Abstract
Purpose:
Parallel signaling reduces the effects of receptor tyrosine kinase (RTK) targeted therapies in glioma. We hypothesized that inhibition of protein N-linked glycosylation, an endoplasmic reticulum co- and post-translational modification crucial for RTK maturation and activation, could provide a new therapeutic approach for glioma radiosensitization.
Experimental design:
We investigated the effects of a small molecule inhibitor of the oligosaccharyltransferase (NGI-1) on EGFR family receptors, MET, PDGFR, and FGFR1. The influence of glycosylation state on tumor cell radiosensitivity, chemotherapy induced cell toxicity, DNA damage, and cell cycle arrest were determined and correlated with glioma cell receptor expression profiles. The effects of NGI-1 on xenograft tumor growth were tested using a nanoparticle formulation validated by in vivo molecular imaging. A mechanistic role for RTK signaling was evaluated through the expression of a glycosylation independent CD8-EGFR chimera.
Results:
NGI-1 reduced glycosylation, protein levels, and activation of most RTKs. NGI-1 also enhanced the radiosensitivity and cytotoxic effects of chemotherapy in those glioma cells with elevated ErbB family activation, but not in cells without high levels of RTK activation. NGI-1 radiosensitization was associated with increases in both DNA damage and G1 cell cycle arrest. Combined treatment of glioma xenografts with fractionated radiation and NGI-1 significantly reduced tumor growth compared to controls. Expression of the CD8-EGFR eliminated NGI-1’s effects on G1 arrest, DNA damage, and cellular radiosensitivity, identifying RTK inhibition as the principal mechanism for the NGI-1 effect.
Conclusion:
This study suggests that OST inhibition with NGI-1 is a novel approach to radiosensitize malignant gliomas with enhanced RTK signaling.
Keywords: RTK, radiosensitization, Oligosaccharyltransferase, N-glycosylation, Glioma
INTRODUCTION
Receptor tyrosine kinases (RTK) are transmembrane glycoproteins that regulate downstream signaling involved in cell proliferation and survival. Receptor overexpression or activation caused by mutation is critical for the development and progression of many tumors including glioblastoma (GBM), an incurable malignant brain tumor (1, 2). RTKs such as ErbB2 and EGFR, as well as the RTK ligand, VEGF, have been successfully targeted in multiple tumor types both as monotherapies or combined with cytotoxic chemotherapy. In head and neck squamous cell carcinoma (HNSCC), EGFR inhibition has also been identified as an effective method for tumor cell radiosensitization (3–5). However, targeting RTKs in glioblastoma has produced limited clinical responses (6–8), questioning whether specific receptors are effective targets in this tumor type.
Parallel or bypass kinase signaling has been identified as a mechanism for resistance to therapeutics that target specific RTKs. For example, de-repression of PDGFRβ transcription has been implicated as an alternative signaling pathway that promotes acquired resistance to EGFR tyrosine kinase inhibitors in glioblastoma (9). The co-expression of EGFR and c-Met has also been shown to rescue survival signaling and counteract EGFR signaling blockade (10, 11). In addition, the dependence of downstream signaling on multiple co-expressed RTKs (12, 13), provides a further understanding of the limitations for targeting individual RTKs in GBM and emphasizes the need for therapeutic strategies that disrupt signaling through multiple RTK proteins.
N-linked glycosylation (NLG) is an endoplasmic reticulum (ER) co- and post-translational protein modification that has an important role in the assembly and maturation of cell surface glycoprotein receptors (reviewed in (14)). NLG is a conserved two-phase process in eukaryotic cells. It involves the assembly of an oligosaccharide on a lipid carrier followed by the transfer of the oligosaccharide to asparagine residues within a specific amino acid consensus sequence (NXS/T; where X can be any amino acid other than proline). The sequential assembly of the mature oligosaccharide is initiated on the ER cytoplasmic leaflet, completed in the ER lumen, and requires the activity of multiple enzymes and glycosyltransferases (15). The glycosylation reaction itself is catalyzed in the ER lumen by the oligosaccharyltransferase (OST), a multisubunit enzyme complex that exists in several isoforms (16). The glycosylation pathway therefore represents a potential upstream biosynthetic node for regulating the function of multiple cell surface receptors, including RTKs, and is therefore an attractive target for cancer biology investigations.
NLG is known to be essential for the function for several transmembrane receptors that are targets for cancer therapy, including the ErbB family members, VEGFR, and IGF-1R (17–19). Furthermore, we have previously demonstrated that inhibition of NLG with tunicamycin, an inhibitor of dolichyl-phosphate N-acetylglucosamine-phospho-transferase (DPAGT1), or siRNA/shRNA knockdown of MPI, an enzyme required for glycan precursor biosynthesis, blocks RTK function and enhances glioma cell radiosensitivity (20, 21).
Recently a high throughput screening campaign for small molecule inhibitors of NLG identified a novel aminobenzamidesulfonamide chemical series that disrupts the function of the OST. The lead compound from this group, NGI-1, targets the OST catalytic subunits through a direct and reversible interaction (22). Importantly, NGI-1 does not completely abolish all OST activity, producing incomplete inhibition of glycosylation that is associated with low toxicity. Because NGI-1 is predicted to alter the glycosylation and function of multiple RTKs we sought to investigate the effects of NGI-1 treatment in malignant glioma cells. Our work tests combinatorial effects of NGI-1 with both radiation or cytotoxic chemotherapy with the goal of understanding the underlying cellular mechanisms that are affected. Together this work evaluates the potential of OST inhibition as novel therapeutic strategy for the treatment of malignant glioma.
MATERIALS AND METHODS
Cell lines and pharmacological inhibitors
In this study, we used the D54, SKMG3, U251, T98G and 42-MG glioma cell lines. The source of D54, SKMG3 and U251 cells has been described previously (17, 23). The 42-MG and T98G cells were provided by Todd Waldman (Georgetown University) and Dr. Ranjit Bindra (Yale University), respectively. The cells were cultured in DMEM/F12 (D54 and T98G), DMEM (42-MG and U251) or RPMI 1640 (SKMG3) + 10% FBS supplemented with penicillin and streptomycin (Gibco, Life Technologies, Grand Island, NY, US) in a humidified incubator with 5% CO2, and they were kept in culture no more than 6 months after resuscitation from the original stocks. All cell lines used in the study were authenticated by the American Type Culture Collection (ATCC) STR profiling, other than SKMG3 which does not have a published STR profile but was confirmed to be of human origin and matched no other cell lines in the ATCC or DMSZ databases. Mycoplasma cell culture contamination was routinely checked and ruled out using a biochemical test (MycoAlert Mycoplasma Detection Kit from Lonza, Rockland, ME USA). Tunicamycin was purchased from Calbiochem/EMD-millipore (Burlington, MA, USA). Cetuximab and Erlotinib were purchased at Selleck Chemicals LLC (Houston, TX, USA). Luciferin was supplied by Promega (Madison, WI USA). For in vitro experiments, radiation (XRT) was administered at room temperature using a Precision X-ray 320-kV orthovoltage unit at a dose rate of 2 Gy/45 seconds (PXI X-Ray Systems) with 2 mm aluminum filter. For in vivo studies, radiation was administered at room temperature using a Precision X-ray 250-kV orthovoltage unit at a dose rate of 6.42 Gy/min (PXI X-Ray Systems) with 2 mm aluminum filter. Quality Assurance for both irradiators was peformed monthly using a P.T.W. 0.3cm3 Ionization Chamber calibrated to NIST standards and quarterly dosimetry using thermoluminescent dosimeter (TLD)-based or ferrous sulfate-based dosimeters.
