Abstract
Hyaluronan (HA) is a glycosaminoglycan composed of repeating disaccharide units of glucuronic acid and N-acetylglucosamine. HA is an extremely long, unbranched polymer, which often exceeds 106 Da and sometimes reaches 107 Da. A feature that epitomizes HA is its rapid turnover: one-third of the total body HA is turned over daily. The current model of HA catabolism postulates that high-molecular weight HA in the extracellular space is first cleaved into smaller fragments by a hyaluronidase(s) that resides at the cell surface, followed by internalization of fragments and their degradation into monosaccharides in lysosomes. Over the last decade, considerable research has shown that the HYAL family of hyaluronidases plays significant roles in HA catabolism. Nonetheless, the identity of a hyaluronidase responsible for the initial step of HA cleavage on the cell surface remains to be determined, as biochemical and enzymological properties of HYAL proteins are not entirely consistent with those expected of cell surface hyaluronidases. Recent identification of transmembrane 2 (TMEM2) as a cell surface protein that possesses potent hyaluronidase activity suggests that it may be the “missing” cell surface hyaluronidase, and that novel models of HA catabolism should include this protein.
Introduction
Hyaluronan (HA) is a linear polymer of repeating disaccharide units of glucuronic acid and N-acetylglucosamine. HA is a large molecule with a molecular weight often exceeding 106 Da, sometimes reaching 107 Da. As a major component of the extracellular matrix (ECM), HA exhibits multiple cellular activities through its unique biophysical and biological properties. Because of its sheer size, cells are thought to possess elaborate mechanisms for its production and degradation. Particularly remarkable is the extremely fast turnover of HA: an estimated one-third of the total body HA (~15 g in a person with a 70 kg body weight) is turned over daily (1), and the metabolic half-life of HA in skin is only 1 to 1.5 days (2).
A current model of HA catabolism is that high-molecular weight HA (106–107 Da) in the extracellular space is first degraded into intermediate-size fragments of 10–100 kDa by hyaluronidase(s) present on the cell surface or in the extracellular space. These fragments are then internalized and eventually degraded to monosaccharides by the combined actions of lysosomal hyaluronidases and exoglucosidases (3,4). It should be noted that this model, which assumes that the entire HA degradation process occurs in a single cell, does not necessarily reflect physiological mechanisms relevant to systemic HA catabolism. It remains possible that the initial degradation into intermediate fragments occurs in peripheral or lymphatic tissues, while degradation into monosaccharides could occur in specific organs such as liver (4). In either case, it is essential to define hyaluronidases responsible for each step of HA degradation and identify their tissue or subcellular sites of action. Since molecular cloning of HYAL1 and HYAL2 some 20 years ago (5,6), studies of HA catabolism have focused mainly on these molecules, and the model of HA catabolism has been formulated based on the assumption that HYALs are the key hyaluronidases both on the cell surface and in intracellular compartments. Yet, recent identification of novel proteins that exhibit HA degrading activity, namely transmembrane 2 (TMEM2) and CEMIP/KIAA1199, now prompts a reevaluation of these models. In this article, we focus on TMEM2 and review the current knowledge of this novel cell surface hyaluronidase, highlighting the distinctive features of TMEM2 relative to HYALs and CEMIP/KIAA1199.
HYAL family molecules
There are six HYAL-like genes in the human genome. HYAL1, HYAL2, and HYAL3 are clustered in 3p21.3, whereas HYAL4 and SPAM1 (also known as PH-20) are present in 7p31.3 (2). The latter cluster also harbors the pseudogene HYALP1. The significant homology between different HYAL genes suggests that they are generated by gene duplication (7).
There are multiple lines of evidence that HYAL proteins are associated with lysosomes and related intracellular vesicles. SPAM1/PH-20, the prototype of the family, is associated with the acrosome (8), a lysosome-related organelle in sperm cells (9). HYAL1, a major hyaluronidase in plasma, is associated with lysosomes and endosomes in cells (10), and HA degradation by HYAL1 occurs intracellularly (11). HYAL2 was originally identified as a lysosomal hyaluronidase (6) and has been shown to be present in lysosomes of various cell types (6,12,13). Interestingly, HYALs can also be anchored to the plasma membrane via a glycosylphosphatydylinositol (GPI) linkage (8,14–16). This phenomenon has been reported for SPAM1, HYAL1, and HYAL2, among which HYAL2 has been studied most extensively. Cell surface translocation of HYAL2 requires co-expression of CD44 (11). In C28/I2 chondrocytes, HYAL2 and CD44 were found to co-immunoprecipitate, suggesting that HYAL2 and CD44 interact directly (17).
HYAL1 and HYAL2 favor acidic pH for their hyaluronidase activity. HYAL1 is active only below pH5.5 (18) and the pH optimum for HYAL2 is pH 4 (6). Such low pH optima are consistent with properties of lysosomal rather than extracellular enzymes. It is also noteworthy that the intrinsic hyaluronidase activity of HYAL2 is reported to be weak relative to those of other HYALs; HYAL2 is ~400-fold and ~50-fold less potent than PH-20/SPAM1 and HYAL1, respectively (14,19,20). There is a suggestion that hyaluronidase activity of HYAL2 requires co-expression of CD44; Harada and Takahashi (11) showed that the membrane fraction of HEK293 cells expressing HYAL2, but not CD44, exhibits little hyaluronidase activity. HYAL family proteins degrade not only HA but also chondroitin sulfate (CS) and dermatan sulfate (DS) (21). For example, HYAL1 exhibits robust chondroitinase activity (22), and HYAL4 appears to act physiologically as a chondroitinase (21).
