Abstract
The stimulator of interferon genes (STING) plays a crucial role in the recognition of a viral infection and subsequent stimulation of an immune response. However, it is unclear whether methylation of the STING promoter affects STING transcription and response to antiviral therapy. The present study determined the methylation status of the STING promoter in patients with chronic hepatitis B (CHB).
This study included 198 participants, of which 159 participants had CHB and 39 were healthy controls (HCs). Methylation-specific polymerase chain reaction was performed to detect the methylation status of the STING promoter. Reverse transcription-quantitative polymerase chain reaction was performed to determine STING mRNA level in peripheral blood mononuclear cells.
The methylation frequency of the STING promoter was significantly higher and STING mRNA level was lower in the patients with CHB than in the HCs. Presence of hepatitis B virus (HBV) DNA was independently correlated with an increased risk of STING promoter methylation. Virological response frequency was higher in the patients with CHB receiving entecavir (ETV) than in those receiving adefovir (ADV). In the ETV group, the virological response frequency was evidently lower in the patients with CHB having methylated STING promoters than in those having unmethylated STING promoters. However, there was no significant difference in the virological response frequency between ADV-treated patients having methylated and unmethylated STING promoters.
These results indicate that the hypermethylation of the STING promoter and thus the transcriptional repression of STING weaken the effect of STING in inhibiting HBV replication and decreases the effectiveness of antiviral therapy.
Keywords: antiviral therapy, chronic hepatitis B, DNA methylation, hepatitis B virus DNA, the stimulator of interferon genes
1. Introduction
Hepatitis B virus (HBV) infection is an important global public health problem affecting approximately 2 billion people.[1] Approximately 240 million people are chronically infected with HBV worldwide[2]; hepatitis B and its associated complications, such as cirrhosis and hepatocellular carcinoma, account for 686,000 deaths annually.[3] An immune response induced during the course of HBV infection promotes hepatic injury.[4] HBV-infected hepatocytes function as target cells and are essential players in inducing host immune response.[5] However, it is unclear why patients with chronic hepatitis B (CHB) show an immunocompromised status.
The stimulator of interferon genes (STING) plays a crucial role in the recognition of a viral infection and subsequent stimulation of an immune response. Patients with CHB show the persistent presence of HBV in their hepatocytes or sera.[6,7] STING contributes to the induction of an early immune response against HBV infection, which may help in more effective clearance of the virus by host.[8] Some studies have suggested that cyclic GMP-AMP synthase (cGAS)-STING pathway plays a role in inhibiting HBV replication in hepatocytes.[9–12] Furthermore, STING activation in liver-resident immune regulatory cells and hepatocytes suppresses HBV replication.[5] One study has reported that STING deficiency in hepatocytes promotes HBV infection.[8]
Epigenetics defines all heritable changes in gene expression occurring during meiosis and mitosiswith without altering the DNA sequence.[13] DNA methylation is an epigenetic modification that regulates the transcription of genes and affects the development of common diseases.[13,14] DNA methylation involves the addition of a methyl group to the cytosine residue in CpG dinucleotides and leads to gene silencing if it occurs in the promoter region of a gene.[15] Hypermethylation of CpG dinucleotides in gene promoters interferes with gene transcription and downregulates gene expression.[16,17] Patients with CHB do not express appropriate levels of STING.[18] Therefore, it is reasonable to suspect that the STING promoter is hypermethylated in patients with CHB. However, it is unclear whether the methylation of the STING promoter affects STING transcription and is associated with response to antiviral therapy in patients with CHB.
At present, bisulfite modification and methylation-specific polymerase chain reaction (MSP) are commonly used methods for detecting DNA methylation.[19] In the present study, we performed MSP to investigate the methylation status of the STING promoter in peripheral blood mononuclear cells (PBMCs) of patients with CHB and healthy controls (HCs). Moreover, we performed reverse transcription-quantitative polymerase chain reaction (RT-qPCR) to assess STING mRNA expression in the PBMCs of the patients with CHB and HCs. Furthermore, we examined whether the methylation status of the STING promoter affected the regulation of STING mRNA expression and response to antiviral therapy.
2. Methods
2.1. Study population
The study enrolled 218 participants, of which 179 participants had CHB, which was confirmed by performing clinical and laboratory evaluations, and 39 participants were HCs. Patients with CHB were enrolled according to the practice guidelines for managing CHB established in the 2018 update of the American Association for the Study of Liver Diseases. Patients were diagnosed with CHB based on the presence of HBsAg for at least 6 months and at elevated alanine aminotransferase (ALT) levels.[20] All the patients with CHB had increased serum ALT levels (>40 U/L). These patients visited the Department of Hepatology, Qilu Hospital of Shandong University, from January 2016 to January 2018. However, 20 patients with CHB were excluded from the study because they had co-existing hepatocellular carcinoma (14 patients), fatty liver disease (4 patients), and hepatitis C infection (2 patients). Thus, the present study included 159 patients with CHB. Of these, 83 patients received antiviral therapy and 76 patients did not receive antiviral treatment. Of the 83 patients who received the antiviral therapy, 51 patients received entecavir and 32 patients received adefovir. Moreover, 22, 32, 16, and 13 patients out of the 83 patients who received the antiviral therapy were previously treated for 1 to 6, 6 to 12, 12 to 24, and >24 months, respectively. Written informed consent was obtained from all the study participants in accordance with the Declaration of Helsinki,[21] and study protocols were approved by the local research and ethics committee of the Qilu Hospital of Shandong University.
2.2. Genomic DNA extraction and bisulfite conversion
Genomic DNA was extracted from the PBMCs of the study participants by using QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany), according to the manufacturer's protocol, and was stored at −20°C until further use for performing bisulfite conversion. The concentration and purity of the extracted DNA were measured using Eppendorf Biophotometer (Brinkmann Instruments, Westbury, NY). Bisulfite conversion was performed by treating the genomic DNA with EZ DNA Methylation-Gold Kit (Zymo Research Corp, Orange, CA), according to the manufacturer's instructions. Finally, 20 μL modified DNA was immediately used as a template for MSP or was stored at −20°C.