Generation of CD8-EGFR cell lines
The CD8 cDNA was a gift from Paula Kavathas (Yale University). The CD8-EGFR was constructed according to a strategy that generated a constitutively active IGF-1R (24). The extracellular domain of CD8 was PCR amplified with a 5’ XbaI and 3’ SalI restriction site and cloned in-frame with the intracellular kinase domain of EGFR using the pDCB5-EGFR plasmid. SKMG3-CD8-EGFR cells were generated by Lipofectamine (Life Technologies) transfection of the plasmid followed by selection with 500 μg/ml G418. Expression of CD8-EGFR was validated by western blot analysis of the wild type and CD8-EGFR.
Radiation survival and Proliferation Assays
Clonogenic survival assays were performed with cells treated in the presence or absence of 10 μM NGI-1 48 hours before radiation and maintained until cells were re-plated. Radiation (XRT) doses of either 0, 2, 4, or 6 Gy were delivered with a Precision X-ray 320-kV orthovoltage unit at a dose rate of 2 Gy/45 seconds (PXI X-Ray Systems). Twenty-four hours following XRT cells were washed, trypsinized, and re-plated in triplicate wells to determine clonogenic survival. Cultures were grown for 14 days, washed once with 1x PBS, and stained with 0.25% crystal violet in 80% methanol. Colonies with >50 cells were counted and clonogenic cell survival differences for each treatment were compared using survival curves generated from the linear quadratic equation using GraphPad Prism7 (GraphPad Software Inc.). Growth rates were determined by CellTiter 96 Non-Radioactive Cell Proliferation Assay (Promega; Madison, WI, USA) according to the manufacturer’s directions. For each experiment one thousand cells were seeded in triplicate wells in 96-wells plates. The following day, cell cultures were treated with NGI-1 (1 μM), Temozolomide (TMZ, 10 μM), Etoposide (VP-16, 0.1 μM) or combinations for 5 days. Absorbance at 540 nm was measured using the spectrophotometric reading (BioTek Synergy 2; Winooski, VT USA).
Western blot analysis
Western blot analyses were performed as previously described (25). We used the following primary antibodies (See Supplementary Table 1). The nitrocellulose-bound primary antibodies, were detected with anti–rabbit IgG horseradish peroxidase–linked antibody or anti–mouse IgG horseradish peroxidase–linked antibody (EMD Millipore; Temecula, CA USA), and were detected by the enhanced chemoluminescence staining ECL (GE Healthcare–Amersham Pharmacia, Buckinghamshire, U.K.).
Immunofluorescence and Cell Cycle Analysis
To determine histone H2AX phosphorylation (γH2AX), cells were cultured on glass cover slips and pre-treated with either vehicle or NGI-1 for 48 hours followed by radiation treatment. Samples were fixed with 4% neutral-buffered formaldehyde, washed (0.1% triton in PBS for 30 minutes) and incubated for 1 hour with protein-blocking solution (PBS containing 10% normal goat–horse serum; Sigma-Aldrich, Saint Louis, MI, US). Next, the slides were incubated with primary anti-phospho-histone γH2AX S139 (1:500, Millipore-Upstate, Billerica, MA, US) followed by incubation with secondary antibody Alexa Fluor 555-conjugated goat anti-mouse IgG (1:750, Molecular probes/Invitrogen, Carlsbad, CA, US), for 1 hour at room temperature. Nuclei were stained using DAPI containing vectashield mounting solution (Vector Labs). Confocal cellular images were captured with an inverted Zeiss LSM 510 Pascal laser confocal microscope (C. Zeiss, Jenna, Germany), using a 63/1.4 plan-apochromat objective. Five randomly selected fields s per slide were analyzed. Cells were counted using the ImageJ program, public domain Java image processing software (http://rsb.info.nih.gov/ij/).
Cell cycle distribution was determined following treatment with vehicle, radiation and/or 10uM NGI-1. Cells were trypsinized and centrifuged, washed once with ice-cold PBS, fixed with ice-cold 70% ethanol and stored overnight at −20 °C. After washing twice with PBS, they were incubated for 30 minutes at room temperature in 200 μL of Guava Cell Cycle Reagent (Guava Technologies). Cytofluorometric acquisitions were performed on a LSRII cytometer (BD Biosciences). First-line analysis was performed with FlowJo software, upon gating of the events characterized by normal forward and side scatter parameters and discrimination of doublets in a FSC-A vs. FSC-H bivariate plot. Approximately 50,000 cells were analyzed per experiment.
NGI-1 nanoparticle (NP) preparation and Evaluation
Polyethylene glycol (PEG)-b-Polylactic acid (PLA) nanoparticles were synthesized using diblock polymer (Mw PEG = 5 kDa, Mw PLA = 10 kDa; Polysciences, Inc. Warrington, PA, USA) and Polyethyeleneimine (PEI; branched – average Mw ~800, average Mn ~600; Sigma-Aldrich, USA) using a nanoprecipitation technique, similar to one previously reported (26). For control NPs 100 mg of polymer was dissolved in 5 ml DMSO at RT for 2 hours and a 200 ul aliquot was added drop-wise to 1 mL deionized (DI) water under strong vortex to create a NP suspension. These suspensions were immediately diluted 5x with DI wàter and transferred to an Amicon Ultracell 100k centrifugal filter unit, and centrifuged at 4000 g, 4°C for 30 minutes. NPs were washed three times with DI water to achieve a final concentration of 100 mg NP/mL DI wàter and snap-frozen at − 80°C until use. For drug-loaded NPs, NGI-1 was dissolved in DMSO at a concentration of 50 mg/ml, and PEI was dissolved in DMSO at 50 mg/ml. The NGI-1 and PEI solutions were then mixed at a 6:1 ratio (by weight) of PEI:drug. The solution was vortexed for ~10 seconds and then incubated at room temperature for 15 minutes. After the incubation period, the PEI/NGI-1 solution was then added to the PLA-PEG solution at a 10% ratio of NGI-1:PLA-25 PEG by weight. This combined solution was briefly vortexed and then water-bath sonicated to ensure uniform mixing. The solution was then added dropwise to DI water under vortex at a final ratio of 1:5 organic:aqueous phase. All NP preparations were tested for particle size distribution by dynamic light scattering (DLS) using a Malvern Nano-ZS (Malvern Instruments).
All experimental procedures were approved in accordance with IACUC and Yale University institutional guidelines for animal care and ethics and guidelines for the welfare and use of animals in cancer research (27). NGI-1 delivery to glioma xenografts was evaluated using a bioluminescent imaging platform that detects inhibtion of NLG (17). Ten days after subcutaneous injection of 1 ×107 gliomas cells, mice bearing palpable tumors were treated with control or NGI-1 NPs (20 mg/kg), or tunicamycin 1mg/kg and imaged 5–30 minutes after delivery of i.p. luciferin (150 mg/kg). Signal intensity was quantified for a region of interest (ROI) encompassing each tumor and induction of bioluminscence was calculated by comparing peak bioluminescent activity from pre- and post-treatment imaging at 24 and 48 hours.
NGI-1 Therapeutic Studies in Glioma Xenografts:
D54 and SKMG3 bilateral xenografts were established in nude mice by subcutaneous injection of 1×106 cells into hind limb. Four days after injection, mice were randomized to one of four treatment groups. They received either control or NGI-1 NPs i.v. (20mg/kg) every other day for a total of 3 doses and either sham irradiation or a total of 10 Gy administered in daily 2 Gy fractions using a Precision X-ray 250-kV orthovoltage unit. Tumor size was measured two times per week and calculated according to the formula π/6 × (large diameter) × (small diameter)2.