Mutations in HYAL1 reportedly cause the mucopolysaccharidosis type IX (MPS IX), a lysosomal storage disease (23). In affected patients, serum HA concentrations increase 38–90-fold, and macrophages and fibroblasts of these patients display lysosomal HA accumulation (24). Hyal1−/− mice are viable and exhibit no gross developmental defects, but develop osteoarthritis as they age (25). Hyal2−/− mice on a C57Bl6 background are also viable and show mild developmental abnormalities, namely shortening of the nose, widened interorbitary space, and slightly deformed cervical vertebrae (20). On a mixed background, approximately half of Hyal2−/− mice exhibit heart defects, including expanded heart valves and cardiac hypertrophy, as well as lung fibrosis (26). Both Hyal1−/− and Hyal2−/− mice show HA accumulation in peripheral tissues. Hyal2−/− mice exhibit lymphadenopathy and buildup of high molecular weight HA in lymph and serum (27). Interestingly, Hyal2 ablation also impairs HA internalization by non-parenchymal cells in the liver (such as sinusoidal endothelial cells and Kupffer cells), suggesting that in these cells, HYAL2 functions as an endocytic receptor for HA (27). More recently, Muggenthaler et al. (28) reported that mutations in HYAL2 found in Amish pedigrees cause orofacial clefting and cor triatriatus sinister in humans and mice, and a whole exome sequencing study identified association of rare human HYAL2 variants with platelet reactivity (29). While it is not entirely clear whether all these phenotypes are direct consequences of loss of hyaluronidase activity of HYAL2, these results are consistent with the notion that HYAL2 physiologically participates in HA catabolism.
CEMIP/KIAA1199
In 2013, Yoshida and colleagues reported that a protein of unknown function encoded by the transcript KIAA1199 has HA binding and degrading activities (30). An independent line of studies identified KIAA1199 as encoding a protein that induces cancer cell migration and named it cell migration inducing protein or CEMIP (31–33). CEMIP, which bears no homology to HYAL family proteins, is a putative secretory protein containing an N-terminal signal sequence. Curiously, neither conditioned media of CEMIP-transfected cells nor recombinant CEMIP show HA degrading activity (30,33). From knockdown experiments, Yoshida concluded that CEMIP-mediated HA degradation requires participation of the clathrin-coated pit pathway (30), suggesting that CEMIP itself does not act as an extracellular hyaluronidase. Recent knockout studies show that systemic Cemip deletion in mice results in mild shortening of long bones (34) and decreased memory function (35).
Several lines of evidence suggest that CEMIP may be involved in human diseases. Mutations in CEMIP have been identified in cases of non-syndromic hearing loss (36). Moreover, a CEMIP point mutant at one of the deafness-associated residues (Arg-187) abrogates hyaluronidase activity (30). However, Cemip−/− mice show no apparent hearing loss (35). In addition, several reports indicate a strong association of CEMIP overexpression with progression and poor prognosis of various human cancers (37–40). CEMIP knockdown in breast cancer cells reportedly reduces cell motility in vitro and lung metastasis in an orthotopic xenograft model (33,38).
TMEM2
As noted above, knockout studies of HYAL family hyaluronidases support their participation in HA catabolism in vivo (20,26). It remains unclear, however, whether HYAL2 is the main player for the initial step of HA degradation on the cell surface, as its enzymatic property and subcellular localization are not entirely consistent with those expected of cell surface hyaluronidases (see above). The observation that Hyal2−/− mice exhibit elevated serum and tissue HA could be explained by reduced lysosomal degradation of HA and does not necessarily support the idea that HYAL2 is the primary hyaluronidase that functions on the cell surface.
In search of a hyaluronidase that is present and might function on the cell surface, we identified TMEM2 as a strong candidate (41). The TMEM family is a heterogeneous collection of more than 300 open reading frames that are grouped based only on the presence of at least one putative transmembrane domain (42). The TMEM2 amino acid sequence predicts a type II transmembrane protein with an extracellular domain showing 48% amino acid identity with CEMIP (41). Interestingly, zebrafish tmem2 mutants reportedly exhibit a phenotype related to endocardial cushion defect (43,44) (see below), a finding that resonates with the fact that HA is the main component of cardiac jelly and plays a critical role in endocardial cushion development (45,46).
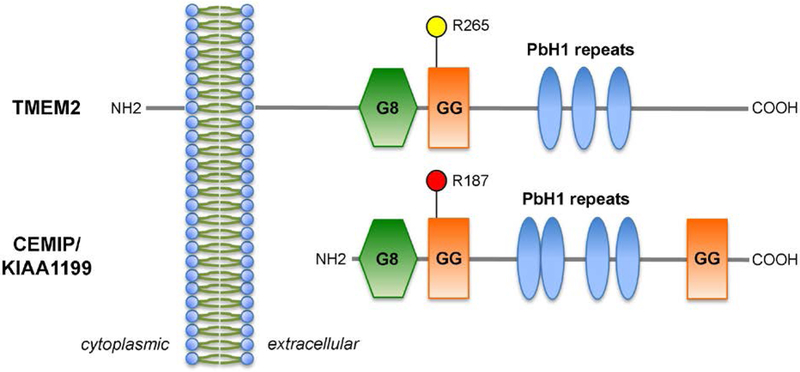
The domain composition of TMEM2 in comparison with CEMIP is shown as a cartoon in Figure 1. The type II transmembrane topology was confirmed by immunocytochemistry of live (unpermeabilized) cells and by a cell surface biotinylation assay (41). The N-terminal domain, which resides in the cytoplasm, is 82 amino acids long, whereas the extracellular C-terminal domain is 1278 amino acids and contains one G8 domain (47), and one GG domain (48), and three parallel β-helix (PbH1) repeats.
