Abstract
The slow pace of discovery of new effective drugs against multi-drug resistant pathogens and largely unsuccessful combinatorial chemistry has resulted in shifting the focus back to natural products as sources of lead molecules for antimicrobial drugs, mainly due to their structural diversity. Investigation of under-explored habitats for potentially novel microorganisms provides for wider chemodiversity. In this study, four actinomycetes, namely UK-274, UK-281, UK-282 and UK-285, which showed broad-spectrum antibacterial and antifungal activities, were isolated from Timli forest range of the biodiversity-rich Himalayan region. 16S rRNA gene sequence analysis showed that the nearest neighbours of the isolates were Actinomadura nitrigenes, Streptomyces niveiscabiei, and Kitasatospora psammotica with similarity values ranging between 97 and 98% suggesting their potential as new isolates. Further morphological and phenotypic characterization strengthened this assumption. Isolate UK-282, of the rare actinomycetes Kitasatospora group, was found to produce antimicrobial activity. Metabolite fingerprinting of ethyl acetate fraction of isolate UK-282 by GC–MS and 1H NMR analysis showed the presence of three novel compounds. The study underlines that a combination approach of bioprospecting of under-studied habitats and focus on rare actinomycetes may result in the identification of novel chemodiversity.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1556-9) contains supplementary material, which is available to authorized users.
Keywords: Multi-drug resistant, Natural products, Forest, Actinomycete, Antimicrobial, Metabolite fingerprinting
Introduction
Increasing incidences of multi-drug resistant pathogens causing infections has led to an increase in life threatening diseases and has necessitated global drug discovery programs. Combinatorial chemistry, with all its promise of high throughput discovery of novel compounds, has proved to be largely unsuccessful resulting in only one commercially approved compound—Nexavar—an anticancer drug (Wilhelm et al. 2006) the others being 2-(4-fluoro-phenyl)-3-(4-methyl-5,6,7,8-tetrahydro-quinazolin-2-yl)-thiazolidin-4-one and 3-(4,6-dimethyl-pyrimidin-2-yl)-2-(2-methoxy-phenyl)-thiazolidin-4-one (Gupta et al. 2016) exhibiting broad-spectrum antimicrobial activity, but yet to be commercialised.
Natural products have provided consistent backbones for drugs with various bioactivities, including treatment of infectious diseases (Berdy 2012). Microorganisms are known to be a rich source of natural bioactive compounds. Actinomycetes are well known producers of bioactive compounds and account for two-third of the approximately 60–70% approved drugs, which have been formulated with skeletons based on natural compounds (Berdy 2012). Among various classes of actinomycetes, the rare actinomycetes groups have been shown to be sources of novel bioactive compounds (Ibeyaima et al. 2017a). The antimicrobial compounds such as Diazepinomicin from Micromonospora sp. (Charan et al. 2004) and Mutactimycin PR from Saccharothrix sp. SA 103 (Zitouni et al. 2004), substantiate this postulate. While nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) biosynthetic pathways have been documented in actinomycetes, there are many cryptic biosynthetic gene clusters, which need to be unravelled to elicit production of potentially novel metabolites (Becerril et al. 2018).
To obtain novel compounds, there is a need to screen un/under-explored niche habitats (Ibeyaima et al. 2017b). Forests are largely under-explored habitats, though in recent times a few studies have focussed on them for bioprospection. For instance, a novel species, Streptomyces pluripotens, from Malaysian mangrove forest soil was reported to produce novel antimicrobial compounds (Lee et al. 2014), as was S. gilvifuscus, from Korean forest (Nguyen and Kim 2015). Sharma et al. (2016) have reported the antimicrobial compounds [(Z)-3-tetradecene, dodecyl acrylate, 2, 4-di-t-butyl-6-nitrophenol, (E)-5-eicosene, (E)-9-octadecene] obtained from actinomycetes of the Himalayan forests of Assam (India).
In the present study, the Timli forest range, a subtropical deciduous Himalayan forest in India has been studied for its cultivable microbial population, with the objective of identifying and characterizing novel actinomycetes. With the twin hypotheses of using a combination of unexplored habitats and focussing on rare actinomycete genera, the current study has been formulated with the following objectives: (i) to characterize microbial diversity for the potential to produce antimicrobial compounds (ii) taxonomically identify selected isolates showing antimicrobial activity (iii) metabolite fingerprinting of organic solvent extract of a selected rare actinomycete isolate (UK-282).
Materials and methods
Selective isolation
Soil samples were collected from the Timli forest range, Dehradun (30°23′N; 77°42′E), India (Ibeyaima et al. 2017a) using standard techniques. The samples were stored at 4 °C for a maximum of 4 days till analysis. Soil samples were pooled, suspended in ¼ strength Ringer’s solution and dilution-plated on Nutrient Agar (NA), Glucose Yeast Extract agar (GYE) and Potato Dextrose Agar (PDA) media for isolation of bacteria, actinomycetes and fungi, respectively. The GYE medium was amended with rifampicin (20 µg ml−1) and actidione (25 µg ml−1) to inhibit the growth of fast-growing bacteria and fungi, respectively.
