The protein ProQ has recently been discovered as the centerpiece of a previously overlooked “third domain” of small RNA-mediated control of gene expression in bacteria. As in vitro work continues to reveal molecular mechanisms, it is also important to understand how ProQ affects the life cycle of bacterial pathogens as these pathogens infect eukaryotic cells. Here, we have determined how ProQ shapes Salmonella virulence and how the activities of this RNA-binding protein compare with those of Hfq, another central protein in RNA-based gene regulation in this and other bacteria. To this end, we apply global transcriptomics of pathogen and host cells during infection. In doing so, we reveal ProQ-dependent transcript changes in key virulence and host immune pathways. Moreover, we differentiate the roles of ProQ from those of Hfq during infection, for both coding and noncoding transcripts, and provide an important resource for those interested in ProQ-dependent small RNAs in enteric bacteria.
KEYWORDS: Hfq, noncoding RNA, ProQ, RNA-seq, bacterial pathogen, posttranscriptional control
ABSTRACT
FinO domain proteins such as ProQ of the model pathogen Salmonella enterica have emerged as a new class of major RNA-binding proteins in bacteria. ProQ has been shown to target hundreds of transcripts, including mRNAs from many virulence regions, but its role, if any, in bacterial pathogenesis has not been studied. Here, using a Dual RNA-seq approach to profile ProQ-dependent gene expression changes as Salmonella infects human cells, we reveal dysregulation of bacterial motility, chemotaxis, and virulence genes which is accompanied by altered MAPK (mitogen-activated protein kinase) signaling in the host. Comparison with the other major RNA chaperone in Salmonella, Hfq, reinforces the notion that these two global RNA-binding proteins work in parallel to ensure full virulence. Of newly discovered infection-associated ProQ-bound small noncoding RNAs (sRNAs), we show that the 3′UTR-derived sRNA STnc540 is capable of repressing an infection-induced magnesium transporter mRNA in a ProQ-dependent manner. Together, this comprehensive study uncovers the relevance of ProQ for Salmonella pathogenesis and highlights the importance of RNA-binding proteins in regulating bacterial virulence programs.
INTRODUCTION
RNA-binding proteins (RBPs) are at the heart of central cellular processes in all living organisms. In bacteria, their functions range from being structural components of essential ribonucleoprotein complexes, such as the ribosome, to facilitate posttranscriptional control of mRNAs, often in conjunction with small regulatory RNAs (sRNAs) (1). RBPs have also increasingly been found to play roles in regulating virulence and stress response genes, helping to ensure the full infectivity and survival of bacterial pathogens inside their host.
Salmonella enterica serovar Typhimurium (henceforth Salmonella) has served as an important model for studying the functions of virulence-associated RBPs. This major Gram-negative bacterium is closely related to Escherichia coli and infects both humans and livestock. Salmonella virulence is predominantly mediated by effector proteins that are translocated into eukaryotic host cells via two type III secretion systems (T3SSs) encoded on the major Salmonella pathogenicity islands (SPIs): SPI-1, required for host cell invasion (2), and SPI-2, for intracellular survival (3). Other SPIs as well as Salmonella motility and chemotaxis loci also contribute, for example, by enabling the bacteria to sense and reach the proper host niche (4). Importantly, these infection-relevant genes are not only controlled transcriptionally by various DNA-binding proteins, they are also targeted at the RNA level by the global RBPs Hfq, CsrA, and CspC/E, each of which interacts with hundreds of different Salmonella transcripts (5–8). Furthermore, Hfq, CsrA, and CspC/E are essential for Salmonella virulence in mouse models of infection (6, 9, 10).
To date, most efforts to understand RBP functions in Salmonella have focused on Hfq, an RNA chaperone that facilitates the base pairing of ∼100 different sRNAs with target mRNAs (11, 12). Early reports that hfq deletion impairs many virulence-related functions (9) motivated mechanistic studies on numerous sRNAs (from both the Salmonella core genome and the SPIs) that directly repress or activate mRNAs of infection-relevant genes. However, the observation that many sRNAs are Hfq independent (5, 7) hinted at the existence of another sRNA-related RBP in enteric pathogens. Indeed, the protein ProQ was recently reported to be a previously unrecognized global RBP in Salmonella and E. coli (13).
ProQ is a ∼25-kDa protein that was originally noted in E. coli for its positive effect on the cellular levels of ProP (a proline transporter) and thus osmolyte accumulation (14, 15). The first hints of RNA-binding activity came from similarities in the protein sequence to FinO, a plasmid-encoded RBP required for cis-antisense RNA-mediated control of conjugation (16). Proteins containing ProQ/FinO domain(s) are present in many alpha-, beta-, and gammaproteobacteria on both chromosomes and mobile elements (13, 17–19). Biochemical and structural analyses of E. coli ProQ have fully demonstrated its ability to bind RNA in vitro (20, 21). More importantly, profiling of ProQ ligands in Salmonella identified an unexpectedly large suite of RNA targets, which includes both hundreds of mRNAs and dozens of sRNAs (13, 22). Two molecular functions for ProQ have been established. First, translational mRNA repression by a trans-encoded sRNA that requires ProQ for its function (23), a mechanism resembling that of the FinO-like protein RocC in sRNA-mediated competence regulation in Legionella pneumophila (24). Second, ProQ stabilizes mRNAs by binding at their 3′ ends, protecting them against decay by exonuclease RNase II (22).
