOTrE. Both working solutions of calibrators and QCs were aliquoted. All stock and working solutions were stored in amber-glass vials at − 80 °C.
A mixture of nine deuterated working internal standard (IS) solutions were similarly prepared by each individual stock (100 ng/μL) of internal standard to give a final concentration of 20 ng/mL in methanol. To simulate the extraction of the endogenous compounds, nine deuterated compounds were used as internal standard (IS) and added to samples before centrifugation. For each analyte, a suitable IS was selected based on structural similarities and retention time (Table 1).
Preparation of calibration standards and QC PMC samples
Oxylipins are endogenously present in plasma, where they can reach relatively high concentrations (10–20 μM for certain species) and no analyte-free matrix is available. Therefore, phosphate buffer saline (PBS)-methanol containing BHT/EDTA was used as a surrogate matrix (referred to as PMC). The PMC matrix was freshly prepared by mixing 200 μL of PBS (100 mM) and 500 μL of cold methanol containing 20 mg/mL BHT/EDTA in an ice bath. The calibration standards were prepared by spiking with 20 μL of the appropriate calibrator working solution into 700 μL of PMC in Eppendorf tube kept in ice, resulting in the calibration concentrations of 0.02, 0.05, 0.1, 0.2, 0.5, 1, 2, 5, 10, 20, 50, 100 ng/mL for all analytes except 0.2, 0.5, 1, 2, 5, 10, 20, 50, 100, 200, 500, and 1000 ng/mL for 9-HODE and 9-HOTrE in the final solution. The QC samples in PMC were prepared in the same way as the calibrators. The final concentrations of QC samples at five levels were 0.2, 0.5, 1, 10, and 80 ng/mL for all analytes except 2, 5, 10, 100, and 800 for 9-HODE and 9-HOTrE in the final solution. A volume of 50 μL of the IS working solution was added resulting in 20 ng/mL in the final solution. The samples were subjected to solid-phase extraction (SPE) as described in Section 1.7.
Preparation of plasma samples and QC plasma samples
All QC plasma samples used for the evaluation of quantitative properties were prepared from the pooled plasma to ensure the same basal levels of analytes in question.
On the day of sample analysis, the frozen plasma samples (stored at − 80 °C) were thawed on ice. The thawed samples were vortexed to ensure complete mixing of contents. A volume of 200 μL of sample was pipetted into 1.5 mL Eppendorf tubes and 500 μL of cold methanol containing 20 mg/mL BHT/EDTA was immediately added. The samples were vortex-mixed followed by placing them at − 80 °C for 30 min. After removing from the − 80 °C freezer, 50 μL of the IS working solution and 20 μL of methanol were added to all samples. For QC plasma samples, 50 μL of the IS working solution and 20 μL of QC solutions were added in QC plasma samples. Samples were then centrifuged at 14,000 rpm for 10 min at 4 °C and were further processed for solid-phase extraction (SPE) as described in Section 1.7.
Solid-phase extraction
Oxylipins were extracted using polymer-based reversed-phase Strata-X cartridges (33 μm, 200 mg/6 mL; Phenomenex, PA). All SPE steps were performed via gravity flow. The cartridge was preconditioned with 6 mL methanol and 6 mL water. Just before loading each sample, the supernatant from plasma sample obtained after centrifugation or standards in the final solution was transferred with glass Pasteur pipette to pre-cooled Kimble glass centrifuge tubes containing 6 mL of cold water in an ice bath and were vortexed briefly. The diluted supernatant was immediately decanted into a SPE cartridge and passed through the cartridge. The cartridge was then washed with 6 mL of 10% methanol in water and air-dried for approximately 2 min. The remaining washing solution in the cartridge tip was removed. Afterwards, the metabolites were eluted with 6 mL methanol containing 0.0004% w/v BHT into a glass tube containing 10 μL of 30% glycerol in methanol. The eluted solutions were evaporated under a stream of nitrogen to dryness at 30 °C. The residues were reconstituted with 40 μL of methanol, transferred to amber autosampler vials, and an aliquot (10 μL) was injected into the LC-MS/MS system.
Container transfer loss test
Due to the lipophilic nature of oxylipins and lack of protein in PMC matrix, there may be non-specific bindings which result in the loss of analytes through adsorption, especially to the hydrophobic surface of the polypropylene materials during transfer with tubes, vials, and tips in sample preparation. The experiments to check for analyte loss during container transfer were performed in the PMC matrix at concentration levels of 0.1 and 0.2 ng/mL and 1 ng/mL for all analytes and 0.5 ng/mL for RvD2, 11-H-9-E LAs, and 11-H-12-E LAs (n = 4 for each). The sample preparation procedure was the same as that detailed in Sections 1.5 and 1.7. Briefly, 700 μL of the freshly prepared cold PMC solution was added in an Eppendorf tube placed in ice, followed by the addition of 20 μL of the standard solution and 50 μL of the IS working solution without mixing. The solution was transferred with a glass Pasteur pipette into a pre-cooled Kimble glass centrifuge tube containing 6 mL of cold water in an ice bath just prior to SPE loading, then vortexed by setting the speed control to the position 8 (approximately 2600 rpm) with touch function on for ~ 5 s using a VORTEX GENIE 2 mixer. The solution was decanted into the preconditioned cartridge with 6 mL of methanol and 6 mL of water. After the samples had passed through the cartridges, they were washed with 6 mL of 10% methanol and eluted with 6 mL of methanol containing 0.0004% w/v BHT into a glass tube that had 10 μL of 30% glycerol in methanol. Samples were dried under a stream of nitrogen. The analyte to IS peak area ratios of pre-spiked PMC samples at these concentrations were compared with those of the spiked samples after the extraction procedure. The results were expressed as percent recoveries:
Quantitation characteristics
The method was evaluated for limit of detection, limit of quantitation, lower limit of quantitation, linearity, precision, accuracy, extraction recovery, matrix effects, and autosampler stability. All samples were processed as described in Sections 1.4 to 1.7.
Limit of detection and quantitation and lower limit of quantitation
The limits of detection (LODs), limits of quantitation (LOQs), and lower limits of quantitation (LLOQs) of the method were determined from a series of calibration standard solutions in PMC matrix going through SPE procedure. LODs were defined as the concentration at which the analytes had signal-to-noise ratios S/N > 3 and LOQs were defined as the lowest point in the calibration curve that have an S/N > 5 and intraday RSD < 25% (n = 4). LLOQs were set to be concentrations in calibration curve at which the analyte peaks are reproducible with RSD of < 20% and accuracy of 70–130%.
Linearity and range
The linearity of the method was assessed with calibration samples of 7 to 12 different concentrations. The calibration curves were constructed using 12 calibration standards (0.02–100 ng/mL) in PMC as described in Sections 1.4 and 1.6. Not all levels were included for each analyte. The calibration curves were constructed by plotting area ratio of each analyte/IS against the corresponding concentration of the analyte and fitting the data using linear regression with the weighting factor of 1/x. The range of the assay was established as the section of the calibration curve where the curve was linear.
In cases of two H-E-LAs (11-H-9-E-LA and 11-H-12-ELA), each standard was composed of two major isomer peaks. Quantitative results of these compounds were expressed as the sum of its related components since the amount in the calibration standard was provided as total of the peaks. The calibration curve was prepared in Microsoft Excel by sum peak area ratios of its related peak area component/peak area IS generated from Analyst 1.6.2 and plotted the best fit of total peak area ratios of analyte/peak area IS vs concentration, and fitted to the equation y = ax + b.
Autosampler stability in PMC solution and plasma extract
Stability of analytes was determined by injecting processed QC samples (n = 4) on three levels (QC3, QC4, and QC5) from PMC or pooled plasma. These QC samples were retained in the autosampler at 4 °C for 24 h prior to reanalysis. The analyte stability was calculated as: (mean peak area ratio of samples at T = 24 h/mean peak area ratio of samples at T = 0 h)*100%.
Precision and accuracy
Method accuracy and precision were evaluated using QC samples processed in PMC solution and pooled plasma. Intraday precision and accuracy was determined by analyzing QC standards at QC concentration levels (n = 4) of four (QC1, QC3, QC4, and QC5) and five (QC1, QC2, QC3, QC4, and QC5) in PMC and plasma, respectively. Interday precision and accuracy were carried out in four replicates by running the same four levels of QC standards in PMC matrix and five levels in plasma on four or five different days. The precision was expressed as the RSD of the intraday and interday replicate analysis for each QC sample. The accuracy was expressed as (mean calculated concentration)/(nominal concentration) × 100. For accuracy of plasma oxylipin measurements, oxylipin concentrations were corrected by subtracting the endogenous concentration from each sample.
Recovery
The recovery of the analytes was performed in three or four replicate samples of four QC levels (QC1, QC3, QC4, and QC5) in PMC and five QC levels (QC1, QC2, QC3, QC4, and QC5) in plasma. For recovery in plasma, three sets of samples were prepared and concentrations (C1 to C3) were measured: set 1 consisted of pooled plasma samples (pre-extraction non-spiked, C3), set 2 consisted of post-extraction spiked samples (plasma was extracted and then spiked with standards, C1), and set 3 consisted of pre-extraction spiked samples (C2). The recovery was calculated according to the following equation:
For determining recovery in PMC, two sets of samples were prepared and concentrations (C1 and C2) were measured: set 1 consisted of post-extraction spiked samples (PMC matrix was extracted and then spiked with standards, C1) and set 2 consisted of pre-extraction spiked PMC samples (C2). The recovery was calculated according to the above Eq. (C3 = 0).
Matrix effect
The matrix effect was evaluated at three QC concentrations (QC3, QC4, and QC5) by comparing the concentration calculated from post-extraction spiked samples prepared in plasma that corrected with endogenous concentration with the concentration calculated post-extraction spiked samples prepared in PMC (n = 3 or 4).
Results and discussion
Method development
The aim of this study was to develop a sensitive method for the quantitation of 57 oxylipins derived from LA (n = 19), AA (n = 19), ALA (n = 2), EPA (n = 3), and DHA (n = 14) from plasma in a single UPLC-MS/MS run. Despite recent advances in LC-MS, development of analytical methods for lipidomic profiling of oxylipins from biological matrices has been challenging due to low concentrations, wide dynamic range of their physiological concentrations, interference from complex matrices, high structural similarity, and high number of isomeric and isobaric species of oxylipins. The combination of selective mass detection and chromatographic separation is crucial to achieve the necessary selectivity and sensitivity of the method and to ensure reliable and accurate quantitation.