Figure 1. Domain structures of TMEM2 and CEMIP.
A red paddle in CEMIP/KIAA1199 indicates the position of mutations (R187C, R187H) identified in cases of non-syndromic hearing loss (36). A yellow paddle in TMEM2 indicates the position (R265) corresponding to R187 in CEMIP. For the sequence similarity in this region, refer to Figure 3 in Yamamoto et al. (41).
PbH1 sequences are the basis for the formation of the right-handed parallel β-helix folds (49). Several bacterial polysaccharide degrading enzymes exhibit this structure (50), among them, the pectate lyase PelC from Erwinia chrysanthemi (51) and chondroitinase B from Flavobacterium heparinum (52). Other than TMEM2 and CEMIP, fibrocystin, which is encoded by the PKHD1 gene, is a notable mammalian protein containing PbH1 repeats. PKHD1 is the causative gene for autosomal recessive polycystic kidney disease (53). None of the HYAL family hyaluronidases contain the PbH1 repeats. Most PbH1-containing prokaryotic polysaccharide degrading enzymes are lyases, whereas HYAL family hyaluronidases are hydrolases (50), suggesting that, unlike HYAL family proteins, TMEM2 may degrade HA by a lytic mechanism.
Characteristics of TMEM2 hyaluronidase activity
In an assay with transfected 293T cells and gel filtration analysis of degraded HA products, TMEM2 depolymerizes high-molecular mass HA (1500 kDa average; ~7500 monosaccharides in length) into fragments of ~5 kDa (~25 monosaccharides in length) (41). Neither increasing TMEM2 expression levels nor use of smaller HA as substrates resulted in generation of HA fragment smaller than ~5 kDa (41). By comparison, HYAL2 has been reported to cleave HA into ~20 kDa fragments, whereas PH-20/SPAM1 degrades them into even smaller fragments (6). As noted above, HYAL family proteins degrade not only HA but also CS and DS (21). In contrast, the GAG degrading activity of TMEM2 is specific for HA: TMEM2 does not degrade CS-A, CS-C, CS-D, or DS (41).
Generally, bacterial polysaccharide lyases containing parallel β-helix folds utilize divalent cations as cofactors. This is also the case with TMEM2: in the absence of Ca2+, TMEM2 does not exhibit HA depolymerizing activity (41). As for HYALs, there are no reports on a requirement for divalent cations in HA depolymerization activities. TMEM2 contains a short segment of amino acid sequence that bears a similarity to the calcium coordination sites in Erwinia pectate lyase (41). Interestingly, this segment also contains an arginine residue (Arg-265), which positionally corresponds to one of the sites (Arg-187) of the deafness mutation in human CEMIP gene (30,36). Mutagenesis experiments revealed that Arg-265 as well as two aspartic acid residues (Asp-273 and Asp-286) in this segment of TMEM2 are important for HA depolymerization activity (41).
Recombinant soluble TMEM2 comprising only its extracellular domain shows a pH optimum in the range of pH 5–8, with the highest activity at pH 6 (41). TMEM2 loses activity below pH 5 and is totally inactive at pH 4. Such a pH optimum is consistent with the possibility that TMEM2 is enzymatically active in the extracellular environment but not in lysosomes. By comparison, HYALs generally exhibit more acidic pH optima, with an exception of Xenopus HYAL2, which is active at both acidic and physiological pH (54). An ancillary implication of the TMEM2 pH optimum experiments mentioned above is that TMEM2 can exert hyaluronidase activity by itself without participation of cellular endocytic pathways, as soluble TMEM2 can degrades HA in a cell free assay (41). This is in contrast to CEMIP, which requires participation of the cellular endocytic machinery for hyaluronidase activity (30,55).
Cells transfected with TMEM2 can degrade not only exogenous HA added to culture media but also substrate-bound HA at cell-substrate contact sites. This activity can be detected by in situ HA degradation assay, in which cells are plated on a substrate of fluorescein-labeled HA immobilized to amino-silanized glass (41). This mode of action is somewhat similar to that of transmembrane ECM-degrading proteinases, such as MT1-MMP (56,57).
TMEM2 expression
TMEM2 mRNA expression in various tissues and cells has been determined by absolute quantification of transcript copy numbers (41). Tmem2 is expressed in essentially all organs in adult mice, with copy numbers greater than 2 ×105 copies per μg total RNA. For example, Tmem2 mRNA is expressed at 3.0 ×105, 2.3 ×105, and 16.0 ×105 copies per μg total RNA in the heart, liver, and lung, respectively. In comparison, Hyal2 mRNA is expressed at 4.1 ×105, 7.3 ×105, and 1.4 ×105 copies per μg total RNA in these organs. In mice, Tmem2 expression levels are much higher than those of Cemip in most organs (41).
