Abstract
Hepatitis B virus (HBV) infects more than 400 million humans Worldwide. Currently, development of new anti-HBV agents is focused on inhibiting of HBV DNA polymerase activity. The natural components of medicinal plant have a broad spectrum of biological activities with therapeutic properties which can be exploited in various steps of drug discovery. Currently, in silico analyses have been introduced as alternative or supplements methods for drug discovery. This study was planned to in silico screening novel HBV DNA polymerase inhibitor(s) from R. palmatum, R. coreanus and S. officinalis. For this purpose, a set of dominant phytochemicals from mentioned plants were retrieved from PubChem database and primary screening was performed with molecular docking method using iGemdock 2.1 software. SwissADME and MedChem Designer 3.0 were used to calculate the drug-likeness parameters of the ligands. Furthermore, the genotoxicity of the studied ligands was predicted using Toxtree 2.6.6 software. Final analysis of screened compounds was done using Autodock 4 software. Result confirmed that Frangulosid and Lindleyin acid have most and least efficacy in HBV DNA polymerase inhibition with the inhibition constant of 2.97 and 53.83 µM, respectively. Results also showed that, the amino acids, involved in interaction, were different for each compound. In this regards, results revealed that the main amino acids residues of the receptor, involved in interaction with Quercetin-3-glucuronide, Frangulosid and Lindleyin separately, located in 420–424, 606–615 and 512–542 spectra, respectively. In conclusion, Frangulosid can be considered as a good candidate for more investigation of its anti-HBV activity.
Electronic supplementary material
The online version of this article (10.1007/s40203-018-0047-3) contains supplementary material, which is available to authorized users.
Keywords: Hepatitis B virus, Medical plant, In silico analysis, Molecular docking
Background
Hepatitis B virus (HBV) infects more than 400 million humans Worldwide. HBV is a common cause of liver cancer and about one million die each year from HBV-infected liver diseases (Lok and McMahon 2009; Michel et al. 2011; Noordeen 2015). More than 80% of HBV carriers have different levels of hepatocellular disease including cirrhosis and liver cancer (Ganem and Prince 2004). HBV is a small-enveloped DNA virus and the prototype member of Hepadnaviridae family. The genome of HBV is a relaxed circular DNA (rcDNA), partially duplexed DNA genome is converted into a covalently closed circular molecule (cccDNA)in HBV life cycle (Nassal and Schaller 1993; Neuveut et al. 2010). The HBV genome is composed of four overlapping open reading frames (ORFs). The P ORF encodes a HBV multifunctional polymerase (HP), which replicates DNA genome via a RNA intermediate termed pregenomic RNA (pgRNA). The main domains of HBV DNA polymerase are including the reverse transcriptase (RT), the terminal protein (TP) and the RNaseH domains. The two main motif of DNA polymerase including YMDD active site and D-E-D-D motif are located in the RT and RNaseH domains, respectively (Jones et al. 2014). The HBV DNA polymerase has been shown to play an essential role in the virus infection.
Due to key role of HBV DNA polymerase in the virus life cycle and progression of HBV infection, currently many efforts have been done to introduce novel HBV DNA polymerase inhibitors (Kim et al. 2010; Langley et al. 2007; Mak et al. 2016; Shaw et al. 1996). Because of development of drug resistance of HBV to common drugs such as nucleotide analogues as well as side effects of available anti-HBV drugs recently new drugs with natural resource especially plant origin compounds are highly regarded. The phytocompounds have broad spectrum of biological activities with therapeutic properties that can be exploited in various steps of drug discovery and design (Meng et al. 2015). Some studies have reported the inhibitory effects of both crud extracts and pure compounds from herbal medicines on some virus such as herpes simplex viruses (HSV), human immunodeficiency virus (HIV) and HBV (Khan et al. 2005; Mukhtar et al. 2008).
Experimental screening of anti-viral compounds and determinate mechanism of action is time and cost consuming. Therefore, currently in silico analysis such as molecular docking, molecular dynamics, target prediction and toxicity prediction have been introduced as alternative or supplements methods (Dibyajyoti et al. 2013; Kandagalla et al. 2016; Muzaffer et al. 2016). In this study, we introduced some HBV DNA polymerase inhibitor(s) form Rheum palmatum, Rubus coreanus and Sanguisorba officinalis which reported in previous studies as the source of anti-HBV agents(Kim et al. 2001). For this purpose, firstly a database of active compounds from mentioned plants was prepared. In the next step, ligand optimization and physicochemical properties prediction were done. Finally, active compounds in HBV DNA polymerase inhibition was determined using primary and final molecular docking studies.