Both Hfq and ProQ have been found to target many mRNAs from pathogenicity loci (5, 7, 9, 13, 22). Therefore, as in vitro work continues to reveal molecular mechanisms, it is also important to understand how ProQ affects the most important phase of the life cycle of Salmonella as a pathogen, i.e., the infection of eukaryotic cells. However, in contrast to the established central role of Hfq in Salmonella virulence (25), the proQ gene was not disrupted in previous transposon-based virulence screens in various animal models of salmonellosis (26, 27), precluding an assessment of the importance of ProQ for pathogenesis. Consequently, we sought to determine whether and how ProQ shapes Salmonella virulence and how the activities of these two major RBPs compare with each other in the infection process.
In this study, we apply global transcriptomics of pathogen and host cells during infection. In doing so, we reveal ProQ-dependent transcript changes in key virulence and host immune pathways. Moreover, we differentiate the roles of ProQ from those of Hfq during infection for both coding and noncoding transcripts.
RESULTS
Attenuated virulence of a Salmonella ΔproQ mutant in cultured HeLa cells.
To identify a cell culture-based model to test for a putative role of ProQ in Salmonella virulence, we moved the previously described proQ deletion (13) into a Salmonella strain that constitutively expresses GFP (28) and infected several established host cell types for the study of Salmonella pathogenesis (see Fig. S1 and S2 in the supplemental material). This strain (ΔproQ/gfp+) was complemented using plasmid pZE12-ProQ (13) which yields a mild (∼2- to 3-fold) overexpression of ProQ (Fig. S1b). Invasion and intracellular replication rates were quantified using flow cytometry to measure the bacterial GFP signal inside host cells. Of the three cell types tested, Salmonella ΔproQ bacteria exhibited a ∼2-fold reduced invasion compared to the wild-type strain into HeLa cells (Fig. S1d). This invasion defect was only partially restored in the complemented strain, suggesting that ProQ levels must be tightly controlled for successful host invasion. In contrast, ProQ deficiency or overexpression only led to a subtle decrease in intracellular replication (Fig. S1e). When testing human phagocytic cells, we found only mild differences between the above mutant strains with respect to uptake or intracellular replication in human monocytic or macrophage-like THP-1 cells (Fig. S2a and b).
Virulence parameters of Salmonella proQ mutants in HeLa cell culture assays. (a) proQ deletion strategy. The 5′ portion of proQ (including its own promoter; red arrow) is deleted from the Salmonella genome, while the 3′ portion is kept as it contains the promoter of the downstream gene prc (as deduced from the respective site in the E. coli genome [85]; blue arrow with the asterisk). The remaining TSS annotations were obtained from reference 64 (black arrows). The read coverage plots show RNA-seq data for two biological replicates of wild-type or ΔproQ Salmonella and confirm the deletion of the respective genomic region. Note that the downstream prc gene is slightly induced in the proQ deletion background. (b) Wild-type Salmonella harboring an empty control vector, a ΔproQ strain with the same empty vector, or a ΔproQ strain overexpressing proQ from that plasmid (strain proQ++) in either the wild-type background (GFP-negative; black) or Ptet::gfp background (green) were grown under SPI-1- or SPI-2-inducing conditions (see Materials and Methods), and the levels of ProQ protein were determined by Western blotting using an antibody against the endogenous protein. GroEL served as a loading control. (c) Assessment of the risk of plasmid loss during infection. HeLa cells were infected with wild-type Salmonella containing an ampicillin resistance-mediating plasmid (MOI of 10) in the presence or absence of ampicillin in the infection medium, and after 24 h, host cells were lysed, and intracellular bacteria were recovered and plated in parallel on LB only or LB-Amp plates. (d) proQ deletion causes a ∼2-fold reduction in invasion of Salmonella into HeLa cells. Salmonella input suspensions were derived from bacteria in late exponential phase (OD600 of 2.0). The fraction of infected cells at 0 h and 24 h p.i. is shown. Significance was evaluated using a two-tailed Student’s t test, and P values of <0.05 were considered statistically significant and are denoted by an asterisk (*). P values of <0.01 were considered very significant (**), and P values of <0.001 were considered extremely significant (***). n.s., not significant. (e) Intracellular replication inside HeLa cells is not affected by proQ deletion or overexpression. In panels d and e, HeLa cells were infected with the indicated Salmonella strains (GFP-expressing background) at an MOI of 5. The proportion of invaded (GFP-positive) cells and the rate of intracellular replication (as deduced from the increase in GFP signal intensity per infected HeLa cell over time) were determined by flow cytometry. Data in panels c to e represent the means ± SD from at least three biological replicate experiments. Download FIG S1, PDF file, 0.1 MB (125.6KB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Assessment of an effect of ProQ on Salmonella virulence in monocyte and macrophage models. (a) Rate of infected (GFP-positive) THP-1 cells at 30 min and 24 h p.i. (MOI of 10) for the indicated Salmonella strains. THP-1 cells were either nondifferentiated (monocytes) or differentiated in vitro by adding phorbol 12-myristate 13-acetate (PMA) into the cell medium 3 days prior to infection (macrophages). Salmonella input suspensions were either derived from exponentially growing cultures (OD600 of 2.0) (exponential) or from overnight cultures opsonized for 20 min in mouse serum (stationary). Significance was evaluated using a two-tailed Student’s t test, and P values of <0.05 were considered statistically significant and are denoted by an asterisk. n.s., not significant. In differentiated THP-1 cells, none of the ProQ-dependent changes were statistically significant. (b) Fold replication of the same strains in the same host cell types between 30 min and 24 h p.i. (MOI of 10). Panels a and b report flow cytometry data (mean ± SD) from at least three biological replicate experiments. Note that for fully differentiated THP-1 cells (but not monocytic THP-1), the rate of Salmonella-containing cells increased over time, because macrophages but not monocytic cells engulf bacteria released from neighboring cells. (c) THP-1 cell death after infection with the indicated Salmonella strains (MOI of 10). Cell death was determined by measuring the rate of lactate dehydrogenase (LDH) released into the culture supernatant at 24 h p.i. The data indicate the means ± SD from at least three biological replicates. Download FIG S2, PDF file, 0.04 MB (46.3KB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ProQ effects on host gene expression in Salmonella-infected HeLa cells.
