Abstract
The importance of gut microbiota in gastrointestinal (GI) physiology was well described, but our ability to study gut microbial ecosystems in their entirety was limited by culture-based methods prior to the sequencing revolution. The advent of high-throughput sequencing opened new avenues, allowing us to study gut microbial communities as an aggregate, independent of our ability to culture individual microbes. Early studies focused on association of changes in gut microbiota with different disease states which was necessary to identify a potential role for microbes and generate novel hypotheses. Over the past few years the field has moved beyond associations to better understand the mechanistic implications of the microbiome in the pathophysiology of complex diseases. This movement also has resulted in a shift in our focus towards therapeutic strategies which rely on better understanding the mediators of gut microbiota-host crosstalk. It is not surprising the gut microbiome has been implicated in pathogenesis of functional gastrointestinal disorders (FGIDs) given its role in modulating physiological processes such as immune development, GI motility and secretion, epithelial barrier integrity, and brain-gut communication. In this review, we focus on the current state of knowledge and future directions in microbiome research as it pertains to FGIDs. We summarize the factors which help shape the gut microbiome in humans. We discuss data from animal models and human studies to highlight existing paradigms regarding the mechanisms underlying microbiota-mediated alterations in physiological processes and their relevance in human interventions. While translation of microbiome science is still in its infancy, the outlook is optimistic and we are advancing in the right direction towards precise mechanism based microbiota therapies.
INTRODUCTION
The human gut is home to a complex microbial ecosystem with bacteria, fungi, viruses, and archaea which exist in a mutualistic relationship with the host in homeostatic conditions. The microbial members along with their genetic content are often referred to as the gut microbiome and can be viewed as a “dynamic organ” capable of mediating a wide variety of biochemical transformations that directly impact host physiology in health and disease1, 2. However, a disruption in this equilibrium can lead to alteration of host physiology resulting in disease states such as functional gastrointestinal disorders (FGIDs).
The role for gut bacteria in FGIDs such as irritable bowel syndrome (IBS) has been well described. An estimated 10% of IBS cases begin after an episode of infectious gastroenteritis3. However, the study of intestinal microbial ecosystems was limited by our inability to identify bacteria without cultivating them in the laboratory. At the turn of the century, ground-breaking advances in the genomics era and sequencing technologies4, 5 gave way to culture-independent molecular approaches allowing us to not only identify and characterize microbial communities based on similarities in DNA sequences, but also provide knowledge that has significantly improved our ability culture bacteria that were previously considered unculturable6.
These advances have led to extensive characterization of microbial communities in FGIDs over the past decade. While no consistent “microbial signature” has been associated with FGIDs, several lines of evidence support a role for gut microbes in the development of FGID symptoms7. There has been a significant effort to move beyond describing associations between the gut microbiome and FGIDs to defining mechanisms underlying microbial contributions to the pathophysiology of FGIDs.
In this review, as a part of our effort to define a path from bench to bedside, we will summarize factors affecting the gut microbiome and describe a conceptual framework for the role of the gut microbiome in FGIDs. This foundation will allow us to identify gaps in our current body of knowledge and develop strategies to translate microbiome science into improved diagnosis, prognosis, and management of FGIDs.
Factors that shape the gut microbiota
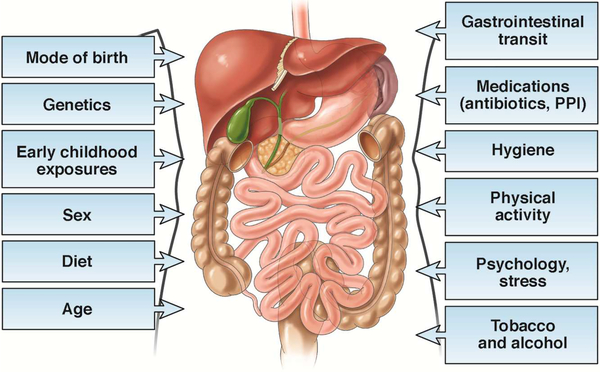
Gut microbial composition and diversity is largely the consequence of host selection pressures such as genetics, habits, sex, and location within the gastrointestinal (GI) tract as well as environmental factors including diet (Figure 1). Gut microbial diversity varies with age, and substantial differences are seen at the extremes of life8. At birth, assembly of the gut microbiota begins with colonization from environmental microbes (e.g., maternal vaginal, fecal, skin microbiota). In the subsequent months to years, gut microbial communities continue to shift in response to key life events (e.g., exposure to solid foods, illnesses, antibiotics) with gradual increases in diversity and convergence to an “adult-like” microbiota9–11. The adult gut microbiota is relatively stable over time and surprisingly resilient to temporary perturbations, changing as we get older to a distinct and less diverse microbiome12.
Figure 1:
Factors that shape the gut microbiota
Sex associations (Figure 1) with the gut microbiota have been characterized by increased relative abundance of Firmicutes and lower Bacteroidetes in women compared to men and may be further influenced by body mass index13, 14. Host genetic influence15,16,17,18, 19,20 on the gut microbiome is apparent from studies of monozygotic and dizygotic twin pairs that demonstrate shared community structures between related individuals21 and temporally stable heritable taxa17. However, the effect size is likely small given recent microbial-genetic association studies showing environmental factors to have substantially greater impact on the gut microbiome than genetics22. The impact of both short23 and long term dietary patterns24 on the gut microbiome cannot be overstated25, 26. The role of diet27 in microbial alterations is of significant interest in FGIDs as dietary intolerances are commonly reported in FGIDs and patients may alter or restrict their diets based on perceived associations between symptoms and food28, 29. The interaction of diet, gut microbiome, and symptoms in FGIDs (reviewed in 30) has not been well studied and the long term consequences of current dietary interventions with reported benefit in IBS, such as supplementation with psyllium fiber and the low FODMAP (fermentable oligosaccharides, disaccharides, monosaccharides, and polyols) diet31, 32, on the gut microbiome remain to be seen.
Other modifying factors (Figure 1) include psychological stress33,34, physical activity35, tobacco use36, alcohol consumption37, and antibiotic exposure38, 39,40. In one population-level analysis of gut microbiome variation, 69 factors were shown to correlate with microbiome community variation, with stool consistency emerging as the most influential covariate 41.
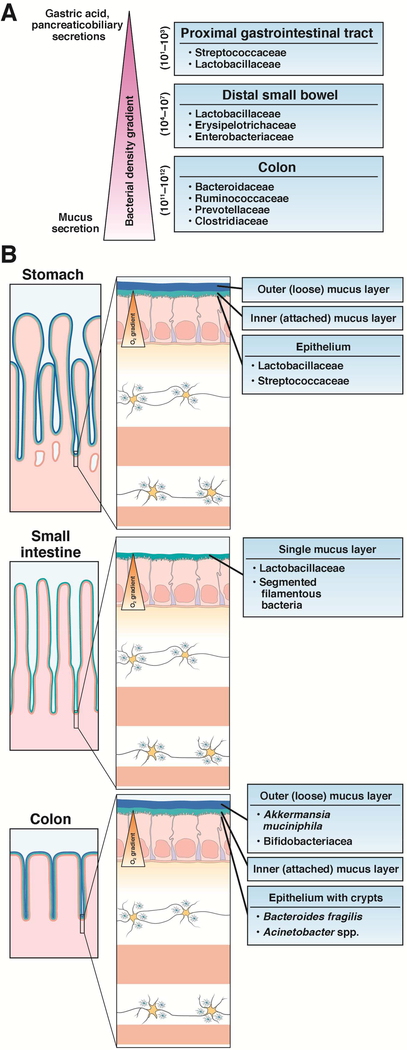
The distribution and composition of the gut microbiota changes along the length of the GI tract (Figure 2) and across the lumen, mucus layer, and the epithelium42, 43. Bacterial density increases from more proximal to distal sites44. Microbial abundance and community structure in the proximal intestine is affected by gastric acid, pancreaticobiliary secretions, and fast transit45. Spatial niche partitioning of microbial populations can also be a result of mucus from goblet cells 46 and differential oxygen tolerance47. Together, these factors lead to distinct microenvironments driving the biogeographical stratification of microbes across the GI tract.
Figure 2:
Distribution of the gut microbiota within the gastrointestinal tract along its longitudinal and radial axes
EFFECT OF THE GUT MICROBIOTA ON HOST PHYSIOLOGY
Gut microbiota and gastrointestinal motility
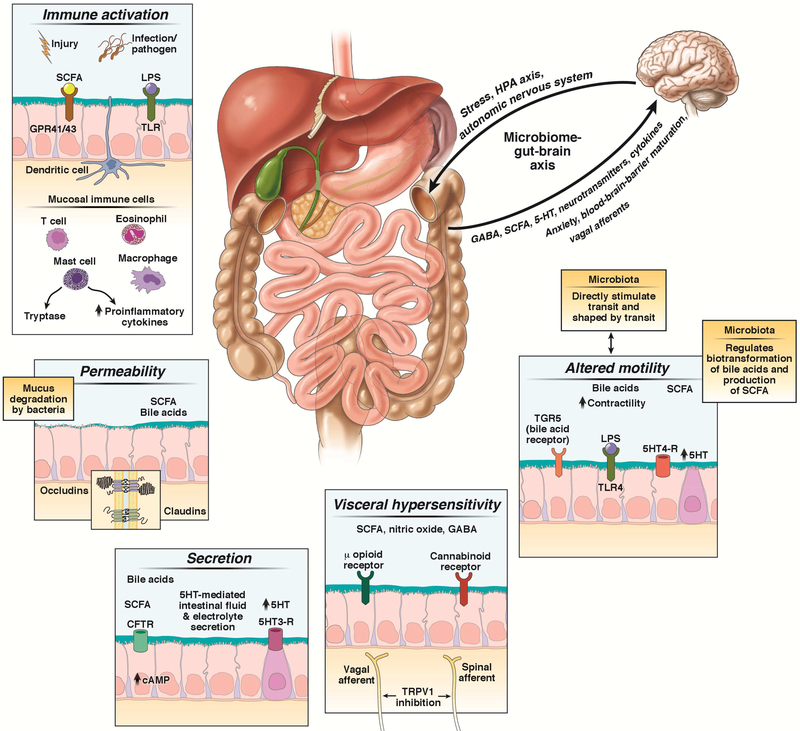
Altered GI motility and transit have long been recognized in the pathobiology of FGIDs such as IBS48 and functional dyspepsia (FD)49, 50. GI motility and the gut microbiota have reciprocal effects (reviewed in 1), highlighting a bidirectional relationship (Figure 3). Gut microbes can accelerate GI transit51, 52. In turn, accelerated GI transit can alter the composition and spatial organization of microbial communities by creating luminal conditions suited for the growth of specific bacterial taxa or by affecting bacterial adherence1. Interestingly, GI motility related changes in the gut microbiome can further perpetuate the alteration in GI motility as a positive feedback effect53. Several microbial mediators (Table 1) of GI motility have been identified (Figure 3), including short chain fatty acids (SCFAs) and bile acids. SCFAs are produced by fermentation of dietary starches or complex carbohydrates by gut bacteria54, while bile acid deconjugation and dehydroxylation by gut bacteria regulates the quantity and derivatives of bile acids in the colon. SCFAs and bile acids may affect gut motility (Table 1). Prokinetic effects of bile acids can be mediated by the G protein-coupled bile acid receptor TGR5 (or GPBAR1), expressed by enteric neurons and enteroendocrine cells based on findings from animal models55. Interestingly, microbial mediators of GI motility can vary by diet56, 57. For example, turmeric, a commonly used spice in Asian dishes, stimulates gallbladder contraction and increases intraluminal bile acids through its active ingredient, curcumin. Similarly, SCFA concentrations can vary based on dietary carbohydrate and protein intake. Other microbial products or metabolites (Table 1) that have been identified as potentially relevant in microbial regulation of GI motility include: bacterial lipopolysaccharide, which can improve survival of enteric neurons by activation of toll-like receptor 4 (TLR4)58. Preliminary studies support the roles of other microbial metabolites such as hydrogen sulfide59,60, tryptamine61, and hydrogen gas62 in regulation of human GI motility by their putative effects on GI smooth muscle and the enteric nervous system1.
