Abstract
Fc gamma receptors (FCGRs) are immune cell surface proteins that bind IgG and facilitate cytokine production, phagocytosis, and antibody-dependent cell-mediated cytotoxicity. FCGRs play a critical role in immunity; variation in these genes is implicated in autoimmunity and other diseases. Cynomolgus macaques are an excellent animal model for many human diseases, and Mauritian cynomolgus macaques (MCMs) are particularly useful due to their restricted genetic diversity. Previous studies of MCM immune gene diversity have focused on the major histocompatibility complex (MHC) and killer cell immunoglobulin-like receptor (KIR). Here we characterize FCGR diversity in 48 MCMs, using PacBio long-read sequencing to identify novel alleles of each of the four expressed MCM FCGR genes. We also developed a high-throughput FCGR genotyping assay, which we used to determine allele frequencies and identify FCGR haplotypes in more than 500 additional MCMs. We found three alleles for FCGR1A, seven each for FCGR2A and FCGR2B, and four for FCGR3A; these segregate into eight haplotypes. We also assessed whether different FCGR alleles confer different antibody binding affinities by surface plasmon resonance, and found minimal difference in binding affinities across alleles for a panel of wild type and Fc-engineered human IgG. This work suggests that although MCMs may not fully represent the diversity of FCGR responses in humans, they may offer highly reproducible results for monoclonal antibody therapy and toxicity studies.
Introduction
Fc gamma receptors (FCGRs) are cell surface glycoproteins that bind to the Fc region of IgG antibodies. Expressed on monocytes, macrophages, neutrophils, natural killer cells, dendritic cells, mast cells, and B cells, FCGRs couple the specificity of antibody responses with cell-mediated immunity. Antigen-specific IgG binds to FCGRs, which trigger a variety of responses, including cytokine production, phagocytosis, and release of cytotoxic granules (1). Antibody-dependent cell-mediated cytotoxicity (ADCC) is an FCGR-facilitated process in which NK cells destroy antibody-bound cells, and is thought to be the mechanism of action for monoclonal antibodies (mAbs) such as rituximab and trastuzumab used to treat cancer (2–4). FCGRs expressed on monocytes and macrophages also facilitate phagocytosis of tumor cells in the presence of certain monoclonal antibodies (5).
Humans have six classical FCGRs encoded by genes on chromosome 1 – the activating receptors FCGR1, FCGR2A, FCGR2C, FCGR3A, and FCGR3B, and the inhibitory receptor FCGR2B. Also included within the broader FCGR family of receptors are the neonatal FCGR (FCGRT), and FCRL5 and TRIM21 (6). All bind IgG at the cell surface, except for FCGRT and TRIM21, which bind IgG intracellularly (6); for this reason we focus on FCGR1, FCGR2, and FCGR3 as they have been shown to interact with and facilitate the effects of mAbs (4). The activating receptors trigger the pro-inflammatory responses described above. The inhibitory FCGR2B dampens activating FCGR responses by inhibiting cytokine release and antibody production, inhibiting T cell stimulation and migration, and increasing the B cell activation threshold (1).
Variation in human FCGR genes alters receptor affinity for IgG. Each FCGR allotype has different affinities for the different IgG subclasses, which has implications for monoclonal antibody therapy design (7, 8). Genetic variation within FCGR genes can also result in altered susceptibility to infection and autoimmunity. Single nucleotide polymorphisms (SNPs) that increase activating receptor affinity for IgG tend to decrease susceptibility to infection, as do SNPs that reduce FCGR2B affinity for IgG (1). Variations in FCGRs are also associated with several specific disease phenotypes. The FCGR2A H131R polymorphism has been associated with an increased rate of progression to AIDS, increased risk of developing systemic lupus erythematosus, and a decreased risk of contracting malaria (9). The FCGR3A F158V polymorphism has been associated with a decreased risk for bloodstream infections following liver transplantation as well as an improved response to rituximab in follicular lymphoma patients (3, 10).
Macaque monkeys are crucial in modeling human disease processes, as they are anatomically and genetically very similar to humans, and mount similar immune responses. Macaque models for HIV infection and progression to AIDS, using simian immunodeficiency virus (SIV), are widely used in research, as are macaque models for organ transplantation (11). Macaques are also used in preclinical and clinical studies assessing toxicity of mAb therapies and other drugs (4, 12). Given the known role of immune gene polymorphisms in disease and treatment outcomes, well-controlled animals studies must take into account immune gene diversity.
FCGR variation has been previously characterized in rhesus macaques (Macaca mulatta, “Mamu”), but only limited studies have been published for cynomolgus macaques (Macaca fascicularis, “Mafa”) (13–15). Mauritian cynomolgus macaques (MCMs) are particularly important for studies where immunogenetics may impact outcome, as they are relatively recently descended from a small founder population, which has led to limited genetic diversity (16, 17). Other immune genes, including the major histocompatibility complex (MHC) (18) and killer cell immunoglobulin-like receptor (KIR) (19) genes, have been well-characterized in this population.
Macaques have four expressed FCGR genes – FCGR1A, FCGR2A, FCGR2B, and FCGR3A (13, 20). In the cynomolgus macaque reference genome Mafa5, FCGR2A, 3A, and 2B are located within 110 kilobases of each other on chromosome 1; FCGR1A is located over 12,000 kb downstream (Figure 1). Cynomolgus macaques express higher levels of FCGR2B on granulocytes, but expression patterns of these genes on different cell types are otherwise consistent between macaques and humans (20). Cynomolgus macaque FCGR2B also has been found to bind IgG2 with greater affinity than human FCGR2B, thought to be due to the presence of a histidine, rather than arginine as seen in humans, at amino acid 131.
Figure 1.
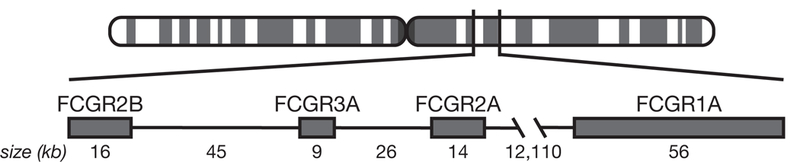
Macaque FCGR family. Due to the proximity of FCGR2A, 3A, and 2B, linkage disequilibrium occurs; FCGR1A is located further away and is not frequently inherited with the others. Locations are based on the Mafa5 reference genome.
Here, we present the first comprehensive characterization of FCGR alleles in MCMs. Using Pacific Biosciences (PacBio) long-read sequencing, we identified full-length alleles of each FCGR gene. We then used the PacBio data to infer haplotypes across FCGR2A, FCGR2B, and FCGR3A. To assess allele frequency in the MCM population, we genotyped 553 individuals using a high-throughput sequencing assay and found that the majority of our animals fell into one of eight common haplotypes. Interestingly, an antibody binding screen revealed that the different allele variants exhibit relatively similar affinities for a variety of IgG constructs. The screening panel included both wild-type and Fc-engineered human IgG used in drug discovery investigations of therapeutic antibodies, particularly biologics used in immuno-oncology research. While these findings may not reflect the variability of human FCGR responses attributed to allele polymorphisms, our data suggest that a single panel of representative FCGRs should be sufficient to represent the MCM population when evaluating FCGR interactions in vitro and lessens the need for complex study groups when using MCMs for preclinical animal model studies.
