Abstract
Protein p68 is a prototype constituent of DEAD-box protein family, which is involved in RNA metabolism, induced during abiotic stress conditions. In order to address the salinity stress faced by economically important soybean crop, we have transformed soybean cv. PUSA 9712 via direct organogenesis with marker free construct of p68 gene by Agrobacterium-mediated genetic transformation. The putative transgenic plants were screened by Polymerase chain reaction (PCR), Dot blot analysis and Southern blot hybridization. Reverse transcriptase-PCR (RT-PCR) and Quantitative real-time PCR (qRT-PCR) established that the p68 gene expressed in three out of five southern positive (T1) plants. The transformed (T1) soybean plants survived irrigation upto 200 mM of NaCl whereas the non-transformed (NT) plants could not survive even 150 mM NaCl. The transgenic soybean (T1) plants showed a higher accumulation of chlorophyll, proline, CAT, APX, SOD, RWC, DHAR and MDHAR than the NT plants under salinity stress conditions. The transformed (T1) soybean plants also retained a higher net photosynthetic rate, stomatal conductance and CO2 assimilation as compared to NT plants. Further analysis revealed that (T1) soybean plants accumulated higher K+ and lower Na+ levels than NT plants. Yield performance of transformed soybean plants was estimated in the transgenic green house under salinity stress conditions. The transformed (T1) soybean plants expressing the p68 gene were morphologically similar to non-transformed plants and produced 22–24 soybean pods/plant containing 8–9 g (dry weight) of seeds at 200 mM NaCl concentration. The present investigation evidenced the role of the p68 gene against salinity, by enhancing the tolerance towards salinity stress in soybean plants.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1553-z) contains supplementary material, which is available to authorized users.
Keywords: Soybean, Agrobacterium tumefaciens, EHA105, p68 gene, Salinity, CaMV 35S promoter, Binary vector, qRT-PCR
Introduction
Soybean [Glycine max (L.) Merrill] is an annual food crop native to East Asia, Japan and China that belongs to the family Leguminosae or Fabaceae. It is widely cultivated as a source of edible oil and protein in tropical and subtropical regions. USA is the leading producer of soybean (82.11 million metric tonnes) followed by Brazil, Argentina and China. India holds the fifth position for soybean production in the world and produces 8.6 million metric tonnes per year (FAOSTAT 2016). It has a high nutrient content and mature dry soybean seeds consist of 36.49% of protein, 19.94% of fat, 4.87% of ash, 30.16% of carbohydrate, 9.3% of fiber, 7.33% of sugars, minerals, vitamins, amino acids, isoflavones and lipids (USDA 2018). The protein and fiber content in soybean can be of help in preventing high blood sugar levels (Birt et al. 2004). Soybean seeds are also rich in isoflavones such as genistein and daidzein, which were used in the treatment of pancreatic cancer (McCue and Shetty 2004). Soybean oil is used for culinary purposes in margarine, bread, salad dressing and a few other food products like soy milk, soy meal and soy flour. Apart from food products, it has also been used in numerous industrial applications such as paints, hydraulic fluids, printing inks, building materials, pharmaceuticals, cosmetics and plastics (Soy Stats 2011).
Abiotic stresses such as salinity, drought, high or low temperature, cold, radiation, and heavy metal contamination of soil limit agricultural productivity. Amongst them, salinity stress adversely affects plant growth, reducing both crop productivity and quality. Around the world, soil salinity occurs generally in semi-arid and arid regions, due to natural build up or by irrigation with saline water (Meloni et al. 2004). The incidence of excess salts in soil interferes with nutrient uptake and water required by the plant in the crop root zone, thereby decreasing crop development and yield (Bauder and Brock 2001; Hanson et al. 1999). The higher levels of sodium and chloride ions, in saline soil exert adverse conditions in plants such as nutrition disorders, ion toxicity, oxidative stress, membrane degradation and alteration of metabolic processes (Hasegawa et al. 2000; Munns 2002). Globally, high salinity occurrence in soil has damaged more than 45 million hectares of irrigated land, and 1.5 million hectares are becoming uncultivable every year (Munns and Tester 2008).
Soybean is classified as a moderately tolerant crop to salinity stress (Munns and Tester 2008; Guan et al. 2014), with a threshold of 0.5 S m− 1, beyond which growth is markedly reduced (Maas and Hoffman 1977). Guan et al. (2014) have identified that GmSALT3 confers the observed moderate tolerance to salinity in soybean. Salinity affects the root nodulation and reduces the number and biomass of roots, which in turn reduces the nitrogen-fixing ability of soybean (Elsheikh and Wood 1995). However, under increased salinity stress conditions, various biochemical and metabolic alterations are evidenced, which includes ionic imbalance, alterations in the synthesis of specific proteins, accumulation of osmotically active compounds and induction of a series of biochemical and physiological responses in plants, such as repression of cell growth, stomatal closure, loss in photosynthesis, activation of respiration, and reduced yield (Subramanyam et al. 2012). In such adverse conditions, free-radical scavengers, osmoprotectants, heat shock proteins, late embryogenesis abundant (LEA) proteins and chaperones activate important stress response mechanisms in alleviating plant stress (Khan et al. 2015).
The p68 helicase is one of the prototype members of ‘DEAD-box’ protein (Linder et al. 1989). Helicase enzymes are known to unwind energetically stable duplex DNA and RNA secondary structures (DNA and RNA helicases), respectively. The p68 enzyme is an RNA helicase that efficiently unwinds dsRNA in both 3′–5′ and 5′–3′ directions (Tuteja and Tuteja 2004a, b; Tuteja and Pradhan 2006; Huang and Liu 2002). RNA helicases play essential roles in RNA metabolism, including mRNA repair, transcription, pre-mRNA and pre-rRNA processing, gene splicing, editing, regulation of RNA stability and translation, which regulates plant growth and development (Tuteja 2003; Hoi and Tuteja 2012; Guan et al. 2013; Tuteja et al. 2014). It belongs to the largest family of RNA helicases with a molecular mass of 68 kDa protein (Hoi and Tuteja 2012). Pradhan et al. (2005b) reported that the helicase and the ATPase activity of p68 RNA helicase is stimulated following stimulation by protein kinase C in Plasmodium falciparum, a human malaria parasite.
The p68 DEAD-box helicase confers tolerance to salinity stress by reducing oxidative stress, controlling the reactive oxygen species (ROS) and improving photosynthesis machinery in tobacco (Tuteja et al. 2014). Pea DNA helicases (PDH47 and PDH45) are reportedly induced by a range of abiotic stress, including salinity stress in shoots and roots, ABA treatment in roots, dehydration and wounding (Vashisht et al. 2005; Hoi and Tuteja 2012). Expression of p68 DNA helicase (PDH45) reportedly enhanced salinity stress tolerance in tobacco and rice, respectively (Sanan-Mishra et al. 2005; Gill et al. 2013). A DEAD-box RNA helicase (OsBIRH1) showed increased tolerance against pathogens and oxidative stresses in rice (Li et al. 2008). DEAD-box RNA helicase family member, OsSUV3, was found to play a key role in the enhancement of salinity stress tolerance (Tuteja et al. 2013). A p68 DEAD-box RNA helicase (AtDRH1) transcript from Arabibopsis thaliana was described to be stored at an immense level, nearly in all the parts of the plant (Okanami et al. 1998). Proteins ZmDRH1 (Z. mays DEAD-box RNA helicase 1) and MA16 (maize RNA-binding protein) were found to be involved in ribosomal RNA (rRNA) metabolism and are speculated to be a part of the ribonucleoprotein complex (Gendra et al. 2004).
Hence, for the first time, in the present study, p68 gene (marker free construct) from Pea (Pisum sativum), has been transferred into the soybean with the help of Agrobacterium tumefaciens strain EHA105, to confer enhanced tolerance to salinity stress, following which the plant characteristics and yield analysis have been carried out for transformed and non-transformed plants comparatively.
Materials and methods
Plant material and seed source
Seeds of soybean cultivar cv. Pusa 9712 (Fig. 1a) were used in the present investigation. The cv. Pusa 9712 is appropriate for growing under normally sown irrigated conditions and has an average yield of 20.5 q/ha. It matures in 116 days and shows significant resistance to soybean mosaic virus, yellow mosaic virus, charcoal rot, bacterial pustule, stem fly and myrothecium leaf spot. The seeds were procured from the Indian Agricultural Research Institute (IARI), Pusa Campus, New Delhi, India. The seeds were germinated and maintained in the greenhouse facility, Department of Biotechnology, Bharathidasan University, Tiruchirappalli, Tamil Nadu, India during the suitable season.
Fig. 1.
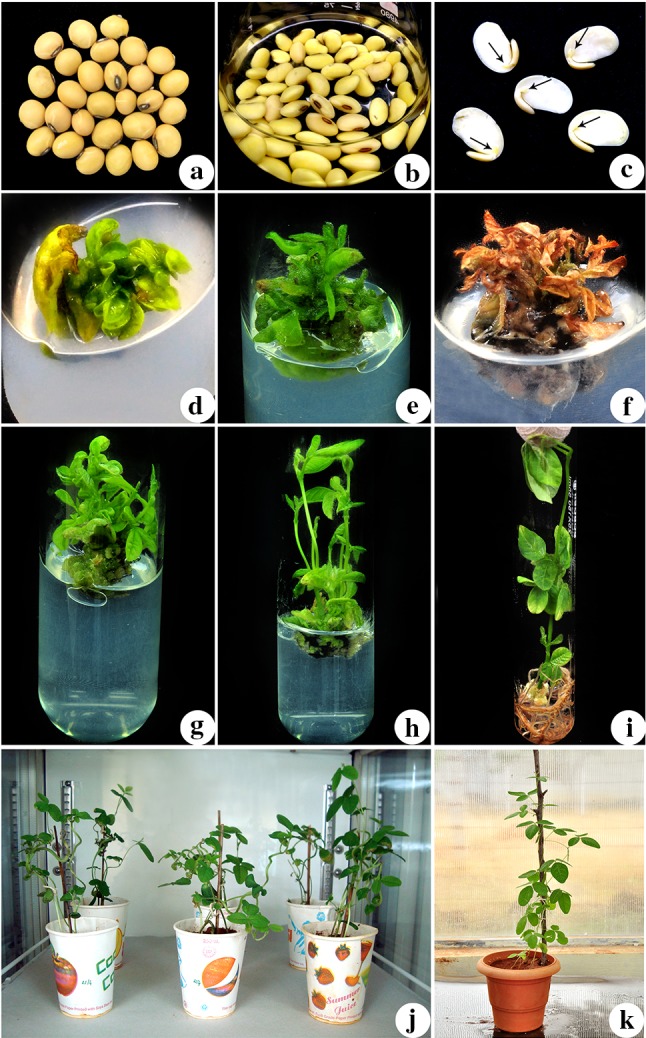
Transformation of cv.Pusa 9712 half seed explants by Agrobacterium tumefaciens strain EHA105 harboring binary vector pCAMBIA1300-p68 to enhance salinity stress tolerance. a mature dry seeds of soybean cv. Pusa 9712 used for preparing the half seed explants; b 1-day old imbibed seeds; c half seed explants made from imbibed seeds (black arrows direct the embryonic area); d explant after infection with A. tumefaciens EHA105 carrying pCAMBIA1300-p68 plasmid and co-cultivated for 5 days in the dark; e shoot induction from explant in SIM supplemented with cefotaxime (200 mg l− 1) without NaCl (after 15 days of culture); f, g selection of regenerated shoots in SIM containing cefotaxime (200 mg l− 1) and NaCl (75 mM) [after 30 days of culture]; h elongated shoots in SEM supplemented with cefotaxime (100 mg l− 1) and NaCl (75 mM) [after 30 days of culture]; i rooted shoots in RIM supplemented with cefotaxime (100 mg l− 1) and NaCl (50 mM) [after 30 days of culture]; j putatively transformed (To) plants maintained in growth chamber; k acclimatization of survived (To) plants under green house condition
Seed surface sterilization and preparation of explants
The seeds of cv. Pusa 9712 were surface sterilized by exposing them to chlorine gas for 16 h (Di et al. 1996), which was achieved by combining 3.5 ml of 12N HCl (Qualigens, Mumbai, India) and 100 ml of 5.25% sodium hypochlorite (Qualigens, Mumbai, India) in a tightly sealed vacuum desiccator along with seeds (Tarsons Product Pvt. Ltd, Kolkata, India). The surface sterilized seeds were then imbibed in an Erlenmeyer flask containing sterile distilled water (Fig. 1b), incubated in total darkness at 25 ± 2 °C on an orbital shaker (Orbitek-Scigenics Biotech Pvt.Ltd, Chennai, India) at 120 rpm for one day. Following, one-half of the imbibed seed cotyledons with attached embryonic axis (Fig. 1c) were used as explants for genetic transformation mediated by Agrobacterium tumefaciens, and the transgenic plants were regenerated through direct organogenesis (Arun et al. 2016).
