The symbiotic N2-fixing cyanobacterium UCYN-A, which is closely related to Braarudosphaera bigelowii, and its eukaryotic algal host have been shown to be globally distributed and important in open-ocean N2 fixation. These unique cyanobacteria have reduced metabolic capabilities, even lacking genes for oxygenic photosynthesis and carbon fixation. Cyanobacteria generally use energy from photosynthesis for nitrogen fixation but require mechanisms for avoiding inactivation of the oxygen-sensitive nitrogenase enzyme by ambient oxygen (O2) or the O2 evolved through photosynthesis. This study showed that symbiosis between the N2-fixing cyanobacterium UCYN-A and its eukaryotic algal host has led to adaptation of its daily gene expression pattern in order to enable daytime aerobic N2 fixation, which is likely more energetically efficient than fixing N2 at night, as found in other unicellular marine cyanobacteria.
KEYWORDS: cyanobacteria, diel cycle, marine microbiology, nitrogen fixation, symbiosis, whole-genome expression
ABSTRACT
Symbiosis between a marine alga and a N2-fixing cyanobacterium (Cyanobacterium UCYN-A) is geographically widespread in the oceans and is important in the marine N cycle. UCYN-A is uncultivated and is an unusual unicellular cyanobacterium because it lacks many metabolic functions, including oxygenic photosynthesis and carbon fixation, which are typical in cyanobacteria. It is now presumed to be an obligate symbiont of haptophytes closely related to Braarudosphaera bigelowii. N2-fixing cyanobacteria use different strategies to avoid inhibition of N2 fixation by the oxygen evolved in photosynthesis. Most unicellular cyanobacteria temporally separate the two incompatible activities by fixing N2 only at night, but, surprisingly, UCYN-A appears to fix N2 during the day. The goal of this study was to determine how the unicellular UCYN-A strain coordinates N2 fixation and general metabolism compared to other marine cyanobacteria. We found that UCYN-A has distinct daily cycles of many genes despite the fact that it lacks two of the three circadian clock genes found in most cyanobacteria. We also found that the transcription patterns in UCYN-A are more similar to those in marine cyanobacteria that are capable of aerobic N2 fixation in the light, such as Trichodesmium and heterocyst-forming cyanobacteria, than to those in Crocosphaera or Cyanothece species, which are more closely related to unicellular marine cyanobacteria evolutionarily. Our findings suggest that the symbiotic interaction has resulted in a shift of transcriptional regulation to coordinate UCYN-A metabolism with that of the phototrophic eukaryotic host, thus allowing efficient coupling of N2 fixation (by the cyanobacterium) to the energy obtained from photosynthesis (by the eukaryotic unicellular alga) in the light.
INTRODUCTION
Nitrogen (N2)-fixing microorganisms (diazotrophs), which reduce atmospheric N2 to biologically available ammonium, are critical components of aquatic and terrestrial ecosystems because they supply fixed inorganic N (1). Cyanobacteria are particularly important in N2 fixation because they can fuel the energy-intensive N2 reduction reaction using energy supplied by oxygenic photosynthesis. In the oceans, the filamentous, non-heterocyst-forming cyanobacterium Trichodesmium and the heterocyst-forming symbiont of diatoms (Richelia and related cyanobacteria) were believed to be the major N2-fixing microorganisms until the discovery of the unicellular cyanobacteria Crocosphaera, Cyanothece, and “Candidatus Atelocyanobacterium thalassa” (UCYN-A) in the open ocean. Crocosphaera and Cyanothece are free-living marine cyanobacteria, but UCYN-A is unusual in that it lacks oxygenic photosynthesis and is a symbiont of a haptophyte alga (related to Braarudosphaera bigelowii). UCYN-A symbiosis is geographically widespread and is important in oceanic N2 fixation (2 – 5). The UCYN-A genome has been greatly reduced, with massive metabolic streamlining, including the loss of oxygen-evolving photosystem II (PSII), the carbon-fixing enzyme RuBisCO, and the entire tricarboxylic acid (TCA) cycle (6). UCYN-A has been shown to supply fixed N to the haptophyte in exchange for fixed carbon (4, 7), but it is not known how these two single-celled organisms coordinate metabolism and cell growth over the daily division cycle.
N2 fixation requires energy and reductant, but the nitrogenase enzyme is inactivated by oxygen (O2). Cyanobacteria generally have access to sufficient energy from photosynthesis but require mechanisms for avoiding inactivation of nitrogenase and N2 fixation by ambient oxygen (O2) or by the O2 evolved through photosynthesis. Trichodesmium and heterocyst-forming cyanobacteria such as Richelia and Nostoc fix N2 during the day, whereas the free-living unicellular genera Crocosphaera and Cyanothece fix N2 at night. Interestingly, the symbiotic UCYN-A strain appears to fix N2 during the day (8 – 10), in contrast to most other unicellular marine N2-fixing cyanobacteria, such as Crocosphaera and Cyanothece.
The processes of N2 fixation and photosynthesis in cyanobacteria are regulated daily to increase cellular fitness and ecological competitiveness (11 – 13). Most cyanobacteria have circadian rhythms (11, 14, 15) that are involved in controlling daily cycles of gene transcription and protein synthesis by signal transduction pathways involving the circadian clock kai genes. UCYN-A lacks two (kaiA and kaiB) of the three kai genes known in most other cyanobacteria, whereas the non-N2-fixing cyanobacterium Prochlorococcus lacks only kaiA. Thus, the daily whole-genome expression pattern in UCYN-A is of interest to determine if there are daily patterns similar to those in all other cyanobacteria compared to evolutionarily related unicellular cyanobacteria.
We used a whole-genome transcription array that targets two genetically distinct uncultivated sublineages of UCYN-A (UCYN-A1 and UCYN-A2), which have similar but genetically distinct hosts. We compared the UCYN-A whole-genome diel transcription patterns to those of Cyanothece sp. ATCC 51142 (16) and Crocosphaera watsonii WH 8501 (17) (both unicellular nighttime N2 fixers) and of Trichodesmium erythraeum IMS101 (a filamentous non-heterocyst-forming daytime N2 fixer). We also compared their expression levels to whole-genome expression of Prochlorococcus sp. MED4 (18) (a marine non-N2 fixer) in order to determine how UCYN-A gene expression levels compare to the general cyanobacterial gene expression levels in a sympatric open-ocean species. We found that many genes in UCYN-A have distinct diel expression patterns and that UCYN-A has unusual gene expression patterns in comparison to unicellular N2-fixing cyanobacteria that fix N2 in the dark; however, it shares some general patterns with daytime N2-fixing cyanobacteria, with heterocysts of heterocyst-forming cyanobacteria, and with unicellular non-N2-fixing cyanobacteria. Results suggest that optimal metabolism for open-ocean cyanobacteria is aligned to the light period and that symbiosis has enabled the unicellular UCYN-A to shift N2 fixation to the daylight period.
RESULTS AND DISCUSSION
UCYN-A has a daily rhythm of gene transcription.
UCYN-A has clear diel patterns of gene transcription, with a large fraction of genes that had periodicity of transcript levels over the dark and light periods (27%).
About a third (31%) of the genes in the UCYN-A genome targeted by the array were transcribed at detectable levels (365 of 1,194 total genes in UCYN-A1 and 394 of 1,244 total genes in UCYN-A2) (see Table S1 in the supplemental material). Approximately 85% of these genes had differences in transcript levels between the dark and light periods, accounting for 27% of the total genes in each strain (Tables S1 and S2). C. watsonii, Cyanothece sp., and Trichodesmium cultures also had a large fraction of genes with changes in transcript levels between the dark and light periods (39% in C. watsonii, 20% in Cyanothece sp., and 34% in Trichodesmium) (Tables S1 and S2).
Genes targeted and detected in the transcriptomic analysis for each organism. The first line shows the total number of genes targeted for the microarray analysis. The second line shows the number of the genes transcribed at detectable levels and the third line the number of diel genes. Percentages of genes determined by comparing the total genes targeted in the microarray in each organism are shown at the bottom. Download Table S1, DOCX file, 0.1 MB (106.4KB, docx) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of genes with periodic transcriptional patterns for all studied cyanobacteria based on Fourier scores. Genes with a FDR of <0.25 were selected and are represented in this table for further comparison. Download Table S2, XLS file, 1.3 MB (1.3MB, xls) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The UCYN-A transcription values (log2 transformed) ranged from 2 to 13.5, with a median of 6.0. In both sublineages, the genes coding for nitrogenase (nif), F0F1-ATP synthase (atpA and atpB) and the cytochrome b6f complex (petB, petC, petF, and petL) and the photosynthetic gene psaC were the most highly transcribed among all of the detected genes (Table S5). The transcript levels of the same genes were also high for both sublineages in metatranscriptomes collected during the TARA expedition in the South Atlantic Ocean (19).
Detailed lists of normalized transcription values and description for all genes in all studied cyanobacteria. There are two spreadsheets for each organism: “Data” and “Description.” The “Data” spreadsheet shows the normalized transcription values for each time point and the mean expression. Eight points were selected for UCYN-A (L6, L9, D3, D6, 2D12, 2L3, 2L9, and 2L12), 9 points for T. erythraeum (D12, L3, L6, L9, L12, D3, D6, D9, and 2D12), 6 points for Cyanothece sp. ATCC 51142 (L2, L6, L10, D2, D6, and D10), 8 points for C. watsonii WH 8501 (D11, L1, L6, L11, D1, D6, 2D11, and 2L1) and 19 points for Prochlorococcus sp. MED4 (D12 to 2L12 [representing time points occurring every 2 h]). “L” and “D” stand for the light period and the dark period, respectively, “2L” and “2D” for the second light-dark cycle, and the number that follows for the hour corresponding to the time of entry into the light or dark period. The “Description” spreadsheet shows the annotation and pathways of each gene for each organism. Download Table S5, XLSX file, 2.6 MB (2.6MB, xlsx) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The two UCYN-A sublineages were similar with respect to the periodicity of their transcript levels, despite divergence in gene sequences at the amino acid level (genome-wide average of 14%), in cell morphology (19), and in genome size (Fig. 1). There were four gene clusters as determined based on the time of day exhibiting the highest relative transcript level (Fig. 1). Cluster I had the highest relative transcript level during the day (with the maximum seen 10 h into the light period) and included genes involved in cell division (e.g., ftsZ, murG, minE, and murB), DNA replication (e.g., topA, rpoE, and DPO3B), ABC transportation (e.g., nikA, nikB, pstC, and cbiO), and carbohydrate and lipid metabolism (e.g., pdhA, pgi, fabG, and fabH) and a few photosynthesis genes (petL, psaD, and ccsB). The transcription levels for the petL gene, encoding subunit 6 of the cytochrome b6f complex, and for the only nitrogen fixation-related gene in this cluster (nifK) showed a substantial (more than 3-fold) change at that time.
FIG 1.