Colony morphology (aerial and reverse substrate mycelia colour and diffusible pigment production) of the isolates was noted by visual observation. Standard chromatographic procedures were used to detect the isomers of 2,6 diaminopimelic acid (A2pm) as per Becker et al. (1964). Fatty acid methyl esters (FAMEs) were extracted, saponified and methylated as described by Miller (1982) with minor modifications from Kuykendall et al. (1988). The FAMEs were separated by gas chromatography and the peaks established using the Standard Microbial Identification (MIDI) system version 6.2 by Sasser (1990), using the Aerobe TSBA 60 version 6.0R database.
Antimicrobial activity
The isolates obtained from NA and GYE plates were evaluated for antimicrobial activity using agar culture plugs as per Bauer et al. (1966) against Gram-positive [Bacillus subtilis (MTCC-121), Micrococcus luteus (MTCC-106)] and Gram-negative bacteria [Escherichia coli (MTCC-1679), Pseudomonas fluorescens (MTCC-2421)] and fungus [Saccharomyces cereviseae (MTCC-747)]. Tetracycline (10 µg ml−1) was used as positive control. The plates were incubated for 24–48 h at 37 °C and zones of inhibition were measured subsequently.
Four isolates (UK-274, UK-281, UK-282, and UK-285) that showed maximum zones of inhibition and broad-spectrum antibacterial and antifungal activity were used for further studies.
Molecular identification and phylogenetic analysis
Bacterial DNA genome of UK-274, UK-281, UK-282, and UK-285 was isolated (Conn and Franco 2004) and quality-checked by agarose gel electrophoresis and spectrophotometrically using NanoDrop1000 (Thermo-Scientific). The universal primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTTC-3′) were used for amplifying the 16S rRNA gene using the following conditions: initial denaturation temperature was set at 95 °C for 5 min, followed by 35 cycles at same temperature for 1 min, primer annealing at 54 °C for 1 min and primer extension at 72 °C for 2 min. The reaction mixture was kept at 72 °C for 10 min subsequently and then cooled to 4 °C (Ibeyaima et al. 2017a). The PCR product obtained was purified using Exosap-IT, and sequenced using the same primers (8F and 1492R) on Applied Biosystems 3130 Genetic Analyzer. Consensus sequence was obtained by alignment of sequences and deleting overlaps, quality-checked using DECIPHER and aligned with sequences of representative type strains obtained from the EzTaxon server (http://eztaxon-e.ezbiocloud.net) (Kim et al. 2012). Evolutionary distances were computed as per Jukes and Cantor (1969) and the bootstrap consensus tree inferred from 1000 replicates was evaluated. MEGA 6.0 software was used to construct the phylogenetic tree based on the neighbour-joining tree algorithm (Tamura et al. 2013). Phylogeny trees were also inferred using maximum-likelihood and maximum-parsimony algorithms. The nearly complete 16S rDNA consensus sequences were deposited in the GenBank database.
Further antimicrobial compound purification and metabolite fingerprinting studies were carried out with the isolate UK-282, which was identified as a rare actinomycete belonging to the genus Kitasatospora.
Purification of antimicrobial compound
Isolate UK-282 was inoculated in 200 ml Bennett’s broth in separate experiments to prepare each type of extract, and incubated in a platform shaker (Kuhner–Therm LT-X) at 200 rpm for 7 days. Subsequently, it was centrifuged at 4000 rpm for 20 min and the supernatant was used as the aqueous extract. Organic solvent extracts were prepared by individually mixing equal volumes of culture supernatant and the solvents ethyl acetate (EA), butanol (BT), hexane (HE), dichloromethane (DCM) and petroleum ether (PE), kept on a vortex mixer for 2 h and the organic layer collected. Each extract was evaporated to dryness, the weight of the residue was measured and dissolved in methanol to give a final concentration of 1 mg ml−1.
The aqueous and organic solvent extracts of UK-282 were used for investigating the antimicrobial activity against an expanded panel of target organisms, with inclusion of Gram-positive Staphylococcus epidermidis (MTCC-435) and Brevibacterium linens (MTCC-268), in addition to the earlier mentioned panel, by well diffusion method. Other conditions were similar as in the culture plug method (Bauer et al. 1966). The inhibition zones were measured after 24–48 h. Tetracycline (10 µg ml−1) was used as positive control. All experiments were performed in triplicates and the result presented as the standard deviation from the mean. The EA extract showed the largest zones of inhibition against all the target organisms. Hence, EA extract was used for determining antimicrobial activity against multi-drug resistant pathogenic clinical isolates [E. coli (26423, 26437 and 26479) and P. aeruginosa (26460 and 26418) from urine samples and P. aeruginosa (26484) from throat swab] (antimicrobial profiles of the clinical isolates are shown in Supplementary Table 2) and for further purification and metabolite fingerprinting studies.
Metabolite fingerprinting
The EA extract was initially subjected to preparatory thin layer chromatography (TLC) on silica gel plates (Silica gel 60F254, 20 cm × 20 cm, Merck). Various organic solvents individually and in combinations (ethyl acetate, hexane, butanol and ethanol) and in various ratios were used as mobile phases to optimise separation of antimicrobial compounds. Ethyl acetate:hexane (2:1) provided best separation of compounds as evaluated by UV visualization. Each well-separated band was further evaluated for antimicrobial activity by bioautography.
Simultaneously, the EA extract was subjected to column chromatography wherein the column was packed with silica gel G (60–120 mesh) slurry. Fractions were collected using similar combinations of organic solvents as for TLC. The fractions obtained using the mobile phase combination of ethyl acetate:ethanol (2:1) showed antimicrobial activity. The fraction that showed maximum zone of inhibition was analysed by GC–MS analysis.