Agrobacterium strain and construction of binary vector
Agrobacterium strain EHA105 (kindly provided by Rafael Perl Treves, Bar-Ilan University, Israel) accommodating binary vector pCAMBIA 1300-p68 was used in the present study. The binary vector (Supplementary Fig. 1) contains p68 gene expression cassette composed of p68 gene under the regulation of CaMV 35S promoter and the nopaline synthase (nos) terminator within the T-DNA, and neomycin phosphotransferase gene (npt II) cassette for bacterial selection in the backbone. The hygromycin phosphotransferase gene (hpt II) fragment of T-DNA, was removed from the transformation vector (Supplementary Fig. 1) by restricting with XhoI so as to achieve marker free transformation. The EHA105 strain harboring pCAMBIA 1300-p68 was maintained on solid AB agar medium (Agrobacterium minimal medium) supplemented with 50 mg l− 1 kanamycin (SRL, Mumbai, India) and 50 mg l− 1 rifampicin (SRL, Mumbai, India).
Sensitivity of half seed explants and seed germination to NaCl
Half seed explants were inoculated on shoot induction medium [SIM: MS salts, MSIII iron, B5 vitamins (Murashige and Skoogs 1962; Gamborg et al. 1968), 3 mM 2-(N-morpholino) ethanesulfonic acid (MES), 87.65 mM sucrose, 4.44 µM N6-benzylaminopurine (BA) and 0.2% phytagel (pH 5.8)] and incubated for 15 days at 25 ± 2 °C with 16-h photoperiod at a light intensity of 50 µmol m− 2 s− 1. After 15 days the half seed explants with microshoots were inoculated into SIM containing different concentrations (0, 25, 50, 75, 100 and 125 mM) of NaCl. Half seed explants were sub cultured into the same medium twice at 15 days interval. After 45 days the multiple shoots were inoculated into shoot elongation medium [SEM: MS salts, MSIII iron, B5 vitamins, 3 mM MES, 87.65 mM sucrose, 1.45 µM gibberellic acid (GA3) and 0.2% phytagel (pH 5.8)] with different concentrations of NaCl (0, 25, 50, 75, 100 and 125 mM) and sub-cultured twice at 15 days interval. Elongated shoots were inoculated into root induction medium [RIM: MS salts, MSIII iron, B5 vitamins, 3 mM MES, 87.65 mM sucrose, 4.93 µM indole-3-butyric acid (IBA) and 0.2% phytagel (pH 5.8)] supplemented with different concentration of NaCl (0, 25, 50, 75, 100 and 125 mM). The sensitivity concentration of NaCl was determined at the stage of shoot induction, elongation and rooting period to lessen the number of escapes. The control explants were simultaneously maintained in the respective medium devoid of NaCl.
Matured and dried soybean seeds were inoculated on MS basal medium containing different concentration of NaCl (0, 25, 50, 75, 100 and 125 mM). The particular concentration of NaCl, wherein the seeds failed to develop into well rooted plantlets was considered as minimum inhibitory concentration (MIC), and this concentration of NaCl was used for screening the seeds harvested from transformed plants. All the cultures with respective medium were incubated at 25 ± 2 °C with 16-h photoperiod at a light intensity of 50 µmol m− 2 s− 1 provided by cool white fluorescent lamps (Philips, Delhi, India).
Overexpression of p68 gene for production of transformed soybean plants
The polyamines-assisted plant transformation was performed by Agrobacterium-mediated genetic transformation methodology developed by Arun et al. (2016), with minor modifications. Half-seed explants following agroinfection were cultured in co-cultivation medium [CCM: MS salts, MSIII iron, B5 vitamins, 3 mM MES, 87.65 mM sucrose, 200 µM acetosyringone and 0.2% phytagel (pH 5.4)]. After 5 days of co-cultivation with EHA105 carrying pCAMBIA 1300-p68, the explants were washed, blot dried and inoculated on SIM supplemented with 200 mg l− 1 cefotaxime (Duchefa, Haarlem, Netherlands), for initiation of shoot buds. To eliminate chimeric transformants the explants were sub-cultured twice at 15 days interval onto selection medium (75 mM NaCl). The explants with surviving shoots were transferred into SEM supplemented with 100 mg l− 1 cefotaxime and 75 mM NaCl, and subcultured for 30 days at 15 days interval. The elongated shoots from explants were subcultured to RIM supplemented with 100 mg l− 1 cefotaxime and 50 mM NaCl incubated for 30 days at 25 ± 2 °C with a 16-h photoperiod at a light intensity of 50 µmol m− 2 s− 1 delivered by cool white fluorescent lamps (Philips, Delhi, India). The surviving plants with adequate roots were transplanted to paper cups filled with sand and peat (1:1 v/v) and allowed to harden in the growth chamber. The hardened plants after 12 days were transferred to earthen pots and grown to maturity in the greenhouse under controlled conditions. Similarly, non-transformed plants were maintained in the respective medium. Transformed (T0) and non-transformed (NT) plants were allowed to undergo self-pollination and set T1 seeds. Following the seeds were harvested, sterilized and inoculated on MS basal medium containing 100 mM NaCl. After 20 days, the seedlings with well-developed rooted plantlets were hardened and acclimatized in the greenhouse for further analysis.
Molecular confirmation of p68 gene
One-month-old transformed (T1) and NT plants were transplanted to paper cups filled with sand and peat (1:1 v/v). The plants in cups were covered with plastic covers, so as to maintain high humidity. Two weeks later, the plants were transferred to green house, wherein they were transplanted to plastic pots containing sand and peat (1:1 v/v). The established plants in the green house, were subjected to molecular analysis.
Polymerase chain reaction (PCR)
Genomic DNA was isolated from the young leaf tissues collected from seventeen transformed lines (T1) and non-transformed (NT) plant using the CTAB method as earlier reported by Dellaporta et al. (1983). The primer pairs (FP-p68: 5′-GGATCCATGTCGTATGTTCCTCCACAC-3′; RP-p68: 5′-GGATCCCCATTACCTACAAACATGACTGAT-3′) was used to amplify 1.8 kb fragment of p68 gene. The NT plant genomic DNA and binary vector pCAMBIA 1300-p68 were used as negative and positive controls, respectively. PCR was performed in the PTC-100™ thermal cycler (MJ research Inc, Waltham, Mass, USA) using the following conditions: Initial denaturation of DNA template at 95 °C for 5 min, followed by 30 cycles of 94 °C (denaturation) for 40 s, 58 °C (annealing) for 40 s and 72 °C (extension) for 2 min, followed by a final extension at 72 °C for 10 min. The amplified products were analyzed by electrophoresis on a 1.0% agarose (Sigma, St. Louis, USA) gel stained with 0.1% ethidium bromide (Sigma, St. Louis, USA) and photographed using the Gel Documentation system (UVITEC, Cambridge, UK).
Dot blot analysis
Five µg of genomic DNA extracted from the transformed plants (T1), non-transformed (NT) plant and 5 µg of binary vector pCAMBIA 1300-p68 were dotted into 1 cm square grids on a nylon membrane (Hybond N+, GE Health care Limted, Buckinghamshire, UK). The membrane was then dried for 5 min and rinsed with Neutralization buffer (0.5 M Tris–HCl (pH 7.4) and 1.5 M NaCl) for 3 min followed by a wash with 2× SSC (saline sodium citrate buffer). The membrane was then UV cross linked and hybridized with probe prepared by labeling the PCR purified fragment of p68 gene, according to the kit manufacturer’s instructions (AlkPhos Direct Labeling and Detection System with CDP Star, GE Healthcare Limited, Buckinghamshire, UK).
Southern hybridization
Southern hybridization analysis was carried out to confirm the transgene integration and determine the copy number of p68 gene in transformed soybean plants. Ten µg of genomic DNA from Dot-positive (T1) plants (named as S1, S2, S12, S13 and S15), NT plant and 5 µg of pCAMBIA 1300-p68 plasmid were digested with XhoI, which recognizes a single site within the T-DNA. The digested DNA samples and plasmid DNA were resolved on a 1% agarose gel and transferred to a nylon membrane (Hybond N+, GE Health care Limted, Buckinghamshire, UK). The membrane was hybridized at 55 °C with ALP-labeled 1.8 kb PCR purified fragment of p68 gene for 8 h and the probe was detected according to manufacturer’s instructions (AlkPhos Direct Labeling and Detection System with CDP Star, GE Healthcare Limited, Buckinghamshire, UK).
Reverse transcriptase-PCR (RT-PCR) and Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from Southern hybridization positive (T1) and NT plants using a RNAqueous kit (Ambion Inc., Austin, USA) and the DNA contamination were eliminated by treatment with DNase I. RT-PCR was performed by a one-step RT-PCR kit (Qiagen, USA) as instructed by the manufacturer. The p68 gene was amplified using the above mentioned gene-specific primers. According to Hu et al. (2009) the GmUKN1 gene (FP: 5′-TGGTGCTGCCGCTATTTACTG-3′ and RP: 5′-GGTGGAAGGAACTGCTAACAATC-3′) was used as an internal control. The PCR amplification profile consisted of an initial denaturation of cDNA at 95 °C for 5 min and followed by 28 cycles for 40 s at 94 °C, 40 s at 58 °C and 20 s at 72 °C, followed by a final extension at 72 °C for 5 min. The amplified products were determined on a 1.0% agarose gel (Sigma, St. Louis, USA) stained with 0.1% ethidium bromide (Sigma, St. Louis, USA) and photographed using the Gel Documentation system (UVITEC, Cambridge, UK).
qRT-PCR analysis was performed in LightCycler® 480 Real-time PCR system (Roche, Germany). The expression analysis of the RT-PCR positive (T1) and NT plants were conducted using Prime Script™ RT Reagent Kit (Takara Bio Inc, Japan) and p68 gene-specific primer set (FP: 5′-CCTCGCATTCTCTTCCTCGTA-3′ and RP: 5′-CGACGAGAACCATTGGCTAGA-3′) (Banu et al. 2015). The GmUKN1 gene was used as the housekeeping gene (NCBI Accession No: BU578186; Unigene ID: Gma.32694) for normalizing the expression values. The experiments were carried out in triplicates. The expression level of p68 was normalized against GmUKN1gene expression level, calculated from cycle threshold (Ct) values, using 2− ΔΔCT method (Livak and Schmittgen 2001).