2.3. Methylation-specific polymerase chain reaction
The stimulator of interferon genes promoter-specific methylated and unmethylated primers were designed using MethPrimer according to recommended criteria.[22] The sequences of these primers are as follows: methylated forward primer: 5′-TTAGGTTGGAGTGTAATGGTACG-3′, methylated reverse primer: 5′-AAATTAACTAAACGTAATAACGCA-3′, unmethylated forward primer: 5′-TAGGTTGGAGTGTAATGGTATGA-3′, and unmethylated reverse primer: 5′-AAAAATTAACTAAACATAATAACACAT-3′. Amplification was performed in a 25-μL reaction mixture containing 1 μL bisulfite-modified DNA, 0.5 μL of each primer (10 μM), 10.5 μL nuclease-free water, and 12.5 μL Premix Taq (Zymo Research Corp). PCR was performed using the following conditions: initial denaturation at 95°C for 10 minutes; 40 cycles of denaturation at 94°C for 30 seconds, annealing at 60.5°C (for methylated STING primers) or 58°C (for unmethylated STING primers) for 30 seconds, primer extension at 72°C for 30 seconds; and final extension at 72°C for 10 minutes. Water without the modified DNA was used as a negative control. PCR products were electrophoresed on 2% agarose gels stained with Gelred (Biotium, CA) and visualized under UV illumination. Each reaction was replicated 3 times.
2.4. RNA extraction from PBMCs and RT-qPCR
Total RNA was extracted from the PBMCs of the study participants by using phenol-chloroform-isopropanol method, according to a recommended protocol, and was suspended in 20 μL RNase-free water. Next, cDNA was synthesized using a first-strand cDNA synthesis kit (Fermentas, Vilnius, Lithuania), according to the manufacturer's instructions. RT-qPCR to measure STING mRNA level was performed using SYBR Green (Toyobo, Osaka, Japan) and Agilent Technologies Stratagene Mx3005P instrument (Stratagene, La Jolla, CA), with the β-actin gene as an internal control and the following condition: initial denaturation at 95°C for 30 seconds, followed by 45 cycles of denaturation at 95°C for 5 seconds, annealing at 60°C for 30 seconds, and a final extension at 72°C for 30 seconds. The RT-qPCR was performed using the following primers: STING forward: 5′-GAGATCTCTGCAGTGTGTGAA-3′, STING reverse: 5′-GCCGCAGATATCCGATGTAATA-3′, β-actin forward: 5′-CACCATTGGCAATGAGCGGTTC-3′, and β-actin reverse 5′-AGGTCTTTGCGGATGTCCACGT-3′.[5] Each reaction was conducted in triplicate, and mRNA expression was evaluated using a comparative (2-DDCt) method.
2.5. Clinical features of the study participants
The levels of serum biochemical markers, including aspartate aminotransferase (AST), ALT, γ-glutamyl transferase (GGT), alkaline phosphatase (AKP), total bilirubin (TBIL), albumin (ALB), blood urea nitrogen (BUN), and creatine (Cr), were determined using COBAS Integra 800 (Roche Diagnostics, Penzberg, Germany). Serum HBV DNA load was determined using ABI 7300 PCR System (Applied Biosystems, Foster City, CA), and HBsAg and HBeAg levels were determined using COBAS 6000 analyzer series (Roche Diagnostics, Basel, Switzerland). Prothrombin time (PT) and prothrombin activity (PTA) were quantified using ACL TOP 700 (Instrument Laboratory, Lexington, MA). All clinical features were determined at the Department of Laboratory Medicine, Qilu Hospital of Shandong University.
2.6. Statistical analysis
Quantitative variables are expressed as median (centile 25; centile 75), and categorical variables are expressed as number (%). All the data were analyzed using SPSS 21 software (SPSS, Inc., Chicago, IL). The baseline characteristics of the participants were assessed using Mann–Whitney U test and chi-square test. The relationship between the methylation status of the STING promoter and the clinical features of the patients with CHB was investigated using multivariate logistic regression analysis. Univariate and multivariate logistic regression analyses were used to investigate the relationship of the virological response of the patients with CHB with clinical features, STING promoter methylation status, and drug treatment. Spearman correlation was used to analyze the association between the quantitative and categorical variables. Biochemical response was defined as the normalization of serum ALT level. Serological response was defined as the loss of HBeAg and seroconversion to anti-HBe in patients with HBeAg-positive CHB infection or loss of HBsAg and seroconversion to anti-HBs. Virological response was defined as the absence of serum HBV DNA during therapy or reduction in serum HBV DNA level to >1 log IU/mL after 24 weeks of oral antiviral therapy in adherent patients. All the responses to antiviral therapy were recorded after 12 weeks of therapy.[2]
3. Results
3.1. General characteristics of the study participants
This study included 159 patients with CHB and 39 HCs. The basic demographic and clinical characteristics of the study participants are shown in Table 1. Significant differences were observed between the patients with CHB and the HCs with respect to ALT (P < .001), AST (P < .001), GGT (P < .001), AKP (P = .001), TBIL (P = .003), ALB (P < .001), BUN (P = .003), and Cr (P = .001) levels; PT (P < .001); and PTA (P < .001). However, no difference was observed between the patients with CHB and the HCs with respect to gender (P = .297) and age (P = .230).
Table 1.
General characteristics of the enrolled participants.