Materials and Methods
Animals and samples
Pacific Biosciences (PacBio) sequencing was performed for 48 MCMs – 22 using whole blood samples, and 26 using purified splenocytes or peripheral blood mononuclear cells. Sixteen of these animals were housed at the Wisconsin National Primate Research Center (WNPRC, Madison, WI), and 32 were housed by Bristol-Myers Squibb. Whole-genome sequencing (WGS) or whole-exome sequencing (macaque exome sequencing, “MES”) was previously performed for the 16 WNPRC animals used for PacBio sequencing (19, 21). MiSeq genotyping was validated with the 48 animals used for allele discovery, and was performed for 553 additional animals provided by Bioculture (Mauritius) and other primate centers. All animals were cared for according to the regulations and guidelines of the Animal Care and Use Committees at their respective institutions (WNPRC, Bioculture, AlphaGenesis, Oregon National Primate Center).
Sample preparation for long-read PacBio sequencing of FCGR cDNA transcripts
RNA purification was performed on each sample using the Maxwell® 16 LEV simplyRNA Purification Kit (Promega, Madison, WI, USA). cDNA synthesis was performed from RNA using the SuperScript® First-Strand Synthesis System (ThermoFisher Scientific, Waltham, MA, USA).
PCR primers were designed to anneal to highly conserved untranslated regions of each FCGR gene (Supplemental table I). A post-hoc analysis of the priming sites was performed using WGS of 18 MCM. Of the eight priming sites, six showed no polymorphisms in the genome/exome sequencing dataset. There was a C/T polymorphism seen in the FCGR2A reverse primer (second base from the 5’ end) and a G/C polymorphism in the FCGR1 reverse primer (fourth base from the 5’ end). As these SNPs are located far from the 3’ end of the primer, it is unlikely that they would have biased amplification. Standard 16 base-pair PacBio multiplex barcodes were added to the 5’ end of each primer (Pacific Biosciences, Menlo Park, CA, USA).
In addition to the amplicons listed in Table I, we also attempted to amplify, but did not find, a macaque FCGR2C by using the forward primer for FCGR2B and the reverse primer for FCGR2A, as FCGR2C in humans is a fusion of the 5’ end of FCGR2B and the 3’ end of FCGR2A.
Table I – Accession numbers for FCGR alleles.
Accession numbers for each FCGR allele. Full sequences of each FCGR allele can be found in GenBank (ncbi.nlm.nih.gov/genbank/). Only 2 of the 21 listed here had been reported previously. One FCGR2B allele (NM_001284131/AF485814) was found in GenBank but not in our samples – this allele was named FCGR2B:03:01 in our naming scheme and was included in the antibody binding assays. Additional alleles found in GenBank but not our samples (and not included in the antibody binding assay) were one FCGR1A allele (NM_001284040), three FCGR2A alleles (NM_001283669/AF485813, XM_005541169, and AB169139), and one FCGR3A allele (NM_001283192/DQ423377).
| Transcript isoform |
||||||
|---|---|---|---|---|---|---|
| Mafa allele | X01 | X02 | X03 | X04 | Splice variant | Previous accession # |
| FCGR1A:01:01 | KU852968 | KU852969 | del18V in X01 vs X02 |
- | ||
| FCGR1A:01:02 | KU852970 | KU852971 | del18V in X01 vs X02 |
AF485812 = X01 | ||
| FCGR1A:02:01 | KU852972 | KU852973 | del18V in X01 vs X02 |
- | ||
| FCGR2A:01:01 | KU852974 | KU852975 | del34A in X04 vs X02 |
- | ||
| FCGR2A:02:01 | KU852976 | KU852977 | del34A in X03 vs X01 |
- | ||
| FCGR2A:03:01 | KU852978 | KU852979 | del34A in X03 vs X01 |
- | ||
| FCGR2A:04:01 | KU852980 | KU852981 | del34A in X04 vs X02 |
- | ||
| FCGR2A:05:01 | KU852982 | KU852983 | del34A in X04 vs X02 |
- | ||
| FCGR2A:06:01 | KU852984 | KU852985 | del34A in X04 vs X02 |
- | ||
| FCGR2A:07:01 | KU852986 | - | ||||
| FCGR2B:01:01 | KU852987 | KU852988 | delEx5 in X02 vs X01 | - | ||
| FCGR2B:01:02 | KU852989 | KU852990 | delEx5 in X02 vs X01 | - | ||
| FCGR2B:01:03 | KU852991 | KU852992 | delEx5 in X02 vs X02 | - | ||
| FCGR2B:01:04 | KU852993 | KU852994 | delEx5 in X02 vs X03 | - | ||
| FCGR2B:01:05 | KU852995 | KU852996 | delEx5 in X02 vs X04 | - | ||
| FCGR2B:02:01 | KU852997 | KU852998 | delEx5 in X02 vs X05 | - | ||
| FCGR2B:02:02 | KU852999 | KU853000 | delEx5 in X02 vs X05 | - | ||
| FCGR3A:01:01 | KU853001 | KU853002 | del21E in X02 vs X01 |
- | ||
| FCGR3A:01:02 | KU853003 | KU853004 | del21E in X02 vs X01 |
- | ||
| FCGR3A:02:01 | KU853005 | KU853006 | del21E in X02 vs X01 |
AF485815 = X01 | ||
| FCGR3A:02:02 | KU853007 | KU853008 | del21E in X02 vs X01 |
- | ||
PCR amplification was performed using the Phusion® High-Fidelity PCR Master Mix (New England BioLabs, Ipswich, MA, USA). PCR products were purified using two rounds of the Agencourt AMPure XP PCR purification system (Beckman Coulter, Brea, CA, USA) and quantified using Quant-iT dsDNA HS Assay Kit (Life Technologies, Grand Island, NY, USA) and a DTX 800 plate reader (Beckman Coulter, Brea, CA, USA). Equimolar ratios of PCR samples were pooled, and two additional rounds of AMPure purification were performed to further concentrate the samples.
Two separate pools were submitted to the University of Washington PacBio Sequencing Services. SMRTbell™ libraries were created from the samples using the PacBio 2-kb Template Preparation protocol for circular consensus sequencing (CCS), which allows each insert to be sequenced multiple times to ensure accurate base calling. Using the PacBio DNA Template Prep Kit 2.0 (Pacific Biosciences, Menlo Park, CA, USA), the PCR products were end-repaired and hairpin adapters were added via blunt end ligation to create SMRTbell™ templates. Failed ligation products were removed using Exonucleases III and IV. The SMRTbell™ products were purified using AMPure PB beads (Pacific Biosciences, Menlo Park, CA, USA). The two libraries were sequenced on the PacBio RSII system using P6C4 sequencing chemistry.
Long-read PacBio sequencing data analysis
PacBio reads of insert were generated using the PacBio SMRT Analysis Pipeline (version 3.1.1), and were quality filtered using the USEARCH (version 9.2) fastq_filter command. Reads were clustered into operational taxonomic units (OTUs) using USEARCH and open reading frames were found using the USEARCH fastx_findorfs command. USEARCH-based OTU clustering creates representative sequences by collapsing reads that are 97% identical. Parameters were set so that the ORF contained the terminating stop codon (orfstyle=12) and the size was above the minimum number of codons necessary for each locus (350, 267, 300, and 230 for FCGR1A, 2A, 2B, and 3A respectively). Clusters that had fewer than two reads were removed, and chimeras and sequencing errors were identified and removed using the USEARCH unoise2 and fastx_uniques commands.