Statistical analysis
Results are expressed as mean ± standard error (S.E.) unless otherwise indicated. The Statistical Package for Social Sciences (SPSS, version 20.0) was used for data analysis. Statistically significant differences in between-group comparisons were defined at a significance level of P-value ≤ 0.05 in the Mann-Whitney test.
RESULTS
NGI-1 disrupts RTK signaling in glioblastoma cells.
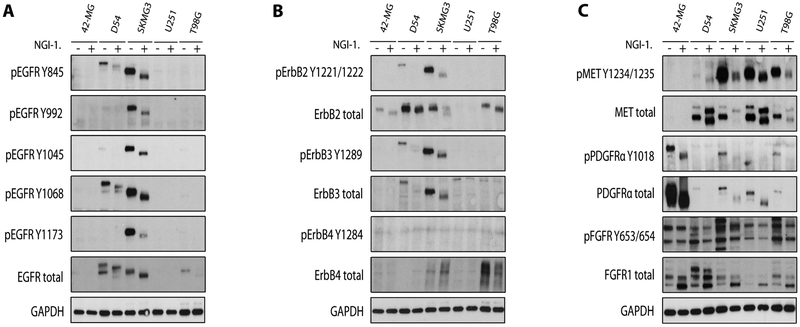
Molecular studies of glioblastoma tumors have identified increased RTK expression (28, 29). Because these receptors are highly glycosylated, we tested the effects of a novel small molecule inhibitor of glycosylation (NGI-1) on RTK glycosylation and activation. We observed that SKMG3 and D54 cell lines had high expression and activation levels of ErbB family members including EGFR, ErbB2, and ErbB3 (Figure 1A and 1B). NGI-1 reduced glycosylation, determined by increased protein gel mobility on western blot, as well as phosphorylation of EGFR, ErbB2, and ErbB3. The reduced EGFR phosphorylation was not caused by decreases in EGFR protein levels, contrary to our prior observations with tunicamycin (20). However, we did observe a reduction in ErbB2 protein levels in SKMG3, as well as a reduction of ErbB3 protein levels in both D54 and SKMG3 cell lines.
Figure 1. NGI-1 reduces RTK glycosylation and phosphorylation in glioma cell lines.
(a.) Western blots demonstrating changes in EGFR molecular weight and phosphorylation after 10 μM NGI-1 treatment for 48 hours in 42-MG, D54, SKMG3, T98G and U251 cell lines. (b.) ErbB2, ErbB3, and ErbB4 phosphorylation and expression, and (c.) MET, PDGFR, and FGFR1 phosphorylation and expression. GADPH was used as a loading control. Representative data from two independent experiments are shown.
To further characterize the effect of NGI-1 on these glioma cell lines, we investigated the phosphorylation levels of MET, PDGFR and FGFR RTKs (Figure 1C). We found that 42-MG, D54, SKMG3, U251 and T98G cell lines each had a distinct profile of receptor expression and activation. Unlike the ErbB family of RTKs, expression levels did not directly correlate with the phosphorylation of these receptors. Regardless, NGI-1 uniformly reduced phosphorylation of MET in D54, SKMG3, U251, and T98G cells, and phosphorylation of PDGFR in 42-MG, SKMG3, and T98G cells. This reduction of phosphorylation was observed to be independent of the effects of NGI-1 on RTK protein levels. Surprisingly FGFR1 levels were increased by NGI-1, although this was not accompanied by an increase in receptor phosphorylation.
NGI-1 radiosensitizes gliobastoma cell lines with activated ErbB receptors.
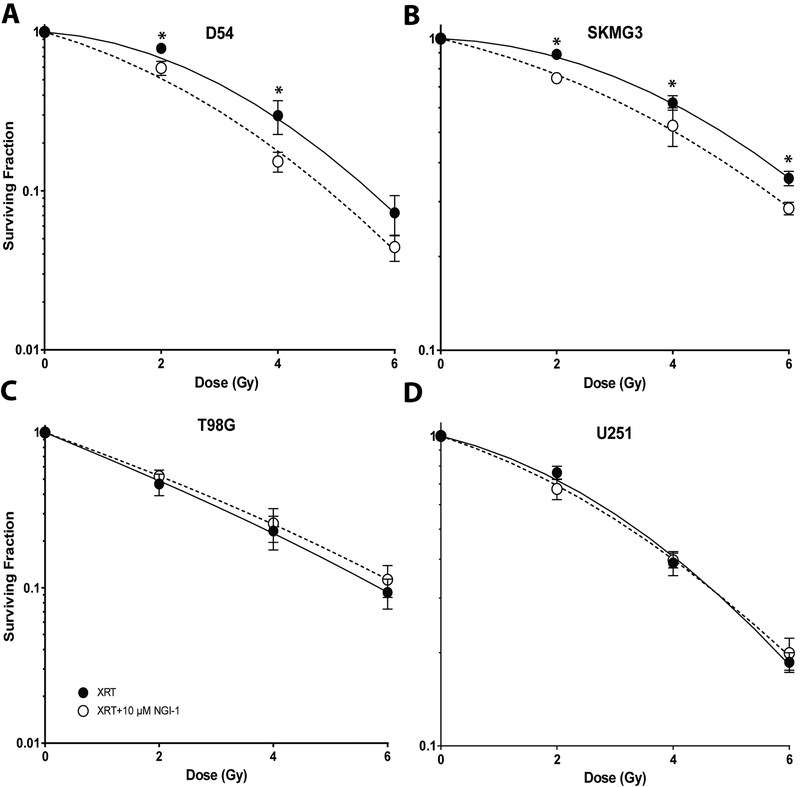
RTK activation protects tumor cells from radiation-induced cell death (30–32). We therefore investigated the effects of NGI-1 on glioma cell radiosensitivity using clonogenic survival analysis (Figure 2). We found that D54 and SKMG3 cell lines, both with significant ErbB family RTK activation, were significantly radiosensitized by NGI-1 at each dose tested (Figure 2A and 2B; p < 0.05). The survival fraction at 2 Gy (SF2Gy) was reduced from 69% to 51% for D54 cells and from 87% to 77% for SKMG3 cells. The dose enhancement factors (DEF) at a surviving fraction of 0.4 for D54 and SKMG3 were 1.3 and 1.2, respectively. In contrast, for both T98G and U251 cells the SF2Gy was unaffected by NGI-1 (Figure 2C and 2D). The correlation of ErbB family RTK signaling with the radiation response suggests that NGI-1 radiosensitizes glioma cells by disrupting ErbB RTK signaling.
Figure 2. NGI-1 radiosensitization in glioma cell lines.
Clonogenic survival of (a.) D54, (b.) SKMG3, (c.) T98G, and (d.) U251 cells treated with vehicle or 10 μM NGI-1. The results represent data from three independent experiments for each cell line. Data are represented as the mean ± standard error. An * indicates a significant difference (p≤ 0.05) compared to radiation alone.
