Abstract
The gut microbiome, a diverse microbial community in the gastrointestinal tract, plays a pivotal role in the maintenance of health. The gut microbiome metabolizes dietary and host-derived molecules to produce bioactive metabolites, which have a wide array of effects on host metabolism and immunity. ‘Dysbiosis’ of the gut microbiome, commonly considered as perturbation of microbiome diversity and composition, has been associated with intestinal and extra-intestinal diseases, including nonalcoholic fatty liver disease (NAFLD). A number of endogenous and exogenous factors, such as nutritional intake and xenobiotic exposure, can alter the gut microbiome. We will review the evolving methods for studying the gut microbiome and how these profiling techniques have been utilized to further our understanding of the gut microbial community composition and functional potential in the clinical spectrum of NAFLD. We will highlight microbiome-host interactions that may contribute to the pathogenesis of NAFLD, with a primary focus on mechanisms related to the metabolic output of the gut microbiome. Finally, we will discuss potential therapeutic implications of the gut microbiome in NAFLD.
Keywords: Microbiota, Nonalcoholic Fatty Liver
The gut microbiota is a diverse microbial community comprised of bacteria, fungi, viruses, and archaea that encodes several orders of magnitude more functional genes than the human genome.1 The collective genetic material of the microbiota is often referred to as the “gut microbiome” and encodes pathways that produce a wide array of bioactive small molecules that are derived from dietary or metabolic precursors and may alter human health.1 While under normal circumstances, the relationship between the human host and gut microbiome is mutually beneficial, perturbations of the gut microbiome, often referred to as “dysbiosis,” have been associated with a number of chronic diseases, including obesity, metabolic syndrome, and nonalcoholic fatty liver disease (NAFLD).1
Pre-clinical studies have provided the strongest evidence for a causal role of the gut microbiome in NAFLD. Several pivotal studies established that mice lacking gut microbiota are resistant to the development of diet-induced hepatic steatosis and that hepatic steatosis is transmissible via fecal microbiota transplantation (FMT) and ameliorated by probiotics and antibiotics in murine models.2 More recent studies suggest that the manipulation of the gut microbiome, either with antibiotics or FMT, also suppresses liver tumorigenesis and reduces portal hypertension in murine models.3, 4
Given this compelling pre-clinical evidence, the gut-liver axis is a rapidly developing area of investigation and new insights are emerging from a growing number of human studies. This review will highlight current methods for studying the microbiome and human gut microbial profiles associated with the clinical spectrum of NAFLD, stratified by community composition and function. We will also review postulated mechanisms linking the gut microbiome to the pathogenesis of NAFLD. Finally, we will discuss potential therapeutic implications of the gut microbiome in NAFLD.
Methods for gut microbiome profiling
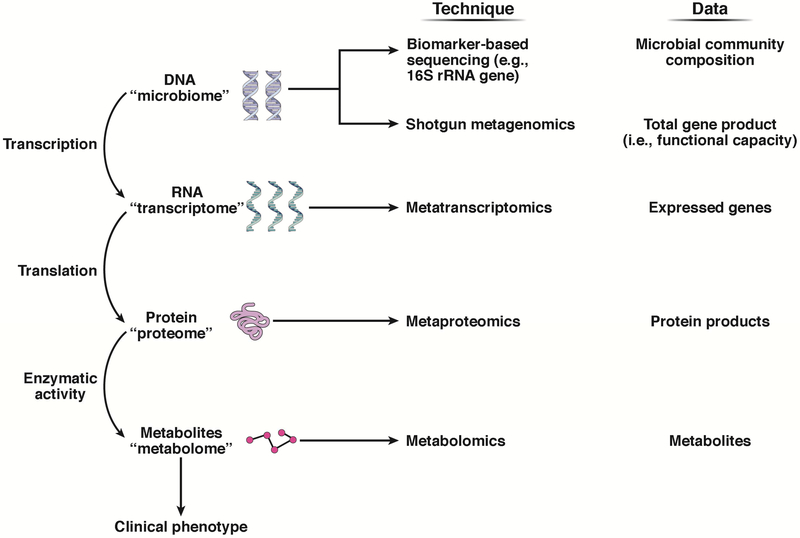
Advances in profiling and analytic techniques are transforming microbiome research and have been recently reviewed elsewhere5, 6, so we will limit our discussion to an overview of methods that have been utilized in human studies in NAFLD. (Figure 1) To date, the majority of studies have utilized culture-independent, biomarker-based profiling techniques. This method involves sequencing a ubiquitous gene, which is represented by the 16S ribosomal RNA (16S rRNA) gene in bacteria. Biomarker-based profiling techniques provide a relatively accurate fingerprint of microbial community composition (i.e. taxonomic relative abundance); however, little can be learned about the microbial community’s functional properties.1 While inferential algorithms based on reference genome databases enable predictions of functional capacities from 16S rRNA sequences, there are limitations to functional predictions.1 Moreover, this sequencing approach lacks the resolution needed to identify bacteria on a species or strain level, and different strains of the same bacterial species can exert different effects on the human host.5
Figure 1.
Methods for characterization of the gut microbiome.
Recent advances in computational biology have improved the feasibility of systems-level “omics” approaches, which allow for microbial community characterization beyond compositional states.5, 6 These approaches include next generation sequencing approaches to determine the functional genes encoded (metagenomics) or expressed (metatranscriptomics) by a microbial community, and mass spectrometry platforms to identify proteins (metaproteomics) and bioactive small molecules (metabolomics) collectively produced by a microbial community.1
Shotgun metagenomic sequencing characterizes the DNA library from a microbial community to obtain the entire gene complement (“metagenome”), although this method cannot assess the activity of microbial gene expression, which is regulated at the transcriptional and translational level.1 Even with metagenomic sequencing data, predicted function should be interpreted with caution because pathway presence does not reveal information about activity or directionality. Nevertheless, when compared to biomarker-based sequencing, metagenomics allows for more accurate characterization of microbial functional properties, in addition to taxonomical resolution to the species level.1 Metabolomics facilitates the identification and quantification of small molecule metabolic products (“metabolome”) through use of complementary analytical chemistry techniques and can include both targeted and untargeted approaches.1
For the purpose of this review, we will use the term “functional potential” to represent the gene content and/or metabolic output of the gut microbiome, as measured by one or more “omic” approaches.