Physiological and biochemical analysis of the transgenic soybean plants
One-month-old qRT-PCR positive (T1) and NT plants, irrigated with different concentration of NaCl (0, 25, 50, 100, 150 and 200 mM) for 2 weeks were subjected to physiological and biochemical analysis.
Total chlorophyll content was measured as described by Arnon (1949). Proline was estimated, as reported by Bates et al. (1973). Catalase activity (CAT) and Ascorbate peroxidase (APX) activity was determined as previously detailed by Aebi (1984) and Chen and Asada (1989) respectively. Monodehydroascorbate reductase (MDHAR) and Dehydroascorbate reductase (DHAR) activities were calculated as previously described by Doulis et al. (1997) and Miyake and Asada (1992), respectively. Superoxide dismutase (SOD) activity was calculated as previously reported by McCord and Fridovich (1969). Malondialdehyde (MDA) content was calculated as previously detailed by Heath and Packer (1968). Relative water content (RWC) was evaluated as described by Turner (1981). Hydrogen peroxide (H2O2) level was evaluated as earlier described by Velikova et al. (2000). All the above mentioned analyses were carried out in triplicates.
Measurement of photosynthetic characteristics
Fully expanded leaves from qRT-PCR positive (T1) and NT plants were analyzed for gas exchange parameters using infra-red gas analyzer (CIRAS-1-PP systems, Hitchin, Herts, UK). The net photosynthetic rate (Pn), stomatal conductance (Gs) and CO2 assimilation rate (A) were measured under the following conditions. The relative humidity, air temperature, photosynthetic photon flux density (PPFD) and CO2 concentration were sustained at 80–90%, 25 °C, 1000 µmol m− 2 s− 1 and 400 µl l− 1, respectively, and the analysis was carried out between 09:00 and 11:00 h. The experiments were performed in triplicates.
Measurement of Na+ and K+ ion content
Na+ and K+ ion content was determined as described earlier by Munns et al. (2010). Fresh leaves were harvested from 1 month old qRT-PCR positive (T1) and NT plants which were treated with different concentrations of NaCl (0, 25, 50, 100, 150 and 200 mM). Harvested leaf material was washed thoroughly with distilled water and then dried at 70 °C for 48 h. Dried leaf material was made into fine powder and used for Na+ and K+ ion estimation. A known weight of finely grounded leaf material was digested overnight in HNO3/H2O2 solutions. After digestion the extract was filtered through a 0.22-µM filter membrane. The Na+ and K+ ion content in the digested sample was determined by inductively coupled plasma emission spectrometry (ICP trace analyzer, Labtam, Braeside, Australia). The analysis was carried out in triplicates.
Yield performance
Three plants of each qRT-PCR positive (T1) transgenic lines (S1, S12 and S15) and NT plants were transplanted into plastic pots in green house containing sand and peat (1:1 v/v). The freshly potted plants were irrigated with water for 1 week, following which they were irrigated with water containing varying concentrations of NaCl (0, 100, 150 and 200 mM) once every 2 days till the pods matured. The matured pods were harvested and the number of seeds per plant, as well as the dry weight of the seeds, was evaluated and noted. The analysis was carried out in triplicates.
Statistical analysis
One-way ANOVA was used to analyze the data, and Duncan’s multiple range test (DMRT) was used to contrast the differences. SPSS 16.0 (SPSS Inc.USA) has been used to perform the statistical analysis at the level of P < 0.05.
Results and discussion
Sensitivity analysis of matured and half-seed explants to NaCl
The proper selection pressure is a vital step in plant genetic transformation to select the transformed cells/tissues/plants from non-transformed counterparts. Hence, to minimize the number of non-transformed plant escapes following transformation and regeneration of half-seed explant, an efficient selection pressure was determined by inoculating and incubating the half-seed explants and seeds on MS basal medium containing varying concentrations of NaCl (0, 25, 50, 75, 100 and 125 mM) for 15 days. Among the various NaCl concentrations evaluated, shoot induction medium supplemented with 75 mM NaCl barred the development of shoot and 50 mM NaCl arrested root formation with dried leaves from the elongated shoots of half seed explants, whereas the MS basal medium supplemented with 100 mM NaCl prohibited seed germination. Hence, the 75, 50 and 100 mM of NaCl was considered as a minimum inhibitory concentration (MIC) in selecting the transformed multiple shoot induction, elongation, rooting and germinating seeds, respectively. Similarly, Zhang et al. (2009) reported that 200 mM NaCl was able to eliminate the non-transformants and thereby was helpful in selecting transformed tobacco plants carrying the rstB gene. Arabidopsis seeds transformed with OsMSR2 gene were successfully screened on 100 mM NaCl supplemented medium (Xu et al. 2011). Transformed somatic embryos and soybean seeds were selected in a medium containing 75 and 100 mM of NaCl, respectively (Subramanyam et al. 2012).
Development of transformed soybean plants by overexpressing p68 gene
Half-seed explants with the embryonic axis were infected with Agrobacterium tumefaciens strain EHA105 harboring the binary vector pCAMBIA1300-p68 and co-cultivated for 5 days in the dark (Fig. 1d). After co-cultivation, the half-seed explants were cultured on SIM incorporated with 200 mg l− 1 cefotaxime for 15 days (Fig. 1e). After 15 days, the half seed explants with initiated shoots were sub-cultured in the same medium incorporated with 75 mM NaCl, for multiplication and selection of putatively transformed shoots (Fig. 1f, g). The actively multiplying and surviving shoots were subcultured to SEM containing 100 mg l− 1 cefotaxime and 75 mM NaCl, wherein they elongated successfully (Fig. 1h). The elongated shoots were subcultured to RIM containing 100 mg l− 1 cefotaxime and 50 mM NaCl developed roots (Fig. 1i). Hardening and acclimatization of transformed plantlets were accomplished in the environmental growth chamber (Fig. 1j) and in the greenhouse, respectively (Fig. 1k). The putative transgenic seeds, collected and labeled as T1 seeds, were screened on MS basal medium incorporated with 100 mM NaCl.
Molecular and expression analysis of transformed plants to confirm p68 gene
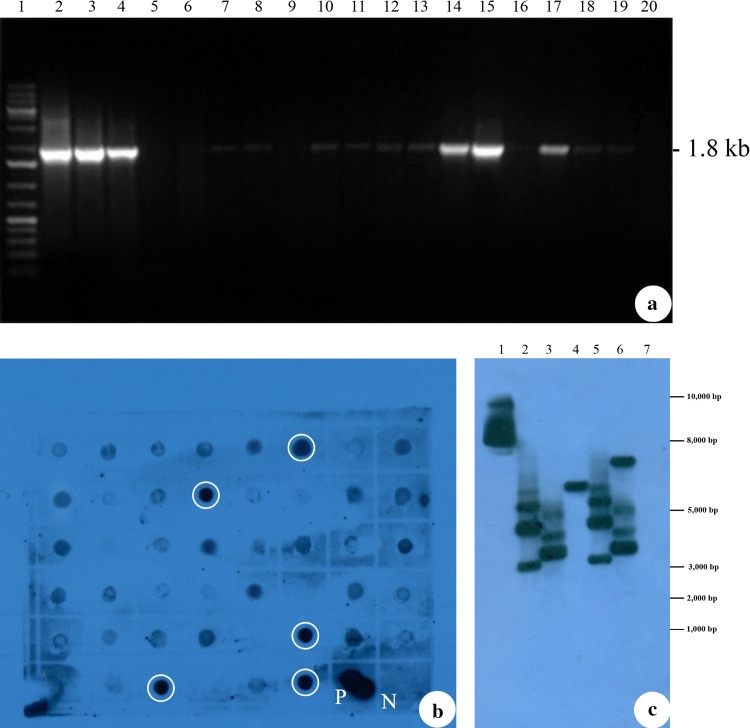
The transformed (T1) and NT plants were analyzed for the presence of p68 gene using PCR. The PCR analysis revealed a 1.8-kb amplicon of p68 gene fragment from the genomic DNA of (T1) soybean plants (Fig. 2a, lanes 3–19) which is similar to the amplicon from the pCAMBIA 1300-p68 (Fig. 2a, lane 2). No amplification was observed from the genomic DNA isolated from NT soybean plants (Fig. 2a, lane 20).
Fig. 2.
Detection of p68 gene integration in transformed soybean plants. a PCR analysis of transformed soybean plants (T1) for the presence of p68 gene. Lane 1, Gene Ruler 1 kb DNA ladder marker, ready to use (#SM0313); lane 2, pCAMBIA1300-p68 as a positive control; lanes 3, 4,14, 15 and 17, transformed soybean plants (S1, S2, S12, S13 and S15) lane 20, non-transformed (NT) soybean plants genomic DNA as a negative control. b Dot blot analysis of transformed soybean plants (T1). Five transgenic lines showing strong positive signal marked in white circles (S1, S2, S12, S13 and S15) genomic DNA; P-pCAMBIA1300-p68 as a positive control; N-non-transformed (NT) soybean genomic DNA. c Southern blot analysis of transformed soybean plants (T1). lane 1, pCAMBIA1300-p68 gene as a positive control; lanes 2–6, transformed soybean plants (S1, S2, S12, S13 and S15) genomic DNA; lane 7, non-transformed (NT) soybean genomic DNA
Dot blot analysis was performed to screen a large number of PCR-positive lines. Samples which were exhibiting strong signal in dot blot (Fig. 2b) were further analyzed by southern blotting to validate the integration of the transgene (p68) and its copy number in (T1) soybean plants. The Genomic DNA was digested with XhoI and hybridized with the probe prepared using the PCR-amplified product of p68 gene fragment as there is a single Xho I site between left TDNA border and the p68 gene cassette with in the T-DNA region of the pCAMBIA1300-p68. Therefore, probing with p68 gene fragment would give us an indication of the number copies of p68 gene integrated into the plant genome. The positive control (pCAMBIA1300-p68) generated hybridization signal (Fig. 2c, lane 1) while the negative control (DNA from NT plant) did not exhibit hybridization signal (Fig. 2c, lane 7). The transformed plants were found to have up to four copies of p68 gene integration (Fig. 2c, lanes 2, 5 and 6), with non-identical hybridization patterns evidencing their individuality.