Figure 3:
Gut microbiota effects on host physiology including gastrointestinal motility, sensation, secretion, epithelial barrier integrity, immune activation, and brain-gut communication
Table 1:
Microbial metabolites as mediators of gastrointestinal physiology
| Metabolite | Microbial regulation | Effect | Mechanism |
|---|---|---|---|
| *SCFA | Fermentation of dietary starches to SCFA | Motility | Butyrate and acetate increase #5-HT biosynthesis193 SCFAs provoke proximal colonic contractions via 5-HT in rats194 |
| Sensation | Butyrate increases colonic hypersensitivity in rats75, decreases visceral sensitivity in healthy humans76 | ||
| Permeability | Butyrate role in maintenance of intestinal barrier195 may vary depending on local concentrations, pH, and cellular differentiation | ||
| Immune activation | SCFAs and n-butyrate regulate neutrophil function, increased tight junction protein expression, reduce cytokine and chemokine release196 | ||
| Secretion | SCFAs promote fluid and electrolyte absorption within the gut99 Acetate effects both small intestine and colon through increased duodenal bicarbonate secretion100 and effects on colonic epithelial 5-HT3 receptor expression101 |
||
| Bile acids | Deconjugation and dehydroxylation | Motility |
$CDCA promotes propagating and non-propagating colonic contractions in humans197 ^DCA-induced intestinal peristalsis and contractions mediated by TGR555 |
| Permeability | Increased permeability associated with bile acids with two hydroxyl groups in α configuration198 Bile acid receptor, TGR5, modulates intestinal barrier function in mice199 |
||
| Secretion | DCA and CDCA stimulate chloride and water secretion via inhibition of Cl/OH- exchange (Alrefai 2007) and activation of CFTR via cAMP98 | ||
| Immune activation | CDCA regulates intestinal antimicrobial environment in mice via Paneth cell α-defensins and C-type lectins200 | ||
| Methane | Gaseous by-product | Motility | Methane augments small bowel contractility and slows intestinal transit152 |
| Hydrogen sulfide | Gaseous by-product | Motility | Sulfate-reducing bacteria slow intestinal transit in mice60 |
| Hydrogen gas | Gaseous by-product | Motility | Hydrogen gas shortens transit in guinea pig colon62 |
SCFA=short chain fatty acid
5-HT=serotonin; Irritable bowel syndrome
CDCA=chenodeoxycholic acid
DCA=deoxycholic acid
Gut microbiota and gastrointestinal sensation
Abdominal pain in IBS48 and other FGIDs63 such as FD64 and functional abdominal bloating/distention65 has been attributed to visceral hypersensitivity to mechanical and chemical stimuli. Evidence for a role of the gut microbiome in regulating GI sensation (Figure 3) comes from gnotobiotic studies showing transfer of the visceral hypersensitivity phenotype following transplantation of gut microbiota from patients with IBS into germ-free (GF) mice 66. A recent study by Riba et al.67 demonstrated a correlation between visceral hypersensitivity and increase of Escherichia coli abundance followed by induction of hypersensitivity in response to E. coli gavage in mice. Disruption of the gut microbiota in early-life also has been associated with longterm changes in visceral sensitivity, emphasizing the importance of the gut microbiome in neurodevelopment of pain pathways68. The exact mechanisms by which bacteria affect visceral perception and sensation still need to be determined. A few putative mechanisms include: microbial induction of epithelial μ-opioid and cannabinoid receptors as shown with oral administration of Lactobacillus strains in rodents69; regulation of central70 and peripheral neuronal pathways71; anti-nociceptive effects from inhibition of transient receptor potential vanilloid as shown with administration of Lactobacillus reuteri72 in rats; microbial metabolites (e.g., organic acids) or byproducts (e.g. nitric oxide)73 altering sensation; and microbially-derived bioactive molecules such as γ-aminobutyric acid (GABA) as shown with administration of GABA-producing Bifidobacterium dentium74. The translation of findings from animal models to humans can, however, be challenging. For instance, rectal administration of butyrate increases colonic hypersensitivity in rats75
Gut microbiota and intestinal permeability
The intestinal epithelium and the overlaying mucus layer serve a key role in protecting the host by providing a physical and immunological barrier against potentially harmful pathogens while also regulating fluid and nutrient absorption77. Increased permeability or disruption of the epithelial barrier has been implicated in the pathophysiology of FGIDs78, 79. Microbes(Figure 3) can directly alter expression of tight junction proteins such as claudin-380 and zonula occludens-181 or enhance expression of genes involved in tight junction signaling82. Microbial metabolites such as bile acids83,84 and SCFAs can also regulate intestinal permeability (Table 1). The mucus layer overlaying the epithelium is a reservoir of antimicrobial peptides and immunoglobulins and provides the first line of defense against gut bacteria85. The mucus layer is compositionally rich in polysaccharides which can serve as a nutrient source for subsets of bacteria. Hence microbial starvation such as with decreased consumption of fiber can increase microbial reliance on the mucus polysaccharides resulting in degradation of the mucus layer and increasing susceptibility to opportunistic pathogens and inflammation27, 86.
Gut microbiota, immune activation, and inflammation
Inflammation or immune activation involving both the innate and adaptive immune systems has been described in subsets of patients with FGIDs87,88. There are several lines of evidence in support of the activation of mucosal and systemic immune responses by gut microbiota (Figure 3). Post-infectious FD is associated with increased numbers of duodenal CD68+ cells and eosinophils when compared with other subtypes of FD (epigastric pain syndrome, post-prandial distress syndrome), and healthy states89. Increased expression of proinflammatory cytokines may be elicited through interactions between bacterial components and pattern recognition receptors including TLRs such as TLR2 and TLR4 that have been associated with IBS90. The gut microbiota can also influence immune activation via effects on lineage differentiation of T-cell subsets91, host-receptor mediated signaling as seen with L. reuteri activation of histamine H2 receptor signaling92, and production of microbial metabolites (Table 1).
Gut microbiota and intestinal secretion
Changes in small intestinal93 and colonic secretion represents another pathophysiologic disturbance in FGIDs that may be influenced by the gut microbiome48 (Figure 3). Secretory mechanisms are common therapeutic targets94, 95 of medications used to treat FGIDs. Microbial mediators of altered intestinal secretion96 include metabolites from breakdown of dietary polysaccharides as well as bile acids (Table 1). Specific bile acids, such as deoxycholate and chenodeoxycholate, can stimulate intestinal chloride secretion97, 98 which is accompanied by water. SCFAs, like bile acids, are important intraluminal determinants of mucus and water secretion through effects on sodium and water influx99, duodenal bicarbonate secretion100, and colonic epithelial 5-HT3 receptor expression101.
Gut microbiota and gastric function
Disturbances in gastric motor and sensory function, including impaired gastric accommodation and increased intragastric pressure, may underlie FGIDs and are often related to food intake102. There is a paucity of data in support of microbial regulation of gastric function. The administration of the prebiotic arabinoxylooligosaccharide in healthy volunteers was not associated with changes in gastric sensitivity, compliance, or accommodation despite increased colonic fermentation102. Reported associations between the presence of small intestinal bacterial overgrowth (SIBO) and delayed gastric emptying103 have not discerned whether associations are a result of microbial mechanisms or merely representative of underlying impairment in small intestinal motility and other confounding factors including chronic acid suppression and opioid analgesics104. Recent evidence showing a similar gastric emptying time among patients with and without SIBO suggests that bacterial overgrowth does not necessarily predispose to impaired gastric emptying105, 106.
Gut microbiota and central nervous system function
The bidirectional microbiome-gut-brain axis (Figure 3) represents the reciprocal regulation of the gut microbiome and the central nervous system (CNS). Recent studies highlight the role of the gut microbiome in modulating brain-gut communication, which may significantly affect the pathophysiology of symptoms associated with FGIDs107,108. Signals from the CNS can influence GI physiology while simultaneously shaping the gut microbial fingerprint as seen in early life stress rodent models which exhibit alterations in gut microbial community composition109. Similar findings have been described in other rodent stress models33. Conversely, microbial colonization and community composition are critical to development of the hypothalamus–pituitary–adrenal axis as evidenced by the exaggerated adrenocorticotropic hormone and corticosterone release in germ-free (GF) mice. This exaggeration is attenuated following colonization with Bifidobacterium infantis110. A different strain within the same genus, Bifidobacterium longum NCC3001, decreases anxiety-like behavior in mice through vagally mediated pathways111. Microbial metabolites such as SCFAs and microbially-derived neurotransmitters such as GABA and 5-HT may further impact brain function and mental health108.
FUNCTIONAL GI DISORDERS
Role of gut microbiota in pathophysiology of irritable bowel syndrome
The gut microbiota of patients with IBS is an area of considerable interest, and has been the most extensively studied among the various FGIDs (reviewed in 44). Despite the lack of a uniform “IBS-microbiota” pattern, key observations include a decrease in α-diversity and alterations in relative abundance of specific taxonomic groups including an increased ratio of Firmicutes to Bacteroidetes, decreased Lactobacillus and Bifidobacterium, and increased Streptococcus and Ruminococcus spp.112. Cross-sectional analysis of extensively phenotyped cohorts also has revealed that while stool consistency is a significant contributor to gut microbiome compositional variation, the contribution imparted by IBS was much less41. These data highlight the importance of investigating specific pathophysiologic disturbances, beyond merely providing descriptive analyses of a heterogeneous patient populations in elucidating the role of the gut microbiome in IBS. Correlative associations between the gut microbiome and IBS have been followed by efforts to better characterize the mechanistic link between the microbiome and pathophysiology of symptoms associated with IBS (Table 2). Among the various aforementioned aspects of gut physiology that are affected by the gut microbiome, many are directly implicated in the pathophysiology of IBS.