GC–MS was performed using QP2010Ultra (SHIMADZU) with internal dimension 30 m × 0.25 mm × 0.25 µm (length × internal diameter × film thickness) as per Nielsen et al. (2014) under the following conditions: GC column oven temperature 50 °C, injector temperature 260 °C at split mode ratio 10 with a flow rate of 16.3 ml min−1; MS with ion source temperature 230 °C, interface temperature: 270 °C, scan range: 40–650 m/z, event time: 0.20 s, solvent cut time: 4.50 min, MS start time: 4 (min), MS end time: 50.32 (min).
The mass spectrum data obtained were further analysed and interpreted using the collated retention time (RT), similarity index (SI), mass ion spectra (MS), and retention indices (RI-observed and calculated) data. The calculated RI was obtained using the Kovats linear index (Babushok et al. 2011) and subsequently was further compared with National Institute of Standards and Technology (NIST11), WILEY8 libraries, PubChem and ChemSpider.
Structure characterizations
1H NMR spectra of the two fractions—one obtained after TLC and other obtained after column chromatography—were recorded on a DRX 400 spectrometer (Bruker, Karlsruhe, Germany) using deuterated methanol as solvent and tetramethylsilane (TMS) as internal standard (TMS, δ = 0.0 ppm). The chemical shifts are expressed in δ values (ppm). The obtained data were preliminarily analysed for functional groups (Silva Sa et al. 2017; Baluja et al. 2017).
Results and discussion
Isolates were selectively isolated in NA, GYE, and PDA. The colony counts were 1.35 × 104 ± 4.08, 1.3 × 102 ± 2.49 and 6.0 × 101 ± 1.70 in NA, GYE and PDA, respectively. A maximum of 55% distinct isolates were obtained from NA as compared to GYE (40%) and PDA (5%).
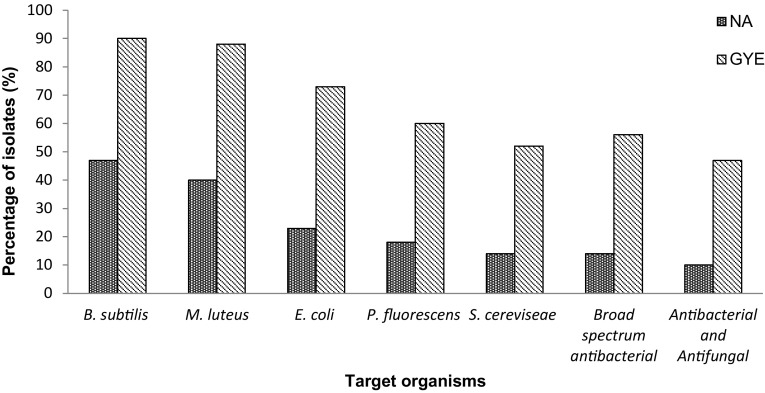
Subsequent to screening for antimicrobial activity, it was observed that around 40% of the isolates obtained on NA exhibited antimicrobial activity against B. subtilis and M. luteus, ~ 20% against E. coli and P. fluorescens, 14% showed broad-spectrum antibacterial activity and against S. cereviseae, while 10% showed both antibacterial and antifungal activity (Fig. 1). The isolates from GYE exhibited best antimicrobial activity (90% against B. subtilis, 88% against M. luteus, 73% against E. coli, 60% against P. fluorescens, 52% against S. cereviseae, 56% showed broad-spectrum activity and 47% both antibacterial and antifungal activities). The isolates from PDA were identified as belonging to Aspergillus and were not studied further.
Fig. 1.
Antimicrobial activity of the isolates from the Timli forest range
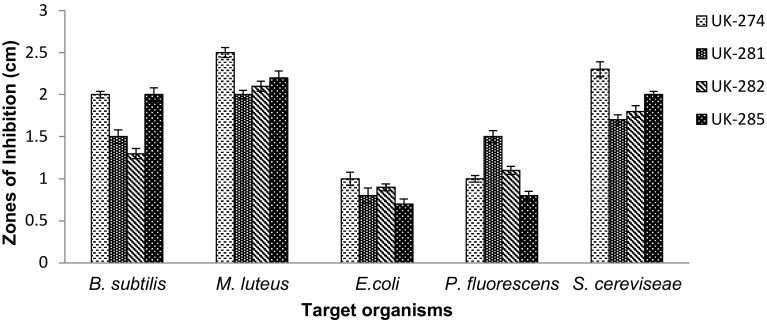
Isolates UK-274, UK-281, UK-282 and UK-285, that showed broad-spectrum antibacterial activity as well as antifungal activity (Fig. 2) were further identified by 16S rDNA sequencing and phylogenetic analysis. The isolates were provided the following accession numbers: MH244352 (UK-274), MH244353 (UK-281), MH244354 (UK-282) and MH244355 (UK-285).
Fig. 2.