Absolute quantification of transcript copy numbers has recently been applied to various human cell types in our laboratory. As observed in adult mice, TMEM2 is generally more highly expressed than CEMIP in most cells tested (Table 1). One exception is primary skin fibroblasts, in which CEMIP is more highly expressed than TMEM2. In contrast, endothelial cells express TMEM2 robustly, whereas CEMIP expression is negligible. Strong TMEM2 expression in endothelial cells appears consistent with the fact that endothelial cells are the major site of HA degradation (4).
Table 1.
Transcript copy numbers of TMEM2, CEMIP, HYAL1, and HYAL2 in various human cells.
| Cell line | Origin/Property | TMEM2 | CEMIP | HYAL1 | HYAL2 |
|---|---|---|---|---|---|
| HT1080 | Fibrosarcoma | 1.66 × 107 | 2.02 × 106 | 1.61 × 104 | 3.52 × 106 |
| RWPE1 | Normal prostate epithelium; non-invasive | 2.31 × 107 | 3.12 × 105 | 3.22 × 105 | 1.85 × 107 |
| RWPE2 | Normal prostate epithelium; invasive | 2.91 × 107 | 1.49 × 106 | 5.56 × 105 | 2.72 × 107 |
| LNCAP | Prostate cancer; low metastatic | 3.44 × 107 | 2.22 × 105 | 1.70 × 105 | 3.72 × 107 |
| PC3 | Prostate cancer; high metastatic | 2.65 × 107 | 4.15 × 106 | 3.25 × 105 | 5.36 × 107 |
| MDA-MB-157 | Mammary medullary carcinoma | 2.02 × 107 | 2.07 × 106 | 1.18 × 104 | 3.73 × 106 |
| MDA-MB-231 | Mammary ductal carcinoma | 3.10 × 107 | 1.69 × 107 | 2.71.× 104 | 7.85 × 106 |
| Primary skin fibroblast | 1.91 × 106 | 1.09 × 107 | ND | ND | |
| Primary dermal microvascular endothelial cells (HDMEC) | 6.96 × 106 | 1.64 × 104 | ND | ND |
Transcript copy numbers were determined by TaqMan gene expression assay with standard curves generated from reference plasmids (41).
Data represent transcript copy numbers per μg total RNA.
ND, not determined.
Thus far, the analysis of spatial expression of TMEM2 in mammalian tissues is based on in situ hybridization. For this, we have used the RNAscope in situ hybridization method (58) and observed strong Tmem2 expression in brain, heart, liver, limb bud, and branchial arch of E10.5 mouse embryos (Fig. 2). While in situ hybridization is informative, there is an acute need for high quality TMEM2 antibodies that can be used in various applications, including immunocytochemistry, immunohistochemistry, and cell sorting, to further define the function and regulation of TMEM2.
Figure 2. Expression of Tmem2 in E10.5 mouse embryos.
In situ hybridization was performed using a custom probe for mouse Tmem2 and the RNAscope 2.5 HD Reagents Kit-RED (Advanced Cell Diagnostics, Newark, CA). This image is a montage of four microphotographs assembled manually. FB, forebrain; MB, midbrain; HB, hindbrain; BA, first branchial arch; Ht, heart; Lv, liver; LB, hindlimb bud.
Developmental roles of TMEM2.
Developmental roles of TMEM2 have been studied using zebrafish, and these studies have provided some intriguing insights into its physiological functions. In 2011, two groups reported simultaneously the identification of zebrafish tmem2 mutants. Totong et al. (43) isolated the wickham (wkm) mutant from an ENU (N-ethyl-N-nitrosourea) mutagenesis screen for defective heart looping. Smith et al. (44) identified the frozen ventricle (frv) allele as a recessive lethal mutant through routine intercrosses. Positional cloning showed that both wkm and frv encode the zebrafish ortholog of TMEM2. Both wkm and frv mutants show abnormal heart looping accompanied by diminished constriction of the atrioventricular canal, suggesting that zebrafish TMEM2 protein functions in endocardial and myocardial morphogenesis (43,44). TMEM2 also functions in skeletal muscle morphogenesis. Zebrafish tmem2 mutants exhibit muscle fiber detachment, plus impaired laminin organization and ineffective fibronectin degradation at the myotendinous junction (59), suggesting that TMEM2 regulates cell-matrix interactions. Since these studies were conducted prior to demonstration of TMEM2 as a hyaluronidase, it will be interesting to see if some or all of these phenotypes can be explained by the premise that TMEM2 is a cell surface hyaluronidase.
More recently, tmem2 was shown to be required for sprouting angiogenesis in zebrafish, and angiogenesis-associated phenotypes were rescued by injection of Streptomyces hyaluronidase or HA oligosaccharides (60). Zebrafish tmem2 mutants also exhibit HA accumulation in heart chambers and surrounding developing blood vessels (44,60), observations consistent with the idea that TMEM2 protein is a hyaluronidase. With regard to the angiogenesis phenotype, De Angelis et al. (60) suggest that HA oligosaccharides, which are presumably generated as degradation products of TMEM2, act upstream of the VEGF receptor and enhance VEGF signaling. This implies that TMEM2 may exert a dual effect in this developmental context, namely to dissolve high molecular weight HA that has acted as space-filling substance and to generate bioactive HA oligomers that promote VEGF signaling.