Human gut microbiome profiles in clinical phenotypes of NAFLD: community composition
Numerous human studies have demonstrated an association between gut dysbiosis and the spectrum of NAFLD in children7–10 and adults11–23. All except one of these studies were cross-sectional12, and the majority utilized biomarker-based sequencing to profile the gut microbiome. We will review gut microbiome profiles, with a focus on genus-level differences, in the following clinical phenotypes: nonalcoholic fatty liver (NAFL), nonalcoholic steatohepatitis (NASH), NAFLD-related advanced fibrosis, and NAFLD-related hepatocellular carcinoma (HCC) (Table 1).
Table 1.
Characterization of the gut microbiome in clinical phenotypes of nonalcoholic fatty liver disease (NAFLD), stratified by genus-level taxonomic composition and “functional potential”
| Clinical Phenotypes of NAFLD |
Community Composition* | Functional Potential+ | ||
|---|---|---|---|---|
| Phylum | Genus | Fecal Metabolites | Serum Metabolites | |
| NAFL± | Firmicutes | ↑Blautia16 ↑Dorea11 ↓↑Roseburia15;11 ↓↑Lactobacillus 15;11,10,18,23 ↑Streptococcus 23 ↑Clostridium 23 ↓Oscillospira9 ↓Coprococcus 21,18,15 ↓Faecalibacterium 18 ↓Moryella 15 ↓Flavonifractor 19 ↓↑Oscillobacter 11,23,15;21 ↓↑Ruminococcus 18,16,15;9 |
↑Butanoic acid11 ↑Propanoic acid11 ↑Acetic acid11 ↑Isobutyric acid18 ↑Propionate18 ↑Unconjugated cholic acid13 ↑Ethanol8 ↓2-butanone11 |
↑2-butanone9 ↑1 pentanol9 ↑2-hydroxy-butyrate18 ↑L-lactic acid18 ↑Phenylacetic acid21 ↑Valine21 ↑Leucine21 ↑Isoleucine21 |
| Bacteroidetes | ↓↑Prevotella
16,23;8 ↓Odoribacter 23 ↓Alistipes23 |
|||
| Proteobacteria | ↓Escherichia 21,16,23 | |||
| Actinobacteria | ↓↑Bifidobacterium10;21 | |||
| NASH± | Firmicutes | ↑Blautia9,16 ↑Dorea9 ↑Lactobacillus10,19 ↑Clostridium 13 ↑Allisonella 12 ↑Oscillospira9 ↓Coprococcus18 |
↑Chenodeoxycholic acid13 ↓Unconjugated cholic acid13 ↑Lithocholic acid13 |
↑Ethanol7 ↑2-butanone9 ↑4-methyl-2-pentanone9 |
| ↓Faecalibacterium
12,18,19 ↓↑Ruminococcus 18,19;9 |
||||
| Bacteroidetes | ↑Bacteroides
24 ↑Parabacteroides 12 ↓Prevotella24 |
|||
| Proteobacteria | ↑Escherichia7 | |||
| Actinobacteria | ↓Bifidobacterium10,19 | |||
| NAFLD- related advanced fibrosis |
Firmicutes Bacteroidetes Proteobacteria |
↑Blautia
20 ↑Roseburia20 ↑Streptococcus20 ↑Lactobacillus20 ↓↑Enterococcus20 ↑Ruminococcus17;20,24 ↑Bacteroides20,17,22,24 ↑Parabacteroides20 ↑Escherichia 17, 22,16 ↑Klebsiella20 ↓↑Prevotella 24;20 |
↑3 phenylpropanoate17 ↑3-(4-hydroxyphenyl)lactate22 |
|
| Verrucomicrobia | ↓Akkermansia20 | |||
| NAFLD- related HCC |
Firmicutes | ↑Enterococcus20 ↑Oscillospira20 ↓Blautia20 |
||
| Bacteroidetes | ↑Bacteroides20 | |||
| Actinobacteria | ↓Bifidobacterium20 | |||
Increased in clinical phenotype of NAFLD; ↓Decreased in clinical phenotype of NAFLD; ↑↓Discordant findings among studies.
Comparison groups differed amongst studies. In NAFL, comparison groups included healthy controls9,8,10,11,15,16,18,23 and obese controls21. In NASH, comparison groups included healthy controls9,10,12,13,18,19, obese controls7, and NAFL24,16. In NAFLD-associated advanced fibrosis, comparison groups included healthy controls20 and NAFLD without advanced fibrosis (stage<2)17,24,22,16. In NAFLD-related HCC, the comparison group included NAFLD-related cirrhosis without HCC20.
The fecal and serum metabolites listed are postulated to be derived from the gut microbiome.
Two studies enrolled only non-obese subjects, in order to examine compositional changes in non-obese (“lean”) NAFL15 and NASH19.
Abbreviations: nonalcoholic fatty liver disease (NAFLD), nonalcoholic fatty liver (NAFL), nonalcoholic steatohepatitis (NASH), and hepatocellular carcinoma (HCC).
NAFL
Studies comparing the bacterial taxonomic composition between patients with NAFL and controls have yielded variable and often contradictory findings.8–11, 15, 16, 18, 21, 23 A common finding in NAFL patients is an increase in Lactobacillus and Escherichia and decrease in Coprococcus. However, studies have demonstrated contradictory findings in the relative abundance of Ruminococcus, Prevotella, and Bifidobacterium. Only one study has evaluated the gut microbiota in non-obese adults and found a decrease in Lactobacillus15, contrary to findings in obese NAFL.