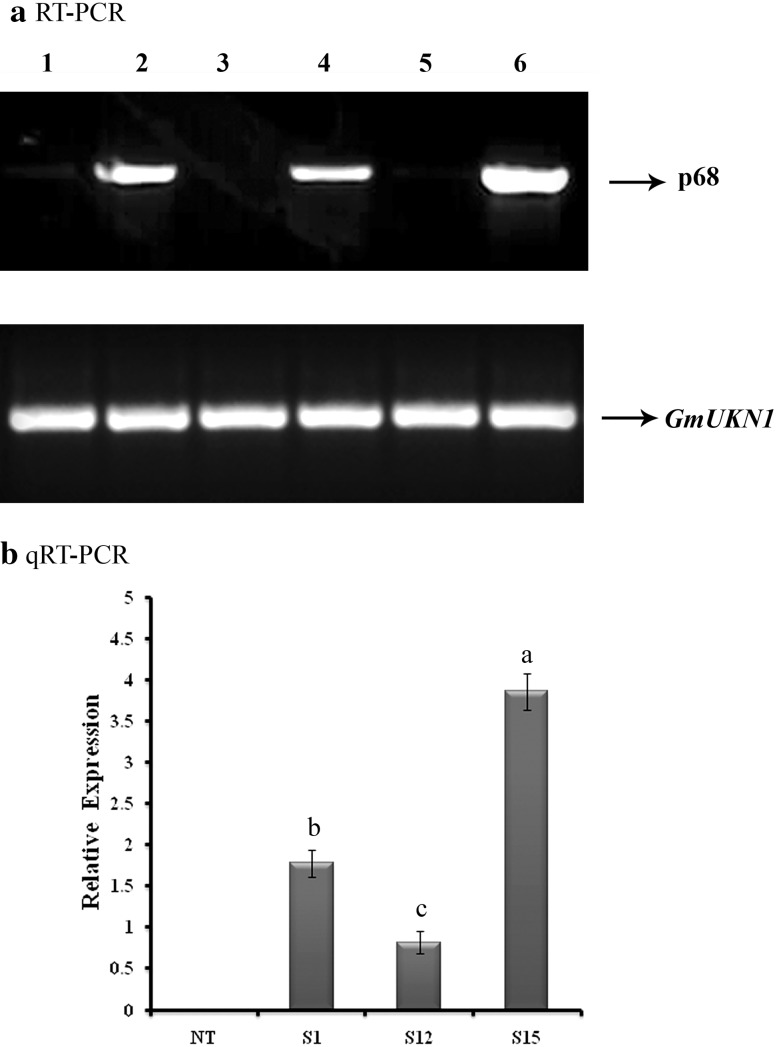
In RT-PCR, a 1.8-kb amplified fragment, specifically amplified by the gene-specific primer, confirmed the expression of p68 gene in three (S1, S12 and S15) of the five Southern-positive (T1) plants (Fig. 3a, lanes 2, 4, 6), and no amplification was observed in remaining two Southern-positive (S2 and S13) (Fig. 3a, lanes 3, 5) and NT plants (Fig. 3a, lane 1). This might be due to gene silencing by the position effect on p68 gene. Similary, Maghuly et al. (2006) observed no expression of GFLV gene in few lines of transformed grapevine plants, suggesting that there is no correlation between the T-DNA copy number and mRNA expression level of transgene.
Fig. 3.
Expression analysis of transformed soybean plants. a RT-PCR expression analysis of p68 gene in transformed soybean plants. Lane 1, non-transformed (NT) soybean plant RNA as a negative control; lanes 2, 4 and 6, transformed (T1) soybean (S1, S12 and S15) plant RNA samples; lanes 3 and 5, failed to amplify p68 gene from transformed (T1) soybean (S2 and S13) plant RNA samples. b qRT-PCR analysis to analyze the expression level of p68 gene in transformed soybean plants. Relative expression of three transgenic lines (S1, S12 and S15) and non-transformed (NT) soybean plant; data were analyzed according to the 2− ΔΔCt method. Mean of three individual experiments with standard errors. Different letters denote significantly different values according to Duncan’s multiple range test (DMRT) at a 5% level
The qRT-PCR analysis confirmed the expression of p68 in three lines of RT-PCR-positive (T1) plants (Fig. 3b) and no expression was observed in non-transformed (NT) plants (Fig. 3b). In the present study, highest level of p68 transcript expression was recorded in S15 and the lowest was observed in line S12 (Fig. 3b). Similarly, Banu et al. (2015) employed qRT-PCR assay to evaluate the expression of p68 gene in transformed rice which conferred salt tolerance.
Assessment of transgenic soybean plants under salt stress
One-month-old qRT-PCR positive (T1) plants were irrigated with 200 mM of NaCl along with non-transformed (NT) control plants. Both (T1) and NT plants exhibited no phenotypic differences under normal growth conditions (0 mM NaCl) (Fig. 4a). However, under increased salinity stress the NT plants exhibited chlorosis, necrosis, leaf burning, defoliation, and completely withered at 200 mM of NaCl (Fig. 4b). Moreover, three (S1, S12 and S15) out of five southern blot positive (T1) plants showed healthy growth at 200 mM NaCl and produced soybean pods (Fig. 4c). This clearly indicates that the expression of p68 gene in (T1) plants played an important role in the salinity tolerance.
Fig. 4.
Phenotypic appearance of transformed (T1) and non-transformed (NT) soybean plants under salinity stress condition (200 mM NaCl). a Phenotype before salt treatment; b phenotype after two weeks of salt treatment; c green house grown (T1) generation transgenic soybean plants produced soybean pods
It has been confirmed that p68 has potentiality towards cell injury recovery. p68 gene in particular stressed condition plays a vital role in preventing aggregation and stabilizing non-native proteins. Thus, considering the aforesaid statements, we presume that p68 expression in soybean played an important role in salinity tolerance. Similarly, earlier reports also support the role of p68 in salinity stress alleviation in rice and tobacco (Banu et al. 2015; Tuteja et al. 2014).
Biochemical analysis of transformed soybean plants under salt stress
The NT plants, showed tolerance upto 100 mM NaCl, beyond which they perished, whereas the transformed plants showed tolerance towards salinity stress of upto 200 mM. Hence, the NT and transformed (T1) plants exposed to salinity stress of 100 mM and 200 mM, respectively, were subjected to physiological and biochemical analysis.
Effect of salinity stress on chlorophyll content
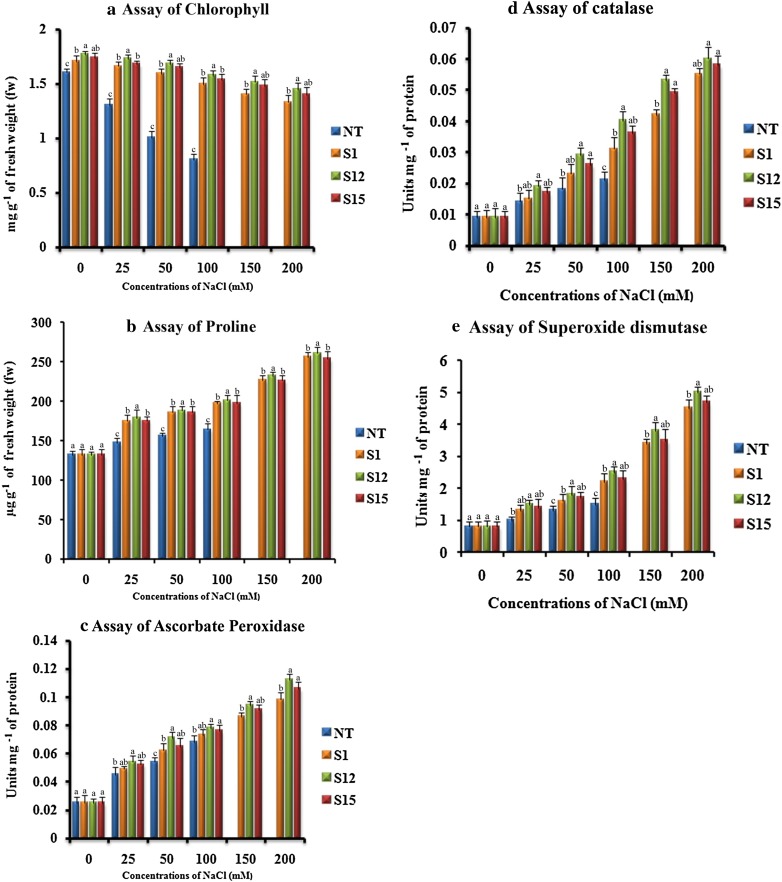
The salinity stress induces photo-oxidative reaction which in turn results in degradation of cell organelle membranes, especially of chloroplast thylakoids which ultimately leads to chlorophyll degradation (Husaini and Abdin 2008). Change in chlorophyll content is fundamental to understand the plant response to the salinity stress. In the present study, the leaf chlorophyll content gradually decreased with an increased concentration of NaCl. However, under same concentrations of NaCl, the chlorophyll loss in (T1) plants was significantly less noticeable than that of the NT plant. At 100 mM of NaCl, the chlorophyll loss was (52.24%) in NT plant, whereas (11.03%) in ‘S12’ (T1) plant (Fig. 5a). A possible explanation is that, overexpression of p68 gene in (T1) plants, induced the expression of downstream genes that inhibited chlorophyll disintegration, thereby increasing the chlorophyll concentration (Zhang et al. 2004; Gao et al. 2009). In accordance with the current study, the expression of p68 gene in tobacco plant resulted in a higher chlorophyll content (Tuteja et al. 2014) which surpassed the WT plants. Similarly, the expression of stress responsive genes such as alfalfa WRKY11 gene in soybean (Wang et al. 2018), Tbosm gene in chilli pepper (Subramanyam et al. 2011), AhBADH gene in trifoliate orange (Fu et al. 2011), Rab 16A gene in tobacco plants (Roychowdhury et al. 2007) and ATHK1 gene in Lyceum barbarum (Chen et al. 2009) resulted in higher chlorophyll content than NT plants.
Fig. 5.
Response of transformed (T1) and non-transformed (NT) soybean plants subjected to different concentrations of NaCl for 12 days under greenhouse conditions. a Chlorophyll concentration (mg g− 1 of fw); b proline concentration (µg g− 1of fw); c APX concentration (Units mg− 1 of protein); d CAT activity (Units mg− 1 of protein); e SOD activity (Units mg− 1 of protein). Mean of three individual experiments with standard errors. Different letters denote significantly different values according to Duncan’s multiple range test (DMRT) at a 5% level
Effect of salinity stress on proline
Increase in the osmotic pressure is caused due to the excessive loss of intracellular water in plants which directs them to accumulate compatible solutes to tolerate the osmotic pressure (Turkan and Demiral 2009). As like all other abiotic stresses, salt stress also affects the flow of metabolic pathways, resulting in accumulation of ROS (Gill and Tuteja 2010). Salinity affected plants require free proline for osmotic adjustment, cellular macromolecules protection, and scavenging hydroxyl radicals (Singh et al. 2000; Chen et al. 2011). Hence, the accumulation of proline in plants during abiotic stress is vital, compared to other compatible solutes (Gao et al. 2009). In the present investigation, under normal growth conditions, the concentration of free proline in leaves of (T1) plants was similar to that of NT plants. When the (T1) and NT plants were treated with increased concentrations of NaCl, the (T1) plants accumulated higher proline than NT plant and (T1) plant ‘S12’ showed highest proline content at all tested NaCl concentrations (Fig. 5b). It indicates that the expression of p68 gene might activate the key enzymes of the proline biosynthetic pathway, which in turn enhance the tolerance to salinity. Similarly, the expression of Tbosm gene in cotton, soybean (Parkhi et al. 2009; Subramanyam et al. 2012) Hva1 gene in mulberry (Checker et al. 2012), ATHK1 gene in Lycium barbarum (Chen et al. 2009) and CgDREBa gene in Chrysanthemum (Chen et al. 2011), enhanced the level of proline content in transgenic plants than that of its counterparts. In addition, overexpression of p68 gene in rice (Banu et al. 2015) enhanced the proline content in transgenic plant than WT plant under salinity stress condition.