Antimicrobial activity of selected isolates (UK-274, UK-281, UK-282 and UK-285) from the Timli forest range
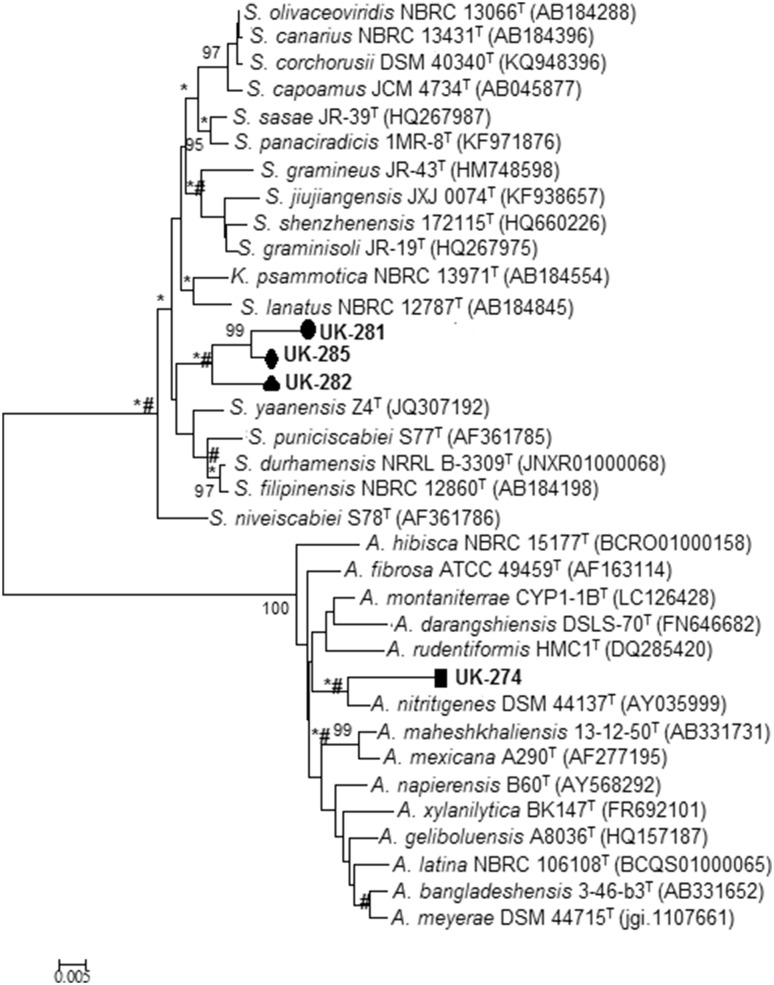
The consensus 16S rDNA sequence of UK-274 showed 97.71% similarity to Actinomadura nitrigenes (Fig. 3) with 29 nucleotide differences in 1269 sites. The aerial and substrate mycelium of this isolate were cream with a crystalline texture. It did not produce diffusible pigments. In contrast, the aerial mycelium of its close relative A. nitrigenes was reported to be white with brown substrate mycelium with no diffusible pigments (Lipski and Altendorf 1995).
Fig. 3.
Neighbour-joining phylogenetic tree based on 16S rDNA sequence showing relationship between isolates UK-274, UK-281, UK-282 and UK-285 with related type strains of Actinomadura sp., Streptomyces sp. and Kitasatospora sp. Bootstrap values at the nodes indicate collated values based on 1000 resampled datasets; only values above > 95% are given. Bar 0.005 indicates substitutions per nucleotide. *And # at nodes indicate branches that were also recovered using the maximum-likelihood and maximum-parsimony algorithms. A Actinomadura, K Kitasatospora, S Streptomyces, NBRC NITE Biological Resource Centre, DSM DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, JCM Japan Collection of Microorganisms, NRRL Agriculture Research Service Culture Collection, ATCC American Type Culture Collection
UK-281 showed 97.97% similarity to Streptomyces niveiscabiei (Fig. 3). There were 24 nucleotide differences in 1182 sites. UK-285 showed 98.14% similarity to S. niveiscabiei, with 24 nucleotide differences in 1288 sites. The aerial and substrate mycelia of isolates UK-281 and UK-285 were light brown and dark brown, respectively, with crystalline textures. Both the isolates secreted brown diffusible pigments. The aerial mycelium of the closely related S. niveiscabiei has been documented to be white with no diffusible pigment secretion (Park et al. 2003).
UK-282 showed 97.29% similarity to Kitasatospora psammotica (Fig. 3) with 35 nucleotides differences in 1290 sites. The aerial and substrate mycelia of this isolate were pinkish brown and light brown, respectively, with a crystalline colony texture, with diffusible brown pigment production. Its closely related genus K. psammotica has been reported to show green-to-yellow aerial mycelium and colourless-to-pale yellow substrate mycelium. No diffusible pigment production was documented. Cell wall composition showed the presence of LL and meso-A2pm, indicating that the isolate does not belong to Streptomyces lineage, and is more closely linked to Kitasatospora sp. (Takahashi 2017). FAME analysis showed that major cellular fatty acids of UK-282 were iso-C16:0 (22.43%), anteiso-C15:0 (18.94%), iso-C15:0 (13.60%) and anteiso-C17:0 (13.38%) while those for the nearest match K. psammotica were iso-C15:0, C16:0, anteiso-C15:0 (Nguyen and Kim 2015) and that for S. shenzhenensis were iso-C16:0 (33.1%), anteiso-C15:0 (12.2%), C14:0 (10.7%), iso- C15:0 (10.6%) (Hu et al. 2011). A low similarity index of 0.6 matching with S. violaceusniger was obtained as per the MIDI system. Collated data analysis shows marked differences in FAME profiles between UK-282 and its nearest matches (Supplementary Table 1). Based on the phylogenetic analysis and the colony characteristics, all the four isolates can be considered novel, at least at the species level.