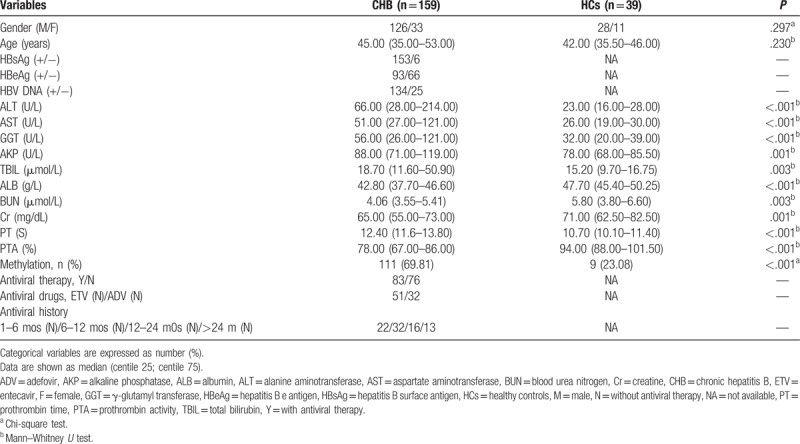
3.2. Methylation status of the STING promoter in the patients with CHB and HCs
The methylation status of the STING promoter was determined by performing MSP. Hypermethylated STING promoter was detected in the PBMCs of 111 out of 159 (69.81%) patients with CHB and 9 out of 39 (23.08%) HCs. The methylation frequency of the STING promoter was significantly higher in the patients with CHB than in the HCs (P < .001; Fig. 1A). STING promoter methylation was detected in 64 of the 83 patients with CHB who received the antiviral therapy and in 47 of the 76 patients with CHB who did not receive the antiviral therapy. Moreover, the methylation frequency of the STING promoter was higher in the patients who received the antiviral therapy than in those who did not receive the antiviral therapy (P = .036; Fig. 1B). However, no significant difference in the methylation frequency of the STING promoter was observed between entecavir (ETV) and adefovir (ADV)-treated patients with CHB and among patients with a history of antiviral therapy (Fig. 1C and D). Fig. 1E shows a typical representative MSP assay of STING promoter methylation.
Figure 1.
(A) The methylation frequency of the STING promoter in the PBMCs of the patients with CHB and the HCs; ∗∗∗P < .001. (B) The methylation frequency of the STING promoter in the patients treated with or without the antiviral therapy; ∗P < .05. (C and D) The methylation frequency of the STING promoter in the patients receiving the different antiviral drugs and with a different antiviral treatment history. (E) A representative image showing the methylation of the STING promoter by performing MSP. A 50-bp marker was used. CHB = chronic hepatitis B, HCs = healthy controls, M = methylated sequence, MSP = methylation-specific polymerase chain reaction, NC = negative control, PBMCs = peripheral blood mononuclear cells, STING = the stimulator of interferon genes, U = unmethylated sequence.
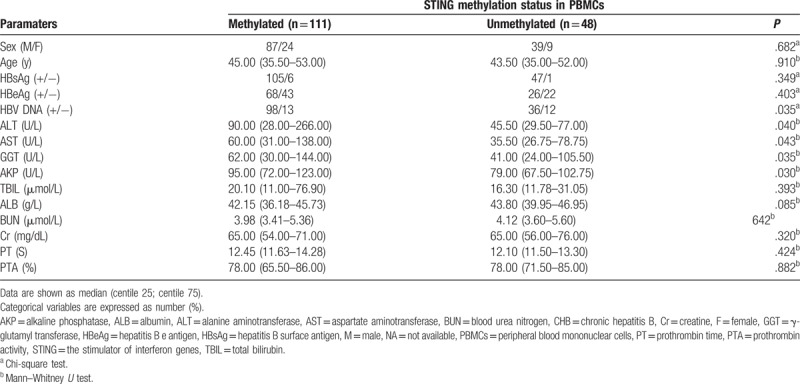
3.3. Correlation between the methylation of the STING promoter and the clinical features of patients with CHB
Table 2 shows the association between the methylation of the STING promoter and the clinical features of the patients with CHB. STING promoter methylation was significantly correlated with HBV DNA (P = .035), ALT (P = .040), AST (P = .043), GGT (P = .035), and AKP (P = .030) levels. HBV DNA positivity was higher in the patients with CHB having methylated STING promoters than in those having unmethylated STING promoters (P = .035). However, no correlation was observed between STING promoter methylation and sex; age; HBeAg, TBIL, ALB, BUN, and Cr levels; PT; and PTA (P > .05 for all).
Table 2.
The association of STING promoter methylation and clinical features of the patients with CHB patients.
