Abstract
Social challenges like territorial intrusions evoke behavioral responses in widely diverging species. Recent work has revealed that evolutionary “toolkits” – genes and modules with lineage-specific variations but deep conservation of function – participate in the behavioral response to social challenge. Here, we develop a multi-species computational-experimental approach to characterize such a toolkit at a systems level. Brain transcriptomic responses to social challenge was probed via RNA-seq profiling in three diverged species – honey bees, mice, and three-spined stickleback fish – following a common methodology, allowing fair comparisons across species. Data were collected from multiple brain regions and multiple time points after social challenge exposure, achieving anatomical and temporal resolution substantially greater than previous work. We developed statistically rigorous analyses equipped to find homologous functional groups among these species at the levels of individual genes, functional and coexpressed gene modules, and transcription factor sub-networks. We identified six orthogroups involved in response to social challenge, including groups represented by mouse genes Npas4 and Nr4a1, as well as common modulation of systems such as transcriptional regulators, ion channels, G-protein coupled receptors, and synaptic proteins. We also identified conserved coexpression modules enriched for mitochondrial fatty acid metabolism and heat shock that constitute the shared neurogenomic response. Our analysis suggests a toolkit wherein nuclear receptors, interacting with chaperones, induce transcriptional changes in mitochondrial activity, neural cytoarchitecture, and synaptic transmission after social challenge. It reveals systems-level mechanisms that have been repeatedly co-opted during evolution of analogous behaviors, thus advancing the genetic toolkit concept beyond individual genes.
Keywords: Comparative Genomics, Animal Behavior, Transcriptomics, Social Behavior, Social Challenge, Systems Biology, Coexpression, Honey Bee, Mouse, Three-Spined Stickleback
INTRODUCTION
A pivotal idea arising from evolutionary developmental biology is that across the bilaterian clade, the same signaling and transcription factor genes, known as “toolkit” genes, underlie the patterning of basic morphological features such as the body plan and the eye (Carroll et al. 2005). This provides a conceptual framework for increasingly detailed explanations of developmental patterning in specific model organisms (Wilkins 2002). Moreover, its success has motivated researchers to ask if the toolkit framework, where common genetic programs coordinate fundamental processes and undergird shared phenotypes, is also applicable to studies of behavior (Rittschof & Robinson 2016; Toth & Robinson 2007).
Identification of behavioral toolkits can enrich both ethology and neurobiology. The idea of the toolkit concept as applied to behavior is that multiple taxa have independently adapted the same gene sets to embed similar social experiences and encode similar behavioral responses. The existence of such toolkits can motivate novel research on a range of topics including: evolutionary research on the origin of diverse behavioral phenotypes derived from a core set of conserved genes, mechanistic research on how the gene systems themselves drive behavioral phenotypes, and genetic research ascertaining the processes governing conservation of toolkit genes across disparate animal taxa (Rittschof & Robinson 2016). However, studying toolkits for behavior poses numerous challenges including: the relative paucity of detailed and directly comparable genetics and genomics datasets for behaviorally relevant phenotypes in most animal species, difficulties in defining correspondence between behaviorally relevant phenotypes in diverged species from different ecological contexts, and ambiguity regarding brain regions and other tissues where shared behaviorally relevant molecular mechanisms may manifest. Further, behaviors, being transitory and directly observable only while an animal is living, cannot be as readily gleaned from fossils as developmental phenotypes (Chen et al. 2013), giving us little evidence from the distant past that contextualizes what we observe in extant species.
In an example of an evolutionary approach, our group recently studied whether shared gene expression correlates constitute a toolkit for the neural response to a territorial intrusion by a conspecific – more generally referred to as a social challenge – in the mouse, the three-spined stickleback fish, and the honey bee (Rittschof et al. 2014). These three highly diverged model social species have well-assembled genomes, providing ample technical resources for detailed comparisons of functional genomic correlates. Further, each of these species displays a robust response to territorial intrusion. Though a toolkit would be derived within each individual species, it would contain a common core of conserved components important for coordinating brain response to social contexts, and an argument for such a toolkit requires the identification of shared functional correlates. Accordingly, we found robust transcriptional responses in brain gene expression profiles across these three species 20–30 minutes after exposure to the intruder and discovered several common molecular mechanisms associated with the intruder response. Though similar to evolutionary development studies in its pursuit of a “toolkit”, our earlier study was notably different for its use of gene expression as the primary means to identify toolkit genes rather than direct or indirect measures of gene sequence. Similarly, other groups have discussed conservation in the transcriptional correlates of aggressive behavior within the vertebrate subphylum (Freudenberg et al. 2016; Malki et al. 2016) and in arthropods (Asahina et al. 2014).
The success of the above studies in identifying shared mechanisms motivates a concerted effort towards more comprehensive and rigorous descriptions of behaviorally relevant evolutionary toolkits. However, further progress has been limited due to two factors. First, the prior studies measured expression at only a single time point after animals were exposed to the social challenge and relatively soon after exposure. Such a design cannot capture longer-acting genetic programs. This simple design also limits the power of this previous study to detect responses whose anatomical and temporal profiles are shaped by the unique cell biology, neuroendocrine, and metabolic properties of brains in these three species (Bukhari et al. 2017; Saul et al. 2017; Shpigler et al. 2017b).
Second, evolutionarily shared mechanisms are likely to be found at various levels of organization that are not reducible to single genes, which have been the main level of comparative analyses thus far. Shared mechanisms embodied by gene orthogroups comprising multiple orthologs and paralogs, coexpressed modules, groups of genes dedicated to specific known biological processes, or regulatory sub-networks (Rittschof & Robinson 2016) have eluded discovery so far. Analytic tools that can identify such higher order functional entities across multiple species, brain regions, and time points in the face of complex gene orthology relationships among highly diverged species have been lacking.
We report here the results of a detailed computational investigation of the shared molecular roots of social behavior, specifically neural response to social challenge, that remedies the above issues. We utilized a powerful analytical design, integrating for the first time three previously published large time series datasets of brain gene expression from mice (Saul et al. 2017), three-spined sticklebacks (Bukhari et al. 2017), and honeybees (Shpigler et al. 2017b). To interrogate these datasets at a systems-level, we developed a suite of computational methods that allowed us to ascertain not only individual genes, but to overcome complex orthology and incomplete annotations in order to identify coordinately expressed Gene Ontology terms, coexpression networks, and transcriptional regulatory cascades commonly associated with a behaviorally relevant stimulus across these distantly related species.
Our work goes beyond analyses of previously published cross-species studies of tissue-specific (Lin et al. 2014) or developmental (Gerstein et al. 2014) time-course transcriptomes; here, we not only identify shared transcriptomic patterns at a systems level but also rigorously test and quantify their associations with brain responses while accounting for complex homology relationships between and within species. For simplicity we will refer to these conserved systems as homologous functional groups (HFGs, see Figure 1). Altogether, this analytical approach permitted the discovery of common molecular correlates of social behavior at varying levels of molecular organization and the aggregation of the discoveries at multiple levels of abstraction from genes to whole systems.
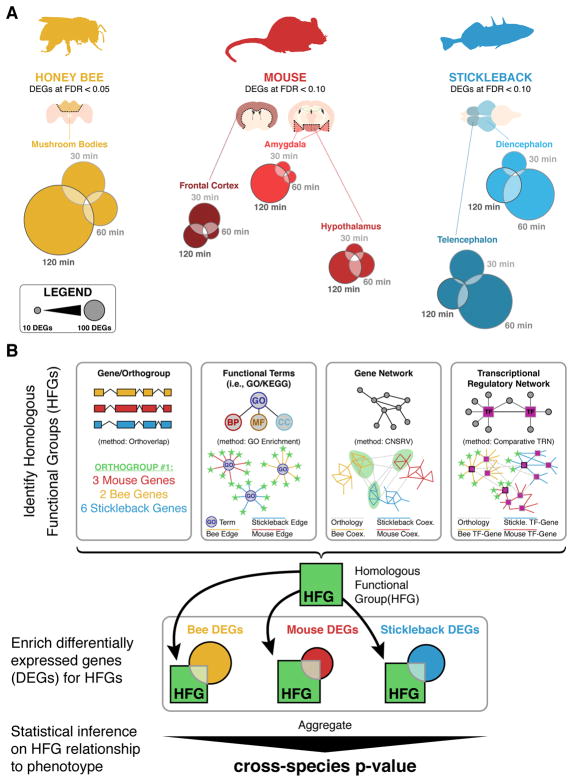
Figure 1.
Summary of (A) dataset and (B) methods used within the paper to assess homologous function groups (HFGs). A) Description of dataset constituting multiple calls of differentially expressed genes (DEGs) within each brain region assayed for each species. Honey bee DEGs are called at FDR < 0.05 while mouse and stickleback DEGs are called at FDR < 0.10. B) Schematic of HFG analysis. Multiple methods of HFG identification utilized included: genes and orthogroups, functional terms, gene networks, and transcriptional regulatory networks (TRNs) and enrichment. HFGs are highlighted in green. Once HFGs were identified, enrichment of DEGs swithin each species was assessed prior to calculation of cross-species simultaneous enrichment.