TMEM2 and cancer
Strong evidence supports an association of CEMIP overexpression with cancer progression and poor prognosis (37–40,61,62). On the other hand, much less is known about TMEM2 involvement in human cancer. Nevertheless, it is interesting that TMEM2 is a SOX4-regulated genes in breast cancer (63). SOX4 is overexpressed in various human cancers and is thought to play a key role in their metastasis (64). Lee et al. (63) found that TMEM2 is one of the 24 direct transcriptional targets of SOX2 in human breast cancer, and that TMEM2 expression positively correlates with reduced overall survival. TMEM2 knockdown significantly decreases metastasis of MDA-LM2 breast cancer cells in an orthotopic xenograft model (63). It is not yet known whether the metastasis-promoting effect of TMEM2 is requires hyaluronidase activity, and this possibility remains to be investigated.
Questions to be resolved and future directions
Biochemically, a key remaining question is the catalytic mechanism of TMEM2. As noted, TMEM2 may depolymerize HA by a lytic mechanism, as do bacterial polysaccharide lyases. Determination of the crystal structure of TMEM2 should address this issue and shed light on structure-function relationships. Also, it remains unknown whether the TMEM2 extracellular domain has a function other than that of a hyaluronidase. In theory, it could serve as a receptor for extracellular ligands, such as growth factors and cytokines, although little evidence currently supports this possibility. There is also little known about the function of the TMEM2 cytoplasmic domain or whether it interacts with intracellular protein(s), which might modulate TMEM2 hyaluronidase activity by inside-out signaling.
Given the identification of TMEM2 as a novel cell surface hyaluronidase, it would be necessary to revise the current model of cellular HA degradation, which assumes that HYAL2 is the primary cell surface hyaluronidase. Regarding the relationship between TMEM2 and HYAL2, several possibilities can be envisioned: (i) TMEM2 is solely responsible for cell surface cleavage of large extracellular HA, while HYAL2 functions in endosomes and lysosomes; (ii) TMEM2 and HYAL2 both function as cell surface hyaluronidases but in different membrane domains (such as apical and basal membranes, lipid rafts, caveolae, etc.); (iii) TMEM2 and HYAL2 both function as cell surface hyaluronidases but under different biological/pathological contexts (e.g., proliferation, migration, cell division, tumor, or inflammation) or in different cell types; (iv) TMEM2 and HYAL2 function in HA degradation on the cell surface sequentially — for example, large HA is first cleaved by TMEM2 and then further degraded into smaller fragments by HYAL2, or vice versa. These hypotheses are not mutually exclusive, and other models are possible. A critical experiment would be a direct comparison of intrinsic HA degrading activities of TMEM2 and HYAL2 in a cell-free system. Another informative experiment would be to knockdown (or knockout) TMEM2 and HYAL2 individually in cells that express both proteins at a similar level and examine its effect on HA degradation. Ultimately, physiological roles of TMEM2 and HYAL2 in various tissues and developmental processes should be defined by comparing phenotypes of Tmem2 and Hyal2 mutant mice and their compound mutants.
The functional relationship of TMEM2 and CEMIP is another issue that needs to be addressed. Considering their structural similarity, it is perplexing that CEMIP protein itself does not exhibit hyaluronidase activity, although transfection of CEMIP confers a significant ability to cleave HA to transfected cells (30). The question emerges: what hyaluronidase molecule cleaves HA in these cells? Is it TMEM2, HYAL2, or different factor? Alternatively, CEMIP could undergo structural changes to an enzymatically active form when associated with an unknown cellular “cofactor.”
In conclusion, the biochemical and cell biological properties of TMEM2 suggest that it could be the long sought-after hyaluronidase that cleaves extracellular HA on the cell surface, and its identification could resolve numerous unanswered questions regarding not only HA catabolism but also HA research in general.
Acknowledgments
This work was supported in part by National Institutes of Health Grant R01 AR062692.