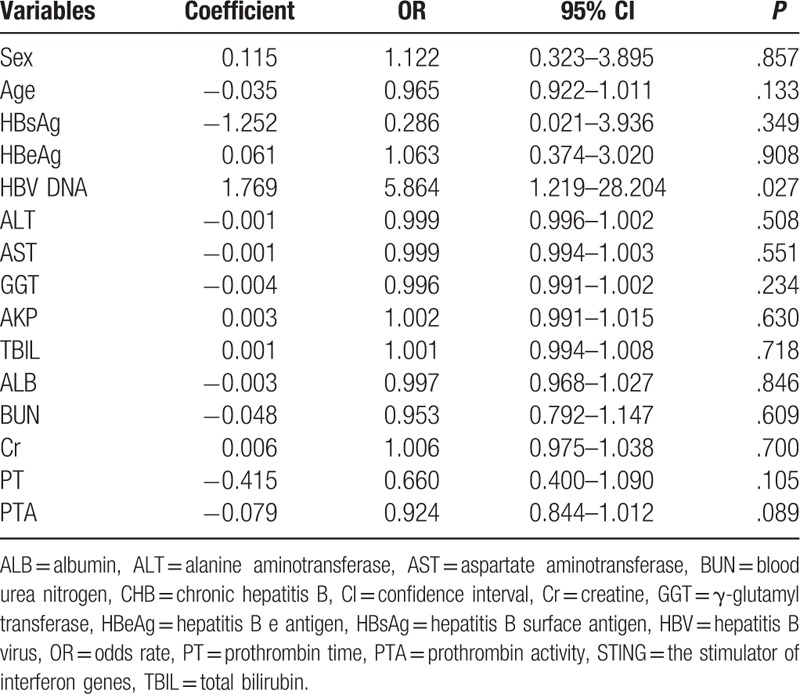
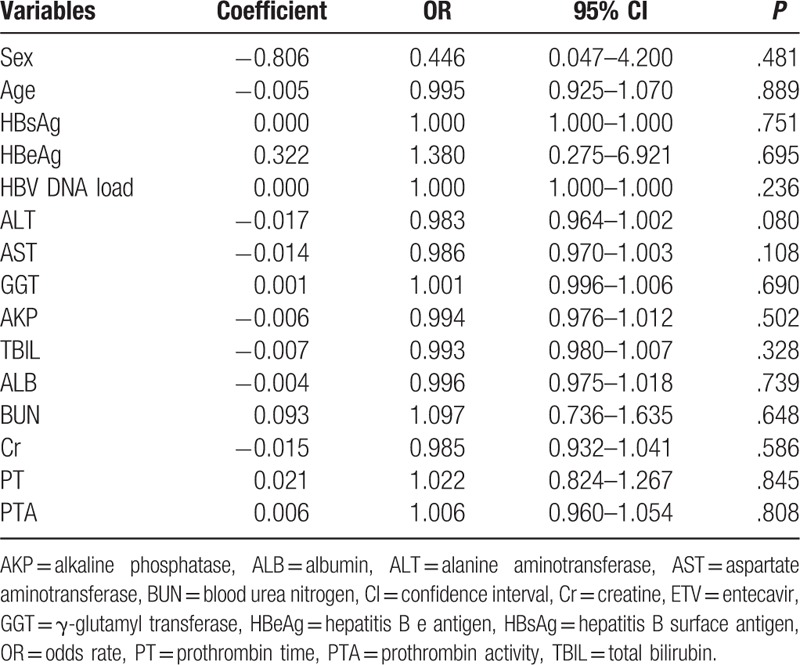
Results of multivariate logistic regression analysis indicated that only HBV DNA was independently correlated with an increased risk of STING promoter methylation (P = .027), whereas sex; age; HBeAg, ALT, AST, GGT, AKP, TBIL, ALB, BUN, and Cr levels; PT; or PTA were not correlated with an increased risk of STING promoter methylation (Table 3).
Table 3.
Multivariate logistic regression analysis of clinicopathological parameters with STING promoter methylation in CHB.
3.4. STING mRNA level in PBMCs
The STING mRNA level was evidently lower in the patients with CHB than in the HCs (P < .001; Fig. 2A). Meanwhile, STING mRNA level was significantly lower in both the patients with CHB (P = .023; Fig. 2B) and HCs (P < .001; Fig. 2C) having methylated STING promoters than in those having unmethylated STING promoters. Moreover, STING mRNA level was lower in the patients showing HBV DNA positivity than in those showing HBV DNA negativity (P = .046; Fig. 2D). However, no obvious difference in STING mRNA level was observed between the patients showing HBsAg positivity and negativity, and between the patients showing HBeAg positivity and negativity (Fig. 2E and F, respectively). The methylation status of the STING promoter was negatively correlated with the STING mRNA level in patients with CHB (r = −0.204, P = .010). We also analyzed the correlation between the STING mRNA level and the clinical features of the patients with CHB to determine the possible correlation between STING promoter methylation and CHB severity. No evident correlation was observed between the STING mRNA level and HBsAg level (r = 0.101, P = .203); HBeAg level (r = 0.075, P = .350); AST, GGT, AKP, TBIL, and ALB levels; PT; and PTA. However, the STING mRNA level was negatively correlated with ALT level (r = −0.218, P = .006) and HBV DNA (r = −0.159, P = .046), and was positively correlated with BUN (r = 0.214, P = .017) and Cr level (r = 0.191, P = .033) (Fig. 2G–P).
Figure 2.
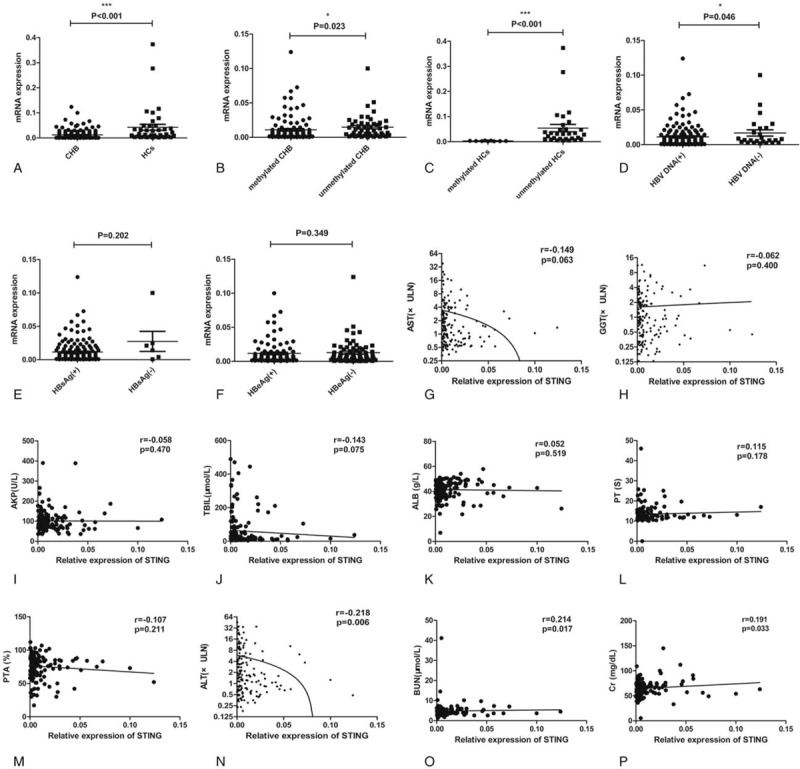
(A–D) STING mRNA level in the different study groups. A significant difference in the STING mRNA level was observed between the patients with CHB and the HCs (P < .001), between the patients with CHB having methylated and unmethylated STING promoters (P = .023), between the HCs having methylated and unmethylated STING promoters (P < .001), and between the patients with CHB showing HBV DNA positivity and negativity (P = .046). (E and F) No obvious difference was observed in the STING mRNA level between the patients with CHB showing HBsAg positivity and negativity (P = .202) and those showing HBeAg positivity and negativity (P = .349). (G–M) No significant correlations were observed between the STING mRNA level and AST (Spearman r = −0.149, P = .063), GGT (Spearman r = −0.062, P = .400), AKP (Spearman r = −0.058, P = .470), TBIL (Spearman r = −0.143, P = .075), and ALB (Spearman r = 0.052, P = .519) levels; PT (Spearman r = 0.115, P = .178); and PTA (Spearman r = −0.107, P = .211). (N–P) Significant correlations were observed between the STING mRNA level and ALT (Spearman r = −0.218, P = .006), BUN (Spearman r = 0.214, P = .017), and Cr (Spearman r = 0.191, P = .033) levels. ∗P < .05; ∗∗∗P < .001. AKP = alkaline phosphatase, ALB = albumin, ALT = alanine aminotransferase, AST = aspartate aminotransferase, BUN = blood urea nitrogen, CHB = chronic hepatitis B, Cr = creatine, GGT = γ-glutamyl transferase, HBeAg = hepatitis B e antigen, HBsAg = hepatitis B surface antigen, HBV = hepatitis B virus, HCs = healthy controls, PT = prothrombin time, PTA = prothrombin activity, STING = the stimulator of interferon genes, TBIL = total bilirubin.