Materials and methods
Retrieval of receptor and ligands from database
The set of phytocompounds selected for this study are present in Table 1. Three-dimensional structures of all the phytochemicals and lamivudine as positive control were retrieved from Pubchem database (http://pubchem.ncbi.nlm.nih.gov). In addition, an amino acid sequence of HBV DNA polymerase as receptor was obtained from NCBI (https://www.ncbi.nlm.nih.gov/) with an accession number of CAA48354.1.
Table 1.
The set of phytochemicals selected for the study
| Plant | Compound | Pubchem CID | Molecular formula |
|---|---|---|---|
| R. palmatium | 2-Methylbutanol | 8723 | C5H12O |
| 4beta-Phorbol | 24832117 | C20H28O6 | |
| 4-Methylhexanol | 98346 | C7H16O | |
| 4alpha-Phorbol | 114708 | C20H28O6 | |
| Aloe-emodin | 10207 | C15H10O5 | |
| Anthocyanin | 145858 | C15H11O6Cl | |
| Anthraquinone | 6780 | C14H8O2 | |
| Calcium oxalate | 33005 | C2CaO4 | |
| Catechin | 9064 | C15H14O6 | |
| Chrysophanic acid | 10208 | C15H10O4 | |
| Chrysophanol-anthron | 68111 | C15H12O3 | |
| Cinnamic acid | 444539 | C9H8O2 | |
| Emodin | 3220 | C15H10O5 | |
| Frangulosid | 196979 | C21H20O9 | |
| Gallic acid | 370 | C7H6O5 | |
| Lindleyin | 42994 | C23H26O11 | |
| Oxalic acid | 971 | C2H2O4 | |
| Procyanidin | 107876 | C30H26O13 | |
| Rhaponticin | 637213 | C21H24O9 | |
| Rhapontigenin | 5320954 | C15H14O4 | |
| Rhein | 10168 | C15H8O6 | |
| Rheochrysidin | 10639 | C16H12O5 | |
| R. coreanus | 2,3-(S)-Hexahydroxydiphenoyl-d-glucose | 14035453 | C20H18O14 |
| 23-Hydroxytormentic acid | 490367 | C30H48O6 | |
| Anthraquinones | 6780 | C14H8O2 | |
| Caffeic acid | 689043 | C9H8O4 | |
| Catechin | 9064 | C15H14O6 | |
| Chlorogenic acid | 1794427 | C16H18O9 | |
| Chrysanthemin | 44256715 | C21H21O11+ | |
| Cinnamic acid | 444539 | C9H8O2 | |
| Ellagic acid | 5281855 | C14H6O8 | |
| Ethyl 3,4-dihydroxybenzoate | 77547 | C9H10O4 | |
| Ferulic acid | 445858 | C10H10O4 | |
| Furan-2-ol | 179383 | C4H4O2 | |
| Gallic acid | 370 | C7H6O5 | |
| Kaempferol | 5280863 | C15H10O6 | |
| Luteolin | 5280445 | C15H10O6 | |
| p-Coumaric | 637542 | C9H8O3 | |
| Pelargonidin-3-rutinoside | 44256626 | C27H31O14+ | |
| Quercetin | 5280343 | C15H10O7 | |
| Salicylic acid | 338 | C7H6O3 | |
| Syringic acid | 10742 | C9H10O5 | |
| Vanillic acid | 8468 | C8H8O4 | |
| β-Carotene | 5280489 | C40H56 | |
| S. officinalis | (+)-Catechin | 9064 | C15H14O6 |
| 3,3′,4′-Tri-O-methylellagic acid | 11674590 | C17H12O8 | |
| Beta-glucogallin | 124021 | C13H16O10 | |
| Ellagic acid | 5281855 | C14H6O8 | |
| Fisetinidol-(4alpha-8)-catechin | 11731408 | C30H26O11 | |
| Gallic acid | 370 | C7H6O5 or C6H2(OH)3COOH | |
| Pomolic acid 28-O-beta-d-glucopyranosyl ester | 76322845 | C36H58O9 | |
| Pomolic acid | 382831 | C30H48O4 | |
| Quercetin | 5280343 | C15H10O7 | |
| Quercetin-3-glucuronide | 11655911 | C21H18O13 | |
| Sanguic acid | 101365640 | C30H46O7 | |
| Ziyuglycoside II | 71773126 | C35H56O8 |
Homology modeling and model assessment
Due to high degree of conservancy between HBV DNA polymerase and HIV-1 polymerases (Das et al. 2001), therefore HIV-1 reverse transcriptase with PDB entry 1RTD was selected as template for modeling of HBV DNA polymerase. Homology modeling was performed using Swiss model server (https://swissmodel.expasy.org/). Quality of the predicted model was evaluated using QMEAN tool in Swiss model server and Ramachandran plot in PROCHECK server (http://services.mbi.ucla.edu/PROCHECK/).