Further studies after purification showed that the aqueous extract of UK-282 presented antimicrobial activity against B. subtilis, S. epidermidis, M. luteus, and P. fluorescens (Table 1). The EA extract showed antimicrobial activity against all the target organisms, while the BT extract showed activity only against S. epidermidis and M. luteus. The other organic solvent extracts (HE, DCM and HE) did not produce inhibition zones against any of the tested microorganisms. EA extract also exhibited antimicrobial activity against pathogenic clinical isolates—E. coli (26437 and 26479), and P. aeruginosa (26460 and 26418) from urine samples and P. aeruginosa (26484) from throat swab (Table 2). Hence the EA extract was further used for metabolite fingerprinting.
Table 1.
Antimicrobial activity of aqueous and organic solvent extracts of UK-282
| Aqueous/organic solvent extracts and positive control | Zones of inhibition (cm) Mean ± SD |
||||||
|---|---|---|---|---|---|---|---|
| B. subtilis | S. epidermidis | B. linens | M. luteus | P. fluorescens | E. coli | S. cereviseae | |
| Aqueous | 0.7 ± 0.06 | 0.8 ± 0.04 | – | 0.9 ± 0.16 | 0.8 ± 0.04 | – | – |
| EA | 1.5 ± 0.08 | 1.8 ± 0.16 | 1.7 ± 0.06 | 2 ± 0.04 | 1 ± 0.16 | 0.9 ± 0.04 | 1.9 ± 0.06 |
| BT | – | 1.1 ± 0.04 | 0.9 ± 0.16 | – | – | – | – |
| HE | – | – | – | – | – | – | – |
| DCM | – | – | – | – | – | – | – |
| PE | – | – | – | – | – | – | – |
| Tetracycline | 2.7 ± 0.08 | 2.3 ± 0.04 | 2.5 ± 0.08 | 3.0 ± 0.08 | 2.6 ± 0.12 | 1.8 ± 0.04 | 2.8 ± 0.20 |
EA ethyl acetate extract, BT butanol extract, HE hexane extract, DCM dichloromethane extract, PE petroleum ether extract
Table 2.
Antimicrobial activity of ethyl acetate extract of UK-282 against clinical isolates
| EA extract | Zones of inhibition (cm) Mean ± SD |
|||||
|---|---|---|---|---|---|---|
| Urine samples | Throat swab sample | |||||
| E. coli (26423) | E. coli (26437) | E. coli (26479) | P. aeruginosa (26460) | P. aeruginosa (26418) | P. aeruginosa (26484) | |
| EA | 0 | 1.2 ± 0.04 | 1.1 ± 0.08 | 1.3 ± 0.12 | 1.0 ± 0.09 | 1.13 ± 0.04 |
| Tetracycline | 2.8 ± 0.04 | 2.7 ± 0.12 | 2.0 ± 0.04 | 2.1 ± 0.06 | 0 | 2.8 ± 0.08 |
EA ethyl acetate extract
Purification and metabolite fingerprinting
Metabolite fingerprinting (by GC–MS) of two distinct components was carried out—the separated component obtained after bioautography (TLC-separated fractions) and the column-purified fraction. Antimicrobial activity measurements of these fractions have been shown in Table 3. Two compounds, hexadecanoic acid and N-(2-hydroxyethyl) dodecanamide, were obtained from the TLC-separated fraction and a single compound 3-hexen-2-one from the column-purified fraction (Table 4).
Table 3.
Antimicrobial activity of metabolite fractions against non-pathogenic and pathogenic microorganisms
| Target organisms | Metabolite fractions Zones of inhibition (cm) Mean ± SD |
Tetracycline (10 µg ml−1) | |
|---|---|---|---|
| Active fraction (TLC) | Active fraction (column chromatography) | ||
| B. subtilis | 1.3 ± 0.04 | 1.2 ± 0.08 | 2.7 ± 0.08 |
| S. epidermidis | 1.65 ± 0.04 | 1.5 ± 0.12 | 2.3 ± 0.04 |
| B. linens | 1.5 ± 0.08 | 1.6 ± 0.06 | 2.5 ± 0.08 |
| M. luteus | 1.8 ± 0.12 | 1.7 ± 0.10 | 3.0 ± 0.08 |
| P. fluorescens | 0.95 ± 0.04 | 0.8 ± 0.08 | 2.6 ± 0.12 |
| E. coli | 0.8 ± 0.04 | 0.63 ± 0.04 | 1.8 ± 0.04 |
| S. cereviseae | 0 | 1.8.0 ± 0.12 | 2.8 ± 0.20 |
| Clinical isolates | |||
| E. coli (26423) | 0 | 0 | 2.8 ± 0.04 |
| E. coli (26437) | 1.2 ± 0.04 | 1.0 ± 0.08 | 2.7 ± 0.12 |
| E. coli (26479) | 0.8 ± 0.06 | 0.95 ± 0.04 | 2.0 ± 0.04 |
| P. aeruginosa (26460) | 1.0 ± 0.12 | 0.8 ± 0.14 | 2.1 ± 0.06 |
| P. aeruginosa (26418) | 1.1 ± 0.22 | 1.0 ± 0.12 | 0 |
| P. aeruginosa (26484) | 1.2 ± 0.26 | 0.96 ± 0.04 | 2.8 ± 0.08 |
Table 4.