3.5. Response of the patients with CHB to antiviral therapy
Because some patients missed their follow-up visits, the outcomes of only 63 patients with CHB (41 ETV-treated patients and 22 ADV-treated patients) were used to evaluate responses to the antiviral therapy. A marked difference in the virological response was observed between the ETV and ADV-treated patients with CHB (P = .023; Table 4); however, no difference was observed in the biochemical and serological responses of these patients. The virological response frequency was higher in the ETV-treated patients than in the ADV-treated patients (P = .023). However, the virological response frequency was lower in the ETV-treated patients with methylated STING promoters than in those with unmethylated STING promoters (P = .046). However, no significant difference in the virological response frequency was observed between the ADV-treated patients with methylated and unmethylated STING promoters (P = .127). Moreover, no significant difference in the virological response frequency was observed in the 63 patients with CHB with methylated and unmethylated STING promoters (P = .464; Fig. 3).
Table 4.
The response to antiviral therapy in patients with CHB.
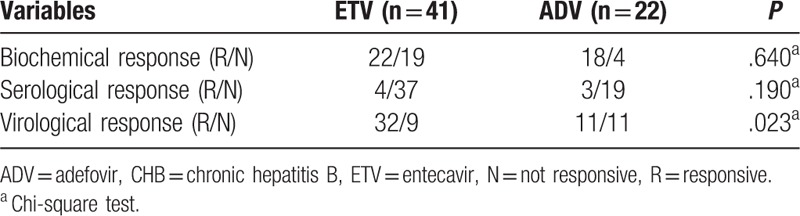
Figure 3.
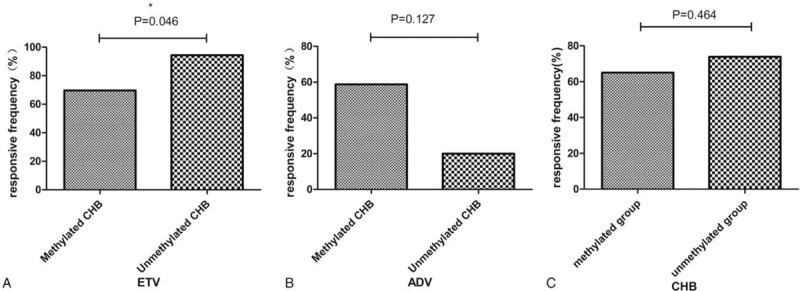
The virological response frequency about methylation in ETV and ADV-treated patients. ∗P < .05. ADV = adefovir, ETV = entecavir.
Results of the univariate analysis showed no association between the virological response and clinical characteristics of the patients with CHB, except for ALT (P = .011) and AST (P = .012) levels (Table 5). Multivariate logistic regression analysis was performed to assess the association of virological response with some clinical characteristics of the patients with CHB (P < .2), STING mRNA level, STING promoter methylation status, and antiviral therapy. Results of the multivariate logistic regression analysis showed that only ALT level (P = .038) and antiviral therapy (P = .011) were associated with the virological response (Table 6).
Table 5.
Univariate analysis of clinicopathological parameters with virological responsive in patients with CHB.
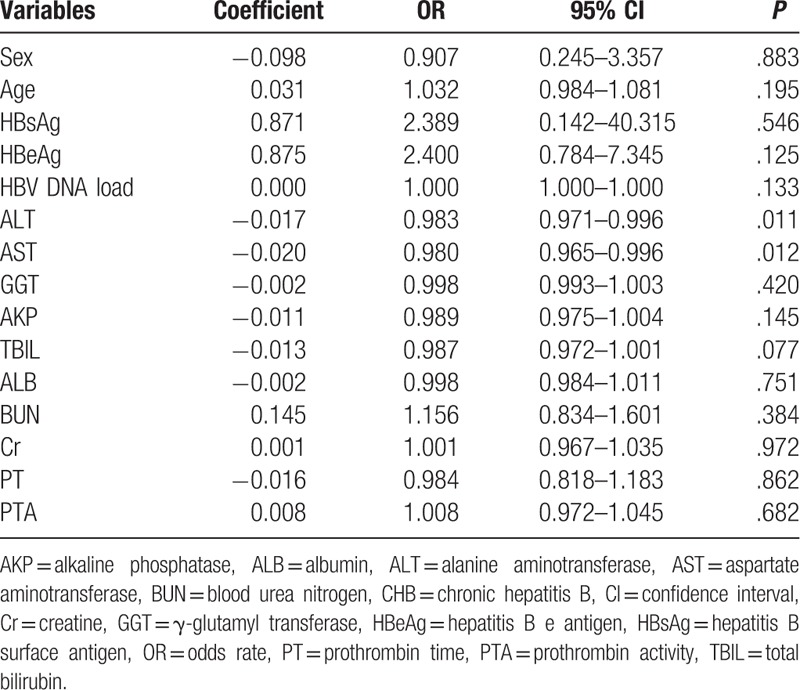
Table 6.