Receptor and ligands preparation and optimization
The raw structure of HBV DNA polymerase and the ligands were further prepared for in silico analysis. For this, the receptor was initially prepared by removing all water molecules and none polar hydrogens, followed by subsequent energy minimization to remove the bad steric clashes using the UCSF Chimera-1.11.2 software. Also all ligand molecules were minimized in Steepest Descent followed by Conjugate Gradient method using Accelrys Discovery Studio (Version 1.7).
Drug likeness and toxicity prediction
SwissADME (http://www.swissadme.ch/index.php) and MedChem Designer 3.0 were used to calculate drug-likeness parameters of the selected compounds including absorption, distribution, metabolism and elimination. Furthermore, the genotoxicity of the studied ligands in Salmonella typhimurium TA 100 was predicted using Toxtree 2.6.6 software.
Primary screening
All dominant phytochemicals from R. palmatum, R. coreanus and S. officinalis (Table 1) which reported in previous studies were subjected to molecular docking as ligands and HBV DNA polymerase was selected as a receptor. Molecular docking was performed using iGemdock 2.1 software with following parameters: population size 200, generation 70, number of solutions 2 and standard docking mode.
Final molecular docking
In this step, the compounds, which showed high affinity to the receptor in primary screening as well as have appropriate physicochemical properties without toxicity potential, were selected for more study. For this, final molecular docking was performed using Autodock4 (version 1.5.6) with the Lamarckian genetic algorithm. Docking parameters which selected for AutoDock4 runs were as follows: 200 docking runs, population size of 200, random starting position and conformation, translation step ranges of 2A, mutation rate of 0.02, cross-over rate of 0.8, local search rate of 0.06 and 2.5 million energy evaluations. Docked conformations were clustered by a tolerance of 2 Å root mean square deviations (RMSD).
Results
Homology modeling and model assessment
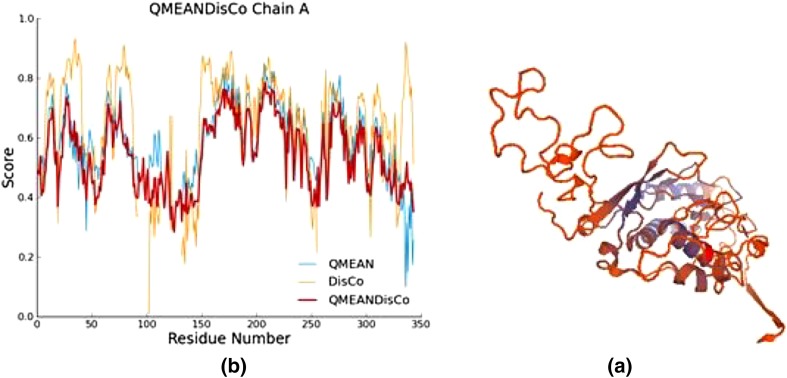
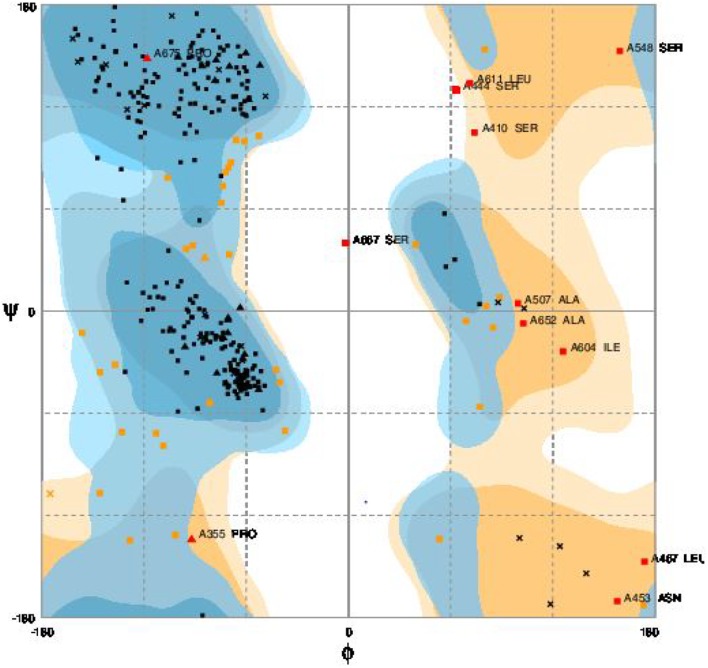
Three-dimension structure of modeled HBV DNA polymerase is depicted in Fig. 1a. Assessment of predicted model showed QMEAN4 value of − 5.92. Furthermore, high conservation level in functional residues of HBV DNA polymerase with the selected template confirmed by local quality validation (Fig. 1b). The Ramachandran plot of predicted model is showed in Fig. 2. Assessment of the Ramachandran plot showed 85.9% of residues in favored region, 10.6% in allowed region and 3.5% in outlier region.