NASH
Although more consistent findings have been reported in studies comparing microbial composition in NASH with NAFL and/or controls, there is still discrepancy in findings.7, 9, 10, 12–14, 16, 18, 19 Several studies found that patients with NASH had a decreased abundance of Faecalibacterium 12, 18 and Ruminococcus18, whereas NASH was associated with an increase in Ruminococcus in a separate study9. Meanwhile, Zhu et al. noted a significant difference only in Escherichia, which was not appreciated in other studies.7 To date, only one study has examined the gut microbiota in non-obese adults and noted a significant reduction in Faecalibacterium 19, which is consistent with findings in obese NASH.12, 18 However, Lactobacillus and Ruminococcus were reduced in non-obese NASH, similar to findings in non-obese NAFL, and consistent with findings that these genera may have a role in weight regulation.24, 25
NAFLD-related advanced fibrosis
NAFLD-related advanced fibrosis (stage>2) is associated with an overall decrease in microbial diversity, secondary to an increase in gram-negative bacteria.14, 16, 17, 20, 22 Multiple studies have identified associations between Bacteroides 14, 17, 20, 22 and Escherichia16, 17, 22 and advanced fibrosis. Similar perturbations of the gut microbiome have been noted in adults with cirrhosis attributable to etiologies other than NAFLD.2 Surprisingly, only one study identified a reduction in Akkermansia, despite evidence that Akkermansia muciniphila is associated with other metabolic diseases.26, 27
Loomba et al. utilized shotgun metagenomic sequencing, thus allowing for species level resolution, and noted that Bacteroides vulgatus and Escherichia coli were the most abundant species in patients with NAFLD-related advanced fibrosis.17 This finding is concordant with a prior observation that Bacteroides vulgatus is a contributor to insulin resistance.28 Due to utilization of 16S rRNA sequencing in other human studies examining advanced fibrosis, comparisons with these species-level findings are not feasible.
NAFLD-related HCC
Human studies examining the role of the gut microbiota in NAFLD-related hepatocarcinogenesis are lacking. Ponziana et al. were the first to examine adults with NAFLD-related cirrhosis with and without HCC, as compared to healthy controls.20 Bacteroides, Oscillospira, and Enterococcus were more abundant in HCC, when compared to cirrhosis without HCC.20 Only one other human study has profiled the gut microbiota in HCC (not limited to NAFLD-related HCC) and found higher levels of Escherichia coli through a culture-based approach.29
Limitations of compositional studies
Contradictory results between compositional studies could be attributed to various study-specific factors such as differences in patient cohorts (e.g. age, race/ethnicity, geographic location, body mass index [BMI]), definition of fatty liver disease (lack of histopathologic diagnosis in some studies), and comparison groups. Moreover, the majority of studies did not measure or adjust for endogenous or exogenous factors that are known to influence the gut microbiome. Although the gut microbiome is comprised of a stable ‘core’, a dynamic component exists that can be influenced by host and environmental factors.1 Age, host genetics, sex and hormonal cycles, diurnal variation, geographic location, illness, physical activity, xenobiotic exposure (including antibiotics), and nutritional intake may all impact the gut microbiome.1 Prior studies have identified country-specific microbial signatures, suggesting that geography and culture (including diet) significantly impact the gut microbiome.30 While antibiotic treatment is known to alter the gut microbiome1, we are increasingly recognizing that non-antibiotic drugs also modulate the gut microbiome. For example, a recent study examined 1,000 non-antibiotic drugs against 40 bacterial strains, and found that 24% of drugs with human targets inhibited growth of at least one strain in vitro.31 These results are consistent with studies that have demonstrated microbiome alterations associated with the use of common medications such as proton pump inhibitors32 and metformin33. While it remains unknown to what degree each of these factors modulate the microbiome-host interaction in NAFLD, these potential confounders must be measured in human studies.5
Of note, all studies to date in patients with NAFLD have focused exclusively on bacterial communities, and nothing is known of changes in co-existing fungal and viral communities. Although bacteria dominate the gut microbiota, fungal and viral communities are increasingly recognized as integral members of the community, and trans-kingdom interactions are likely to be in part responsible for ecological balance.1, 34 Perturbations in fungal community composition have now been associated with several chronic diseases, including obesity35, and warrants further investigation in NAFLD.
Ultimately, gut microbiome studies in larger, well-characterized, multi-ethnic, international cohorts are needed to better profile the gut microbiota in the clinical spectrum of NAFLD. However, as opposed to the presence of a single microbial species or pathogenic microbiome as the mediator of NAFLD, it’s plausible that several pathogenic microbiome states exist and exert differential effects on the human host based on environmental factors and/or genetic susceptibility for NAFLD. For example, a re-analysis of combined data from gut microbiome studies in obesity refuted prior claims that obesity is associated with one unique taxonomic signature.36 Nonetheless, changes in composition are likely not as important as changes in the functional potential of the gut microbial community. It is increasingly recognized that the metagenome encodes substantial redundancy among microorganisms and that there is incongruence between species abundance and transcriptional activity.37 This was exemplified in a study of lean and obese individuals who were better differentiated based on their gut metagenome, as opposed to their taxonomic profile.38
Human gut microbiome profiles in clinical phenotypes of NAFLD: functional potential
Several human studies have employed “omics” techniques and thus added new perspectives on functional attributes of the gut microbiome in NAFL, NASH, and NAFLD-related advanced fibrosis.7–9, 11, 13, 17, 18, 21, 22 Results from fecal and serum metabolite profiling are listed in Table 1. To date, no human studies have functionally profiled the gut microbiome in NAFLD-related HCC.
NAFL
Two studies have performed integrated analyses of the gut microbiota via 16S rRNA sequencing and targeted metabolomics for fecal metabolites in adults with NAFL. Raman et al. identified 18 fecal metabolites that were differentially abundant in obese adults with NAFL, when compared to healthy controls. The majority of differentially abundant fecal metabolites were esters of short-chain fatty acids (SCFAs), including propanoic acid and butanoic acid.11 Da Silva et al. performed targeted profiling of eight fecal metabolites of interest and found that higher concentration of two fecal SCFAs, propionate and isobutyric acid, differentiated adults with NAFL from healthy controls, corroborating findings from Raman et al.18
Hoyles et al. performed an integrated analysis of the gut metagenome, hepatic transcriptome, and serum and urine metabolomes in a cohort of obese, non-diabetic women.21 NAFL was associated with low microbial gene richness and alterations in branched-chain and aromatic amino acid pathways and endotoxin synthesis. While multiple microbial-derived metabolites were correlated with NAFL, serum phenylacetic acid, a product of amino acid metabolism, had the strongest association.