NGI-1 enhances cytotoxic chemotherapy in glioma
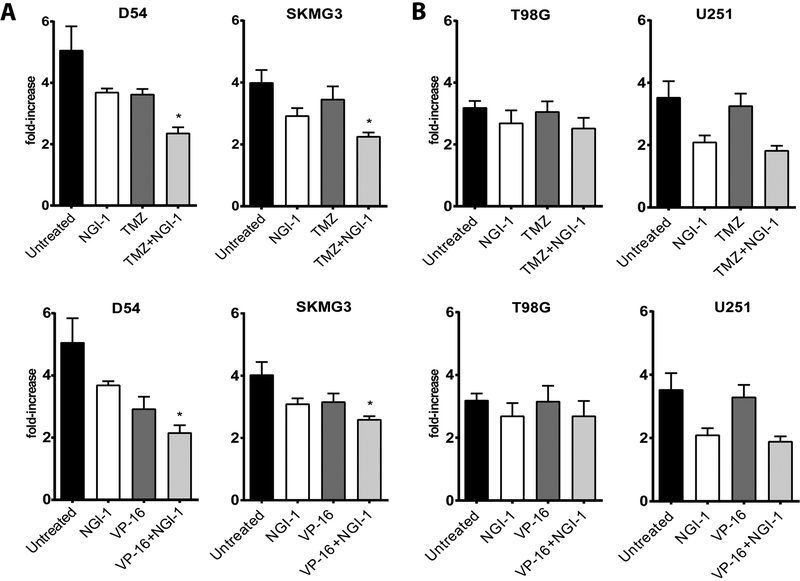
To investigate the effects of NGI-1 in combination with cytotoxic chemotherapy, we first performed dose response experiments with NGI-1, temozolomide (TMZ), or etoposide (VP-16) to determine the effects on proliferation and to select the appropriate concentrations for this line of experimentation (Supplementary Figure 1). Glioma cell line proliferation was sensitive to NGI-1 and we therefore used a dose of 1μM for combination treatment experiments. D54, SKMG3, T98G or U251 cells were then treated for 5 days with NGI-1, with or without TMZ or VP-16. For D54 and SKMG3, we found that the combinations of NGI-1 with TMZ or VP-16 further reduced glioma cell proliferation, although these effects were not synergistic (Figure 3A; p ≤ 0.05, light gray bars). In contrast, NGI-1 treatment did not enhance the anti-proliferative effects of TMZ or VP-16 in T98G or U251 cells (Figure 3B). This data parallels the observations with radiation clonogenic survival and suggests that blockade of RTK signaling may also enhance the effects of cytotoxic chemotherapy in malignant glioma.
Figure 3. Combined effects of NGI-1 and cytotoxic chemotherapy on glioma cell proliferation.
Bar graphs representing fold increases in proliferation for (a.) D54 and SKMG3 or (b.) T98G and U251 after 5 days of drug exposure. Cultures were treated as described in Materials and Methods. The results are mean values ± standard error for three independent experiments for each cell line. An * indicates a significant difference (p≤ 0.05).
NGI-1 enhances cell cycle arrest and DNA damage in glioma
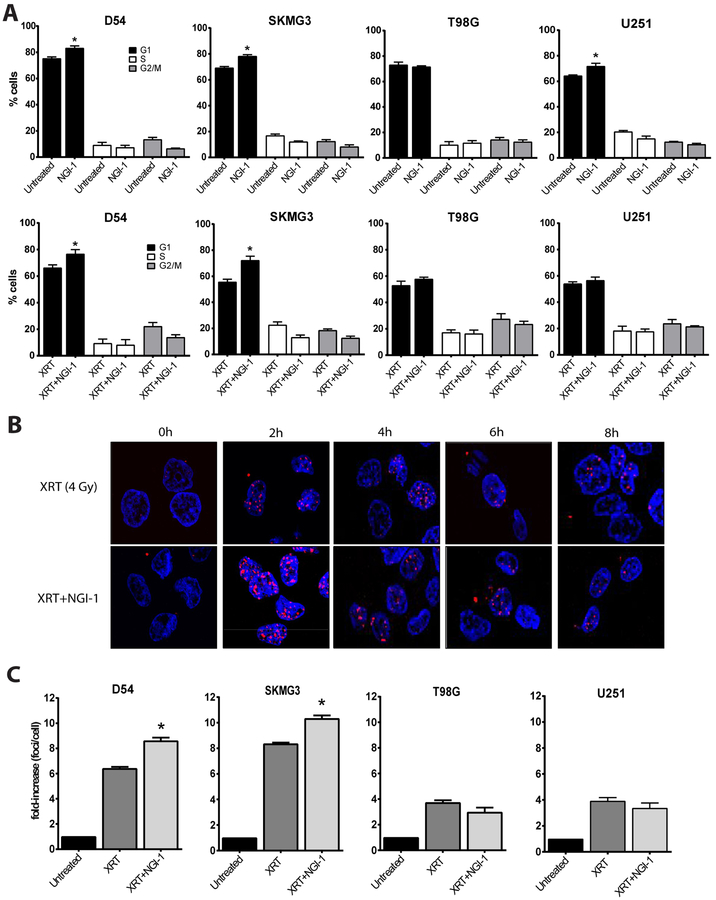
We have previously identified an accumulation of cells in the G1 phase of the cell cycle after NGI-1 treatment in NSCLC (22). We therefore examined the effects of NGI-1 on cell cycle distributions after 48 hours of NGI-1 treatment. Our results demonstrate that NGI-1 caused a significant G1 arrest in D54 (83% vs. 75%), SKMG3 (78% vs. 69%) and U251 (64% vs. 72%) cells (Figure 4A, p ≤ 0.05), but no arrest in the T98G cell line. Six hours after 4 Gy, a significant G1 arrest was again observed with NGI-1 treatment for D54 (77% vs. 66%) and SKMG-3 (72% vs. 56%), but not for either the T98G or U251 cells. This data indicates that NGI-1 reduces progression through the cell cycle after radiation therapy and suggests that inhibition of upstream growth factor receptor glycoproteins alters these downstream cell cycle distributions.
Figure 4. Effects of NGI-1 on cell cycle and γH2AX foci formation.
(a.) Flow cytometry and cell cycle distribution of D54, SKMG3, T98G and U251 cells after vehicle or 10μM NGI-1 treatment. Cells were also treated with 4 Gy under similar conditions and harvested for cell cycle analysis after 6 hours. The percent of cells in G1 (black bars), S (white bars) and G2/M (gray bars) are shown. Data were obtained from three independent experiments and are represented as mean ± standard error. An * indicates a significant difference between NGI-1 treated and control samples (p≤ 0.05). (b.) Time course of γH2AX foci formation after 4 Gy in the presence or absence of 10 μM of NGI-1 in D54 cells. DAPI stained nuclei (blue) and γH2AX fluorescence (red) are shown 0 (pre-radiation), 2, 4, 6 and 8 hours after radiation treatment. Representative images from three independent experiments are shown. (c.) quantification of γH2AX foci in D54, SKMG3, T98G and U251 cells 2 hours after irradiation with 4 Gy in the presence or abscence of 10 μM of NGI-1. Bar graphs represent the fold-increase of total number of foci counted per total number of cells in the picture. Foci of a cell were counted when a nucleus contained >10 foci. Data were obtained from three independent experiments and are represented as mean ± standard error. An * indicates a significant difference between NGI-1 treated and control samples (p≤ 0.05).
To evaluate the effects of NGI-1 on the DNA damage response we performed time course experiments to determine the kinetics of phospho-γH2AX foci formation in the presence or absence of NGI-1. D54 cells were irradiated with 4 Gy and foci were detected with a S139 phospho-specific antibody at 0 (pre-radiation), 2, 4, 6 and 8 hours. Radiation alone induced a 6.4-fold increase of phospho-γH2AX foci formation followed by a rapid decrease at 4 hours and full resolution of foci by 8 hours. NGI-1 treatment increased phospho-γH2AX foci formation significantly up to 8.6-fold (P = 0.03, Figure 4B), followed by a similar time frame for foci resolution as radiation alone (Supplementary Figure 2).
We next quantified the induction of phospho-γH2AX foci at 2 hours in D54, SKMG3, U251 and T98G cell lines irradiated with 4 Gy. NGI-1 increased phospho-γH2AX foci formation (Figure 4C; p ≤ 0.05, light gray bars) by a factor of 1.3 and 1.2 in D54 and SKMG3 cell lines, respectively, compared to radiation alone (p ≤ 0.05; dark gray bars) consistent with an increase in DNA damage. However, no enhancement of phospho-γH2AX foci formation was observed in either T98G or U251 cells. Together these results suggest that in cells with high levels of cytoprotective RTK signaling, NGI-1 enhances DNA damage which contributes to cell cycle arrest and tumor cell death.