Because of the rather subtle phenotypes in infection experiments of cultured cells, we employed the more sensitive Dual RNA-seq technique (29) to assess molecular consequences of ProQ deficiency, using the transcriptomes of the pathogen and host as a readout. This approach has been successfully used in the past to extract molecular phenotypes of, for example, infection-induced sRNAs in the absence of strong macroscopic phenotypes (30). In the present study, we selected HeLa cells to determine ProQ-dependent gene expression changes during the course of Salmonella infection. Following infection with Salmonella wild-type or ΔproQ bacteria, infected HeLa cells (GFP-positive) were enriched at 8 h and 16 h postinfection (p.i.), and subjected to Dual RNA-seq (Fig. S3a and b; Table S1A).
Experimental overview of the Dual RNA-seq screen for ProQ-dependent expression changes during Salmonella infection of HeLa cells. (a) Dual RNA-seq pipeline. HeLa cells were infected with wild-type or ΔproQ Salmonella (GFP-expressing strains), and invaded (GFP-positive) cells were enriched by FACS. Total RNA was extracted, and ribosomal transcripts of host and pathogen were depleted prior to sequencing. (b) Comparative Dual RNA-seq outline. RNA samples were taken before infection (0 h; lysate from Salmonella in exponential growth phase after the medium shift from LB to host cell media, artificially mixed with HeLa lysate prior to joint RNA extraction) or at 8 and 16 h after infection (mixed human-Salmonella infection samples). Each two biological replicates were analyzed. (c) Dual RNA-seq mapping statistics. Of the ∼20 million reads per sample that could be aligned to the reference annotations, between 1% (0 h and 8 h) and 2% (16 h) mapped to Salmonella and the remaining 98 or 99% mapped to the human genome. (d) RNA class distribution of reads mapped to the Salmonella genome. (e) RNA class distribution of reads mapped to the human genome. Abbreviations in panels d and e: rRNA, ribosomal RNA; tRNA, transfer RNA; mRNA, messenger RNA; sRNA, bacterial small regulatory RNA; IGRs, intergenic regions; lncRNA, long noncoding RNA; miRNA, microRNA; sn(o)RNA, small nucle(ol)ar RNA; miscRNA, miscellaneous RNA. The percentage values denote the range of the fraction of reads mapping to the corresponding RNA class (relative to the total fraction of reads assigned to the respective organism) for the two replicate experiments. Download FIG S3, PDF file, 0.1 MB (98.5KB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Compiled data sets of the present study. Access table for detailed descriptions. Download Table S1, XLSX file, 23.0 MB (23.6MB, xlsx) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
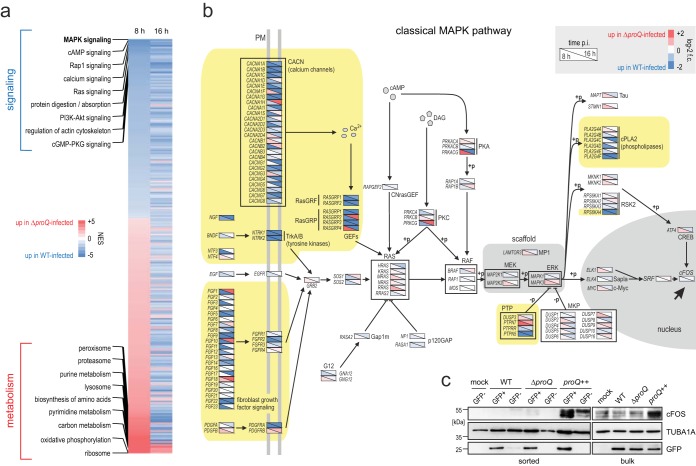
Starting with the host transcriptome, Salmonella has been shown to evoke a proinflammatory response in epithelial cells (31, 32) which is thought to help this pathogen to access otherwise unavailable nutrients and outcompete other members of the gut microbiota (33, 34). A gene set enrichment analysis (GSEA) of differentially expressed host genes (Table S1A) revealed 266 human pathways that were significantly (FDR < 0.1) different upon infection with the ΔproQ mutant relative to wild-type infection (Fig. 1a). Generally, the lack of ProQ-mediated gene regulation in the infecting bacteria caused an upregulation of metabolic processes in the host, whereas host pathways involving immune, calcium, and G-protein signaling were downregulated, with mitogen-activated protein kinase (MAPK) signaling being the most strongly repressed host pathway.
FIG 1.