MS optimization
MS instrument parameters were tuned for optimal sensitivity. The compound specific parameters (DP, CE, and CXP) were tuned for selected precursor-product pairs by syringe pump infusion using individual authentic standards. For each analyte, the selected transition ion for quantification relied on the specificity and intensity of the transition ion. Source parameters (ion spray voltage, gas settings GS1 and GS2, and temperature) were optimized by coupling the UPLC-MS/MS conditions with flow injection at the analytical flow rate, and optimum values were selected for the best overall selectivity and resolution of the most compounds. Additionally, our method implemented scheduled MRM (sMRM) that restricts scans to compound elution time; therefore, the sensitivity of the assay was further enhanced. A complete list of analytes and internal standards together with retention times, MRM transitions, the two diagnostic ions, and the detailed mass spectrometric parameters is summarized in Table 1. The diagnostic ions are used to confirm analyte identity when needed.
LC optimization
Liquid chromatography was optimized to achieve the best possible separation for critical isomeric and isobaric analytes. LC separation of these analytes was investigated by using several reverse-phase C18 columns. The mobile phases were tested with different organic modifiers (acetonitrile and acetonitrile/water mixtures at various concentrations in water) and a low amount of formic acid or acetic acid. The buffer strength of the mobile phases was investigated with ammonium formate and ammonium acetate. Ammonium acetate buffer provided better separation of isomers and was investigated for improving the resolutions of H-E-LAs or K-E-LAs. Four different concentrations of ammonium acetate: 4, 10, 12, and 15 mM were tested in the mobile phase. We found that 12 mM ammonium acetate yielded the best separation. Higher buffer concentrations showed no significant enhancement of resolution for the analytes. Additionally, the gradient was optimized by modulating the mobile phase composition and flow rate. Finally, the optimal UPLC separation of the target oxylipins was achieved using an ZorBAX RRHD Eclipse Plus C18 column filled with 1.8 μm particles and a gradient elution with mobile phases 0.02% aqueous acetic acid and acetonitrile/water/acetic acid (90:10/0.02 v/v/v) both containing 12 mM ammonium acetate. A representative chromatogram of 57 targeted oxylipins injected on the UPLC-MS/MS system is presented in Electronic Supplementary Material (ESM) Fig. S1.
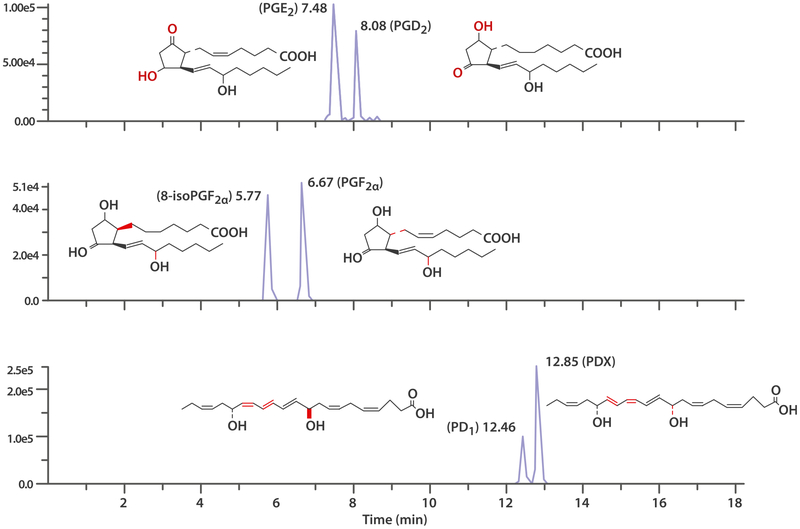
Resolution of isomers to prevent inter channel cross-talk
When highly specific LC-MS/MS MRM with unique transition is used, the chromatographic separation is often not required such as in co-eluting pairs of regioisomers, 5-HETE and 8-HETE, 9-HETE and 15-HETE, 8,9-EET and 12-EET, 7-HDHA and 10-HDHA. For most of the isomers, specific MRM transitions were carefully selected to avoid cross-talk. This has the advantage of shortened run times and increased throughput. However, a significant limitation of commonly deployed mass spectrometric assays is the inability to differentiate closely related isomeric and isobaric compounds with similar fragmentation patterns. The chromatographic separation is necessary for independent quantitation. While these structures may appear quite similar, biological functions of these compounds can be profoundly different, as previously reported for PD1 and PDX [52]. Thus, isomeric pairs need to be resolved and separately characterized in order to answer certain biological questions. Our method achieved excellent chromatographic separation of the target oxylipins with similar MS/MS spectra. For example, PD1 and PDX differ in double bond geometry and stereochemistry of the hydroxyl group at C10 position; PGF2α and 8-isoPGF2α stereoisomers differ in the configuration of the C7–C8 bond; and PGE2 and PGD2 regioisomers bear similar major fragment ions (Fig. 2). Additionally, we also achieved baseline resolution for the isomers sharing the same MRM transition for quantitation such as LXA4 and LXB4 (351 > 115), RvD1 and RvD2 (375 > 215), and RvD3 and RvD4 (375 > 147) with minimal cross-talk. Moreover, isobaric species, such as hydroxy-PUFA, were all baseline resolved from corresponding epoxy-PUFA. Two pairs of regioisomers, 9-HETE and 11-HETE (319 > 167), 7-HDHA and 17-HDHA (343 > 245) were nearly baseline separated. However, 9,10,13-TriHOME and 9,12,13-TriHOME (329 > 171) only partially resolved. Furthermore, for coeluting isomers 9-HODE and 13-HODE, and partially resolved isomers 9-EpOME and 13-EpOME, 9-HODE and 9-EpOME were each found to produce a very minor peak at m/z 195, forming an MRM transition 295 > 195, corresponding to less than 1% of the response at the same concentration level for 13-HODE and 13-EpOME. To our knowledge, few LC-MS/MS assays reported in the literature for the quantitation of PD1 have achieved baseline separation of PD1 and PDX without using a chiral column. We do not yet know whether other isomers AT-NPD1, 10S, 17R-DiHDHA, and 10S/R-17R-DiHDHA can be separated under the current LC condition since the standards are not commercially available. Mas et al. obtained a partial chromatographic resolution for PD1 and PDX by means of LCMS/MS system consisting of a Zorbax Eclipse XDB C18 column and 5 mmol/L ammonium acetate pH 9 and methanol as the mobile phase [53].
Fig. 2.
Chromatographic separation of critical isomer pairs: PGE2 and PGD2 (upper), PGF2α and 8-isoPGF2α (middle), and PD1 and PDX (bottom). Analytes were separated by UPLC using an Agilent ZorBAX RRHD Eclipse Plus C18 UPLC column and detected by MS/MS-MRM based on specific product ion(s) from each of the analytes. Experimental details in Section of Liquid chromatography and mass spectrometry
Resolution of targeted hydroxy-epoxy-octadecenoic acids and keto-epoxy-octadecenoic acids
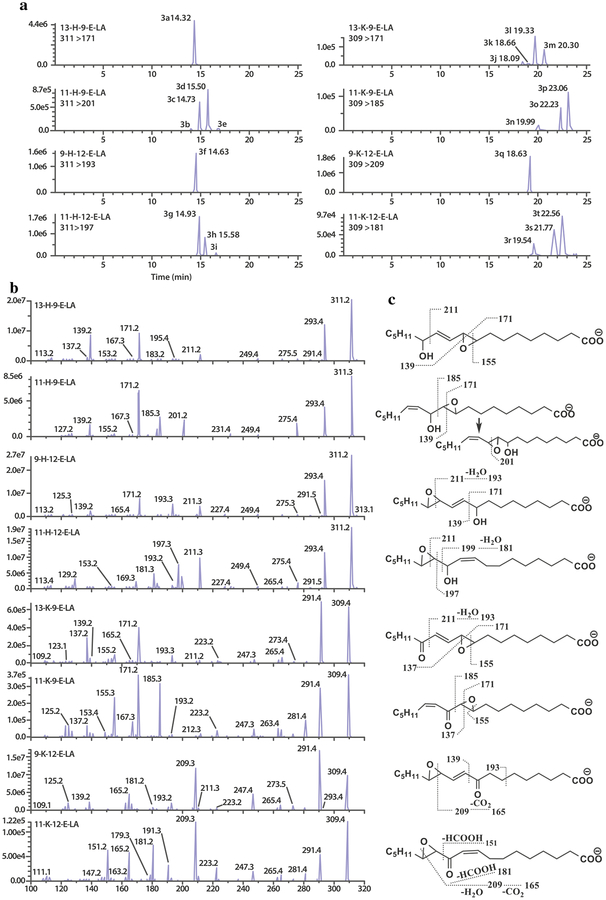
Seven out of eight targeted hydroxy-epoxy-octadecenoic acid (H-E-LAs) and keto-epoxy-octadecenoic acid (K-E-LAs) are not commercially available. To our knowledge, tandem LCMS methods have not been previously used for systematic analysis of these H-E-LAs and K-E-LAs. Two methods have been described for partial analysis of H-E-LAs or K-E-LAs, one for quantifying 9-K-12-E-LA in human plasma [54] and the other for separating four stereo-isomeric 13-H-9-E-LA enantiomers using SP-HPLC analysis on a silica column [55].