Multivariate logistic regression analysis in patients with CHB.
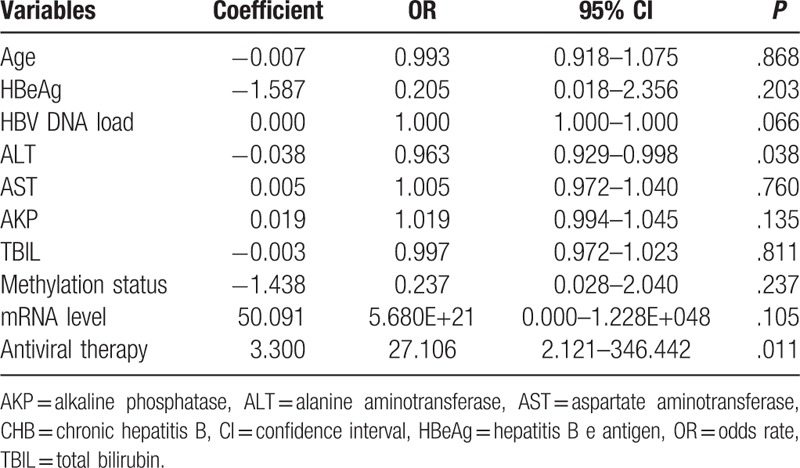
Univariate analysis of the ETV-treated patients with CHB showed no association between the clinical characteristics of these patients and virological response (Table 7). Therefore, ALT and AST levels (P < .2), STING mRNA level, and STING promoter methylation status were used to perform multivariate logistic regression analysis. Results of this analysis showed that only STING promoter methylation status (P = .027) was associated with and that ALT level (P = .091), AST level (P = .912), and STING mRNA level (P = .329) were not associated with the virological response. The number of ADV-treated patients with CHB was very less to perform univariate and multivariable logistic regression analyses for assessing the association of the clinical characteristics, STING mRNA level, STING promoter methylation status, and antiviral therapy with virological response in these patients.
Table 7.
Univariate analysis of clinicopathological parameters with virological responsive in ETV group.
4. Discussion
To our knowledge, this is the first study to investigate the methylation status of the STING promoter in patients with CHB. In the present study, we found that the methylation frequency of the STING promoter was higher in the patients with CHB than in the HCs. Moreover, we found that the methylation frequency of the STING promoter was higher in the patients with CHB receiving antiviral therapy than in those not receiving the antiviral therapy. The STING mRNA level was lower in the patients with CHB than in the HCs. Moreover, the STING mRNA level was lower in the patients with CHB having methylated STING promoters than in those having unmethylated STING promoters. Furthermore, we found that the methylation status of the STING promoter was negatively correlated with the STING mRNA level. HBV DNA positivity was evidently higher in the patients with CHB having methylated STING promoters than in those having unmethylated STING promoters; moreover, presence of HBV DNA was correlated with an increased risk of STING promoter methylation. In addition, HBV DNA positivity was negatively correlated with the STING mRNA level. The virological response was higher in the ETV-treated patients with CHB than in ADV-treated patients with CHB. Among the ETV-treated patients, the virological response frequency was lower in the patients having methylated STING promoters than in those having unmethylated STING promoters. Therefore, we concluded that STING methylation suppressed the effect of STING in inhibiting HBV replication and thus could be a potential therapeutic target for treating patients with CHB.
Hepatitis B virus is an enveloped double-stranded DNA virus, and a complex interaction between the host immune system and the replicating virus leads to HBV infection.[23,24] cGAS-cGAMP-STING pathway is essential for DNA-mediated immune response.[25] Cytoplasmic DNA activates cGAS to produce cGAMP, which binds to STING to induce type I interferons (IFNs).[26,27] A recent study reported that STING agonists prevented HBV infection by inducing an innate antiviral immune response.[28] Another study reported that STING activation suppressed HBV replication.[5] Several studies have reported that DNA methyltransferases (DNMTs) mRNA level is significantly higher in patients with CHB than in HCs, and that persistent HBV infection stimulates DNMT overexpression.[29,30] The present study showed that STING mRNA level was negatively correlated with STING promoter methylation status and HBV DNA positivity in the patients with CHB. Gene promoter methylation is usually associated with transcriptional silencing and may result in gene function loss.[31] Therefore, we hypothesized that the hypermethylation of the STING promoter reduced STING mRNA expression by decreasing gene transcription in patients with CHB, thus weakening the effect of STING on the inhibition of HBV replication.
A previous study suggested that some therapies influence DNA methylation status.[32] Similarly, the present study showed that the methylation frequency of the STING promoter was higher in the patients receiving the antiviral therapy than in those not receiving the antiviral therapy. However, no difference in the methylation frequency of the STING promoter was observed between the ETV and ADV-treated patients with CHB. Therefore, we hypothesized that these antiviral drugs increased the DNA methylation status by themselves or by affecting HBV DNA replication. Interestingly, we found that the methylation frequency of the STING promoter increased with time; however, this increase was not statistically significant. Therefore, additional studies involving a large sample size are needed to investigate whether methylation frequency of the STING promoter is associated with the history of antiviral therapy.