Effect of salinity stress on activities of APX, CAT, and SOD
ROS are continuously produced in plants, during salinity and drought stress conditions as by-products of various metabolic pathways, which interact with the macromolecules making them malfunction (Gill and Tuteja 2010). The chloroplasts and mitochondria are prime locations for the production of ROS owing to their active electron transport activities (Tambussi et al. 2000; Bartoli et al. 2004). To overcome oxidative stress induced by drought and salinity, plants up-regulate antioxidative enzymes such as APX, CAT, and SOD to detoxify their system (Turkan and Demiral 2009). In the present investigation, under standard growth conditions, the (T1) and NT soybean plants accumulated APX, CAT, and SOD similarly. The antioxidative enzyme activities gradually increased with the increased concentration of NaCl in both (T1) and NT soybean plants (Fig. 5c–e). However, the (T1) plants maintained a higher APX, CAT, and SOD activity than their counterparts (Fig. 5c–e). Among the three (T1) plants analyzed, plant ‘S12’ showed the highest APX, CAT, and SOD activities than ‘S15’ at all tested NaCl concentrations (Fig. 5c–e). The results obtained from the current study are relatable to earlier reports where soybean plants expressing Tbosm showed elevated levels of SOD, CAT, and APX (Subramanyam et al. 2012). Similarly, SOD, CAT, and APX levels were elevated in the transformed chilli and tomato expressing Tbosm and BcZAT12 respectively, under stressed salinity and drought conditions (Subramanyam et al. 2011; Shah et al. 2013). Tobacco plant expressing p68 gene showed an improved level of APX, CAT and SOD activity under salinity stress conditions (Tuteja et al. 2014). Transgenic rice plants expressing p68 gene also displayed a higher APX and CAT activity upon exposure to salt stress as compared to NT plants (Banu et al. 2015).
Effect of salinity stress on activities of DHAR and MDHAR
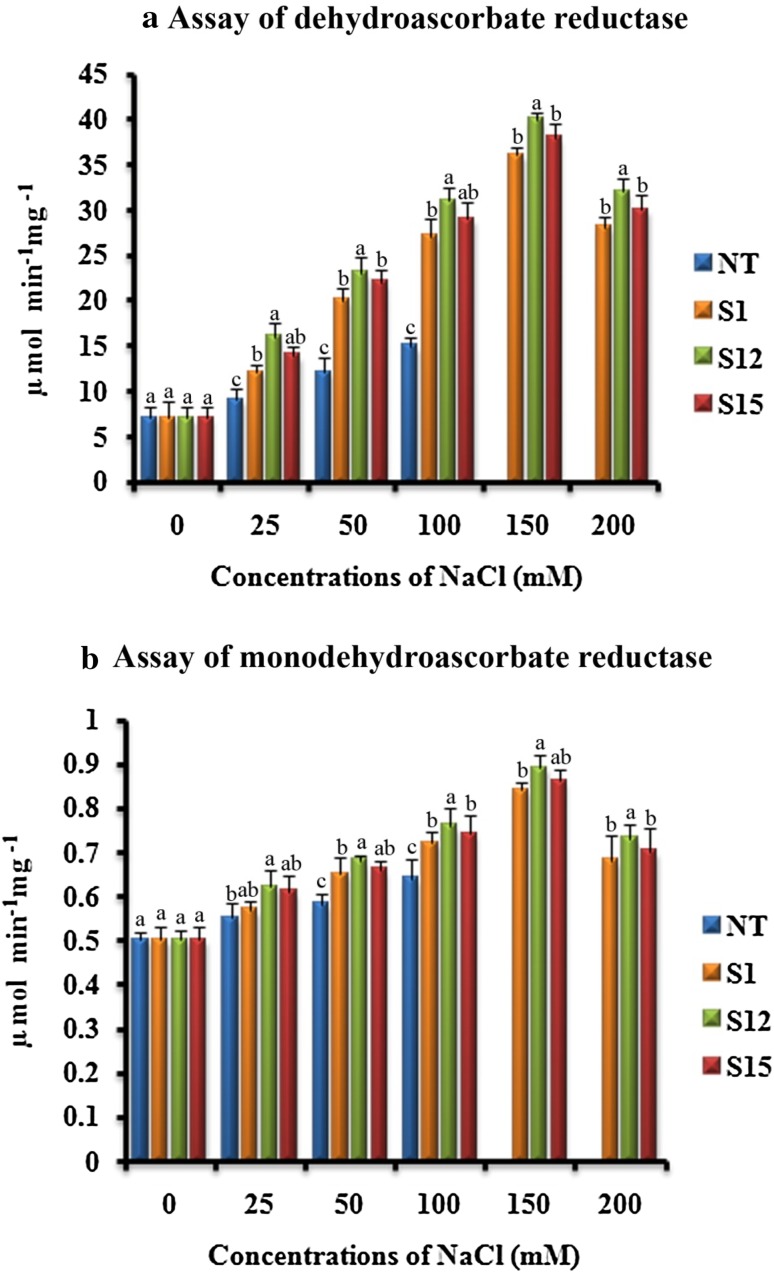
Noctor and Foyer (1998) reported that the two enzymes DHAR and MDHAR involved in the production of reducing agents such as ascorbate and glutathione during several abiotic and biotic stress conditions, helps in foraging the increased levels of H2O2. In our studies, the activities of DHAR and MDHAR were similar in (T1) and NT plants under normal growth conditions (Fig. 6a, b). When the (T1) and NT soybean plants were irrigated with different concentrations of NaCl, the activities of DHAR and MDHAR increased up to 150 mM and thereafter decreased. However, the (T1) soybean plants accumulated DHAR and MDHAR higher than NT soybean plants (Fig. 6a, b). Among the three (T1) soybean plants, ‘S12’ showed higher activity than the rest at all tested NaCl concentrations. The results obtained were in accordance with the earlier reports where glutathione S-transferase expression in cotton (Light et al. 2005) and Tbosm gene expression in chilli pepper and soybean (Subramanyam et al. 2011, 2012) showed enhanced levels of DHAR and MDHAR for effective elimination of H2O2 under the salinity conditions.
Fig. 6.
Response of transformed (T1) and non-transformed (NT) soybean plants subjected to different concentrations of NaCl for 12 days under greenhouse conditions. a DHAR activity (µ mol min− 1 mg− 1); b MDHAR activity (µ mol min− 1 mg− 1). Mean of three individual experiments with standard errors. Different letters denote significantly different values according to Duncan’s multiple range test (DMRT) at a 5% level
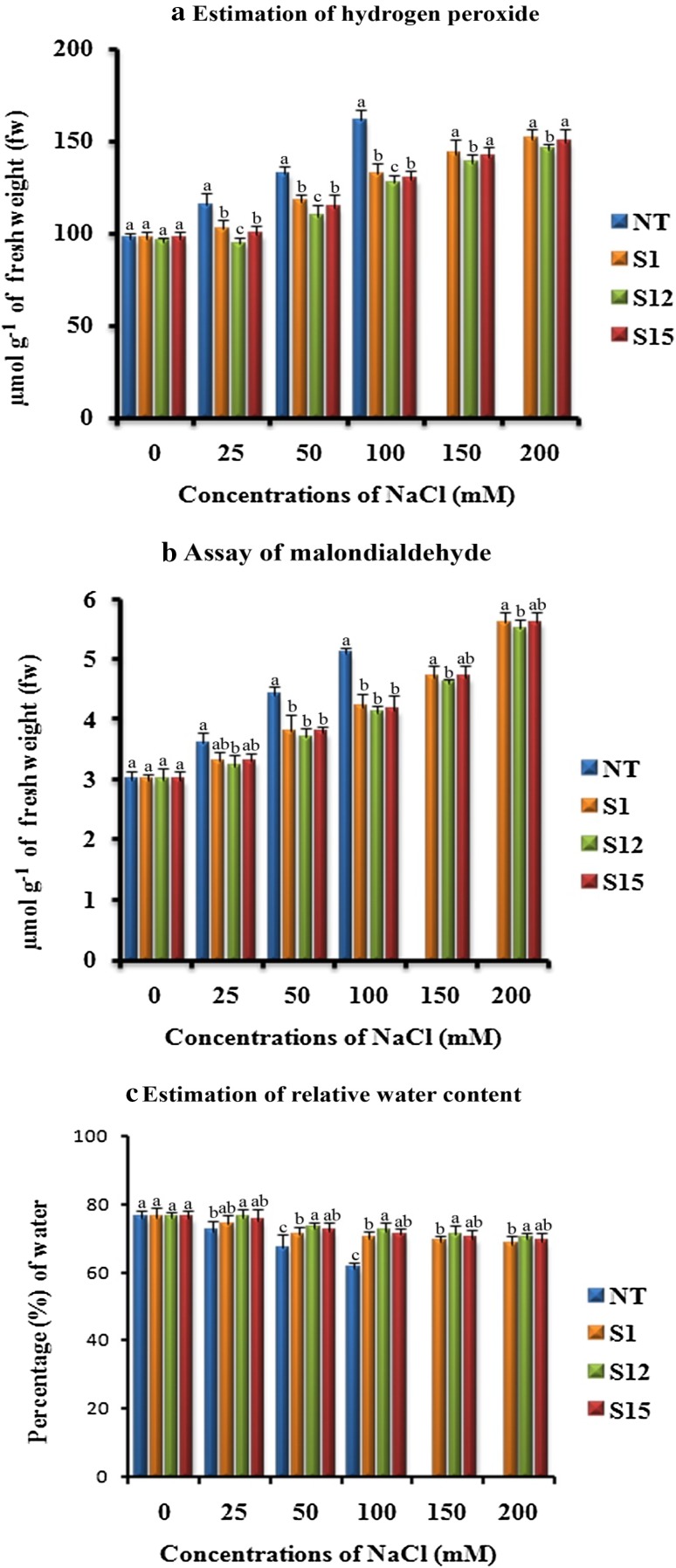
Estimation of H2O2 concentration
It is a well-known fact that excessive accumulation of H2O2 leads to the prevalence of oxidative stress in plants. This is a cause for apoptosis, DNA fragmentation, chromatin condensation as well as cell shrinkage (Houot et al. 2001). A variety of enzymatic and non-enzymatic anti-oxidants play crucial role in eliminating H2O2 (Gill and Tuteja 2010). Foyer et al. (1994) reported that CAT or APX, which converts H2O2 to water molecules, prevents the accumulation of H2O2. In our studies, there were no prominent differences in H2O2 content between the (T1) and NT soybean plants under normal growth conditions (Fig. 7a). However, when (T1) and NT soybean plants were irrigated with different concentrations of NaCl, the (T1) soybean plants accumulated less amount of H2O2 than NT plant (Fig. 7a). As we noticed earlier, higher accumulation of antioxidative enzymes in the transgenic soybean plants might have lowered the amount of H2O2 under stressed conditions. In a similar fashion, p68 gene expressed in tobacco and rice, respectively, maintained a decent level of H2O2 content, even under stressed condition (Tuteja et al. 2014; Banu et al. 2015).
Fig. 7.
Response of transformed (T1) and non-transformed (NT) soybean plants subjected to different concentrations of NaCl for 12 days under greenhouse conditions. a H2O2 concentration (µmol g− 1 of fw); b MDA content (µmol g− 1 of fw); c relative water content (%). Mean of three individual experiments with standard errors. Different letters denote significantly different values according to Duncan’s multiple range test (DMRT) at a 5% level
Effect of salinity stress on MDA
Membranes are the main targets of lipid peroxidation degradative process due to the accumulation of ROS induced by salinity. In plant cells, MDA content is an indicator of lipid peroxidation levels (Shalata and Tal 1998). In the present study, under normal growth conditions, the (T1) and NT soybean plants displayed similar concentrations of MDA (Fig. 7b). The MDA concentration gradually increased in both (T1) and NT plants with increased concentrations of NaCl (Fig. 7b). However, under all tested NaCl concentrations, the NT plant showed higher MDA content than (T1) plants (Fig. 7b). This implies that the degree of lipid peroxidation in (T1) plants is lower than that in the NT plant. Therefore, suggesting that the expression of p68 gene in (T1) soybean plants protected the cell membrane from damage caused by salinity stress. In accordance with the present study, p68 gene expressed in rice recorded a less amount of MDA under salinity condition (Banu et al. 2015). Similar results were also observed during the expression of BcZAT12 gene in tomato plants under heat stress (Shah et al. 2013) and osmotin gene in strawberry plants under salt stress (Husaini and Abdin 2008).