Analysis of the two groups of H-E-LAs and K-E-LAs, four regio-, geometric and stereoisomers for each group, presents a challenge due to not only structural similarity within the group, but also being racemic mixtures for each synthesized standard. Each synthesized compound was prepared selectively with respect to double bond and epoxide geometry; however, each chiral center was prepared non-selectively so that hydroxyls were racemic and absolute configuration of epoxides was not determined. The structures, extracted ion chromatograms, MS/MS spectra and fragmentation patterns are displayed in Fig. 3. H-E-LAs contain one chiral alcohol and both hydroxy- and keto-epoxide LAs have an epoxy ring as well as a double bond. Therefore, each H-E-LAs and K-ELAs can theoretically have eight or four structural isomers, respectively, although based on our predictions, there would be expected to be only four isomers of hydroxy-epoxides and two for each keto epoxide [56, 57]. Under our UPLC-MS/MS system, five out of seven synthesized standards were observed to contain a mixture of the related isomers (Fig. 3A): four peaks for 11-H-9-E-LA (3b, 3c, 3d, 3e), three for 11-H-12-ELA (3g, 3h, 3i), four for 13-K-9-E-LA (3j, 3k, 3l, 3m), three for 11-K-9-E-LA (3n, 3o, 3p), and three for 11-K-12-E-LA (3r, 3s, 3t), and one peak for both 13-H-9-E-LA (3a) and 9-H-12-E-LA (3f). These single peaks could be the sum of contributions from more than one respective isomer. Additional (>2) peaks that were observed for certain keto-epoxides could potentially be introduced through double bond isomerization during sample preparation and storage. Each peak of H-E-LAs and K-E-LAs in Fig. 3A showed a deprotonated molecular ion of 311 and 309, respectively. The peaks within each compound yielded identical MS/MS spectra; thus, only one representative spectrum for each compound is shown in Fig. 3B along with fragmentation patterns of these compounds shown in Fig. 3C. As noted, MS/MS spectra of H-E-LAs or K-E-LAs indicate that compounds within each group share many identical product ions (Fig. 3B). For instance, the major product ions at m/z 139, 171, and 211 of 13-H-9-E-LA (3a) are the identical major ions shown in 9-H-12-E-LA spectrum (3f). Quantitation of these two isomers requires chromatographic resolution as shown in Fig. 3A of 3a and 3f. In the cases of closely eluted peaks 3c (11-H-9-E-LA) and 3f (9-H-12-ELA); and 3d, 3e (11-H-9-E-LA) and respective 3h, 3i (11-H-12-E-LA), the corresponding characteristic ions at m/z 201, 193, and 197, were chosen as MRM transition ions for 11-H-9-E-LA, 9-H-12-E-LA, and 11-H-12-E-LA, respectively, to allow their differentiation and quantitation.
Fig. 3.
(a) Representative UPLC-MS/MS MRM chromatograms of H-E-▶LAs and K-E-LAs from a standard mixture solution using a ZorBAX RRHD Eclipse Plus C18 UPLC column with established gradient elution. (b) Representative MS/MS spectra of H-E-LAs and K-E-LAs. The H-ELAs (m/z 311) and K-E-LAs (m/z 309) undergo collision-induced fragmentation to produce product ion mass spectra. Only one spectrum for each compound is shown since peaks within each compound yielded identical MS/MS spectra. Experimental details refer to Section of Liquid chromatography and mass spectrometry. (c) Fragmentation patterns of H-E-LAs and K-E-Las
Similarly, for the K-E-LA group, the three major fragment ions of 13-K-9-E-LA (m/z 137, 155, 171) and of 9-K-12-ELA (m/z 165, 193, 209) are all present in the fragment patterns of 11-K-9-E-LA and 11-K-12-E-LA, respectively. Therefore, chromatographic separations of peaks between 13-K-9-E-LA and 11-K-9-E-LA, and 9-K-12-E-LA and 11-K-12-E-LA are required for quantitation. In the present method, each isomer peak of K-E-LAs was separated from others as indicated in Fig. 3A from peak 3j to 3t. Overall, good chromatographic separations are essential in this work for assuring accuracy in analyses of these isomers. With one standard containing several possible isomers, it is possible that each may have different biological activity. The mass spectra analyses do not allow for discrimination between these conformational isomers of related peaks. Future studies are required to isolate the individual isomers and to characterize each isomer by NMR to obtain absolute configuration.
Sample preparation
Many common sample preparation methods including protein precipitation, liquid-liquid extraction, and SPE have been used for analysis of oxylipins. Protein precipitation (PPT) does not provide enough sensitivity for low level of analytes in the pg/mL range. Liquid-liquid extraction (LLE) with organic solvents provides poor extraction of hydrophilic compounds such as prostaglandins and leukotrienes. SPE has been shown to provide a better purification and enrichment than LLE [58].
The most common sample preparation method used to minimize matrix effects and increase sensitivity in oxylipin profiling is methanol precipitation followed by SPE. C18-bonded silica or polymeric sorbent (Strata-X and Oasis HLB) SPEs are used most often. Sample loading solutions are usually slightly acidified to prevent ionization of these acidic analytes, which would reduce their retention [43, 59, 60]. During method development, we observed that acidification of the loading solution to pH 4 with 1 M HCL resulted in a loss of approximately 70% of 13-H-9-E-LA (data not shown). This observed loss is likely due to the instability of epoxyalcohols under acidic conditions, with hydrolysis to vicinal diols [61]. By contrast, neutral loading without acidification of the loading solution greatly improved recovery of 13-H-9-E-LA (86.9 to 107.1% across concentration range) and achieved good recoveries for all other analytes except 5,6-EET (Table 2), although these acidic analytes would likely be charged at a neutral condition. Good analyte retention could be attributed to properties of hydrophilic-lipophilic-balanced and higher capacity of polymeric sorbent cartridges. Thus, the final SPE extraction was performed under neutral conditions.
Table 2.
Recovery of targeted oxylipins from quality control samples at five different concentration levels (n = 3 or 4)
| Compound | LQC2 (0.5 ng/mL)* | LQC2 (0.5 ng/mL)* | QC3 (1 ng/mL)* | QC4 (10 ng/mL)* | QC5 (80 ng/mL)* | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PMC | Plasma | Plasma | PMC | Plasma | PMC | Plasma | PMC | Plasma | ||||||||||
| RC | RSD% | RC | RSD% | RC | RSD% | RC | RSD% | RC | RSD% | RC | RSD% | RC | RSD% | RC | RSD% | RC | RSD% | |
| 9-HODE | 98.4 | 1.8 | 89.8 | 15.1 | 93.3 | 13.4 | 98.9 | 12.4 | 95.1 | 7.7 | 93.4 | 5.6 | 99.6 | 10.5 | 102.6 | 4.6 | 88.4 | 4.8 |
| 13-HODE | NQ | NA | NQ | NA | 97.3 | 11.0 | 103.5 | 6.8 | 99.9 | 14.5 | 94.9 | 7.8 | 104.0 | 4.1 | 103.7 | 7.5 | 87.1 | 9.3 |
| 9-oxoODE | 109.5 | 17.0 | 87.3 | 8.2 | 95.9 | 7.7 | 102.8 | 15.7 | 99.4 | 9.8 | 99.3 | 2.2 | 101.7 | 11.9 | 103.9 | 11.7 | 80.8 | 14.3 |
| 13-oxoODE | 90.1 | 21.9 | 103.2 | 10.9 | 99.6 | 11.1 | 101.7 | 14.0 | 89.6 | 15.1 | 98.1 | 3.7 | 100.0 | 10.1 | 104.6 | 11.5 | 82.0 | 11.0 |
| 9,10-EpOME | NQ | NA | NQ | NA | NQ | NA | 96.0 | 7.9 | 114.2 | 19.2 | 92.5 | 2.6 | 101.7 | 12.7 | 104.1 | 8.8 | 86.0 | 4.0 |
| 12,13-EPOME | NQ | NA | NQ | NA | NQ | NA | 99.4 | 15.2 | 101.9 | 13.4 | 91.9 | 2.9 | 100.0 | 18.1 | 103.0 | 10.4 | 83.8 | 8.3 |
| 9,10-DiHOME | 102.7 | 4.6 | 82.4 | 12.9 | 90.7 | 10.4 | 102.6 | 7.3 | 94.6 | 4.9 | 100.9 | 7.2 | 97.7 | 13.5 | 107.9 | 3.8 | 88.9 | 10.2 |
| 12,13-DiHOME | 99.8 | 7.4 | 90.3 | 12.6 | 73.3 | 12.0 | 101.3 | 10.7 | 90.1 | 6.8 | 99.8 | 5.6 | 99.6 | 15.0 | 108.6 | 6.3 | 88.1 | 7.5 |
| 9,12,13-TriHOME | NQ | NA | NQ | NA | NQ | NA | 100.8 | 2.5 | 112.6 | 13.9 | 100.4 | 3.2 | 102.6 | 13.1 | 105.5 | 8.1 | 89.1 | 11.0 |
| 9,10,13-TriHOME | NQ | NA | NQ | NA | 100.4 | 9.9 | 101.0 | 7.4 | 100.7 | 9.2 | 99.5 | 4.0 | 101.0 | 12.3 | 104.9 | 6.0 | 90.7 | 9.9 |
| 9,10,11-TriHOME | 116.2 | 8.3 | 97.0 | 10.6 | 98.6 | 11.4 | 96.5 | 5.0 | 94.8 | 14.1 | 99.8 | 5.4 | 100.0 | 11.0 | 105.0 | 11.0 | 90.8 | 12.9 |