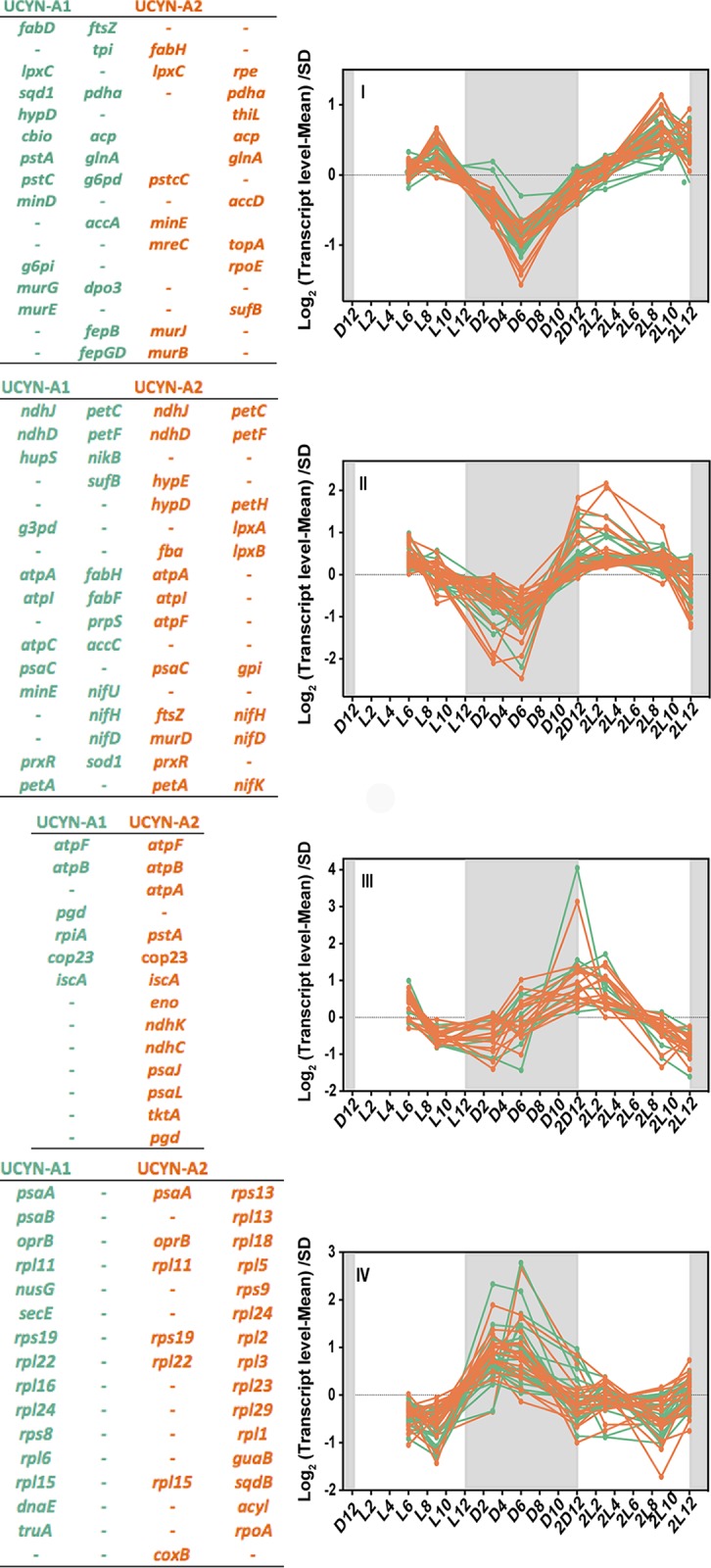
Four different clades (indicated by I-IV) based on Pearson correlation of the transcription profiles of UCYN-A1 and UCYN-A2 genes over light-dark cycles. The transcription value of each gene at each time point was normalized to the mean at all time points and divided by the standard deviation (SD) (y axis, log 2 scale). The x axis represents time points where “D” and “L” stand for dark and light, respectively, followed by a number corresponding to the hour of entry into the light or dark period. The second light-dark cycle is indicated as “2D” followed by a number corresponding to the hour of entry into the light or dark period. The shaded area represents the dark period. In each cluster, the most representative genes are listed in the table attached to the plot. UCYN-A1 genes are coded in green and UCYN-A2 genes are coded in orange.
The transcript abundances of genes from clusters II and III had similar patterns, with an increase before sunrise and a decrease during the dark period. The highest relative transcript levels for the genes in clusters II and III were seen at 4 h and 1 h after sunrise, respectively, and included genes involved in nitrogen fixation (nifHDK operon) that increased 4-fold in transcription during the light period. However, these clusters also included genes involved in oxidative phosphorylation (e.g., NADH dehydrogenase subunit-related genes and ATP synthase-related genes) and in carbohydrate catabolism such as those involved in glycolysis (e.g., gap1, fbaA, pgi, and eno), in the pentose phosphate pathway (opcA and zwf), and in photosynthesis (e.g., cytochrome b6f complex subunit genes). In most cyanobacteria, genes encoding proteins involved in carbohydrate catabolism are highly transcribed during the night and are essential for survival under dark conditions.
The gene with the most dramatic difference in transcript levels between the light and dark periods encoded membrane protein COP23 (23-kDa circadian oscillating protein), which had a change in transcript abundance of more than 5-fold in both UCYN-A strains (Fig. 1). COP23, a protein which may have a critical role in membrane function, has been detected only in nitrogen-fixing cyanobacteria (20).
Cluster IV had genes with the highest transcript level during the night and the lowest during the day and included genes encoding photosystem I (PSI) subunits and a carbohydrate porin (oprB) and also genes encoding ribosomal proteins with 2- and 4-fold changes during the night period. Cluster IV had the lowest number of genes compared with the other clusters. Surprisingly, the PSI genes (psaA and psaB) were expressed during the night, as seen in many anoxygenic phototrophic bacteria (21), whereas these genes are expressed during the day in most oxygenic cyanobacteria (including mats) (22).
The results show that UCYN-A has a daily rhythm of gene expression with strong periodicities of transcript levels over the diel cycle. Daily patterns of gene transcription in cyanobacteria are typically regulated by a circadian rhythm mediated by kai gene products (11). Rhythmic daily transcription patterns are still possible without the full suite of kai genes; for example, the marine cyanobacterium Prochlorococcus sp. MED4 lacks one of the circadian genes, kaiA, and yet it maintains strong diel gene transcription patterns (18). However, Prochlorococcus sp. PCC 9511 loses the typical periodicities of the circadian clock under conditions of continuous light (23). In the case of UCYN-A, it lacks two of the three kai genes (24), which is unique among cyanobacteria; furthermore, the kaiC gene was not transcribed at detectable levels. It is unclear what controls the UCYN-A diel gene expression pattern, but it could be that (i) there are unidentified components of a clock and signal transduction pathway or (ii) the pattern could be driven by the physiological differences between light and dark conditions and might primarily be driven by energy supplied by the eukaryotic partner. It is possible that the diel transcription patterns in UCYN-A are primarily regulated by the daily host metabolism, which itself is likely to be circadian. However, it is not yet known whether the UCYN-A diel cycle is maintained under constant conditions in UCYN-A or whether the diel pattern is maintained in the absence of the partner alga.
UCYN-A transcription patterns are similar to those of aerobic marine daytime N2-fixers and non-N2-fixers.
UCYN-A had diel whole-genome expression patterns that were different from those of phylogenetically closely related unicellular cyanobacteria (17). Only a few genes (such as those encoding ATP synthase) had the same daily pattern among all cyanobacteria, presumably differing because of physiology (e.g., N2-fixing or not). The unicellular cyanobacteria C. watsonii WH 8501 and Cyanothece sp. ATCC 51142, which fix N2 during the night, expressed many genes in a pattern opposite that seen with daytime N2-fixing T. erythraeum and UCYN-A (Fig. 2; see also Tables S3, S4, and S5). Interestingly, the diel transcription patterns of N2 fixation and PSI genes in UCYN-A were opposite those in Cyanothece sp. ATCC 51142 and C. watsonii WH 8501 and similar to those in T. erythraeum (Fig. 2; see also Tables S3, S4, and S5).
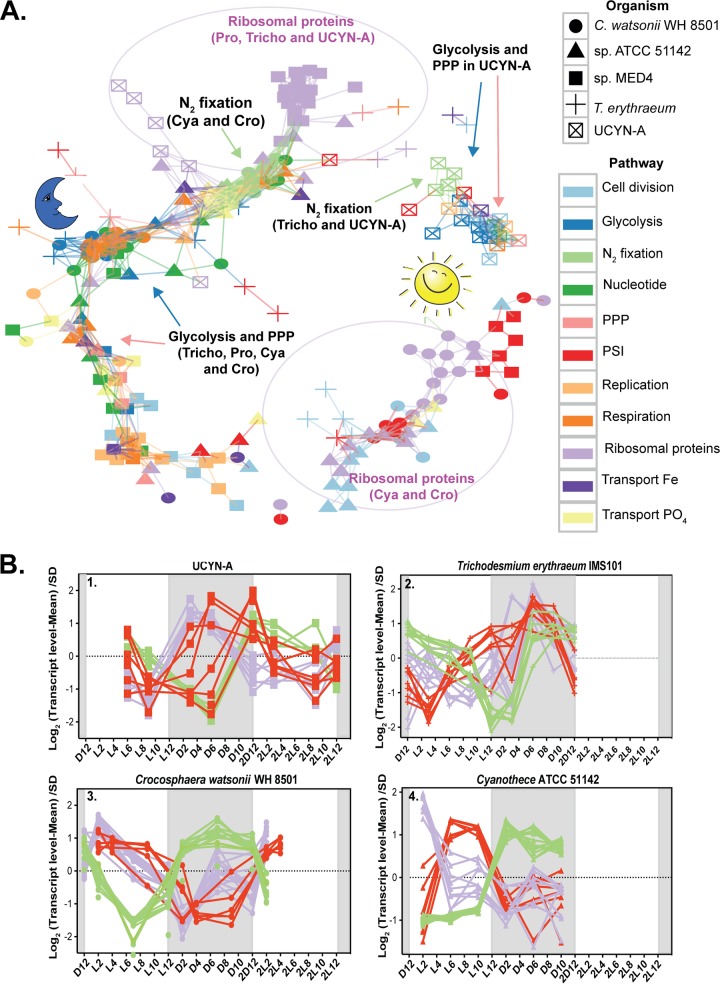
FIG 2.