Assessment of liver disease severity is crucial and is based on physical examination and evaluation of biochemical parameters such as ALT, AST, GGT, AKP, and BUN level, and PT.[33] In the present study, STING promoter methylation was significantly associated with ALT (P = .040), AST (P = .043), GGT (P = .035), and AKP (P = .030) levels. STING mainly activates type I IFN expression.[10] We observed that the patients with CHB having high ALT levels showed high STING methylation frequency. These results suggest that methylation of the STING promoter inhibits type I IFN secretion and induces a weak innate anti-HBV response during the immune clearance of CHB. These results are consistent with those of previous studies that reported that IFN-γ rather than type I IFN mainly contributed to immune clearance. CD8+ T cells and IFN-γ secretion play critical roles in HBV clearance in patients of CHB.[34] The early stage of acute HBV infection is characterized by the induction of interleukin-10 rather than of type I IFNs.[35] Therefore, we believe that the hypermethylation of the STING promoter contributes to the severity of CHB.
Entecavir and ADV are broadly used for treating patients with CHB. In the present study, the virological response frequency was higher among the ETV-treated patients with CHB than among the ADV-treated patients with CHB (P = .023). Moreover, we found that the antiviral therapy was associated with the virological response (P = .011). Similar to the results of some previous studies,[36,37] the results of the present study suggest that ETV induces a more potent virological response than ADV in patients with CHB. Moreover, our results suggest that the methylation of the STING promoter affects the efficacy of antiviral therapy. We found that the virological response frequency was evidently lower in the ETV-treated patients with CHB having methylated STING promoters than in those having unmethylated STING promoters (P = .046). Moreover, we found that the virological response was evidently associated with the methylation status of the STING promoter (P = .027). A separate analysis of the 63 patients with CHB and of the ADV-treated patients with CHB showed no obvious difference in the virological response frequency between the patients with methylated and unmethylated STING promoters. This may be associated with the very less number of patients in the ADV treatment group. However, we still believe that the methylation of the STING promoter reduces the effect of antiviral drugs (at least ETV).
However, the present study has some limitations. First, MSP is a qualitative method for detecting DNA methylation, and other methods such as direct sequencing may provide detailed information on DNA methylation status. However, because MSP is easy to perform, it can be used for the primary screening of patients suspected of having methylated DNA. Second, HBV replication occurs in hepatocytes. However, it is out of reality that all the patients accept the liver biopsy in real world. The present study was aimed to determine whether STING promoter methylation in PBMCs was a promising noninvasive biomarker in the clinical setting. Therefore, we investigated STING promoter methylation in PBMCs rather than in hepatocytes. Third, the number of patients receiving ADV was very less, which may have affected some study results. Therefore, additional studies involving a large sample size are needed to confirm the findings of the present study.
5. Conclusions
In conclusion, the present study is the first to report the elevated methylation frequency of the STING promoter in patients with CHB. We found that the methylation of the STING promoter and transcriptional repression of STING were associated with HBV replication and CHB severity. Moreover, we observed that in patients with ETV therapy, there was a decreased virological response frequency in the patients with CHB having methylated STING promoters. Thus, the hypermethylation of the STING promoter can be used to develop potential new prevention and treatment strategies for patients with CHB, especially for patients who are sensitive to ETV therapy.
Author contributions
Data curation: Chen-Si Wu, Qian Zhao, Jun Zhang.
Formal analysis: Chen-Si Wu, Jing-Wen Wang, Yu Qian.
Funding acquisition: Kai wang.
Investigation: Chen-Si Wu.
Methodology: Chen-Si Wu.
Supervision: Kai wang.
Writing – original draft: Kai wang, Chen-Si Wu.
Writing – review & editing: Kai wang, Yu-Chen Fan.
Footnotes
Abbreviations: ADV = adefovir, AKP = alkaline phosphatase, ALB = albumin, ALT = alanine aminotransferase, AST = aspartate aminotransferase, BUN = blood urea nitrogen, CHB = chronic hepatitis B, Cr = creatine, DNMTs = DNA methyltransferases, ETV = entecavir, GGT = γ-glutamyl transferase, HBeAg = hepatitis B e antigen, HBsAg = hepatitis B surface antigen, HBV = hepatitis B virus, HCs = healthy controls, MSP = methylation-specific polymerase chain reaction, PBMCs = peripheral blood mononuclear cells, PT = prothrombin time, PTA = prothrombin activity, RT-qPCR = reverse transcription-quantitative polymerase chain reaction, STING = the stimulator of interferon genes, TBIL = total bilirubin.
Funding: This work was supported by the Key Project of Chinese Ministry of Science and Technology (2018ZX10301406 and 2017ZX10202202) and the Fundamental Research Funds of Shandong University-Clinical Research Project of Qilu Hospital (2014QLKY11).
The authors declare that they do not have any competing financial interests related to this study.