Fig. 1.
Three dimension structure of modeled HBV DNA polymerase (a) and model assessment results (b)
Fig. 2.
Ramachandran plot of modeled HBV DNA polymerase
Drug likeness and toxicity prediction
The results of drug likeness and toxicity prediction of studied ligands are shown in (Supplementary data, Tables 1–3). Results confirmed that any tested ligand has genotoxicity but some of them showed moderate cytotoxicity because of inhibition essential cytochromes. Results also revealed that most tested ligands have high gastrointestinal (GI) absorption. In term of water solubility, phytochemicals from S. officinalis showed higher solubility in compared to other compounds. In addition, comparison of logP and logD value of the phytochemicals indicated that phytochemicals from S. officinalis have higher permeation (logP) and distribution (logD) than other compounds.
Primary screening
The results of primary screening are shown in Table 2. Results showed that three compounds including Quercetin-3-glucuronide, Frangulosid and Lindleyin acid have appropriate and strong interactions to the receptor in compared to other ligands and lamivudine, so these compounds were selected for more evaluation. Results also revealed that H-bond and Van der Waals interactions were main inter molecular forces between studied ligands and the receptor. Comparison analysis showed that phytochemicals from R. palmatium had stronger interaction against the receptor in compared to other studied plants.
Table 2.
Results of primary screening of the phytochemicals for introducing effective HBV DNA polymerase inhibitors
| Compound | Total energy | Van der Waals | H-bond | Electrostatic |
|---|---|---|---|---|
| 2-Methylbutanol | − 41.25 | − 34.25 | − 7 | 0 |
| 4-Beta-Phorbol | − 80.12 | − 65.9 | − 14.22 | 0 |
| 4-Methylhexanol | − 47.91 | − 43.45 | − 4.46 | 0 |
| 4-Alpha-Phorbol | − 74.94 | − 61.38 | − 13.56 | 0 |
| Aloe-Emodin | − 82.03 | − 68.25 | − 13.79 | 0 |
| Anthocyanin | − 73.6 | − 65.1 | − 8.5 | 0 |
| Anthraquinone | − 51.01 | − 23.9 | − 27.91 | 0 |
| Catechin | − 82.68 | − 71.43 | − 11.25 | 0 |
| Chrysophanic acid | − 77.68 | − 72.84 | − 4.84 | 0 |
| Chrysophanol-anthron | − 76.03 | − 67.58 | − 9.49 | 0 |
| Cinnamic acid | − 63.1 | − 52.6 | − 10.5 | 0 |
| Emodin | − 82.99 | − 68.99 | − 13.99 | 0 |
| Frangulosid | − 106.54 | − 88.59 | − 17.95 | 0 |
| Gallic acid | − 67.53 | − 49.41 | − 18.12 | 0 |
| Lindleyin | − 99.88 | − 74.72 | − 25.16 | 0 |
| Oxalic acid | − 51.11 | − 23.61 | − 27.5 | 0 |
| Procyanidin | − 93.77 | − 79.67 | − 14.1 | 0 |
| Rhaponticin | − 94.95 | − 79.91 | − 15.04 | 0 |
| Rhapontigenin | − 77.07 | − 67.58 | − 9.49 | 0 |
| Rhein | − 98.61 | − 73.76 | − 24.85 | 0 |
| Rheochrysidin | − 87.59 | − 67.42 | − 20.17 | 0 |
| 2,3-(S)-Hexahydroxydiphenoyl-d-glucose | − 99.15 | − 82.12 | − 17.03 | 0 |
| 23-Hydroxytormentic acid | − 89.25 | − 64.25 | − 25 | 0 |
| Caffeic acid | − 76.06 | − 61.75 | − 14.13 | 0 |
| Chlorogenic acid | − 87.82 | − 58.67 | − 29.15 | 0 |