Effect of salinity stress on RWC
Flower and Ludlow (1986) reported that maintenance of plants in high water level helped them to survive under salt stress. Hence, RWC is considered as an important parameter for salinity tolerance (Malatrasi et al. 2002; Rampino et al. 2006; Talame et al. 2007). We noticed a gradual decrease in the RWC in both NT and (T1) plants with an increase in NaCl concentration, however, the (T1) plants maintained a higher RWC than that of its counterparts (Fig. 7c). In a similar fashion, Banu et al. (2015) reported that p68 gene expressed in rice revealed higher amount of RWC under salinity condition when compared to WT plants. RWC was observed to be similar in the NT and (T1) plants under normal growth conditions (Fig. 7c). Likewise, expression of BcZAT12 and Tbosm genes in transgenic tomato and soybean, respectively (Shah et al. 2013; Subramanyam et al. 2012), exhibited high RWC than that of its non-transformed counterparts.
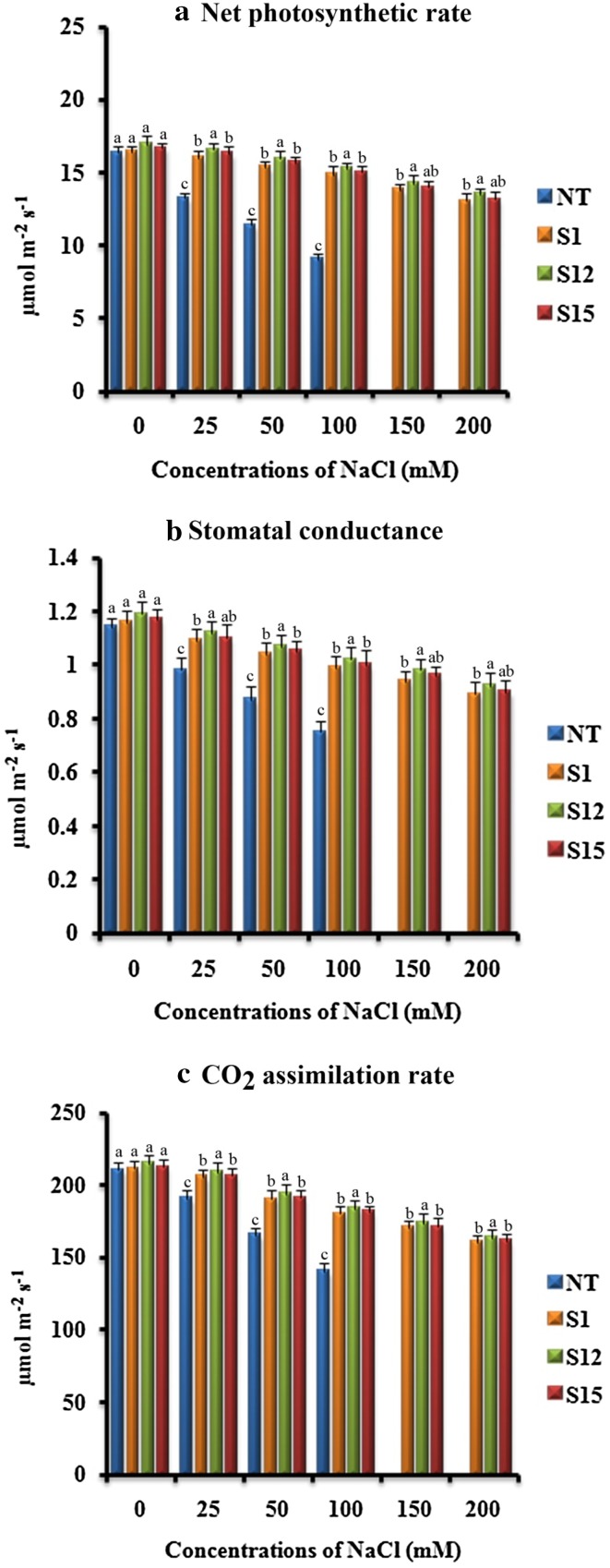
Effect of salinity stress on photosynthetic characteristics
Net photosynthetic rate (Pn), stomatal conductance (Gs) and CO2 assimilation rate (A) are the important parameters to evaluate the plant performance under salinity stress (Subramanyam et al. 2012). The difference in Pn, Gs and A was not significant between the (T1) and NT plants when grown under control conditions (Fig. 8a–c). However, Pn, Gs and A was retained at a higher level in (T1) plants as compared to NT plants under all tested NaCl concentrations (Fig. 8a–c). The observed results suggested that (T1) plants subjected to salinity stress utilized internal carbon dioxide efficiently. Similarly, a higher retention of Pn, Gs and A was observed in mtlD expressing Populus tomentosa (Hu et al. 2005), AVP1 expressing cotton (Pasapula et al. 2011), tbosm expressing soybean (Subramanyam et al. 2012) and p68 expressing tobacco (Tuteja et al. 2014) which displayed higher tolerance to salinity stress.
Fig. 8.
Response of transformed (T1) and non-transformed (NT) soybean plants subjected to different concentrations of NaCl for 12 days under greenhouse conditions. a Net photosynthetic rate (Pn) (µmol m− 2 s− 1); b stomatal conductance (Gs) (µmol m− 2 s− 1); c CO2 assimilation (A) (µmol m− 2 s− 1). Mean of three individual experiments with standard errors. Different letters denote significantly different values according to Duncan’s multiple range test (DMRT) at a 5% level
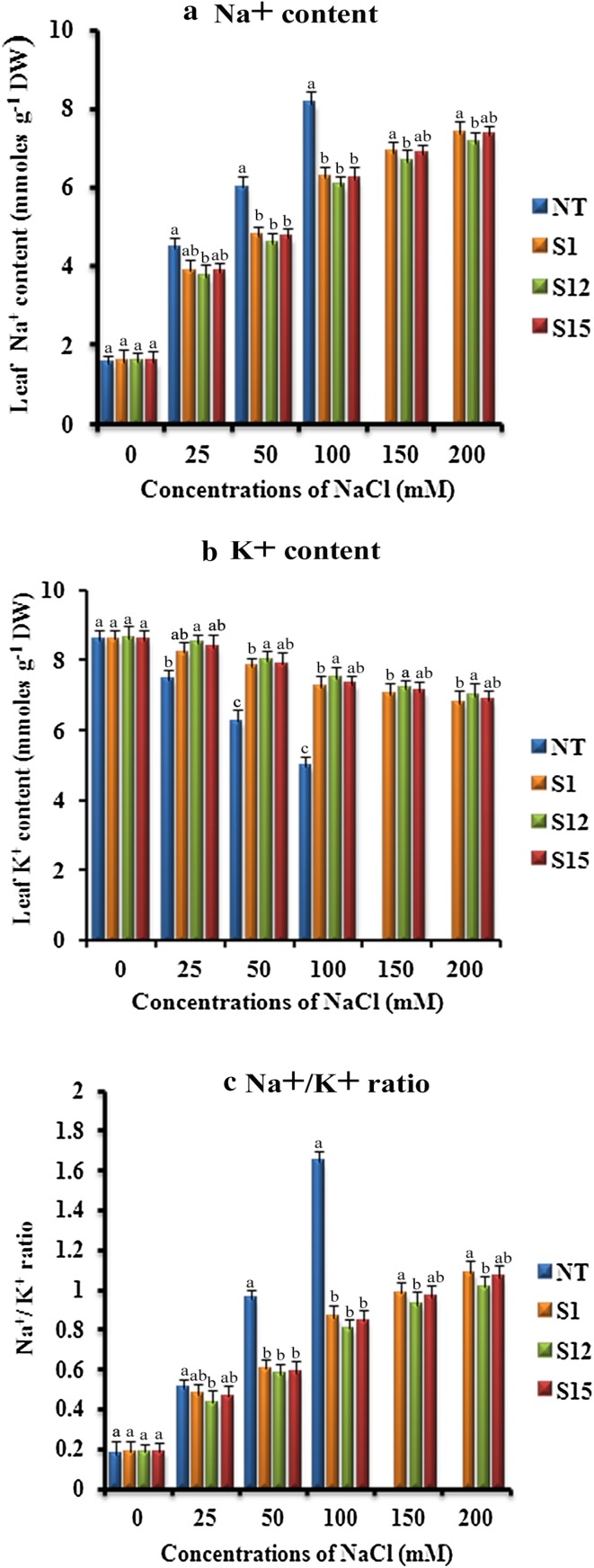
Effect of salinity stress on Na+ and K+ ion content
Ionic balance is necessary for cellular metabolism, plant development, growth and productivity that provides tolerance to salinity stress (Cuin et al. 2008; Conde et al. 2011; Huda et al. 2013a). There was no significant variation observed in the accumulation of Na+ ions between (T1) and NT plants grown under control conditions. However, in the plants subjected to salinity stress the Na+ ion content gradually increased to a significant level in NT plant as compared to the (T1) plants (Fig. 9a). Further analysis of K+ ion content revealed that (T1) and NT plants displayed similar K+ ion content under normal growth conditions, however, when subjected to salinity stress the transgenic plants retained higher K+ ions compared to the NT plant (Fig. 9b). Thereby, we finally observed that Na+/K+ ratio was less in (T1) plants as compared to NT plant (Fig. 9c), which concluded that the transgenic plants have higher potential to tolerate salinity stress. Similarly, Tuteja et al. (2014) reported that p68 expressing tobacco plants accumulated higher K+ and lower Na+ ions as compared to the wild-type plants when subjected to salinity stress. The higher K+ ion content has been reported to delay leaf senescence, whereas lower K+ ion content reportedly regulates caspase-like protease and endonuclease activity resulting in leaf senescence (Huda et al. 2013a; Shabala 2009). Munns et al. (2006) also reported increase of K+ ions and decrease of Na+ ions in wheat as a response to salinity stress. The expression of p68 in tobacco plants reduced Na+/K+ ratio in transgenic plants as compared to control plants, and the restricted entry of Na+ ions into the cells protected the photosynthetic machinery from abiotic stress (Tuteja et al. 2014).
Fig. 9.
Response of transformed (T1) and non-transformed (NT) soybean plants subjected to different concentrations of NaCl for 12 days under greenhouse conditions. a Na+ content (mmoles g− 1 DW); b K+content (mmoles g− 1 DW); c Na+/K+ ratio. Mean of three individual experiments with standard errors. Different letters denote significantly different values according to Duncan’s multiple range test (DMRT) at a 5% level
Yield performance of transformed soybean plants
Under the greenhouse conditions, (T1) and NT soybean plants were phenotypically similar (Fig. 4a), but NT plants were unable to survive beyond 100 mM NaCl, which indicates that beyond 100 mM NaCl is lethal and prevented further development of plants. Whereas (T1) soybean plants grew fairly even at 200 mM NaCl and produced upto 22–24 pods containing 8–9 g dry weight of seeds (Table 1). On the other hand, NT soybean plants produced 19 matured soybean pods containing 6 g of dry weight of seeds at 100 mM NaCl (Table 1).
Table 1.