ProQ-dependent changes in Salmonella-infected HeLa cells. (a) Gene set enrichment analysis (GSEA) heat map showing human pathways that were significantly altered (FDR < 0.1; normalized enrichment score [NES] > 2 or < −2) between the infections of HeLa cells by wild-type (WT) or proQ deletion (ΔproQ) Salmonella strains. Some of the most drastically affected pathways are labeled. The results stem from two biological replicate experiments. (b) Schematic overview of the classical MAPK signaling pathway with comparative Dual RNA-seq data (ΔproQ- versus wild-type-infected HeLa cells; 8 and 16 h postinfection [p.i.]) plotted on top. The scheme was drawn manually using the human KEGG Pathview representation as the template. The yellow-shaded regions highlight branches of the pathway that were most severely affected (on the transcript level) by the absence of ProQ. Many of the downstream regulatory events are mediated by phosphorylation (+p), and thus, differences are not expected to be directly reflected in transcriptome data. Individual branches of the pathway converge at the activation of cFOS (black arrow). log-2 f.c., log2 fold change; PM, plasma membrane. (c) Western blot analysis confirms alterations in cFOS levels in differentially infected host cells. HeLa cells were infected (MOI of 50) with either wild-type Salmonella (WT), or the deletion (ΔproQ) or overexpression (proQ++) strain of ProQ—the two latter strains have highly similar invasion efficiencies and replication kinetics in this system (see Fig. S1d and e in the supplemental material) and are therefore directly comparable. Cells were harvested after 16 h p.i., sorted into invaded (GFP-positive) cells and uninfected (GFP-negative) bystanders or left unsorted (bulk), and total protein samples were separated on a 10% SDS-PAGE gel, transferred to a PVDF membrane, and probed with cFOS-specific antibodies. Tubulin (TUBA1A) serves as a human loading control, and detection of bacterial GFP confirms the purity of the sorted fractions.
Detailed inspection of human MAPK signaling component transcripts (Fig. 1b) pinpointed the most affected branches of the pathway and suggested that Salmonella ProQ is required to activate host cFOS, a constituent of the activator protein 1 transcription factor. On the protein level, we detected similar amounts of cFOS after infection with wild-type or ΔproQ mutant bacteria (Fig. 1c). However, overproduction of ProQ in the Salmonella proQ++ strain, which shows invasion efficiency and replication kinetics similar to those of the Salmonella ΔproQ strain in HeLa cells (Fig. S1d and e), led to a substantial increase in cFOS levels (Fig. 1c). Therefore, enhanced ProQ activity in infecting Salmonella increases MAPK signaling in infected epithelial host cells.
ProQ effects in invading Salmonella involve motility, chemotaxis, and SPI-1 gene expression.
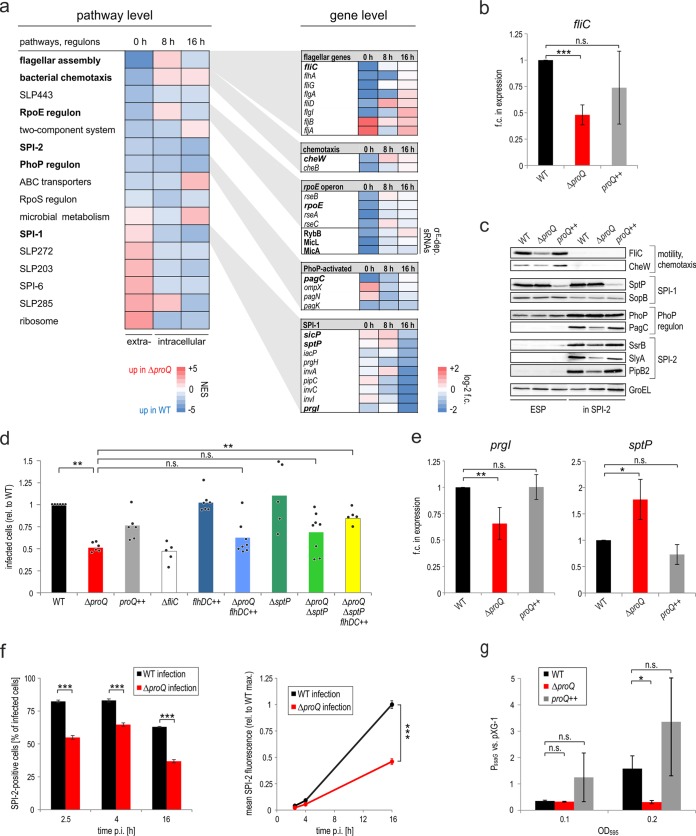
Upon analyzing the impact of ProQ on Salmonella genes during host infection, we observed ∼200 mRNAs that were significantly (FDR < 0.05) differentially expressed between wild-type and ΔproQ Salmonella, at least at one of the three sampled time points (Table S1A). GSEA was used to identify the Salmonella pathways most severely affected by the absence of ProQ (Fig. 2a). At the time of invasion (0 h), the ΔproQ mutation caused downregulation of the genes encoding components of the motility and chemotaxis pathways as well as of the σE regulon and its associated sRNAs, whereas invasion (SPI-1) and ribosomal genes showed higher expression levels, despite a similar growth rate of the strains (Fig. 3a). Some of the most prominent ProQ-dependent expression changes during the invasion stage were independently validated. For example, the Dual RNA-seq results predicted a downregulation of the major flagellin FliC mRNA in the ΔproQ strain (Fig. S4a), and this was confirmed by qRT-PCR measurements (Fig. 2b) and Western blot analysis (Fig. 2c). Importantly, differential expression is likely due to a posttranscriptional effect of ProQ on fliC mRNA and not the result of genomic inversion of this phase-variable gene (Fig. S4b). The changes were dependent on ProQ since proQ complementation partially rescued fliC expression (Fig. 2b). Similar results were obtained for the transcripts of the chemotaxis gene cheW (Fig. S4c and d) and the rpoE operon (Fig. S4e, f, and g), including a slight regulation of CheW on the protein level (Fig. 2c). The reduced expression of motility and chemotaxis genes provided a possible explanation for the invasion defect observed for the ΔproQ mutant (Fig. S1d). However, attempts to restore flagellar gene expression through overexpression of the flagellar master regulator FlhDC (ΔproQ/flhDC++ strain) failed to restore wild-type invasion efficiency (Fig. 2d).