(A) Transcriptional network based on Pearson correlation of gene transcription over the diel cycle in all studied cyanobacteria. The genes are connected if correlation coefficient for their transcription patterns is higher than 0.5. The genes shown are diel genes with variable transcription patterns among the studied cyanobacteria. The arrows point to genes for glycolysis, the pentose phosphate pathway, and N2 fixation in the studied diazotrophs. The purple circles demarcate genes for ribosomal proteins included in the analysis. Abbreviations: Prochlorococcus sp. MED4 (Pro), Cyanothece sp. ATCC 51142 (Cya), C. watsonii WH 8501 (Cro), T. erythraeum (Tricho), pentose phosphate pathway (PPP), photosystem I (PSI). (B) Four time course plots for the N2-fixing cyanobacteria, showing the diel transcription patterns of photosystem I genes, N2 fixation genes, and genes for ribosomal proteins.
Gene orthologs among all studied cyanobacteria. The orthologs were identified as the best hits in reciprocal blastn searches for each pair of genomes. Orthologs of UCYN-A diel genes detected in the other cyanobacteria are listed in sheets 2 to 5. The percentages of diel genes shared between UCYN-A and the respective cyanobacterial species were calculated relative to the total gene orthologs identified by blastn for the two cyanobacterial species. Download Table S3, XLSX file, 0.4 MB (407.6KB, xlsx) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genes selected for network analysis. A minimum of 50% of genes of UCYN-A with diel transcription patterns is represented in the figure. The same genes or genes in the same pathway were represented for the rest of cyanobacteria. The last column (“Genes selected for network”) indicates the genes that were used to build the network analysis whose results are presented in Fig. 2 and 6. Download Table S4, XLS file, 1.2 MB (1.2MB, xls) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
As observed for the activity of nitrogenase, it has been demonstrated that levels of nif transcripts and the biosynthesis of different components of the nitrogenase complex are very sensitive to O2 (22, 25 – 27), most likely as a consequence of the need to avoid energy losses associated with the degradation of this enzyme by O2. Thus, the different patterns observed in the genes involved in N2 fixation in the cyanobacteria studied here presumably are due to the different mechanisms used to protect the nitrogenase complex from the O2 produced by photosynthesis. T. erythraeum and UCYN-A showed maximum transcript levels of the nitrogenase and PSI genes just prior to dawn but maintained high levels of transcripts for both sets of genes during the day. The peak of transcript levels just before dawn was likely due to the advantage of synthesizing nitrogenase in preparation for N2 fixation in the early hours of the day (28).
The diel expression patterns of genes that are unrelated to N2 fixation in the aerobic daytime N2 fixers (T. erythraeum and UCYN-A) were also more similar to those of non-N2-fixing sympatric cyanobacteria of the genus Prochlorococcus and of heterocysts of heterocyst-forming cyanobacteria than to those of the nighttime N2-fixing cyanobacteria (C. watsonii and Cyanothece sp.). The transcript levels of genes encoding ribosomal proteins in both UCYN-A and T. erythraeum were higher during the night, probably because the reduced nitrogen required for the synthesis of new proteins was obtained during the day (Fig. 2; see also Tables S4 and S5). Similar patterns were observed in Prochlorococcus, with higher transcript levels seen during the night (Fig. 2; see also Tables S4 and S5), while genes encoding ribosomal proteins in C. watsonii WH 8501 and Cyanothece sp. ATCC 41142 had maximum transcript levels during the day (Fig. 2; see also Tables S4 and S5). Intriguingly, these results imply that both UCYN-A and T. erythraeum have adopted daytime gene transcription patterns for the main metabolic pathways, minimizing cellular processes in the dark. The nighttime patterns of the transcript levels of the ribosomal proteins (genes) would make it possible to have proteins synthesized in order to make the most efficient use of the light period, as seen in Prochlorococcus. Because UCYN-A and Trichodesmium are likely the two most abundant N2-fixing cyanobacteria in the open ocean, it appears that direct coupling of N2 fixation to photosynthesis is important in the oligotrophic environment (as long as low oxygen concentrations are maintained in the cell).
Phosphorus is a vital element for cellular energetics and growth and is acquired by oceanic bacterioplankton primarily as phosphate (29 – 31). The UCYN-A phosphate ABC transporter had the same diel pattern as that in Trichodesmium for genes involved in DNA replication, with higher transcript levels during the day (Table S5) but with maximum transcript abundances during the late afternoon in Crocosphaera and Cyanothece (17, 32). The presence of high levels of phosphate transporters during the day could meet the increased demand for inorganic phosphate (33, 34) during DNA replication, which occurs during the day in UCYN-A and Trichodesmium. Similar patterns were observed in the heterocyst-forming Richelia species, with peak expression of P acquisition genes at approximately 15:00 h, suggesting that the apparent rhythmicity of P acquisition could be a common feature of daytime N2 fixers (35).
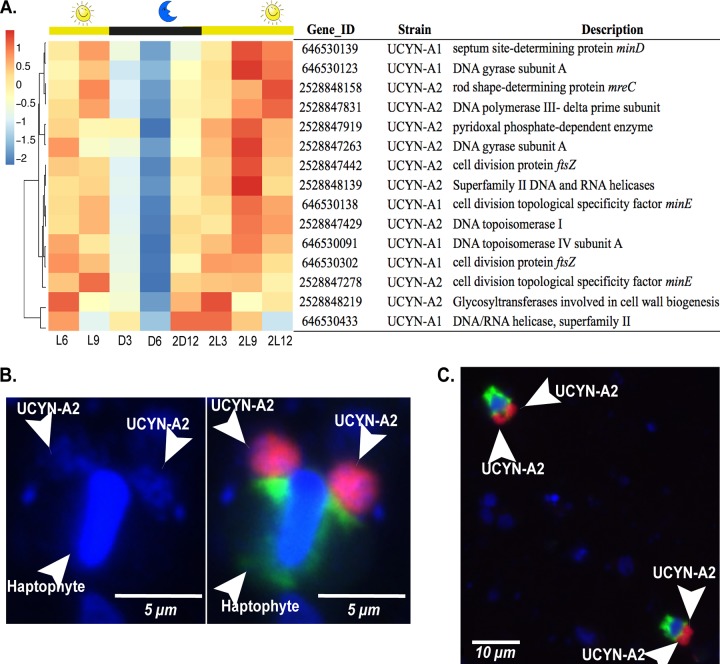
The factor initiating DNA replication, DnaA, is a protein that is highly conserved in prokaryotes although it is absent in red algae, the cyanobacterial symbiont Nostoc azollae (36), and also the spheroid bodies of diatoms (37). The genome of UCYN-A lacks the dnaA gene as well. Recent studies suggested that DnaA is not essential for DNA replication and that the lack of dnaA could suggest a preadaptation of the genome to enable the symbiosis (38). In UCYN-A and T. erythraeum, genes for DNA replication (dnaE and the RNase H1 gene), DNA topoisomerases, DNA gyrases, and cell division (ftsZ, mre, and min) showed maximum transcript levels during the day (i.e., after midday) and minimum levels at night (Fig. 3A; see also Fig. S1 in the supplemental material). In contrast, the nighttime N2-fixing Cyanothece sp. ATCC 51142 and C. watsonii WH 8501 confined cell division to the period of transition from dark to light at sunrise. The temporal delay in cell division in Cyanothece and Crocosphaera has been suggested to reflect the need to recover energy reserves with light-derived energy after nighttime metabolic activity (39). The similarity of the pattern in UCYN-A to that in Trichodesmium is consistent with UCYN-A shifting metabolism to the daytime.
FIG 3.
Transcription of genes for replication and cell division in UCYN-A. (A) (Upper panel) Diel transcription patterns for cell division and replication genes in UCYN-A1 and UCYN-A2 over the light-dark cycle. Hierarchical clustering of genes was based on Pearson correlations between their transcription profiles. The transcription values of the genes were standardized at each time point, and the blue-red scale shows by how many standard deviations a transcription value was lower (blue) or higher (red) than the mean transcription values over the diel cycle (Z score). The Gene ID and the gene product corresponding to each gene for UCYN-A1 and UCYN-A2 are shown. Time periods are indicated on the x axis as “L” (light) and “D” (dark) followed by a number corresponding to the hour after the sunrise and sunset periods started. The second light-dark cycle is indicated as “2D” followed by a number corresponding to the hour of entry into the light or dark period. (Lower panel) Epifluorescence micrographs of dividing UCYN-A2 detected with CARD-FISH (19). (B) Two big clusters of UCYN-A2 cells and the attached haptophyte host. (Left panel) The nucleus of the host and the UCYN-A2 cells were visualized with DAPI stain (blue). (Right panel) The UCYN-A2 strain (red) and its haptophyte host (green). (C) Two different associations of UCYN-A2 with its haptophyte dividing in samples from Scripps Pier.
Transcription patterns in T. erythraeum for the cell division and replication genes over the light-dark cycle. Hierarchical clustering of genes was based on Pearson correlation of gene transcription profiles. The transcription values fof each gene at each time point were standardized, and the blue-red scale shows by how many standard deviations a transcription value is lower (blue) or higher (red) than the mean transcription values over the diel cycle (Z score). “L” and “D” stand for light and dark, respectively; those notations are followed by the hour corresponding to the time after the light and dark periods started. The second light-dark cycle is indicated as “2D,” and that notation is followed by the number of the hours corresponding to entry into the light or dark period. Download FIG S1, PDF file, 0.8 MB (809.5KB, pdf) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microscopy counts of B. bigelowii–UCYN-A2 symbiosis were performed eight times during two diel cycles in order to observe the timing of cell division (Fig. 3B and C; see also Table S6). In both diel cycles, single host cells with two associated UCYN-A2 cells (or groups of cells), corresponding to approximately 60% of the total cell counts, were present at night between 21:00 and 03:00 h. The delay observed between the time of the higher transcription levels after midday and the time of actual cell division at h 21:00 may be explained by the need of the cell to coordinate the assembly of the cell division machinery prior to cell division.
Abundance of the haptophyte UCYN-A2 association in CARD-FISH samples during two diel cycles at the Scripps Institution of Oceanography, indicating the number of UCYN-A2 clusters per host cell. Download Table S6, DOCX file, 0.2 MB (190.9KB, docx) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Unique UCYN-A transcription patterns.