MATERIALS AND METHODS
Social Challenge Exposure, Sequencing, and Identification of Differentially Expressed Genes
The results described derive from three separate experiments that proceeded in parallel previously in three different species. Briefly, within each species, animals were exposed to an analogous social stimulus or to a non-social control stimulus: either the social challenge of a conspecific resident-intruder (challenged) or a novel non-social stimulus of roughly equal size and shape (control). Challenged female honey bees were exposed to an intruder bee from a different hive while control honey bees were exposed to a microcentrifuge tube; challenged C57BL/6J male mice were exposed to a male territorial intruder of an unrelated strain while control mice were exposed to a paper cup; and challenged male sticklebacks were exposed to an unrelated male in a flask while control sticklebacks were exposed to an empty flask. All animals were exposed to either challenge or control stimulus for five minutes, then the stimulus was removed for the rest of the elapsed time. We waited either 30 min, 60 min, or 120 min after initial stimulus presentation to collect brains for transcriptomic profiling within each species for both challenged and control animals. recording transcriptional events with a time series (30 min, 60 min, and 120 min after the exposure to the social challenge within each species). While measurements approximately 30 min after stimulus exposure were used in previous work (Rittschof et al. 2014), the addition of 60 and 120 min allows us to more broadly accommodate the cross-species variations in the timing of transcriptional trajectory; such variations may arise from differences in molecular mechanisms and may also reflect differences in animal behavior such as the time it takes to recover from a social challenge. For details about the specific experimental paradigms used within each individual species, see our previous work (Bukhari et al. 2017; Saul et al. 2017; Shpigler et al. 2017b).
The experimental design integrated datasets on mice, sticklebacks, and honey bees. Insights into the social neural transcriptomes for each of these individual species have been published (Bukhari et al. 2017; Saul et al. 2017; Shpigler et al. 2017b). These datasets were sequenced in parallel, allowing for comparative analysis and discovery of shared mechanisms with minimal technical variation. Differential gene expression sets compared challenged to control animals at each of the three time points and in each brain region sampled. These brain regions included: amygdala, frontal cortex, and hypothalamus for mouse; diencephalon and telencephalon for three-spined sticklebacks; and mushroom bodies for honey bees (Figure 1A).
In our choices of the tissues we used for sequencing, we limited ourselves to a subset of regions within each species’ brains that are closely tied to social behavior. Though there are both social and non-social regions of the brain that may be of interest in the future, this design generated the well-powered individual species social brain transcriptomes needed for this analysis. Within the vertebrates, the specific brain regions were chosen because they have been shown to have roles in coordination of aggressive behavior (Takahashi and Miczek 2014) and to contain important nodes of the vertebrate social decision-making network (O’Connell and Hofmann 2012). The mushroom bodies, a higher order integration center important in insect learning and memory (Heisenberg 2003; Farris and Sinakevitch 2003), were chosen as a site of honey bee experience-dependent neuroanatomical plasticity (Withers et al. 1993; Farris et al. 2001) and as a well-documented substrate for honey bee social behavior-related neural phenomena (Robinson et al. 1997). We make no specific assumptions here about whether brain regions are homologous across distantly related vertebrates and honey bees; instead, these experiments aimed to probe discrete brain regions relevant to social behavior in each species, which should increase the specificity of RNA-seq signals relative to sequencing whole brains as was done previously (Rittschof et al. 2014).
The data for these three sets are deposited in the GEO under accession numbers: GSE85876 (honey bee, Shpigler et al. 2017b), GSE80346 (mouse, Saul et al. 2017), and GSE96673 (three-spined stickleback, Bukhari et al. 2017). Sample sizes for each experiment are summarized in Supplementary Table 1. The analysis utilized differentially expressed gene (DEG) lists generated by comparing challenged to control experiments within each species. Briefly, after alignment using TopHat and read counting using htseq-count, analysis of differential expression proceeded using edgeR with TMM normalization. Pairwise comparisons performed using either the exactTest() function (mice and three-spined sticklebacks) or a generalized linear model with blocking for inter-hive variance (honey bee). In each species, we used a 1 CPM cutoff for an equivalent of the smallest group size for expression, as proposed in the edgeR documentation (Robinson et al. 2010). DEGs for each time point compare transcriptomes of challenged (experimental) to neutral (control). FDR thresholds from each individual species project were chosen to compile the DEG lists: 5% for bee, 10% for mouse, and 10% for stickleback (Figure 1A). We chose a lower FDR threshold for the honey bee because its experimental design was more powerful and therefore produced more DEGs. Note that DEGs were defined only based on statistical significance, and we did not impose any thresholds on fold-change in expression.
OrthoDB
Comparing DEGs between species requires a reliable orthology map between these three species. Using the raw data from OrthoDB v8 (Kriventseva et al. 2015), we first filtered for the three species of interest. We then identified the metazoan level orthogroups present within all of the three individual species used in this experiment, a total of 4,982 orthogroups. We found all paralogs inside of each orthogroup for the individual species, which brought us to a total of 10,158 genes in mouse, 6,725 genes in bee, and 10,869 genes in stickleback. The scripts used to annotate these orthogroups have been uploaded to GitHub (https://github.com/msaul/three_species_orthology).
Multi-scale characterization of conserved molecular basis for analogous cross-species phenotype
We probed for an evolutionary toolkit for social challenge response at multiple levels of molecular organization in a uniform and systematic manner. For each level of organization – individual genes, cellular processes, coexpression modules, and TF regulons – we first identified HFGs (Figure 1B) in the three species as sets of genes that exhibited intra-species as well as inter-species commonality, e.g., involvement in the same cellular process, being paralogs or orthologs of each other, etc. We then tested each HFG for association with phenotype across all three species (see below). This systematic two-step approach is a novel feature of our work, and while our previous work (Rittschof et al. 2014) reported an initial use of the approach, it is developed fully in this work with a focus on statistical rigor.
Identifying orthogroups with a conserved response to social challenge
We identified a given HFG as associated with phenotype if its constituent species-specific gene sets are simultaneously enriched in phenotype-associated genes. To test for this simultaneous enrichment, we combined enrichment p-values obtained from each gene set, then tested the significance of the combined p-value by simulating a null distribution according to a precisely specified null model.
In each species, for each orthogroup we only considered genes in the orthogroup and in the corresponding species’ gene “universe”, that is, the full complement of genes expressed above a threshold in each species. Under the null hypothesis of no orthogroup activity in response to social challenge, we modeled the number of DEGs contained in an orthogroup as a hypergeometric random variable. We tested if each orthogroup contained more DEGs from that species than expected by chance, using a one-tailed hypergeometric test. We did not separate brain region- and time point-specific DEGs within each species at this stage of analysis. This resulted in three p-values for each orthogroup, pbee, pmouse, and pfish, which we then aggregated using Fisher’s combination test statistic T = −2lnpbee − 2lnpfish.
For each orthogroup triplet, we calculated the p-value of the test statistic T under the reasonable assumption that the pbee, pmouse, and pfish were statistically independent. If they were uniformly distributed, classical theory gives that T would be -distributed under the null hypothesis that none of the three orthogroups was responsive to social challenge. However, due to the discrete nature of the hypergeometric variables from which the pbee, pmouse, and pfish were calculated, we resorted to simulations to calculate the true p-value of T. We simulated 5 million instances of the hypergeometric variables for each orthogroup in each species under the null hypothesis and calculated the p-value of each orthogroup triplet’s T using the simulated distribution.
Technically, the alternative hypothesis of this test is that there is at least one species in which the corresponding orthogroup is enriched in social challenge DEGs. This does not exactly match the conservation hypothesis, which should state that all orthogroups in all three species are enriched in DEGs. However, this latter hypothesis corresponds to a composite null hypothesis, which is difficult to formally test without sacrificing a great deal of statistical power. Here we instead test the simpler sharp null hypothesis where all orthogroups are inactive, it is known that the test statistic T that we have chosen is oriented toward the desired alternative hypothesis where all three orthogroups are enriched in DEGs. Thus, our tests are oriented toward the desired conservation hypothesis.
Identifying Gene Ontology terms and transcription factor orthogroups with a conserved response to social challenge
We downloaded Gene Ontology (GO) annotations for mouse and stickleback from Ensembl Biomart (Ensembl v83, Kinsella et al. 2011) and for bee from Ensembl Metazoa Biomart (v29, Kinsella et al. 2011) and considered only the 341 terms that contained at least 5 genes as HFGs. We used our orthogroup analysis method, described above, to identify terms that were significantly enriched in DEGs in multiple species. Transcriptional regulatory networks (TRNs) were reconstructed for each individual species individually as previously described (Bukhari et al. 2017; Saul et al. 2017; Shpigler et al. 2017b). In each species, for each orthogroup of transcription factors (TFs), we collected the gene targets of all TFs in the orthogroup into a single set. We then used our orthogroup analysis method to identify TF orthogroups whose target sets were enriched in DEGs in multiple species as HFGs.