FIG 2.
ProQ-dependent expression changes in infecting Salmonella. (a) Differential transcriptome analysis. (Left) GSEA pinpoints Salmonella pathways most severely affected by the absence of ProQ. Plotted are all pathways that were significantly altered (FDR < 0.1; normalized enrichment score [NES] > 2 or < −2) between wild-type and ΔproQ Salmonella for at least one time point. (Right) The significantly differentially expressed (FDR < 0.05) genes from several of the affected pathways are plotted. Genes with ProQ-dependent expression changes that were independently validated in this study are shown in bold type. The results stem from two biological replicate experiments. (b) Repression of fliC mRNA in the absence of ProQ. The indicated Salmonella strains were grown in LB to an OD600 of 2.0, total RNA was extracted and served as the template for qRT-PCR measurements using the constitutively expressed gfp mRNA as a reference. Values are means (bars) ± standard deviations (SD) (error bars) from four biological replicate experiments. (c) Western blot analyses demonstrate that ProQ-dependent differential virulence gene expression extends to the protein level. The indicated Salmonella strains were grown in LB to an OD600 of 2.0 (early stationary phase [ESP]) or in minimal SPI-2-inducing medium to an OD600 of 0.3 (in SPI-2), prior to the separation of total protein samples on a 10% SDS-PAGE gel. Membranes were probed with protein-specific (FliC, SopB, PagC, and GroEL) or FLAG-specific (other) antibodies. GroEL serves as loading control (one representative image is shown). (d) Influence of ProQ on Salmonella motility genes and on SptP expression is partially responsible for differential infection rates. HeLa infection rates of the indicated Salmonella strains. The Salmonella ΔfliC strain was included as a nonmotile control strain. The data are derived from flow cytometry measurements of infected (GFP-positive) cells relative to wild-type infection (individual replicate measurements shown as single dots; bars refer to the respective mean). Evaluation of significance was performed using a two-tailed nonparametric Mann-Whitney-Wilcoxon test. P values of ≤ 0.01 were considered very significant (**). n.s., not significant (P value > 0.05). (e) The mRNA of the T3SS-1 tip component PrgI is suppressed and that of the SPI-1 effector SptP is derepressed in the absence of ProQ. qRT-PCR measurements were performed preinfection (0 h; prgI) or at 1 h p.i. of HeLa cells (sptP; MOI of 50) on proQ-expressing versus non-proQ-expressing Salmonella strains. Constitutively expressed gfp mRNA served as a reference transcript. The data refer to the mean ± SD from five (prgI) or three (sptP) biological replicate experiments. (f) ProQ affects SPI-2 expression inside host cells. (Left) Cultured HeLa cells were infected (MOI 50) with Salmonella strains containing a transcriptional reporter of SPI-2 expression (PssaG::gfp), in the respective proQ background. At the indicated time points, the fraction of SPI-2-positive relative to all infected HeLa cells was measured by flow cytometry. (Right) From the same experiment, the mean SPI-2 fluorescence level per SPI-2-positive host cell was quantified. Data refer to the mean ± SD from four independent experiments. (g) ProQ-dependent SPI-2 activation in defined minimal medium mimicking the vacuolar compartment. The indicated Salmonella strains were grown to an OD595 of 0.1 or 0.2, respectively, and the activity of the same transcriptional reporter as in panel f was measured and normalized against a constitutive GFP reporter (pXG-1). Data refer to the means ± SD from three independent experiments. In panels b, e, f, and g, significance was evaluated using a two-tailed Student’s t test and indicated as follows:*, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant.
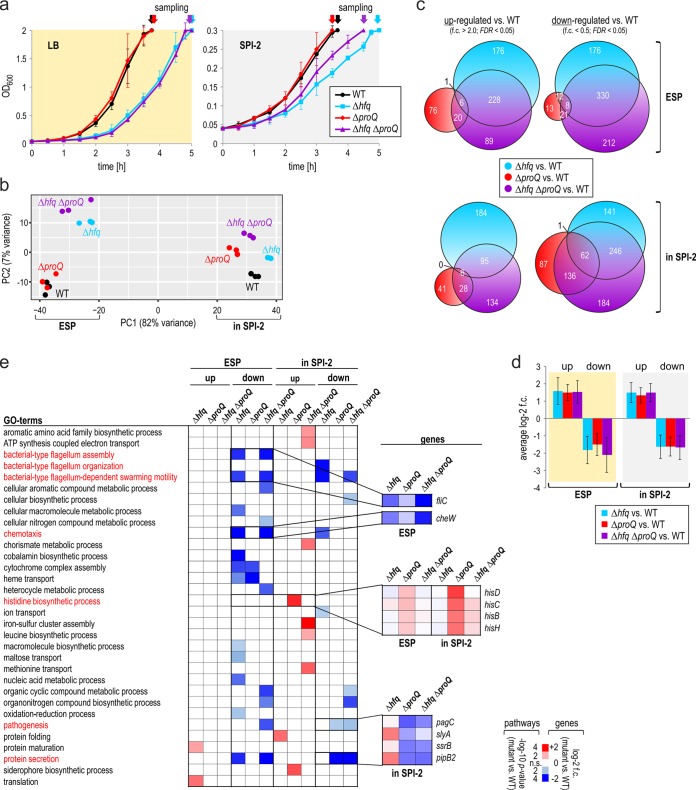
FIG 3.