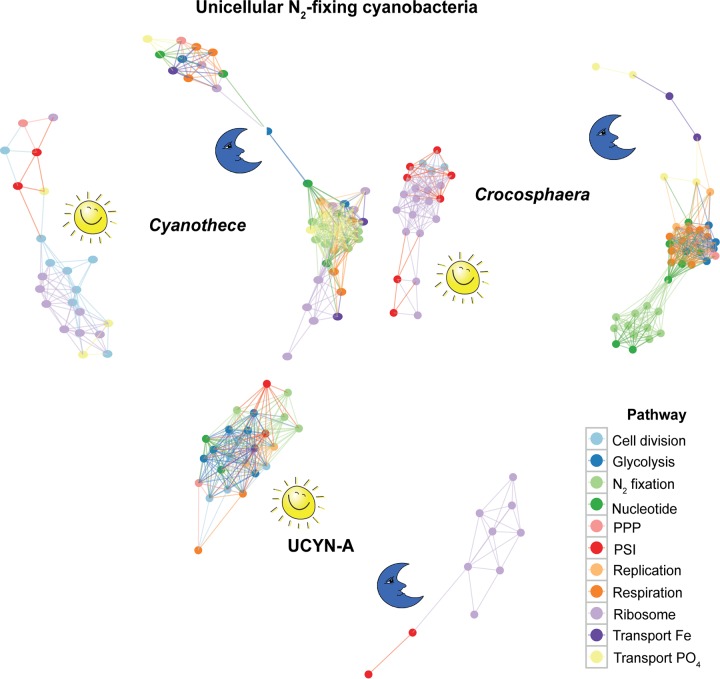
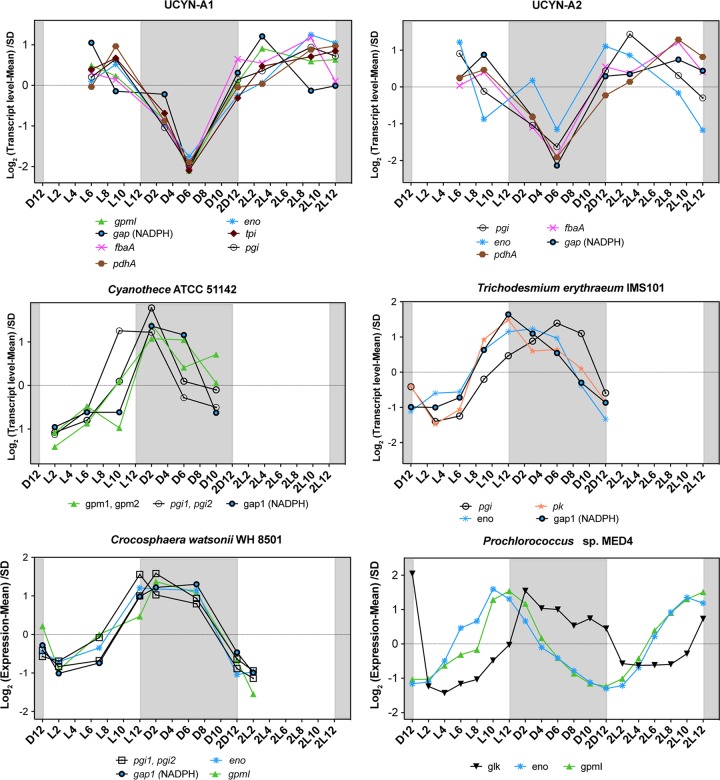
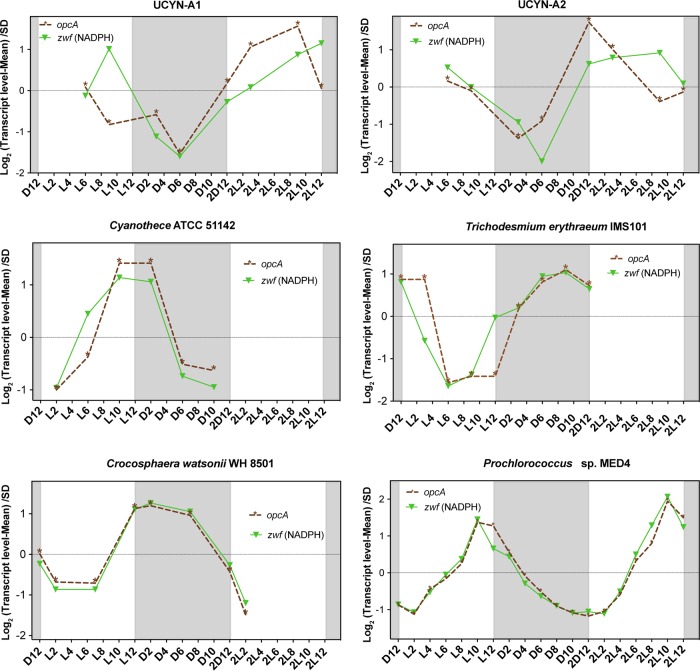
Although many gene transcription patterns in UCYN-A are more similar to those in Trichodesmium than to those in other unicellular N2-fixing cyanobacteria, some of the patterns were unique to UCYN-A. Such unique gene transcription patterns in the UCYN-A symbiosis may provide clues to possible roles of specific genes involved in adaptation to N2-fixing symbiosis, revealing metabolic interdependence between host and symbiont. In order to compare the transcriptomic patterns of these specific genes with those of the rest of the N2-fixers, we performed network analysis of the genes using Pearson correlation. Whereas most of the key genes of the major pathways in UCYN-A had higher transcript levels during the day, those of the other unicellular N2-fixing cyanobacteria had maximum transcript levels at night (Fig. 4). For example, glycolysis genes in UCYN-A had the highest levels of transcripts at sunrise and midday (maximum light conditions) compared to the other cyanobacteria (Fig. 4 and 5). The metabolic pathway that generates reductant for biosynthesis activities (NADPH), i.e., the pentose phosphate pathway (PPP), had similar patterns. The allosteric effector opcA, which redirects carbon flow to the first enzyme of the PPP (glucose-6-P dehydrogenase [zwf]) (18, 40), had a periodic transcript level pattern in UCYN-A (Fig. 4 and 6) that was different from that previously reported in other cyanobacteria (41, 42).
FIG 4.
Network showing the Pearson correlation for gene transcriptions in the unicellular N2-fixing cyanobacteria Cyanothece sp. ATCC 51142 (Cyanothece), C. watsonii WH 8501 (Crocosphaera), and UCYN-A. Shown here are key genes in major metabolic pathways with distinct diel transcription patterns. The genes are shown as connected if their correlation coefficient for transcription patterns is higher than 0.2. PPP, pentose phosphate pathway; PSI, photosystem I.
FIG 5.
Transcriptional profiles of the genes for glycolysis over light-dark cycles in the cyanobacteria studied here. The transcription value of each gene at each time point was normalized to the mean at all time points and divided by the standard deviation (SD) (y axis, log scale). The x axis represents time points where “D” and “L” stand for dark and light, respectively, followed by a number corresponding to the hour of entry into the light or dark period. The second light-dark cycle is indicated as “2D” followed by a number corresponding to the hour of entry into the light or dark period. The shaded area represents the dark period.
FIG 6.
Transcriptional profiles of opcA (allosteric effector) and zwf (glucose-6-P dehydrogenase) over light-dark cycles in the cyanobacteria studied here. The transcription value of each gene at each time point was normalized to the mean at all time points and divided by the standard deviation (SD) (y axis, log scale). The x axis represents time points where “D” and “L” stand for dark and light, respectively, followed by a number corresponding to the hour of entry into the light or dark period. The second light-dark cycle is indicated as “2D” followed by a number corresponding to the hour of entry into the light or dark period. The shaded area represents the dark period.
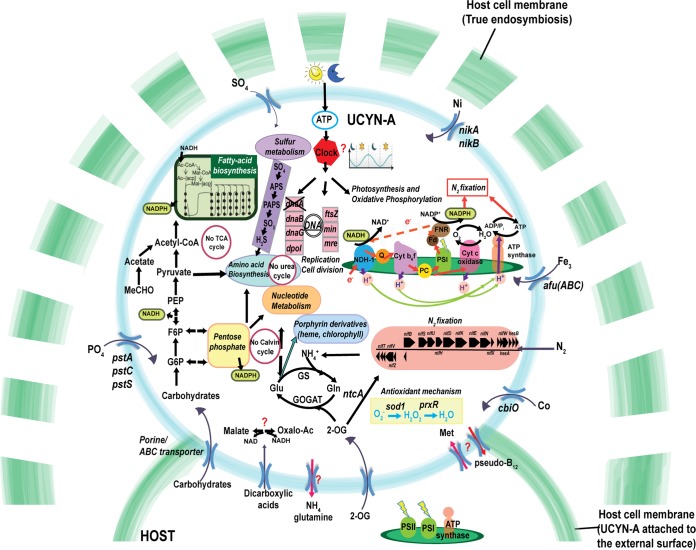
N2 fixation in UCYN-A depends on the light period for the supply of photosynthate from the host during the day, as well as possibly producing ATP by cyclic photophosphorylation with PSI. Because UCYN-A cannot fix carbon dioxide, it has to obtain reduced carbon compounds in the same way. On the basis of the genome and transcriptomic profiles, we propose a pathway of carbon metabolism for the regeneration of reductant and ATP in UCYN-A that is needed for N2 fixation (Fig. 7). Carbohydrate porins or ABC transporters could transport the carbohydrates from the host to the cyanobacteria during the day and the carbon compounds metabolized through the oxidative pentose phosphate (OPP) pathway or glycolysis pathway. Pyruvate is required for generation of reductant for nitrogenase and also to generate acetyl-coenzyme A (acetyl-CoA) for synthesis of fatty acids.
FIG 7.
Schematic model of UCYN-A showing the possible main cellular functions, metabolic pathways, and transporters.
Because UCYN-A lacks photosystem II, which normally supplies electrons to photosystem I by splitting water, UCYN-A needs alternative electron donors if it uses PSI to make the reductant NADPH. The NADH generated by the OPP pathway or by glycolysis could reduce the plastoquinone (PQ) pool via the NDH-1 complex and transfer electrons to ferredoxin though the PQ pool, cytochrome b6f plastocyanin, and the action of PSI. Ferredoxin could deliver electrons to the ferredoxin:NADPH oxidoreductase (FNR), which might supply reductant and ATP directly to the dinitrogenase reductase. To increase the ATP/e− ratio, UCYN-A can redirect electrons from PSI to NDH-1 in cyclic phosphorylation. This mechanism to supply nitrogenase with electrons was proposed years ago for heterocysts (43).
Together, the results are consistent with the assumption that UCYN-A uses host-supplied carbohydrates during the day whereas other unicellular cyanobacteria synthesize their own carbohydrates during the day and use them during the evening or at night. The unique distribution of these metabolic processes suggests that UCYN-A has developed the ability to perform light-driven, daytime N2 fixation under oxic conditions as a result of symbiosis.
Apart from fixed carbon, several other compounds may be made available to UCYN-A, which may be endosymbiotic and relies on the host for all of its essential nutrients. Interestingly, UCYN-A has the whole pathway for the synthesis of the cyanobacterial type of vitamin B12, pseudocobalamin, that can be required for the activity of several vital enzymes in central metabolism (44) (Table S8). Transcription of genes involved in B12 synthesis were detected in all cyanobacteria, and some of them had diel patterns (Table S2 and S8). It is unknown if UCYN-A has enzymes that require pseudocobalamin or whether it can be used by the host. However, in order for the host to use pseudocobalamin, it would have to be remodeled in order for it to be accessible to the haptophyte (45). The role of pseudo-B12 biosynthesis in UCYN-A is unclear, but the fact that UCYN-A retains this entire pathway, in such a reduced genome, indicates that it is likely to have an important role, perhaps in symbiosis.