CNSRV
We developed a method to discover homologous gene coexpression modules across divergent species as a method of ab initio discovery of HFGs. This was necessitated by our multi-scale analysis strategy but may also be of independent interest. Our module inference method, called “Common NetworkS ReVealed” (CNSRV), is closest in spirit to the OrthoClust method (Yan et al. 2014), but uses a different score for the quality of cross-species modules. This score helps avoid a bias towards large or small modules that is commonly seen with existing methods of module discovery (Langfelder & Horvath, 2008). We performed systematic assessments to demonstrate that our methods led to less extreme module sizes (Supplementary Figure SM2) and also found the resulting modules to be more statistically enriched for Gene Ontology terms (Supplementary Figure SM3). The code for CNSRV has been deposited in GitHub (https://github.com/weiyangedward/CNSRV). For a full description of CNSRV and its evaluations, see Supplementary Methods. We outline its main steps below.
Construction of coexpression networks
For each species, we first calculated coexpression of gene pairs as the Pearson correlation of their expression values in a specific brain region at different time points after exposure (including intruder-exposed as well as control animals) and retained pairs that had correlation coefficient above 0.7 in all brain regions considered for that species.
Cross-species coexpression module detection
The algorithm partitions the genes in each species’ coexpression network into K = 20 non-overlapping clusters, referred to by identifiers 1, 2, … K, such that cluster i in one species “corresponds to” clusters labeled i in the other species. The algorithm seeks to find partitions such that (1) clusters in each species exhibit “modularity” (Newman 2006) – high density of within-cluster coexpression edges compared to cross-cluster density of such edges, and (2) corresponding clusters in a pair of species exhibit high density of orthology edges (an orthology edge is created for any pair of genes in the same orthogroup from the two species). To meet these two goals, the CNSRV method attempts to maximize the following objective function:
Here S is the number of species, K is the desired number of clusters. ŵk is the normalized count of coexpression edges in cluster k of species s, defined as ŵks = wks/Es, where wks is the number of coexpression edges in cluster k of that species and Es is the total number of edges in that species. Similarly, ν̂ks is the normalized count of coexpression edges connected to nodes in cluster k of species s, defined as ν̂ks = ν ks/Es, where ν ks is the count of coexpression edges incident to nodes in cluster k in that species. (a,b) refers to any pair of orthologous genes from species i and j such that both genes are in cluster k of their respective species. To normalize the number of orthologous edges from many-to-many gene mappings, ω̂ab = 1/2(1/da + 1/db) where da is the number of orthologous edges from gene a in species i to genes in species j. The two terms in this formula, representing the “modularity” and “orthology” goals respectively, are weighted by factors of λ and (1 − λ) respectively. We chose a value of λ = 0.05 to provide a suitable balance between the coexpression modularity and cross-species sharing aspects of our desired gene modules (Supplementary Figure SM1).
The objective function is maximized with a Simulated Annealing algorithm. Initially, genes are assigned random cluster labels from 1 to K and the “temperature” variable is set to 10. In each proposed move, a gene is selected at random and assigned a different cluster label. The objective function is re-evaluated, “good” moves that generate a better score are accepted, whereas “bad” moves are rejected with probability that depends on the score of the proposed reassignment and the temperature variable. Specifically, the probability of accepting a proposed move that generates a new clustering with score Qnew, assuming the current score is Qcur, is given by min(1, (Qnew/Qold)T), where temperature T changes across iterations according to the cooling schedule Tk+1 = αTk, where α = 0.9, and k is the iteration index. This results in bad moves being rejected with low probability in earlier iterations (when the “temperature” is higher), and with higher probability in later iterations. The iterative procedure stops once no good move can be found after certain amount of attempts, or a pre-determined number of iterations have been performed.
Identifying and annotating gene coexpression modules with a conserved response to social challenge
We used 19-df chi-square tests of independence to test if the DEGs in each species/brain region/time point combination were distributed randomly across the 20 modules. A non-random distribution indicates that exposure to social challenge results in certain modules being more activated than others. To identify the active ones, we used post-hoc hypergeometric enrichment tests in the species/brain region/time point combinations with significant chi-square tests.
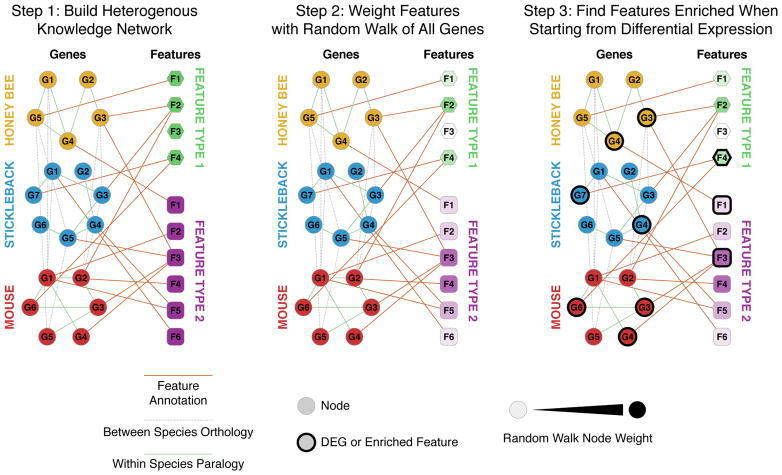
It is not clear immediately clear how to annotate these active modules in a way that also accounts for the available orthology information across the species. This is because annotations for the same module can change depending which species is considered. To address this issue, we employed a multi-species extension of our previously reported DRaWR tool (Blatti & Sinha 2016, see Supplemenatary Methods for details). DRaWR takes a heterogeneous biological network with gene and annotation nodes and ranks all annotation nodes in the network for their proximity to a set of gene nodes of interest by using random walks (Figure 2). We constructed a network containing “gene nodes” representing genes from all three species and “annotation nodes” that represent Gene Ontology annotations (obtained from Biomart for Ensembl v83, Kinsella et al. 2011) and Pfam domains (whose presence was predicted using HMMER, Finn et al. 2011). Edges connected genes with their properties (GO annotations and Pfam domains), and also connected homologous pairs of genes from the same or different species. For a given module, we executed the DRaWR random walk with restarts from module genes from all three species, so that the method is also able to “walk” from a gene to its ortholog(s) in other species. Separately, we also executed the random walk with restarts from module genes of each species individually, and selected annotation nodes that were ranked highest across all four restart configurations. As such, an annotation that is highly ranked by our multi-species DRaWR technique is either enriched in module genes from multiple species or enriched in orthologs of those genes (even if it is not enriched in the module genes themselves), or both. We also required that the reported annotations be significantly enriched (p-value < 0.05 using one-sided Fisher exact test) in at least one of the three species. To our knowledge, the resulting “multi-species DRaWR” algorithm is the first method capable of functional annotation of gene sets in a cross-species manner, making it ideal for identifying HFGs across distantly related species.
Figure 2.
Schematic of heterogeneous knowledge network enrichment using cross-species DRaWR. In brief, the algorithm operates in 3 steps: 1) Construct a heterogeneous network consisting of edges connecting gene nodes with one another within species (paralogy relationships in this dataset), with one another between species (cross-species orthology information in this dataset), and with annotations features (Gene Ontology terms and Pfam domains in this dataset). 2) Using a random walk with restarts starting on the whole gene set, identify features that are frequently traversed and weight them appropriately. 3) Using a random walk with restarts on the differential expression gene set, identify features that are more frequently traversed than in all genes.
RESULTS
Brain transcriptomic response to social challenge in three diverged species shares several orthologous gene groups
We profiled gene expression by sequencing mRNA at 30 min, 60 min, or 120 min after exposure to an intruder from discrete brain regions chosen for each species: the mushroom bodies in honey bee; the amygdala, frontal cortex, and hypothalamus in mouse; and the diencephalon and telencephalon in stickleback. We considered only genes that were sufficiently expressed in these RNA-seq experiments for downstream analysis 10,701 in honey bee, 15,388 in mouse, and 17,435 in stickleback. Differentially expressed genes (DEGs) were obtained by comparison of intruder-exposed animals to control animals in matched conditions (see Materials and Methods), providing three sets of DEGs in honey bee, nine sets in mouse, and six sets in stickleback; these results have been reported elsewhere as individual species studies (Bukhari et al. 2017; Saul et al. 2017, Shpigler et al. 2017b) and are summarized in Figure 1A, but this is the first time that these data have been analyzed and discussed in a comparative context. These DEG sets varied in size between 36 genes (mouse amygdala, 60 min) to 1,151 (honey bee mushroom bodies, 120 min).
We were first interested in whether the same (orthologous) genes were associated with social challenge responses across these three species. However, the great evolutionary divergence of these species precludes unambiguous ortholog assignments at the gene level. We instead used orthologous groups (“orthogroups”) of genes as our fundamental unit of analysis. A resulting major analytical challenge is that most orthogroups contain different numbers of paralogs in the genomes of each individual species, and furthermore different numbers of brain regions were assessed in each species. These challenges make it difficult to ensure a fair comparison across species. Overcoming these issues requires carefully designed statistics, and existing approaches to this type of analysis, such as the one we previously employed in ref. (Rittschof et al. 2014), cannot be applied. To address this problem, we developed a method to identify orthogroups with the strongest evidence for activity in multiple species, where activity was measured by the proportion of DEGs, at any time point and brain region, within the orthogroup in each species (see Materials and Methods). Our procedure is based on another algorithm we recently developed called Orthoverlap (Shpigler et al. 2017a) and offers stringent control of false positives.