To filter out reads matching known FCGR alleles, we used bbmap (version 36.86) to map reads to the FCGR reference database. This database contains existing GenBank sequences for cynomolgus macaque FCGRs, including two FCGR1A, three FCGR2A, two FCGR2B, and two FCGR3A alleles. Reads that exactly matched alleles in the database (100% sequence identity) were removed. The remaining unmapped reads were manually validated by aligning them to known sequences and identifying mismatches and splice variants (based on insertions/deletions compared to known alleles). Reads for individual animals were mapped against known and novel alleles using Geneious R9 to count the number of reads mapping perfectly to putative novel alleles. Alleles were provisionally deemed legitimate if they appeared as one of the two most prevalent alleles in at least one MCM, with the support of at least three reads, and with read counts roughly in proportion with the other most common allele for that subject, should one exist. Additionally, some provisional alleles were deemed PCR artifacts and discarded if they appeared to have insertions or deletions compared to legitimate alleles, only appeared in MCMs that had those particular legitimate alleles, and represented what appeared to be “third” alleles for some of those MCMs.
Establishing a standardized nomenclature for macaque FCGR sequences
Existing macaque FCGR sequences have an inconsistent nomenclature that complicates comparisons of sequences identified across different studies. To address this, we established a simple syntax for describing FCGR alleles in MCMs, which can be applied more broadly across all macaque species.
Nucleotide-distinct transcript sequences were named in the form Mafa-FCGR1A:01:02|X01.
Mafa – indicates that the species in question is Macaca fascicularis.
FCGR1Α – indicates to which of the four FCGR genes the allele belongs.
:01 – identifies which protein the allele belongs to; each protein is differentiated from the rest by at least one amino acid difference.
:02 – identifies alleles for the same protein that are differentiated by synonymous nucleotide changes.
X01 – refers to the splice variant/isoform, typically numbered from most to least abundant.
There are six additional cynomolgus macaque FCGR alleles listed in GenBank that were not found in our allele discovery sampling. These additional alleles include one FCGR1A allele (NM_001284040), three FCGR2A alleles (NM_001283669/AF485813, XM_005541169, and AB169139), one FCGR2B allele (NM_001284131/AF485814), and one FCGR3A allele (NM_001283192/DQ423377).
Whole genome sequencing (WGS) of MCMs
In this study, we used 18 previously-published MCM WGS datasets to aid in primer design, validate PacBio allele discovery, add to our allele frequency estimation, and evaluate an intron for an alternative splice site. The WGS datasets were generated by Ericsen et al. (21), using 18 male MCMs housed at the WNPRC. The macaque exome sequencing (MES) data was produced by Prall et al. (22). Variants were then called against the Macaca_fascicularis_5.0 reference genome (23).
High-throughput FCGR genotyping using the Illumina MiSeq
High-throughput deep sequencing of diagnostic gene fragments is a cost-effective method for genotyping complex immune genes, such as the MHC (18). We developed an analogous method for FCGR genotyping using the MiSeq platform (Illumina, San Diego, CA, USA). The genotyping assay was developed to sequence FCGR1A exon 4, FCGR2A exons 3 and 4, FCGR2B exon 4, and FCGR3A exon 3 (using PCR amplicons prepared from genomic DNA), as our PacBio allele discovery results indicated that these amplicons were sufficient to distinguish between known non-synonymous alleles in MCMs.
We attempted to select PCR priming sites in highly conserved regions of the introns to minimize the potential for biased amplification (Supplemental table I). To avoid intronic SNP sites, the forward primer for FCGR1 exon 4 ends 1 bp into the exon, and the reverse primer ends 4 bp into the exon. According to an analysis of the 18 MCM WGS datasets, there were no polymorphisms in any of the genotyping primer sites. Fluidigm Access Array CS1 and CS2 adapters were added to the 5’ ends of each primer to enable the addition of barcode identifiers during PCR (Fluidigm, South San Francisco, CA, USA).
Genomic DNA was purified from blood and PBMC samples using Promega Maxwell 16 LEV Blood DNA kits. PCR amplification of the gDNA was performed using the Phusion® High-Fidelity PCR Master Mix. Reactions for each animal contained a final concentration of 0.0125 µM of each site-specific FCGR primer with CS1 or CS2 adapters attached (Supplemental table I) and 0.1 µM of two unique Illumina barcode primers. During PCR cycling, the barcode IDs are added as tags to the ends of the FCGR amplicons, serving as a unique identifier for each MCM. After validation studies with individual PCR reactions for animals with known FCGR genotypes, multiplex PCR reactions were performed using the Fluidigm Access Array system. The resulting amplicons were pooled and purified with Agencourt XP SPRI beads (Beckman Coulter, Brea, CA, USA) prior to deep sequencing on an Illumina MiSeq.
Data analysis for high-throughput genotyping assay
Illumina MiSeq read data was binned by barcode identifier and then merged using the flash merger tool (24). Geneious was used to map the reads for each sample against the database of known FCGR alleles (including both known sequences found in GenBank and new alleles found in this study), requiring exact matches to the database sequences. A Python script was applied to the Geneious mapping output to generate a table listing the number of reads for each sample corresponding to each known allele. This table was manually evaluated to assign genotypes to each individual for each gene.
Haplotyping
We inferred haplotypes across FCGR2A, FCGR2B, and FCGR3A using an algorithm which assigns haplotypes to each individual in a population such that the lowest number of total haplotypes (and ancestral recombinations) is required (25). The algorithm is traditionally applied to genotype data composed of a series of unphased SNPs for each individual. Our genotype data has phased the SNPs within each gene, so we instead applied the algorithm across the three genes in order to phase the full-length alleles for each locus into unified haplotypes.
The algorithm first identifies individuals who are homozygous at all three loci. For these individuals, only one possible haplotype can explain both chromosomes, and so that haplotype is considered established and is assigned to each of those individuals. The same process is next applied to individuals who are heterozygous at a single locus. These individuals’ genotypes can also be explained by a single pair of possible haplotypes, and these are added to the list of established haplotypes.
The remaining individuals are heterozygous at two or more loci, and their genotypes are then evaluated using the list of established haplotypes. If an established haplotype can explain one chromosome of an individual, it is assumed that they have that haplotype plus one novel haplotype explaining the remaining alleles. The novel haplotype is added to the list of haplotypes, and the process is repeated until all individuals have haplotype assignments.
Immunoglobulin binding assay
Surface plasmon resonance (SPR) studies were performed on a Biacore T100 and/or T200 instrument (GE Healthcare, Chicago, IL, USA) at 25°C in 10 mM NaPO4, 130 mM NaCl, 0.05% p20 (PBS-T) (pH 7.1). To study the binding of IgG to FCGR variants, Fab fragments from a murine anti-6xHis Ab (generated in-house) were immobilized on a CM5 sensor chip using ethyl(dimethylaminopropyl) carbodiimide/NHS to a density of ∼3800 RU. His-tagged constructs for the extracellular domains of 14 distinct FCGR1A, FCGR2A, FCGR2B, and FCGR3A sequences were expressed in human epithelial kidney (HEK) cells. The FCGR-containing supernatants were concentration-normalized and captured via the his-tag using a contact time of 30 seconds at 10µl/min and to a total capture level of 200–600 RU. The binding of a diverse panel of wild type and engineered recombinantly produced human IgG isotypes were screened at 1 µM concentration with a 120-second injection at 30 µl/min and regeneration back to baseline between binding cycles was performed by 2 × 15 second injections of 10 mM glycine-HCL pH1.5. The screening assay was analyzed by determining the percent of theoretical maximum binding response for each antibody based on the level of captured FCGR from the supernatant, assuming 100% fractional activity and only taking into account protein mass without glycosylation. Apparent KD values were determined for representative IgG isotypes binding to select FCGR2A proteins by analyzing the steady state binding level of a 2-fold serially diluted titration of antibody starting at 10 µM in the same capture assay format. The data were analyzed using the Biacore T200 Evaluation software (GE Healthcare). Replicates of select IgG/FCGR interactions were performed to ensure the reproducibility of our reported data; however, the collective SPR dataset was not acquired in a manner that would enable robust statistical analysis.