Detailed list of genes involved in B12 metabolism in all studied cyanobacteria. Download Table S8, XLSX file, 0.04 MB (42.1KB, xlsx) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
It is still unclear how N2 fixation in UCYN-A avoids the oxygen evolved by the photosynthetic host alga. There are only two possible pathways for consuming O2 in UCYN-A, including aerobic (cytochrome-dependent) respiration and the photocatalyzed reduction of O2 to H2O in PSI such as occurs in the heterocysts of cyanobacteria like Nostoc sp. PCC 7120 (46 – 48). The latter, called the Mehler reaction, results in the production of the superoxide radical O2–, which is subsequently reduced to water (46, 49).
In UCYN-A, the cytochrome c oxidase coxA gene was transcribed during the night (cluster IV) but also (rarely) during the day, along with a few N2 fixation genes (cluster I) (Fig. 1). Moreover, we also found higher transcript levels during the day for the antioxidant enzyme superoxide dismutase (sod1) and two peroxiredoxins (prxR), which have the ability to detoxify peroxide (Fig. 1 and 7). Both antioxidants would protect the nitrogenase against the reactive oxygen species produced by UCYN-A or the haptophyte host (Fig. 7).
It is not currently possible to directly determine the oxygen protection mechanisms in this uncultured microorganism because (i) transcription cannot definitively be related to function and (ii) it is not possible to perform physiological experiments with this low-abundance microorganism, which has yet to be obtained in an axenic culture. Consequently, the issue of protection from O2 cannot be directly addressed experimentally, but our results suggest that some of the proteins in UCYN-A could help to protect nitrogenase from the O2 generated by host photosynthesis.
Because UCYN-A shows genome reduction normally associated with endosymbiosis (e.g., in Paulinella chromatophora [50]), the unique gene transcription patterns of UCYN-A may provide insights into the evolution of endosymbiosis and organellar evolution. Future studies are needed to determine if the rhythm of these patterns is maintained under constant conditions as in the case of a circadian rhythm, whether the host has a circadian rhythm, and/or whether the daily cycle in UCYN-A simply responds to metabolite availability from the host. It will also be interesting to determine how PSI is involved in supporting the energy or reductant requirements of N2 fixation. Such experiments will have to await the establishment of a pure culture.
MATERIALS AND METHODS
Diel sampling of UCYN-A.
Surface seawater samples for UCYN-A transcription and catalyzed reported deposition-fluorescence in situ hybridization (CARD-FISH) analyses were collected using a bucket from the end of the Scripps Institution of Oceanography (SIO) Ellen Browning Scripps Memorial Pier in La Jolla, CA, USA. Two replicates were collected from the bucket at each time point within 48 h between 28 July and 1 August 2014 for transcriptomic analysis and between 3 and 8 May 2016 for CARD-FISH analysis. A total of 16 samples were collected every 3 to 6 h (with two replicates taken at each of eight time points) as follows: 12:00-L6, 15:00-L9, 21:00-D3, 00:00-D6, 06:00-2D12, 09:00-2L3, 15:00-2L9, and 18:00-2L12. (“L” and “D” stand for the light period and the dark period, respectively, “2L” and “2D” for the second set of light-dark cycles, and the final number for the hour corresponding to entry into the light or dark period.)
For the CARD-FISH assay, 190 ml of seawater from each seawater replicate was fixed with 10 ml 37% formaldehyde (final concentration, 1.87% [vol/vol]) at 4°C in the dark for 1 h. After fixation, 100 ml was filtered at a maximum vacuum pressure of 100 mm Hg onto a 0.6-μm-pore-size, 25-mm-diameter polycarbonate membrane filter (Millipore Isopore; EMD Millipore, Billerica, MA, USA) with a support filter consisting of a 0.8-μm-pore-size, 25-mm-diameter polycarbonate cellulose acetate membrane (Sterlitech Corporation, Kent, WA, USA). The filters were kept at −80°C until processing.
Samples for RNA extraction were collected by filtering a total of 500 ml from each seawater replicate through 0.22-μm-pore-size, 47-mm-diameter Supor filters (Pall Corporation, Port Washington, NY, USA) using a peristaltic pump. Filters were placed in sterile 2-ml bead-beating tubes with sterile glass beads, flash-frozen in liquid nitrogen, and stored at −80°C until extraction.
Double CARD-FISH assay.
The double CARD-FISH assay was carried out following the protocol designed by Cabello et al. (3) and Cornejo-Castillo et al. (19). Details for all of the probes, competitors, and helpers used in this work are compiled in Table S7 in the supplemental material. More details are provided in Text S1 in the supplemental material. Microscopic evaluation and counting was performed with a Carl Zeiss Axioplan-2 imaging fluorescent microscope (Zeiss, Berlin, Germany) in 3 transects (8.0 by 0.1 mm2 each) across the filter piece. Cell dimensions were estimated using AxioVision 4.8 and Image J software (51).
Extended description of materials and methods described in the main text. Download Text S1, DOCX file, 0.06 MB (57.6KB, docx) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of probes utilized in the visualization of UCYN-A associations by double CARD-FISH. Bold letters represent the mismatches between the sequences designed to distinguish UCYN-A1 and UCYN-A2 and their hosts with high specificity. Download Table S7, DOCX file, 0.07 MB (67.1KB, docx) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Diel sampling of Trichodesmium erythraeum IMS101 cultures.
Biological triplicate cultures of T. erythraeum were grown in rectangular canted neck polycarbonate cell culture flasks with a 0.2-μm-pore-size vent cap and 225-cm2 surface area (Corning Inc., Corning, NY, USA). The cultures were maintained at 26°C on a 12h/12h light/dark cycle at 50 μmol quanta m−2 s−1 in YBCII media (52) supplemented with 2.8 μmol liter−1 ferric ammonium citrate. The light was turned on at h 7:00 and off at h 19:00. The cultures were diluted 10-fold from the inoculum and were verified to be axenic by staining with DAPI (4′,6-diamidino-2-phenylindole) and visualizing cells under an epifluorescence microscope (Carl Zeiss, Thornwood, NY, USA). Growth and cell density were monitored until the cultures reached the exponential phase (∼10 to 14 days after inoculation), during which the cells were harvested for the diel transcription assay. Samples were taken at 3-h intervals starting at the onset of the light period and continuing until the end of the dark period for a total of 24 h. A total of 27 samples were collected from the following nine time points: 7:00-D12, 10:00-L3, 13:00-L6, 16:00-L9, 19:00-L12, 22:00-D3, 1:00-D6, 4:00-D9, and 7:00-2D12 (where “L” and “D” stand for the light period and the dark period, respectively, “2D” for the second light-dark cycle, and the final number for the hour corresponding to entry into the light or dark period). At each time point, 200 ml of each of triplicate cultures (replicates from different flasks) was filtered using a 5-μm-pore-size, 47-mm-diameter polycarbonate membrane filter (Osmonics, Minnetonka, MN, USA). The filters were immediately frozen in liquid nitrogen and stored at −80°C until processing.
RNA extraction and processing for hybridization to the microarray.
Environmental RNA containing transcripts from UCYN-A cells was extracted using an Ambion RiboPure Bacteria kit (Ambion, Thermo Fisher), with modifications that included mechanical lysis using glass beads (Biospec, Bartlesville, OK). The extracted RNA was treated with a Turbo-DNA-free DNase kit (Ambion, Thermo Fisher) to remove genomic DNA. Sufficient environmental RNA was obtained for two replicates at 4 sampling times (L6, L9, D3, and 2L12) as follows: L6-1, L6-2, L9-1, L9-2, D3-1, D3-2, 2L12-1, and 2L12-2.
Total RNA for T. erythraeum was extracted using an Ambion RiboPure Bacteria kit (Ambion, Thermo Fisher), followed by in-solution DNase digestion with an RNase-free DNase kit and on-column cleanup with an RNeasy MiniElute kit (Qiagen, Valencia, CA, USA).
RNA purity, concentration, and quality were determined using a NanoDrop 1000 instrument (Thermo Scientific, Waltham, MA, USA), a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), and an RNA 6000 Nano kit (Agilent Technologies). Only samples with RNA integrity values of >7.0 and ratios of A260/A230 and A260/A280 of ≥1.8 were processed further.
Double-stranded cDNA (ds-cDNA) was synthesized from environmental RNA samples that contained UCYN-A and amplified following the procedure previously described by Shilova et al. (53). Briefly, 400 ng RNA from each sample was used, and 1 μl of a 1:100 dilution (corresponding to 4.7 aM of ERCC-0016) of RNA spike-in mix 1 (External RNA Control Consortium [54]) (Ambion) was added before amplification was performed to monitor the technical performance of the assay (54).
Double-stranded cDNA was synthesized and amplified using a TransPlex whole-transcriptome amplification kit (WTA-2; Sigma-Aldrich, St. Louis, MO, USA) and antibody-inactivated hot-start Taq DNA polymerase (Sigma-Aldrich). The amplified cDNA was purified with a GenElute PCR cleanup kit (Sigma-Aldrich), and the quality and quantity of ds-cDNA were determined using a NanoDrop 1000 instrument, a 2100 Bioanalyzer, and an Agilent DNA 7500 kit (Agilent Technologies). Total RNA volumes of 400 ng yielded on average 12 μg of ds-cDNA. The labeling and hybridization of cDNA samples (1.0 μg of ds-cDNA) to the microarray were done at the Roy J. Carver Center for Genomics (CCG) Facility (University of Iowa, Iowa City, IA, USA) according to the Agilent Technology protocol for arrays.
For T. erythraeum, at least 30 μg of unamplified total RNA with a concentration of 1.0 μg μl−1 per sample was provided for 27 samples. A control sample was generated by mixing equal amounts of total RNA, based on NanoDrop-measured concentrations, from each of the 27 samples, resulting in 28 samples in total. Reverse transcription of the total RNA, labeling of cDNA, and hybridization to the array were performed at the Roche NimbleGen facility according to the protocol of the manufacturer (Roche NimbleGen, Inc., Madison, WI, USA).
Design of the UCYN-A array.