| 9-HOTrE | 96.3 | 1.6 | 97.1 | 5.7 | 95.4 | 7.1 | 102.4 | 10.9 | 98.2 | 11.0 | 96.8 | 10.0 | 99.0 | 7.8 | 103.3 | 7.1 | 88.9 | 4.2 |
| 13-HOTrE | 107.0 | 3.4 | 79.8 | 25.5 | 85.6 | 5.1 | 101.1 | 14.8 | 98.1 | 9.4 | 96.8 | 7.1 | 99.8 | 12.1 | 106.1 | 6.7 | 84.9 | 12.1 |
| PGE2 | 102.4 | 10.7 | 92.5 | 14.0 | 104.9 | 6.7 | 107.9 | 6.7 | 97.1 | 13.8 | 104.5 | 9.3 | 100.9 | 7.0 | 113.4 | 7.7 | 93.1 | 10.1 |
| PGF2α | NQ | NA | NQ | NA | NQ | NA | 103.7 | 8.5 | 92.9 | 7.5 | 99.0 | 8.9 | 98.1 | 8.1 | 104.8 | 8.9 | 89.1 | 13.1 |
| 8-IsoPGF2α | NQ | NA | NQ | NA | 104.6 | 12.3 | 107.7 | 6.0 | 94.8 | 10.7 | 100.6 | 6.7 | 101.6 | 10.4 | 105.2 | 8.2 | 89.4 | 12.7 |
| TXB2 | 95.6 | 4.7 | 102.5 | 5.2 | 97.0 | 4.0 | 101.8 | 9.3 | 95.1 | 12.0 | 99.8 | 4.4 | 97.4 | 13.6 | 103.5 | 13.7 | 89.4 | 12.6 |
| LTB4 | 90.7 | 5.1 | 110.4 | 10.4 | 94.4 | 10.4 | 100.2 | 9.0 | 96.4 | 5.4 | 97.1 | 2.7 | 97.3 | 13.0 | 104.7 | 7.7 | 85.8 | 13.9 |
| LXA4 | 98.6 | 9.6 | 69.6 | 7.4 | 92.1 | 6.3 | 102.8 | 9.7 | 93.8 | 6.3 | 97.4 | 6.5 | 98.3 | 9.6 | 107.5 | 7.6 | 87.9 | 14.9 |
| LXB4 | 104.1 | 13.3 | NQ | NA | 98.8 | 10.8 | 102.6 | 8.9 | 96.9 | 2.1 | 99.0 | 5.5 | 98.9 | 8.5 | 104.2 | 6.2 | 87.9 | 15.0 |
| 5-HETE | 86.4 | 4.2 | 85.1 | 13.5 | 92.3 | 1.8 | 89.0 | 11.5 | 96.2 | 12.4 | 86.7 | 2.0 | 98.3 | 10.3 | 97.4 | 8.9 | 84.1 | 12.5 |
| 8-HETE | 96.5 | 7.6 | 104.3 | 12.1 | 92.6 | 7.0 | 89.5 | 8.3 | 90.6 | 7.9 | 87.9 | 4.9 | 98.2 | 9.4 | 100.9 | 12.9 | 81.2 | 12.8 |
| 9-HETE | 93.5 | 13.0 | 91.9 | 11.7 | 94.9 | 8.6 | 97.0 | 7.8 | 95.1 | 13.1 | 89.0 | 6.8 | 99.6 | 10.3 | 99.4 | 14.1 | 83.3 | 7.0 |
| 11-HETE | 94.1 | 1.9 | 100.1 | 9.8 | 92.7 | 10.5 | 92.9 | 11.7 | 95.2 | 11.1 | 89.7 | 2.4 | 99.3 | 12.6 | 102.1 | 12.1 | 85.0 | 9.5 |
| 12-HETE | 90.9 | 15.6 | 88.2 | 13.9 | 85.7 | 5.7 | 96.4 | 12.5 | 96.1 | 12.0 | 94.5 | 3.0 | 99.1 | 9.2 | 101.6 | 11.7 | 81.6 | 12.0 |
| 15-HETE | 96.2 | 9.6 | 95.3 | 17.9 | 96.9 | 9.7 | 89.6 | 14.8 | 101.1 | 6.7 | 92.0 | 4.3 | 102.4 | 7.7 | 104.0 | 12.9 | 84.9 | 11.7 |
| 20-HETE | 121.2 | 21.8 | 86.7 | 10.7 | 96.7 | 9.4 | 103.0 | 12.8 | 103.6 | 17.4 | 94.0 | 12.2 | 103.6 | 3.0 | 104.0 | 11.2 | 83.6 | 15.6 |
| 5-oxoETE | 85.2 | 10.2 | 87.4 | 12.7 | 95.5 | 8.5 | 86.3 | 12.7 | 97.2 | 12.1 | 99.0 | 11.2 | 96.5 | 18.5 | 99.9 | 7.8 | 88.8 | 10.8 |
| 5,6-EET | − | − | − | − | − | − | 38.3 | 13.4 | 61.3 | 11.7 | 43.4 | 5.1 | 66.2 | 7.9 | 53.1 | 13.8 | 63.3 | 13.8 |
| 8.9-EET | 89.3 | 5.0 | 89.8 | 9.6 | 97.3 | 8.8 | 86.9 | 7.4 | 97.0 | 10.1 | 83.0 | 2.9 | 97.4 | 12.2 | 95.8 | 10.2 | 79.3 | 11.6 |
| 11,12-EET | 82.3 | 12.0 | 94.4 | 4.8 | 91.6 | 6.9 | 91.5 | 5.1 | 93.7 | 5.8 | 81.9 | 4.7 | 95.6 | 10.7 | 96.0 | 9.4 | 81.6 | 11.8 |
| 14,15-EET | 92.1 | 7.4 | 93.7 | 3.0 | 97.9 | 5.7 | 92.0 | 13.2 | 92.7 | 5.8 | 90.9 | 6.9 | 99.7 | 12.0 | 99.5 | 5.7 | 86.1 | 12.6 |
| 18-HEPE | 98.6 | 7.6 | 99.8 | 6.3 | 92.4 | 6.1 | 98.0 | 12.1 | 98.2 | 10.7 | 95.3 | 6.4 | 98.5 | 11.3 | 101.9 | 7.3 | 84.8 | 10.6 |
| 14,15-EEQ | 93.6 | 4.9 | 92.3 | 7.9 | 93.6 | 7.7 | 95.8 | 7.9 | 92.0 | 9.5 | 91.7 | 5.9 | 100.5 | 11.3 | 103.1 | 12.7 | 82.7 | 10.9 |
| 17,18-EEQ | 103.8 | 5.8 | 103.8 | 11.1 | 91.8 | 8.1 | 104.6 | 14.1 | 96.8 | 5.8 | 94.8 | 2.7 | 100.4 | 9.2 | 104.6 | 11.9 | 80.4 | 14.8 |
| 4-HDHA | 87.8 | 5.5 | 109.5 | 15.4 | 92.3 | 10.7 | 86.3 | 10.9 | 96.3 | 6.5 | 77.7 | 2.7 | 95.5 | 10.7 | 90.7 | 13.5 | 83.6 | 10.6 |
| 7-HDHA | 101.0 | 9.8 | 99.5 | 7.2 | 95.7 | 5.9 | 94.6 | 6.7 | 97.2 | 13.6 | 86.0 | 5.9 | 98.3 | 11.3 | 97.7 | 11.5 | 83.4 | 11.5 |
| 10-HDHA | 88.2 | 6.8 | 95.2 | 6.2 | 91.6 | 14.6 | 86.0 | 6.9 | 97.5 | 13.6 | 88.2 | 5.4 | 98.3 | 10.4 | 98.8 | 13.2 | 83.0 | 10.8 |
| 14-HDHA | 106.7 | 17.8 | 92.5 | 17.4 | 93.2 | 7.2 | 90.5 | 11.4 | 99.4 | 9.7 | 85.1 | 4.9 | 98.3 | 12.6 | 98.3 | 12.5 | 82.2 | 12.7 |
| 17-HDHA | 93.8 | 14.1 | 73.7 | 11.4 | 88.1 | 12.7 | 99.5 | 8.9 | 104.2 | 11.8 | 88.3 | 2.1 | 104.1 | 7.7 | 98.4 | 11.9 | 86.3 | 12.7 |
| PDX | 97.9 | 6.3 | 100.8 | 10.2 | 96.1 | 8.9 | 98.0 | 11.3 | 96.7 | 4.5 | 96.5 | 3.9 | 98.7 | 12.1 | 107.0 | 9.9 | 85.9 | 12.3 |
| MaR1 | 89.2 | 5.7 | 93.4 | 2.3 | 91.0 | 4.9 | 100.3 | 13.7 | 95.4 | 5.5 | 96.2 | 6.3 | 96.1 | 11.3 | 107.1 | 7.9 | 86.8 | 12.1 |
| RvD1 | 92.4 | 8.8 | 72.1 | 4.8 | 93.2 | 10.1 | 98.8 | 15.7 | 87.3 | 11.3 | 101.6 | 4.0 | 100.0 | 7.3 | 103.0 | 9.6 | 90.8 | 5.3 |
| RvD2 | NQ | NA | NQ | NA | 97.2 | 14.3 | 87.9 | 15.0 | 100.0 | 5.5 | 98.5 | 6.6 | 97.4 | 7.7 | 102.1 | 7.7 | 88.4 | 13.3 |
| 16,17-EDP | 86.4 | 7.9 | 88.5 | 13.7 | 82.9 | 9.2 | 81.9 | 7.2 | 103.3 | 15.0 | 84.6 | 4.0 | 101.3 | 15.8 | 94.6 | 11.5 | 80.5 | 9.9 |
| 19,20-EDP | 91.6 | 8.3 | 97.4 | 10.8 | 87.1 | 8.2 | 81.2 | 12.1 | 99.1 | 6.8 | 83.4 | 5.1 | 98.3 | 10.9 | 96.1 | 10.3 | 79.5 | 14.1 |
| 11-H-9-E-LA | NQ | NA | NQ | NA | 99.3 | 16.1 | 92.4 | 13.9 | 80.1 | 17.2 | 101.3 | 6.6 | 100.3 | 14.0 | 108.4 | 8.1 | 87.8 | 12.8 |
| 13-H-9-E-LA | 104.7 | 17.7 | 91.0 | 9.7 | 91.7 | 14.9 | 91.0 | 20.0 | 94.6 | 8.4 | 102.1 | 7.2 | 98.5 | 13.5 | 107.1 | 10.2 | 86.9 | 13.1 |
| 9-H-12-E-LA | 111.8 | 7.4 | 99.3 | 11.4 | 86.2 | 14.2 | 94.4 | 18.3 | 92.3 | 8.4 | 102.4 | 5.0 | 96.0 | 12.3 | 108.4 | 7.4 | 86.7 | 14.5 |
| 11-H-12-E-LA | NQ | NA | NQ | NA | NQ | NA | 71.3 | 20.3 | 66.8 | 10.4 | 104.3 | 4.7 | 95.0 | 13.6 | 107.8 | 8.6 | 92.0 | 14.0 |
Recoveries (RC) of oxylipins were evaluated using four or five spiked concentrations for PMC and plasma, respectively, and oxylipin levels in plasma were adjusted for endogenous level subtraction. Experimental details in Section 1.9.5. For EHODEs and EKODEs with two or three peaks, recoveries were determined by sum of each peaks in Excel. NQ: the concentration is below the LOQ or out of the linear range or not detected, NA: not available,
: for 9-HODE and 9-HOTrE, concentration levels of QC1, QC2, QC3, QC4, and QC5 were 2, 5, 10,100, and 800, respectively. Experimental details in Recovery Section under Method
Container transfer loss test
Container transfer loss of analytes in the PMC matrix was assessed at low concentration levels of 0.1 or 0.2 ng/mL and a higher level of 1 ng/mL for all analytes and 0.5 ng/mL for RvD2, 11-H-9-E LAs, and 11-H-12-E LAs (n = 4) during sample preparation and results were expressed as percent recoveries. ESM Table S1 gave the overall recovery of analytes at these concentration levels ranging from 68.9 to 114.6% with RSD less than 15% for majority of analytes except low recovery for 5,6-EET that has been discussed in the section of Recovery and matrix effects below. Results showed no significant adsorptive loss for analytes at these concentrations, so the container absorption was not an issue in PMC matrix within the calibration range. The container adsorption may be concentration-dependent, and may become an issue at lower concentration levels than tested. Interestingly, other investigators found that using water as the surrogate matrix led to a significant loss, with only approximately 10% recoveries of oxylipins following extraction [3]. In our case, the surrogate matrix PMC in the Eppendorf tube consisted of greater than 71% methanol. The solution was then transferred to a Kimble borosilicate glass tube which is designed to minimize the surface adsorption. The process transferring the solution to SPE loading is rapid. It is likely that the greater than 70% methanol concentration facilitated the disruption of container surface adsorption. Blanc et al. [62] reported that container surface adsorption to cannabinoid calibrators can be reversed by ethanol extraction. Additionally, the recoveries we measured were determined by comparing the pre- and post-spiked samples in PMC using calibration curve going through the same process of sample work-up. Therefore, the losses during sample preparation have been compensated by calibration curve. Yang et al. reported good recovery for the 39 oxylipins spiked in PBS solution going through SPE [44]. It would be worthwhile to determine factors that affect these processes such as material used (tubes, vials, and tips) and the transfer times, etc., in sample preparation.