Comparison of the ProQ and Hfq regulons under infection-relevant conditions. (a) Growth curves of the four indicated Salmonella strains in LB or SPI-2-inducing medium. The graphs show the means ± SDs from three biological replicate experiments. The arrows indicate the sampling times of the cultures for total RNA extraction. (b) Principal-component analysis (PCA) plot of the RNA-seq data of the strains grown under the conditions shown in panel a (three replicates/condition). (c) Venn diagrams display the overlap of the ProQ and Hfq regulons. Plotted are all transcripts that were significantly differentially expressed (FDR < 0.05; log2 fold change [f.c.] > 1 or < −1) in the respective mutant compared to the wild-type background. (d) Average magnitude of the fold changes of the significantly differentially expressed genes (plotted in panel c) in the indicated mutants compared to the wild-type strain. (e, left) Gene Ontology (GO) term enrichment analysis of Salmonella pathways significantly affected (P value < 0.05; Bonferroni correction for multiple testing) in the respective RBP deletion backgrounds. (Right) Significantly differentially expressed (FDR < 0.05; log2 f.c. > 1 or < −1) genes from several of the affected pathways are plotted. The results stem from three biological replicate experiments.
RNA-seq coverage plots and validation data for ProQ-dependent effects on Salmonella motility, chemotaxis, and the σE response. (a) Read coverage over the flagellar locus of Salmonella and zoom-in on the fliC gene. At the time of infection (0 h), fliC mRNA is downregulated in ΔproQ Salmonella (red) compared to wild-type Salmonella (black). (b) Orientation-specific PCR of the fliC locus of the indicated Salmonella strains using genomic DNA as the template. Information about the invertible region comes from reference 81. While all three strains contain populations of both fliC-ON and fliC-OFF bacteria, the majority express fliC (ON state). Importantly, the ratio (fliC-ON/fliC-OFF) is unchanged in the three strains. (c) Read coverage over the chemotaxis regulon and close-up view of the cheW locus. (d) qRT-PCR data for cheW mRNA. The indicated Salmonella strains were grown in LB to an OD600 of 2.0, total RNA was extracted and served as the template for qRT-PCR measurements. gfp mRNA was the reference transcript. Bars and error bars indicate the means ± SD from four biological replicate experiments. Significance was evaluated using a two-tailed Student’s t test (**, P < 0.01; ***, P < 0.001). (e) Read coverage over the rpoE operon and close-up view of the rpoE gene. (f) qRT-PCR data for rpoE mRNA. Data shown as described above for panel d. (g) Northern blot shows the expression of the σE-dependent sRNAs RybB and MicA upon growth in LB to an OD600 of 2.0 (ESP) or in the minimal SPI-2-inducing medium to an OD600 of 0.3 (in SPI-2), respectively. Since both sRNAs were expressed at low levels in LB at an OD600 of 2.0, the respective membrane was reexposed for an extended time period (shown to the right). 5S rRNA serves as a loading control. Download FIG S4, PDF file, 0.3 MB (315.5KB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The absence of ProQ also had divergent effects on individual SPI-1-associated transcripts. For example, the prg operon encoding the T3SS-1 was downregulated in invading ΔproQ versus wild-type Salmonella (i.e., the 0-h time point), whereas the sicP-sptP transcriptional unit encoding the SPI-1 effector SptP and its chaperone SicP were expressed at higher levels than in wild-type Salmonella (Fig. 2a; Fig. S5a). These Dual RNA-seq-based predictions for the prgI, sicP, and sptP mRNAs were independently confirmed by qRT-PCR (Fig. 2e; Fig. S5b). Notably, the mRNA for the major SPI-1 transcriptional activator HilD was largely unaffected by ProQ status (Fig. S5b), indicating that ProQ acts directly on the prgI and sicP-sptP mRNAs. This is further supported by our previous observation that the prgHIJK and sicP-sptP mRNAs, but not the hilD mRNA, can be cross-linked to ProQ (22) (Table S1B).
RNA-seq coverage plots and validation data for ProQ-dependent expression changes of Salmonella pathogenicity islands. (a) Read coverage over the SPI-1 locus of Salmonella and zoom-in onto the TSS of the sicP-sptP transcript. TSS annotation (black arrow) obtained from reference 64. (b) qRT-PCR data on the expression of selected Salmonella SPI-1 virulence genes (the sicP-sptP operon and the mRNA of the SPI-1 master regulator, hilD) in proQ-expressing versus non-proQ-expressing strains during the infection of HeLa cells (MOI of 50). gfp mRNA was used as a reference. The plot displays the mean ± SD from three biological replicate experiments. (c) Read coverage over the SPI-2 locus. (d) qRT-PCR data on the expression of selected SPI-2 genes (a gene for a secreted effector, pipB2; the gene for the SPI-2 master regulator, ssrB) in proQ-expressing versus non-proQ-expressing strains during the infection of HeLa cells (MOI of 50). gfp mRNA was the reference. The data are the means ± SD from three biological replicate experiments. Download FIG S5, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Upon their translocation, certain SPI-1 effectors (namely, SopE, SopE2, and SopB) cooperate to reorganize the host actin cytoskeleton, enabling Salmonella to invade epithelial cells (35–37). Translocated SptP protein subsequently counteracts the activities of these effectors, helping to reorganize the cytoskeleton to preinfection conditions (36, 38). This suggested that elevated SptP secretion could, at least in part, be the basis for the invasion defect of the ΔproQ strain (Fig. S1d). To test this, we first deleted the sptP gene alone (ΔproQ/ΔsptP strain) yet failed to see significant improvement of the invasion efficiency (Fig. 2d). However, combining sptP deletion with flagellar overexpression (ΔproQ/ΔsptP/flhDC++ strain) did significantly increase the invasion rate of the ΔproQ strain, albeit not to the level of wild-type Salmonella (Fig. 2d). Therefore, the impaired invasion rate of the Salmonella ΔproQ strain is a multifactorial phenotype and at least partially due to the reduced expression of motility genes and enhanced levels of the SptP effector.