The abbreviations used are:
- HA
hyaluronan
- CS
chondroitin sulfate
- DS
dermatan sulfate
- ECM
extracellular matrix
References
- 1.Fraser JR, and Laurent TC (1989) Turnover and metabolism of hyaluronan. Ciba Found Symp 143, 41–53; discussion 53–49, 281–285 [PubMed] [Google Scholar]
- 2.Stern R (2003) Devising a pathway for hyaluronan catabolism: are we there yet? Glycobiology 13, 105R–115R [DOI] [PubMed] [Google Scholar]
- 3.Stern R, Kogan G, Jedrzejas MJ, and Soltes L (2007) The many ways to cleave hyaluronan. Biotechnol Adv 25, 537–557 [DOI] [PubMed] [Google Scholar]
- 4.Lepperdinger G, Fehrer C, and Reitinger S (2004) Biodegradation of hyaluronan in Chemistry and biology of hyaluronan (Garg HG, and Hales CA eds.), Elsevier; pp 71–82 [Google Scholar]
- 5.Frost GI, Csoka AB, Wong T, and Stern R (1997) Purification, cloning, and expression of human plasma hyaluronidase. Biochem Biophys Res Commun 236, 10–15 [DOI] [PubMed] [Google Scholar]
- 6.Lepperdinger G, Strobl B, and Kreil G (1998) HYAL2, a human gene expressed in many cells, encodes a lysosomal hyaluronidase with a novel type of specificity. J Biol Chem 273, 22466–22470 [DOI] [PubMed] [Google Scholar]
- 7.Csoka AB, and Stern R (2013) Hypotheses on the evolution of hyaluronan: a highly ironic acid. Glycobiology 23, 398–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherr GN, Yudin AI, and Overstreet JW (2001) The dual functions of GPI-anchored PH-20: hyaluronidase and intracellular signaling. Matrix Biol 20, 515–525 [DOI] [PubMed] [Google Scholar]
- 9.Moreno RD, and Alvarado CP (2006) The mammalian acrosome as a secretory lysosome: new and old evidence. Mol Reprod Dev 73, 1430–1434 [DOI] [PubMed] [Google Scholar]
- 10.Puissant E, Gilis F, Dogne S, Flamion B, Jadot M, and Boonen M (2014) Subcellular trafficking and activity of Hyal-1 and its processed forms in murine macrophages. Traffic 15, 500–515 [DOI] [PubMed] [Google Scholar]
- 11.Harada H, and Takahashi M (2007) CD44-dependent intracellular and extracellular catabolism of hyaluronic acid by hyaluronidase-1 and −2. J Biol Chem 282, 5597–5607 [DOI] [PubMed] [Google Scholar]
- 12.Chow G, Knudson CB, and Knudson W (2006) Expression and cellular localization of human hyaluronidase-2 in articular chondrocytes and cultured cell lines. Osteoarthritis Cartilage 14, 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow G, Knudson CB, and Knudson W (2006) Human hyaluronidase-2 is localized intracellularly in articular chondrocytes and other cultured cell lines. Osteoarthritis Cartilage 14, 1312–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rai SK, Duh FM, Vigdorovich V, Danilkovitch-Miagkova A, Lerman MI, and Miller AD (2001) Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc Natl Acad Sci U S A 98, 4443–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andre B, Duterme C, Van Moer K, Mertens-Strijthagen J, Jadot M, and Flamion B (2011) Hyal2 is a glycosylphosphatidylinositol-anchored, lipid raft-associated hyaluronidase. Biochem Biophys Res Commun 411, 175–179 [DOI] [PubMed] [Google Scholar]
- 16.Csoka AB, Frost GI, and Stern R (2001) The six hyaluronidase-like genes in the human and mouse genomes. Matrix Biol 20, 499–508 [DOI] [PubMed] [Google Scholar]
- 17.Hida D, Danielson BT, Knudson CB, and Knudson W (2015) CD44 knock-down in bovine and human chondrocytes results in release of bound HYAL2. Matrix Biol 48, 42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Afify AM, Stern M, Guntenhoner M, and Stern R (1993) Purification and characterization of human serum hyaluronidase. Arch Biochem Biophys 305, 434–441 [DOI] [PubMed] [Google Scholar]
- 19.Vigdorovich V, Strong RK, and Miller AD (2005) Expression and characterization of a soluble, active form of the jaagsiekte sheep retrovirus receptor, Hyal2. J Virol 79, 79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jadin L, Wu X, Ding H, Frost GI, Onclinx C, Triggs-Raine B, and Flamion B (2008) Skeletal and hematological anomalies in HYAL2-deficient mice: a second type of mucopolysaccharidosis IX? FASEB J 22, 4316–4326 [DOI] [PubMed] [Google Scholar]
- 21.Stern R, and Jedrzejas MJ (2006) Hyaluronidases: their genomics, structures, and mechanisms of action. Chem Rev 106, 818–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada S (2015) Role of hyaluronidases in the catabolism of chondroitin sulfate. Adv Exp Med Biol 842, 185–197 [DOI] [PubMed] [Google Scholar]
- 23.Triggs-Raine B, Salo TJ, Zhang H, Wicklow BA, and Natowicz MR (1999) Mutations in HYAL1, a member of a tandemly distributed multigene family encoding disparate hyaluronidase activities, cause a newly described lysosomal disorder, mucopolysaccharidosis IX. Proc Natl Acad Sci U S A 96, 6296–6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Natowicz MR, Short MP, Wang Y, Dickersin GR, Gebhardt MC, Rosenthal DI, Sims KB, and Rosenberg AE (1996) Clinical and biochemical manifestations of hyaluronidase deficiency. N Engl J Med 335, 1029–1033 [DOI] [PubMed] [Google Scholar]
- 25.Martin DC, Atmuri V, Hemming RJ, Farley J, Mort JS, Byers S, Hombach-Klonisch S, Csoka AB, Stern R, and Triggs-Raine BL (2008) A mouse model of human mucopolysaccharidosis IX exhibits osteoarthritis. Hum Mol Genet 17, 1904–1915 [DOI] [PubMed] [Google Scholar]