Table 2:
Summary of studies investigating pathophysiologic mechanisms and gut microbiota in patients with functional gastrointestinal disorders
| Study | Study population | Intervention | Sample | Mechanism studied | Role of microbiota |
|---|---|---|---|---|---|
| Shin et al. 2018201 | 60 IBS*-D | L. gasseri BNR17 vs. pcbo& | Fecal | Transit | Transit significantly ↑ during 8 weeks with L. gasseri BNR17 |
| Tap et al. 20177 | 110 IBS, 39 HV+ | NA | Fecal, mucosal | Transit, GBA** | ↑Transit with Clostridiales vs. Prevotella and Bacteroides enterotypes. No association between HADS# and enterotype |
| Acosta et al. 2016119 |
24 nonconstipated IBS | Rifaximin vs. pcbo |
Fecal | Transit, permeability, SCFAΔ and bile acid production | No significant effects of rifaximin on permeability, bile acids, SCFAs. Rifaximin associated with ↑ascending colon emptying, and colonic transit at 48H |
| Dior et al. 2016202 | 15 HV, 15 IBS-C, 16 IBS-D | NA | Fecal | Fecal bile acids | ↓bacterial deconjugation of bile acids in IBS-D and IBS-C feces vs. HV |
| Le Neve et al. 2016203 | 100 IBS | NA | Fecal | Sensation, transit | Response to lactulose challenge associated with rectal sensitivity but not with fecal microbiota or transit |
| Chumpitazi et al. 2014131 | 12 IBS children | LFSFD@ | Fecal | Transit, metabolite composition | LFSD response associated with ↑abundance of Sporobacter and Subdoligranulum and ↓Bacteroides, but not with transit. Stool metabolites (L-urobilin, cholate) associated with response and microbiome composition |
| Jeffery et al. 2012156 | 37 IBS, 20 HV | NA | Fecal | Sensation, transit, GBA | Proteobacteria associated with ↑mental component and pain threshold; Actinomycetales inversely associated with depression. Desulfohalobiaceae and Methanobacteriaceae associated with transit |
| Labus et al. 2017204 | 29 IBS, 23 HV | NA | Fecal | GBA | No correlations between anxiety or depression symptom scores and microbial parameters; Clostridia and Bacteroidia correlated with sensory integration regions |
| Liu et al. 2016205 | 40 IBS, 15 Depression, 25 IBS and Depression, 20 HV |
NA | Fecal | GBA, immune | ↑Bacteroidetes and ↓Firmicutes in IBS-D, depression, and IBS-D with depression; Colonic mucosa inflammation associated with ↑Bacteroides or Prevotella |
| Azpiroz et al. 2017206 |
79 IBS | scFOS^ vs. pcbo | Fecal | GBA, sensation | scFOS reduced anxiety scores and increased fecal Bifidobacteria; No significant difference in rectal sensory threshold foe scFOS vs. pcbo |
| Le Gall et al. 2011207 | 10 IBS, 13 UC, 22 HV |
NA | Fecal | Fecal metabolites | Correlation between gut microbiota profile and metabolite composition |
| Heitkemper et al. 2018208 |
93 IBS | NA | Fecal | Permeability | Higher stool δTFF3 associated with lower permeability and microbial diversity. Christensenellaceae inversely related to stool TFF3. |
| Bednarska et al. 2017209 |
32 IBS, 15 HV | NA | Mucosal | Immune, Permeability |
Increased permeability to E. coli strain HS and S. typhimurium in IBS biopsies vs. controls; ↑plasma VIP in IBS vs. HV; ↑tryptase and mast cells in IBS biopsies vs. HV |
| Valentin et al. 2017210 |
15 IBS-D | SBI$ | Duodenal brushing, fecal | Immune, permeability, metabolism |
Bile acid synthesis, tryptophan metabolism, permeability and stool microbiome not significantly different with SBI. Changes in β diversity analysis, increased ↑Proteobacteria Burkholderiales, Firmicutes Catonella, and unclassified genus organisms with SBI in duodenal microbiome. |
| Ko et al. 2013211 | 53 IBS-D | Herbal (GJS), Probiotic (Duolac7S), pcbo |
Fecal | Permeability | GJS with DuoLac7 ↑B. lactis, L. rhamnosus, L. plantarum. No significant difference observed in permeability |
| Crouzet et al. 201366 |
3 IBS-C, 2 HV | NA | Fecal | Rectal sensitivity | IBS with rectal hypersensitivity have ↓bifidobacteria, ↑Enterobacteriaceae, and ↑H2-utilizing sulfide-producing bacteria vs. HV |
| Shulman et al. 2017136 | 103 IBS children | Fiber vs. placebo | Fecal | GBA, permeability |
No differences in psychological symptoms, permeability, or microbiome between groups |
| Compare et al. 2017212 |
10 IBS-D, 10 HV (ex vivo) |
LC-DG!, postbiotic | Mucosal | Immune | ↑IL-1α, IL-6 and IL-8 mRNA, TLR-4 protein expression with ↓IL-10 mRNA levels in PI-IBS D vs. HV. LC-DG and PB ↓mRNA levels of proinflammatory cytokines and TLR-4 but ↑IL-10 after LPS⌘ stimulation |
| Hustoft et al. 2017213 | 20 IBS-D or IBS- M |
low FODMAP diet, FOS vs. pcbo |
Fecal | Immune, SCFA | ↓IL-6 and IL-8, fecal bacteria (Actinobacteria, Bifidobacterium, Faecalibacterium prausnitzii), total SCFAs, and n-butyric acid on LFD. FOS supplement then ↑levels of these bacteria, but cytokines and SCFAs unchanged. |
| McIntosh et al. 2017214 | 37 IBS | low vs. high FODMAP |
Fecal | Urinary metabolites | Significant correlations between relative bacterial abundance and symptoms and urinary metabolites (histamine, p-hydroxybenzoic acid) |
| Sundin et al. 2015215 | 11 PI-IBS, 10 HV (ex vivo) |
NA | Mucosal | Immune | IL-1β ↑ in PI-IBS vs. HV after stimulation with Subdoligranulum variabile; IL-10 ↓ in HV vs. PIIBS after stimulation with Eubacterium limosum. |
| Sundin et al. 2015216 | 13 PI-IBS, 19 IBS, 16 HV |
NA | Fecal, mucosal | Immune, GBA | Naive CD8+ CD45RA+ intraepithelial lymphocytes and lamina propria lymphocytes negatively correlated with mucosal microbial diversity. Fecal microbial diversity negatively correlated with HADS |
| Pinto-Sanchez et al. 2017127 | 44 IBS | BL∞ vs. pcbo | Fecal | GBA, immune, urinary metabolites, neurotransmitte rs, and neurotrophins. | BL ↓depression and associated with ↓limbic reactivity. No difference in fecal microbiota, serum markers of inflammation, neurotrophins and neurotransmitters. Reduced urine methylamines and aromatic amino acids metabolites with BL. |
| Parthasarathy et al. 2017217 | 25 CC, 25 HV | NA | Fecal | Transit | Reproducibility of fecal microbiota lower in normal transit vs. slow transit constipation |
| Parthasarathey et al. 2016157 | 25 CC, 25 HV | NA | Fecal, mucosal | Transit | Fecal microbiota profile associated with colonic transit; genera from Firmicutes correlated with faster colonic transit. |
| Tian et al. 2017218 |
60 STC | Fecal microbiota transplantation (FMT) |
NA | Transit | FMT associated with faster transit vs. control treatment |
IBS=Irritable bowel syndrome (IBS-D=diarrhea-predominant IBS, IBS-C=constipation-predominant IBS, PI-IBS=post-infectious IBS)
pcbo=Placebo
HV=Healthy volunteer
GBA=Gut brain axis
HADS=Hospital Anxiety and Depression Scale
SCFA=short chain fatty acid
LFSD= Low fermentable substrate diet
scFOS= Short-chain fructooligosaccharide
TFF3= urine trefoil factor 3
SBI=Serum-derived bovine immunoglobulin/protein isolate
LC-DG=Lactobacillus casei DG
LPS=lipopolysaccharide
BL=Bifidobacterium longum NCC3001
Summary of findings from animal studies
Animal models, although imperfect correlates to IBS pathophysiology in humans, have allowed us to explore putative interactions between the gut microbiome and mechanisms implicated in IBS such as altered motility, visceral hypersensitivity, increased permeability, immune activation, intestinal secretion, and disturbances in central mechanisms. De Palma et al.52 recently demonstrated that GF mice colonized with the fecal microbiota of diarrheapredominant IBS (IBS-D) patients exhibited faster GI transit, increased colonic permeability, increased anxiety-like behavior, and increased infiltration by CD3+ T lymphocytes compared to those colonized by microbiota from healthy controls. Study of specific IBS pathways include reports describing microbiota-induced hypersensitivity to colonic distension in GF rats inoculated with the fecal microbiota from IBS patients66. Microbial regulation of host immune responses may be further relevant to IBS. An increase in mucosal immune cells including mast cells, macrophages or monocytes, T-cells, and eosinophils has been reported in both pediatric and adult FGID populations79, 113, 114. Mast cells contain biologically active substances including histamine, tryptase, cytokines, and membrane-derived arachidonic acid metabolites (e.g., prostaglandins) that are released upon their activation. These mediators may alter nociceptive pathways in IBS115 or increase intestinal permeability116. Macrophages and monocytes are important in modulating the adaptive immune responses and producing proinflammatory cytokines such as IL-6 and IL-8 which in some studies are increased in IBS patients117. The role of gut microbes in these immune pathways remains unknown.
Summary of findings from studies in adult IBS patients
Numerous studies have examined microbiome-related effects on pathophysiological changes in IBS, building upon work performed in animal models. Interventional studies investigating the use of probiotic and antibiotic therapy in IBS have led to identification of potential microbial effects on transit (Table 2). Treatment with a probiotic containing Bifidobacterium lactis118 accelerates whole gut transit and improves symptoms in patients with constipation-predominant IBS (IBS-C), while treatment with the non-absorbable antibiotic rifaximin is associated with increases in both ascending colonic emptying and overall colonic transit rate at 48 hours119 in non-constipated IBS patients. A role for the gut microbiome in immune modulation was suggested by findings from the clinical trial wherein B. infantis 35624 alleviated symptoms and was associated with normalization of abnormal IL-10/IL-12 ratios in IBS patients120. Colonic mucosal gene expression profiling of IBS patients also has found differential expression of genes associated with host immune responses against microbial invasion, further suggesting that immune activation may be shaped by microbial interactions121. Alterations in mucus-associated bacteria that may influence mucus integrity and intestinal secretion (e.g. Akkermansia muciniphila, Ruminococcus gnavus and Ruminococcus torques) also have been associated with IBS122. Microbially-mediated effects on intestinal secretion in IBS may be a consequence of differential bile acid biotransformation by the gut microbiome96. This concept is supported by the decreased concentrations of fecal unconjugated bile acids known to stimulate colonic secretion (deoxycholate and chenodeoxycholate) in IBS-C123. Regarding the role of the microbiome-gut-brain axis in IBS (reviewed in 124 and 125), probiotic therapy has been shown to modulate CNS function in healthy volunteers126 through effects on brain regions controlling processing of emotion and sensation. More recently, in a recent placebo controlled trial in IBS patients, treatment with the probiotic B. longum NCC3001 was associated with improved symptoms of depression and changes in brain activation patterns measured by functional magnetic resonance imaging127.
Summary of findings from studies in pediatric IBS patients
Similar to adult studies, the composition of the gut microbiome differs between children with IBS and age-matched healthy controls, despite lack of a uniform “IBS-microbiota” signature across studies. One study enrolling children ages 7–12 years found pediatric IBS to be associated with decreased relative abundance of Bacteroides spp. and increased relative abundance of the class Gammaproteobacteria, including Haemophilus parainfluenzae, along with increased abundance of novel taxa related to the genus Ruminococcus. In this cohort, microbiota composition correlated with abdominal pain severity and frequency, and could be used to distinguish IBS-C from unsubtyped IBS128. Another study of children ages 11–18 years found IBS-D to be associated with increased abundance of the genera Veillonella, Prevotella, Lactobacillus, and Parasporobacterium, and with decreased abundance of Bifidobacterium and Verrucomicrobium129. By adding fecal metabolomic profiling to microbiome signatures, stool from children with IBS-D could be more accurately discriminated from that of healthy controls, with formate, pyruvate, and glucose being the most predictive metabolites130. Fecal microbial community composition also might be used to predict which children with IBS are more likely to respond to a low-FODMAP diet: in two separate studies, responders had distinct baseline microbiome signatures compared to non-responders131, 132.
Among the most studied probiotics in pediatric IBS is Lactobacillus rhamnosus GG, which was found in a meta-analysis of three randomized, placebo-controlled trials (RCTs) to confer a modest but significantly increased rate of treatment response versus placebo133. Two multicenter, randomized, double-blind, placebo-controlled crossover studies provide further evidence of microbiota involvement in pediatric IBS. One study found that VSL#3 improved GI symptoms134, while the other reported that a combination of three bifidobacteria resolved abdominal pain and improved quality of life to a greater extent than placebo135. On the other hand, psyllium fiber reduced pain episodes in an RCT enrolling children with IBS without altering the composition of the gut microbiota based on 16S ribosomal RNA analysis136.
RCTs in children with functional abdominal pain (FAP) have revealed that L. reuteri DSM 17938 is effective in treating abdominal symptoms. Jadrešin et al.137 demonstrated a reduction in days with pain and pain severity in children with IBS and FAP. In studies focused specifically on FAP, Romano et al.138 reported reduced pain severity and Weizman et al.139 and Maragkoudaki et al.140 both reported reduced pain severity and frequency in those treated with the probiotic compared with placebo.
In summary, both animal and human studies underscore the importance of the gut microbiome in mediating peripheral and central mechanisms implicated in IBS. Moreover, factors affecting gut microbiota composition are akin to the etiological factors in IBS and probiotic interventions have a generally beneficial effect. However, given the heterogenous nature of the disease with multiple putative mechanisms, our broad nontargeted approach without consideration for the underlying physiological disturbance likely dilutes the overall impact and makes it difficult to ascertain the precise benefit of microbiota modulation. As we move forward, it will be important to phenotype patients based on the underlying physiological alterations so that we can develop targeted approaches directed towards specific microbes driving the host phenotype.
Role of gut microbiota in pathophysiology of functional dyspepsia
Similar to IBS, multiple pathogenic mechanisms including altered gastric function, visceral hypersensitivity, low grade inflammation or immune activation, increased duodenal permeability, and abnormal CNS function have been postulated to contribute to symptoms in FD141. As summarized above, gut microbiota have been shown to modulate the majority of these physiological functions. Although data on the gastroduodenal microbiome and its particular role in FD are sparse, there are a few studies that lay the groundwork for future work investigating the role of microbial community alterations in FD.