References
- [1].Hoan NX, Van Tong H, Giang DP, et al. SOCS3 genetic variants and promoter hypermethylation in patients with chronic hepatitis B. Oncotarget 2017;8:17127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sarin SK, Kumar M, Lau GK, et al. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatology international 2016;10:1–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mortality GBD. Causes of Death C Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015;385:117–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rehermann B, Bertoletti A. Immunological aspects of antiviral therapy of chronic hepatitis B virus and hepatitis C virus infections. Hepatology 2015;61:712–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Guo F, Tang L, Shu S, et al. Activation of stimulator of interferon genes in hepatocytes suppresses the replication of hepatitis B virus. Antimicrob Agents Chemother 2017;61: doi: 10.1128/AAC.00771-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sajadi SM, Mirzaei V, Hassanshahi G, et al. Decreased expressions of Toll-like receptor 9 and its signaling molecules in chronic hepatitis B virus-infected patients. Arch Pathol Lab Med 2013;137:1674–9. [DOI] [PubMed] [Google Scholar]
- [7].Assar S, Arababadi MK, Mohit M, et al. T helper and B cell escape mutations within the HBc gene in patients with asymptomatic HBV infection: a study from the South-Eastern region of Iran. Clin Lab 2012;58:53–60. [PubMed] [Google Scholar]
- [8].Dickson I. Viral hepatitis: a lack of hepatocyte STING favours HBV infection. Nat Rev Gastroenterol Hepatol 2016;13:438. [DOI] [PubMed] [Google Scholar]
- [9].Liu S, Zhao K, Su X, et al. MITA/STING and its alternative splicing isoform MRP restrict hepatitis B virus replication. PloS One 2017;12:e0169701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].He J, Hao R, Liu D, et al. Inhibition of hepatitis B virus replication by activation of the cGAS-STING pathway. J Gen Virol 2016;97:3368–78. [DOI] [PubMed] [Google Scholar]
- [11].Dansako H, Ueda Y, Okumura N, et al. The cyclic GMP-AMP synthetase-STING signaling pathway is required for both the innate immune response against HBV and the suppression of HBV assembly. FEBS J 2016;283:144–56. [DOI] [PubMed] [Google Scholar]
- [12].Cui X, Clark DN, Liu K, et al. Viral DNA-dependent induction of innate immune response to hepatitis B virus in immortalized mouse hepatocytes. J Virol 2016;90:486–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Egger G, Liang G, Aparicio A, et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004;429:457–63. [DOI] [PubMed] [Google Scholar]
- [14].Feinberg AP. Epigenetics at the epicenter of modern medicine. JAMA 2008;299:1345–50. [DOI] [PubMed] [Google Scholar]
- [15].Feng Q, Stern JE, Hawes SE, et al. DNA methylation changes in normal liver tissues and hepatocellular carcinoma with different viral infection. Exp Mol Pathol 2010;88:287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yu J, Ni M, Xu J, et al. Methylation profiling of twenty promoter-CpG islands of genes which may contribute to hepatocellular carcinogenesis. BMC Cancer 2002;2:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhong C, Lu H, Han T, et al. CpG methylation participates in regulation of hepatitis B virus gene expression in host sperm and sperm-derived embryos. Epigenomics 2017;9:123–5. [DOI] [PubMed] [Google Scholar]
- [18].Karimi-Googheri M, Daneshvar H, Khaleghinia M, et al. Decreased expressions of STING but not IRF3 molecules in chronic HBV infected patients. Arch Iran Med 2015;18:351–4. [PubMed] [Google Scholar]
- [19].Yang AS, Estecio MR, Doshi K, et al. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucl Acids Res 2004;32:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Terrault NA, Lok ASF, McMahon BJ, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 2018;67:1560–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].General Assembly of the World Medical Association World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. J Am Coll Dentists 2014;81:14–8. [PubMed] [Google Scholar]
- [22].Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002;18:1427–31. [DOI] [PubMed] [Google Scholar]
- [23].Leong CR, Oshiumi H, Suzuki T, et al. Nucleic acid sensors involved in the recognition of HBV in the liver-specific in vivo transfection mouse models: pattern recognition receptors and sensors for HBV. Med Sci (Basel) 2015;3:16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wieland S, Thimme R, Purcell RH, et al. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci USA 2004;101:6669–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell 2014;54:289–96. [DOI] [PubMed] [Google Scholar]
- [26].Gao D, Wu J, Wu YT, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 2013;341:903–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008;455:674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Guo F, Han Y, Zhao X, et al. STING agonists induce an innate antiviral immune response against hepatitis B virus. Antimicrob Agents Chemother 2015;59:1273–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fan XP, Ji XF, Li XY, et al. Methylation of the glutathione-S-transferase P1 gene promoter is associated with oxidative stress in patients with chronic hepatitis B. Tohoku J Exp Med 2016;238:57–64. [DOI] [PubMed] [Google Scholar]
- [30].Li H, Yang F, Gao B, et al. Hepatitis B virus infection in hepatocellular carcinoma tissues upregulates expression of DNA methyltransferases. Int J Clin Exp Med 2015;8:4175–85. [PMC free article] [PubMed] [Google Scholar]
- [31].Zhao J, Fan YC, Liu XY, et al. Hypermethylation of the galectin-3 promoter is associated with poor prognosis of acute-on-chronic hepatitis B liver failure. Digest Liver Dis 2017;49:664–71. [DOI] [PubMed] [Google Scholar]
- [32].Madeddu G, Ortu S, Garrucciu G, et al. DNMT1 modulation in chronic hepatitis B patients and hypothetic influence on mitochondrial DNA methylation status during long-term nucleo(t)side analogs therapy. J Med Virol 2017;89:1208–14. [DOI] [PubMed] [Google Scholar]
- [33].European Association for the Study of the Liver Electronic addressee, European Association for the Study of the L. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol 2017;67:370–98. [DOI] [PubMed] [Google Scholar]
- [34].Lan P, Zhang C, Han Q, et al. Therapeutic recovery of hepatitis B virus (HBV)-induced hepatocyte-intrinsic immune defect reverses systemic adaptive immune tolerance. Hepatology 2013;58:73–85. [DOI] [PubMed] [Google Scholar]
- [35].Dunn C, Peppa D, Khanna P, et al. Temporal analysis of early immune responses in patients with acute hepatitis B virus infection. Gastroenterology 2009;137:1289–300. [DOI] [PubMed] [Google Scholar]
- [36].Shi H, Han Z, Liu J, et al. Comparing efficacy of lamivudine, adefovir dipivoxil, telbivudine, and entecavir in treating nucleoside analogues naive for HBeAg-negative hepatitis B with medium hepatitis B virus (HBV) DNA levels. Med Sci Monit 2017;23:5230–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Leung N, Peng CY, Hann HW, et al. Early hepatitis B virus DNA reduction in hepatitis B e antigen-positive patients with chronic hepatitis B: a randomized international study of entecavir versus adefovir. Hepatology 2009;49:72–9. [DOI] [PubMed] [Google Scholar]