NGI-1 reduces tumor growth of glioblastoma with activated ErbB receptors in vivo.
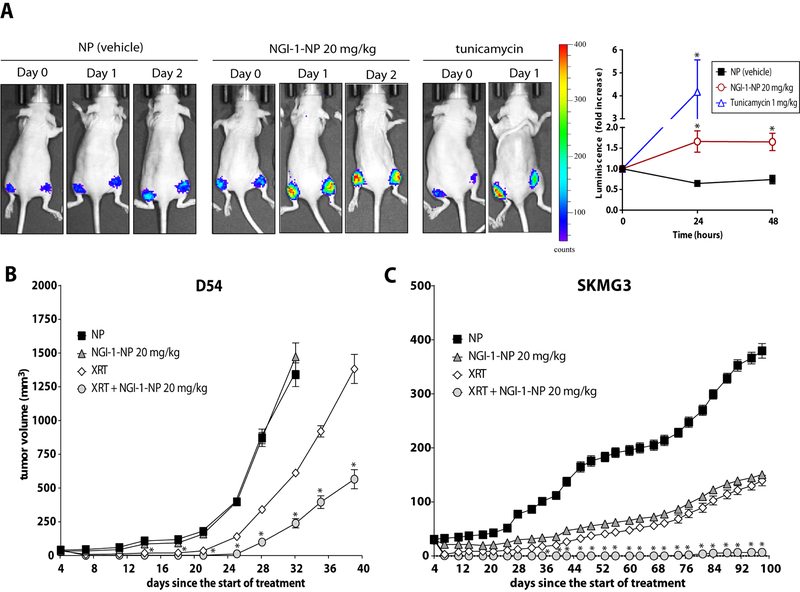
To assess the effect of NGI-1 on xenograft tumor growth, we used an NGI-1 nanoparticle formulation that overcomes the low solubility of this compound. First the effect of NGI-NPs were tested using D54-ERLucT xenografts, which increase biolouminescence after inhibition of glycoyslation (17). We found a significant induction of bioluminescence in mice that received NGI-1 at both 24 (1.7 fold, p =.03; Fig. 5A) and 48 hour (1.7 fold, p =.03; Fig. 5A) time points. Tunicmaycin, another inhibidor of N-linked glycosylation was used as a positive control and induced bioluminescence (4.2 fold at 24 hours (p =.007)). These results confirmed the ability of NGI-1 NPs to inhibit glycosylation in D54 tumors in vivo.
Figure 5. NGI-1 Radiosensitizes Glioma Xenografts.
(a.) In vivo imaging of D54 ER-LucT xenografts treated with i.v. control NPs, NGI-1 NP (20mg/kg), or tunicmaycin (1mg/kg). Representative images over 48 hours demonstrate induction of bioluminescence. Tumor average changes in luminescence with standard error for control NP (n=4), NGI-1-NP (n=8) or Tn (n=4) over 48 hours are also reported. (b.) D54 and (c.) SKMG3 averge xenograft tumor growth following treatment with control NP, NGI-1 NP, RT, or RT + NGI-1 NP. NPs were delivered 24 hours before the first fraction of radiotherapy (day 3) and on days 5 and 7 before radiation. An * indicates a significant difference between radiation+NGI-1 treated and radiation tumors (p≤ 0.05).
To evaluate the therapeutic potential of NGI-1 in vivo, we tested the effect of NGI-1 NPs on glioma tumor growth both alone and in combination with radiation for both D54 and SKMG3 cell lines. In these experiments mice were randomly assigned to receive treatment in one of four groups: control NPs, control NPs + RT, NGI-1NPs, and NGI-1 NPs + RT. NGI-1 NPs (20mg/kg) were delivered every other day for a total of 3 doses and RT was delivered in 5 daily doses of 2Gy. In D54 xenografts tumor growth was significantly delayed by radiation alone or radiation + NGI-NP treatment. The addition of NGI-NPs to RT significantly reduced tumor growth compared to those treated with radiation alone. At 39 days median tumor volumes for the NGI-1 NP + RT group were 566 ± 200 mm3 compared to 1383 ± 305 mm3 for the RT alone group (p = .001; Figure 5B). Similar results favoring combined treatment with NGI-1 NPs and RT were observed in the SKMG3 xenografts. In this cell line, both radiation and NGI-NPs reduced tumor growth when administered as a single therapy. The combination of NGI-1 NPs + RT prodcued significantly larger reductions in tumor growth. The mean tumor volume at day 98 for the radiation + NGI-1-NP group was nearly undetectable. In comparison tumor volumes for blank NPs (379 ± 38 mm3; p = .001), radiation (139 ± 27 mm3; p = .001) and NGI-1-NP (151 ± 7 mm3; p = .001) were all significantly greater (Figure 5C). For both in vivo xenograft experiments there was no evidence for significant weight loss or other toxicity in animals treated with the NGI-NP. Taken together, these results indicate that the combination of NGI-1 + RT could be a therapeutic approach for the treatment of glioblastoma.
CD8-EGFR rescues glioma cells from radiosensitization.
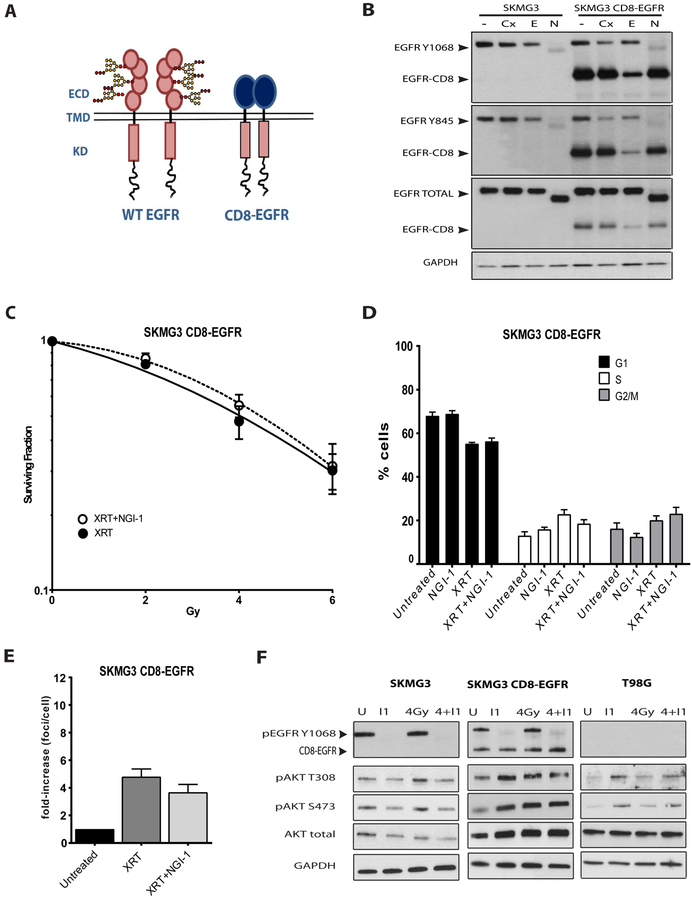
NGI-1 disrupts glycosylation of multiple cell surface glycoproteins, but our data suggests that the mechanism of radiosensitization is the blockade of ErbB RTK family signaling. To test this hypothesis, we designed a glycosylation-independent EGFR transgene that is resistant to the effects of NGI-1. This EGFR construct combines the extracellular domain of the CD8 protein, which contains no N-linked glycosylation sites, with the intracellular domain of the EGFR. Because CD8 spontaneously dimerizes, the CD8-EGFR produces constitutive EGFR kinase activation (Figure 6A). To validate the construct, we analyzed phospho-Y1068, phospho-Y845 and total EGFR levels in SKMG3 parental cells and SKMG3 cells with stable expression of the CD8-EGFR. CD8-EGFR phosphorylation is insensitive to cetuximab, an antibody that recognizes the EGFR extracellular domain, but sensitive to erlotinib, a tyrosine kinase inhibitor. CD8-EGFR phosphorylation is also resistant to NGI-1, while phosphorylation of the wild type receptor continues to be significantly reduced by this inhibitor (Figure 6B). We next evaluated the effects of NGI-1 on radiation clonogenic survival and found that CD8-EGFR prevented radiosensitization of SKMG3 by NGI-1 (Figure 6C). We also found that CD8-EGFR eliminated the G1 cell cycle arrest caused by NGI-1 both alone and after exposure to radiation (Figure 6D). This result coincided with no effect of NGI-1 on phospho-γH2AX foci formation in SKMG3-CD8-EGFR cells (Figure 6E). Radiation stimulates RTK-dependent downstream signaling, and we therefore compared AKT activation in the parental SKMG3 and SKMG3-CD8-EGFR cells. The results show that NGI-1 blocks radiation-induced activation of AKT in parental but not in the CD8-EGFR expressing SKMG3 cells. This result is similar to the effect of NGI-1 on T98G cells, where NGI-1 does not block AKT activation (Figure 6F). In summary, the CD8-EGFR model system provides additional evidence that NGI-1 radiosensitizes SKMG3 through inhibition of ErbB family RTK signaling.