The UCYN-A oligonucleotide expression array was designed using UCYN-A1 and UCYN-A2 genes and the eArray Web-based tool (Agilent Technology Inc.; https://earray.chem.agilent.com/earray/) similarly to the array design previously described by Shilova et al. (53). The gene sequences were obtained from the National Center of Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov). Briefly, six probes of 60 nucleotides (nt) in length were designed for each gene, and a total of 6,618 probes (1,199 genes) and 6,862 probes (1,246 genes) were designed for UCYN-A1 and UCYN-A2, respectively. These probes were replicated 4 times in the 8 × 60K array slides and 13 times in the 4 × 180K array slide, which allowed internal evaluation of signals. The sequences of all oligonucleotide probes were tested in silico for possible cross-hybridization as described below. The probe sequences were used as queries in the BLASTN against the following available nt databases in June 2014: Marine microbes, Microbial Eukaryote Transcription, and Non-redundant Nucleotides in the Community Cyberinfrastructure for Advanced Microbial Ecology Research and Analysis (CAMERA; https://www.imicrobe.us [55]). Agilent technology allows 5% nt mismatch in the whole probe region; thus, sequences with a range of 95% to 100% nt identity to the target probe are detected. Therefore, all probes with BLASTN hits with ≥95% over 100% of the nt length were deleted. Next, the probe sequences that passed the cross-hybridization filter were clustered using CD-HIT-EST (56, 57) at 95% nt similarity to select unique probes for UCYN-A1 and unique probes for UCYN-A2. Finally, to select probes specific for each strain, the probes with ≥95% nt identity to the genes in the other strain were deleted. However, a few probes that showed cross-hybridization between the two strains for highly conserved genes (such as the nifH nitrogenase gene) were retained. In summary, 6,120 probes for 1,194 genes of UCYN-A1 and 6,324 probes for 1,244 genes of UCYN-A2 were chosen.
In addition, standard control probes (IS-62976-8-V2_60Kby8_GX_EQC_201000210 with ERCC control probes added) were included randomly as part of the Agilent Technology array to feature locations on the microarray slide. The final design of the microarray was synthesized on two platforms: ca. 62,976 experimental probes and 1,319 control probes on the 8 × 60K array slide and ca. 180,880 experimental probes and 4,854 control probes on the 4 × 180K array slide. The probe sequences are available at NCBI Gene Expression Omnibus (GEO) under accession number GSE100124.
Design of the T. erythraeum IMS101 array.
A custom oligonucleotide array for T. erythraeum was designed using a Roche NimbleGen platform (NimbleGen design identifier [ID]: 080610_Trich_erth_UCSC_TS_expr) according to the complete genome assembly of T. erythraeum IMS101 (NC_008312). The genome sequence is publicly available via on-line gateways, including GenBank (https://www.ncbi.nlm.nih.gov/nuccore/NC_008312), IMG (https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=637000329), and the genome browser at the University of California, Santa Cruz (UCSC) (http://microbes.ucsc.edu/cgi-bin/hgGateway?db=tricEryt_IMS101). Up to six 60-nt-long tiling probes were designed to target each of the 4,788 genes in the genome, resulting in a total of 28,235 probes. The probes were duplicated on the array to allow internal evaluation of hybridization signals. Moreover, 60-nt oligonucleotide tiling probes were also designed to target the intergenic regions that were >60 bp in length at 150-bp intervals, leading to a total of 11,175 probes targeting 3,877 intergenic regions (average, 2.9 probes per intergenic region); however, hybridization data for intergenic probes are not presented here. All the probes were rank ordered and selected based on the following criteria: (i) a minimum annealing temperature of 68°C; (ii) no cross-contamination among the probes for different genes and for different intergenic regions. In addition to the experimental probes, standard control probes were also included on the microarray for quality assessment of the sample preparation, the hybridization process, and the intensity measurements (see Fig. S3 in the supplemental material). The final microarray slides were printed in 4-plex (4 × 72K), format with 67,645 experimental probe features and 7,454 control probe features on one array. The full microarray platform descriptions and data for T. erythraeum are available at NCBI GEO under accession number GSE99896. Microarray hybridization signals were quantified using a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA, USA) at the Roche NimbleGen facility.
Box plot showing that quantile normalization of gene hybridization signals for the T. erythraeum IMS101 arrays resulted in similar signal distribution profiles. Raw hybridization data (A) and normalized data (B) are shown for a total of 28 samples. The different colors correspond to different samples. All samples for T. erythraeum IMS101 hybridization passed the quality control testing. Download FIG S3, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microarray data analysis.
All data analyses were performed with R (www.R-project.org) and the Bioconductor Project (58), specifically, using the Biobase (59), Linear Models for Microarray LIMMA (60), arrayQualityMetrics (61), affyPLM (62, 63), and genefilter packages.
(i) UCYN-A microarray. Transcription values for each gene were obtained using median polish summarization, and values were normalized using quantile normalization (62, 63) (see Fig. S2). The transcription values for UCYN-A at L6, L9, D3, and 2L12 represent the mean transcription values for the two replicates (L6-1, L6-2, L9-1, L9-2, D3-1, D3-2, 2L12-1, and 2L12-2). Raw and normalized microarray data for UCYN-A were submitted to NCBI GEO under accession number GSE100124. To determine if transcription of a gene was detected, the signal-to-noise ratio (SNR) of each chip was calculated as SNR = (Si – BG)/BG, where Si is the hybridization signal for the gene and BG is the chip background signal determined as the average of the lowest 5% of all signals. Transcription was considered to have been detected if the SNR of a transcript was ≥5 (as described previously by Shilova et al. [53]). Transcription values were centered and scaled across genes and samples, and a distance matrix was calculated using Pearson’s correlation coefficient. The distance matrix was then used in hierarchical clustering by a complete agglomeration method to identify clusters of genes with similar patterns of transcription during the diel transcription.
Box plot showing that normalization of gene hybridization signals for the three UCYN-A arrays resulted in similar signal distribution profiles. All samples used for UCYN-A hybridization passed the quality control testing. Raw hybridization probe data (A, B, and C) and quantile-normalized data (D, E, and F) are shown for a total of 12 samples hybridized to three different slides. The distribution of hybridization signals is shown for 10,000 randomly sampled probes (including experimental and control probes) for raw data and for 14,000 probes (experimental) for normalized data. Y-axis data represent the hybridization signal shown as a log2 exponent. Download FIG S2, PDF file, 0.9 MB (967.1KB, pdf) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(ii) T. erythraeum microarray. The raw microarray data for T. erythraeum were subjected to robust multichip average (RMA) analysis (64) and quantile normalization (62, 63) (see Fig. S3 in the supplemental material). Transcription values for each gene were obtained using median polish summarization (53). The final transcription value for each sample represented the mean of data from up to 12 technical replicates (blocks 1 and 2, with up to six replicate probes in each block in the T. erythraeum microarray design). A gene was selected for further analysis if it had a log2 transcription value above 64 in at least 25% of samples and an interquartile range across all samples on the log2 scale of at least 0.5. This filtering resulted in 4,128 genes, which were used in further analysis.
(iii) Comparison of diel transcription patterns for all cyanobacteria. Transcription data for Prochlorococcus sp. MED4, Cyanothece sp. ATCC 51142, and Crocosphaera watsonii WH 8501 were collected from previously published data (16 – 18). Cyanothece sp. ATCC 51142 and C. watsonii WH 8501 microarray data were downloaded from ArrayExpress (http://www.ebi.ac.uk/aerep/) using accession no. E-TABM-386 and E-TABM-737, respectively. The genes with periodic transcriptional patterns for all cyanobacteria studied here (Prochlorococcus sp. MED4, Cyanothece sp. ATCC 51142, C. watsonii WH 8501, T. erythraeum, and UCYN-A) were identified using the R package “cycle” based on Fourier analysis, and the genes with a false-discovery rate (FDR) of <0.25 were selected for further comparison (65) (Table S2). To compare the diel transcription patterns among the cyanobacteria, gene transcription values for each cyanobacterium were selected for over 36 h. Eight points were selected for UCYN-A (L6, L9, D3, D6, 2D12, 2L3, 2L9, and 2L12), 9 points for T. erythraeum (D12, L3, L6, L9, L12, D3, D6, D9, and 2D12), 6 points for Cyanothece sp. ATCC 51142 (L2, L6, L10, D2, D6, and D10), 8 points for C. watsonii WH 8501 (D11, L1, L6, L11, D1, D6, 2D11, and 2L1) and 19 points for Prochlorococcus sp. MED4 (D12 to 2L12 [every 2 h]). “L” and “D” stand for the light period and the dark period, respectively, “2L” and “2D” for the second light-dark cycle, and the final number for the hour corresponding to entry into the light or dark period. Because the studies had a few dissimilar sampling times, the missing values were interpolated using the Stineman algorithm implemented in the imputeTS package (66). A network was constructed based on the Pearson correlation and the ‘make_network’ function in phyloseq (67). The maximum distance between connecting nodes was selected as 0.5 unless otherwise noted in the figure legends.
Accession number(s). Microarray data have been deposited at the NCBI Gene Expression Omnibus (GEO) under accession numbers GSE100124 and GSE99896.
ACKNOWLEDGMENTS
We thank J.C. Meeks (University of California, Davis [UC Davis]) for discussions, F. Azam (Scripps Institution of Oceanography, UC San Diego) for access to Scripps facilities, and K. Turk-Kubo and M. Hogan for laboratory and field support. We thank J. M. García-Fernández and J. Díez-Dapena (University of Córdoba, Córdoba, Spain) and Marine Landa (University of Santa Cruz, Santa Cruz, CA, USA) for helping us to improve the manuscript.
M.C.M.-M. designed the UCYN-A array, designed and performed the research, and analyzed the data. I.N.S. analyzed the T. erythraeum array data and aided with the design of the UCYN-A array and with the comparison of transcription levels among cyanobacteria. T.S. designed the T. erythraeum array and performed the diel sampling of T. erythraeum cultures. H.F. aided the sampling of the diel UCYN-A samples and performed the phylogenetic tree analysis. A.M.C. carried out the CARD-FISH experiments and counted the CARD-FISH diel samples. J.P.Z. conceptualized the study. M.C.M.-M., I.N.S., T.S., H.F., and J.P.Z. drafted and edited the manuscript and figures. All of us read and approved the final manuscript.
We declare that we have no competing financial interest.
Footnotes
Citation Muñoz-Marín MC, Shilova IN, Shi T, Farnelid H, Cabello AM, Zehr JP. 2019. The transcriptional cycle is suited to daytime N2 fixation in the unicellular cyanobacterium “Candidatus Atelocyanobacterium thalassa” (UCYN-A). mBio 10:e02495-18. https://doi.org/10.1128/mBio.02495-18.
Contributor Information
Douglas G. Capone, University of Southern California.
Donald Bryant, Pennsylvania State University.
Dave Scanlan, University of Warwick.