We obtained 4,982 orthogroups common to the three species from the OrthoDB database (Kriventseva et al. 2015), and our method identified six orthogroups that were responsive to a social challenge in all three species at FDR ≤ 0.10 (Figure 3, Supplementary Table 2). Three of the six contained at least one DEG in each of the species. Group EOG80K992 (p-value ≤ 1 × 10−7) – which includes the mouse genes F5, Nrp2, Sned1, and Vwf – is potentially involved in a deeply conserved immune response (Chang et al. 2012), but is also related to neurite outgrowth (Hey-Cunningham et al. 2013) and axon guidance (Klagsbrun & Eichmann, 2005). Group EOG8THX4X (p-value = 8.2 × 10−6), which includes the mouse gene Npas4, is a gene that is involved in activity-dependent development of synapses (Lin et al. 2008) and that regulates the balance between GABA and glutamate in neural circuits (Spiegel et al. 2014). This finding is consistent with our previous work (Rittschof et al. 2014; Saul et al. 2017), which also identified Npas4 as a central gene in the shared response to social challenge based on transcriptomic analysis. Finally, group EOG8TMSCQ (p-value = 1.6 × 10−5) includes subunits of the heat shock protein 70 family, which is nominally associated with stressors like heat shock that require protein refolding and that often acts in concert with co-chaperones in the heat shock protein 90 family (Mayer & Bukau, 2005). Heat shock proteins from the Hsp70/Hsp90 complex have an additional documented but less discussed role, being necessary for ligand binding and subsequent signal transduction of nuclear receptors and other signaling molecules (Pratt & Toft, 2003).
Figure 3.
Orthogroups with significant conservation of differential expression across all three species.
The remaining three statistically significant orthogroups include DEGs in two out of the three species. We still considered these orthogroups of potential importance. For example, group EOG8M934T (p-value = 9.6 × 10−6), which includes the mouse gene Nr4a1, only contained DEGs in honey bee and mouse. However, one of the stickleback orthologs was detected at an FDR of 0.1012 (uncorrected p-value = 0.0189) in telencephalon at 30 min, only slightly higher than the 10% FDR cutoff used for that species. This group of Nr4a orthologs, orphan nuclear receptors with unknown ligands, thus appears to have common socially regulated activity. These receptors, which are known to regulate glucose metabolism and homeostasis (Close et al. 2013), have documented roles in memory and in object recognition (McNulty et al. 2012) and have been documented as related to social aggression in vertebrates previously (Malki et al. 2016). Additionally, group EOG8F4TSP (p-value = 2 × 10−7), which contains “zinc finger of the cerebellum” (Zic) proteins, contains at least one DEG in both mouse and stickleback, but not in honey bee. This group of C2H2 zinc finger proteins is known for their evolutionarily conserved roles in neural development (Aruga, 2004; Fujimi et al. 2006).
Several of these findings rose to significance only because we collected RNA-seq data in three species. For example, Npas4 and Nr4a1, transcription factors involved in neural function and/or development, had not been identified as central molecules in the response to social challenge in each individual species (but see Rittschof et al. 2014; Shpigler et al. 2017b), but our comparative analysis showed that these genes were consistently involved in the brain’s response to challenge in all three of our species. The multiple brain region/time point resolution of these RNA-seq data also allowed us to identify shared genes that are transiently expressed, and/or expressed in a brain-region specific manner. For example, the heat shock orthogroup, which contains the chaperone gene Hspa1a, a potential cofactor with nuclear receptors like Nr4a1, was only active at 120 min in the mouse and in the diencephalon in the stickleback and would have been missed if not for the time course design.
Social challenge triggers shared hormone-dependent neuronal signaling
Shared mechanisms of the response to social challenge may emerge at higher levels of organization than that of individual genes. We asked if the same cellular processes (e.g., Gene Ontology terms) are transcriptionally active in response to social challenge, even if specific genes exhibiting differential expression are not strictly orthologs of each other. This allows us to be more sensitive to cellular mechanisms that may have evolved by convergence through repeated coopting the same biological pathways.
We considered gene sets defined by 341 Gene Ontology (GO) terms that contained at least 5 genes in each of the species studied here. Using the method that we developed for our analysis of shared gene orthogroups above, we identified those GO terms that had the strongest evidence for enrichment of DEGs in each of the three species. We identified 66 GO terms at FDR ≤ 0.10 and 37 GO terms at a more stringent threshold of FWER ≤ 0.10 (Table 1). These terms centered on five major categories: hormone activity, transmembrane transport, G-protein coupled receptor (GPCR) signal transduction, synaptic activity, and extracellular matrix components. This analysis thus identified a set of processes that correspond to the same general functions, though they have a slightly different complement of genes between distantly related taxa. Specifically, these results suggest that hormone receptors, acting as nuclear receptors, signaling molecules and transcription factors, are essential in the coordination of the large-scale social challenge induced transcriptional responses that potentially cause remodeling of axons and dendrites, which lead to differences in synapse-related proteins, extracellular matrix proteins, transmembrane transporters, and the modulation of GPCRs for neural signaling.
Table 1.
Multiple cross-species-mapped GO terms show conserved activity in response to social challenge (includes Biological Processes, Cellular Components, and Molecular Functions).
| Biological Process GO ID – Term | P Sim. | Ratio (Hits/Total Genes in GO Term) | ||
|---|---|---|---|---|
| Honey Bee | Mouse | Stickleback | ||
| GO:0007186 – G-protein coupled receptor signaling pathway | < 1 × 10−7 | 30/139 | 52/344 | 58/399 |
| GO:0007218 – neuropeptide signaling pathway | < 1 × 10−7 | 5/17 | 20/70 | 2/6 |
| GO:0007601 – visual perception | < 1 × 10−7 | 1/6 | 7/71 | 12/16 |
| GO:0055085 – transmembrane transport | < 1 × 10−7 | 36/246 | 42/338 | 67/425 |
| GO:0007155 – cell adhesion | 2.0 × 10−7 | 8/54 | 40/308 | 20/120 |
| GO:0006836 – neurotransmitter transport | 4.0 × 10−7 | 4/16 | 11/28 | 3/28 |
| GO:0043401 – steroid hormone mediated signaling pathway | 5.4 × 10−7 | 11/21 | 9/52 | 5/61 |
| GO:0007169 – transmembrane receptor protein tyrosine kinase signaling pathway | 5.8 × 10−6 | 1/7 | 14/81 | 11/45 |
| GO:0006811 – ion transport | 6.6 × 10−6 | 14/108 | 17/176 | 39/190 |
| GO:0006366 – transcription from RNA polymerase II promoter | 7.2 × 10−6 | 2/22 | 41/314 | 0/6 |
| GO:0007165 – signal transduction | 1.8 × 10−4 | 50/285 | 60/633 | 50/506 |
| GO:0007166 – cell surface receptor signaling pathway | 2.9 × 10−4 | 3/16 | 17/132 | 11/58 |
|
| ||||
| Cellular Component GO ID – Term | P Sim. | Ratio (Hits/Total Genes in GO Term) | ||
| Honey Bee | Mouse | Stickleback | ||
|
| ||||
| GO:0005576 – extracellular region | < 1 × 10−7 | 27/113 | 85/481 | 31/176 |
| GO:0005578 – proteinaceous extracellular matrix | < 1 × 10−7 | 2/20 | 40/193 | 3/14 |
| GO:0005615 – extracellular space | < 1 × 10−7 | 6/17 | 107/712 | 5/18 |
| GO:0005886 – plasma membrane | < 1 × 10−7 | 8/58 | 203/2146 | 23/109 |
| GO:0005887 – integral component of plasma membrane | < 1 × 10−7 | 3/7 | 83/562 | 1/9 |
| GO:0016021 – integral component of membrane | < 1 × 10−7 | 119/785 | 245/3266 | 153/1212 |
| GO:0016459 – myosin complex | < 1 × 10−7 | 8/23 | 2/39 | 21/53 |
| GO:0031012 – extracellular matrix | 2.0 × 10−7 | 2/11 | 28/158 | 6/40 |
| GO:0016020 – membrane | 4.8 × 10−6 | 123/813 | 147/2521 | 174/1229 |
| GO:0045202 – synapse | 6.7 × 10−5 | 1/25 | 31/236 | 1/6 |
|
| ||||
| Molecular Function GO ID – Term | P Sim. | Ratio (Hits/Total Genes in GO Term) | ||
| Honey Bee | Mouse | Stickleback | ||
|
| ||||
| GO:0003774 – motor activity | < 1 × 10−7 | 8/23 | 2/51 | 21/53 |
| GO:0004930 – G-protein coupled receptor activity | < 1 × 10−7 | 26/124 | 37/247 | 51/360 |
| GO:0005179 – hormone activity | < 1 × 10−7 | 3/8 | 12/41 | 10/40 |
| GO:0005509 – calcium ion binding | < 1 × 10−7 | 29/169 | 57/488 | 90/535 |
| GO:0043565 – sequence-specific DNA binding | 2.0 × 10−7 | 32/133 | 44/385 | 48/365 |
| GO:0005515 – protein binding | 1.2 × 10−6 | 279/1635 | 534/8766 | 417/3710 |
| GO:0003707 – steroid hormone receptor activity | 1.4 × 10−6 | 10/18 | 9/45 | 5/62 |
| GO:0005216 – ion channel activity | 2.4 × 10−6 | 5/65 | 11/109 | 30/119 |
| GO:0005198 – structural molecule activity | 4.8 × 10−6 | 2/24 | 12/86 | 20/79 |
| GO:0005201 – extracellular matrix structural constituent | 5.6 × 10−6 | 4/7 | 6/26 | 7/25 |
| GO:0020037 – heme binding | 3.3 × 10−5 | 10/63 | 15/77 | 11/72 |
| GO:0004714 – transmembrane receptor protein tyrosine kinase activity | 3.4 × 10−5 | 1/6 | 8/36 | 8/28 |
| GO:0005215 – transporter activity | 4.6 × 10−5 | 13/95 | 19/110 | 16/122 |
| GO:0005506 – iron ion binding | 2.6 × 10−4 | 8/60 | 16/95 | 11/82 |
| GO:0003700 – transcription factor activity, sequence-specific DNA binding | 2.7 × 10−4 | 40/168 | 51/637 | 38/346 |
Shared gene coexpression modules responding to social challenge
In addition to defining sets of genes using GO terms, we sought to identify coordinately expressed sets of genes, often called gene modules, ab initio, without the need for prior knowledge. Coexpressed gene modules have become a mainstay of systems-level analysis of transcriptional programs (Langfelder & Horvath 2008). Studies in evolutionary developmental biology have noted that modules underlying development are deeply conserved and are an important facet of the genetic “toolkit” concept (Peter & Davidson 2011; Rittschof & Robinson 2016; Toth & Robinson 2007). Coexpressed modules are also conserved in other biological contexts across evolutionary spans as great as humans, flies, worms, and yeast (Stuart et al. 2003), but typically have not been explicitly examined as an HFG representing shared responses to social stimuli.