NASH
Del Chierico et al. identified increased abundance of two serum metabolites, 2-butanone and 4-methyl-2-pentanone, in children with NASH.39 When combined with taxonomic differences (decreased Oscillospira and increased Dorea and Ruminococcus), these fecal metabolites differentiated children with NASH from healthy controls. Intriguingly, 2-butanone was also increased in the serum of adults with NAFL21, but the functional significance of this metabolite is unknown.
NAFLD-related advanced fibrosis
Loomba et al. performed shotgun metagenomic sequencing in adults with NAFLD (with a focus on NAFLD-related advanced fibrosis), which was integrated with serum metabolomics.17 Thirty-seven bacterial species, of which Escherichia coli was the most abundant, were differentially represented in the gut of patients with advanced fibrosis, compared to those without fibrosis. When these species were incorporated into a model that also included patient age, BMI, and a microbial diversity index, the model possessed high accuracy for detecting advanced fibrosis. However, the differential abundance of serum metabolites and gut microbial gene pathways between groups did not achieve statistical significance. A more recent study by Caussy et al. identified a shared genetic determination of serum 3-(4-hydroxyphenyl)lactate, a microbial-derived metabolite involved in amino acid metabolism, with NAFLD-related fibrosis.22 Interestingly, 3-(4-hydroxyphenyl)lactate was strongly correlated with the abundance of 7 bacterial species that were previously associated with advanced fibrosis, including Bacteroides caccae, Escherichia coli, and Clostridium sp.17 Moreover, the finding of dysregulation in amino acid metabolism is consistent with metagenomic shifts noted in NAFL.21
Postulated mechanisms linking the gut microbiome to NAFLD
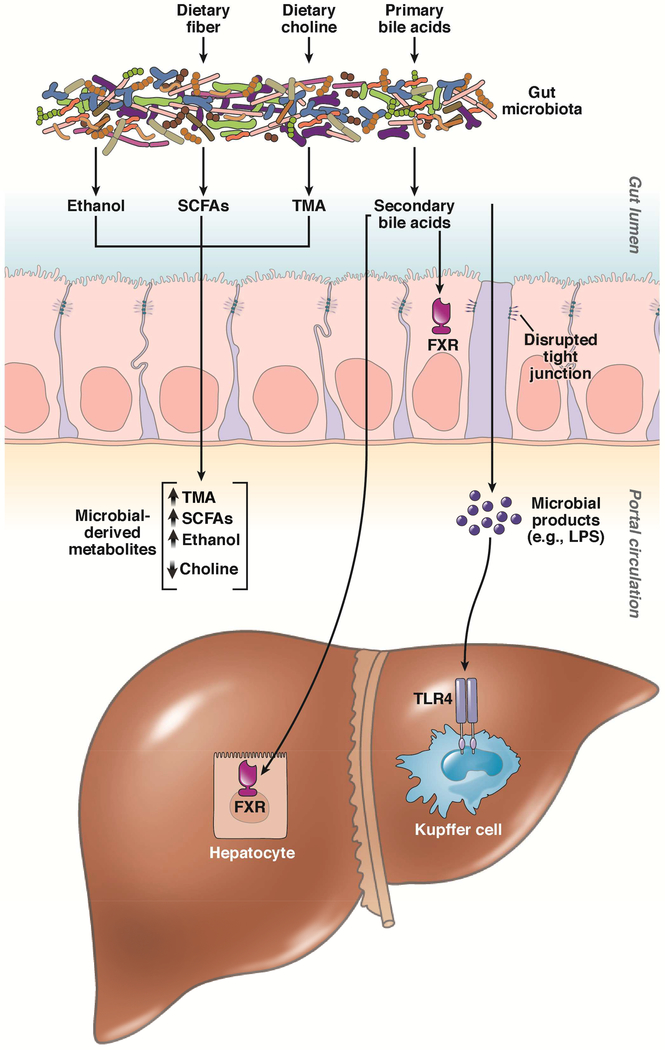
Although human studies have yielded insight into functional attributes of the gut microbiota in NAFLD, much of the mechanistic evidence linking the gut microbiome and NAFLD pathogenesis has been obtained from experiments in animal models. We will summarize the current evidence for postulated microbiome-associated mechanisms contributing to the pathogenesis of NAFLD (Figure 2).
Figure 2. Postulated mechanisms linking the gut microbiome to nonalcoholic fatty liver disease.
Impairment of intestinal epithelial function, via disrupted tight junctions, leads to endotoxemia and induction of toll-like receptor 4 (TLR4). Dietary choline is metabolized by the gut microbiome to produce trimethylamine (TMA), which reduces choline bioavailability. Indigestable carbohydrates (e.g. dietary fiber) undergo fermentation by the gut microbiome and give rise to short-chain fatty acids (SCFAs). The gut microbiome metabolizes bile acids, thus regulating the bile acid pool and leading to alterations in farnesoid X receptor (FXR) signaling. Ethanol can be generated by the gut microbiome. The gut microbiome may contribute to disruptions in amino acid homeostasis (not pictured).