Summary of findings from animal studies
In general, animal studies investigating microbial effects on putative pathophysiologic mechanisms in FD are lacking given the absence of reliable models. The described effects are attributed to fermentative end products such as SCFAs. Bacterially-derived or ingested SCFAs can alter duodenal bicarbonate secretion100. In addition, the absorption of SCFAs can also influence the luminal bacterial population which may be relevant in FD100.
Summary of findings from studies in adult patients with functional dyspepsia
There are few human studies describing the gut microbiome in patients with FD, hence the precise role of the microbiota remains unknown. SIBO has been proposed to trigger symptoms in FD142, although studies examining the role of SIBO in FD are limited by the relative inaccessibility of the more distal regions of the small intestine and concerns regarding accuracy and interpretation of available testing methods for the diagnosis of SIBO143. Recently, Zhong et al.144 found the relative abundance of the anaerobic genera Prevotella, Veillonella and Actinomyces were significantly decreased in the duodenal mucosa of nine patients with FD compared to controls. Interestingly, severity of symptom responses to a standardized meal was positively correlated with mucosal bacterial load, which in turn was inversely correlated with bacterial diversity. Igarashi et al145 found that gastric fluid samples from patients with FD were characterized by an increased Bacteroidetes to Proteobacteria ratio and absence of Acidobacteria. In contrast, healthy volunteers had a decreased Bacteroidetes to Proteobacteria ratio and presence of Acidobacteria. Non-blinded probiotic therapy with Lactobacillus gasseri OLL2716 was subsequently associated with shifts in gastric fluid microbial community composition similar to that found in healthy controls. In another RCT among patients with FD, rifaximin treatment was associated with significant improvement in global dyspeptic symptoms, belching, and postprandial fullness/bloating, further suggesting a potential role for the microbiome in FD146.
Summary of findings from studies in pediatric patients with functional dyspepsia
Relatively little is known regarding the gut microbiome in pediatric FD. Although the previously highlighted multicenter, randomized, double blind, placebo controlled crossover study reported that a combination of three probiotic bifidobacteria improved pain scores and quality of life among 48 children with IBS, no benefit was observed among the 25 enrolled children who had FD, perhaps owing to the small number of patients treated135. Likewise, the moderate overall benefit associated with L. rhamnosus GG treatment in an RCT of children with IBS or FD was not observed in the subset of children with FD147. However, it would be premature to make definitive conclusions given the small sample size (n=20 with FD versus n=37 with IBS).
In summary, the gut microbiome can affect mechanisms underlying FD similar to IBS, but the microbial community composition of the stomach and small bowel remain elusive and much work is needed before we can target specific microbial mediators that drive symptoms in FD. The overall positive impact of probiotics is encouraging and highlights the need for better mechanistic understanding in order to develop more precise microbiota-based therapeutics.
Role of gut microbiota in pathophysiology of functional abdominal bloating
Abdominal bloating and distension are common complaints among patients suffering from FGIDs, and are among the most challenging symptoms to treat. The pathophysiologic mechanisms contributing to bloating are poorly understood, although SIBO and alterations in gut microbial communities have been hypothesized148 to play a role through microbial fermentation of dietary nutrients. As this is predominantly a subjective sensation, there are no animal models to mimic these symptoms.
Summary of findings from adult patients with functional abdominal bloating
The majority of clinical studies investigating symptoms of bloating have been performed in IBS patients, with bloating and distension evaluated as secondary endpoints148. A recent study showed depletion of operational taxonomic units within Subdoligranulum and Anaerovorax, belonging to the families Ruminococcaceae and Eubacteriaceae, respectively, in IBS patients without bloating compared to those with bloating and to healthy controls149. Placebo-controlled studies of antibiotic (rifaximin) treatment in FGIDs and IBS have demonstrated significant reduction in bloating scores with rifaximin compared to placebo150–152. Efficacy of probiotic administration for symptoms of bloating have been less consistently reported153 although some studies in IBS patients have suggested benefit with specific probiotic strains including B. lactis DN-173118, Bifidobacterium animalis DN-173 010154, and VSL#3155.
Summary of findings from pediatric studies
Little is known regarding the microbiome in functional abdominal bloating in children. The trial noted previously by Weizman and colleagues139, which reported benefit for abdominal pain with the probiotic L. reuteri DSM 17938, also reported a lower incidence of perceived abdominal distention and bloating. Similarly, patients in the VSL#3 trial had decreased abdominal bloating/gassiness compared to placebo134.
In summary, while gut microbes can potentially impact these symptoms both via fermentative end products and by their effect on visceral sensation, we need to better characterize the potential microbial mediators in order to develop relevant therapeutics.
Role of gut microbiota in pathophysiology of functional constipation
There is evidence supporting an association between the altered mucosal and fecal microbiota and chronic constipation156, 157. Most of our knowledge regarding the effects of the gut microbiota on peripheral mechanisms associated with constipation, such as GI motility, comes from animal studies. However, in recent years, several studies have been published exploring the gut microbiome in patients with constipation (Table 2).
Summary of findings from animal studies
Investigation of the causal relationship between alterations in gut microbial communities and constipation has been described in a recent study158 reporting upregulation of 5-HT transporter and decreased 5-HT content in the colonic tissue of germ-free mice that received fecal microbiota from constipated patients. 5-HT was negatively correlated with transit time and changes were accompanied by decreased relative abundance of the phylum Firmicutes and increased Bacteroidetes in mice receiving fecal microbiota from constipated patients. Genus level analyses further showed decreased relative abundance of Clostridium, Lactobacillus, Desulfovibrio and Methylobacterium and increased relative abundance of Bacteroides and Akkermansia. The findings suggest a potential role for gut microbiota in the pathogenesis of chronic constipation via increased expression of 5-HT transporter158. Interestingly, gut microbiota changes resulting from constipation can further impact GI motility, suggesting a more complex interaction with feedforward regulation rather than a simple cause-effect relationship53.
The potential role of microbially-derived metabolites is further supported by findings of delayed GI transit and altered SCFA and bile acid profiles following transfer of fecal microbiota from patients with slow transit constipation to antibiotic-treated mice159.
Summary of findings from adult patients with functional constipation
Several studies have reported a positive relationship between prolonged colon transit times, with increased richness and diversity of the fecal microbiome in adults without prior history of GI disorders160,161. However, the association between constipation and the gut microbiome may involve mechanisms beyond that of slow transit. In a study of adults with chronic constipation, overall composition of the colonic mucosa-associated microbiota could discriminate patients with constipation from control subjects independent of transit time157. Taxonomic profiling of the fecal microbiome from patients with functional constipation (FC) and healthy volunteers has shown decreased abundance of Bacteroides, Roseburia, and Coprococcus in FC patients. Furthermore, healthy volunteers were found to have a gut microbiome enriched in genes involved in carbohydrate, fatty acid, and lipid metabolism while FC patients harbored a high abundance of genes involved in methanogenic pathways, hydrogen production, and glycerol162. Analysis of functional gene targets in constipated and healthy females also has demonstrated increased abundance of hydrogenogenic (hydrogen producing) and hydrogenotrophic (hydrogen utilizing) genes by qPCR in colonic mucosa of constipated individuals163.
Summary of findings from pediatric studies
In a cross-sectional study of 8 constipated obese children and 14 non-constipated obese children, FC was associated with decreased abundance of the phylum Bacteroidetes, including a significant reduction of the genus Prevotella, and increased abundance of multiple genera within the phylum Firmicutes, including Blautia, Coprococcus, and Ruminococcus164. A recent systematic review included seven RCTs enrolling a total of 515 children that investigated the effects of probiotics in pediatric FC. Although two of the included studies, those evaluating L. reuteri DSM 17938165 and B. longum166, reported significantly increased defecation frequency in the treatment arm, the meta-analysis concluded that there is currently insufficient evidence to support the use of probiotics for pediatric FC167. Finally, although a low-fiber diet is a known risk factor for FC in children168, there is currently little evidence to support the use of fiber for pediatric FC. Multiple systematic reviews note the sparse data and high risk of bias among the current evidence base169–172.
In summary, the reciprocal interactions between GI transit and gut microbiota suggest that even if changes in gut microbiota are initiated by a change in transit, the altered microbial community can perpetuate the alteration in GI transit, highlighting the adaptability of the gut microbial community. Consequently, we need to think beyond the simple cause-effect paradigm as irrespective of the inciting event that alters the microbial community, these changes can still perpetuate a disease phenotype. The effect of gut microbiota on the host serotonergic system provides a plausible target for altering GI transit.
Role of gut microbiota in pathophysiology of infant colic
Infant colic, a characteristic group of behaviors featuring prolonged crying, is present in up to 25% of infants at 6 weeks of life173 and is associated with increased risk of recurrent abdominal pain and allergic disorders later in childhood174. Underlying mechanisms are unclear, due in part to a lack of small animal models. Multiple pathophysiologies, including gut microbiome alterations, have been proposed to promote abdominal pain. Early culture-dependent studies by Savino et al. revealed that colicky infants were more frequently colonized by anaerobic gram-negative proinflammatory bacteria and less frequently colonized by lactobacilli when compared to non-colicky infants175, 176. Subsequent molecular studies confirmed enrichment of proinflammatory and gas producing taxa within Proteobacteria in stool from colicky infants177–179.
Given these observations, the probiotic L. reuteri, one of the few endogenous lactobacilli in the human GI tract, was proposed as a means of normalizing these gut microbial community alterations and potentially reducing crying times in infant colic and has become the most extensively studied microbiome-targeting therapy for colic. L. reuteri has been tested in six prospective, RCTs: two meta-analyses that included more than 400 infants found that L. reuteri significantly reduced crying time in formula fed infants by a mean of nearly one hour per day180, 181. Of note, the only other therapy to demonstrate efficacy in infant colic was fennel oil while often-recommended interventions including simethicone and maternal diet manipulation produced mixed results181. Interestingly, L. reuteri also has shown benefit in prevention trials, reducing the risk of developing colic at three months of life182, 183. Finally, a number of small studies have tested other microbiome-targeting therapies, including L. rhamnosus GG184, 185 and a synbiotic combination of fructooligosaccharide and seven probiotics186; these small studies generated mixed results.
Modulating the gut microbiota for treatment of functional gastrointestinal disorders
Targeting the gut microbiota for therapeutic intervention in FGIDs remains an area of significant interest for patients and clinicians. Probiotics have been studied extensively in adult and pediatric FGID populations as previously discussed and summarized in Table 2. A prior systematic review of probiotics in IBS suggested evidence for efficacy on global IBS symptoms, abdominal pain, bloating, and flatulence187; however, there remain many unanswered questions regarding strain-specific effects, mechanisms of action, mode of administration and dosing, and patient selection. Despite their relative accessibility and general safety, clinical recommendations regarding specific probiotic use in FGIDs are limited by a lack of rigorous clinical trial data. Rifaximin has been studied in functional dyspepsia146, abdominal bloating, and flatulence150, and is approved for treatment of adults with IBS-D151. The exact mechanisms by which rifaximin exerts its effects in IBS, however, remain uncertain, with a recent study of patients with nonconstipated IBS showing borderline effects on microbial richness and increased rates of proximal colonic emptying but no clear effects on bowel function, permeability, or production of intraluminal metabolites119. More recently, results of several trials investigating the efficacy of fecal microbiota transplantation (FMT) for IBS have been reported. In one RCT among patients with moderate-to-severe IBS, higher response (p=0.049) rates at three months, defined as a 75point improvement in the IBS severity scoring system, were observed in patients receiving FMT (65%) compared to those receiving placebo (43%). However, differences were no longer significant at 12 months followup188. On the other hand, a separate multicenter RCT189 comparing FMT capsules to placebo in patients with diarrhea-predominant IBS was unable to demonstrate significant symptom relief at three months with FMT, although subgroup analysis suggested patients with post-infectious IBS experienced greater improvement with FMT compared to placebo (p=0.09). The role of FMT in IBS needs to be better defined as there may be specific features in the donor microbiome as well as additional recipient characteristics that predict clinical outcomes. FMT, however, represents a stop-gap measure and it is imperative that we determine which specific microbes, microbial consortia, or microbial products yield benefit in FGIDs to provide precision care without unwanted effects.
TRANSLATING MICROBIOME RESEARCH: Where are we, and what do we need?