Using the new developed CNSRV method for cross-species analysis, we sought to discover broadly shared gene modules (see schematic representation of coexpressed shared modules in Figure 4A) from our multi-species brain transcriptomic data, then query if any of these are regulated by social challenge commonly across the three species. Our experimental design allowed us to characterize modules with coordinated anatomic and temporal expression profiles in bee, mouse, and stickleback. We then combined these results with our DEGs to identify modules that were highly responsive to social challenge (see Materials and Methods). These represent shared core regulatory programs, where conservation is identified at the level of coexpressed modules rather than individual genes.
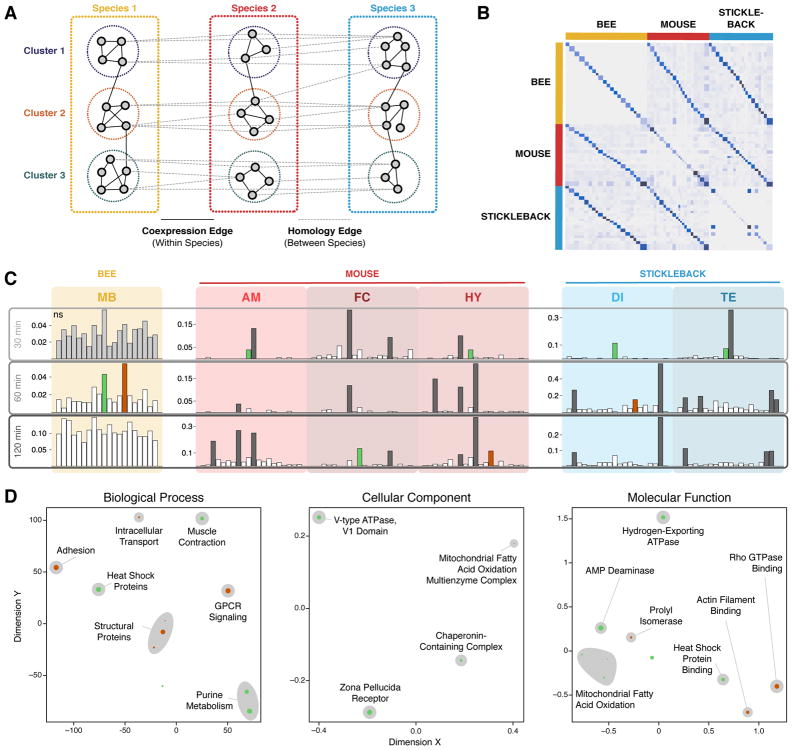
Figure 4.
Cross-species coexpression module algorithm (A) conceptual schematic and (B–D) results. A) Schematic of CNSRV, the cross-species clustering algorithm used to find conserved gene modules, which uses evidence derived from both coexpression and conservation to find gene modules enriched in conservation. B) Clustering results from CNSRV show that conserved modules, shown by the ancillary diagonals off the main diagonal, cluster better between species than do unmatched modules. C) Enrichment results for DEGs for CNSRV modules within each species reveal significant differences among clusters in all but honey bee mushroom body at 30 min (light gray). Multiple CNSRV clusters were enriched in individual species (dark gray), but two modules – 10 and 14, shown in green and dark orange respectively – show simultaneous enrichment for differential expression across all 3 species. D) Multidimensional scaling on semantic distances for GO terms enriched in the multi-species DRAWR results show clusters of GO terms commonly related to clusters 10 and 14 across all 3 species. Larger points associated with each GO term correspond to stronger p-values. Gray clouds correspond to a high-level biological description of the GO terms within each cluster annotated by the authors.
With CNSRV, we identified 20 homologous modules (Figure 4B), each ranging between 140 and 523 genes in size (see Materials and Methods and Supplementary Table 3). These modules show both dense coexpression within modules in individual species (Figure 2B, central diagonal) and elevated frequency of orthology relationships between corresponding modules (Figure 2B, ancillary diagonals). Next, for each combination of brain region and time point in each species, we tested if DEGs were differentially distributed among the modules (see Materials and Methods) and found this to be the case (FDR ≤ 0.10) for all but one of the 18 species/brain region/time point combination (Figure 4C). Within these 17 significant combinations of region, time, and species, we then conducted post-hoc tests at FWER ≤ 0.10 to identify significantly enriched modules.
This analysis revealed that two gene coexpression modules, numbered 10 and 14, have shared social challenge-specific activity across all three species (Figure 4C). Specifically, module 10 is significantly associated with DEGs in honey bee mushroom body (60 min), mouse frontal cortex (120 min) and hypothalamus (30 min), as well as stickleback diencephalon (30 min) and telencephalon (30 min). Similarly, module 14 is enriched for DEGs in honey bee mushroom body (60 min), mouse hypothalamus (120 min), and stickleback diencephalon (60 min). We note that though coexpression module enrichment showed limited evidence of a conserved order – green module enrichment was identified earlier than orange module enrichment in both mice and sticklebacks – the time points where the orthologous modules were observed often did not match between the species, which may have resulted from differences in the timing of the neurobiological responses of each species, underscoring the importance of multiple time points in the study design.
While it was instructive to observe shared modules apparently regulated by social challenge, it was not as clear what biological functions these modules might be involved with. Functional annotation of these modules is difficult because a module in our context is not a single list of genes but a set of three different species-specific gene lists with strong mutual orthology, and standard gene set enrichment tests do not take into account the multi-species nature of the modules or their orthology relationships. To solve this problem, we adapted our previously developed tool DRaWR (Blatti & Sinha, 2016), which considers a network whose nodes are genes and annotations (e.g., Gene Ontology terms) and edges connect a gene to each of its annotations. It annotates a gene set by performing a random walk starting from nodes in the gene set and recording the annotation nodes that are visited most frequently. We extended this approach here to annotate the orthologous CNSRV modules by constructing a network using module genes, GO annotations, and orthology edges from all three of our species (see Materials and Methods).
Figure 4D shows the top functional annotations for modules 10 and 14, as revealed by a high DRaWR percentile score in every species, and with additional support from standard enrichment tests (hypergeometric test nominal p-value ≤ 0.05) in at least two of the three species. Module 14 comprises genes involved in cell-matrix adhesion, a process involved in neural development and plasticity (Murase & Schuman 1999); Rho GTPase binding, a process implicated in several aspects of neuronal development as well as neurological diseases (Govek et al. 2005); and actin binding, a process associated with function and plasticity of dendritic spines and synapses (Lin & Webb, 2009). Module 10 includes genes annotated for AMP deaminase activity and IMP biosynthesis, processes associated with purine balance in the brain. Purine balance and purinergic reception play well-known roles in neuronal repair and protection, acting as a bridge between neural signaling and the neural immune system in mammals (Skaper et al. 2010; Thauerer et al. 2012). Further, the enrichment of enoyl-CoA hydratase activity found in Module 10, as a step of fatty acid metabolism found in the Cellular Component and Molecular Function results, potentially bridges neural signaling and the metabolic processes previously observed in response to social challenge both across species and within individual species (Chandrasekaran et al. 2015; Rittschof et al. 2014). These coexpression modules bolster evidence from the shared DEGs and from the GO results in support of a shared transcriptomic response that includes structural proteins, heat shock proteins, and GPCR signaling proteins.