Results
FCGR allele discovery
To identify novel MCM FCGR alleles, we analyzed PacBio sequence data for 48 MCMs to identify the common alleles for the FCGR1A, FCGR2A, FCGR2B, and FCGR3A genes. All alleles are listed in Table I and can be found in GenBank (ncbi.nlm.nih.gov/genbank/).
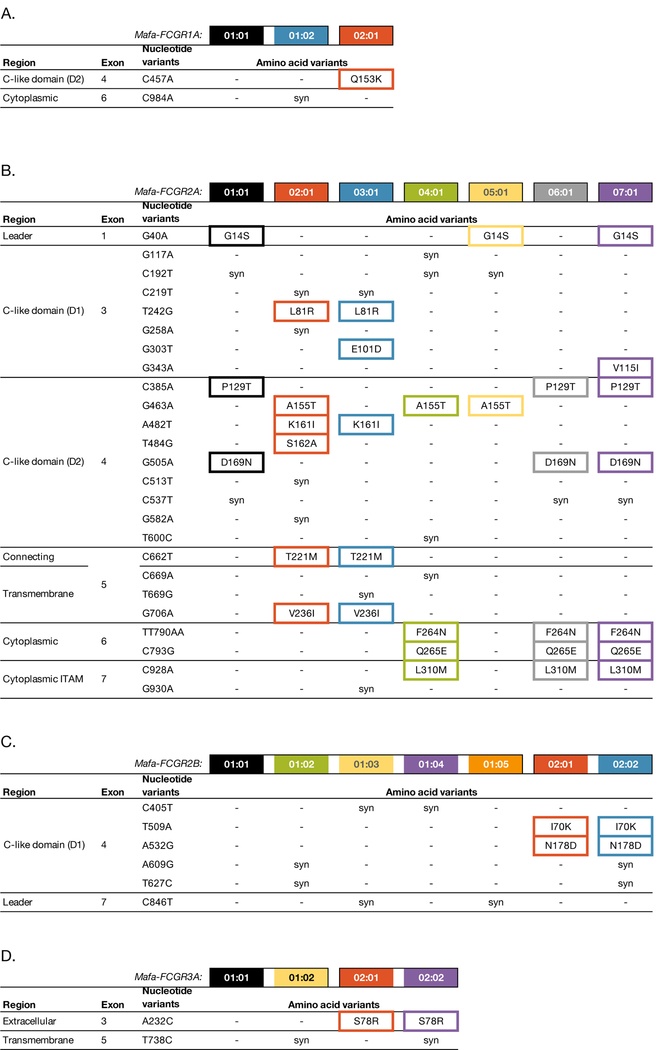
We identified three full-length alleles for FCGR1A (Figure 2A), which encode two distinct proteins. One of these alleles, FCGR1A:01:02, matched an existing cynomolgus macaque GenBank sequence. The other two alleles were novel, with one encoding the known FCGR1A protein with a synonymous variant and one encoding a distinct FCGR1A protein with a single amino acid change located in the extracellular domain of the receptor.
Figure 2.
FCGR polymorphisms by allele. Every possible nucleotide variant for each region is listed, and the presence or absence of that variant for each allele is noted. If the variant is present and causes a change in an amino acid, it is indicated; otherwise, if it is present but does not change the amino acid at that position, “syn” is used to indicate a synonymous variant. Nucleotides are numbered from the start of the leader peptide sequence using the longest isoform. (A) FCGR1A polymorphisms; longest isoform is X02. (B) FCGR2A polymorphisms; longest isoform is X02. (C) FCGR2B polymorphisms; longest isoform is X01. (D) FCGR3A polymorphisms; longest isoform is X01.
We identified seven novel full-length alleles for FCGR2A (Figure 2B), each of which encodes a distinct protein. Across the transcript, we found fourteen polymorphic amino acid sites, including one in the peptide leader sequence, eight in the extracellular domain, one in the connecting region, one in the transmembrane domain, and three in the cytoplasmic region. Among the MCM FCGR genes, FCGR2A was found to have the greatest degree of polymorphism.
We also identified seven novel full-length alleles for FCGR2B (Figure 2C). Together, these encode two distinct novel proteins. FCGR2B has two polymorphic amino acids, both of which are located in the extracellular region. This pair of FCGR2B variants is analogous to a pair of SNPs identified previously in cynomolgus macaques (14).
Finally, we identified four full-length alleles for FCGR3A (Figure 2D), encoding two distinct proteins. Three of these alleles, and one of the proteins, were novel; the allele FCGR3A:02:01 matched the GenBank sequence AF485815. The polymorphic sites differentiating the two proteins are located in the extracellular domain, and the nonsynonymous variant corresponds to a SNP that was also identified previously (14).
Splice variants
We next used the PacBio cDNA transcript sequences to evaluate splice variants for each gene and assess isoform abundance for each allele. In most cases, splice variants in each FCGR gene are not closely associated with particular alleles. For FCGR1A and 3A, the X01 isoform comprised over 90% of reads. However, the two most common isoforms of FCGR2B are expressed in roughly equal proportion for all alleles. The X01 isoform has an additional 19 amino acids in its cytoplasmic region compared to the X02 isoform, corresponding to presence or absence of exon 5 sequences in the mature transcript. The X01 isoform represented about one third of all FCGR2B isoform reads, while the X02 isoform represented about half.
Notably, the FCGR2A:01, 04, 05, 06, and 07 alleles predominantly expressed isoforms X02, X04, and X07, which comprised 99% of all isoform reads for these alleles. These isoforms include an additional 5 amino acids prior to the connecting region compared to the FCGR2A:02 and 03 alleles that exclusively expressed isoforms X01, X03, and X05, which lack those 5 amino acids. To determine why this dichotomy exists, we looked to our WGS/MES datasets, which revealed a T to G nucleotide substitution in the intron sequence just prior to exon 5 of FCGR2A (Supplemental figure 1). This mutation is in perfect linkage disequilibrium with the SNPs in exon 5 that differentiate the alleles. In our datasets, the FCGR2A:01, 04, 05, 06 and 07 alleles always have an AG dinucleotide 15 base pairs upstream from the typical start site of exon 5. This creates an early splice site not seen in the FCGR2A:02 and 03 alleles, which have an AT at this locus. The lack of an early splice site makes these alleles more similar to known rhesus macaque and human FCGR2A alleles.
FCGR genotyping assay
To streamline FCGR genotyping we developed a high-throughput assay that can be performed on a large number of animals. We determined that SNPs sufficient to differentiate all MCM FCGR alleles with nonsynonymous variation can be identified by sequencing only five exons across all four genes. We used WGS/MES data from our MCMs to develop primers (Supplemental table I) in the introns flanking FCGR1A exon 4, FCGR2A exons 3 and 4, FCGR2B exon 4, and FCGR3A exon 3, and incorporated these into our standard MHC Fluidigm panel in order to generate FCGR amplicons for sequencing (26). These were then sequenced on an Illumina MiSeq, and the genotyping assay was validated using the same 48 animals that we characterized by PacBio sequencing.