- 26.Chowdhury B, Hemming R, Hombach-Klonisch S, Flamion B, and Triggs-Raine B (2013) Murine hyaluronidase 2 deficiency results in extracellular hyaluronan accumulation and severe cardiopulmonary dysfunction. J Biol Chem 288, 520–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bourguignon V, and Flamion B (2016) Respective roles of hyaluronidases 1 and 2 in endogenous hyaluronan turnover. FASEB J 30, 2108–2114 [DOI] [PubMed] [Google Scholar]
- 28.Muggenthaler MM, Chowdhury B, Hasan SN, Cross HE, Mark B, Harlalka GV, Patton MA, Ishida M, Behr ER, Sharma S, Zahka K, Faqeih E, Blakley B, Jackson M, Lees M, Dolinsky V, Cross L, Stanier P, Salter C, Baple EL, Alkuraya FS, Crosby AH, Triggs-Raine B, and Chioza BA (2017) Mutations in HYAL2, Encoding Hyaluronidase 2, Cause a Syndrome of Orofacial Clefting and Cor Triatriatum Sinister in Humans and Mice. PLoS Genet 13, e1006470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eicher JD, Chen MH, Pitsillides AN, Lin H, Veeraraghavan N, Brody JA, Metcalf GA, Muzny DM, Gibbs RA, Becker DM, Becker LC, Faraday N, Mathias RA, Yanek LR, Boerwinkle E, Cupples LA, and Johnson AD (2017) Whole exome sequencing in the Framingham Heart Study identifies rare variation in HYAL2 that influences platelet aggregation. Thromb Haemost 117, 1083–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida H, Nagaoka A, Kusaka-Kikushima A, Tobiishi M, Kawabata K, Sayo T, Sakai S, Sugiyama Y, Enomoto H, Okada Y, and Inoue S (2013) KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc Natl Acad Sci U S A 110, 5612–5617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evensen NA, Li Y, Kuscu C, Liu J, Cathcart J, Banach A, Zhang Q, Li E, Joshi S, Yang J, Denoya PI, Pastorekova S, Zucker S, Shroyer KR, and Cao J (2015) Hypoxia promotes colon cancer dissemination through up-regulation of cell migration-inducing protein (CEMIP). Oncotarget 6, 20723–20739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shostak K, Zhang X, Hubert P, Goktuna SI, Jiang Z, Klevernic I, Hildebrand J, Roncarati P, Hennuy B, Ladang A, Somja J, Gothot A, Close P, Delvenne P, and Chariot A (2014) NF-kappaB-induced KIAA1199 promotes survival through EGFR signalling. Nat Commun 5, 5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evensen NA, Kuscu C, Nguyen HL, Zarrabi K, Dufour A, Kadam P, Hu YJ, Pulkoski-Gross A, Bahou WF, Zucker S, and Cao J (2013) Unraveling the role of KIAA1199, a novel endoplasmic reticulum protein, in cancer cell migration. J Natl Cancer Inst 105, 1402–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimoda M, Yoshida H, Mizuno S, Hirozane T, Horiuchi K, Yoshino Y, Hara H, Kanai Y, Inoue S, Ishijima M, and Okada Y (2017) Hyaluronan-Binding Protein Involved in Hyaluronan Depolymerization Controls Endochondral Ossification through Hyaluronan Metabolism. Am J Pathol 187, 1162–1176 [DOI] [PubMed] [Google Scholar]
- 35.Yoshino Y, Ishisaka M, Tsuruma K, Shimazawa M, Yoshida H, Inoue S, Shimoda M, Okada Y, and Hara H (2017) Distribution and function of hyaluronan binding protein involved in hyaluronan depolymerization (HYBID, KIAA1199) in the mouse central nervous system. Neuroscience 347, 1–10 [DOI] [PubMed] [Google Scholar]
- 36.Abe S, Usami S, and Nakamura Y (2003) Mutations in the gene encoding KIAA1199 protein, an inner-ear protein expressed in Deiters’ cells and the fibrocytes, as the cause of nonsyndromic hearing loss. J Hum Genet 48, 564–570 [DOI] [PubMed] [Google Scholar]
- 37.Fink SP, Myeroff LL, Kariv R, Platzer P, Xin B, Mikkola D, Lawrence E, Morris N, Nosrati A, Willson JK, Willis J, Veigl M, Barnholtz-Sloan JS, Wang Z, and Markowitz SD (2015) Induction of KIAA1199/CEMIP is associated with colon cancer phenotype and poor patient survival. Oncotarget 6, 30500–30515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jami MS, Hou J, Liu M, Varney ML, Hassan H, Dong J, Geng L, Wang J, Yu F, Huang X, Peng H, Fu K, Li Y, Singh RK, and Ding SJ (2014) Functional proteomic analysis reveals the involvement of KIAA1199 in breast cancer growth, motility and invasiveness. BMC Cancer 14, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuzaki S, Tanaka F, Mimori K, Tahara K, Inoue H, and Mori M (2009) Clinicopathologic significance of KIAA1199 overexpression in human gastric cancer. Ann Surg Oncol 16, 2042–2051 [DOI] [PubMed] [Google Scholar]