Figure 6. Glycosylation-indpendent EGFR signaling rescues NGI-1 radiosensitization.
(a.) Overview of the strategy for generating NGI-1 resistant EGFR signaling. The EGFR is sensitive to NGI-1 because it’s extracellular domain has 11 consensus and one atypical N-linked gflycosylation site. The CD8 protein has no N-linked glycoyslation sites and spontaneously dimerizes, thus providing EGFR signaling that is resistant to NGI-1. (b.) Western blots of parental and CD8-EGFR expressing SKMG3 cell lines treated with vehicle, 30 nM cetuximab (Cx), 500 nM erlotinib (E), or 10 μM of NGI-1 (N) for 48 hours. GAPDH was used as a loading control. Arrows denote either the wild type EGFR or CD8-EGFR. (c.) Radiation dose response clonogenic survival in SKMG3-CD8-EGFR. Data represented the mean ± S.E. for two independent experiments (d.) Quantification of γH2AX foci formation and (e.) cell cycle distribution in control and irradiated cells (4 Gy) in the presence or abscence of 10 μM of NGI-1. Experimental conditions were identical to figure 4. (f.) Western blot analyis of SKMG3, SKMG3-CD8-EGFR, and T98G cells after 4 Gy radiation with or without 10 μM NGI-1 treatment. Western blots are representative of two independent experiments.
DISCUSSION
RTK glycoproteins have been established as important cellular targets for modifying radiosensitivity (3, 4). However, the co-expression of multiple pro-survival cell surface receptors by a single cell or heterogeneous tumor populations complicates therapeutic strategies for blocking these signals and enhancing the effects of radiation therapy (12, 31, 33). In this work, we report the effects of a new small molecule inhibitor of N-linked glycosylation (NGI-1) on RTK expression and function in malignant glioma; one of the most radioresistant tumor types. NGI-1 partially reduces glycosylation of most RTKs, in some instances also affecting RTK stability, and enhances radiosensitivity of glioma cells that have upregulated ErbB family RTK signaling. NGI-1 also enhances the anti-proliferative effects of cytotoxic chemotherapy suggesting that a global reduction in RTK signaling combines favorably with standard anti-tumor therapeutic approaches. Because NGI-1 is a first in class small molecule inhibitor of the OST, our data also identifies the OST as an enzyme that can be targeted to enhance standard cancer therapies.
The OST is a multisubunit complex that exists in several isoforms and contains one of two individually encoded catalytic subunits; STT3A and STT3B. NGI-1 impairs the activity of the OST through a direct and reversible interaction with both catalytic subunits, but unlike tunicamycin does not cause complete inhibition of glycosylation (22). Because NGI-1 does not block all glycosylation, we have hypothesized that highly glycosylated proteins with complex secondary structure and protein folding requirements, such as the cysteine rich domains found in ErbB family RTKs (34), would be preferentially affected by this inhibitor. However, our present data indicate that the effects of OST inhibition on RTK function are likely multifactorial and depend upon both the target protein that is observed, as well as the cellular context. We have previously reported that NGI-1 disrupts EGFR localization and reduces membrane expression (22), and in the current work we observe that RTK protein levels can also be affected in a cell-type specific manner. For example, NGI-1 reduces total EGFR protein levels in D54 vs SKMG3, total ErbB2 in SKMG3 vs D54, and total MET in SKMG3 and T98G vs D54 and U251 (See Figure 1). In contrast, the reduction of PDGFR and ErbB3 receptor levels appear consistent across cell lines, and surprisingly FGFR1 levels were increased (although not activated) in several cell lines. Together our data therefore suggest that both protein-intrinsic and broader cell-intrinsic factors may govern cell surface expression levels of RTKs in the setting of aberrant N-linked glycosylation. These cellular factors are likely to include both downstream transcriptional and post-translational regulation of RTK protein levels, the effect of ER stress responses, and the contributions of other cytoplasmic chaperone interactions; all of which will require further investigation. Despite differences in receptor fate, however, the consequences of aberrant N-linked glycosylation are similar for RTKs with consistent reduction of phosphorylation for individual receptors across the glioma cell lines.
Molecular studies of glioblastoma tumors have identified frequent dysregulation of growth factor receptor signaling via amplification or mutational activation of RTK genes like EGFR, ErbB2, PDGFR or MET (28, 35). In large scale sequencing investigations ~70% of GBMs showed RTK genetic abnormalities that could potentially lead to receptor activation, with the majority involving EGFR (36). In GBM patient samples and in most GBM cancer cell lines, ErbB2 protein expression has been shown to be elevated (37). Similarly, PDGFRα overexpression is associated with glioma transformation and proliferation and contributes to tumor progression and therapeutic resistance (38, 39). MET receptor overexpression and amplification has also been found in GBM and can mediate cellular reprogramming, aberrant vascularization, and chemoresistance (40–42). Other receptors such as FGFR, INSR, or IGF-1R have also been found to be overexpressed or activated and contribute to tumor progression in glioblastoma (33, 43–45). Therefore, the impaired function of ErbB family members, MET, and PDGFR produced by OST inhibition provides a potentially important targeted approach for disrupting multiple co-expressed and functionally redundant oncogenic RTKs in malignant glioma.