Haplotypes
We sought to infer haplotypes across FCGR2A, FCGR2B, and FCGR3A, which are located close to one another on chromosome 1, within a span of ~110 kb (Figure 3). We anticipated that the proximity of these genes would lead to a high degree of linkage disequilibrium – that is, they should be inherited together, and a relatively small number of allele combinations, or haplotypes, should exist. Although FCGR1A is also on chromosome 1, it is ~12,120 kb away from FCGR2A. Over approximately 100 generations since the original cynomolgus macaques were introduced to Mauritius, recombination between the FCGR1A and the rest of the FCGR cluster makes it difficult to discern the ancestral haplotypes for the extended FCGR region.
Figure 3.
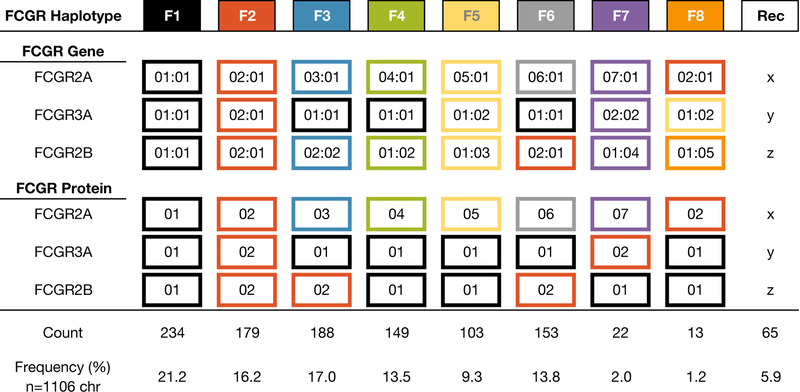
Summary of the alleles, and the proteins they form, comprising each FCGR haplotype. “Rec” indicates recombinant haplotypes.
We applied our haplotyping algorithm, described above, to 553 MCMs for which we had data covering FCGR2A, FCGR2B, and FCGR3A. Most of this data was from the MiSeq genotyping assay.
We began by identifying animals in the PacBio dataset that were homozygous at each gene; their haplotypes were unambiguous and therefore served as a launching point for determining the haplotypes of the others. Two animals (cy0333 and B5_075) carried identical homozygous alleles, and we designated this combination the F3 haplotype. Another animal (cy0320) was also homozygous at each gene, and we designated this combination the F6 haplotype. Subsequently, we identified animals that were heterozygous for either the F3 or F6 haplotype – one chromosome could be explained by these haplotypes, and based on this the other could be inferred. Based on the F3 haplotype, we were able to identify three additional haplotypes (F1, F2, and F4); based on F6 we were able to identify one additional haplotype (F5). Repeating the process using the F1–6 haplotypes as our established haplotypes, we identified two more (F7 and F8). The remaining haplotypes identified were consistent with recombination events between the eight ancestral haplotypes.
With the exception of FCGR2A:02:01, which is found in two of the eight ancestral haplotypes, each FCGR2A allele is overwhelmingly associated with a unique combination of FCGR2B and FCGR3A alleles (Figure 3). For example, among the eight haplotypes, the FCGR3A:01:01 and FCGR2B:01:01 combination is only associated with the FCGR2A:01:01 allele; all other instances of this pair co-occurring with a different FCGR2A allele were deemed to be recombinant haplotypes. Of the 553 haplotypes we analyzed, 94% were consistent with the eight most prevalent combinations of proteins, with the remainder consistent with simple recombination events.
Immunoglobulin binding assays
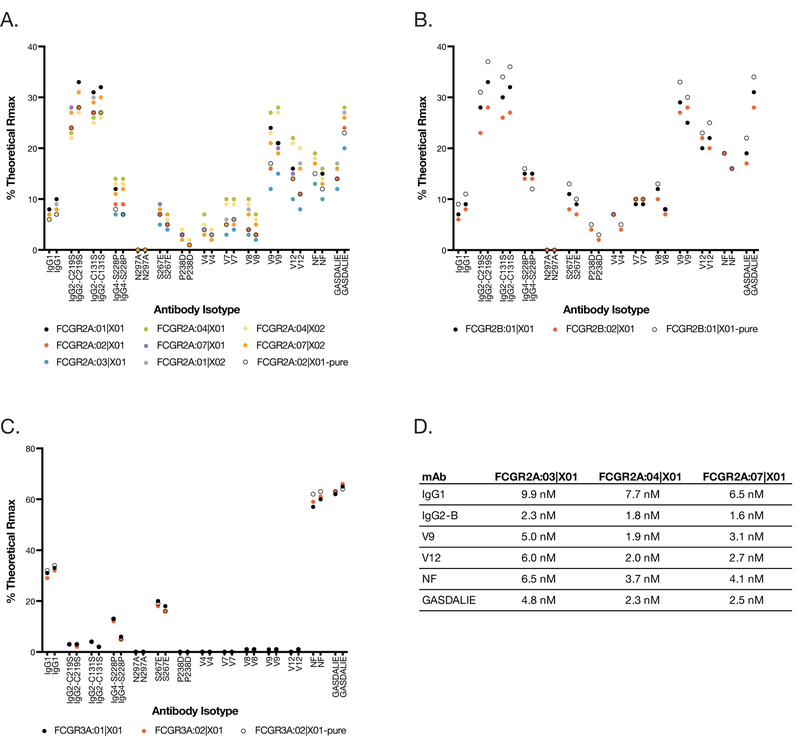
To determine whether FCGR variants demonstrate different binding affinities for IgG antibodies, an antibody binding screen was performed. Structure-based modeling to identify FCGR/Fc contact and proximal regions indicated that none of the polymorphic FCGR amino acids are direct FCGR/Fc contact residues, although some are proximal to the predicted contact residues (data not shown). Based on the allele discovery studies described above, His-tagged constructs for the extracellular domains of 14 distinct FCGR1A, FCGR2A, FCGR2B, and FCGR3A sequences were expressed in HEK cells, and tested for IgG binding by SPR.
The SPR screening assay captured His-tagged FCGRs out of cell culture supernatant and onto Biacore sensor chip surfaces containing immobilized anti-His-Fab fragments; purified IgG analytes were then bound to the FCGR molecules (Supplemental figure 2). We used a diverse panel of wild-type and engineered human IgG isotypes to represent the diversity of therapeutic IgGs that are currently in exploratory and clinical development for a variety of disease indications (Table II). Representative FCGR1A, FCGR2A, FCGR2B, and FCGR3A proteins were also expressed at a larger scale and were purified for comparison. The expression and purification of FCGR2A, FCGR2B, and FCGR3A were typical for well-behaved His-tagged proteins, and SPR data demonstrated comparable binding for purified and unpurified receptors (Figure 4, Supplemental figure 2). In contrast, FCGR1A demonstrated low expression and the purified protein was prone to aggregation and had low fractional activity (Supplemental figure 3).