The role of the gut microbiome in FGIDs must be considered in the context of the environment, the host, and host-specific factors. In order for us to advance the field and develop novel microbiota-based diagnostic and therapeutic targets in FGIDs, we will need to move from simple taxonomic associations to functional phenotypes and mechanism-based studies. In animal studies, we need to determine the specific microbes or microbial products as well as the mechanisms that alter host physiology. The use of gnotobiotic models allows inclusion of heterogeneity among gut microbiome and diet similar to human subjects, phenotype transfer to better understand cause-effect relationships, and complex reciprocal interactions among the host and microbiome. In terms of human studies, we need well controlled longitudinal studies incorporating functional genomic, transcriptomic, metagenomic, and metabolomic analyses as well as robust clinical metadata for the evaluation of “mechanism-based phenotypes.”. There are several factors that can affect the gut microbiome including diet, demographics, body mass index, medication etc. as described above and hence these should be controlled before linking the microbiome with host outcomes. In addition to understanding the role of microbiome in the pathophysiology of symptoms in FGIDs, assessing the impact of the microbiome on efficacy of dietary and pharmacologic therapy in conjunction with host features will allow for better treatment stratification compared to the current one size fits all approach. Finally, we need to move the needle from empirically selected prebiotic and probiotic therapies to the next generation of precise mechanism-based diagnostic and therapeutic interventions. The use of genetically engineered bacterial strains to assess the gut environment, release metabolites of interest at specific locations within the GI tract, and optimize drug metabolism appears to be on the horizon190, 191. Rapid advances in these areas provide an optimistic outlook for microbiotabased interventions in FGIDs.
PERSPECTIVE
It is now apparent that the gut microbiome is an integral player in the pathophysiology of FGIDs through its effects on host physiological processes even though the precise mechanisms underlying microbial regulation remain an area of active investigation. The improved understanding of factors that shape the gut microbiome allow us to better identify confounding effects in human studies, including physiological development through childhood and adolescence to adulthood30, 128, 11, and at the same time, appreciate the adaptation of this resilient microbial ecosystem to short- and long-term perturbations in host environment. A comprehensive view of the gut microbiome in both pediatric and adult FGIDs is important in order to account for the dynamics of the gut microbiome as it exhibits a continuum across the lifespan, with hallmark characteristics in different phases of life192.
The expanding ecosystem of microbiome-based startups and industry funding, the shift away from compositional changes towards functional products of the microbiome, better integration of clinical metadata, and genetic engineering and synthetic biology tools to make designer probiotics targeting specific host functions, together instill confidence in our ability to move microbiome science from bench to the bedside.
Acknowledgments:
Dr. AS is supported by grants KL2TR001106, and UL1TR001108 (A. Shekhar, PI) from the National Institutes of Health (NIH), National Center for Advancing Translational Sciences (NCATS), Clinical and Translational Sciences Award (CTSA). Dr. GAP is supported by NIH grants K08 DK113114 and 30 DK056338, the AGA-Rome Foundation Functional GI and Motility Pilot Research Award, the American Neurogastroenterology and Motility Society Research Grant, and the Ting Tsung and Wei Fong Chao Foundation. Dr. RS is supposed by NIH grant R01 NR013497 and the Daffy’s Foundation. Dr. P.K. is supported by NIH DK111850 and DK114007
Abbreviations:
- CNS
central nervous system
- FC
functional constipation
- FD
functional dyspepsia
- FGIDs
functional gastrointestinal disorders
- FODMAP
fermentable oligosaccharides, disaccharides, monosaccharides, and polyols
- GABA
γ-aminobutyric acid
- GF
germfree
- GI
gastrointestinal
- IBS
irritable bowel syndrome
- IBS-C
constipation predominant IBS
- IBS-D
diarrhea predominant IBS
- SCFA
short chain fatty acid(s)
- SIBO
small intestinal bacterial overgrowth
- TLR
toll-like receptor
- 5-HT
5 hydroxytryptamine
Footnotes
Disclosures: Dr. AS and PK have no disclosures to declare. Dr. GP was supported by a Career Development award from NASPGHAN Foundation/Nestlé Nutrition Research. Dr. RS has served as a consultant for Nutrinia.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- 1.Reigstad CS, Kashyap PC. Beyond phylotyping: understanding the impact of gut microbiota on host biology. Neurogastroenterol Motil 2013;25:358–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Preidis GA, Versalovic J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology b2009;136:2015–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jalanka J, Salonen A, Fuentes S, et al. Microbial signatures in post-infectious irritable bowel syndrome--toward patient stratification for improved diagnostics and treatment. Gut Microbes 2015;6:364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860–921. [DOI] [PubMed] [Google Scholar]
- 5.Human Microbiome Project C. A framework for human microbiome research. Nature 2012;486:215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaughan EE, Schut F, Heilig HG, et al. A molecular view of the intestinal ecosystem. Curr Issues Intest Microbiol 2000;1:1–12. [PubMed] [Google Scholar]
- 7.Tap J, Derrien M, Tornblom H, et al. Identification of an Intestinal Microbiota Signature Associated With Severity of Irritable Bowel Syndrome. Gastroenterology 2017;152:111123 e8. [DOI] [PubMed] [Google Scholar]
- 8.Biagi E, Nylund L, Candela M, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One 2010;5:e10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 2011;108 Suppl 1:4578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer C, Bik EM, DiGiulio DB, et al. Development of the human infant intestinal microbiota. PLoS Biol 2007;5:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollister EB, Riehle K, Luna RA, et al. Structure and function of the healthy preadolescent pediatric gut microbiome. Microbiome 2015;3:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claesson MJ, Cusack S, O’Sullivan O, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A 2011;108 Suppl 1:4586–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominianni C, Sinha R, Goedert JJ, et al. Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS One 2015;10:e0124599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haro C, Rangel-Zuniga OA, Alcala-Diaz JF, et al. Intestinal Microbiota Is Influenced by Gender and Body Mass Index. PLoS One 2016;11:e0154090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 2011;9:279–90. [DOI] [PubMed] [Google Scholar]
- 16.Kolde R, Franzosa EA, Rahnavard G, et al. Host genetic variation and its microbiome interactions within the Human Microbiome Project. Genome Med 2018;10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodrich JK, Davenport ER, Beaumont M, et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016;19:731–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKnite AM, Perez-Munoz ME, Lu L, et al. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS One 2012;7:e39191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petnicki-Ocwieja T, Hrncir T, Liu YJ, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A 2009;106:15813–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blekhman R, Goodrich JK, Huang K, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol 2015;16:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018;555:210–215. [DOI] [PubMed] [Google Scholar]
- 23.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 2010;107:14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta VK, Paul S, Dutta C . Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front Microbiol 2017;8:1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desai MS, Seekatz AM, Koropatkin NM, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016;167:1339–1353 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monsbakken KW, Vandvik PO, Farup PG. Perceived food intolerance in subjects with irritable bowel syndrome-- etiology, prevalence and consequences. Eur J Clin Nutr 2006;60:667–72. [DOI] [PubMed] [Google Scholar]
- 29.Chumpitazi BP, Weidler EM, Lu DY, et al. Self-Perceived Food Intolerances Are Common and Associated with Clinical Severity in Childhood Irritable Bowel Syndrome. Acad Nutr Diet 2016;116:1458–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajilic-Stojanovic M, Jonkers DM, Salonen A, et al. Intestinal microbiota and diet in IBS: causes, consequences, or epiphenomena? Am J Gastroenterol 2015;110:278–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halmos EP, Christophersen CT, Bird AR, et al. Diets that differ in their FODMAP content alter the colonic luminal microenvironment. Gut 2015;64:93–100. [DOI] [PubMed] [Google Scholar]
- 32.Staudacher HM, Lomer MCE, Farquharson FM, et al. A Diet Low in FODMAPs Reduces Symptoms in Patients With Irritable Bowel Syndrome and A Probiotic Restores Bifidobacterium Species: A Randomized Controlled Trial. Gastroenterology 2017;153:936–947. [DOI] [PubMed] [Google Scholar]
- 33.Bangsgaard Bendtsen KM, Krych L, Sorensen DB, et al. Gut microbiota composition is correlated to grid floor induced stress and behavior in the BALB/c mouse. PLoS One 2012;7:e46231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karl JP, Margolis LM, Madslien EH, et al. Changes in intestinal microbiota composition and metabolism coincide with increased intestinal permeability in young adults under prolonged physiological stress. Am J Physiol Gastrointest Liver Physiol 2017;312:G559–G571. [DOI] [PubMed] [Google Scholar]
- 35.Codella R, Luzi L, Terruzzi I. Exercise has the guts: How physical activity may positively modulate gut microbiota in chronic and immune-based diseases. Dig Liver Dis 2018;50:331–341. [DOI] [PubMed] [Google Scholar]
- 36.Biedermann L, Zeitz J, Mwinyi J, et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One 2013;8:e59260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mutlu EA, Gillevet PM, Rangwala H, et al. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol 2012;302:G966–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Cobas AE, Gosalbes MJ, Friedrichs A, et al. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut 2013;62:1591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jernberg C, Lofmark S, Edlund C, et al. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 2007;1:56–66. [DOI] [PubMed] [Google Scholar]
- 40.Hernandez E, Bargiela R, Diez MS, et al. Functional consequences of microbial shifts in the human gastrointestinal tract linked to antibiotic treatment and obesity. Gut Microbes 2013;4:306–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falony G, Joossens M, Vieira-Silva S, et al. Population-level analysis of gut microbiome variation. Science 2016;352:560–4. [DOI] [PubMed] [Google Scholar]
- 42.McCracken VJ, Lorenz RG. The gastrointestinal ecosystem: a precarious alliance among epithelium, immunity and microbiota. Cell Microbiol 2001;3:1–11. [DOI] [PubMed] [Google Scholar]
- 43.Sartor RB. Gut microbiota: Optimal sampling of the intestinal microbiota for research. Nat Rev Gastroenterol Hepatol 2015;12:253–4. [DOI] [PubMed] [Google Scholar]
- 44.Simren M, Barbara G, Flint HJ, et al. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut 2013;62:159–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep 2006;7:688–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swidsinski A, Weber J, Loening-Baucke V, et al. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol 2005;43:3380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albenberg L, Esipova TV, Judge CP, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 2014;147:1055–63 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Camilleri M Peripheral mechanisms in irritable bowel syndrome. N Engl J Med 2012;367:1626–35. [DOI] [PubMed] [Google Scholar]