ProQ affects SPI-2 expression and the σE response in intracellular Salmonella.
To understand the role of ProQ after successful host cell invasion, we analyzed the two intracellular stages (8 h and 16 h p.i.) of the Dual RNA-seq data (Table S1A). Host cell invasion is generally accompanied by widespread expression changes in the infecting Salmonella, including the upregulation of metal ion uptake systems and induction of the envelope stress response (30, 39–41). Interestingly, envelope stress was differentially affected by the absence of ProQ with lower steady-state levels of σE-dependent sRNAs, RybB and MicA, in ΔproQ Salmonella (Fig. S4g). Additionally, the SPI-2 locus was exclusively activated once Salmonella reached its intracellular niche (Fig. S5c). Interestingly, SPI-2 expression was slightly reduced in the ΔproQ strain compared to the wild-type strain (Fig. 2a; Fig. S5c). qRT-PCR measurements during the infection of HeLa cells revealed only marginal differences in the expression of selected SPI-2 genes (the gene for the secreted effector PipB2 and that for the SPI-2 master regulator SsrB) in the absence of ProQ. However, overproduction of ProQ substantially enhanced intracellular SPI-2 activation. Likewise, a ProQ-dependent effect on PipB2 protein levels was observed in vitro by Western blot analysis (Fig. 2c).
To further support the positive effect of ProQ on SPI-2 activity, cultured HeLa cells were infected with Salmonella strains containing a robust transcriptional reporter of SPI-2 expression (PssaG::gfp) in the respective proQ background (Fig. 2f). In this reporter, GFP expression is driven by the promoter of the SsrB-activated gene ssaG (42), and thus, GFP intensity serves as a proxy for SPI-2 activity. At all three time points measured (2.5, 4, and 16 h p.i.), the fraction of SPI-2-positive HeLa cells was lower for ΔproQ bacterial infections than for wild-type infections (Fig. 2f, left). Likewise, mean GFP intensity (corresponding to mean SPI-2 activity) per SPI-2-positive host cell was reduced in the absence of ProQ (Fig. 2f, right). A similar attenuation in SPI-2 induction in the ΔproQ background as well as an (over)complementation in the proQ++ background were observed in an infection-relevant in vitro assay. Here, Salmonella bacteria were grown to defined densities in minimal SPI-2-inducing medium (42), and ssaG promoter activity was measured as a proxy for SPI-2 induction (Fig. 2g).
In summary, ProQ affects the expression of Salmonella motility and virulence genes during infection. Of note, fliC and cheW mRNAs as well as selected SPI-1 and SPI-2 transcripts have previously been identified as direct ProQ ligands in cross-linking experiments (22) (Table S1B), arguing for a direct effect of ProQ binding on the steady-state levels of these transcripts.
Comparative analysis of the ProQ and Hfq regulons under infection-relevant conditions.
To understand the extent of the ProQ-dependent regulations, we compared the identified genes with the regulon of the other major RNA chaperone of Salmonella, Hfq. Importantly, several of the flagellar and virulence genes affected by ProQ have previously been shown to be dysregulated in a Salmonella Δhfq strain (9, 10). To avoid secondary effects caused by the different invasion efficiencies of ΔproQ and Δhfq bacteria, we performed RNA-seq analysis of bacteria grown in medium that mimics invasion (early stationary phase [ESP] in LB) or intracellular replication (defined minimal SPI-2-inducing medium [42]). Under these conditions, the proQ deletion mutant grew indistinguishably from wild-type Salmonella, whereas the Δhfq strain displayed a considerably longer lag phase (in LB medium) or slightly reduced growth (SPI-2-inducing conditions) (Fig. 3a).
Principal-component analysis of the RNA-seq data (Fig. 3b) revealed two major clusters, splitting the samples along principal component 1 (PC1), reflecting the two growth conditions (ESP versus “in SPI-2”). Additionally, within the ESP cluster, the ΔproQ bacteria colocalized with the wild-type Salmonella but separated from Δhfq and the ΔproQ Δhfq double mutant. In contrast, in SPI-2-inducing medium, the wild-type and ΔproQ transcriptomes were segregated from each other, suggesting that ProQ may exert stronger effects on the Salmonella transcriptome in intracellular bacteria compared to invading bacteria. Likewise, the absolute numbers of significantly (FDR < 0.05) differentially expressed genes in ΔproQ compared to wild-type Salmonella were higher in SPI-2 than in ESP, albeit the total number of differentially expressed genes was lower than the number of Hfq-dependent regulated genes (Fig. 3c; Table S1C). Notwithstanding this, the average magnitude of the fold changes of differentially expressed genes in the respective mutant backgrounds was highly similar between the Hfq and ProQ regulon (Fig. 3d). Of note, the set of genes differentially expressed in the ΔproQ Δhfq double mutant was not simply the sum of genes deregulated in the two single mutant strains (Fig. 3c), suggesting some degree of synergy between these two major RBPs.