Evaluation of quantitation characteristics
The present method was evaluated with human plasma samples to demonstrate the limit of detection and quantitation, linearity, accuracy, precision, recovery, matrix effects, and autosampler stability of the method. A detailed summary of these data is shown in the ESM Tables S2–S6 and Table 2. For plasma samples, calculated concentrations were adjusted with the determined endogenous concentration levels where necessary. Despite having 57 targeted oxylipins included in the current method development, seven of them, PD1, RvD3, RvD4, and four K-E-LAs have not been included in the evaluation due to availability of only small amounts of analytical standards for spiking.
Sensitivity and linearity
The LODs, LOQs, and LLOQs of the method were determined from a series of calibration standard solutions following SPE. LODs and LOQs were defined as the concentration that had S/N > 3 and LOQs were the lowest value on the standard curve for each analyte with S/N > 5 and resulted in intraday RSDs ≤ 25% (n = 4). The LODs and LOQs ranged from 0.02 to 0.2 and 0.05 to 0.5 ng/mL, respectively (ESM Table S2). The LLOQs were the concentrations of each analyte in the calibration curve that resulted in a precision RSD of < 20% and accuracy of 70–130%. Values for LLOQs were in the range of 0.1 to 1 ng/mgL and are presented in ESM Table S2, and representative MRM chromatograms of oxylipins at the LLOQ concentrations are shown in ESM Fig. S2.
The calibration was carried out with SPE extraction of the analytes and internal standards. Linearity was tested by using a 12-point calibration curve of standard extracts. This high number of levels was selected to achieve the optimal concentration range for each target analyte, considering the large differences in sensitivity between the single substances. A linear regression model with 1/x weighting was applied to all calibration curves. The results of correlation coefficients (R) and calibration ranges for all targeted 50 oxylipins are listed in ESM Table S2. The correlation coefficient ranged from 0.990 to 0.999 for all analytes, which indicated good linearity within the considered concentration ranges.
Autosampler stability
The stability of analytes in both PMC and plasma extracted reconstituted solution was evaluated relative to freshly prepared samples. QC samples at three levels (Q3 to Q5), four replicates each, were left in the autosampler at 4 °C for 24 h and reinjected. Stability of 5,6-EET and deuterated 5,6-EET-d11 decreased to 61 and 57%, respectively, at this storage condition for plasma extract at 24 h relative to 0 h. This decrease may be due to instability of 5,6-EET in acidic and neutral conditions [63, 64]. Loss of 5,6-EET during sample storage was compensated by the use of deuterated 5,6-EET-d11 as internal standard during quantitation of 5,6-EET as shown in ESM Table S2. The stability results are summarized in ESM Table S3. No significant degradation (89.3–113.4%) was observed for all other analytes, and the RSD of stability ranged from 0.3 to 14.5% for all, except 17-HDHA (RSD of 18% at 1 ng/mL) for plasma extract. The results revealed all analytes except 5,6-EET are stable in the reconstitute solution at 4 °C within the duration of 24 h. Recently, Wang et al. examined autosampler stability of 104 oxylipins including 41 of oxylipins in our assay and found less than 20% loss in plasma extract when stored at 4 °C for 8 h in the autosampler loading tray [48]. It is worth to mention that Wolfer et al. have reported stabilities of long-term storage (− 80 °C for 20 weeks), undergoing three freeze-thaw cycles, for 43 oxylipins generated from LA, AA, EPA, and DHA across concentration levels ranging from 0.5 to 250 ng/mL in MeOH/H2O (1:1) and found no significant loss (< 15%) over the storage period [47]. We did not conduct the analyte long-term storage stability (− 80 °C) in the methanol working solution. It may be necessary to do so in the future. Additionally, since our working solutions were aliquoted before their storage at − 80 °C for single use only, our methodology involved only one freeze-thaw cycle.
Precision and accuracy
Evaluation of intraday and interday accuracy and precision was performed with four QC levels from PMC and five levels from plasma. Intraday accuracy and precision were evaluated in four replicates at each concentration on the same day. Interday accuracy and precision were carried out in four replicates on four or five different days of analysis. The accuracy and precision results for intraday and interday PMC and plasma samples are shown in ESM Tables S4 and S5, respectively. A total of 41 of the 50 oxylipins quantified in PMC showed accuracy in the range of 80.5–118.5% across four QC levels and 69.3–127.1% for all except 11-H-9-E-LA and 11-H-12-ELA that had interday accuracy of 143.1 and 139.6% at LLOQ level. The intraday RSDs in the PMC ranged from 0.4 to 15.3% for all QC samples. The interday RSDs ranged from 4.3 to 15.0% for 44 out of 50 across the concentration levels and 16.7% for 12,13-EpOME at LLOQ level except five analytes (13-oxoODE, 9,12,13-TriHOME, 13-H-9-E-LA, 9-H-12-E-LA, and 11-H-12-E-LA) with RSDs of 21.7–27.7% at LLOQ or medium QC levels (ESM Table S4). For all five QC levels, 42 out of 50 oxylipins quantified in the plasma samples showed accuracy in the range of 70.3 and 120.5%. Overall, all 50 oxylipins had accuracy between 67.8 and 129.3%. The intraday RSDs for 48 of 50 in the plasma were ≤ 15% across the concentration range and for 2 of 50 in the range of 17.3–20.3% at LOQ or LLOQ levels. The interday RSDs for 35 out of 50 were ≤ 15% across the concentration range, for 10 of 50 analytes between 15 and 20% at LOQ or LLOQ levels, and except for 5 of 50 analytes (9-oxoODE, 13-oxoODE, 13-HOTrE, 18-HEPE, and 17-HDHA) that had RSDs ranging between 23.3 and 30.1% at LOQ, or LLOQ levels. A significant batch-to-batch variation has been observed for 13-HOTrE and 18-HEPE (as high as 70%). It is worth mentioning that four analytes 9,10-EpOME, 12,13-EpOME, 9,10,13-TriHOME and 9,12,13-TriHOME showed good sensitivity, but large variations were observed for the concentrations between LOQs and LLOQs (data not shown), which will require further investigation. In future studies, the relatively high RSDs of these analytes might be improved by using their corresponding deuterated analogs if available, the use of a type I internal standard to correct for instrument and volume variability, and separation of co-eluting or endogenous compounds in the samples. Nevertheless, these results demonstrate that the method is reproducible and reliable for the majority of oxylipins analyzed across these concentration ranges.
Recovery and matrix effects
The recoveries of the target analytes spiked in PMC and plasma were evaluated using three or four replicates in four and five levels of QC samples respectively, by comparing analyte concentrations calculated from samples spiked with analytes before extraction to samples spiked with analytes after extraction. For each pooled plasma sample, calculated concentrations were adjusted with the determined endogenous concentration levels before recoveries were calculated. Recoveries for all target analytes are shown in Table 2. Recoveries of 80 to 110% were achieved for most oxylipins. Forty-nine out of the of 50 analytes showed recoveries in the range of 66.8 to 121.2% except 5,6-EET that showed recoveries of 38.3–66.2% across three QC concentration levels (QC3 to QC5) in PMC and plasma. Since no matrix suppression was observed (ESM Table S6), low recoveries of 5,6-EET most likely resulted from degradation and losses during sample preparation, which were compensated by the use of deuterated 5,6-EET-d11 as indicated in its accuracy determination (ESM Tables S4 and S5). Extraction recovery experiments displayed satisfactory repeatability, with RSD ≤ 15% for the majority of analytes, and RSD ≤ 20% for all analytes except for 13-oxoODE (21.9% in PMC), 13-HOTrE (25.5% in plasma), and 20-HETE (21.8% in PMC) at the lowest QC1level. Wang et al. [48] also used the Strata-X cartridge SPE for sample preparation and compared the peak areas before extraction with the corresponding peak areas of pure internal standard solutions. Their results showed slightly lower recoveries for 17 deuterated internal standards (62.1–92.8%) compared to the same 17 unlabeled oxylipins. In the present study, the observed recovery range of 86.9–107.9% at 1 ng/mL concentration in plasma indicates the effectiveness of the procedures to extract the targeted oxylipins from PMC and plasma samples.
Matrix effects assessments have not been reported in many LC-MS/MS profiling manuscripts. In the present study, we measured matrix effects as a matrix factor percent by comparing concentrations of extracts of post-spiked plasma corrected by endogenous concentration levels with those of extracts of post-spiked PMC at three QCs levels (QC3 to QC5). Results show very low matrix effects for the majority of samples across the tested concentration ranges (ESM Table S6) with greater than 73.9% and RSD < 20 for 96% of all analytes, indicating that no significant ion suppression was observed across assayed concentration levels. Among the 50 oxylipins evaluated, 14 observed signal enhancement effects, with some results ranging from 123.3 to 144.5% across the concentration range, while others fell within that range for only one or two concentration levels and RSD values were < 17.2% suggesting that these matrix effects are reproducible. Despite the low observed matrix effects for these compounds across these concentration levels, it should be noted that matrix effect is concentration-dependent, the analytes at lower concentration may experience suppression and enhancement of matrix effect different from higher concentration and their analytical responses would be expected to be different from matrix to matrix. The matrix effects may be mitigated by modifying chromatographic and mass spectrometric conditions, or using the respective isotope-labeled internal standard whenever available.
Overall, the method here provided comprehensive quantitative evaluation, including limit of detection and quantitation, linearity, precision and accuracy, recovery, stability and matrix effects.