1. Altered gut barrier function, endotoxemia, and activation of toll-like receptor mediated pathways
Impairment of the gut barrier, predominantly caused by disruption of intracellular tight junctions, is more common in adults with NAFLD and can even occur in healthy subjects transitioned to a Western diet.40, 41 Intestinal epithelial barrier disruption leads to increased translocation of microbial products such as lipopolysaccharide (LPS) into the portal circulation, and resultant endotoxemia can induce hepatic inflammation. Rahman et al. demonstrated that administering a high fat, fructose and cholesterol diet to knockout mice for the gene encoding junctional adhesion molecule A resulted in severe fibrotic steatohepatitis compared to only modest steatosis in control mice, and administration of oral antibiotics or sequestration of bacterial endotoxins resulted in improvement of liver histology.42 While impairment in gut permeability appears to contribute to NAFLD pathogenesis, it remains unclear if patients with NAFLD are predisposed to altered gut barrier function, if dietary changes directly affect intestinal permeability, or if a Western diet leads to deleterious changes in the microbiota that mediate impairments in gut barrier function.43
Interestingly, knockout mice for mucin 2, with a resulting decrease in intestinal mucus, appear to be protected from high-fat diet induced NAFLD mediated by an increase in IL-22, suggesting an important relationship between the intestinal immune system, the gut barrier, and NAFLD.44 Modulation of the gut immune system in beta 7 integrin-deficient mice through use of 5-aminosylicylclic acid as a topical anti-inflammatory improved metabolic parameters and reduced gut permeability and endotoxemia.45 Downstream effects of endotoxin translocation may include induction of toll-like receptors (TLR) in the liver, specifically TLR4, with downstream activation of transcription factors inducing an inflammatory response, and TLR4 knockout may mitigate hepatic inflammation.46 However, a recent phase 2 clinical trial demonstrated no benefit from a TLR4 antagonist in patients with biopsy-proven NASH, and the ability to manipulate this pathway to ameliorate NAFLD remains uncertain.47
2. Choline metabolism
The link between reduced choline bioavailability and the development of NAFLD was established decades ago.48 Choline deficiency leads to abnormal phospholipid synthesis, defective very-low-density lipoprotein secretion, and alterations in the enterohepatic circulation of bile acids.49 A number of factors affect choline bioavailability, including dietary intake, estrogen status, and single-nucleotide polymorphisms (SNPs) in genes for de novo choline metabolism.49 However, it was more recently discovered that dietary choline is metabolized by the gut microbiome to produce a variety of metabolites such as trimethylamine (TMA), and thus can reduce choline bioavailability.50
At least 8 human gut microbes are avid choline metabolizers, and only low levels of colonization with TMA-producing species are required to significantly reduce host choline levels.51, 52 A high-fat diet leads to an increase in gut microbes that metabolize choline and subsequent development of hepatic steatosis in mice.53 Manipulation of dietary intake of choline in human subjects resulted in variations in the abundance of Gammaproteobacteria and Erysipelotrichi, which were directly associated with the degree of liver fat accumulation during periods of choline depletion. Abundance of these two classes of bacteria, along with SNPs in choline metabolism, accurately predicted the degree to which subjects developed hepatic steatosis while on a choline-deficient diet.54
TMA diffuses into the bloodstream and is metabolized by the liver to generate trimethylamine-N-oxide (TMAO). Elevated circulating TMAO levels have been associated with cardiovascular disease55 and chronic kidney disease33. Intriguingly, these are extrahepatic manifestations of NAFLD, which is suggestive of a microbialderived mechanistic link between inter-related cardiometabolic diseases. However, the association between circulating TMAO and NAFLD, in addition to specific microbe-host interactions, is not well delineated.
3. Production of short-chain fatty acids
Indigestable carbohydrates (e.g. dietary fiber) undergo fermentation by the gut microbiota and give rise to SCFAs, which are amongst the most abundant of microbialderived metabolites. The Bacteroidetes phylum is a major contributor to the production of acetate and propionate, whereas the Firmicutes phylum predominantly produces butyrate.56 While SCFAs provide the majority of energy needs for intestinal epithelial cells, they also cross the intestinal epithelial barrier and mediate a diverse array of biological activities, including regulation of energy expenditure, appetite, and satiety hormone production, through G-protein coupled receptors in multiple tissue sites.57, 58
Overweight adults, as compared to lean adults, have increased total gut SCFAs.59 Additionally, total gut SCFAs decrease in obese adults with anti-obesity treatment and prebiotic administration.60, 61 Altogether, these data suggest that increased energy extraction from dietary intake, manifested by an increase in SCFAs, is a hallmark of the obesity phenotype. However, in contrast to these findings, increasing colonic propionate prevents weight gain in overweight adults62, and the beneficial metabolic effects of FMT from lean donors to obese men was associated with an increased abundance of butyrate-producing gut microbes.63
Supplementation of SCFAs improves diet-induced hepatic steatosis in murine models64; however, in contrast to these findings, human studies have noted increased fecal concentrations of SCFAs in adults with NAFL.11, 18 (Table 1) In both obesity and NAFLD, incongruous findings for the association between clinical phenotypes and SCFAs are likely attributable to the differential abundance of individual SCFAs, each of which may have different effects on host metabolism.
4. Metabolism of bile acids
Bile acids are increasingly recognized as important signaling molecules that activate a number of host receptors, including farnesoid X receptor (FXR), with effects on host metabolism and immunity.65 Bile acids prevent intestinal bacterial overgrowth, both directly (via membrane damaging effects) and indirectly (via production of antimicrobial proteins). As such, bile acids modulate the composition of the gut microbiome. On the other hand, the gut microbiota deconjugate and convert primary bile acids to secondary bile acids, thus regulating the bile acid pool.66 While a wide array of gut microbes have the capacity to deconjugate bile acids, only small number of bacteria (predominantly Clostridium species) facilitate conversion of primary bile acids to secondary bile acids.67
Inhibition of intestinal FXR ameliorates high-fat diet induced hepatic steatosis in murine models.68 Recent pre-clinical studies have also supported a role for microbial alterations in bile acid metabolism, specifically an overabundance of deoxycholic acid (a secondary bile acid), in the development of obesity-related liver cancer.69 However, it is imperative to highlight that differences in bile acid composition between humans and mice must considered when extrapolating these findings to humans.70
FXR agonists have been studied in the management of NASH in humans. A clinical trial of obeticholic acid, a potent FXR agonist, in patients with non-cirrhotic NASH showed that 72-week treatment improved histological NASH.71 Adults with NAFLD have elevated total serum bile acids and alterations in the ratio of secondary to primary bile acids72, which was subsequently found to be associated with a shift in abundance of gut microbes associated with bile acid deconjugation.73 Moreover, fecal bile acid composition distinguishes adults with NASH from healthy controls.13 (Table 1) Overall, while it is likely that altered bile acid metabolism and ensuing effects on FXR signaling contribute to NAFLD pathogenesis, there is still much to be gleaned from the complex, reciprocal relationship between the gut microbiome and bile acids.