REFERENCES
- 1.Postgate JR. 1982. The fundamentals of nitrogen fixation. Cambridge University Press, London, United Kingdom. [Google Scholar]
- 2.Montoya JP, Holl CM, Zehr JP, Hansen A, Villareal TA, Capone DG. 2004. High rates of N2 fixation by unicellular diazotrophs in the oligotrophic Pacific Ocean. Nature 430:1027–1031. doi: 10.1038/nature02824. [DOI] [PubMed] [Google Scholar]
- 3.Cabello AM, Cornejo-Castillo FM, Raho N, Blasco D, Vidal M, Audic S, de Vargas C, Latasa M, Acinas SG, Massana R. 2016. Global distribution and vertical patterns of a prymnesiophyte-cyanobacteria obligate symbiosis. ISME J 10:693–706. doi: 10.1038/ismej.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martínez-Pérez C, Mohr W, Löscher C, Dekaezemacker J, Littmann S, Yilmaz P, Lehnen N, Fuchs BM, Lavik G, Schmitz R, LaRoche J, Kuypers MM. 2016. The small unicellular diazotrophic symbiont, UCYN-A, is a key player in the marine nitrogen cycle. Nat Microbiol 1:16163. doi: 10.1038/NMICROBIOL.2016.163. [DOI] [PubMed] [Google Scholar]
- 5.Farnelid H, Turk-Kubo K, Muñoz-Marín MC, Zehr JP. 2016. New insights into the ecology of the globally significant uncultured nitrogen-fixing symbiont UCYN-A. Aquat Microb Ecol 77:125–138. doi: 10.3354/ame01794. [DOI] [Google Scholar]
- 6.Tripp HJ, Bench SR, Turk KA, Foster R, Desany BA, Niazi F, Affourtit JP, Zehr JP. 2010. Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature 464:90–94. doi: 10.1038/nature08786. [DOI] [PubMed] [Google Scholar]
- 7.Thompson AW, Foster RA, Krupke A, Carter BJ, Musat N, Vaulot D, Kuypers MMM, Zehr JP. 2012. Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 337:1546–1550. doi: 10.1126/science.1222700. [DOI] [PubMed] [Google Scholar]
- 8.Church M, Short C, Jenkins B, Karl D, Zehr J. 2005. Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl Environ Microbiol doi: 10.1128/AEM.71.9.5362-5370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krupke A, Musat N, LaRoche J, Mohr W, Fuchs BM, Amann RI, Kuypers MMM, Foster RA. 2013. In situ identification and N2 and C fixation rates of uncultivated cyanobacteria populations. Syst Appl Microbiol 36:259–271. doi: 10.1016/j.syapm.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Thompson A, Carter BJ, Turk-Kubo K, Malfatti F, Azam F, Zehr JP. 2014. Genetic diversity of the unicellular nitrogen-fixing cyanobacteria UCYN-A and its prymnesiophyte host. Environ Microbiol 16:3238–3249. doi: 10.1111/1462-2920.12490. [DOI] [PubMed] [Google Scholar]
- 11.Cohen SE, Golden S. 2015. Circadian rhythms in cyanobacteria. Microbiol Mol Biol Rev 79:373–385. doi: 10.1128/MMBR.00036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. 2005. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reimers AM, Knoop H, Bockmayr A, Steuer R. 2017. Cellular trade-offs and optimal resource allocation during cyanobacterial diurnal growth. Proc Natl Acad Sci U S A 2017:201617508. doi: 10.1073/pnas.1617508114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golden SS, Ishiura M, Johnson CH, Kondo T. 1997. Cyanobacterial circadian rhythms. Annu Rev Plant Physiol Plant Mol Biol 48:327–354. doi: 10.1146/annurev.arplant.48.1.327. [DOI] [PubMed] [Google Scholar]
- 15.Kondo T, Ishiura M. 2000. The circadian clock of cyanobacteria. Bioessays 22:10–15. doi:. [DOI] [PubMed] [Google Scholar]
- 16.Toepel J, Welsh E, Summerfield TC, Pakrasi HB, Sherman LA. 2008. Differential transcriptional analysis of the cyanobacterium Cyanothece sp. strain ATCC 51142 during light-dark and continuous-light growth. J Bacteriol 190:3904–3913. doi: 10.1128/JB.00206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi T, Ilikchyan IN, Rabouille S, Zehr JP. 2010. Genome-wide analysis of diel gene expression in the unicellular N2-fixing cyanobacterium Crocosphaera watsonii WH 8501. ISME J 4:621–632. doi: 10.1038/ismej.2009.148. [DOI] [PubMed] [Google Scholar]
- 18.Zinser ER, Lindell D, Johnson ZI, Futschik ME, Steglich C, Coleman ML, Wright MA, Rector T, Steen R, McNulty NP, Thompson LR, Chisholm SW. 2009. Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS One 4:e5135. doi: 10.1371/journal.pone.0005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornejo-Castillo FM, Cabello AM, Salazar G, Sanchez-Baracaldo P, Lima-Mendez G, Hingamp P, Alberti A, Sunagawa S, Bork P, de Vargas C, Raes J, Bowler C, Wincker P, Zehr JP, Gasol JM, Massana R, Acinas SG. 2016. Cyanobacterial symbionts diverged in the late Cretaceous towards lineage-specific nitrogen fixation factories in single-celled phytoplankton. Nat Commun 7:11071. doi: 10.1038/ncomms11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen HM, et al. 1996. Regulation and molecular structure of a circadian oscillating protein located in the cell membrane of the prokaryote Synechococcus RF-1. Planta 199:520–527. [DOI] [PubMed] [Google Scholar]
- 21.Klatt CG, Liu Z, Ludwig M, Kühl M, Jensen SI, Bryant DA, Ward DM. 2013. Temporal metatranscriptomic patterning in phototrophic Chloroflexi inhabiting a microbial mat in a geothermal spring. ISME J 7:1775–1789. doi: 10.1038/ismej.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steunou AS, Jensen SI, Brecht E, Becraft ED, Bateson MM, Kilian O, Bhaya D, Ward DM, Peters JW, Grossman AR, Kühl M. 2008. Regulation of nif gene expression and the energetics of N2 fixation over the diel cycle in a hot spring microbial mat. ISME J 2:364–378. doi: 10.1038/ismej.2007.117. [DOI] [PubMed] [Google Scholar]
- 23.Holtzendorff J, Partensky F, Mella D, Lennon J-F, Hess WR, Garczarek L. 2008. Genome streamlining results in loss of robustness of the circadian clock in the marine cyanobacterium Prochlorococcus marinus PCC 9511. J Biol Rhythms 23:187–199. doi: 10.1177/0748730408316040. [DOI] [PubMed] [Google Scholar]
- 24.Axmann I, Hertel S, Wiegard A, Dörrich A, Wilde A. 2014. Diversity of KaiC-based timing systems in marine Cyanobacteria. Mar Genomics 14:3–16. doi: 10.1016/j.margen.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Fay P. 1992. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol Rev 56:340–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staal M, Rabouille S, Stal LJ. 2007. On the role of oxygen for nitrogen fixation in the marine cyanobacterium Trichodesmium sp. Environ Microbiol 9:727–736. doi: 10.1111/j.1462-2920.2006.01195.x. [DOI] [PubMed] [Google Scholar]
- 27.Bürgmann H, Widmer F, Sigler WV, Zeyer J. 2003. mRNA extraction and reverse transcription-PCR protocol for detection of nifH gene expression by Azotobacter vinelandii in soil. Appl Environ Microbiol 69:1928–1935. doi: 10.1128/AEM.69.4.1928-1935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen YB, Dominic B, Mellon MT, Zehr JP. 1998. Circadian rhythm of nitrogenase gene expression in the diazotrophic filamentous nonheterocystous cyanobacterium Trichodesmium sp. strain IMS 101. J Bacteriol 180:3598–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zubkov MV, Martin AP, Hartmann M, Grob C, Scanlan D. 22 July 2015. Dominant oceanic bacteria secure phosphate using a large extracellular buffer. Nat Commun doi: 10.1038/ncomms8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zubkov MV, Mary I, Woodward EMS, Warwick PE, Fuchs BM, Scanlan DJ, Burkill PH. 2007. Microbial control of phosphate in the nutrient-depleted North Atlantic subtropical gyre. Environ Microbiol 9:2079–2089. doi: 10.1111/j.1462-2920.2007.01324.x. [DOI] [PubMed] [Google Scholar]
- 31.Michelou VK, Lomas MW, Kirchman DL. 2011. Phosphate and adenosine-5´-triphosphate uptake by cyanobacteria and heterotrophic bacteria in the Sargasso Sea. Limnol Oceanogr 56:323–332. doi: 10.4319/lo.2011.56.1.0323. [DOI] [Google Scholar]
- 32.Stockel J, Welsh EA, Liberton M, Kunnvakkam R, Aurora R, Pakrasi HB. 2008. Global transcriptomic analysis of Cyanothece 51142 shows a robust diurnal oscillation of central metabolic processes. Proc Natl Acad Sci U S A 105:6156–6161. doi: 10.1073/pnas.0711068105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davey M, Tarran G, Mills M, Ridame C, Geider R, Roche JL. 2008. Nutrient limitation of picophytoplankton photosynthesis and growth in the tropical North Atlantic. Limnol Oceanogr 53:1722–1733. doi: 10.4319/lo.2008.53.5.1722. [DOI] [Google Scholar]
- 34.Klausmeier C, Litchman E, Levin S. 2004. Phytoplankton growth and stoichiometry under multiple nutrient limitation. Limnol Oceanogr 49:1463–1470. doi: 10.4319/lo.2004.49.4_part_2.1463. [DOI] [Google Scholar]
- 35.Harke MJ, Frischkorn KR, Haley ST, Aylward FO, Zehr J, Dyhrman ST. 16 August 2018. Periodic and coordinated gene expression between a diazotroph and its diatom host. ISME J doi: 10.1038/s41396-018-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ran L, Larsson J, Vigil-Stenman T, Nylander J, Ininbergs K, Zheng W, Lapidus A, Lowry S, Haselkorn R, Bergman B. 2010. Genome erosion in a nitrogen-fixing vertically transmitted endosymbiotic multicellular cyanobacterium. PLoS One 5:e11486. doi: 10.1371/journal.pone.0011486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakayama T, Kamikawa R, Tanifuji G, Kashiyama Y, Ohkouchi N, Archibald JM, Inagaki Y. 21 July 2014. Complete genome of a nonphotosynthetic cyanobacterium in a diatom reveals recent adaptations to an intracellular lifestyle. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1405222111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohbayashi R, Watanabe H, Ehira S, Kanesaki Y, Chibazakura T, Yoshikawa H. 2016. Diversification of DnaA dependency for DNA replication in cyanobacterial evolution. ISME J 10:1113–1121. doi: 10.1038/ismej.2015.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson ST, Aylward FO, Ribalet F, Barone B, Casey JR, Connell PE, Eppley JM, Ferrón S, Fitzsimmons JN, Hayes CT, Romano AE, Turk-Kubo KA, Vislova A, Armbrust EV, Caron DA, Church MJ, Zehr JP, Karl DM, DeLong EF. 2017. Coordinated regulation of growth, activity and transcription in natural populations of the unicellular nitrogen-fixing cyanobacterium Crocosphaera. Nat Microbiol 2:17118. doi: 10.1038/nmicrobiol.2017.118. [DOI] [PubMed] [Google Scholar]
- 40.Hagen KD, Meeks JC. 2001. The unique cyanobacterial protein OpcA is an allosteric effector of glucose-6-phosphate dehydrogenase in Nostoc punctiforme ATCC 29133. J Biol Chem 276:11477–11486. doi: 10.1074/jbc.M010472200. [DOI] [PubMed] [Google Scholar]
- 41.Min HT, Golden SS. 2000. A new circadian class 2 gene, opcA, whose product is important for reductant production at night in Synechococcus elongatus PCC 7942. J Bacteriol 182:6214–6221. doi: 10.1128/JB.182.21.6214-6221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S, Ryu JY, Kim SY, Jeon JH, Song JY, Cho HT, Choi SB, Choi D, de Marsac NT, Park YI. 2007. Transcriptional regulation of the respiratory genes in the cyanobacterium Synechocystis sp. PCC 6803 during the early response to glucose feeding. Plant Physiol 145:1018–1030. doi: 10.1104/pp.107.105023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maldener I, Muro-Pastor AM. 2010. Cyanobacterial heterocysts In Sons JW. (ed), Encyclopedia of life sciences (ELS). John Wiley & Sons Ltd, Chichester, United Kingdom. doi: 10.1002/9780470015902.a0000306.pub2. [DOI] [Google Scholar]