Yield assay of transformed (T1) and non-transformed (NT) soybean plants subjected to 0, 100, 150 and 200 mM NaCl induced salinity stress conditions
| S. no | Soybean plant | Yield in the absence of NaCl | Yield (100 mM of NaCl) | Yield (150 mM of NaCl) | Yield (200 mM of NaCl) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Number of matured (pods/plant) | Dry weight of seeds (g/plant) | Number of matured (pods/plant) | Dry weight of seeds (g/plant) | Number of matured (pods/plant) | Dry weight of seeds (g/plant) | Number of matured (pods/plant) | Dry weight of seeds (g/plant) | ||
| 1 | NT | 31.3 ± 0.27a,b | 11.3 ± 0.28b | 19.6 ± 0.20c | 6.3 ± 0.34c | – | – | – | – |
| 2 | S1 | 30.6 ± 0.20b | 10.0 ± 0.25c | 27.3 ± 0.24b,a | 12.6 ± 0.24a,b | 25.6 ± 0.20b | 10.3 ± 0.28c | 22.0 ± 0.25b | 8.6 ± 0.15a,b |
| 3 | S12 | 32.0 ± 0.25a | 12.3 ± 0.23a | 28.6 ± 0.20a | 13.3 ± 0.14a | 26.6 ± 0.28a | 12.6 ± 0.24a | 24.3 ± 0.26a | 9.0 ± 0.25a |
| 4 | S15 | 29.3 ± 0.19c | 11.6 ± 0.21a,b | 27.6 ± 0.27b | 12.0 ± 0.21b | 24.0 ± 0.21c | 11.3 ± 0.28b | 23.6 ± 0.18a,b | 8.3 ± 0.24b |
Mean of three individual experiments (±) with standard errors. Different letters inside the same column denote significantly different values according to Duncan’s multiple range test (DMRT) at a 5% level
Conclusion
In conclusion, the p68 gene was transferred and expressed in soybean plants. The expression of the p68 gene in three transgenic lines of soybean exhibited enhanced salinity tolerance. The morphological, physiological and biochemical evidence revealed that three transgenic lines are more tolerant to salinity stress as compared to NT plants. These results suggest the role of p68 in conferring tolerance to salinity stress without affecting yield, photosynthesis, and by controlling the reactive oxygen species (ROS) through modulating antioxidative defence machinery. The obtained results pioneered the efficacy of the p68 gene against salinity and paved way to understand its role under abiotic stress conditions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1 Schematic representation of binary vector pCAMBIA1300 with pea p68 gene (marker free construct). (DOCX 325 KB)
Acknowledgements
Jawaharlal Nehru Scholarship (Ref no: SU-1/88/2016-17/79) for doctoral studies awarded by Jawaharlal Nehru Memorial Fund, New Delhi, India is thankfully acknowledged by the first author (Sivabalan Karthik).
Abbreviations
- NaCl
Sodium chloride
- CaMV 35S
Cauliflower mosaic virus 35S promoter
- APX
Ascorbate peroxidase
- CAT
Catalase
- SOD
Superoxide dismutase
- DHAR
Dehydroascorbate reductase
- MDHAR
Monodehydroascorbate reductase
- MDA
Malondialdehyde
- RWC
Relative water content
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Aebi H. Catalase in vitro. Methods Enzymol. 1984;105:21–126. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- Arnon DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24(1):1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun M, Chinnathambi A, Subramanyam K, Karthik S, Sivanandhan G, Theboral J, Alharbi SA, Kim CK, Ganapathi A. Involvement of exogenous polyamines enhances regeneration and Agrobacterium-mediated genetic transformation in half-seeds of soybean. 3 Biotech. 2016;6(2):148. doi: 10.1007/s13205-016-0448-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banu MS, Huda KM, Sahoo RK, Garg B, Tula S, Islam SS, Tuteja R, Tuteja N. Pea p68 imparts salinity stress tolerance in rice by scavenging of ROS-mediated H2O2 and interacts with argonaute. Plant Mol Biol Rep. 2015;33(2):221–238. [Google Scholar]
- Bartoli CG, Gomez F, Martinez DE, Guiamet JJ. Mitochondria are the main target for oxidative damage in leaves of wheat (Triticum aestivum L.) J Exp Bot. 2004;55:1663–1669. doi: 10.1093/jxb/erh199. [DOI] [PubMed] [Google Scholar]
- Bates LS, Waldren RP, Teare JD. Rapid determination of free proline for water stress studies. Plant Soil. 1973;39:205–207. [Google Scholar]
- Bauder JW, Brock TA. Irrigation water quality, soil amendment, and crop effects on sodium leaching. Arid Land Res Manag. 2001;5(2):101–113. [Google Scholar]
- Birt DF, Hendrich S, Anthony M, Alkel DL. Soybeans and the prevention of chronic human disease. In: Specht J, BOerma R, editors. Soybeans: improvement, production and uses. 3. Madison: American society of Agronomy; 2004. pp. 1047–1117. [Google Scholar]
- Checker VG, Chhibbar AK, Khurana P. Stress-inducible expression of barley Hva1 gene in transgenic mulberry displays enhanced tolerance against drought, salinity and cold stress. Transgenic Res. 2012;21(5):939–957. doi: 10.1007/s11248-011-9577-8. [DOI] [PubMed] [Google Scholar]
- Chen GX, Asada K. Ascorbate peroxidase in tea leaves:occurrence of two isozymes and the differences in their enzymatic and molecular properties. Plant Cell Physiol. 1989;30:987–998. [Google Scholar]
- Chen N, Liu Y, Liu X, Chai J, Hu Z, Guo G, Liu H. Enhanced tolerance to water deficit and salinity stress in transgenic Lycium barbarum L. plants ectopically expressing ATHK1, an Arabidopsis thaliana histidine kinase gene. Plant Mol Biol Rep. 2009;27:321–333. [Google Scholar]
- Chen S, Cui X, Chen Y, Gu C, Miao H, Gao H, Chen F, Liu Z, Guan Z, Fang W. CgDREBa transgenic Chrysanthemum confers drought and salinity tolerance. Environ Exp Bot. 2011;74:255–260. [Google Scholar]
- Conde A, Chaves MM, Geros H. Membrane transport, sensing and signaling in plant adaptation to environmental stress. Plant Cell Physiol. 2011;52:1583–1602. doi: 10.1093/pcp/pcr107. [DOI] [PubMed] [Google Scholar]
- Cuin TA, Betts SA, Chalmandrier R, Shabala S. A root’s ability to retain K+ correlates with salt tolerance in wheat. J Exp Bot. 2008;59:2697–2706. doi: 10.1093/jxb/ern128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA mini preparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Di R, Purcell V, Collins GB, Ghabrial SA. Production of transgenic soybean lines expressing the bean pod mottle virus coat protein precursor gene. Plant Cell Rep. 1996;15(10):746–750. doi: 10.1007/BF00232220. [DOI] [PubMed] [Google Scholar]
- Doulis AG, Debian N, Kingston-Smith AH, Foyer CH. Differential localization of antioxidants in maize leaves. Plant Physiol. 1997;114:1031–1037. doi: 10.1104/pp.114.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsheikh EA, Wood M. Nodulation and N2 fixation by soybean inoculated with salt-tolerant rhizobia or salt-sensitive bradyrhizobia in saline soil. Soil Biol Biochem. 1995;27(4–5):657–661. [Google Scholar]
- FAOSTAT (2016) Agricultural data. http://www.fao.org/faostat/en#data/QC/visualize. Accessed 3 Aug 2018
- Flower DJ, Ludlow MM. Contribution of osmotic adjustment to the dehydration tolerance of water-stressed pigeon pea [Cajanus cajan (L.) Mill sp.] leaves. Plant Cell Environ. 1986;9:33–40. [Google Scholar]
- Foyer CH, Descourvieres P, Kunert KJ. Protection against oxygen radicals: an important defense mechanism studied in transgenic plants. Plant Cell Environ. 1994;17:507–523. [Google Scholar]
- Fu XZ, Khan EU, Hu SS, Fan QJ, Liu JH. Overexpression of the betaine aldehyde dehydrogenase gene from Atriplex hortensis enhances salt tolerance in the transgenic trifoliate orange (Poncirus trifoliata L. Raf.) Environ Exp Bot. 2011;74:106–113. [Google Scholar]
- Gamborg OL, Miller RA, Ojiama K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Gao SQ, Chen M, Xia LQ, Xiu HJ, Xu ZS, Li LC, Zhao CP, Chen XG, Ma YZ. A cotton (Gossypium hirsutum) DREbinding transcription factor gene, GhDREB, confers enhanced tolerance to drought, high salt, and freezing stresses in transgenic wheat. Plant Cell Rep. 2009;28:301–311. doi: 10.1007/s00299-008-0623-9. [DOI] [PubMed] [Google Scholar]
- Gendra E, Moreno A, Alba MM, Pages M. Interaction of the plant glycine-rich RNA-binding protein MA16 with a novel nucleolar DEAD box RNA helicase protein from Zea mays. Plant J. 2004;38:875–886. doi: 10.1111/j.1365-313X.2004.02095.x. [DOI] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48:909–939. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Gill SS, Tajrishi M, Madan M, Tuteja N. A DESD-box helicase functions in salinity stress tolerance by improving photosynthesis and antioxidant machinery in rice (Oryza sativa L. cv. PB1) Plant Mol Biol. 2013;82:1–22. doi: 10.1007/s11103-013-0031-6. [DOI] [PubMed] [Google Scholar]
- Guan Q, Wu J, Zhang Y, Jiang C, Liu R, Chai C, Zhu J. A DEAD box RNA helicase is critical for pre-mRNA splicing, cold responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell. 2013;25:342–356. doi: 10.1105/tpc.112.108340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R, Qu Y, Guo Y, Yu L, Liu Y, Jiang J, Chen J, Ren Y, Liu G, Tian L, Jin L. Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 2014;80(6):937–950. doi: 10.1111/tpj.12695. [DOI] [PubMed] [Google Scholar]
- Hanson B, Grattan SR, Fulton A. Agricultural salinity and drainage. Davis: University of California Irrigation Program. University of California; 1999. [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- Heath RL, Packer L. Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys. 1968;125:189–198. doi: 10.1016/0003-9861(68)90654-1. [DOI] [PubMed] [Google Scholar]
- Hoi PX, Tuteja N. Identification and sequencing analysis of P68 DEAD-box RNA helicase from Pisum sativum. Nat Sci Technol. 2012;28:28–36. [Google Scholar]
- Houot V, Etienne P, Petitot AS, Barbier S, Blein JP, Suty L. Hydrogen peroxide induces programmed cell death features in cultured tobacco BY-2 cells, in a dose- dependent manner. J Exp Bot. 2001;52:1721–1730. [PubMed] [Google Scholar]
- Hu L, Lu H, Liu Q, Chen X, Jiang X. Overexpression of mtlD gene in transgenic Populus tomentosa improves salt tolerance through accumulation of mannitol. Tree Physiol. 2005;25:1273–1281. doi: 10.1093/treephys/25.10.1273. [DOI] [PubMed] [Google Scholar]
- Hu R, Fan C, Li H, Zhang Q, Fu YF. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol. 2009;10:93. doi: 10.1186/1471-2199-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Liu ZR. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J Biol Chem. 2002;277:12810–12815. doi: 10.1074/jbc.M200182200. [DOI] [PubMed] [Google Scholar]
- Huda KM, Banu MS, Garg B, Tula S, Tuteja R, Tuteja N. OsACA6, a P-type IIB Ca2+ATPase promotes salinity and drought stress tolerance in tobacco by ROS scavenging and enhancing the expression of stress-responsive genes. Plant J. 2013;76:997–1015. doi: 10.1111/tpj.12352. [DOI] [PubMed] [Google Scholar]