Common transcription factor regulatory activities underlie social challenge
The previous sections provide insight into the common biological processes and gene modules underlying the response to social challenge. We next sought to identify transcription factors (TFs) that act as master regulators of those processes and modules, using state-of-the-art tools for reconstruction of transcriptional regulatory networks in each species. In particular, we queried if the same TFs (or their paralogs) regulate the brain transcriptomic response to social challenge across species. TF-gene relationships are among the best studied and most widely accepted conception of gene networks, and they have been explored in the context of genetic toolkit studies in evo-devo (Rittschof & Robinson, 2016). The gene orthogroup analysis reported above (Figure 3) identified multiple TF orthogroups containing social challenge DEGs; however, there may be TFs which do not detectably change in transcript expression upon social challenge but may for example be activated by post-transcriptional modifications. Regulatory targets for these TFs may nevertheless be socially regulated and the TFs reasonably speculated to have a role in the transcriptional response to social challenge. We reasoned that even if the TFs are not differentially expressed on social challenge, their regulatory targets may still be identifiable from a covariance between TF and gene expression across the many brain regions and time points assayed here, allowing us to test enrichment between each TF’s targets and social challenge DEGs.
To explore this idea, we constructed transcriptional regulatory networks (TRNs) for each species using the previously developed tool ASTRIX (Chandrasekaran et al. 2011), which uses the ARACNE algorithm (Margolin et al. 2006) to identify putative TFs for a gene, then employs Least Angle Regression (Efron et al. 2004) to identify those TFs that best predict expression levels of that gene target in multiple experiments. In this case, each TRN had been reconstructed from different brain regions and time points within each individual species previously (Bukhari et al. 2017; Saul et al. 2017; Shpigler et al. 2017b). We used these previously reconstructed TRNs to identify TF orthogroups whose gene targets were enriched in DEGs in all three species, using the same method as described above for identifying the HFGs of shared gene orthogroups and GO terms. We considered only orthogroups that contained at least one TF with at least one gene target in each species. This analysis detects TFs important to social challenge even if the TFs themselves are not significantly differentially expressed.
We detected six TF orthogroups (FDR ≤ 0.10) that are likely to be conserved regulators of the transcriptomic response to social challenge (Table 2). For instance, the orthogroup EOG8KWM99 comprises the mouse TF genes Pbx1 and Pbx3, for which the ASTRIX-derived TRN included 2 target genes in mouse, both of which are social challenge DEGs, 45 targets (including 3 DEGs) in stickleback and 25 targets (including 9 DEGs) in honey bee. Further, one orthogroup that includes the mouse neural development transcription factor genes Rax and Pax6 may be involved in the conserved regulation of the formation of new neurons from a neural stem cell lineage (Davis et al. 2003; Pak et al. 2014). Rax was identified as a transcriptional regulator in our earlier work (Rittschof et al. 2014). One particularly interesting TF, the orphaned nuclear receptor mouse gene Nr2e1, has been implicated in our previous cross-species work (Rittschof et al. 2014), in aggression in mice (Abrahams et al. 2005), and in aggression in flies (Davis et al. 2014). These results identify specific transcriptional regulators that appear to be important central regulators of the processes described in the above sections and therefore constitute potential key shared master regulators of the transcriptional response to social challenge.
Table 2.
Conserved transcription factor expressed in the brain implicated as regulators of response to social challenge.
| Orthogroup | Ratio (Hits/Total Targets) | p | Mouse Gene Names | ||
|---|---|---|---|---|---|
| Honey Bee | Mouse | Stickleback | |||
| EOG8JT1HM | 1/3 | 12/84 | 30/128 | 0 | Sp3, Klf16, Klf13, Klf10, Sp4, Klf4, Klf9, Sp1, Sp7, Klf5, Klf12, Klf7 |
| EOG873R3N | 0/1 | 8/42 | 11/34 | 0.000004 | Barx2, Nkx2-1, Hmx2, Hhex |
| EOG8KWM99 | 9/25 | 2/2 | 3/45 | 0.0003072 | Pbx3, Pbx1 |
| EOG86DNH2 | 5/9 | 0/1 | 1/1 | 0.0012816 | Nr2e1 |
| EOG8JWWWP | 4/5 | 1/6 | 0/6 | 0.0055148 | Gsx1 |
| EOG81RRB5 | 5/105 | 1/2 | 9/42 | 0.0171374 | Arx, Pax6, Rax |
DISCUSSION
The evolution of gene regulatory programs is a subject of long-standing interest (Halfon & Michelson 2002) and has been studied by cross-species comparisons of cis-regulatory sequences (Sinha et al. 2004), TF-DNA binding (ENCODE Project Consortium 2012), as well as gene expression measurements in matched tissues and organs (Breschi et al. 2017; Gerstein et al. 2014; Lin et al. 2014). An important feature of our study was its explicit coupling of gene and gene network comparisons with objectively defined and analogous phenotypic states measured experimentally to identify shared mechanisms that may constitute a behavioral toolkit. We note that such an evolutionary toolkit, consisting of a non-unitary system of genes, does not require framing in the dichotomy of common descent versus convergence. It is instead possible that shared functions derive from redeployment and elaboration of ancestral gene modules into new contexts as has been observed in insect gene regulatory network evolution (Kazemian et al. 2014, Suryamohan et al. 2016).
Our approach utilizes an array of novel tools with a common theme of studying different tiers of organization for evidence of a shared genetic program: each test assays if groups of related genes – orthogroups, functional systems, coexpressed modules, or transcription factor regulons – have a non-random association with socially responsive genes expressed in the brain in multiple species. Our methodology for cross-species associations between HFGs and phenotypes can also be used in other contexts to identify similar broadly shared molecular systems in association with other phenotypes of interest. Moreover, the scope of such transcriptome-wide comparisons distinguishes this work from more directed studies of regulatory evolution where expression and cis-regulatory divergence of individual genes was linked to morphological differences between species (Wray 2007). Our goal is similar to the work of (Malki et al. 2016), who compared aggression-related DEGs in brains of mouse and zebrafish, but our study pursues the goal through an experiment design wherein data were collected from multiple species in a parallel manner, addressing several key technical and statistical challenges in the process.
One technical challenge observed in these data is the difficulty in matching gene expression sets across such long evolutionary distances. We note that time points where the orthologous modules were observed often did not match between the species, which may have resulted from differences in metabolic rates between the species. This observation underscores the importance of time series in the study design. Furthermore, it demonstrates that experimental designs in future studies must proceed carefully to identify matching expression sets across species. We were able to go beyond identification of differentially expressed genes and rigorously analyze coexpression relationships only because our experimental design included multiple brain regions and time points. Thus, the design gave us access to the higher order systems mechanisms mentioned above, significantly elaborating upon our earlier work (Rittschof et al. 2014).
Integrating results from these multiple analytical levels, we propose a system of genes acting commonly in the adult brains of these diverged species to transduce social challenge stimuli into transcriptional and epigenomic responses. This is graphically summarized in Figure 5. It involves the integration of nuclear receptor signaling to drive the transcriptional regulatory events that result in changes in neural signaling observed after a social challenge. We speculate that because nuclear receptors are both liganded receptors and transcription factors, they act as key drivers of the large-scale transcriptional changes seen across all of these species. We further speculate that these transcriptional changes occur in concert with transcription factors commonly associated with neural development to drive neural signaling modulation, which likely take place through alterations in dendritic architecture, axon architecture, signaling molecules like GPCRs and ion channels, mitochondrial metabolism, or all of these processes simultaneously.
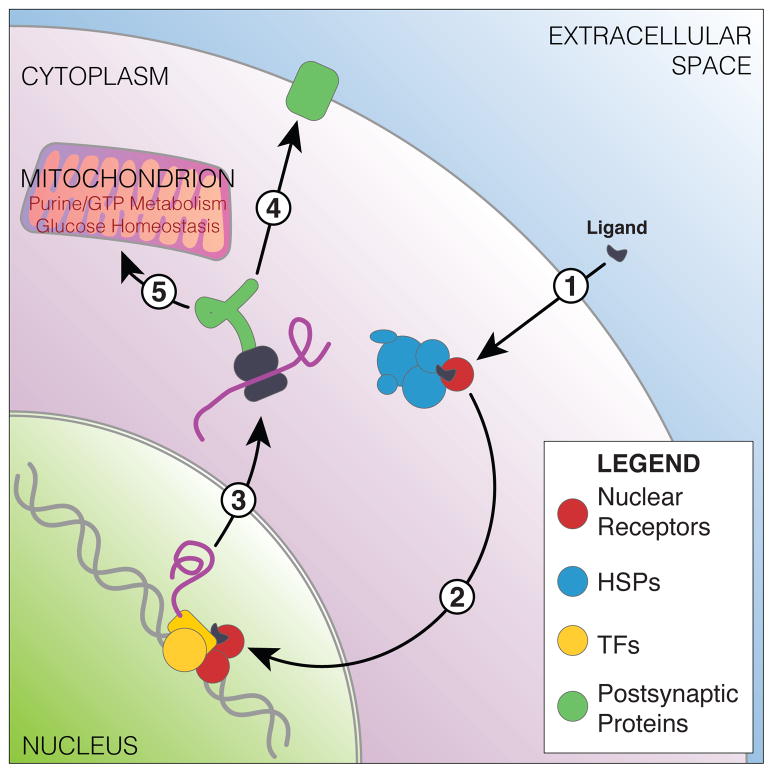
Figure 5.