Application of the method
The method was successfully applied to determine basal concentrations of the targeted oxylipins in human plasma and serum. In healthy human control subjects, we were able to quantitate 25 metabolites in plasma samples and 31 in serum samples corresponding to four enzymatic and non-enzymatic pathways (Table 3). A large numbers of analytes were found in basal plasma and serum in our method, demonstrating that our method is sufficiently sensitive. Table 3 shows that basal levels of oxylipins in plasma and serum obtained from our studies are in agreement with those previously reported by others [47–49, 65–69]. Our results of higher 12-HETE levels in serum than in plasma corroborate earlier investigations [66, 68, 69]. Higher levels of the 12-LOX products 12-HETE and 14-HDHA in serum are likely due to activation of platelet-derived 12-LOX during the clotting cascade [69]. Similarly, remarkable differences were also observed in basal levels of oxylipins for human serum [69]. The analytes we determined in Table 3 had the concentration above LOQ although the concentrations were measured potentially with less than or equivalent accuracy than those greater than LLOQ. The identities of novel incorporated compounds, H-E-LAs, K-E-LAs, and 9,10,11-TriHOME, in plasma were confirmed by comparing retention times and product ion spectra with authentic standard. The novel methodology is capable of detecting oxylipin changes from clinical and preclinical studies and has now been used to quantify these eight lipid mediators in human and rodent skin and in human serum and plasma. Moreover, we found that these lipids cause pain and itch-related behaviors in rodent models. In addition, we used this assay to show that these novel lipids can be altered by changing the amounts of their precursor fatty acids in the diet [70]. It has also been successfully applied for the profiling of these lipid mediators in human psoriasis skin and blood and showed alterations of the targeted mediators by comparing to the healthy individuals [71]. In future studies, we plan to synthesize additional standards of K-E-LAs to expand method performance evaluation, and to incorporate targeted PUFAs in order to investigate how parent PUFA levels relate to their corresponding oxylipin derivatives.
Table 3.
Comparison of endogenous levels of oxylipins in human plasma and serum determined by our method with those previously reported measurement [47–4,9, 66–6,9]
| Compound | Current work*1 | Reported in Ref [66]a | Reported in Ref [48]b | Reported in Ref [65]c | Reported in Ref [47]d | Reported in Ref [49]e | Reported in Ref [67]f | Current work*2 | Reported in Ref [68]g | Reported in Ref [47]d | Reported in Ref [69]h |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma concentration (ng/mL) | Serum concentration (ng/mL) | ||||||||||
| 9-HODE | 0.667–5.28 | 0.865–4.132 | 27.20 | 11.961–20.776 | 0–26.969 | 2.80–5.64 | 0.744–3.270 | 14.05–21.43 | 1.748 ±0.151 | ||
| 13-HODE | 0.506–6.76 | 1.286–5.826 | 38.30 | 9.933–19.933 | 0–21.094 | 1.72–9.67 | 1.290–8.370 | 11.93–24.61 | 2.281 ±0.178 | ||
| 9-oxoODE | NQ-0.578 | 4.11 | 0.23–0.53 | ||||||||
| 13-oxoODE | NQ-0.755 | 0.347–2.136 | 2.05 | 0.39–0.87 | 0–1.270 | ||||||
| 9,10-EpOME | NQ-0.772 | 0.308–1.368 | 0.06 | NQ-2.08 | 0^4.500 | 1.688 ±0.195 | |||||
| 12,13-EPOME | NQ −11.6 | 0.181–0.684 | 0.07 | 2.82–7.63 | 4.230–23.90 | 1.925 ± 0.190 | |||||
| 9,10-DiHOME | 0.304–3.13 | 0.644–2.130 | 0.10 | 0.59–1.00 | 0.221–1.730 | 1.100 ± 0.167 | |||||
| 12,13-DiHOME | 0.238–2.88 | 1.053–3.739 | 0.09 | 0.77–2.45 | 0.536–2.630 | 1.948 ± 0.151 | |||||
| 9,12,13-TriHOME | NQ-2.24* | 0.168–0.429 | − | 0.218–0.932 | |||||||
| 9,10,13-TriHOME | NQ* | 0.248–0.528 | − | 0.047–0.257 | |||||||
| 9,10,11-TriHOME | ND* | − | |||||||||
| 9-HOTrE | NQ-0.212 | 0.10–0.24 | 0.118 ±0.012 | ||||||||
| 13-HOTrE | NQ-0.452 | 1.38 | 0.20–0.47 | 0.115 ±0.011 | |||||||
| PGE2 | NQ | 0.053–0.106 | 0.07 | 0.029 ±0.013 | NQ | NQ | NQ-0.54 | 0.043–0.462 | 0.135–0.454 | ||
| PGF2α | NQ | 0.078–0.184 | NQ | <0.05 | 0.175–0.533 | NQ | NQ-0.70 | 0.231–6.090 | 0.210–0.537 | ||
| 8-IsoPGF2α | NQ | NQ | 0.052 ±0.026 | 0.162–0.567 | NQ | NQ | NQ | ||||
| TXB2 | NQ-0.249 | 0.096–0.481 | NQ | 0.179 ±0.065 | 0.057–0.324 | NQ | 0.12–68.10 | 0.108–37.10 | 0.908–4.762 | ||
| LTB4 | NQ | 0.55 | <0.01 | 0.284–0.552 | NQ | 0.20–0.71 | 0.008–0.213 | 0.282–0.544 | |||
| LXA4 | NQ | 0.15 | 0.016±0.019 | NQ | NQ | NQ | 0.251–0.738 | ||||
| LXB4 | NQ | 0.26 | 0.284–0.553 | NQ | NQ | 1.020–1.519 | |||||
| 5-HETE | 0.127–0.866 | 0.026–0.138 | 16.66 | 0.399 ±0.148 | 0.287–0.529 | 0.442–0.707 | 1.09–2.10 | 0.158–0.569 | 0.613–0.993 | 0.512 ±0.042 | |
| 8-HETE | NQ-0.265 | 0.022–0.090 | 1.52 | 0.280 ±0.092 | 0.041–0.075 | 0.25–1.28 | 0–21.200 | 0.003–0.291 | 0.112 ±0.004 | ||
| 9-HETE | NQ-0.182 | 0–0.032 | 3.13 | 0.328±0.122 | NQ | NQ −0.45 | 0.054 ± 0.003 | ||||
| 11-HETE | NQ-0.270 | 0.019–0.102 | 2.54 | 0.316 ± 0.148 | 0.110–0.166 | 0.172–0.284 | 0.25^.78 | 0.036–1.050 | 0.160–0.381 | 0.179 ±0.023 | |
| 12-HETE | NQ-0.627 | 0.051–0.416 | 2.87 | 3.12 ±2.502 | 0.260–0.482 | NQ | 8.9–524.00 | 2.610–38.60 | 0.912–4.610 | 4.963 ± 0.929 | |
| 15-HETE | NQ-0.614 | 0.061–0.311 | 3.39 | 1.307 ±0.356 | 0.268–0.532 | 0.435–0.756 | 0.54–6.65 | 0.182–1.80 | 0.396–0.497 | 0.352 ± 0.035 | |
| 20-HETE | NQ-0.761 | 0.016–0.118 | NQ | 1.308 ±0.212 | NQ | 0.74–1.46 | 0–1.900 | ||||
| 5-oxoETE | NQ-0.247 | 0–0.051 | 0.263–0.388 | NQ | 0.10–0.18 | NQ | |||||
| 5,6-EET | NQ | 0–019-0.096 | 0.109–0.195 | NQ | NQ | 0.333–1.820 | NQ | ||||
| 8.9-EET | NQ | 0.006–0.054 | NQ | 0.147±0.104 | 0.181–0.346 | NQ | NQ-0.70 | 0.109–0.791 | NQ | 0.202 ± 0.020 | |
| 11,12-EET | NQ | 0–0.058 | NQ | 0.165 ±0.225 | NQ | NQ | NQ-0.21 | 0.191–0.870 | NQ | 0.448 ± 0.045 | |
| 14,15-EET | NQ | 0–0.026 | NQ | 0.125 ± 0.136 | NQ | 0.188–0.220 | 0.15–0.30 | 0.081–0.803 | NQ | 0.416 ± 0.042 | |
| 18-HEPE | NQ | 0.068–0.119 | 0.11–0.30 | ||||||||
| 14(15)-EEQ | NQ | NQ | NQ | 0.067 ± 0.008 | |||||||
| 17(18)-EEQ | NQ | NQ | NQ | 0.102 ±0.009 | |||||||
| 4-HDHA | NQ-0.114 | 2.73 | 0.15–0.35 | ||||||||
| 7-HDHA | NQ | 0.13 | NQ −0.15 | ||||||||
| 10-HDHA | NQ | 0.20 | NQ | ||||||||
| 14-HDHA | NQ-0.659 | 0.56 | 0.237–1.415 | 0.169–0.780 | 2.65–71.4 | 0.400–1.314 | |||||
| 17-HDHA | NQ-0.387 | 0–0.145 0.40 | 0–0.145 0.40 | 0.227–0.670 | 0.047–0.157 | 0.23–3.48 | 0.029–0.513 | ||||
| PDX | NQ | 0.591–1.718 | NQ | NQ | |||||||
| MaR1 | NQ | NQ | NQ | ||||||||
| RvD1 | NQ | NQ | NQ | NQ | 0.028–0.568 | NQ | 0–0.51 | NQ | |||
| RvD2 | NQ | NQ | 0.047–0.084 | NQ | 0–0.470 | NQ | |||||
| 16,17-EDP | NQ | NQ | NQ | 0.165 ±0.018 | |||||||
| 19(20)-EDP | NQ-0.243 | NQ | 0.16–0.31 | 0.379 ± 0.038 | |||||||
NQ: not quantifiable, the concentration is below the LOQ or out of the linear range or not detected from the matrix, −: not quantified
n = 40,
n = 5,
n = 4
Eicosanoid profiles in control human plasma (n = 3), concentrations were converted from nM reported in the article to ng/mL
Eicosanoid profiles in control human plasma (mean value, n = 3)
Eicosanoid profiles in human plasma from healthy volunteers (n = 6)
Eicosanoid profiles in non-fasting control human plasma (n = 9)
Eicosanoid profiles in control human plasma (n = 50)
Eicosanoid profiles in control human plasma (n = 21)
Eicosanoid profiles in control human serum (n = 16)
Oxylipin profiles in control human plasma (n = 20), concentrations were converted from pM (±SE) reported in the article to ng/mL (±SE)
Summary and conclusion
We developed a sensitive and selective UPLC-MS/MS method for the simultaneous profiling of 57 targeted oxylipins derived from five major n-6 and n-3 PUFAs including LA, AA, ALA, EPA, and DHA. The method achieved good separation performance on a UPLC reversed-phase column and allows in-depth analysis of isomeric and isobaric species of oxylipin extracts in biological samples. To our knowledge, this work is the first report to integrate four H-E-LAs and four K-E-LAs, as well as 9,10,11-TriHOME in a targeted oxylipin profiling platform. The surrogate matrix PMC was used for calibration and calibrators were prepared through SPE. The method was evaluated in human plasma for 50 oxylipins. The LODs, LOQs, and LLOQs of the method ranged from 0.02 to 0.2, 0.05 to 0.5 ng/mL, and 0.1 to 1 ng/mL, respectively. The method demonstrated good linearity (R = 0.990–0.999), 24 h autosampler stability (89.3–113.4%), intra- and interday precision as relative standard deviation (RSD%) < 20% for the majority of analytes (45 out of 50) across concentration levels, accuracy between 67.8 and 129.3% for all analytes, and recoveries in the range of 66.8 to 121.2% for all except 5,6-EET that has been compensated by the use of its deuterated internal standard. Ion enhancement in plasma was observed for 28% of analytes in tested concentrations but the results were still reproducible (RSD < 17.2). The method has been successfully employed in clinical and preclinical applications.