| Chrysanthemin | − 88.9 | − 72.2 | − 16.7 | 0 |
| Cinnamic acid | − 69.15 | − 53.19 | − 15.95 | 0 |
| Ellagic acid | − 85.86 | − 70.32 | − 15.54 | 0 |
| Ethyl 3,4-dihydroxybenzoate | − 65.98 | − 42.66 | − 23.3 | 0 |
| Ferulic acid | − 64.82 | − 56.32 | − 8.5 | 0 |
| Furan-2-Ol | − 46.2 | − 35.75 | − 10.45 | 0 |
| Kaempferol | − 79.94 | − 51.41 | − 28.52 | 0 |
| Luteolin | − 76.36 | − 68.08 | − 8.28 | 0 |
| p-Coumaric | − 61.36 | − 52.1 | − 9.26 | 0 |
| Pelargonidin-3-Rutinoside | − 94.31 | − 83.81 | − 10.5 | 0 |
| Quercetin | − 81.48 | − 73.05 | − 8.43 | 0 |
| Salicylic acid | − 60.87 | − 36.02 | − 24.85 | 0 |
| Syringic acid | − 79.33 | − 65.88 | − 13.45 | 0 |
| Vanillic acid | − 68.5 | − 56.58 | − 11.91 | 0 |
| Beta-carotene | − 80.89 | − 80.89 | 0 | 0 |
| 3,3′,4′-Tri-O-methylellagic acid | − 79.13 | − 68.24 | − 10.89 | 0 |
| B-Glucogallin | − 79.76 | − 60.82 | − 18.94 | 0 |
| Fisetinidol-(4alpha-8)-catechin | − 97.98 | − 76.91 | − 21.07 | 0 |
| Pomolic acid 28-O-beta-d-glucopyranosyl ester | − 72.4 | − 10.2 | − 62.2 | 0 |
| Pomolic acid | − 78.35 | − 59.36 | − 18.99 | 0 |
| Quercetin-3-glucuronide | − 108.06 | − 78.01 | − 30.05 | 0 |
| Sanguic acid | − 93.75 | − 78.14 | − 15.61 | 0 |
| Ziyuglycoside II | − 85.66 | − 67.82 | − 17.84 | 0 |
| Lamivudine | − 89.73 | − 75.16 | − 14.57 | 0 |
Final molecular docking
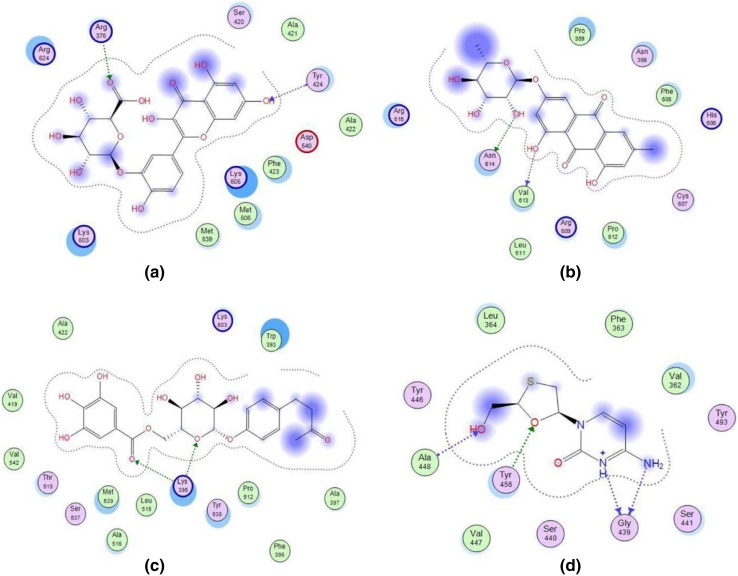
The results of final molecular docking study are presented in Table 3. Result confirmed that Frangulosid and Lindleyin acid have most and least efficacy in HBV DNA polymerase inhibition with the inhibition constant of 2.97 and 53.83 µM, respectively. Results also showed that, the amino acids, involved in the interactions, were different for each compound. In this regards, results revealed that the main amino acids residues of receptor, involved in interaction with Quercetin-3-glucuronide, Frangulosid and Lindleyin separately, located in 420–424, 606–615 and 512–542 spectra. respectively (Fig. 3a–c). In addition, active amino acids for interaction with lamivudine were located in two different spectra consist of 362–364 and 439–448 (Fig. 3d).
Table 3.
The results of final molecular docking study. Frangulosid and Quercetin-3-glucuronide have more effective interaction with HBV DNA polymerase rather than lamivudine
| Compound | Binding energy (kcal/mol) | Inhibition constant or Ki (µM) | Amino acid involved in interaction |
|---|---|---|---|
| Quercetin-3-glucuronide | − 7.13 | 5.96 | Arg376, Ser420, Ala421, Ala422, Phe423, Tyr424, Met506, Met539, Lys603, Lys605, Arg624 |
| Frangulosid | − 7.54 | 2.97 | Asn368, Pro369, His606, Cys607, Phe608, Arg609, Leu611, Pro612, Val613, Asn614, Arg615 |
| Lindleyin acid | − 5.82 | 53.83 | Trp393, Lys395, Phe396, Ala397, Val419, Ala422, Pro512, Leu515, Ala516, Thr519, Ser537, Tyr538, Met539, Val542, Lys603 |
| Lamivudine | − 6.79 | 6.80 | Val362, Phe363, Leu364, Gly439, Ser440, Ser441, Tyr446, Val447, Ala448, Tyr493 |
Fig. 3.