- 49.Quartero AO, de Wit NJ, Lodder AC, et al. Disturbed solid-phase gastric emptying in functional dyspepsia: a meta-analysis. Dig Dis Sci 1998;43:2028–33. [DOI] [PubMed] [Google Scholar]
- 50.Bredenoord AJ, Chial HJ, Camilleri M, et al. Gastric accommodation and emptying in evaluation of patients with upper gastrointestinal symptoms. Clin Gastroenterol Hepatol 2003;1:264–72. [PubMed] [Google Scholar]
- 51.Husebye E, Hellstrom PM, Sundler F, et al. Influence of microbial species on small intestinal myoelectric activity and transit in germ-free rats. Am J Physiol Gastrointest Liver Physiol 2001;280:G368–80. [DOI] [PubMed] [Google Scholar]
- 52.De Palma G, Lynch MD, Lu J, et al. Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci Transl Med 2017;9. [DOI] [PubMed] [Google Scholar]
- 53.Touw K, Ringus DL, Hubert N, et al. Mutual reinforcement of pathophysiological hostmicrobe interactions in intestinal stasis models. Physiol Rep 2017;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roy CC, Kien CL, Bouthillier L, et al. Short-chain fatty acids: ready for prime time? Nutr Clin Pract 2006;21:351–66. [DOI] [PubMed] [Google Scholar]
- 55.Alemi F, Poole DP, Chiu J, et al. The receptor TGR5 mediates the prokinetic actions of intestinal bile acids and is required for normal defecation in mice. Gastroenterology 2013;144:145–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dey N, Wagner VE, Blanton LV, et al. Regulators of gut motility revealed by a gnotobiotic model of diet-microbiome interactions related to travel. Cell 2015;163:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kashyap PC, Marcobal A, Ursell LK, et al. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology 2013;144:967–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anitha M, Vijay-Kumar M, Sitaraman SV, et al. Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology 2012;143:1006–16 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jimenez M, Gil V, Martinez-Cutillas M, et al. Hydrogen sulphide as a signalling molecule regulating physiopathological processes in gastrointestinal motility. Br J Pharmacol 2017;174:2805–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ritz NL, Lin DM, Wilson MR, et al. Sulfate-reducing bacteria slow intestinal transit in a bismuth-reversible fashion in mice. Neurogastroenterol Motil 2017;29. [DOI] [PubMed] [Google Scholar]
- 61.Takaki M, Mawe GM, Barasch JM, et al. Physiological responses of guinea-pig myenteric neurons secondary to the release of endogenous serotonin by tryptamine. Neuroscience 1985;16:223–40. [DOI] [PubMed] [Google Scholar]
- 62.Jahng J, Jung IS, Choi EJ, et al. The effects of methane and hydrogen gases produced by enteric bacteria on ileal motility and colonic transit time. Neurogastroenterol Motil 2012;24:185–90, e92. [DOI] [PubMed] [Google Scholar]
- 63.Simren M, Tornblom H, Palsson OS, et al. Visceral hypersensitivity is associated with GI symptom severity in functional GI disorders: consistent findings from five different patient cohorts. Gut 2018;67:255–262. [DOI] [PubMed] [Google Scholar]
- 64.Oustamanolakis P, Tack J. Dyspepsia: organic versus functional. J Clin Gastroenterol 2012;46:175–90. [DOI] [PubMed] [Google Scholar]
- 65.Burri E, Barba E, Huaman JW, et al. Mechanisms of postprandial abdominal bloating and distension in functional dyspepsia. Gut 2014;63:395–400. [DOI] [PubMed] [Google Scholar]
- 66.Crouzet L, Gaultier E, Del’Homme C, et al. The hypersensitivity to colonic distension of IBS patients can be transferred to rats through their fecal microbiota. Neurogastroenterol Motil 2013;25:e272–82. [DOI] [PubMed] [Google Scholar]
- 67.Riba A, Olier M, Lacroix-Lamande S, et al. Paneth Cell Defects Induce Microbiota Dysbiosis in Mice and Promote Visceral Hypersensitivity. Gastroenterology 2017;153:1594–1606 e2. [DOI] [PubMed] [Google Scholar]
- 68.O’Mahony SM, Felice VD, Nally K, et al. Disturbance of the gut microbiota in early-life selectively affects visceral pain in adulthood without impacting cognitive or anxiety-related behaviors in male rats. Neuroscience 2014;277:885–901. [DOI] [PubMed] [Google Scholar]
- 69.Rousseaux C, Thuru X, Gelot A, et al. Lactobacillus acidophilus modulates intestinal pain and induces opioid and cannabinoid receptors. Nat Med 2007;13:35–7. [DOI] [PubMed] [Google Scholar]
- 70.Ait-Belgnaoui A, Eutamene H, Houdeau E, et al. Lactobacillus farciminis treatment attenuates stress-induced overexpression of Fos protein in spinal and supraspinal sites after colorectal distension in rats. Neurogastroenterol Motil 2009;21:567–73, e18–9. [DOI] [PubMed] [Google Scholar]
- 71.Kunze WA, Mao YK, Wang B, et al. Lactobacillus reuteri enhances excitability of colonic AH neurons by inhibiting calcium-dependent potassium channel opening. J Cell Mol Med 2009;13:2261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perez-Burgos A, Wang L, McVey Neufeld KA, et al. The TRPV1 channel in rodents is a major target for antinociceptive effect of the probiotic Lactobacillus reuteri DSM 17938. Physiol 2015;593:3943–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ait-Belgnaoui A, Han W, Lamine F, et al. Lactobacillus farciminis treatment suppresses stress induced visceral hypersensitivity: a possible action through interaction with epithelial cell cytoskeleton contraction. Gut 2006;55:1090–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pokusaeva K, Johnson C, Luk B, et al. GABA-producing Bifidobacterium dentium modulates visceral sensitivity in the intestine. Neurogastroenterol Motil 2017;29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bourdu S, Dapoigny M, Chapuy E, et al. Rectal instillation of butyrate provides a novel clinically relevant model of noninflammatory colonic hypersensitivity in rats. Gastroenterology 2005;128:1996–2008. [DOI] [PubMed] [Google Scholar]
- 76.Vanhoutvin SA, Troost FJ, Kilkens TO, et al. The effects of butyrate enemas on visceral perception in healthy volunteers. Neurogastroenterol Motil 2009;21:952–e76. [DOI] [PubMed] [Google Scholar]
- 77.Bischoff SC, Barbara G, Buurman W, et al. Intestinal permeability--a new target for disease prevention and therapy. BMC Gastroenterol 2014;14:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Camilleri M, Madsen K, Spiller R, et al. Intestinal barrier function in health and gastrointestinal disease. Neurogastroenterol Motil 2012;24:503–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vanheel H, Vicario M, Vanuytsel T, et al. Impaired duodenal mucosal integrity and lowgrade inflammation in functional dyspepsia. Gut 2014;63:262–71. [DOI] [PubMed] [Google Scholar]
- 80.Patel RM, Myers LS, Kurundkar AR, et al. Probiotic bacteria induce maturation of intestinal claudin 3 expression and barrier function. Am J Pathol 2012;180:626–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ukena SN, Singh A, Dringenberg U, et al. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS One 2007;2:e1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anderson RC, Cookson AL, McNabb WC, et al. Lactobacillus plantarum MB452 enhances the function of the intestinal barrier by increasing the expression levels of genes involved in tight junction formation. BMC Microbiol 2010;10:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun Y, Fihn BM, Sjovall H, et al. Enteric neurones modulate the colonic permeability response to luminal bile acids in rat colon in vivo. Gut 2004;53:362–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Forsgard RA, Korpela R, Stenman LK, et al. Deoxycholic acid induced changes in electrophysiological parameters and macromolecular permeability in murine small intestine with and without functional enteric nervous system plexuses. Neurogastroenterol Motil 2014;26:1179–87. [DOI] [PubMed] [Google Scholar]
- 85.Johansson ME, Gustafsson JK, Holmen-Larsson J, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 2014;63:281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Llewellyn SR, Britton GJ, Contijoch EJ, et al. Interactions Between Diet and the Intestinal Microbiota Alter Intestinal Permeability and Colitis Severity in Mice. Gastroenterology 2018;154:1037–1046 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ohman L, Simren M. Pathogenesis of IBS: role of inflammation, immunity and neuroimmune interactions. Nat Rev Gastroenterol Hepatol 2010;7:163–73. [DOI] [PubMed] [Google Scholar]
- 88.Fan K, Talley NJ. Functional dyspepsia and duodenal eosinophilia: A new model. J Dig Dis 2017;18:667–677. [DOI] [PubMed] [Google Scholar]
- 89.Futagami S, Shindo T, Kawagoe T, et al. Migration of eosinophils and CCR2-/CD68double positive cells into the duodenal mucosa of patients with postinfectious functional dyspepsia. Am J Gastroenterol 2010;105:1835–42. [DOI] [PubMed] [Google Scholar]
- 90.Belmonte L, Beutheu Youmba S, Bertiaux-Vandaele N, et al. Role of toll like receptors in irritable bowel syndrome: differential mucosal immune activation according to the disease subtype. PLoS One 2012;7:e42777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Powell N, Walker MM, Talley NJ. The mucosal immune system: master regulator of bidirectional gut-brain communications. Nat Rev Gastroenterol Hepatol 2017;14:143159. [DOI] [PubMed] [Google Scholar]
- 92.Gao C, Major A, Rendon D, et al. Histamine H2 Receptor-Mediated Suppression of Intestinal Inflammation by Probiotic Lactobacillus reuteri. MBio 2015;6:e01358–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Peters SA, Edogawa S, Sundt WJ, et al. Constipation-Predominant Irritable Bowel Syndrome Females Have Normal Colonic Barrier and Secretory Function. Am J Gastroenterol 2017;112:913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Acosta A, Camilleri M. Elobixibat and its potential role in chronic idiopathic constipation. Therap Adv Gastroenterol 2014;7:167–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shah ED, Kim HM, Schoenfeld P. Efficacy and Tolerability of Guanylate Cyclase-C Agonists for Irritable Bowel Syndrome with Constipation and Chronic Idiopathic Constipation: A Systematic Review and Meta-Analysis. Am J Gastroenterol 2018;113:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ridlon JM, Kang DJ, Hylemon PB, et al. Bile acids and the gut microbiome. Curr Opin Gastroenterol 2014;30:332–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Alrefai WA, Saksena S, Tyagi S, et al. Taurodeoxycholate modulates apical Cl-/OH- exchange activity in Caco2 cells. Dig Dis Sci 2007;52:1270–8. [DOI] [PubMed] [Google Scholar]
- 98.Ao M, Sarathy J, Domingue J, et al. Chenodeoxycholic acid stimulates Cl(−) secretion via cAMP signaling and increases cystic fibrosis transmembrane conductance regulator phosphorylation in T84 cells. Am J Physiol Cell Physiol 2013;305:C447–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Murray RD, McClung HJ, Li BU, et al. Stimulatory effects of short-chain fatty acids on colonic absorption in newborn piglets in vivo. J Pediatr Gastroenterol Nutr 1989;8:95101. [DOI] [PubMed] [Google Scholar]
- 100.Kaji I, Iwanaga T, Watanabe M, et al. SCFA transport in rat duodenum. Am J Physiol Gastrointest Liver Physiol 2015;308:G188–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bhattarai Y, Schmidt BA, Linden DR, et al. Human-derived gut microbiota modulates colonic secretion in mice by regulating 5-HT3 receptor expression via acetate production. Am J Physiol Gastrointest Liver Physiol 2017;313:G80–G87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scarpellini E, Deloose E, Vos R, et al. The effect of arabinoxylooligosaccharides on gastric sensory-motor function and nutrient tolerance in man. Neurogastroenterol Motil 2016;28:1194–203. [DOI] [PubMed] [Google Scholar]
- 103.Reddymasu SC, McCallum RW. Small intestinal bacterial overgrowth in gastroparesis: are there any predictors? J Clin Gastroenterol 2010;44:e8–13. [DOI] [PubMed] [Google Scholar]
- 104.George NS, Sankineni A, Parkman HP. Small intestinal bacterial overgrowth in gastroparesis. Dig Dis Sci 2014;59:645–52. [DOI] [PubMed] [Google Scholar]
- 105.Chander Roland B, Mullin GE, Passi M, et al. A Prospective Evaluation of Ileocecal Valve Dysfunction and Intestinal Motility Derangements in Small Intestinal Bacterial Overgrowth. Dig Dis Sci 2017;62:3525–3535. [DOI] [PubMed] [Google Scholar]
- 106.Roland BC, Ciarleglio MM, Clarke JO, et al. Small Intestinal Transit Time Is Delayed in Small Intestinal Bacterial Overgrowth. J Clin Gastroenterol 2015;49:571–6. [DOI] [PubMed] [Google Scholar]