Table II –
IgG isotypes tested in SPR binding assay.
| Isotype | Mutations | Description | References |
|---|---|---|---|
| IgG1 | none | Wild type IgG1 FcR binding affinity. | |
| IgG2-A | IgG2-C219S | C219S mutation stabilizes A-form disulfide bonding pattern. Has wild type IgG2 FcR binding activity. |
(29–32) |
| IgG2-B | IgG2-C131S | C131S mutation stabilizes B-form disulfide bonding pattern. Has wild type IgG2 FcR binding activity. | (29, 32) |
| IgG4 | IgG4-S228P | IgG4-S228P, where S228P mutation stabilizes hinge structure. Has wild type IgG4 FcR binding activity. |
(33, 34) |
| N297A | IgG1-N297A | N297A mutation results in aglycosylated Fc which dramatically reduces FcR affinity. |
(35, 36) |
| S267E | IgG1-S267E | S267E mutation alters human FCGR2 selectivity. |
(37, 38) |
| P238D | IgG1-P238D | Mutations alter human FCGR2 selectivity. |
(28, 39) |
| V4 | IgG1-P238D, P271G | ||
| V7 | IgG1-E233D, P238D, P271G, A330R | ||
| V8 | IgG1-G237D, P238D, H268D, P271G | ||
| V9 | IgG1-G237D, P238D, P271G, A330R | ||
| V12 | IgG1-E233D, G237D, P238D, H268D, P271G, A330R | ||
| NF | none | Wild type IgG1 with Fc glycan lacking fucose, increases human FCGR3A affinity. |
(40–42) |
| GASDALIE | G236A, S239D, A330L, I332E | G236A, S239D, A330L, and I332E mutations increase human FCGR3A affinity. |
(40, 43) |
Figure 4.
FCGR protein binding to fourteen different IgG isotypes, with two different monoclonal antibodies per isotype, was tested by surface plasmon resonance assay. SPR screening data showing percentage of theoretical Rmax is shown for (A) FCGR2A, (B) FCGR2B, and (C) FCGR3A. The sequences used to generate the 2A:01 protein were the same as the 2A:06 protein; likewise for 2A:04 and 2A:05. (D) KDapparent values are shown for representative IgG isotypes binding to select FCGR2A proteins, obtained from 2-fold serially diluted IgG titrations on captured FCGR surfaces, fit to a 1:1 steady-state model.
Because FCGR2A, FCGR2B, and FCGR3A all demonstrate fast association/dissociation kinetics and steady state binding, the relative binding of different IgG isotypes can be compared by analysis of the binding response as a percentage of the mass-normalized theoretical maximum binding response (%Rmax). As expected, the different wild-type and engineered IgG isotypes demonstrated different FCGR selectivities (Figure 4, Supplemental figure 2). However, the differences in binding among the allelic variants for a given FCGR were modest. To further quantitate any differences in binding, antibody titrations were performed for some of the FCGR2A interactions that demonstrated the largest differences in %Rmax in the screening data. For these titrations, the apparent KD values for wild type IgG1 and IgG2-B varied by 1.5-fold or less among the different FCGR2A variants (Figure 4D). The apparent KD values for the engineered IgG, such as V9, V12, and GASDALIE, showed slightly larger differences, but still varied by less than 3-fold for a given IgG. Collectively, these data show that the allelic variations in the MCM FCGR proteins have a very modest impact on the binding affinity to a wide range of wild type and Fc-engineered IgG, which is in contrast to human FCGR variants which can have IgG binding affinities that differ by five-fold or more for wild type IgG, and as much as 20-fold for some engineered variants such as V12 (27, 28).
Discussion
Due to their restricted genetic diversity, MCMs offer a more reproducible model for biomedical research than non-Mauritian cynomolgus macaques. Here we characterized FCGR allelic diversity in MCMs, using a combination of PacBio long-read sequencing, Illumina MiSeq genotyping, and WGS/MES to define full-length allele sequences and haplotypes as well as determine allele frequencies.
Variation in FCGR genes can produce receptors with varying affinities for IgG, leading to different susceptibilities and responses to disease. We identified polymorphic sites in each of the four FCGR proteins’ extracellular domains; these regions are the most likely to directly impact receptor binding to IgG.
FCGRs are likely responsible for the effects of monoclonal antibody (mAb) cancer therapies, such as rituximab and trastuzumab, due to their role in triggering antibody-dependent cell-mediated cytotoxicity (ADCC). Polymorphisms in human FCGRs, particularly FCGR2A H131R and FCGR3A F158V, have been found to impact the effectiveness of mAb therapies (4, 44). We identified just eight ancestral haplotypes across the FCGR2A, FCGR2B, and FCGR3A genes, which could simplify efforts to design balanced experimental cohorts of MCMs. Analogous control of experimental groups with other more diverse macaque species, especially rhesus macaques, is virtually impossible. However, we found that MCM FCGRs showed minimal differences in antibody binding affinity, and substantially less variability than reported for human FCGR polymorphisms (27, 45). In vitro mAb studies using MCM FCGRs can therefore be simplified by testing antibody binding to a standard panel of representative FCGR proteins rather than a full panel representing all MCM FCGRs. Furthermore, while MCMs may not reflect the full diversity of human FCGR binding affinities, they may serve as highly reproducible models for in vivo mAb therapy studies. Our work suggests that for preclinical and toxicology studies, time-consuming and costly FCGR haplotyping required for creating balanced study cohorts may not be necessary when using MCMs, which would simplify and streamline experimental design.
Our use of PacBio long-read sequencing to sequence full-length alleles allowed us to identify phased SNPs within each gene, a difficult task with shorter sequence reads that require assembly. To confirm that our PacBio sequencing was unbiased and comprehensive, we compared our data to whole-genome sequences. For FCGR1A and FCGR3A, the PacBio data encompassed all alleles and SNPs found in the WGS data. For FCGR2A and FCGR2B, however, we could not map WGS reads uniquely to either gene in exons 3 and 4 due to the homology between the extracellular domains of the two genes. Long-read sequencing that spans entire alleles eliminates this problem.
While our sample size was large, the majority of our animals originated from a single vendor (Bioculture Mauritius) and allele frequencies may differ somewhat for MCMs from different suppliers. Our dataset also may not encompass the entirety of all MCM FCGR variation since PacBio long-read sequencing was only performed for 48 individuals (96 chromosomes), and our MiSeq genotyping assay is designed to only identify known, not novel, alleles. However, because of the restricted genetic diversity of MCMs, any additional FCGR alleles are likely to be exceedingly rare.
Although identifying copy number variations or polymorphisms in FCGR promoter regions is beyond the scope of this work, this is an avenue for future research. The human G(–286)C SNP found in the promoter region of FCGR2B is associated with higher levels of transcription, and copy number variations have been found in the human FCGR3B, FCGR2C, and FCGR3A genes (46, 47). Our methods also were biased against identifying nonsense mutations that would lead to truncated proteins, either due to nonsense-mediated mRNA decay or removal during our open reading frame analyses.
Future directions include PacBio sequencing of FCGRs from additional animals, which will allow for both genotyping and ongoing novel allele discovery. We also plan to sequence several whole macaque genomes using PacBio and Oxford Nanopore long-read sequencing platforms, which will allow us to identify non-coding variation. Further research will also focus on characterizing the binding affinities of each allele and the ability of each allele to trigger ADCC (15), and our allele database will provide the basis for generating protein constructs to study this. Additionally, it will be important to study the effects of transmembrane and intracellular polymorphic residues on function. Studies of FCGR expression level differences and their presence on difference cell subsets were beyond the scope of this work, but will be important in confirming whether, as our work suggests, FCGR haplotyping is not essential when using MCMs in preclinical and toxicology studies. This research advances the development of MCMs as an important animal model for disease modeling and for drug toxicity studies, and shows that their use can streamline preclinical testing of biological therapies.