Schematic representation of genes and gene sets found enriched in the brain’s response to social challenge across honey bees, stickleback fish and mice. Hypothesized pathway includes 1) nuclear receptor signaling interacting with heat shock/chaperones. 2) These nuclear receptors translocate across the membrane, interacting with well-known neurally active transcription factors to cause 3) alterations in transcription. These induce changes in 4) postsynaptic proteins and 5) mitochondrial function.
In this pathway, we call specific attention to the signaling molecules, transcription factors, and nuclear receptors that can act as both. Specifically, the various homologs of Npas4, Nr2e1, and Nr4a1 are transcription factor genes well-known in neural response to stimuli (Abrahams et al. 2005, Kim et al. 2010; Maxwell & Muscat 2006). We speculate that the ancestral versions of these genes, which were likely present in the most recent common ancestor of all living bilaterians, were potentially already active in the response to social challenge stimuli that was exhibited by their contemporaries around the time of the Cambrian explosion (Carbone & Narbonne 2014). Translating these gene expression patterns into knowledge about how the cellular systems inside the brain change in response to social challenge is an important next step. Such research will require careful work across species to identify important points of similarity as well as how these systems diverge.
Ascertainment of the conserved components of a behavioral toolkit permit translational work in the future, such as the use of “bottom-up” approaches for identifying similarities in behaviors across widely diverged species through similarities in functional genomic profiling (Rittschof & Robinson 2016). This approach already shows promise; recent work reported similarities in the gene sets related to social responsiveness in humans and honey bees (Shpigler et al. 2017a). To that point, though we discussed the role and neurobiological relevance of some of the above-mentioned systems in detail in our previous work– we described hormone receptors in sticklebacks (Bukhari et al. 2017), developmental transcription factors in mice (Saul et al. 2017), dendritic architecture in honey bees (Shpigler et al. 2017b), and GPCRs in all three species (Bukhari et al. 2017; Saul et al. 2017; Shpigler et al. 2017b) – the present work identifies core systems relating to social behavior. The genes and systems-level mechanisms we proposed here as drivers of the response to social challenge constitute real, testable connections about a possibly conserved toolkit for the response to a social challenge, something that was lacking before this analysis. However, we note that these mechanisms may not be specific to social contexts, but may instead coordinate information from multiple contexts, and thus, the specificity of these gene sets for social challenge response also needs rigorous testing. The analytical pipeline we lay out in this work is suited to address this question in the future with further data.
Supplementary Material
Acknowledgments
We would like to thank Yoshitsugu Oono and Jian Ma for their critical comments during the compilation of this manuscript. This research was primarily funded by grant #SFLife 291812 from the Simons Foundation. In addition, this research was supported in part by grant 1U54GM114838 awarded by NIGMS through funds provided by the trans-NIH Big Data to Knowledge (BD2K) initiative and by NSF grant DMS-1613005 to SDZ.
Footnotes
Supplementary Methods (Supplementary_Methods.pdf): Detailed materials and methods for the CNSRV cross-species coexpression module discovery algorithm (includes Supplementary Figures SM1–3).
Supplementary Table 1 (Supplementary_Table_1.pdf): Sample sizes of RNA-seq datasets utilized in this manuscript.
Supplementary Table 2 (Supplementary_Table_2.xls): Orthogroups displaying significant cross-species enrichment in DEG lists.
Supplementary Table 3 (Supplementary_Table_3.xls): CNSRV-derived cross-species coexpression modules.
References
- Abrahams BS, Kwok MC, Trinh E, Budaghzadeh S, Hossain SM, Simpson EM. Pathological aggression in “fierce” mice corrected by human nuclear receptor 2E1. J Neurosci. 2005;25:6263–6270. doi: 10.1523/JNEUROSCI.4757-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruga J. The role of Zic genes in neural development. Mol Cell Neurosci. 2004;26:205–221. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Asahina K, Watanabe K, Duistermars BJ, Hoopfer E, Gonzalez CR, Eyjolfsdottir EA, Perona P, Anderson DJ. Tachykinin-expressing neurons control male-specific aggressive arousal in Drosophila. Cell. 2014;156:221–235. doi: 10.1016/j.cell.2013.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatti C, Sinha S. Characterizing gene sets using discriminative random walks with restart on heterogeneous biological networks. Bioinformatics. 2016;32:2167–2175. doi: 10.1093/bioinformatics/btw151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breschi A, Gingeras TR, Guigo R. Comparative transcriptomics in human and mouse. Nat Rev Genet. 2017;18:425–440. doi: 10.1038/nrg.2017.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari SA, Saul MC, Seward CH, Zhang H, Bensky M, James N, Zhao SD, Chandrasekaran S, Stubbs L, Bell AM. Temporal Dynamics of Neurogenomic Plasticity in Response to Social Interactions in Male Threespined Sticklebacks. PLoS Genetics. 2017;13:e1006840. doi: 10.1371/journal.pgen.1006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone C, Narbonne GM. When life got smart: the evolution of behavioral complexity through the Ediacaran and early Cambrian of NW Canada. Journal of Paleontology. 2014;88:309–330. [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to diversity: molecular genetics and the evolution of animal design. 2. Blackwell; Malden, MA: 2005. [Google Scholar]
- Chandrasekaran S, Ament SA, Eddy JA, Rodriguez-Zas SL, Schatz BR, Price ND, Robinson GE. Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states. Proc Natl Acad Sci U S A. 2011;108:18020–18025. doi: 10.1073/pnas.1114093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran S, Rittschof C, Djukovic D, Gu H, Raftery D, Price N, Robinson G. Aggression is associated with aerobic glycolysis in the honey bee brain1. Genes Brain Behav. 2015;14:158–166. doi: 10.1111/gbb.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HJ, Dhanasingh I, Gou X, Rice AM, Dushay MS. Loss of Hemolectin reduces the survival of Drosophila larvae after wounding. Dev Comp Immunol. 2012;36:274–278. doi: 10.1016/j.dci.2011.04.009. [DOI] [PubMed] [Google Scholar]
- Chen Z, Zhou C, Meyer M, Xiang K, Schiffbauer JD, Yuan X, Xiao S. Trace fossil evidence for Ediacaran bilaterian animals with complex behaviors. Precambrian Res. 2013;224:690–701. [Google Scholar]
- Close AF, Rouillard C, Buteau J. NR4A orphan nuclear receptors in glucose homeostasis: a minireview. Diabetes Metab. 2013;39:478–484. doi: 10.1016/j.diabet.2013.07.005. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ, Tavsanli BC, Dittrich C, Walldorf U, Mardon G. Drosophila retinal homeobox (drx) is not required for establishment of the visual system, but is required for brain and clypeus development. Dev Biol. 2003;259:272–287. doi: 10.1016/s0012-1606(03)00201-x. [DOI] [PubMed] [Google Scholar]
- Davis SM, Thomas AL, Nomie KJ, Huang L, Dierick HA. Tailless and Atrophin control Drosophila aggression by regulating neuropeptide signalling in the pars intercerebralis. Nat Commun. 2014;5:3177. doi: 10.1038/ncomms4177. [DOI] [PubMed] [Google Scholar]
- Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Ann Stat. 2004;32:407–499. [Google Scholar]
- Farris SM, Robinson GE, Fahrbach SE. Experience- and age-related outgrowth of intrinsic neurons in the mushroom bodies of the adult worker honeybee. J Neurosci. 2001;21:6395–6404. doi: 10.1523/JNEUROSCI.21-16-06395.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris SM, Sinakevitch I. Development and evolution of the insect mushroom bodies: towards the understanding of conserved developmental mechanisms in a higher brain center. Arthropod Struct Dev. 2003;32:79–101. doi: 10.1016/S1467-8039(03)00009-4. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenberg F, Carreno Gutierrez H, Post AM, Reif A, Norton WH. Aggression in non-human vertebrates: Genetic mechanisms and molecular pathways. Am J Med Genet B Neuropsychiatr Genet. 2016;171:603–640. doi: 10.1002/ajmg.b.32358. [DOI] [PubMed] [Google Scholar]