5. Ethanol production
Alcohol is mainly metabolized in the liver by alcohol dehydrogenase (ADH), and hepatic gene transcription of ADH is increased in NAFLD.74 Ethanol production in the gut can be altered by manipulation of the gut microbiome75, and a common finding in multiple NAFLD phenotypes is enrichment in Escherichia and Lactobacillus, genera which can produce ethanol.2 (Table 1) While increased serum and fecal ethanol has been described pediatric patients with NAFL8 and NASH7, this finding has not been replicated in adult cohorts.18 Recent evidence suggests that insulin-dependent impairments of liver ADH activity, as opposed to endogenous alcohol synthesis from the gut microbiome, could explain elevated serum ethanol levels in NAFLD, pointing to the existence of distinct disease mechanisms that may be related to differences in gut microbiome function.76 Ultimately, additional studies are needed to confirm these findings.
6. Amino acid biosynthesis and metabolism
The gut microbiome can exert effects on amino acid homeostasis, in part due to biosynthesis and metabolism of aromatic (AAA) and branched-chain amino acids (BCAA). Cohort studies have identified elevated serum BCAAs as a biomarker for insulin resistance.77, 78 In adults with insulin resistance, Prevotella copri and Bacteroides vulgatus are associated with enriched biosynthetic potential for BCAAs and a reduced potential for BCAA transport into bacterial cells.28
Perturbations in microbial metabolism of AAAs and BCAAs, and ensuing alterations in the serum metabolite profile, have more recently been identified in adults with NAFL.21 (Table 1) Phenylacetic acid, an AAA-derived microbial metabolite strongly correlated with hepatic steatosis in humans, was found to induce hepatic steatosis in both a primary culture of human hepatocytes and in rodents, suggesting a causal role in NAFL pathogenesis.21 This study provides a proof-of-concept of how findings from integrated multi-omic analyses in patient cohorts can be complemented by in vitro and in vivo mechanistic studies, in order to facilitate the identification of microbial-driven causal pathways.
The gut microbiome and NAFLD: from research to bedside
Microbiome-targeted therapy (MTT) is considered to include antibiotics, probiotics (culture of living microorganisms which could have health benefits for the human host), prebiotics (fermentable dietary fibers that stimulate the growth and survival of probiotics), synbiotics (combination of probiotics and prebiotics), and FMT.1 High-quality, large-scale clinical interventional trials examining MTT in NAFLD are lacking. Several randomized controlled trials have examined the use of non-FMT MTT in NAFLD but yielded mixed results.79 To date, FMT trials in human subjects have been limited to obese adults with metabolic syndrome (without defined NAFLD), but at least two clinical trials examining FMT in adults with biopsy-confirmed NASH are actively recruiting subjects.81, 82 FMT from lean to obese donors was shown to improve insulin sensitivity, albeit with only short-term improvement.63, 80 Given NAFLD is commonly associated with insulin resistance, these results suggest that FMT could be efficacious in the management of NAFLD, however, improvement may be short-lived and limit the feasibility of this approach.
Ultimately, until we better understand the intricacies of microbial metabolism and microbe-host interactions in the pathogenesis of NAFLD, it’s unlikely that MTT will be optimized for use in clinical practice. Nonetheless, even without elucidation of exact mechanistic pathways, stool and/or serum microbial biomarkers will likely yield diagnostic and/or prognostic utility. This is especially relevant in NAFLD given the need for non-invasive approaches to evaluate for progressive forms of disease.
Conclusions
In summary, pre-clinical evidence supports a causal role of gut microbiome in liver disease progression in NAFLD. However, there is much that we do not understand about the gut-liver axis. Due to the cross-sectional nature of published human data, along with methods utilized for microbial profiling, the majority of clinical evidence supports an association between dysbiosis and NAFLD, but mechanistic links have not been well established. Further well-designed, longitudinal, prospective cohort studies with multi-omic profiling techniques, and complemented by mechanistic studies in animal models, are needed to decipher the complex (and likely multifactorial) microbiome-host interactions in NAFLD. Nevertheless, it is becoming increasingly apparent that the gut microbiome may play a role as a biomarker of disease severity in NAFLD and provide novel insights into the pathogenesis of NAFLD in the coming years.
Acknowledgments
Grant Support: Dr. Sharpton is supported by the National Research Service Award (2T32DK060414–16) from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). Dr. Ajmera is supported by the Advanced/Transplant Hepatology Fellowship and the Alan Hofmann Clinical and Translational Research Award from the AASLD Foundation. Dr. Loomba is supported by NIH/R01-DK106419–02.