- 40.Tiwari A, Schneider M, Fiorino A, Haider R, Okoniewski MJ, Roschitzki B, Uzozie A, Menigatti M, Jiricny J, and Marra G (2013) Early insights into the function of KIAA1199, a markedly overexpressed protein in human colorectal tumors. PLoS One 8, e69473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto H, Tobisawa Y, Inubushi T, Irie F, Ohyama C, and Yamaguchi Y (2017) A mammalian homolog of the zebrafish Transmembrane Protein 2 (TMEM2) is the long-sought-after cell surface hyaluronidase. J Biol Chem [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wrzesinski T, Szelag M, Cieslikowski WA, Ida A, Giles R, Zodro E, Szumska J, Pozniak J, Kwias Z, Bluyssen HA, and Wesoly J (2015) Expression of pre-selected TMEMs with predicted ER localization as potential classifiers of ccRCC tumors. BMC Cancer 15, 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Totong R, Schell T, Lescroart F, Ryckebusch L, Lin YF, Zygmunt T, Herwig L, Krudewig A, Gershoony D, Belting HG, Affolter M, Torres-Vazquez J, and Yelon D (2011) The novel transmembrane protein Tmem2 is essential for coordination of myocardial and endocardial morphogenesis. Development 138, 4199–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith KA, Lagendijk AK, Courtney AD, Chen H, Paterson S, Hogan BM, Wicking C, and Bakkers J (2011) Transmembrane protein 2 (Tmem2) is required to regionally restrict atrioventricular canal boundary and endocardial cushion development. Development 138, 4193–4198 [DOI] [PubMed] [Google Scholar]
- 45.Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, Calabro A Jr., Kubalak S, Klewer SE, and McDonald JA (2000) Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest 106, 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camenisch TD, Biesterfeldt J, Brehm-Gibson T, Bradley J, and McDonald JA (2001) Regulation of cardiac cushion development by hyaluronan. Exp Clin Cardiol 6, 4–10 [PMC free article] [PubMed] [Google Scholar]
- 47.He QY, Liu XH, Li Q, Studholme DJ, Li XW, and Liang SP (2006) G8: a novel domain associated with polycystic kidney disease and non-syndromic hearing loss. Bioinformatics 22, 2189–2191 [DOI] [PubMed] [Google Scholar]
- 48.Guo J, Cheng H, Zhao S, and Yu L (2006) GG: a domain involved in phage LTF apparatus and implicated in human MEB and non-syndromic hearing loss diseases. FEBS Lett 580, 581–584 [DOI] [PubMed] [Google Scholar]
- 49.Jenkins J, Mayans O, and Pickersgill R (1998) Structure and evolution of parallel beta-helix proteins. J Struct Biol 122, 236–246 [DOI] [PubMed] [Google Scholar]
- 50.Garron ML, and Cygler M (2014) Uronic polysaccharide degrading enzymes. Curr Opin Struct Biol 28, 87–95 [DOI] [PubMed] [Google Scholar]
- 51.Scavetta RD, Herron SR, Hotchkiss AT, Kita N, Keen NT, Benen JA, Kester HC, Visser J, and Jurnak F (1999) Structure of a plant cell wall fragment complexed to pectate lyase C. Plant Cell 11, 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang W, Matte A, Li Y, Kim YS, Linhardt RJ, Su H, and Cygler M (1999) Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution. J Mol Biol 294, 1257–1269 [DOI] [PubMed] [Google Scholar]
- 53.Ward CJ, Hogan MC, Rossetti S, Walker D, Sneddon T, Wang X, Kubly V, Cunningham JM, Bacallao R, Ishibashi M, Milliner DS, Torres VE, and Harris PC (2002) The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat Genet 30, 259–269 [DOI] [PubMed] [Google Scholar]
- 54.Lepperdinger G, Mullegger J, and Kreil G (2001) Hyal2--less active, but more versatile? Matrix Biol 20, 509–514 [DOI] [PubMed] [Google Scholar]
- 55.Yoshida H, Nagaoka A, Nakamura S, Tobiishi M, Sugiyama Y, and Inoue S (2014) N-Terminal signal sequence is required for cellular trafficking and hyaluronan-depolymerization of KIAA1199. FEBS Lett 588, 111–116 [DOI] [PubMed] [Google Scholar]
- 56.Galvez BG, Matias-Roman S, Albar JP, Sanchez-Madrid F, and Arroyo AG (2001) Membrane type 1-matrix metalloproteinase is activated during migration of human endothelial cells and modulates endothelial motility and matrix remodeling. J Biol Chem 276, 37491–37500 [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, and McNiven MA (2012) Invasive matrix degradation at focal adhesions occurs via protease recruitment by a FAK-p130Cas complex. J Cell Biol 196, 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, and Luo Y (2012) RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14, 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ryckebusch L, Hernandez L, Wang C, Phan J, and Yelon D (2016) Tmem2 regulates cell-matrix interactions that are essential for muscle fiber attachment. Development 143, 2965–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Angelis JE, Lagendijk AK, Chen H, Tromp A, Bower NI, Tunny KA, Brooks AJ, Bakkers J, Francois M, Yap AS, Simons C, Wicking C, Hogan BM, and Smith KA (2017) Tmem2 Regulates Embryonic Vegf Signaling by Controlling Hyaluronic Acid Turnover. Dev Cell 40, 123–136 [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y, Jia S, and Jiang WG (2014) KIAA1199 and its biological role in human cancer and cancer cells (review). Oncol Rep 31, 1503–1508 [DOI] [PubMed] [Google Scholar]
- 62.Kuscu C, Evensen N, Kim D, Hu YJ, Zucker S, and Cao J (2012) Transcriptional and epigenetic regulation of KIAA1199 gene expression in human breast cancer. PLoS One 7, e44661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee H, Goodarzi H, Tavazoie SF, and Alarcon CR (2016) TMEM2 is a SOX4-regulated gene that mediates metastatic migration and invasion in breast cancer. Cancer Res 76, 4994–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J, Ju HL, Yuan XY, Wang TJ, and Lai BQ (2016) SOX4 is a potential prognostic factor in human cancers: a systematic review and meta-analysis. Clin Transl Oncol 18, 65–72 [DOI] [PubMed] [Google Scholar]