- 107.Drossman DA, Tack J, Ford AC, et al. Neuromodulators for Functional Gastrointestinal Disorders (Disorders of Gut-Brain Interaction): A Rome Foundation Working Team Report. Gastroenterology 2018;154:1140–1171 e1. [DOI] [PubMed] [Google Scholar]
- 108.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 2012;13:701–12. [DOI] [PubMed] [Google Scholar]
- 109.O’Mahony SM, Marchesi JR, Scully P, et al. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry 2009;65:263–7. [DOI] [PubMed] [Google Scholar]
- 110.Sudo N, Chida Y, Aiba Y, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol 2004;558:263–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bercik P, Park AJ, Sinclair D, et al. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterol Motil 2011;23:1132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bhattarai Y, Muniz Pedrogo DA, Kashyap PC. Irritable bowel syndrome: a gut microbiota-related disorder? Am J Physiol Gastrointest Liver Physiol 2017;312:G52G62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barbara G, Stanghellini V, De Giorgio R, et al. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology 2004;126:693–702. [DOI] [PubMed] [Google Scholar]
- 114.Barbara G, Feinle-Bisset C, Ghoshal UC, et al. The Intestinal Microenvironment and Functional Gastrointestinal Disorders. Gastroenterology 2016. [DOI] [PubMed] [Google Scholar]
- 115.Wouters MM, Balemans D, Van Wanrooy S, et al. Histamine Receptor H1-Mediated Sensitization of TRPV1 Mediates Visceral Hypersensitivity and Symptoms in Patients With Irritable Bowel Syndrome. Gastroenterology 2016;150:875–87 e9. [DOI] [PubMed] [Google Scholar]
- 116.Barbara G, Cremon C, Carini G, et al. The immune system in irritable bowel syndrome. J Neurogastroenterol Motil 2011;17:349–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scully P, McKernan DP, Keohane J, et al. Plasma cytokine profiles in females with irritable bowel syndrome and extra-intestinal co-morbidity. Am J Gastroenterol 2010;105:2235–43. [DOI] [PubMed] [Google Scholar]
- 118.Agrawal A, Houghton LA, Morris J, et al. Clinical trial: the effects of a fermented milk product containing Bifidobacterium lactis DN-173 010 on abdominal distension and gastrointestinal transit in irritable bowel syndrome with constipation. Aliment Pharmacol Ther 2009;29:104–14. [DOI] [PubMed] [Google Scholar]
- 119.Acosta A, Camilleri M, Shin A, et al. Effects of Rifaximin on Transit, Permeability, Fecal Microbiome, and Organic Acid Excretion in Irritable Bowel Syndrome. Clin Transl Gastroenterol 2016;7:e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.O’Mahony L, McCarthy J, Kelly P, et al. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology 2005;128:541–51. [DOI] [PubMed] [Google Scholar]
- 121.Aerssens J, Camilleri M, Talloen W, et al. Alterations in mucosal immunity identified in the colon of patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 2008;6:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lyra A, Rinttila T, Nikkila J, et al. Diarrhoea-predominant irritable bowel syndrome distinguishable by 16S rRNA gene phylotype quantification. World J Gastroenterol 2009;15:5936–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shin A, Camilleri M, Vijayvargiya P, et al. Bowel functions, fecal unconjugated primary and secondary bile acids, and colonic transit in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 2013;11:1270–1275 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Quigley EMM. The Gut-Brain Axis and the Microbiome: Clues to Pathophysiology and Opportunities for Novel Management Strategies in Irritable Bowel Syndrome (IBS). J Clin Med 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mayer EA, Savidge T, Shulman RJ. Brain-gut microbiome interactions and functional bowel disorders. Gastroenterology 2014;146:1500–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tillisch K, Labus J, Kilpatrick L, et al. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology 2013;144:1394–401, 1401 e1–4. 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pinto-Sanchez MI, Hall GB, Ghajar K, et al. Probiotic Bifidobacterium longum NCC3001 Reduces Depression Scores and Alters Brain Activity: A Pilot Study in Patients With Irritable Bowel Syndrome. Gastroenterology 2017;153:448–459 e8. [DOI] [PubMed] [Google Scholar]
- 128.Saulnier DM, Riehle K, Mistretta TA, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 2011;141:1782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rigsbee L, Agans R, Shankar V, et al. Quantitative profiling of gut microbiota of children with diarrhea-predominant irritable bowel syndrome. Am J Gastroenterol 2012;107:1740–51. [DOI] [PubMed] [Google Scholar]
- 130.Shankar V, Reo NV, Paliy O. Simultaneous fecal microbial and metabolite profiling enables accurate classification of pediatric irritable bowel syndrome. Microbiome 2015;3:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chumpitazi BP, Hollister EB, Oezguen N, et al. Gut microbiota influences low fermentable substrate diet efficacy in children with irritable bowel syndrome. Gut Microbes 2014;5:165–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chumpitazi BP, Cope JL, Hollister EB, et al. Randomised clinical trial: gut microbiome biomarkers are associated with clinical response to a low FODMAP diet in children with the irritable bowel syndrome. Aliment Pharmacol Ther 2015;42:418–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Horvath A, Dziechciarz P, Szajewska H. Meta-analysis: Lactobacillus rhamnosus GG for abdominal pain-related functional gastrointestinal disorders in childhood. Aliment Pharmacol Ther 2011;33:1302–10. [DOI] [PubMed] [Google Scholar]
- 134.Guandalini S, Magazzu G, Chiaro A, et al. VSL#3 improves symptoms in children with irritable bowel syndrome: a multicenter, randomized, placebo-controlled, double-blind, crossover study. J Pediatr Gastroenterol Nutr 2010;51:24–30. [DOI] [PubMed] [Google Scholar]
- 135.Giannetti E, Maglione M, Alessandrella A, et al. A Mixture of 3 Bifidobacteria Decreases Abdominal Pain and Improves the Quality of Life in Children With Irritable Bowel Syndrome: A Multicenter, Randomized, Double-Blind, Placebo-Controlled, Crossover Trial. J Clin Gastroenterol 2017;51:e5–e10. [DOI] [PubMed] [Google Scholar]
- 136.Shulman RJ, Hollister EB, Cain K, et al. Psyllium Fiber Reduces Abdominal Pain in Children With Irritable Bowel Syndrome in a Randomized, Double-Blind Trial. Clin Gastroenterol Hepatol 2017;15:712–719 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jadresin O, Hojsak I, Misak Z, et al. Lactobacillus reuteri DSM 17938 in the Treatment of Functional Abdominal Pain in Children: RCT Study. J Pediatr Gastroenterol Nutr 2017;64:925–929. [DOI] [PubMed] [Google Scholar]
- 138.Romano C, Ferrau V, Cavataio F, et al. Lactobacillus reuteri in children with functional abdominal pain (FAP). J Paediatr Child Health 2014;50:E68–71. [DOI] [PubMed] [Google Scholar]
- 139.Weizman Z, Abu-Abed J, Binsztok M. Lactobacillus reuteri DSM 17938 for the Management of Functional Abdominal Pain in Childhood: A Randomized, Double-Blind, Placebo-Controlled Trial. J Pediatr 2016;174:160–164 e1. [DOI] [PubMed] [Google Scholar]
- 140.Maragkoudaki M, Chouliaras G, Orel R, et al. Lactobacillus reuteri DSM 17938 and a placebo both significantly reduced symptoms in children with functional abdominal pain. Acta Paediatr 2017;106:1857–1862. [DOI] [PubMed] [Google Scholar]
- 141.Stanghellini V, Chan FK, Hasler WL, et al. Gastroduodenal Disorders. Gastroenterology 2016;150:1380–92. [DOI] [PubMed] [Google Scholar]
- 142.Tziatzios G, Giamarellos-Bourboulis EJ, Papanikolaou IS, et al. Is small intestinal bacterial overgrowth involved in the pathogenesis of functional dyspepsia? Med Hypotheses 2017;106:26–32. [DOI] [PubMed] [Google Scholar]
- 143.Aziz I, Tornblom H, Simren M. Small intestinal bacterial overgrowth as a cause for irritable bowel syndrome: guilty or not guilty? Curr Opin Gastroenterol 2017;33:196–202. [DOI] [PubMed] [Google Scholar]
- 144.Zhong L, Shanahan ER, Raj A, et al. Dyspepsia and the microbiome: time to focus on the small intestine. Gut 2017;66:1168–1169. [DOI] [PubMed] [Google Scholar]
- 145.Igarashi M, Nakae H, Matsuoka T, et al. Alteration in the gastric microbiota and its restoration by probiotics in patients with functional dyspepsia. BMJ Open Gastroenterol 2017;4:e000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tan VP, Liu KS, Lam FY, et al. Randomised clinical trial: rifaximin versus placebo for the treatment of functional dyspepsia. Aliment Pharmacol Ther 2017;45:767–776. [DOI] [PubMed] [Google Scholar]
- 147.Gawronska A, Dziechciarz P, Horvath A, et al. A randomized double-blind placebocontrolled trial of Lactobacillus GG for abdominal pain disorders in children. Aliment Pharmacol Ther 2007;25:177–84. [DOI] [PubMed] [Google Scholar]
- 148.Schmulson M, Chang L. Review article: the treatment of functional abdominal bloating and distension. Aliment Pharmacol Ther 2011;33:1071–86. [DOI] [PubMed] [Google Scholar]
- 149.Ringel-Kulka T, Benson AK, Carroll IM, et al. Molecular characterization of the intestinal microbiota in patients with and without abdominal bloating. Am J Physiol Gastrointest Liver Physiol 2016;310:G417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sharara AI, Aoun E, Abdul-Baki H, et al. A randomized double-blind placebo-controlled trial of rifaximin in patients with abdominal bloating and flatulence. Am J Gastroenterol 2006;101:326–33. [DOI] [PubMed] [Google Scholar]
- 151.Pimentel M, Lembo A, Chey WD, et al. Rifaximin therapy for patients with irritable bowel syndrome without constipation. N Engl J Med 2011;364:22–32. [DOI] [PubMed] [Google Scholar]
- 152.Pimentel M, Lin HC, Enayati P, et al. Methane, a gas produced by enteric bacteria, slows intestinal transit and augments small intestinal contractile activity. Am J Physiol Gastrointest Liver Physiol 2006;290:G1089–95. [DOI] [PubMed] [Google Scholar]
- 153.Hungin AP, Mulligan C, Pot B, et al. Systematic review: probiotics in the management of lower gastrointestinal symptoms in clinical practice -- an evidence-based international guide. Aliment Pharmacol Ther 2013;38:864–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Guyonnet D, Chassany O, Ducrotte P, et al. Effect of a fermented milk containing Bifidobacterium animalis DN-173 010 on the health-related quality of life and symptoms in irritable bowel syndrome in adults in primary care: a multicentre, randomized, doubleblind, controlled trial. Aliment Pharmacol Ther 2007;26:475–86. [DOI] [PubMed] [Google Scholar]
- 155.Kim HJ, Camilleri M, McKinzie S, et al. A randomized controlled trial of a probiotic, VSL#3, on gut transit and symptoms in diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther 2003;17:895–904. [DOI] [PubMed] [Google Scholar]
- 156.Jeffery IB, O’Toole PW, Ohman L, et al. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 2012;61:997–1006. [DOI] [PubMed] [Google Scholar]
- 157.Parthasarathy G, Chen J, Chen X, et al. Relationship Between Microbiota of the Colonic Mucosa vs Feces and Symptoms, Colonic Transit, and Methane Production in Female Patients With Chronic Constipation. Gastroenterology 2016;150:367–79 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cao H, Liu X, An Y, et al. Dysbiosis contributes to chronic constipation development via regulation of serotonin transporter in the intestine. Sci Rep 2017;7:10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ge X, Zhao W, Ding C, et al. Potential role of fecal microbiota from patients with slow transit constipation in the regulation of gastrointestinal motility. Sci Rep 2017;7:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Roager HM, Hansen LB, Bahl MI, et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat Microbiol 2016;1:16093. [DOI] [PubMed] [Google Scholar]
- 161.Vandeputte D, Falony G, Vieira-Silva S, et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 2016;65:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mancabelli L, Milani C, Lugli GA, et al. Unveiling the gut microbiota composition and functionality associated with constipation through metagenomic analyses. Sci Rep 2017;7:9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wolf PG, Parthasarathy G, Chen J, et al. Assessing the colonic microbiome, hydrogenogenic and hydrogenotrophic genes, transit and breath methane in constipation. Neurogastroenterol Motil 2017;29:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhu L, Liu W, Alkhouri R, et al. Structural changes in the gut microbiome of constipated patients. Physiol Genomics 2014;46:679–86. [DOI] [PubMed] [Google Scholar]