- 44.Martens J, Barg H, Warren M, Jahn D. 2002. Microbial production of vitamin B12. Appl Microbiol Biotechnol 58:275–285. doi: 10.1007/s00253-001-0902-7. [DOI] [PubMed] [Google Scholar]
- 45.Helliwell KE, Lawrence AD, Holzer A, Kudahl UJ, Sasso S, Kräutler B, Scanlan DJ, Warren MJ, Smith AG. 2016. Cyanobacteria and eukaryotic algae use different chemical variants of vitamin B12. Curr Biol 26:999–1008. doi: 10.1016/j.cub.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milligan AJ, Berman-Frank I, Gerchman Y, Dismukes GC, Falkowski PG. 2007. Light-dependent oxygen consumption in nitrogen-fixing cyanobacteria plays a key role in nitrogenase protection. J Phycol 43:845–852. doi: 10.1111/j.1529-8817.2007.00395.x. [DOI] [Google Scholar]
- 47.Valladares A, Herrero A, Pils D, Schmetterer G, Flores E. 2003. Cytochrome c oxidase genes required for nitrogenase activity and diazotrophic growth in Anabaena sp. PCC 7120. Mol Microbiol 47:1239–1249. doi: 10.1046/j.1365-2958.2003.03372.x. [DOI] [PubMed] [Google Scholar]
- 48.Jones KM, Haselkorn R. 2002. Newly identified cytochrome c oxidase operon in the nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120 specially induced in heterocysts. J Bacteriol 184:2491–2499. doi: 10.1128/JB.184.9.2491-2499.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Latifi A, Ruiz M, Zhang C-C. 2009. Oxidative stress in cyanobacteria. FEMS Microbiol Rev 33:258–278. doi: 10.1111/j.1574-6976.2008.00134.x. [DOI] [PubMed] [Google Scholar]
- 50.Nakayama T, Archibald JM. 24 April 2012. Evolving a photosynthetic organelle. BMC Mol Biol doi: 10.1186/1741-7007-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins TJ. 2007. Introduction to ImageJ for light microscopy. Microsc Microanal 13:1674–1675. doi: 10.1017/S1431927607072911. [DOI] [Google Scholar]
- 52.Chen Y, Zehr JP, Mellon MT. 1996. Growth and nitrogen fixation of the diazotrophic filamentous nonheterocystous cyanobacterium Trichodesmium sp. IMS 101 in defined media: evidence for a circadian rhythm. J Phycol 32:916–923. doi: 10.1111/j.0022-3646.1996.00916.x. [DOI] [Google Scholar]
- 53.Shilova IN, Robidart JC, James Tripp H, Turk-Kubo K, Wawrik B, Post AF, Thompson AW, Ward B, Hollibaugh JT, Millard A, Ostrowski M, J Scanlan D, Paerl RW, Stuart R, Zehr JP. 2014. A microarray for assessing transcription from pelagic marine microbial taxa. ISME J 8:1476–1491. doi: 10.1038/ismej.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baker SC, et al. 2005. The External RNA Controls Consortium: a progress report. Nat Methods 2:731–734. doi: 10.1038/nmeth1005-731. [DOI] [PubMed] [Google Scholar]
- 55.Sun S, Chen J, Li W, Altintas I, Lin A, Peltier S, Stocks K, Allen EE, Ellisman M, Grethe J, Wooley J. 2011. Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: the CAMERA resource. Nucleic Acids Res 39:D546–D551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang Y, Niu B, Gao Y, Fu L, Li W. 2010. CD-HIT Suite: a Web server for clustering and comparing biological sequences. Bioinformatics 26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 58.Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini A, Sawitzki G, Smith C, Smyth G, Tierney L, Yang J, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleś AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smyth GK. 2005. Limma: linear models for microarray data, p 397–420. In Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W (ed), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY. [Google Scholar]
- 61.Kauffmann A, Gentleman R, Huber W. 2009. arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics 25:415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolstad BM, Collin F, Brettschneider J, Simpson K, Cope L, Irizarry RA, Speed TP. 2005. Quality assessment of Affymetrix GeneChip data, p 33–47. In Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W (ed), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY. [Google Scholar]
- 63.Bolstad B. 2004. Low level analysis of high-density oligonucleotide array data: background, normalization and summarization. University of California, Berkeley, Berkeley, CA. [Google Scholar]
- 64.Irizarry R, Hobbs B, Collin F, Beazer-Barclay Y, Antonellis K, Scherf U, Speed T. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 65.Futschik ME, Herzel H. 2008. Are we overestimating the number of cell-cycling genes? The impact of background models on time series analysis. Bioinformatics 24:1063–1069. doi: 10.1093/bioinformatics/btn072. [DOI] [PubMed] [Google Scholar]
- 66.Moritz S, Sardá A, Bartz-Beielstein T, Zaefferer M, Stork J. 2015. Comparison of different methods for univariate time series imputation in R. arXiv arXiv:151003924 [stat.AP]. https://arxiv.org/abs/1510.03924.
- 67.McMurdie P, Holmes S. 2013. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes targeted and detected in the transcriptomic analysis for each organism. The first line shows the total number of genes targeted for the microarray analysis. The second line shows the number of the genes transcribed at detectable levels and the third line the number of diel genes. Percentages of genes determined by comparing the total genes targeted in the microarray in each organism are shown at the bottom. Download Table S1, DOCX file, 0.1 MB (106.4KB, docx) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of genes with periodic transcriptional patterns for all studied cyanobacteria based on Fourier scores. Genes with a FDR of <0.25 were selected and are represented in this table for further comparison. Download Table S2, XLS file, 1.3 MB (1.3MB, xls) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed lists of normalized transcription values and description for all genes in all studied cyanobacteria. There are two spreadsheets for each organism: “Data” and “Description.” The “Data” spreadsheet shows the normalized transcription values for each time point and the mean expression. Eight points were selected for UCYN-A (L6, L9, D3, D6, 2D12, 2L3, 2L9, and 2L12), 9 points for T. erythraeum (D12, L3, L6, L9, L12, D3, D6, D9, and 2D12), 6 points for Cyanothece sp. ATCC 51142 (L2, L6, L10, D2, D6, and D10), 8 points for C. watsonii WH 8501 (D11, L1, L6, L11, D1, D6, 2D11, and 2L1) and 19 points for Prochlorococcus sp. MED4 (D12 to 2L12 [representing time points occurring every 2 h]). “L” and “D” stand for the light period and the dark period, respectively, “2L” and “2D” for the second light-dark cycle, and the number that follows for the hour corresponding to the time of entry into the light or dark period. The “Description” spreadsheet shows the annotation and pathways of each gene for each organism. Download Table S5, XLSX file, 2.6 MB (2.6MB, xlsx) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene orthologs among all studied cyanobacteria. The orthologs were identified as the best hits in reciprocal blastn searches for each pair of genomes. Orthologs of UCYN-A diel genes detected in the other cyanobacteria are listed in sheets 2 to 5. The percentages of diel genes shared between UCYN-A and the respective cyanobacterial species were calculated relative to the total gene orthologs identified by blastn for the two cyanobacterial species. Download Table S3, XLSX file, 0.4 MB (407.6KB, xlsx) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genes selected for network analysis. A minimum of 50% of genes of UCYN-A with diel transcription patterns is represented in the figure. The same genes or genes in the same pathway were represented for the rest of cyanobacteria. The last column (“Genes selected for network”) indicates the genes that were used to build the network analysis whose results are presented in Fig. 2 and 6. Download Table S4, XLS file, 1.2 MB (1.2MB, xls) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transcription patterns in T. erythraeum for the cell division and replication genes over the light-dark cycle. Hierarchical clustering of genes was based on Pearson correlation of gene transcription profiles. The transcription values fof each gene at each time point were standardized, and the blue-red scale shows by how many standard deviations a transcription value is lower (blue) or higher (red) than the mean transcription values over the diel cycle (Z score). “L” and “D” stand for light and dark, respectively; those notations are followed by the hour corresponding to the time after the light and dark periods started. The second light-dark cycle is indicated as “2D,” and that notation is followed by the number of the hours corresponding to entry into the light or dark period. Download FIG S1, PDF file, 0.8 MB (809.5KB, pdf) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Abundance of the haptophyte UCYN-A2 association in CARD-FISH samples during two diel cycles at the Scripps Institution of Oceanography, indicating the number of UCYN-A2 clusters per host cell. Download Table S6, DOCX file, 0.2 MB (190.9KB, docx) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed list of genes involved in B12 metabolism in all studied cyanobacteria. Download Table S8, XLSX file, 0.04 MB (42.1KB, xlsx) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Extended description of materials and methods described in the main text. Download Text S1, DOCX file, 0.06 MB (57.6KB, docx) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of probes utilized in the visualization of UCYN-A associations by double CARD-FISH. Bold letters represent the mismatches between the sequences designed to distinguish UCYN-A1 and UCYN-A2 and their hosts with high specificity. Download Table S7, DOCX file, 0.07 MB (67.1KB, docx) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Box plot showing that quantile normalization of gene hybridization signals for the T. erythraeum IMS101 arrays resulted in similar signal distribution profiles. Raw hybridization data (A) and normalized data (B) are shown for a total of 28 samples. The different colors correspond to different samples. All samples for T. erythraeum IMS101 hybridization passed the quality control testing. Download FIG S3, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Box plot showing that normalization of gene hybridization signals for the three UCYN-A arrays resulted in similar signal distribution profiles. All samples used for UCYN-A hybridization passed the quality control testing. Raw hybridization probe data (A, B, and C) and quantile-normalized data (D, E, and F) are shown for a total of 12 samples hybridized to three different slides. The distribution of hybridization signals is shown for 10,000 randomly sampled probes (including experimental and control probes) for raw data and for 14,000 probes (experimental) for normalized data. Y-axis data represent the hybridization signal shown as a log2 exponent. Download FIG S2, PDF file, 0.9 MB (967.1KB, pdf) .
Copyright © 2019 Muñoz-Marin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.