- Fujimi TJ, Mikoshiba K, Aruga J. Xenopus Zic4: conservation and diversification of expression profiles and protein function among the Xenopus Zic family. Dev Dyn. 2006;235:3379–3386. doi: 10.1002/dvdy.20906. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Rozowsky J, Yan KK, Wang D, Cheng C, Brown JB, Davis CA, Hillier L, Sisu C, Li JJ, Pei B, Harmanci AO, Duff MO, Djebali S, Alexander RP, Alver BH, Auerbach R, Bell K, Bickel PJ, Boeck ME, Boley NP, Booth BW, Cherbas L, Cherbas P, Di C, Dobin A, Drenkow J, Ewing B, Fang G, Fastuca M, Feingold EA, Frankish A, Gao G, Good PJ, Guigo R, Hammonds A, Harrow J, Hoskins RA, Howald C, Hu L, Huang H, Hubbard TJ, Huynh C, Jha S, Kasper D, Kato M, Kaufman TC, Kitchen RR, Ladewig E, Lagarde J, Lai E, Leng J, Lu Z, MacCoss M, May G, McWhirter R, Merrihew G, Miller DM, Mortazavi A, Murad R, Oliver B, Olson S, Park PJ, Pazin MJ, Perrimon N, Pervouchine D, Reinke V, Reymond A, Robinson G, Samsonova A, Saunders GI, Schlesinger F, Sethi A, Slack FJ, Spencer WC, Stoiber MH, Strasbourger P, Tanzer A, Thompson OA, Wan KH, Wang G, Wang H, Watkins KL, Wen J, Wen K, Xue C, Yang L, Yip K, Zaleski C, Zhang Y, Zheng H, Brenner SE, Graveley BR, Celniker SE, Gingeras TR, Waterston R. Comparative analysis of the transcriptome across distant species. Nature. 2014;512:445–448. doi: 10.1038/nature13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Halfon MS, Michelson AM. Exploring genetic regulatory networks in metazoan development: methods and models. Physiol Genomics. 2002;10:131–143. doi: 10.1152/physiolgenomics.00072.2002. [DOI] [PubMed] [Google Scholar]
- Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neuroscience. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- Hey-Cunningham AJ, Markham R, Fraser IS, Berbic M. Dysregulation of vascular endothelial growth factors and their neuropilin receptors in the eutopic endometrium of women with endometriosis. Reprod Sci. 2013;20:1382–1389. doi: 10.1177/1933719113485299. [DOI] [PubMed] [Google Scholar]
- Kazemian M, Suryamohan K, Chen JY, Zhang Y, Samee MA, Halfon MS, Sinha S. Evidence for deep regulatory similarities in early developmental programs across highly diverged insects. Genome Biol Evol. 2014;6:2301–2320. doi: 10.1093/gbe/evu184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella RJ, Kahari A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, Kersey P, Flicek P. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klagsbrun M, Eichmann A. A role for axon guidance receptors and ligands in blood vessel development and tumor angiogenesis. Cytokine Growth Factor Rev. 2005;16:535–548. doi: 10.1016/j.cytogfr.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Kriventseva EV, Tegenfeldt F, Petty TJ, Waterhouse RM, Simao FA, Pozdnyakov IA, Ioannidis P, Zdobnov EM. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 2015;43:D250–D256. doi: 10.1093/nar/gku1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Lin Y, Nery JR, Urich MA, Breschi A, Davis CA, Dobin A, Zaleski C, Beer MA, Chapman WC, Gingeras TR, Ecker JR, Snyder MP. Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci U S A. 2014;111:17224–17229. doi: 10.1073/pnas.1413624111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WH, Webb DJ. Actin and Actin-Binding Proteins: Masters of Dendritic Spine Formation, Morphology, and Function. Open Neurosci J. 2009;3:54–66. doi: 10.2174/1874082000903020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malki K, Du Rietz E, Crusio WE, Pain O, Paya-Cano J, Karadaghi RL, Sluyter F, de Boer SF, Sandnabba K, Schalkwyk LC, Asherson P, Tosto MG. Transcriptome analysis of genes and gene networks involved in aggressive behavior in mouse and zebrafish. Am J Med Genet B Neuropsychiatr Genet. 2016;171:827–838. doi: 10.1002/ajmg.b.32451. [DOI] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty SE, Barrett RM, Vogel-Ciernia A, Malvaez M, Hernandez N, Davatolhagh MF, Matheos DP, Schiffman A, Wood MA. Differential roles for Nr4a1 and Nr4a2 in object location vs. object recognition long-term memory. Learn Mem. 2012;19:588–592. doi: 10.1101/lm.026385.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase S, Schuman EM. The role of cell adhesion molecules in synaptic plasticity and memory. Curr Opin Cell Biol. 1999;11:549–553. doi: 10.1016/s0955-0674(99)00019-8. [DOI] [PubMed] [Google Scholar]
- Newman ME. Modularity and community structure in networks. Proc Natl Acad Sci U S A. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell LA, Hofmann HA. Evolution of a vertebrate social decision-making network. Science. 2012;336:1154–1157. doi: 10.1126/science.1218889. [DOI] [PubMed] [Google Scholar]
- Pak T, Yoo S, Miranda-Angulo AL, Wang H, Blackshaw S. Rax-CreERT2 knock-in mice: a tool for selective and conditional gene deletion in progenitor cells and radial glia of the retina and hypothalamus. PLoS One. 2014;9:e90381. doi: 10.1371/journal.pone.0090381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. Evolution of gene regulatory networks controlling body plan development. Cell. 2011;144:970–985. doi: 10.1016/j.cell.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Rittschof CC, Bukhari SA, Sloofman LG, Troy JM, Caetano-Anollés D, Cash-Ahmed A, Kent M, Lu X, Sanogo YO, Weisner PA. Neuromolecular responses to social challenge: Common mechanisms across mouse, stickleback fish, and honey bee. Proc Natl Acad Sci U S A. 2014;111:17929–17934. doi: 10.1073/pnas.1420369111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittschof CC, Robinson GE. Behavioral Genetic Toolkits: Toward the Evolutionary Origins of Complex Phenotypes. Curr Top Dev Biol. 2016;119:157–204. doi: 10.1016/bs.ctdb.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Robinson GE, Fahrbach SE, Winston ML. Insect societies and the molecular biology of social behavior. BioEssays. 1997;19:1099–1108. doi: 10.1002/bies.950191209. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul MC, Seward CH, Troy JM, Zhang H, Sloofman LG, Lu X, Weisner PA, Caetano-Anolles D, Sun H, Zhao SD, Chandrasekaran S, Sinha S, Stubbs L. Transcriptional regulatory dynamics drive coordinated metabolic and neural response to social challenge in mice. Genome Res. 2017;27:959–972. doi: 10.1101/gr.214221.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpigler HY, Saul MC, Corona F, Block L, Cash Ahmed A, Zhao SD, Robinson GE. Deep evolutionary conservation of autism-related genes. Proc Natl Acad Sci U S A. 2017a;114:9653–9658. doi: 10.1073/pnas.1708127114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpigler HY, Saul MC, Murdoch EE, Cash-Ahmed AC, Seward CH, Sloofman L, Chandrasekaran S, Sinha S, Stubbs LJ, Robinson GE. Behavioral, transcriptomic and epigenetic responses to social challenge in honey bees. Genes Brain Behav. 2017b;116:579–591. doi: 10.1111/gbb.12379. [DOI] [PubMed] [Google Scholar]
- Sinha S, Schroeder MD, Unnerstall U, Gaul U, Siggia ED. Cross-species comparison significantly improves genome-wide prediction of cis-regulatory modules in Drosophila. BMC Bioinformatics. 2004;5:129. doi: 10.1186/1471-2105-5-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaper SD, Debetto P, Giusti P. The P2×7 purinergic receptor: from physiology to neurological disorders. The FASEB Journal. 2010;24:337–345. doi: 10.1096/fj.09-138883. [DOI] [PubMed] [Google Scholar]
- Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, Harmin DA, Greenberg ME. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- Suryamohan K, Hanson C, Andrews E, Sinha S, Scheel MD, Halfon MS. Redeployment of a conserved gene regulatory network during Aedes aegypti development. Dev Biol. 2016;416:402–413. doi: 10.1016/j.ydbio.2016.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Miczek KA. Neurogenetics of aggressive behavior: studies in rodents. Curr Top Behav Neurosci. 2014;17:3–44. doi: 10.1007/7854_2013_263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauerer B, zur Nedden S, Baier-Bitterlich G. Purine nucleosides: endogenous neuroprotectants in hypoxic brain. J Neurochem. 2012;121:329–342. doi: 10.1111/j.1471-4159.2012.07692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth AL, Robinson GE. Evo-devo and the evolution of social behavior. Trends in Genetics. 2007;23:334–341. doi: 10.1016/j.tig.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Wilkins AS. The evolution of developmental pathways. Sinauer Associates; Sunderland, MA, USA: 2002. [Google Scholar]
- Withers GS, Fahrbach SE, Robinson GE. Selective Neuroanatomical Plasticity and Division-of-Labor in the Honeybee. Nature. 1993;364:238–240. doi: 10.1038/364238a0. [DOI] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Yan KK, Wang D, Rozowsky J, Zheng H, Cheng C, Gerstein M. OrthoClust: an orthology-based network framework for clustering data across multiple species. Genome Biol. 2014;15:R100. doi: 10.1186/gb-2014-15-8-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.