Supplementary Material
Acknowledgments
The authors wish to acknowledge and thank Alan Korman for support of these research efforts, Stanley Krystek for performing structure-based modeling, Michael Doyle for valuable input to the manuscript, and the following individuals for contributions to the production of the purified antibodies used in this study: Michael Pietras, Martin Corbett, Eric Lawrence, Karlene Melero, Yongmi An, Selyna Mai, Brenda Stevens, Claire Noriega, Derek Bond, Karla Henning, Brian Lee and Deepa Sharma.
Footnotes
A portion of this work was supported by Bristol-Myers Squibb and National Library of Medicine Grant 5T15LM007359.
References
- 1.Castro-Dopico T, and Clatworthy MR. 2016. Fcγ Receptors in Solid Organ Transplantation. Current Transplantation Reports 3: 284–293. PMID: 27909648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dahal LN, Roghanian A, Beers SA, and Cragg MS. 2015. FcγR requirements leading to successful immunotherapy. Immunological Reviews 268: 104–122. PMID: 26497516 [DOI] [PubMed] [Google Scholar]
- 3.Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, and Watier H. 2002. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood 99: 754–758. PMID: 11806974 [DOI] [PubMed] [Google Scholar]
- 4.Barnhart BC, and Quigley M. 2017. Role of Fc-FcγR interactions in the antitumor activity of therapeutic antibodies. Immunology and Cell Biology 95: 340–346. PMID: 27974746 [DOI] [PubMed] [Google Scholar]
- 5.Braster R, O’Toole T, and van Egmond M. 2014. Myeloid cells as effector cells for monoclonal antibody therapy of cancer. Methods 65: 28–37. PMID: 23811299 [DOI] [PubMed] [Google Scholar]
- 6.Gillis C, Gouel-Chéron A, Jönsson F, and Bruhns P. 2014. Contribution of human FcγRs to disease with evidence from human polymorphisms and transgenic animal studies. Frontiers in Immunology 5: 1–13. PMID: 24910634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nimmerjahn F, and Ravetch JV. 2005. Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science 310: 1510–1512. PMID: 16322460 [DOI] [PubMed] [Google Scholar]
- 8.Nimmerjahn F, and Ravetch JV. 2006. Fcgamma receptors: old friends and new family members. Immunity 24: 19–28. PMID: 16413920 [DOI] [PubMed] [Google Scholar]
- 9.Forthal DN, Landucci G, Bream J, Jacobson LP, Phan TB, and Montoya B. 2007. FcγRIIa genotype predicts progression of HIV infection. Journal of immunology (Baltimore, Md.: 1950) 179: 7916–7923. PMID: 18025239 [DOI] [PubMed] [Google Scholar]
- 10.Shimizu S, Tanaka Y, Tazawa H, Verma S, Onoe T, Ishiyama K, Ohira M, Ide K, and Ohdan H. 2016. Fc-Gamma Receptor Polymorphisms Predispose Patients to Infectious Complications after Liver Transplantation. American Journal of Transplantation 16: 625–633. PMID: 26517570 [DOI] [PubMed] [Google Scholar]
- 11.Hérodin F, Thullier P, Garin D, and Drouet M. 2005. Nonhuman primates are relevant models for research in hematology, immunology and virology. European Cytokine Network 16: 104–116. PMID: 15941681 [PubMed] [Google Scholar]
- 12.Keler T, Halk E, Vitale L, O’Neill T, Blanset D, Lee S, Srinivasan M, Graziano RF, Davis T, Lonberg N, and Korman A. 2003. Activity and safety of CTLA-4 blockade combined with vaccines in cynomolgus macaques. J Immunol 171: 6251–6259. PMID: 14634142 [DOI] [PubMed] [Google Scholar]
- 13.Nguyen DC, Scinicariello F, and Attanasio R. 2011. Characterization and allelic polymorphisms of rhesus macaque (Macaca mulatta) IgG Fc receptor genes. Immunogenetics 63: 351–362. PMID: 21327607 [DOI] [PubMed] [Google Scholar]
- 14.Wu H, and Adkins K. 2012. Identification of polymorphisms in genes of the immune system in cynomolgus macaques. Mammalian Genome 23: 467–477. PMID: 22527486 [DOI] [PubMed] [Google Scholar]
- 15.Sanford JC, Wu H, Abdiche Y, Harney JA, Chaparro-Riggers J, and Adkins K. 2017. Single nucleotide polymorphisms in the FcγR3A and TAP1 genes impact ADCC in cynomolgus monkey PBMCs. Immunogenetics 241–253. PMID: 28154890 [DOI] [PubMed]
- 16.Sussman RW, and Tattersall I. 1986. Distribution, Abundance, and Putative Ecological Strategy of Macaca fascicularis on the Island of Mauritius, Southwestern Indian Ocean. Folia Primatologica 46: 28–43. [Google Scholar]
- 17.Bonhomme M, Blancher A, Cuartero S, Chikhi L, and Crouau-Roy B. 2008. Origin and number of founders in an introduced insular primate: Estimation from nuclear genetic data. Molecular Ecology 17: 1009–1019. PMID: 18261045 [DOI] [PubMed] [Google Scholar]
- 18.Wiseman RW, Karl JA, Bohn PS, Nimityongskul FA, Starrett GJ, and O’Connor DH. 2013. Haplessly hoping: Macaque major histocompatibility complex made easy. ILAR Journal 54: 196–210. PMID: 24174442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bimber BN, Moreland AJ, Wiseman RW, Hughes AL, and O’Connor DH. 2008. Complete characterization of killer Ig-like receptor (KIR) haplotypes in Mauritian cynomolgus macaques: novel insights into nonhuman primate KIR gene content and organization. Journal of immunology (Baltimore, Md.: 1950) 181: 6301–6308. PMID: 18941221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warncke M, Calzascia T, Coulot M, Balke N, Touil R, Kolbinger F, and Heusser C. 2012. Different adaptations of IgG effector function in human and nonhuman primates and implications for therapeutic antibody treatment. Journal of immunology (Baltimore, Md.: 1950) 188: 4405–4411. PMID: 22461693 [DOI] [PubMed] [Google Scholar]
- 21.Ericsen AJ, Starrett GJ, Greene JM, Lauck M, Raveendran M, Deiros DR, Mohns MS, Vince N, Cain BT, Pham NH, Weinfurter JT, Bailey AL, Budde ML, Wiseman RW, Gibbs R, Muzny D, Friedrich TC, Rogers J, and O’Connor DH. 2014. Whole genome sequencing of SIV-infected macaques identifies candidate loci that may contribute to host control of virus replication. Genome Biology 15: 478 PMID: 25418588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prall TM, Graham ME, Karl JA, Wiseman RW, Ericsen AJ, Raveendran M, Alan Harris R, Muzny DM, Gibbs RA, Rogers J, and O’Connor DH. 2017. Improved full-length killer cell immunoglobulin-like receptor transcript discovery in Mauritian cynomolgus macaques. Immunogenetics 69: 325–339. PMID: 28343239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebeling M, Küng E, See A, Broger C, Steiner G, Berrera M, Heckel T, Iniguez L, Albert T, Schmucki R, Biller H, Singer T, and Certa U. 2011. Genome-based analysis of the nonhuman primate Macaca fascicularis as a model for drug safety assessment. Genome Research 21: 1746–1756. PMID: 21862625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magoč T, and Salzberg SL. 2011. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. PMID: 21903629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark AG 1990. Inference of haplotypes from PCR-amplified samples of diploid populations. Molecular biology and evolution 7: 111–122. PMID: 2108305 [DOI] [PubMed] [Google Scholar]