Compounds/metabolites and their characteristics, obtained by GC–MS after preparatory thin layer chromatography and column chromatography from ethyl acetate extract of UK-282
| Compound source | Chemical group | Name of compound | Retention time (RT) min | Similarity index, SI (%) | Chemical abstracts service (CAS) no. | Retention indices from databases (RI) | Calculated retention indices (RI) |
|---|---|---|---|---|---|---|---|
| TLC | Fatty acid | Hexadecanoic acid | 33.707 | 95 | 57-10-3 | 1968 | 1514 |
| TLC | Fatty acid amide | N-(2-Hydroxyethyl) dodecanamide | 35.923 | 93 | 142-78-9 | 2056 | 1568 |
| Column chromatography | Ketone | 3-Hexen-2-one | 8.597 | 93 | 763-93-9 | 762 | 956 |
The three compounds identified as per the (NIST11), WILEY8 libraries, PubChem and ChemSpider databases belonged to three major chemical groups, namely fatty acid, amide and ketone. Babushok et al. (2011) described a standard approach to identify compounds by comparing the observed retention indices from databases with the calculated retention index. This approach is applicable for the identification of those compounds for which standard reference compounds are not available. For compound identification, Kovats RI is a validated method (Babushok et al. 2011). Kovats RI values were calculated for all these three compounds and compared with the documented RI in the standard databases. The SI values were used for the initial data analysis. Amongst the identified compounds, hexadecanoic acid showed SI value of 95%. The other two compounds showed SI values of 93% each. The calculated RI value for N-hexadecanoic acid and N-(2-hydroxyethyl) dodecanamide were widely different from that reported in NIST11/WILEY8/PubChem/ChemSpider databases. The reported RI value for N-hexadecanoic acid was 1968 and calculated RI 1514. Likewise, the reported RI value for N-(2-hydroxyethyl) dodecanamide was 2056 and calculated RI 1568. The reported RI value of 3-hexen-2-one was 762 and calculated RI 762.
Among these compounds, hexadecanoic acid (palmitic acid) has been reported in plants and known to have both antibacterial and antifungal activities (Huang et al. 2011). N-(2-Hydroxyethyl) dodecanamide is an amide-linked hydroxy fatty acid derivative (amide derivative of lauric acid), also known for both antibacterial and antifungal activity (Kabara et al. 1972). This compound has not been reported as a secretary product from microorganisms or other systems. Some of the amide-linked hydroxy fatty acids have been reported in lipopolysaccharides (LPS) of Gram-negative bacteria (endotoxins) (Bhat and Carlson 1992). However, the compound documented in this study has been obtained from a Gram-positive bacterium and hence presence of LPS is ruled out. The third identified compound (3-hexen-2-one) is also reported to be produced by plants (Malik et al. 2013).
Many species of Kitasatospora have been reported for the production of novel bioactive compounds. For instance, a phenazine-type compound was reported in Kitasatospora sp. MBT66 with antimicrobial activity (Wu et al. 2015). An antifungal compound, Cystragin, was produced by K. cystarginea RK-419 (Kusakabe and Isono 1988). K. psammotica (earlier named S. psammoticus) has been reported to show production of the tetracycline derivative, 2-acetyl-2-decarboxamidotetracycline (Lancini and Sensi 1964). Streptomyces psammoticus KP1404 produces the polyene antifungal compounds Strevertene A and B (Kim et al. 2011). The other matching species, S. graminisoli and S. shenzhenensis have not been documented to show any antimicrobial compound production.
1H NMR
1H NMR spectrum of TLC-separated fraction exhibited a prominent peak at 4.198 ppm suggesting the presence of alkene groups. Other peaks obtained were of alcohols/esters/ethers between 3.320 and 3.336 ppm. A less dense peak was obtained at 2.034 ppm, suggesting the presence of carbonyl groups. While GC–MS analysis showed the presence of hexadecanoic acid and N-(2-hydroxyethyl) dodecanamide, 1H NMR spectrum has not shown chemical shifts in the acid or amide regions.
1H NMR spectrum of the fraction obtained after column chromatography also showed a prominent peak in the alkene region (4.925 ppm) and less dense peaks (possibly corresponding to alcohols/esters/ethers) between 3 and 4 ppm and similar peaks of carbonyl/alkenes/aromatics at around 2 ppm. Overlapped peaks of methyl groups were observed at 1–2 ppm. The presence of alkene and carbonyl functional groups is similar to that of 3-hexen-2-one (from GC–MS analysis). However, in addition, peaks corresponding to alcohols/esters/ethers were also obtained suggesting that the compound may not contain only ketone and alkene groups. Further, the spectrum obtained does not match the standard spectrum of these compounds.
The low similarity indices of hexadecanoic acid (95%), N-(2-hydroxyethyl) dodecanamide (93%) and 3-hexen-2-one (93%) from the GC–MS analysis and the differing functional groups obtained from 1HNMR data, indicate that the obtained compounds are novel. Further studies can confirm the structural configuration of the obtained compounds.