The targeted oxylipin panel offers broad coverage of lipid mediators and pathway markers generated from enzymatic COX, LOX, CYP, and non-enzymatic oxidation pathways. The ability to profile these targeted oxylipins may contribute to better understanding of disease mechanisms and perturbations, and facilitate biomarker identification, and the assessment of nutritional interventions.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Programs of the National Institute on Aging (NIA), National Institute on Alcohol Abuse and Alcoholism (NIAAA), NIH Clinical Center Department of Perioperative Medicine, and the Mayday Fund. The authors would like to thank Dr. Charles N. Serhan for sharing methods and providing resolvin and protectin standards, the research participants who provided plasma samples, the many NIH co-investigators and staff who contributed to collection and processing of these plasma samples.
Compliance with ethical standards
Protocol 11-AA-0028 was reviewed and approved by an NIH Institutional Review Board (FWA# 00005897), and informed consent was obtained from all study participants.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00216-018-1222-4) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that they have no conflicts of interest.
References
- 1.Gabbs M, Leng S, Devassy JG, Monirujjaman M, Aukema HM. Advances in our understanding of oxylipins derived from dietary PUFAs. Adv Nutr 2015;6(5):513–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niki E Lipid peroxidation: physiological levels and dual biological effects. Free Radic Biol Med. 2009;47(5):469–84. [DOI] [PubMed] [Google Scholar]
- 3.Strassburg K, Huijbrechts AM, Kortekaas KA, Lindeman JH, Pedersen TL, Dane A, et al. Quantitative profiling of oxylipins through comprehensive LC-MS/MS analysis: application in cardiac surgery. Anal Bioanal Chem. 2012;404(5):1413–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serhan CN, Dalli J, Colas RA, Winkler JW, Chiang N. Protectins and maresins: new pro-resolving families of mediators in acute inflammation and resolution bioactive metabolome. Biochim Biophys Acta. 2015;1851(4):397–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serhan CN, Petasis NA. Resolvins and protectins in inflammation resolution. Chem Rev. 2011;111(10):5922–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capdevila JH, Falck JR. Biochemical and molecular characteristics of the cytochrome P450 arachidonic acid monooxygenase. Prostaglandins Other Lipid Mediat. 2000;62(3):271–92. [DOI] [PubMed] [Google Scholar]
- 7.Yu Z, Schneider C, Boeglin WE, Marnett LJ, Brash AR. The lipoxygenase gene ALOXE3 implicated in skin differentiation encodes a hydroperoxide isomerase. Proc Natl Acad Sci U S A. 2003;100(16):9162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bui P, Imaizumi S, Beedanagari SR, Reddy ST, Hankinson O. Human CYP2S1 metabolizes cyclooxygenase- and lipoxygenase-derived eicosanoids. Drug Metab Dispos. 2011;39(2):180–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts LJ 2nd, Fessel JP. The biochemistry of the isoprostane, neuroprostane, and isofuran pathways of lipid peroxidation. Chem Phys Lipids. 2004;128(1–2):173–86. [DOI] [PubMed] [Google Scholar]
- 10.Spiteller G Lipid peroxidation in aging and age-dependent diseases. Exp Gerontol. 2001;36(9):1425–57. [DOI] [PubMed] [Google Scholar]
- 11.Blank C, Neumann MA, Makrides M, Gibson RA. Optimizing DHA levels in piglets by lowering the linoleic acid to alpha-linolenic acid ratio. J Lipid Res. 2002;43(9):1537–43. [DOI] [PubMed] [Google Scholar]
- 12.Markworth JF, Kaur G, Miller EG, Larsen AE, Sinclair AJ, Maddipati KR, Cameron-Smith D. Divergent shifts in lipid mediator profile following supplementation with n-3 docosapentaenoic acid and eicosapentaenoic acid. FASEB J. 2016;209(3):192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramsden CE, Zamora D, Majchrzak-Hong S, Faurot KR, Broste SK, Frantz RP, et al. Re-evaluation of the traditional diet-heart hypothesis: analysis of recovered data from Minnesota coronary experiment (1968–73). BMJ. 2016;353:i1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramsden CE, Ringel A, Feldstein AE, Taha AY, MacIntosh BA, Hibbeln JR, et al. Lowering dietary linoleic acid reduces bioactive oxidized linoleic acid metabolites in humans. Prostaglandins Leukot Essent Fat Acids. 2012;87(4–5):135–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramsden CE, Faurot KR, Zamora D, Suchindran CM, Macintosh BA, Gaylord S, et al. Targeted alteration of dietary n-3 and n-6 fatty acids for the treatment of chronic headaches: a randomized trial. Pain. 2013;154(11):2441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramsden CE, Ringel A, Majchrzak-Hong SF, Yang J, Blanchard H, Zamora D, Loewke JD, Rapoport SI, Hibbeln JR, Davis JM, Hammock BD, Taha AY. Dietary linoleic acid-induced alterations in pro- and anti-nociceptive lipid autacoids: Implications for idiopathic pain syndromes? Mol Pain 2016;12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundstrom SL, Yang J, Brannan JD, Haeggstrom JZ, Hammock BD, Nair P, et al. Lipid mediator serum profiles in asthmatics significantly shift following dietary supplementation with omega-3 fatty acids. Mol Nutr Food Res. 2013;57(8):1378–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nording ML, Yang J, Georgi K, Hegedus Karbowski C, German JB, Weiss RH, et al. Individual variation in lipidomic profiles of healthy subjects in response to omega-3 fatty acids. PLoS One. 2013;8(10):e76575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 2001;294(5548):1871–5. [DOI] [PubMed] [Google Scholar]
- 20.Amtul Z, Uhrig M, Wang L, Rozmahel RF, Beyreuther K. Detrimental effects of arachidonic acid and its metabolites in cellular and mouse models of Alzheimer’s disease: structural insight. Neurobiol Aging. 2012;33(4):831 e21–31. [DOI] [PubMed] [Google Scholar]
- 21.Corsinovi L, Biasi F, Poli G, Leonarduzzi G, Isaia G. Dietary lipids and their oxidized products in Alzheimer’s disease. Mol Nutr Food Res. 2011;55(Suppl 2):S161–72. [DOI] [PubMed] [Google Scholar]
- 22.Narumiya S, Furuyashiki T. Fever, inflammation, pain and beyond: prostanoid receptor research during these 25 years. FASEB J. 2011;25(3):813–8. [DOI] [PubMed] [Google Scholar]
- 23.Tessaro FH, Ayala TS, Martins JO. Lipid mediators are critical in resolving inflammation: a review of the emerging roles of eicosanoids in diabetes mellitus. Biomed Res Int. 2015;2015:568408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuck RN, Theken KN, Edin ML, Caughey M, Bass A, Ellis K, et al. Cytochrome P450-derived eicosanoids and vascular dysfunction in coronary artery disease patients. Atherosclerosis. 2013;227(2): 442–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez-Reyes RE, Nava-Mesa MO, Vargas-Sanchez K, Ariza-Salamanca D, Mora-Munoz L. Involvement of astrocytes in Alzheimer’s disease from a neuroinflammatory and oxidative stress perspective. Front Mol Neurosci. 2017;10:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sallam N, Laher I. Exercise modulates oxidative stress and inflammation in aging and cardiovascular diseases. Oxidative Med Cell Longev. 2016;2016:7239639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapiro H, Singer P, Ariel A. Beyond the classic eicosanoids: peripherally-acting oxygenated metabolites of polyunsaturated fatty acids mediate pain associated with tissue injury and inflammation. Prostaglandins Leukot Essent Fat Acids. 2016;111:45–61. [DOI] [PubMed] [Google Scholar]
- 28.Luo P, Wang MH. Eicosanoids, beta-cell function, and diabetes. Prostaglandins Other Lipid Mediat. 2011;95(1–4):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nasrallah R, Hassouneh R, Hebert RL. PGE2, kidney disease, and cardiovascular risk: beyond hypertension and diabetes. J Am Soc Nephrol. 2016;27(3):666–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldman M, Peterson SJ, Arad M, Hochhauser E. The role of 20-HETE in cardiovascular diseases and its risk factors. Prostaglandins Other Lipid Mediat. 2016;125:108–17. [DOI] [PubMed] [Google Scholar]
- 31.Bhat AH, Dar KB, Anees S, Zargar MA, Masood A, Sofi MA, et al. Oxidative stress, mitochondrial dysfunction and neurodegenerative diseases; a mechanistic insight. Biomed Pharmacother. 2015;74: 101–10. [DOI] [PubMed] [Google Scholar]
- 32.Daiber A, Steven S, Weber A, Shuvaev VV, Muzykantov VR, Laher I, Li H, Lamas S, Munzel T. Targeting vascular (endothelial) dysfunction. Br J Pharmacol. 2017;174(12):1591–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraser PA. The role of free radical generation in increasing cerebrovascular permeability. Free Radic Biol Med. 2011;51(5):967–77. [DOI] [PubMed] [Google Scholar]
- 34.Gong Z, Muzumdar RH. Pancreatic function, type 2 diabetes, and metabolism in aging. Int J Endocrinol. 2012;2012:320482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H, Weng J, Wang MH. EETs/sEH in diabetes and obesity-induced cardiovascular diseases. Prostaglandins Other Lipid Mediat. 2016;125:80–89. [DOI] [PubMed] [Google Scholar]