- 26.Karl JA, Heimbruch KE, Vriezen CE, Mironczuk CJ, Dudley DM, Wiseman RW, and O’Connor DH. 2014. Survey of major histocompatibility complex class II diversity in pig-tailed macaques. Immunogenetics 66: 613–623. PMID: 25129472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okazaki A, Shoji-Hosaka E, Nakamura K, Wakitani M, Uchida K, Kakita S, Tsumoto K, Kumagai I, and Shitara K. 2004. Fucose depletion from human IgG1 oligosaccharide enhances binding enthalpy and association rate between IgG1 and FcgammaRIIIa. J Mol Biol 336: 1239–1249. PMID: 15037082 [DOI] [PubMed] [Google Scholar]
- 28.Mimoto F, Katada H, Kadono S, Igawa T, Kuramochi T, Muraoka M, Wada Y, Haraya K, Miyazaki T, and Hattori K. 2013. Engineered antibody Fc variant with selectively enhanced FcγRIIb binding over both FcγRIIa(R131) and FcγRIIa(H131). Protein Eng Des Sel 26: 589–598. PMID: 23744091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dillon TM, Ricci MS, Vezina C, Flynn GC, Liu YD, Rehder DS, Plant M, Henkle B, Li Y, Deechongkit S, Varnum B, Wypych J, Balland A, and Bondarenko PV. 2008. Structural and functional characterization of disulfide isoforms of the human IgG2 subclass. J Biol Chem 283: 16206–16215. PMID: 18339626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wypych J, Li M, Guo A, Zhang Z, Martinez T, Allen MJ, Fodor S, Kelner DN, Flynn GC, Liu YD, Bondarenko PV, Ricci MS, Dillon TM, and Balland A. 2008. Human IgG2 antibodies display disulfide-mediated structural isoforms. J Biol Chem 283: 16194–16205. PMID: 18339624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu YD, Chen X, Enk JZ, Plant M, Dillon TM, and Flynn GC. 2008. Human IgG2 antibody disulfide rearrangement in vivo. J Biol Chem 283: 29266–29272. PMID: 18713741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H, and May K. 2012. Disulfide bond structures of IgG molecules: structural variations, chemical modifications and possible impacts to stability and biological function. MAbs 4: 17–23. PMID: 22327427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Angal S, King DJ, Bodmer MW, Turner A, Lawson AD, Roberts G, Pedley B, and Adair JR. 1993. A single amino acid substitution abolishes the heterogeneity of chimeric mouse/human (IgG4) antibody. Mol Immunol 30: 105–108. PMID: 8417368 [DOI] [PubMed] [Google Scholar]
- 34.Yang X, Wang F, Zhang Y, Wang L, Antonenko S, Zhang S, Zhang YW, Tabrizifard M, Ermakov G, Wiswell D, Beaumont M, Liu L, Richardson D, Shameem M, and Ambrogelly A. 2015. Comprehensive Analysis of the Therapeutic IgG4 Antibody Pembrolizumab: Hinge Modification Blocks Half Molecule Exchange In Vitro and In Vivo. J Pharm Sci 104: 4002–4014. PMID: 26308749 [DOI] [PubMed] [Google Scholar]
- 35.Bolt S, Routledge E, Lloyd I, Chatenoud L, Pope H, Gorman SD, Clark M, and Waldmann H. 1993. The generation of a humanized, non-mitogenic CD3 monoclonal antibody which retains in vitro immunosuppressive properties. Eur J Immunol 23: 403–411. PMID: 8436176 [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Mathieu M, and Brezski RJ. 2018. IgG Fc engineering to modulate antibody effector functions. Protein Cell 9: 63–73. PMID: 28986820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chu SY, Vostiar I, Karki S, Moore GL, Lazar GA, Pong E, Joyce PF, Szymkowski DE, and Desjarlais JR. 2008. Inhibition of B cell receptor-mediated activation of primary human B cells by coengagement of CD19 and FcgammaRIIb with Fc-engineered antibodies. Mol Immunol 45: 3926–3933. PMID: 18691763 [DOI] [PubMed] [Google Scholar]
- 38.Moore GL, Chen H, Karki S, and Lazar GA. 2010. Engineered Fc variant antibodies with enhanced ability to recruit complement and mediate effector functions. MAbs 2: 181–189. PMID: 20150767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dahan R, Barnhart BC, Li F, Yamniuk AP, Korman AJ, and Ravetch JV. 2016. Therapeutic Activity of Agonistic, Human Anti-CD40 Monoclonal Antibodies Requires Selective FcγR Engagement. Cancer Cell 29: 820–831. PMID: 27265505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, Weikert SH, and Presta LG. 2002. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem 277: 26733–26740. PMID: 11986321 [DOI] [PubMed] [Google Scholar]
- 41.Smith P, DiLillo DJ, Bournazos S, Li F, and Ravetch JV. 2012. Mouse model recapitulating human Fcγ receptor structural and functional diversity. Proc Natl Acad Sci U S A 109: 6181–6186. PMID: 22474370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satoh M, Iida S, and Shitara K. 2006. Non-fucosylated therapeutic antibodies as next-generation therapeutic antibodies. Expert Opin Biol Ther 6: 1161–1173. PMID: 17049014 [DOI] [PubMed] [Google Scholar]
- 43.Ahmed AA, Keremane SR, Vielmetter J, and Bjorkman PJ. 2016. Structural characterization of GASDALIE Fc bound to the activating Fc receptor FcγRIIIa. J Struct Biol 194: 78–89. PMID: 26850169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Julià M, Guilabert A, Lozano F, Suarez-Casasús B, Moreno N, Carrascosa JM, Ferrándiz C, Pedrosa E, Alsina-Gibert M, and Mascaró JM. 2013. The role of Fcγ receptor polymorphisms in the response to anti–tumor necrosis factor therapy in psoriasis A pharmacogenetic study. JAMA Dermatol 149: 1033–1039. PMID: 24048425 [DOI] [PubMed] [Google Scholar]
- 45.Richards JO, Karki S, Lazar GA, Chen H, Dang W, and Desjarlais JR. 2008. Optimization of antibody binding to FcgammaRIIa enhances macrophage phagocytosis of tumor cells. Mol Cancer Ther 7: 2517–2527. PMID: 18723496 [DOI] [PubMed] [Google Scholar]
- 46.Su K, Li X, Edberg JC, Wu J, Ferguson P, and Kimberly RP. 2004. A Promoter Haplotype of the Immunoreceptor Tyrosine-Based Inhibitory Motif-Bearing Fc RIIb Alters Receptor Expression and Associates with Autoimmunity. II. Differential Binding of GATA4 and Yin-Yang1 Transcription Factors and Correlated Receptor Expressio. The Journal of Immunology 172: 7192–7199. PMID: 15153543 [DOI] [PubMed] [Google Scholar]
- 47.Bournazos S, Woof JM, Hart SP, and Dransfield I. 2009. Functional and clinical consequences of Fc receptor polymorphic and copy number variants. Clinical and Experimental Immunology 157: 244–254. PMID: 19604264 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.