The compounds (deduced from combined GC–MS and 1H NMR analyses) in the present study have not been reported earlier in K. psammotica or other microorganisms. The results of this study further support the hypothesis that a combination of sampling from under-explored habitats, selective isolation of actinomycetes, and focussed study of rare actinomycetes can result in elaboration of novel bioactive compounds, as also substantiated by our earlier studies (Ibeyaima et al. 2017a, b).
Conclusion
Diversity analysis for antimicrobial activity showed that a good percentage of isolates obtained from the Himalayan biodiversity hotspot, exhibited both antibacterial and antifungal activities. Based on molecular and morphological data, isolates UK-274, UK-281, UK-282 and UK-285 have been shown to be novel taxa (at the species level). GC–MS analysis of the EA extract of UK-282 showed the presence of three compounds [hexadecanoic acid and N-(2-hydroxyethyl) dodecanamide and 3-hexen-2-one]. Combined analysis of GC–MS and 1H NMR revealed that the obtained compounds do not match those present in standard databases and are novel. We conclude that bioprospection of under-studied habitats for rare actinomycetes can elaborate promising novel bioactive compounds.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are thankful to Jaypee Institute of Information Technology, Noida for providing the necessary facilities. We acknowledge, Advanced Institute of Research Facility (AIRF), Jawaharlal Nehru University, New Delhi, for GC–MS analyses. Nidhi Srivastava thanks the Indian Council of Medical Research, Government of India, for providing ICMR fellowship [3/1/3JRF-2013/HRD-136 (30690)].
Conflict of interest
On the behalf of all authors, corresponding author states that there is no conflict of interest.
Compliance with ethical requirements
This article does not contain any studies with human or animal subjects.
References
- Babushok VI, Linstrom PJ, Zenkevich IG. Retention indices for frequently reported compounds of plant essential oils. J Phys Chem Ref Data. 2011;40:43101. doi: 10.1063/1.3653552. [DOI] [Google Scholar]
- Baluja S, Chanda S, Padalia H, Talaviya H. Synthesis, characterization and in vitro antimicrobial screening studies of some pyridyl-coumarin compounds. Rev Colomb Cienc Quím Farmn. 2017;46(1):5–21. doi: 10.15446/rcciquifa.v64n1.67287. [DOI] [Google Scholar]
- Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- Becerril A, Alvarez S, Brana AF, Rico S, Diaz M, Santamaría RI, Salas JA, Mendez C. Uncovering production of specialized metabolites by Streptomyces argillaceus: activation of cryptic biosynthesis gene clusters using nutritional and genetic approaches. PLoS One. 2018;13(5):e0198145. doi: 10.1371/journal.pone.0198145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B, Lechevalier MP, Gordon RA, Lechevalier HA. Rapid differentiation between Nocardia and Streptomyces by paper chromatography of whole-cell hydrolysates. Appl Microbiol. 1964;12:421–423. doi: 10.1128/am.12.5.421-423.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdy J. Thoughts and facts about antibiotics: where we are now and where we are heading. J Antibiot (Tokyo) 2012;65:385–395. doi: 10.1038/ja.2012.27. [DOI] [PubMed] [Google Scholar]
- Bhat UR, Carlson RW. A new method for the analysis of amide-linked hydroxy fatty acids in lipid-As from gram-negative bacteria. Glycobiology. 1992;2:535–539. doi: 10.1093/glycob/2.6.535. [DOI] [PubMed] [Google Scholar]
- Charan RD, Schlingmann G, Janso J, Bernan V, et al. Diazepinomicin, a new antimicrobial alkaloid from a marine Micromonospora sp. J Nat Prod. 2004;67(8):1431–1433. doi: 10.1021/np040042r. [DOI] [PubMed] [Google Scholar]
- Conn VM, Franco CMM. Analysis of the endophytic actinobacterial population in the roots of wheat (Triticum aestivum L.) by terminal restriction fragment length polymorphism and sequencing of 16S rRNA clones. Appl Environ Microbiol. 2004;70:1787–1794. doi: 10.1128/AEM.70.3.1787-1794.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Singh R, Sonar PK, Saraf SK. Novel 4-thiazolidinone derivatives as anti-infective agents: synthesis, characterization, and antimicrobial evaluation. Biochem Res Int. 2016;2016:8. doi: 10.1155/2016/8086762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Lin HP, Xie Q, Li L, Xie XQ, Sun M, Hong K. Streptomyces shenzhenensis sp. nov., a novel actinomycete isolated from mangrove sediment. Antonie Van Leeuwenhoek. 2011;100(4):631–637. doi: 10.1007/s10482-011-9618-6. [DOI] [PubMed] [Google Scholar]
- Huang CB, Alimova Y, Myers TM, Ebersole JL. Short- and medium-chain fatty acids exhibit antimicrobial activity for oral microorganisms. Arch Oral Biol. 2011;56:650–654. doi: 10.1016/j.archoralbio.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibeyaima A, Dwivedi AK, Saini N, Gupta S, Sarethy IP. Saccharothrix sp. TD-093 from the Thar Desert, India: metabolite fingerprinting of antimicrobial compounds and in silico analysis. Curr Microbiol. 2017;74:334–343. doi: 10.1007/s00284-016-1183-9. [DOI] [PubMed] [Google Scholar]