- 165.Coccorullo P, Strisciuglio C, Martinelli M, et al. Lactobacillus reuteri (DSM 17938) in infants with functional chronic constipation: a double-blind, randomized, placebocontrolled study. J Pediatr 2010;157:598–602. [DOI] [PubMed] [Google Scholar]
- 166.Guerra PV, Lima LN, Souza TC, et al. Pediatric functional constipation treatment with Bifidobacterium-containing yogurt: a crossover, double-blind, controlled trial. World J Gastroenterol 2011;17:3916–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wojtyniak K, Szajewska H. Systematic review: probiotics for functional constipation in children. Eur J Pediatr 2017;176:1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Morais MB, Vitolo MR, Aguirre AN, et al. Measurement of low dietary fiber intake as a risk factor for chronic constipation in children. J Pediatr Gastroenterol Nutr 1999;29:132–5. [DOI] [PubMed] [Google Scholar]
- 169.Pijpers MA, Tabbers MM, Benninga MA, et al. Currently recommended treatments of childhood constipation are not evidence based: a systematic literature review on the effect of laxative treatment and dietary measures. Arch Dis Child 2009;94:117–31. [DOI] [PubMed] [Google Scholar]
- 170.Tabbers MM, Boluyt N, Berger MY, et al. Nonpharmacologic treatments for childhood constipation: systematic review. Pediatrics 2011;128:753–61. [DOI] [PubMed] [Google Scholar]
- 171.Tabbers MM, Benninga MA. Constipation in children: fibre and probiotics. BMJ Clin Evid 2015;2015. [PMC free article] [PubMed] [Google Scholar]
- 172.Gordon M, Naidoo K, Akobeng AK, et al. Cochrane Review: Osmotic and stimulant laxatives for the management of childhood constipation (Review). Evid Based Child Health 2013;8:57–109. [DOI] [PubMed] [Google Scholar]
- 173.Wolke D, Bilgin A, Samara M. Systematic Review and Meta-Analysis: Fussing and Crying Durations and Prevalence of Colic in Infants. J Pediatr 2017;185:55–61 e4. [DOI] [PubMed] [Google Scholar]
- 174.Savino F, Castagno E, Bretto R, et al. A prospective 10-year study on children who had severe infantile colic. Acta Paediatr Suppl 2005;94:129–32. [DOI] [PubMed] [Google Scholar]
- 175.Savino F, Cresi F, Pautasso S, et al. Intestinal microflora in breastfed colicky and noncolicky infants. Acta Paediatr 2004;93:825–9. [PubMed] [Google Scholar]
- 176.Savino F, Bailo E, Oggero R, et al. Bacterial counts of intestinal Lactobacillus species in infants with colic. Pediatr Allergy Immunol 2005;16:72–5. [DOI] [PubMed] [Google Scholar]
- 177.Savino F, Cordisco L, Tarasco V, et al. Molecular identification of coliform bacteria from colicky breastfed infants. Acta Paediatr 2009;98:1582–8. [DOI] [PubMed] [Google Scholar]
- 178.Rhoads JM, Fatheree NY, Norori J, et al. Altered fecal microflora and increased fecal calprotectin in infants with colic. J Pediatr 2009;155:823–828 e1. [DOI] [PubMed] [Google Scholar]
- 179.de Weerth C, Fuentes S, Puylaert P, et al. Intestinal microbiota of infants with colic: development and specific signatures. Pediatrics 2013;131:e550–8. [DOI] [PubMed] [Google Scholar]
- 180.Xu M, Wang J, Wang N, et al. The Efficacy and Safety of the Probiotic Bacterium Lactobacillus reuteri DSM 17938 for Infantile Colic: A Meta-Analysis of Randomized Controlled Trials. PLoS One 2015;10:e0141445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Harb T, Matsuyama M, David M, et al. Infant Colic-What works: A Systematic Review of Interventions for Breast-fed Infants. J Pediatr Gastroenterol Nutr 2016;62:668–86. [DOI] [PubMed] [Google Scholar]
- 182.Indrio F, Di Mauro A, Riezzo G, et al. Prophylactic use of a probiotic in the prevention of colic, regurgitation, and functional constipation: a randomized clinical trial. JAMA Pediatr 2014;168:228–33. [DOI] [PubMed] [Google Scholar]
- 183.Savino F, Ceratto S, Poggi E, et al. Preventive effects of oral probiotic on infantile colic: a prospective, randomised, blinded, controlled trial using Lactobacillus reuteri DSM 17938. Benef Microbes 2015;6:245–51. [DOI] [PubMed] [Google Scholar]
- 184.Partty A, Lehtonen L, Kalliomaki M, et al. Probiotic Lactobacillus rhamnosus GG therapy and microbiological programming in infantile colic: a randomized, controlled trial. Pediatr Res 2015;78:470–5. [DOI] [PubMed] [Google Scholar]
- 185.Fatheree NY, Liu Y, Ferris M, et al. Hypoallergenic formula with Lactobacillus rhamnosus GG for babies with colic: A pilot study of recruitment, retention, and fecal biomarkers. World J Gastrointest Pathophysiol 2016;7:160–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kianifar H, Ahanchian H, Grover Z, et al. Synbiotic in the management of infantile colic: a randomised controlled trial. J Paediatr Child Health 2014;50:801–5. [DOI] [PubMed] [Google Scholar]
- 187.Ford AC, Quigley EM, Lacy BE, et al. Efficacy of prebiotics, probiotics, and synbiotics in irritable bowel syndrome and chronic idiopathic constipation: systematic review and meta-analysis. Am J Gastroenterol 2014;109:1547–61; quiz 1546, 1562. [DOI] [PubMed] [Google Scholar]
- 188.Johnsen PH, Hilpusch F, Cavanagh JP, et al. Faecal microbiota transplantation versus placebo for moderate-to-severe irritable bowel syndrome: a double-blind, randomised, placebo-controlled, parallel-group, single-centre trial. Lancet Gastroenterol Hepatol 2018;3:17–24. [DOI] [PubMed] [Google Scholar]
- 189.Aroniadis OB L; Oneto C; Feuerstadt P; Sherman A; Wolkoff A; Downs I; Zanetti-Yabur A; Ramos Y; Cotto C; Kassam Z; Elliott R; Rosenbaum R; Budree S; Sadovsky R; Timberlake S; Swanson P; Kim M; Keller M A Double-Blind, Randomized, Placebo-Controlled Trial of Fecal Microbiota Transplantation Capsules (FMTC) for the Treatment of Diarrhea-Predominant Irritable Bowel Syndrome (IBS-D). Gastroenterology 2018;154:S-154–S-155. [Google Scholar]
- 190.Kalantar-Zadeh K, Berean KJ, Ha N, et al. A human pilot trial of ingestible electronic capsules capable of sensing different gases in the gut. Nature Electronics 2018;1:79–87. [Google Scholar]
- 191.Mimee M, Nadeau P, Hayward A, et al. An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 2018;360:915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kundu P, Blacher E, Elinav E, et al. Our Gut Microbiome: The Evolving Inner Self. Cell 2017;171:1481–1493. [DOI] [PubMed] [Google Scholar]
- 193.Reigstad CS, Salmonson CE, Rainey JF 3rd, et al. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J 2015;29:1395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Fukumoto S, Tatewaki M, Yamada T, et al. Short-chain fatty acids stimulate colonic transit via intraluminal 5-HT release in rats. Am J Physiol Regul Integr Comp Physiol 2003;284:R1269–76. [DOI] [PubMed] [Google Scholar]
- 195.Ploger S, Stumpff F, Penner GB, et al. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann N Y Acad Sci 2012;1258:52–9. [DOI] [PubMed] [Google Scholar]
- 196.Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science 2012;336:1262–7. [DOI] [PubMed] [Google Scholar]
- 197.Bampton PA, Dinning PG, Kennedy ML, et al. The proximal colonic motor response to rectal mechanical and chemical stimulation. Am J Physiol Gastrointest Liver Physiol 2002;282:G443–9. [DOI] [PubMed] [Google Scholar]
- 198.Chadwick VS, Gaginella TS, Carlson GL, et al. Effect of molecular structure on bile acid-induced alterations in absorptive function, permeability, and morphology in the perfused rabbit colon. J Lab Clin Med 1979;94:661–74. [PubMed] [Google Scholar]
- 199.Cipriani S, Mencarelli A, Chini MG, et al. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS One 2011;6:e25637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Tremblay S, Romain G, Roux M, et al. Bile Acid Administration Elicits an Intestinal Antimicrobial Program and Reduces the Bacterial Burden in Two Mouse Models of Enteric Infection. Infect Immun 2017;85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Shin SP, Choi YM, Kim WH, et al. A double blind, placebo-controlled, randomized clinical trial that breast milk derived-Lactobacillus gasseri BNR17 mitigated diarrheadominant irritable bowel syndrome. J Clin Biochem Nutr 2018;62:179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Dior M, Delagreverie H, Duboc H, et al. Interplay between bile acid metabolism and microbiota in irritable bowel syndrome. Neurogastroenterol Motil 2016;28:1330–40. [DOI] [PubMed] [Google Scholar]
- 203.Le Neve B, Brazeilles R, Derrien M, et al. Lactulose Challenge Determines Visceral Sensitivity and Severity of Symptoms in Patients With Irritable Bowel Syndrome. Clin Gastroenterol Hepatol 2016;14:226–33 e1–3. [DOI] [PubMed] [Google Scholar]
- 204.Labus JS, Hollister EB, Jacobs J, et al. Differences in gut microbial composition correlate with regional brain volumes in irritable bowel syndrome. Microbiome 2017;5:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Liu Y, Zhang L, Wang X, et al. Similar Fecal Microbiota Signatures in Patients With Diarrhea-Predominant Irritable Bowel Syndrome and Patients With Depression. Clin Gastroenterol Hepatol 2016;14:1602–1611 e5. [DOI] [PubMed] [Google Scholar]
- 206.Azpiroz F, Dubray C, Bernalier-Donadille A, et al. Effects of scFOS on the composition of fecal microbiota and anxiety in patients with irritable bowel syndrome: a randomized, double blind, placebo controlled study. Neurogastroenterol Motil 2017;29. [DOI] [PubMed] [Google Scholar]
- 207.Le Gall G, Noor SO, Ridgway K, et al. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res 2011;10:4208–18. [DOI] [PubMed] [Google Scholar]
- 208.Heitkemper MM, Cain KC, Shulman RJ, et al. Stool and urine trefoil factor 3 levels: associations with symptoms, intestinal permeability, and microbial diversity in irritable bowel syndrome. Benef Microbes 2018;9:345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bednarska O, Walter SA, Casado-Bedmar M, et al. Vasoactive Intestinal Polypeptide and Mast Cells Regulate Increased Passage of Colonic Bacteria in Patients With Irritable Bowel Syndrome. Gastroenterology 2017;153:948–960 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Valentin N, Camilleri M, Carlson P, et al. Potential mechanisms of effects of serumderived bovine immunoglobulin/protein isolate therapy in patients with diarrheapredominant irritable bowel syndrome. Physiol Rep 2017;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ko SJ, Han G, Kim SK, et al. Effect of korean herbal medicine combined with a probiotic mixture on diarrhea-dominant irritable bowel syndrome: a double-blind, randomized, placebo-controlled trial. Evid Based Complement Alternat Med 2013;2013:824605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Compare D, Rocco A, Coccoli P, et al. Lactobacillus casei DG and its postbiotic reduce the inflammatory mucosal response: an ex-vivo organ culture model of post-infectious irritable bowel syndrome. BMC Gastroenterol 2017;17:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hustoft TN, Hausken T, Ystad SO, et al. Effects of varying dietary content of fermentable short-chain carbohydrates on symptoms, fecal microenvironment, and cytokine profiles in patients with irritable bowel syndrome. Neurogastroenterol Motil 2017;29. [DOI] [PubMed] [Google Scholar]
- 214.McIntosh K, Reed DE, Schneider T, et al. FODMAPs alter symptoms and the metabolome of patients with IBS: a randomised controlled trial. Gut 2017;66:1241–1251. [DOI] [PubMed] [Google Scholar]
- 215.Sundin J, Rangel I, Repsilber D, et al. Cytokine Response after Stimulation with Key Commensal Bacteria Differ in Post-Infectious Irritable Bowel Syndrome (PI-IBS) Patients Compared to Healthy Controls. PLoS One 2015;10:e0134836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sundin J, Rangel I, Fuentes S, et al. Altered faecal and mucosal microbial composition in post-infectious irritable bowel syndrome patients correlates with mucosal lymphocyte phenotypes and psychological distress. Aliment Pharmacol Ther 2015;41:342–51. [DOI] [PubMed] [Google Scholar]
- 217.Parthasarathy G, Chen J, Chia N, et al. Reproducibility of assessing fecal microbiota in chronic constipation. Neurogastroenterol Motil 2017;29:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tian H, Ge X, Nie Y, et al. Fecal microbiota transplantation in patients with slow-transit constipation: A randomized, clinical trial. PLoS One 2017;12:e0171308. [DOI] [PMC free article] [PubMed] [Google Scholar]