- 36.Waddington E, Sienuarine K, Puddey I, Croft K. Identification and quantitation of unique fatty acid oxidation products in human atherosclerotic plaque using high-performance liquid chromatography. Anal Biochem. 2001;292(2):234–44. [DOI] [PubMed] [Google Scholar]
- 37.Tsikas D Application of gas chromatography-mass spectrometry and gas chromatography-tandem mass spectrometry to assess in vivo synthesis of prostaglandins, thromboxane, leukotrienes, isoprostanes and related compounds in humans. J Chromatogr B Biomed Sci Appl. 1998;717(1–2):201–45. [DOI] [PubMed] [Google Scholar]
- 38.Montine TJ, Sidell KR, Crews BC, Markesbery WR, Marnett LJ, Roberts LJ 2nd, et al. Elevated CSF prostaglandin E2 levels in patients with probable AD. Neurology. 1999;53(7):1495–8. [DOI] [PubMed] [Google Scholar]
- 39.Tacconelli S, Capone ML, Patrignani P. Measurement of 8-isoprostaglandin F2alpha in biological fluids as a measure of lipid peroxidation. Methods Mol Biol. 2010;644:165–78. [DOI] [PubMed] [Google Scholar]
- 40.Hughes H, Mitchell JR, Gaskell SJ. Quantitation of leukotriene B4 in human serum by negative ion gas chromatography-mass spectrometry. Anal Biochem. 1989;179(2):304–8. [DOI] [PubMed] [Google Scholar]
- 41.Yang J, Eiserich JP, Cross CE, Morrissey BM, Hammock BD. Metabolomic profiling of regulatory lipid mediators in sputum from adult cystic fibrosis patients. Free Radic Biol Med. 2012;53(1): 160–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song J, Liu X, Wu J, Meehan MJ, Blevitt JM, Dorrestein PC, et al. A highly efficient, high-throughput lipidomics platform for the quantitative detection of eicosanoids in human whole blood. Anal Biochem. 2013;433(2):181–8. [DOI] [PubMed] [Google Scholar]
- 43.Yang R, Chiang N, Oh SF, Serhan CN. Metabolomics-lipidomics of eicosanoids and docosanoids generated by phagocytes. Curr Protoc Immunol. 2011;Chapter 14:Unit 14 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, Schmelzer K, Georgi K, Hammock BD. Quantitative profiling method for oxylipin metabolome by liquid chromatography electrospray ionization tandem mass spectrometry. Anal Chem. 2009;81(19):8085–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia V, Gilani A, Shkolnik B, Pandey V, Zhang FF, Dakarapu R, et al. 20-HETE signals through G-protein-coupled receptor GPR75 (Gq) to affect vascular function and trigger hypertension. Circ Res. 2017;120(11):1776–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jamieson KL, Endo T, Darwesh AM, Samokhvalov V, Seubert JM. Cytochrome P450-derived eicosanoids and heart function. Pharmacol Ther. 2017;179:47–83. [DOI] [PubMed] [Google Scholar]
- 47.Wolfer AM, Gaudin M, Taylor-Robinson SD, Holmes E, Development NJK. Validation of a high-throughput ultrahigh-performance liquid chromatography-mass spectrometry approach for screening of oxylipins and their precursors. Anal Chem. 2015;87(23):11721–31. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Armando AM, Quehenberger O, Yan C, Dennis EA. Comprehensive ultra-performance liquid chromatographic separation and mass spectrometric analysis of eicosanoid metabolites in human samples. J Chromatogr A. 2014;1359:60–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kortz L, Dorow J, Becker S, Thiery J, Ceglarek U. Fast liquid chromatography-quadrupole linear ion trap-mass spectrometry analysis of polyunsaturated fatty acids and eicosanoids in human plasma. J Chromatogr B Anal Technol Biomed Life Sci. 2013;927: 209–13. [DOI] [PubMed] [Google Scholar]
- 50.Le Faouder P, Baillif V, Spreadbury I, Motta JP, Rousset P, Chene G, et al. LC-MS/MS method for rapid and concomitant quantification of pro-inflammatory and pro-resolving polyunsaturated fatty acid metabolites. J Chromatogr B Anal Technol Biomed Life Sci. 2013;932:123–33. [DOI] [PubMed] [Google Scholar]
- 51.Foreman HD, Brown JB. Solubilities of the fatty acids in organic solvents at low temperatures. J Am Oil Chem Soc. 1994;21(7): 183–7. [Google Scholar]
- 52.Balas L, Guichardant M, Durand T, Lagarde M. Confusion between protectin D1 (PD1) and its isomer protectin DX (PDX). An overview on the dihydroxy-docosatrienes described to date. Biochimie. 2014;99:1–7. [DOI] [PubMed] [Google Scholar]
- 53.Mas E, Croft KD, Zahra P, Barden A, Mori TA. Resolvins D1, D2, and other mediators of self-limited resolution of inflammation in human blood following n-3 fatty acid supplementation. Clin Chem. 2012;58(10):1476–84. [DOI] [PubMed] [Google Scholar]
- 54.Goodfriend TL, Ball DL, Egan BM, Campbell WB, Nithipatikom K. Epoxy-keto derivative of linoleic acid stimulates aldosterone secretion. Hypertension. 2004;43(2):358–63. [DOI] [PubMed] [Google Scholar]
- 55.Thomas CP, Boeglin WE, Garcia-Diaz Y, O’Donnell VB, Brash AR. Steric analysis of epoxyalcohol and trihydroxy derivatives of 9-hydroperoxy-linoleic acid from hematin and enzymatic synthesis. Chem Phys Lipids. 2013;167–168:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oliw EH, Garscha U, Nilsson T, Cristea M. Payne rearrangement during analysis of epoxyalcohols of linoleic and alpha-linolenic acids by normal phase liquid chromatography with tandem mass spectrometry. Anal Biochem. 2006;354(1):111–26. [DOI] [PubMed] [Google Scholar]
- 57.Lin D, Zhang J, Sayre LM. Synthesis of six epoxyketooctadecenoic acid (EKODE) isomers, their generation from nonenzymatic oxidation of linoleic acid, and their reactivity with imidazole nucleophiles. J Org Chem. 2007;72(25):9471–80. [DOI] [PubMed] [Google Scholar]
- 58.Quehenberger O, Armando AM, Dennis EA. High sensitivity quantitative lipidomics analysis of fatty acids in biological samples by gas chromatography-mass spectrometry. Biochim Biophys Acta. 2011;1811(11):648–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masoodi M, Mir AA, Petasis NA, Serhan CN, Nicolaou A. Simultaneous lipidomic analysis of three families of bioactive lipid mediators leukotrienes, resolvins, protectins and related hydroxy-fatty acids by liquid chromatography/electrospray ionisation tandem mass spectrometry. Rapid Commun Mass Spectrom. 2008;22(2):75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Balvers MG, Verhoeckx KC, Bijlsma S, Rubingh CM, Meijerink J, Wortelboer HM, et al. Fish oil and inflammatory status alter the n-3 to n-6 balance of the endocannabinoid and oxylipin metabolomes in mouse plasma and tissues. Metabolomics. 2012;8(6):1130–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pace-Asciak CR, Reynaud D, Demin PM. Hepoxilins: a review on their enzymatic formation, metabolism and chemical synthesis. Lipids. 1995;30(2):107–14. [DOI] [PubMed] [Google Scholar]
- 62.Blanc JA, Manneh VA, Ernst R, Berger DE, de Keczer SA, Chase C, et al. Adsorption losses from urine-based cannabinoid calibrators during routine use. Clin Chem. 1993;39(8):1705–12. [PubMed] [Google Scholar]
- 63.Vandernoot VA, VanRollins M. Capillary electrophoresis of cyto-chrome P-450 epoxygenase metabolites of arachidonic acid. 2. Resolution of stereoisomers. Anal Chem. 2002;74(22):5866–70. [DOI] [PubMed] [Google Scholar]
- 64.Fulton D, Falck JR, McGiff JC, Carroll MA, Quilley J. A method for the determination of 5,6-EET using the lactone as an intermediate in the formation of the diol. J Lipid Res. 1998;39(8):1713–21. [PubMed] [Google Scholar]
- 65.Shinde DD, Kim KB, Oh KS, Abdalla N, Liu KH, Bae SK, et al. LC-MS/MS for the simultaneous analysis of arachidonic acid and 32 related metabolites in human plasma: basal plasma concentrations and aspirin-induced changes of eicosanoids. J Chromatogr B Anal Technol Biomed Life Sci. 2012;911:113–21. [DOI] [PubMed] [Google Scholar]
- 66.Gouveia-Figueira S, Spath J, Zivkovic AM, Nording ML. Profiling the Oxylipin and Endocannabinoid Metabolome by UPLC-ESIMS/MS in Human Plasma to Monitor Postprandial Inflammation. Plos One. 2015;10(7):e0132042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barden A, Mas E, Croft KD, Phillips M, Mori TA. Short-term n-3 fatty acid supplementation but not aspirin increases plasma proresolving mediators of inflammation. J Lipid Res. 2014;55(11):2401–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gouveia-Figueira S, Nording ML, Gaida JE, Forsgren S, Alfredson H, Fowler CJ. Serum levels of oxylipins in achilles tendinopathy: an exploratory study. PLoS One. 2015;10(4):e0123114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schuchardt JP, Schmidt S, Kressel G, Dong H, Willenberg I, Hammock BD, et al. Comparison of free serum oxylipin concentrations in hyper- vs. normolipidemic men. Prostaglandins Leukot Essent Fat Acids. 2013;89(1):19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramsden CE, Domenichiello AF, Yuan ZX, Sapio MR, Keyes GS, Mishra SK, Gross JR, Majchrzak-Hong S, Zamora D, Horowitz MS, Davis JM, Sorokin AV, Dey A, LaPaglia DM, Wheeler JJ, Vasko MR, Mehta NN, Mannes AJ, Iadarola MJ. A systems approach for discovering linoleic acid derivatives that potentially mediate pain and itch. Sci Signal. 2017;10(493):eaal5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sorokin AV, Domenichiello AF, Dey AK, Yuan ZX, Goyal A, Rose SM, Playford MP, Ramsden CE, Mehta NN. Bioactive Lipid Mediator Profiles in Human Psoriasis Skin and Blood. J Invest Dermatol. 2018;138(7):1518–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.