Abbreviations:
- (ADH)
alcohol dehydrogenase
- (AAA)
aromatic amino acids
- (BMI)
body mass index
- (BCAAs)
branched-chain amino acids
- (DNA)
deoxyribonucleic acid
- (FXR)
farnesoid X receptor
- (FMT)
fecal microbiota transplantation
- (HCC)
hepatocellular carcinoma
- (LPS)
lipopolysaccharide
- (MTT)
microbiome-targeted therapy
- (NAFL)
nonalcoholic fatty liver
- (NAFLD)
nonalcoholic fatty liver disease
- (NASH)
nonalcoholic steatohepatitis
- (rRNA)
ribosomal ribonucleic acid
- (SCFA)
short-chain fatty acid
- (SNP)
single-nucleotide polymorphism
- (TLR4)
toll-like receptor 4
- (TMA)
trimethylamine
- (TMAO)
trimethylamine-N-oxide
Footnotes
Conflicts of Interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. The New England journal of medicine 2016;375:2369–2379. [DOI] [PubMed] [Google Scholar]
- 2.Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology 2014;146:1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu LX, Schwabe RF. The gut microbiome and liver cancer: Mechanisms and clinical translation. Nature Reviews Gastroenterology and Hepatology 2017;14:527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.García-Lezana T, Raurell I, Bravo M, et al. Restoration of a healthy intestinal microbiota normalizes portal hypertension in a rat model of nonalcoholic steatohepatitis. Hepatology 2018;67:1485–1498. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert JA, Blaser MJ, Caporaso JG, et al. Current understanding of the human microbiome. Nat Med 2018;24:392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knight R, Vrbanac A, Taylor BC, et al. Best practices for analysing microbiomes. Nat Rev Microbiol 2018;16:410–422. [DOI] [PubMed] [Google Scholar]
- 7.Zhu L, Baker SS, Gill C, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis patients: a connection between endogenous alcohol and NASH. Hepatology 2013;57:601–609. [DOI] [PubMed] [Google Scholar]
- 8.Michail S, Lin M, Frey MR, et al. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS microbiology ecology 2015;91:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Chierico F, Nobili V, Vernocchi P, et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 2017;65:451–464. [DOI] [PubMed] [Google Scholar]
- 10.Nobili V, Putignani L, Mosca A, et al. Bifidobacteria and lactobacilli in the gut microbiome of children with non-alcoholic fatty liver disease: which strains act as health players? Arch. Med. Sci 2018;14:81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raman M, Ahmed I, Gillevet PM, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clinical gastroenterology and hepatology 2013;11:868–75.e1–3. [DOI] [PubMed] [Google Scholar]
- 12.Wong VW-S, Tse C-H, Lam TT-Y, et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis--a longitudinal study. PloS one 2013;8:e62885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mouzaki M, Wang AY, Bandsma R, et al. Bile Acids and Dysbiosis in Non-Alcoholic Fatty Liver Disease. PloS one 2016;11:e0151829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boursier J, Mueller O, Barret M, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016;63:764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang B, Jiang X, Cao M, et al. Altered Fecal Microbiota Correlates with Liver Biochemistry in Nonobese Patients with Non-alcoholic Fatty Liver Disease. Scientific reports 2016;6:32002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen F, Zheng RD, Sun XQ, et al. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary and Pancreatic Diseases International 2017;16:375–381. [DOI] [PubMed] [Google Scholar]
- 17.Loomba R, Seguritan V, Li W, et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell metabolism 2017;25:1054–1062.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Da Silva HE, Teterina A, Comelli EM, et al. Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Scientific reports 2018;8:1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duarte SMB, Stefano JT, Miele L, et al. Gut microbiome composition in lean patients with NASH is associated with liver damage independent of caloric intake: A prospective pilot study. Nutrition, metabolism, and cardiovascular diseases: 2018;28:369–384. [DOI] [PubMed] [Google Scholar]
- 20.Ponziani FR, Bhoori S, Castelli C, et al. Hepatocellular Carcinoma is Associated with Gut Microbiota Profile and Inflammation in Non-Alcoholic Fatty Liver Disease. Hepatology 2018. [DOI] [PubMed] [Google Scholar]
- 21.Hoyles L, Fernandez-Real JM, Federici M, et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med 2018;24:1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caussy C, Hsu C, Lo M-T, et al. Novel link between gut-microbiome derived metabolite and shared gene-effects with hepatic steatosis and fibrosis in NAFLD. Hepatology 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang W, Wu N, Wang X, et al. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep 2015;5:8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Million M, Maraninchi M, Henry M, et al. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes 2012;36:817–25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Ottosson F, Brunkwall L, Ericson U, et al. Connection Between BMI-Related Plasma Metabolite Profile and Gut Microbiota. J Clin Endocrinol Metab 2018;103:1491–1501. [DOI] [PubMed] [Google Scholar]
- 26.Schneeberger M, Everard A, Gomez-Valades AG, et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep 2015;5:16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plovier H, Everard A, Druart C, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 2017;23:107–113. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen HK, Gudmundsdottir V, Nielsen HB, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016;535:376–81. [DOI] [PubMed] [Google Scholar]
- 29.Grat M, Wronka KM, Krasnodebski M, et al. Profile of Gut Microbiota Associated With the Presence of Hepatocellular Cancer in Patients With Liver Cirrhosis. Transplant Proc 2016;48:1687–91. [DOI] [PubMed] [Google Scholar]
- 30.Li J, Jia H, Cai X, et al. An integrated catalog of reference genes in the human gut microbiome. Nature biotechnology 2014;32:834–841. [DOI] [PubMed] [Google Scholar]
- 31.Maier L, Pruteanu M, Kuhn M, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018;555:623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson MA, Goodrich JK, Maxan M-E, et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut 2016;65:749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015;528:262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nash AK, Auchtung TA, Wong MC, et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017;5:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mar Rodríguez M, Pérez D, Javier Chaves F, et al. Obesity changes the human gut mycobiome. Scientific reports 2015;5:14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finucane MM, Sharpton TJ, Laurent TJ, et al. A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PloS one 2014;9:e84689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abu-Ali GS, Mehta RS, Lloyd-Price J, et al. Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nature microbiology 2018;3:356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013;500:541–546. [DOI] [PubMed] [Google Scholar]
- 39.Del Chierico F, Nobili V, Vernocchi P, et al. Gut microbiota profiling of pediatric NAFLD and obese patients unveiled by an integrated meta-omics based approach. Hepatology 2016. [DOI] [PubMed] [Google Scholar]