- Ibeyaima A, Rana J, Dwivedi AK, Saini N, Gupta S, Sarethy IP. Pseudonocardiaceae sp. TD-015 from the Thar Desert, India: antimicrobial activity and identification of antimicrobial compounds. Curr Bioact Compd. 2017 doi: 10.2174/1573407213666170104124315. [DOI] [Google Scholar]
- Jukes TH, Cantor CR. Mammalian protein metabolism. Amsterdam: Elsevier; 1969. Evolution of protein molecules; pp. 21–132. [Google Scholar]
- Kabara JJ, Swieczkowski DM, Conley AJ, Truant JP. Fatty acids and derivatives as antimicrobial agents. Antimicrob Agents Chemother. 1972;2:23–28. doi: 10.1128/AAC.2.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JD, Han JW, Lee SC, Lee D, Hwang IC, Kim BS. Disease control effect of strevertenes produced by Streptomyces psammoticus against tomato Fusarium Wilt. J Agric Food Chem. 2011;59(5):1893–1899. doi: 10.1021/jf1038585. [DOI] [PubMed] [Google Scholar]
- Kim O-S, Cho Y-J, Lee K, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- Kusakabe H, Isono K. Taxonomic studies on Kitasatosporia cystarginea sp. nov., which produces a new antifungal antibiotic cystargin. J Antibiot (Tokyo) 1988;41:1758–1762. doi: 10.7164/antibiotics.41.1758. [DOI] [PubMed] [Google Scholar]
- Kuykendall LD, Roy MA, O’Neill JJ, Divine TE. Fatty acids, antibiotic resistance and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int J Syst Evol Microbiol. 1988;38:358–361. [Google Scholar]
- Lancini GC, Sensi P. Isolation of 2-acetyl-2-decarboxamidotetracycline from cultures of Streptomyces psammoticus. Experientia. 1964;20(2):83–84. doi: 10.1007/BF02151254. [DOI] [PubMed] [Google Scholar]
- Lee L-H, Zainal N, Azman A-S, et al. Streptomyces pluripotens sp. nov., a bacteriocin-producing streptomycete that inhibits meticillin-resistant Staphylococcus aureus. Int J Syst Evol Microbiol. 2014;64:3297–3306. doi: 10.1099/ijs.0.065045-0. [DOI] [PubMed] [Google Scholar]
- Lipski A, Altendorf K (1995) Actinomadura nitritigenes sp. nov., isolated from experimental biofilters. Int J Syst Bacteriol 717–723
- Malik AA, Mir SR, Ahmad J. Ruta graveolens L. essential oil composition under different nutritional treatments. Environ Sci. 2013;13:1390–1395. doi: 10.5829/idosi.aejaes.2013.13.10.1124831. [DOI] [Google Scholar]
- Miller LT. Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxyl acids. J Clin Microbiol. 1982;16:584–586. doi: 10.1128/jcm.16.3.584-586.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TM, Kim J. Streptomyces gilvifuscus sp. nov., an actinomycete that produces antibacterial compounds isolated from soil. Int J Syst Evol Microbiol. 2015;65:3493–3500. doi: 10.1099/ijsem.0.000447. [DOI] [PubMed] [Google Scholar]
- Nielsen MT, Ranberg JA, Christensen U, et al. Microbial synthesis of the forskolin precursor manoyl oxide in an enantiomerically pure form. Appl Environ Microbiol. 2014;80:7258–7265. doi: 10.1128/AEM.02301-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Kim JS, Kwon SW, Wilson C, Yu YM, Hur JH, Lim CK. Streptomyces luridiscabiei sp. nov., Streptomyces puniciscabiei sp. nov. and Streptomyces niveiscabiei sp. nov., which cause potato common scab disease in Korea. Int J Syst Evol Microbiol. 2003;53:2049–2054. doi: 10.1099/ijs.0.02629-035. [DOI] [PubMed] [Google Scholar]
- Sasser M. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101. Newark: MIDI Inc; 1990. [Google Scholar]
- Sharma P, Kalita MC, Thakur D. Broad spectrum antimicrobial activity of forest-derived soil Actinomycete, Nocardia sp. PB-52. Artic Front Microbiol. 2016 doi: 10.3389/fmicb.2016.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva Sa FA, de Paula JAM, dos Santos PA, Oliveira LAR, Oliveira GAR, Liao LM, Paula JR, Silva MRR. Phytochemical analysis and antimicrobial activity of Myrcia tomentosa. (Aubl.) DC. leaves. Molecules. 2017;22:1100. doi: 10.3390/molecules22071100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y. Genus Kitasatospora, taxonomic features and diversity of secondary metabolites. J Antibiot. 2017;70:506–513. doi: 10.1038/ja.2017.8. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm S, Carter C, Lynch M, Lowinger T, Dumas J, Smith RA, Schwartz B, Simantov RKS. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat Rev Drug Discov. 2006;5(10):835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- Wu C, van Wezel GP, Hae Choi Y. Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66. J Antibiot (Tokyo) 2015;68:445–452. doi: 10.1038/ja.2015.14. [DOI] [PubMed] [Google Scholar]
- Zitouni A, Mathieu F, Coppel Y, et al. Mutactimycin PR, a new anthracycline antibiotic from Saccharothrix sp. SA 103. II. Physico-chemical properties and structure elucidation. J Antibiot (Tokyo) 2004;57:373–378. doi: 10.7164/antibiotics.57.373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.