Schematic configuration of docking results of Quercetin-3-glucuronide (a), Frangulosid (b), Lindleyin acid (c) and Lamivudine (d) with binding pocket of HBV DNA polymerase. Polar residues are shown in pink. Acidic residues have been circled in red color while basic ones are encircled by blue color. Acceptance or donation of electron is shown by the respective arrow pointing towards or away from the residue. Dashed line shows the proximity contour
Discussion
In the study, in silico screening of novel phytochemical compounds to inhibit of HBV DNA polymerase were performed. These compounds were from three medicinal plants including R. palmatum, R. coreanus and S. officinalis. According to obtained results, two compounds including Quercetin-3-glucuronide and Frangulosid have high potential to inhibition of HBV DNA polymerase in comparison with other mentioned compounds and lamivudine as a positive control. Therefore, these two compounds, which are founds in S. officinalis and R. palmatum, respectively, are the appropriate candidates for in vitro and in vivo studies.
Worldwide, HBV is the most common among other hepatitis viruses and more than 400 million peoples are chronically infected by this virus, caused to 80% of hepatocellular carcinoma. Vaccination is a effective way to reduce the affection of hepatitis, but these preventative approaches are most thoroughly performed only in developed countries (Chemin and Zoulim 2009; Lavanchy 2004).
The widespread of HBV infection with rapid rise in the incidence and mortality rate of liver cancer have led to identification of effective and specific anti-hepatitis treatment strategies over recent years. Due to key role of HBV DNA polymerase in the virus infection cycle, currently treatment of chronic HBV infection using DNA polymerase inhibitors are high considered. Although recently, the new anti-HBV drugs have become available for clinical use, but due to rapid development of drug resistance and financial constraints the use of these drugs are encountered with restrictions (Lu and Zhuang 2009).
In this regards, natural resources are widely considered due to availability, minimizing adverse side effects and cost. Therefore, in the recent years development of new HBV polymerase inhibitor agents is focused on discovering diverse compounds from medicinal plants (Lin et al. 2013; Shin et al. 2005; Siddiqui et al. 2017). Herbal medicine can exhibit strong activity against HBV and other viruses. Moronic and betulonic acids from the herbal ethylacetate extract of Rhus javanica showed anti-HSV (herpes simplex-1 virus) activity (Kurokawa et al. 1999). Glycyrrhizin from Glycyrrhiza glabra has been found to improve the resistance of mice to HSV-1 infection (Utsunomiya et al. 1995). In another study, the antiviral activity of forty-two Egyptian medicinal plants was surveyed on HSV infection, poliomyelitis-1 virus (POLIO) and vesicular stomatitis virus (VSV) (Soltan and Zaki 2009). Antiviral activities for curcumin extracted from Curcuma longa L. were confirmed against HIV (human immunodeficiency virus), Influenza viruses, HSV-1, HSV-2, HPV (human papillomaviruses), HCV (hepatitis C virus) and HBV. Curcumin has a role in suppression of HBV replication by increasing the p53 level (Zorofchian Moghadamtousi et al. 2014). In this regards the antiviral effects of aqueous extracts from four medicinal plants including Terminalis chebula, S. officinalis, R. coreanus and R. palmatum were confirmed against HBV (Kim et al. 2001).
Experimental analyses of wide series of antiviral compounds from different medicinal plants are tedious, costly and time consuming. Therefore, recently computational approaches have been regarded to screening of new compounds with antiviral effects (Ahmad et al. 2015; Byler et al. 2016; Jain et al. 2017). Homology modeling and molecular docking analysis of phytochemical compounds including Ethyl brevifolincarboxylate, Tenofovir, and Quercitrin from Phyllanthus niruri against HBV DNA polymerase were performed and the results clearly demonstrated strong affinity of the ligands with HBV DNA polymerase (Mekha Mohan et al. 2015).
Meng et al. (2015) reported that bentysrepinine (Y101) isolated from Dichondra repens, can inhibits HIV reverse transcriptase, also has the potential to interacts with HBV and HCV DNA polymerase. The validation of antiviral activity of repensine and its over 200 derivatives showed that Y101, as one of repensine derivative, was able to inhibit HBV cccDNA production. Furthermore, they reported that reverse transcription activity of HBV was associated with residues 354–694 and Y101 can interact with this area through van der Waals interactions with lower energy than repensine (Meng et al. 2015). In the present study, similar results were observed about amino acids involved in interaction between HBV DNA polymerase and Quercetin-3-glucuronide as well as Frangulosid.
Prediction of physicochemical properties of Quercetin-3-glucuronide and Frangulosid revealed that the both compounds have low gastrointestinal absorption and solubility but these problems can be resolved using novel drug delivery system such as nano-carrier systems (Bonifácio et al. 2014; Suri et al. 2007). Quercetin-3-glucuronide and Frangulosid do not show Genotoxicity or mutagenicity and therefore can be used as safe anti-HBV candidates.