Although NGI-1 has similar effects on reducing RTK glycosylation and activation across glioma cells, radiosensitization was not observed in each cell line. An attractive explanation for this observation is the presence of mutations that are downstream from EGFR signaling. Examples include PIK3CA and PTEN, which are mutated or deleted, respectively, in ~40% of GBMs and are known to enhance survival signaling and tumor progression (28, 36). In agreement with this hypothesis, the cell lines in this study that were not radiosensitized by NGI-1 and also showed no combined effect of NGI-1 with chemotherapy (U251 and T98G) have PTEN loss. These cell lines also did not display elevated levels of ErbB family RTK activation, suggesting independence from RTK signaling at multiple levels. Another route for eliminating the effect of NGI-1 on radiosensitivity is the reconstitution of glycosylation-independent RTK signaling. Because all RTKs are glycosylated, we accomplished this maneuver through the generation of a CD8-EGFR transgene, which due to the lack of N-linked glycosylation sites and spontaneous dimerization of CD8, leads to constitutive activation of the EGFR kinase domain. Expression of the CD8-EGFR in SKMG3 cells prevented radiosensitization by NGI-1, and was accompanied by a loss of NGI-1’s effect on AKT signaling following radiation treatment. These results demonstrate that sustained RTK signaling is sufficient for eliminating the effect of NGI-1 on glioma radiosensitization and thus provide strong evidence that RTK inhibition is a primary mechanism for this synergistic effect. Taken together these results also imply that a subset of malignant gliomas would be sensitive to OST inhibition but that tumors with PTEN deletion, PI3K mutations, NF1 mutations, and other rare genetic alterations such as FGFR3-TACC3 fusions, may not be ideal candidates for radiosensitization with this strategy.
DNA damage induced by radiation or targeted small molecules can effectively lead to cell cycle arrest and activation of cell death programs (46–48). OST inhibition by NGI-1 caused a significant increase in G1 cell cycle arrest both alone and following treatment with radiation. Although G2/M is considered the most radiosensitive phase of the cell cycle, G1 is relatively radiosensitive as well, and an arrest could contribute to the effects of NGI-1 on radiosensitivity. Notably, however, the G1 cell cycle arrest after NGI-1 treatment plus radiation was only present in cells with high levels of RTK signaling suggesting that OST inhibition causes a more potent G1 arrest, a known indicator of increased clonogenic cell death (49). Follow up experiments measuring DNA damage responses using γ-H2AX foci formation demonstrated that NGI-1 treatment caused significantly more DNA damage accumulation over the first 2 hours after radiation exposure in glioma cells with high levels of RTK activation compared to those that had low levels of RTK activation. This observation is consistent with inhibition of an early RTK-dependent DNA damage response that could be mediated by a downstream pathway such as PI3K signaling. In addition, the expression of the CD8-EGFR reversed both the G1 arrest and the increase in γ-H2AX foci formation. Together these experiments suggest that disrupting the function of multiple RTKs enhances the accumulation of DNA damage and reduces clonogenic survival of glioma cells.
Glioblastoma multiforme has a poor prognosis with a median survival of 12–15 months. Although several clinical trials have investigated targeted inhibition of the EGFR (50, 51) or other RTKs (13, 52) in malignant glioma, this work has not yet translated into significant advances in patient outcomes. Because N-linked glycosylation is a common biosynthetic step for RTKs identified as potential therapeutic targets in malignant glioma (eg EGFR, Met, PDGFR, and VEGFR) we investigated whether inhibition of this post-translational modification with NGI-1 is a new approach for more broadly reducing RTK dependent survival signaling. The results from our work demonstrate that partial reduction of NLG through OST targeting with small molecules can indeed reduce parallel RTK signaling and increase tumor cell radiosensitivity for a subset of malignant gliomas.
Supplementary Material
TRANSLATIONAL RELEVANCE:
Receptor tyrosine kinases are validated therapeutic targets for the treatment of malignant disease. However, redundant signaling through co-expressed RTK networks enables therapeutic resistance and limits the effects of targeting individual receptors. N-linked glycosylation is an endoplasmic reticulum co- and post-translational protein modification that has a vital role in the assembly and maturation of multiple cell surface glycoprotein receptors. Recently we identified a small molecule inhibitor (NGI-1) that partially blocks the function of the oligosaccharyltransferase, a multisubunit enzymatic complex that transfers glycan precursors to elongating proteins in the ER. In the present study we investigate the effect of NGI-1 on multiple RTKs important for the progression and survival of malignant gliomas. We show that NGI-1 radiosensitizes and enhances cytotoxic chemotherapy in vitro, and that delivery of NGI-1 in vivo significantly reduces tumor growth. Together these results suggest that OST inhibition is a novel therapeutic approach for the treatment of malignant glioma.
ACKNOWLEDGEMENTS
This work is supported by US National Institutes of Health (NIH) R01CA172391 and by a Research grant from the American Cancer Society (J.N. Contessa) as well as by NIH RO1 CA149128 (W.M. Saltzman) and NIH F30 CA206386 (A. Quijano).
Footnotes
COMPETING FINANCIAL INTERESTS
JNC, WMS and AQ are listed as inventors on a patent application for the NGI-1 NP formulation reported in this manuscript.
REFERENCES
- 1.Xu AM, Huang PH. Receptor tyrosine kinase coactivation networks in cancer. Cancer Res. 2010;70:3857–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casaletto JB, McClatchey AI. Spatial regulation of receptor tyrosine kinases in development and cancer. Nat Rev Cancer. 2012;12:387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer. 2011;11:239–53. [DOI] [PubMed] [Google Scholar]
- 4.Mahajan K, Mahajan NP. Cross talk of tyrosine kinases with the DNA damage signaling pathways. Nucleic Acids Res. 2015;43:10588–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contessa JN, Reardon DB, Todd D, Dent P, Mikkelsen RB, Valerie K, et al. The inducible expression of dominant-negative epidermal growth factor receptor-CD533 results in radiosensitization of human mammary carcinoma cells. Clin Cancer Res. 1999;5:405–11. [PubMed] [Google Scholar]
- 6.Rich JN, Reardon DA, Peery T, Dowell JM, Quinn JA, Penne KL, et al. Phase II trial of gefitinib in recurrent glioblastoma. J Clin Oncol. 2004;22:133–42. [DOI] [PubMed] [Google Scholar]
- 7.Reardon DA, Rich JN, Friedman HS, Bigner DD. Recent advances in the treatment of malignant astrocytoma. J Clin Oncol. 2006;24:1253–65. [DOI] [PubMed] [Google Scholar]
- 8.Wen PY, Yung WK, Lamborn KR, Dahia PL, Wang Y, Peng B, et al. Phase I/II study of imatinib mesylate for recurrent malignant gliomas: North American Brain Tumor Consortium Study 99–08. Clin Cancer Res. 2006;12:4899–907. [DOI] [PubMed] [Google Scholar]
- 9.Akhavan D, Pourzia AL, Nourian AA, Williams KJ, Nathanson D, Babic I, et al. De-repression of PDGFRbeta transcription promotes acquired resistance to EGFR tyrosine kinase inhibitors in glioblastoma patients. Cancer Discov. 2013;3:534–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi J, McTigue MA, Rogers A, Lifshits E, Christensen JG, Janne PA, et al. Multiple mutations and bypass mechanisms can contribute to development of acquired resistance to MET inhibitors. Cancer Res. 2011;71:1081–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jun HJ, Acquaviva J, Chi D, Lessard J, Zhu H, Woolfenden S, et al. Acquired MET expression confers resistance to EGFR inhibition in a mouse model of glioblastoma multiforme. Oncogene. 2012;31:3039–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–90. [DOI] [PubMed] [Google Scholar]