- 40.Pendyala S, Walker JM, Holt PR. A high-fat diet is associated with endotoxemia that originates from the gut. Gastroenterology 2012;142:1100–1101.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miele L, Valenza V, La Torre G, et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology 2009;49:1877–1887. [DOI] [PubMed] [Google Scholar]
- 42.Rahman K, Desai C, Iyer SS, et al. Loss of Junctional Adhesion Molecule A Promotes Severe Steatohepatitis in Mice on a Diet High in Saturated Fat, Fructose, and Cholesterol. Gastroenterology 2016;151:733–746.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergheim I, Weber S, Vos M, et al. Antibiotics protect against fructose-induced hepatic lipid accumulation in mice: role of endotoxin. J Hepatol 2008;48:983–92. [DOI] [PubMed] [Google Scholar]
- 44.Hartmann P, Seebauer CT, Mazagova M, et al. Deficiency of intestinal mucin-2 protects mice from diet-induced fatty liver disease and obesity. Am J Physiol Gastrointest Liver Physiol 2016;310:G310–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luck H, Tsai S, Chung J, et al. Regulation of obesity-related insulin resistance with gut anti-inflammatory agents. Cell Metab 2015;21:527–42. [DOI] [PubMed] [Google Scholar]
- 46.Henao-Mejia J, Elinav E, Jin C, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diehl AM, Harrison S, Caldwell S, et al. JKB-121 in patients with nonalcoholic steatohepatitis: A phase 2 double blind randomized placebo control study. Journal of Hepatology 2018;68:S103. [Google Scholar]
- 48.Blumberg H, McCollum EV. THE PREVENTION BY CHOLINE OF LIVER CIRRHOSIS IN RATS ON HIGH FAT, LOW PROTEIN DIETS. Sci. 1941;93:598–599. [DOI] [PubMed] [Google Scholar]
- 49.Corbin KD, Zeisel SH. Choline metabolism provides novel insights into nonalcoholic fatty liver disease and its progression. Current opinion in gastroenterology 2012;28:159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stremmel W, Schmidt KV, Schuhmann V, et al. Blood Trimethylamine-N-Oxide Originates from Microbiota Mediated Breakdown of Phosphatidylcholine and Absorption from Small Intestine. PloS one 2017;12:e0170742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romano KA, Vivas EI, Amador-Noguez D, et al. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio 2015;6:e02481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rath S, Heidrich B, Pieper DH, et al. Uncovering the trimethylamine-producing bacteria of the human gut microbiota. Microbiome 2017;5:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dumas M-E, Barton RH, Toye A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proceedings of the National Academy of Sciences of the United States of America 2006;103:12511–12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spencer MD, Hamp TJ, Reid RW, et al. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 2011;140:976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang WHW, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. The New England journal of medicine 2013;368:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinez KB, Leone V, Chang EB. Microbial metabolites in health and disease: Navigating the unknown in search of function. The Journal of biological chemistry 2017;292:8553–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nature Reviews Endocrinology 2015;11:577–591. [DOI] [PubMed] [Google Scholar]
- 58.Ríos-Covián D, Ruas-Madiedo P, Margolles A, et al. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Frontiers in microbiology 2016;7:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwiertz A, Taras D, Schäfer K, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 2010;18:190–195. [DOI] [PubMed] [Google Scholar]
- 60.Patil DP, Dhotre DP, Chavan SG, et al. Molecular analysis of gut microbiota in obesity among Indian individuals. Journal of biosciences 2012;37:647–657. [DOI] [PubMed] [Google Scholar]
- 61.Salazar N, Dewulf EM, Neyrinck AM, et al. Inulin-type fructans modulate intestinal Bifidobacterium species populations and decrease fecal short-chain fatty acids in obese women. Clinical nutrition 2015;34:501–507. [DOI] [PubMed] [Google Scholar]
- 62.Chambers ES, Viardot A, Psichas A, et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 2015;64:1744–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vrieze A, Van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012;143:913–6.e7. [DOI] [PubMed] [Google Scholar]
- 64.Mattace Raso G, Simeoli R, Russo R, et al. Effects of sodium butyrate and its synthetic amide derivative on liver inflammation and glucose tolerance in an animal model of steatosis induced by high fat diet. PloS one 2013;8:e68626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Slijepcevic D, van de Graaf SFJ. Bile Acid Uptake Transporters as Targets for Therapy. Digestive diseases 2017;35:251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ridlon JM, Harris SC, Bhowmik S, et al. Consequences of bile salt biotransformations by intestinal bacteria. Gut microbes 2016;7:22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Urdaneta V, Casadesus J. Interactions between Bacteria and Bile Salts in the Gastrointestinal and Hepatobiliary Tracts. Front Med 2017;4:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang C, Xie C, Li F, et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. The Journal of clinical investigation 2015;125:386–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nature Reviews Gastroenterology and Hepatology 2018;15:111–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wahlström A, Kovatcheva-Datchary P, Ståhlman M, et al. Crosstalk between Bile Acids and Gut Microbiota and Its Impact on Farnesoid X Receptor Signalling. Digestive diseases (Basel, Switzerland) 2017;35:246–250. [DOI] [PubMed] [Google Scholar]
- 71.Neuschwander-Tetri BA, Loomba R, Sanyal AJ, et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet 2015;385:956–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ferslew BC, Xie G, Johnston CK, et al. Altered Bile Acid Metabolome in Patients with Nonalcoholic Steatohepatitis. Digestive diseases and sciences 2015;60:3318–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiao N, Baker SS, Chapa-Rodriguez A, et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut 2017:314307. [DOI] [PubMed] [Google Scholar]
- 74.Baker SS, Baker RD, Liu W, et al. Role of alcohol metabolism in non-alcoholic steatohepatitis. PloS one 2010;5:e9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Elshaghabee FMF, Bockelmann W, Meske D, et al. Ethanol Production by Selected Intestinal Microorganisms and Lactic Acid Bacteria Growing under Different Nutritional Conditions. Frontiers in microbiology 2016;7:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Engstler AJ, Aumiller T, Degen C, et al. Insulin resistance alters hepatic ethanol metabolism: studies in mice and children with non-alcoholic fatty liver disease. Gut 2016;65:1564–1571. [DOI] [PubMed] [Google Scholar]
- 77.Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med 2011;17:448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Menni C, Fauman E, Erte I, et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes 2013;62:4270–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.S Lavekar A V Raje D, Manohar T, et al. Role of Probiotics in the Treatment of Nonalcoholic Fatty Liver Disease: A Meta-analysis. Euroasian journal of hepato-gastroenterology 2017;7:130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kootte RS, Levin E, Salojärvi J, et al. Improvement of Insulin Sensitivity after Lean Donor Feces in Metabolic Syndrome Is Driven by Baseline Intestinal Microbiota Composition. Cell metabolism 2017;26:611–619.e6. [DOI] [PubMed] [Google Scholar]
- 81.Clinicaltrials.gov, Identifier: NCT02721264.
- 82.Clinicaltrials.gov, Identifier: NCT02469272.