Because of highest affinity and stronger interactions between Frangulosid with reverse transcriptase region of HBV DNA polymerase as well as appropriate physicochemical properties of the compounds, it can be concluded that this compounds are good candidates for in vitro and in vivo validation of their anti-HBV activity.
Conclusion
This study was conducted to in silico screening novel HBV DNA polymerase inhibitor(s) from R. palmatum, R. coreanus and S. officinalis. The result confirmed that Frangulosid and Lindleyin acid have most and least efficacy in HBV DNA polymerase inhibition with inhibition constant of 2.97 and 53.83 µM, respectively. In conclusion, Frangulosid can be considered as a good candidate for more investigation of its anti-HBV activity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to acknowledge the University of Isfahan for the financial support of this study.
Compliance with ethical standards
Conflict of interest
The authors have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmad A, Ahad A, Rao AQ, Husnain T. Molecular docking based screening of neem-derived compounds with the NS1 protein of Influenza virus. Bioinformation. 2015;11:359. doi: 10.6026/97320630011359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifácio BV, da Silva PB, dos Santos Ramos MA, Negri KMS, Bauab TM, Chorilli M. Nanotechnology-based drug delivery systems and herbal medicines: a review. Int J Nanomed. 2014;9:1. doi: 10.2217/nnm.13.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byler KG, Ogungbe IV, Setzer WN. In-silico screening for anti-Zika virus phytochemicals. J Mol Graph Model. 2016;69:78–91. doi: 10.1016/j.jmgm.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemin I, Zoulim F. Hepatitis B virus induced hepatocellular carcinoma. Cancer Lett. 2009;286:52–59. doi: 10.1016/j.canlet.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Das K, Xiong X, Yang H, Westland CE, Gibbs CS, Sarafianos SG, Arnold E. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC) J Virol. 2001;75:4771–4779. doi: 10.1128/JVI.75.10.4771-4779.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibyajyoti S, Bin ET, Swati P. Bioinformatics: the effects on the cost of drug discovery. Galle Med J. 2013;18:44–50. doi: 10.4038/gmj.v18i1.5511. [DOI] [Google Scholar]
- Ganem D, Prince AM. Hepatitis B virus infection—natural history and clinical consequences. N Engl J Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- Jain J, Kumari A, Somvanshi P, Grover A, Pai S, Sunil S. In silico analysis of natural compounds targeting structural and nonstructural proteins of chikungunya virus. F1000Research. 2017;6:1601. doi: 10.12688/f1000research.12301.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Clark DN, Cao F, Tavis JE, Hu J. Comparative analysis of hepatitis B virus polymerase sequences required for viral RNA binding, RNA packaging, and protein priming. J Virol. 2014;88:1564–1572. doi: 10.1128/JVI.02852-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandagalla S, Sharath B, Bharath BR, Manjunatha H. Molecular docking analysis of curcumin analogues against kinase domain of ALK5. In Silico Pharmacol. 2016;5:15. doi: 10.1007/s40203-017-0034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MTH, Ather A, Thompson KD, Gambari R. Extracts and molecules from medicinal plants against herpes simplex viruses. Antiviral Res. 2005;67:107–119. doi: 10.1016/j.antiviral.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Kim TG, Kang SY, Jung KK, Kang JH, Lee E, Han HM, Kim SH. Antiviral activities of extracts isolated from Terminalis chebula Retz., Sanguisorba officinalis L., Rubus coreanus Miq. and Rheum palmatum L. against hepatitis B virus. Phytother Res. 2001;15:718–720. doi: 10.1002/ptr.832. [DOI] [PubMed] [Google Scholar]
- Kim K-H, Kim ND, Seong B-L. Discovery and development of anti-HBV agents and their resistance. Molecules. 2010;15:5878–5908. doi: 10.3390/molecules15095878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa M, et al. Anti-herpes simplex virus activity of moronic acid purified from Rhus javanica In vitro and in vivo. J Pharmacol Exp Ther. 1999;289:72–78. [PubMed] [Google Scholar]
- Langley DR, et al. Inhibition of hepatitis B virus polymerase by entecavir. J Virol. 2007;81(8):3992–4001. doi: 10.1128/JVI.02395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepatitis. 2004;11:97–107. doi: 10.1046/j.1365-2893.2003.00487.x. [DOI] [PubMed] [Google Scholar]