- Husaini AM, Abdin MZ. Development of transgenic strawberry (Fragaria × ananassa Dutch.) plants tolerant to salt stress. Plant Sci. 2008;174:446–455. [Google Scholar]
- Khan MS, Ahmad D, Khan MA. Utilization of genes encoding osmoprotectants in transgenic plants for enhanced abiotic stress tolerance. Electron J Biotechnol. 2015;18(4):257–266. [Google Scholar]
- Li D, Zhang H, Wang X, Song F. OsBIRH1, a DEAD box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress. J Exp Bot. 2008;59:2133–2146. doi: 10.1093/jxb/ern072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light GG, Mahan JR, Roxas VP, Allen RD. Transgenic cotton (Gossypium hirsutum L.) seedlings expressing a tobacco glutathione S-transferase fail to provide improved stress tolerance. Planta. 2005;222:346–354. doi: 10.1007/s00425-005-1531-7. [DOI] [PubMed] [Google Scholar]
- Linder P, Lasko PF, Ashburner M, Leroy P, Nielsen PJ, Nishi K, Schneir J, Slonimiski PP. Birth of DEAD-box. Nature. 1989;337:121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2∆∆CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maas EV, Hoffman GJ. Crop salt tolerance–current assessment. J Irrig Drain Div. 1977;103(2):115–134. [Google Scholar]
- Maghuly F, Leopold S, Camara Machado AD, Fernandez EB, Khan MA, Gambino G, Gribaudo I, Schartl A, Laimer M. Molecular characterization of grapevine plants transformed with GFLV resistance genes:II. Plant Cell Rep. 2006;25:546–553. doi: 10.1007/s00299-005-0087-0. [DOI] [PubMed] [Google Scholar]
- Malatrasi M, Close TJ, Marmiroli N. Identification and mapping of a putative stress response regulator gene in barley. Plant Mol Biol. 2002;50:143–152. doi: 10.1023/a:1016051332488. [DOI] [PubMed] [Google Scholar]
- McCord JM, Fridovich I. Superoxide dismutase: an enzymatic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- McCue P, Shetty K. Health benefits of soy isoflavonoids and strategies for enhancement: a review. Crit Rev Food Sci Nutr. 2004;44(5):361–367. doi: 10.1080/10408690490509591. [DOI] [PubMed] [Google Scholar]
- Meloni DA, Gulotta MR, Martínez CA, Oliva MA. The effects of salt stress on growth, nitrate reduction and proline and glycinebetaine accumulation in Prosopis alba. Braz J Plant Physiol. 2004;16(1):39–46. [Google Scholar]
- Miyake C, Asada K. Thylakoid-bound ascorbate peroxidase in spinach chloroplasts and photoreduction of its primary oxidation product monodehydroascorbate radicals in the thylakoids. Plant Cell Physiol. 1992;35:539–549. [Google Scholar]
- Munns R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002;25(2):239–250. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Ann Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Lauchli A. Approaches to increasing the salt tolerance of wheat and other cereals. J Exp Bot. 2006;57:1025–1043. doi: 10.1093/jxb/erj100. [DOI] [PubMed] [Google Scholar]
- Munns R, Wallace PA, Teakle NL, Colmer TD. Measuring soluble ion concentrations (Na+, K+, Cl) in salt-treated plants. In: Sunkar R, editor. Methods in molecular biology Plant stress tolerance: methods and protocols. New York: Humana Press Springer; 2010. pp. 371–382. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Ann Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Okanami M, Meshi T, Iwabuchi M. Characterization of a DEAD box ATPase/RNA helicase protein of Arabidopsis thaliana. Nucleic Acids Res. 1998;26:2638–2643. doi: 10.1093/nar/26.11.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhi V, Kumar V, Sunilkumar G, Campbell LAM, Singh NK, Rathore KS. Expression of apoplastically secreted tobacco osmotin in cotton confers drought tolerance. Mol Breed. 2009;23:625–639. [Google Scholar]
- Pasapula V, Shen G, Kuppu S, Paez-Valencia J, Mendoza M, Hou P, Chen J, Qiu X, Zhu L, Zhang X, Auld D, Blumwald E, Zhang H, Gaxiola R, Payton P. Expression of an Arabidopsis vacuolar H+-pyrophosphatase gene (AVP1) in cotton improves drought-and salt tolerance and increases fiber yield in the field conditions. Plant Biotechnol J. 2011;9:88–99. doi: 10.1111/j.1467-7652.2010.00535.x. [DOI] [PubMed] [Google Scholar]
- Pradhan A, Chauhan VS, Tuteja R. Plasmodium falciparum DNA helicase 60 is a schizont stage specific, bipolar and dual helicase stimulated by PKC phosphorylation. Mol Biochem Parasitol. 2005;144:133–141. doi: 10.1016/j.molbiopara.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Rampino P, Pataleo S, Gerardi C, Mita G, Perrotta C. Drought stress response in wheat: physiological and molecular analysis of resistant and sensitive genotypes. Plant Cell Environ. 2006;29:2143–2152. doi: 10.1111/j.1365-3040.2006.01588.x. [DOI] [PubMed] [Google Scholar]
- Roychowdhury A, Roy C, Sengupta DN. Transgenic tobacco plants overexpressing the heterologous lea gene Rab 16a from rice during high salt and water deficit display enhanced tolerance to salinity stress. Plant Cell Rep. 2007;26:1839–1859. doi: 10.1007/s00299-007-0371-2. [DOI] [PubMed] [Google Scholar]
- Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N. Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci USA. 2005;102:509–514. doi: 10.1073/pnas.0406485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S. Salinity and programmed cell death: unraveling mechanisms for ion specific signalling. J Exp Bot. 2009;60:709–712. doi: 10.1093/jxb/erp013. [DOI] [PubMed] [Google Scholar]
- Shah K, Singh M, Rai AC. Effect of heat-shock induced oxidative stress is suppressed in BcZAT12 expressing drought tolerant tomato. Phytochemistry. 2013;95:109–117. doi: 10.1016/j.phytochem.2013.07.026. [DOI] [PubMed] [Google Scholar]
- Shalata A, Tal M. The effect of salt stress on lipid peroxidation and antioxidants in the leaf of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii. Physiol Plant. 1998;104:169–174. doi: 10.1034/j.1399-3054.2001.1120405.x. [DOI] [PubMed] [Google Scholar]
- Singh SK, Sharma HC, Goswami AM, Datta SP, Singh SP. In vitro growth and leaf composition of grapevine cultivars as affected by sodium chloride. Biol Plant. 2000;43:283–286. [Google Scholar]
- Soystats (2011) American Soybean Association. http://soystats.com/archives/2011/page_06.htm. Accessed 3 Aug 2018
- Subramanyam K, Sailaja KV, Subramanyam K, Muralidhara Rao D, Lakshmidevi K. Ectopic expression of an osmotin gene leads to enhanced salt tolerance in transgenic chilli pepper (Capsicum annum L.) Plant Cell Tissue Organ Cult. 2011;105:181–192. [Google Scholar]
- Subramanyam K, Arun M, Mariashibu TS, Theboral J, Rajesh M, Singh NK, Manickavasagam M, Ganapathi A. Overexpression of tobacco osmotin (Tbosm) in soybean conferred resistance to salinity stress and fungal infections. Planta. 2012;236(6):1909–1925. doi: 10.1007/s00425-012-1733-8. [DOI] [PubMed] [Google Scholar]
- Talame V, Ozturk NZ, Bohnert HJ, Tuberosa R. The dynamics of water loss affects the expression of drought-related genes in barley. J Exp Bot. 2007;58:229–240. doi: 10.1093/jxb/erl163. [DOI] [PubMed] [Google Scholar]
- Tambussi EA, Bartoli CG, Beltrano J, Guiamet JJ, Araus JL. Oxidative damage to thylakoid proteins in water-stressed leaves of wheat (Triticum aestivum) Physiol Plant. 2000;108:398–404. [Google Scholar]
- Turkan I, Demiral T. Recent developments in understanding salinity tolerance. Environ Exp Bot. 2009;67(1):2–9. [Google Scholar]
- Turner NC. Techniques and experimental approaches for the measurement of plant water stress. Plant Soil. 1981;58:339–366. [Google Scholar]
- Tuteja N. Plant DNA helicases: the long unwinding road. J Exp Bot. 2003;54(391):2201–2214. doi: 10.1093/jxb/erg246. [DOI] [PubMed] [Google Scholar]
- Tuteja R, Pradhan A. Unraveling the ‘DEAD-box’ helicases of Plasmodium falciparum. Gene. 2006;376:1–12. doi: 10.1016/j.gene.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N, Tuteja R. Prokaryotic and eukaryotic DNA helicases. Essential molecular motor proteins for cellular machinery. Eur J Biochem. 2004;271:1835–1848. doi: 10.1111/j.1432-1033.2004.04093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N, Tuteja R. Unraveling DNA helicases. Motif, structure, mechanism and function. Eur J Biochem. 2004;271:1849–1863. doi: 10.1111/j.1432-1033.2004.04094.x. [DOI] [PubMed] [Google Scholar]
- Tuteja N, Sahoo RK, Garg B, Tuteja R. OsSUV3 dual helicase functions in salinity stress tolerance by maintaining photosynthesis and antioxidant machinery in rice (Oryza sativa L. cv. IR64) Plant J. 2013;76:115–127. doi: 10.1111/tpj.12277. [DOI] [PubMed] [Google Scholar]
- Tuteja N, Banu MS, Huda KM, Gill SS, Jain P, Pham XH, Tuteja R. Pea p68, a DEAD-box helicase, provides salinity stress tolerance in transgenic tobacco by reducing oxidative stress and improving photosynthesis machinery. PLoS One. 2014;9(5):e98287. doi: 10.1371/journal.pone.0098287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA (2018) Food composition database. https://ndb.nal.usda.gov/ndb/search/list?home=true. Accessed 3 Aug 2018
- Vashisht AA, Pradhan A, Tuteja R, Tuteja N. Cold- and salinity stress-induced bipolar pea DNA helicase 47 is involved in protein synthesis and stimulated by phosphorylation with protein kinase C. Plant J. 2005;44:76–87. doi: 10.1111/j.1365-313X.2005.02511.x. [DOI] [PubMed] [Google Scholar]
- Velikova V, Yordanov J, Edreva A. Oxidative stress and some antioxidant systems in acid rain treated bean plants. Protective role of exogenous polyamines. Plant Sci. 2000;151:59–66. [Google Scholar]
- Wang Y, Jiang L, Chen J, Tao L, An Y, Cai H, Guo C. Overexpression of the alfalfa WRKY11 gene enhances salt tolerance in soybean. PLoS One. 2018;13(2):e0192382. doi: 10.1371/journal.pone.0192382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GY, Rocha PSCF, Wang ML, Xu ML, Cui YC, Li LY, Zhu YX, Xia X. A novel rice calmodulin-like gene, OsMSR2, enhances drought and salt tolerance and increases ABA sensitivity in Arabidopsis. Planta. 2011;234:47–59. doi: 10.1007/s00425-011-1386-z. [DOI] [PubMed] [Google Scholar]
- Zhang HW, Huang ZJ, Xie BY, Chen Q, Tian X, Zhang XL, Zhang HB, Lu XY, Huang DF, Huang RF. The ethylene, jasmonate, abscisic acid and NaCl-responsive tomato transcription factor JERF1 modulates expression of GCC box containing genes and salt tolerance in tobacco. Planta. 2004;220:262–270. doi: 10.1007/s00425-004-1347-x. [DOI] [PubMed] [Google Scholar]
- Zhang WJ, Yang SS, Shen XY, Jin YS, Zhao HJ, Wang T. The salt-tolerance gene rstB can be used as a selectable marker in plant genetic transformation. Mol Breed. 2009;23:269–277. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 Schematic representation of binary vector pCAMBIA1300 with pea p68 gene (marker free construct). (DOCX 325 KB)