- 13.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370:709–22. [DOI] [PubMed] [Google Scholar]
- 14.Aebi M N-linked protein glycosylation in the ER. Biochim Biophys Acta. 2013;1833:2430–7. [DOI] [PubMed] [Google Scholar]
- 15.Freeze HH. Genetic defects in the human glycome. Nat Rev Genet. 2006;7:537–51. [DOI] [PubMed] [Google Scholar]
- 16.Kelleher DJ, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47R–62R. [DOI] [PubMed] [Google Scholar]
- 17.Contessa JN, Bhojani MS, Freeze HH, Ross BD, Rehemtulla A, Lawrence TS. Molecular imaging of N-linked glycosylation suggests glycan biosynthesis is a novel target for cancer therapy. Clin Cancer Res. 2010;16:3205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Croci DO, Cerliani JP, Dalotto-Moreno T, Mendez-Huergo SP, Mascanfroni ID, Dergan-Dylon S, et al. Glycosylation-dependent lectin-receptor interactions preserve angiogenesis in anti-VEGF refractory tumors. Cell. 2014;156:744–58. [DOI] [PubMed] [Google Scholar]
- 19.Itkonen HM, Mills IG. N-linked glycosylation supports cross-talk between receptor tyrosine kinases and androgen receptor. PLoS One. 2013;8:e65016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Contessa JN, Bhojani MS, Freeze HH, Rehemtulla A, Lawrence TS. Inhibition of N-linked glycosylation disrupts receptor tyrosine kinase signaling in tumor cells. Cancer Res. 2008;68:3803–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cazet A, Charest J, Bennett DC, Sambrooks CL, Contessa JN. Mannose phosphate isomerase regulates fibroblast growth factor receptor family signaling and glioma radiosensitivity. PLoS One. 2014;9:e110345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Sambrooks C, Shrimal S, Khodier C, Flaherty DP, Rinis N, Charest JC, et al. Oligosaccharyltransferase inhibition induces senescence in RTK-driven tumor cells. Nat Chem Biol. 2016;12:1023–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez CA, Feng FY, Herman JM, Nyati MK, Lawrence TS, Ljungman M. Phenylbutyrate sensitizes human glioblastoma cells lacking wild-type p53 function to ionizing radiation. Int J Radiat Oncol Biol Phys. 2007;69:214–20. [DOI] [PubMed] [Google Scholar]
- 24.Carboni JM, Lee AV, Hadsell DL, Rowley BR, Lee FY, Bol DK, et al. Tumor development by transgenic expression of a constitutively active insulin-like growth factor I receptor. Cancer Res. 2005;65:3781–7. [DOI] [PubMed] [Google Scholar]
- 25.Baro M, de Llobet LI, Figueras A, Skvortsova I, Mesia R, Balart J. Dasatinib worsens the effect of cetuximab in combination with fractionated radiotherapy in FaDu- and A431-derived xenografted tumours. Br J Cancer. 2014;111:1310–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King AR, Corso CD, Chen EM, Song E, Bongiorni P, Chen Z, et al. Local DNA Repair Inhibition for Sustained Radiosensitization of High-Grade Gliomas. Mol Cancer Ther. 2017;16:1456–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Workman P, Aboagye EO, Balkwill F, Balmain A, Bruder G, Chaplin DJ, et al. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102:1555–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen DJ, Nirodi CS. The epidermal growth factor receptor: a role in repair of radiation-induced DNA damage. Clin Cancer Res. 2007;13:6555–60. [DOI] [PubMed] [Google Scholar]
- 31.De Bacco F, Luraghi P, Medico E, Reato G, Girolami F, Perera T, et al. Induction of MET by ionizing radiation and its role in radioresistance and invasive growth of cancer. J Natl Cancer Inst. 2011;103:645–61. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt-Ullrich RK, Contessa JN, Lammering G, Amorino G, Lin PS. ERBB receptor tyrosine kinases and cellular radiation responses. Oncogene. 2003;22:5855–65. [DOI] [PubMed] [Google Scholar]
- 33.Gouaze-Andersson V, Delmas C, Taurand M, Martinez-Gala J, Evrard S, Mazoyer S, et al. FGFR1 Induces Glioblastoma Radioresistance through the PLCgamma/Hif1alpha Pathway. Cancer Res. 2016;76:3036–44. [DOI] [PubMed] [Google Scholar]
- 34.Dawson JP, Bu Z, Lemmon MA. Ligand-induced structural transitions in ErbB receptor extracellular domains. Structure. 2007;15:942–54. [DOI] [PubMed] [Google Scholar]
- 35.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, Burger MC, Jennewein L, Genssler S, Schonfeld K, Zeiner P, et al. ErbB2/HER2-Specific NK Cells for Targeted Therapy of Glioblastoma. J Natl Cancer Inst. 2016;108. [DOI] [PubMed] [Google Scholar]
- 38.Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K, et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26:288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu F, Chen Y, Zhao C, Wang H, He D, Xu L, et al. Olig2-Dependent Reciprocal Shift in PDGF and EGF Receptor Signaling Regulates Tumor Phenotype and Mitotic Growth in Malignant Glioma. Cancer Cell. 2016;29:669–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joo KM, Jin J, Kim E, Ho Kim K, Kim Y, Gu Kang B, et al. MET signaling regulates glioblastoma stem cells. Cancer Res. 2012;72:3828–38. [DOI] [PubMed] [Google Scholar]
- 41.van den Heuvel C, Navis AC, de Bitter T, Amiri H, Verrijp K, Heerschap A, et al. Selective MET Kinase Inhibition in MET-dependent Glioma Models Alters Gene Expression and Induces Tumor Plasticity. Mol Cancer Res. 2017. [DOI] [PubMed] [Google Scholar]
- 42.Huang M, Liu T, Ma P, Mitteer RA Jr., Zhang Z, Kim HJ, et al. c-Met-mediated endothelial plasticity drives aberrant vascularization and chemoresistance in glioblastoma. J Clin Invest. 2016;126:1801–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Almiron Bonnin DA, Ran C, Havrda MC, Liu H, Hitoshi Y, Zhang Z, et al. Insulin-Mediated Signaling Facilitates Resistance to PDGFR Inhibition in Proneural hPDGFB-Driven Gliomas. Mol Cancer Ther. 2017;16:705–16. [DOI] [PubMed] [Google Scholar]
- 44.Ma Y, Tang N, Thompson RC, Mobley BC, Clark SW, Sarkaria JN, et al. InsR/IGF1R Pathway Mediates Resistance to EGFR Inhibitors in Glioblastoma. Clin Cancer Res. 2016;22:1767–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinn B, Tallen G, Schroeder G, Grassl B, Schulze J, Budach V, et al. Caffeine confers radiosensitization of PTEN-deficient malignant glioma cells by enhancing ionizing radiation-induced G1 arrest and negatively regulating Akt phosphorylation. Mol Cancer Ther. 2010;9:480–8. [DOI] [PubMed] [Google Scholar]
- 47.Cheng Z, Gong Y, Ma Y, Lu K, Lu X, Pierce LA, et al. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19:1748–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.See WL, Tan IL, Mukherjee J, Nicolaides T, Pieper RO. Sensitivity of glioblastomas to clinically available MEK inhibitors is defined by neurofibromin 1 deficiency. Cancer Res. 2012;72:3350–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta N, Vij R, Haas-Kogan DA, Israel MA, Deen DF, Morgan WF. Cytogenetic damage and the radiation-induced G1-phase checkpoint. Radiat Res. 1996;145:289–98. [PubMed] [Google Scholar]
- 50.Chakravarti A, Wang M, Robins HI, Lautenschlaeger T, Curran WJ, Brachman DG, et al. RTOG 0211: a phase 1/2 study of radiation therapy with concurrent gefitinib for newly diagnosed glioblastoma patients. Int J Radiat Oncol Biol Phys. 2013;85:1206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peereboom DM, Shepard DR, Ahluwalia MS, Brewer CJ, Agarwal N, Stevens GH, et al. Phase II trial of erlotinib with temozolomide and radiation in patients with newly diagnosed glioblastoma multiforme. J Neurooncol. 2010;98:93–9. [DOI] [PubMed] [Google Scholar]
- 52.Franceschi E, Stupp R, van den Bent MJ, van Herpen C, Laigle Donadey F, Gorlia T, et al. EORTC 26083 phase I/II trial of dasatinib in combination with CCNU in patients with recurrent glioblastoma. Neuro Oncol. 2012;14:1503–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.