- Lin X, Zhang S, Huang Q, Zheng L, Huang J, Zhang X, Huang R. Anti-hepatitis B virus activity of total saponins isolated from Taraphochlamys affinis in vitro and in vivo. J Med Plants Res. 2013;7:2841–2846. [Google Scholar]
- Lok AS, McMahon BJ. Chronic hepatitis B: update. Hepatology. 2009;50:661–662. doi: 10.1002/hep.23190. [DOI] [PubMed] [Google Scholar]
- Lu F, Zhuang H. Management of hepatitis B in China. Chin Med J (Engl) 2009;122:3–4. [PubMed] [Google Scholar]
- Mak L-Y, Seto W-K, Lai C-L, Yuen M-F. DNA polymerase inhibitors for treating hepatitis B: a safety evaluation. Expert Opin Drug Saf. 2016;15:383–392. doi: 10.1517/14740338.2016.1139573. [DOI] [PubMed] [Google Scholar]
- Mekha Mohan PJ, Valsalan R, Nazeem PA. Molecular docking studies of phytochemicals from Phyllanthus niruri against hepatitis B DNA polymerase. Bioinformation. 2015;11:426. doi: 10.6026/97320630011426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F-C, Xu W-R, Li Y-Z, Huang Z-M, Liang G-Y, Liu C-X. In silico molecular docking study of repensine and bentysrepinine against HBV DNA polymerase. Chin Herb Med. 2015;7:39–44. doi: 10.1016/S1674-6384(15)60018-1. [DOI] [Google Scholar]
- Michel M-L, Deng Q, Mancini-Bourgine M. Therapeutic vaccines and immune-based therapies for the treatment of chronic hepatitis B: perspectives and challenges. J Hepatol. 2011;54(6):1286–1296. doi: 10.1016/j.jhep.2010.12.031. [DOI] [PubMed] [Google Scholar]
- Mukhtar M, Arshad M, Ahmad M, Pomerantz RJ, Wigdahl B, Parveen Z. Antiviral potentials of medicinal plants. Virus Res. 2008;131:111–120. doi: 10.1016/j.virusres.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzaffer U, Paul V, Prasad NR. Molecular docking of selected phytoconstituents with signaling molecules of ultraviolet-B induced oxidative damage. In Silico Pharmacol. 2016;5:17. doi: 10.1007/s40203-017-0035-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal M, Schaller H. Hepatitis B virus replication. Trends Microbiol. 1993;1:221–228. doi: 10.1016/0966-842X(93)90136-F. [DOI] [PubMed] [Google Scholar]
- Neuveut C, Wei Y, Buendia MA. Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol. 2010;52:594–604. doi: 10.1016/j.jhep.2009.10.033. [DOI] [PubMed] [Google Scholar]
- Noordeen F. Hepatitis B virus infection: an insight into infection outcomes and recent treatment options. Virusdisease. 2015;26:1–8. doi: 10.1007/s13337-015-0247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw T, Mok SS, Locarnini SA. Inhibition of hepatitis B virus DNA polymerase by enantiomers of penciclovir triphosphate and metabolic basis for selective inhibition of HBV replication by penciclovir. Hepatology. 1996;24:996–1002. doi: 10.1002/hep.510240504. [DOI] [PubMed] [Google Scholar]
- Shin MS, Kang EH, Lee YI. A flavonoid from medicinal plants blocks hepatitis B virus-e antigen secretion in HBV-infected hepatocytes. Antiviral Res. 2005;67:163–168. doi: 10.1016/j.antiviral.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Siddiqui MH, Alamri SA, Al-Whaibi MH, Hussain Z, Ali HM, El-Zaidy ME. A mini-review of anti-hepatitis B virus activity of medicinal plants. Biotechnol Biotechnol Equip. 2017;31:9–15. doi: 10.1080/13102818.2016.1240593. [DOI] [Google Scholar]
- Soltan MM, Zaki AK. Antiviral screening of forty-two Egyptian medicinal plants. J Ethnopharmacol. 2009;126:102–107. doi: 10.1016/j.jep.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Suri SS, Fenniri H, Singh B. Nanotechnology-based drug delivery systems. J Occup Med Toxicol. 2007;2:16. doi: 10.1186/1745-6673-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya T, Kobayashi M, Herndon DN, Pollard RB, Suzuki F. Glycyrrhizin (20β-carboxy-11-oxo-30-norolean-12-en-3β-yl-2-O-β-d-glucopyranuronosyl-α-d-glucopyranosiduronic acid) improves the resistance of thermally injured mice to opportunistic infection of herpes simplex virus type 1. Immunol Lett. 1995;44:59–66. doi: 10.1016/0165-2478(94)00183-R. [DOI] [PubMed] [Google Scholar]
- Zorofchian Moghadamtousi S, Abdul Kadir H, Hassandarvish P, Tajik H, Abubakar S, Zandi K. A review on antibacterial, antiviral, and antifungal activity of curcumin. BioMed Res Int. 2014;2014:186864. doi: 10.1155/2014/186864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.