Pathway analysis of the differentially expressed genes (Fig. 3e) recapitulated many of the findings from the comparative Dual RNA-seq screen for ProQ (Fig. 2a). Interestingly, however, motility and chemotaxis pathways were even more strongly repressed in the absence of Hfq compared to ProQ, and again more so in the absence of both chaperones, suggesting additive effects of Hfq and ProQ on the respective transcript abundance. In contrast, SPI-2 genes were specifically repressed in the absence of ProQ (ΔproQ and ΔproQ Δhfq strains) but not affected by the deletion of hfq. Another ProQ-specific footprint was revealed with respect to genes involved in histidine biosynthesis. Interestingly, these genes were derepressed in the absence of ProQ which was partially rescued by removing Hfq, suggesting that the two major chaperones might fulfill opposing functions in the regulation of histidine biosynthesis (note, however, that SL1344, the Salmonella strain used in this study, is a histidine auxotroph [43]).
Screen for infection-relevant ProQ-dependent sRNAs.
ProQ not only binds hundreds of different mRNAs but also binds close to 50 sRNAs, most of which are of uncharacterized function. To obtain a better understanding of the expression of these ProQ-associated sRNAs (13, 22), we reanalyzed our previous high-resolution Dual RNA-seq data of intracellular Salmonella (30). Figure 4a summarizes the expression kinetics of these ProQ-associated sRNAs during a 24-h time course of HeLa cell infection. Interestingly, the maximal fold changes in their expression (Fig. 4a, right) are about an order of magnitude lower than for ProQ-independent sRNAs (Fig. 4a, left). We identified three sRNAs—SraL (44, 45), RaiZ (23), and the uncharacterized 3′-derived sRNA STnc540 (7)—for which expression was consistently reduced in ProQ-deficient Salmonella grown in HeLa cells (Fig. 4b; Table S1A).
FIG 4.
Screen for infection-induced ProQ-dependent sRNAs. (a) Dual RNA-seq data on the expression of ProQ-associated sRNAs (as defined in references 13 and 22) during a high-resolution time course of the infection of HeLa cells by wild-type Salmonella. Expression data were retrieved from a previous study (30) (accession number GSE117256). (b) Comparative expression of ProQ-associated infection-induced sRNAs (colored in panel a) between ΔproQ strain and the wild-type Salmonella at the indicated time points after HeLa infection. The results stem from two biological replicate experiments. (c) Northern blot on the expression of the denoted sRNAs. Total RNA samples were taken from wild-type Salmonella grown under the indicated in vitro conditions, separated on a 6% PAA/urea gel, blotted, and probed with sRNA-specific DNA oligonucleotides (Table S1G). OD, optical density (in LB); ON, overnight growth in LB; SPI-2, SPI-2-inducing conditions (see Materials and Methods). (d and e) Northern blot data for the same sRNAs in the presence or absence of ProQ (d) or Hfq (e) in ESP (LB; OD 2.0) or SPI-2-inducing conditions. Detection was performed as described above for panel c. 5S rRNA serves as loading control in panels c to e.
The expression of SraL, RaiZ, and STnc540 as well as six additional intracellularly induced (30) ProQ-associated sRNAs in wild-type Salmonella during various growth stages in vitro (Fig. 4c) was analyzed by Northern blotting. STnc540 levels, for which we detected two main species (∼90 and ∼60 nt), did not necessarily correlate with the expression of its parental transcript, the ihfA mRNA encoding integration host factor α (Fig. 4c). For example, STnc540 expression peaked under SPI-2-inducing conditions, while maximal ihfA expression occurred in early exponential phase (OD of 2.0), suggesting that different factors determine the steady-state levels of the sRNA and mRNA. Confirming previous results (23), however, the 3′-derived sRNA RaiZ accumulated in stationary-growth bacteria as both a long (∼160-nt) and short (∼120-nt) isoform (Fig. 4c). The intergenic sRNA SraL accumulated in both stationary phase and in SPI-2 medium (Fig. 4c), which agrees with SraL being a σS-dependent sRNA (44) and its ∼25-fold induction during intracellular replication (Fig. 4a). Similarly, the antisense-encoded RyjB, the sense-overlapping STnc1680, as well as the antitoxin RNAs IstR-1, SibA, and SibC were induced in stationary phase and under SPI-2 conditions, whereas the candidate sRNA STnc1275 was exclusively detected in SPI-2 medium (Fig. 4c).
We expanded this Northern blot analysis by including the ΔproQ and proQ++ strains (Fig. 4d). This confirmed that ProQ is generally required for full expression of STnc540 and RaiZ, whereas its effect on SraL was limited to growth in LB medium (ESP) (Fig. 4d). This agrees with the RNA-seq data, which suggested an effect of ProQ on SraL levels primarily prior to infection (Fig. 4b). Of the other sRNAs, expression of IstR-1, STnc1275, and SibC positively correlated with ProQ. In contrast, RyjB and SibA showed unchanged steady-state levels in the absence of ProQ, despite them being bona fide ligands of this RBP (22). Finally, STnc1680 levels were negatively affected by both the absence of ProQ and its overexpression. This highlights that for many of the tested sRNAs, the function of ProQ is likely more complex than mediating sRNA stabilization.
Finally, we investigated the potential role of Hfq on the expression of the above-described sRNAs. While Hfq slightly affected the abundance of most of the sRNAs, STnc540 was exclusively affected by ProQ (ΔproQ and ΔproQ Δhfq strains; Fig. 4e). As this suggested STnc540 to be a fully ProQ-dependent and Hfq-independent sRNA, we selected it for further characterization.
Interplay between STnc540 and ProQ affects expression of a magnesium importer.
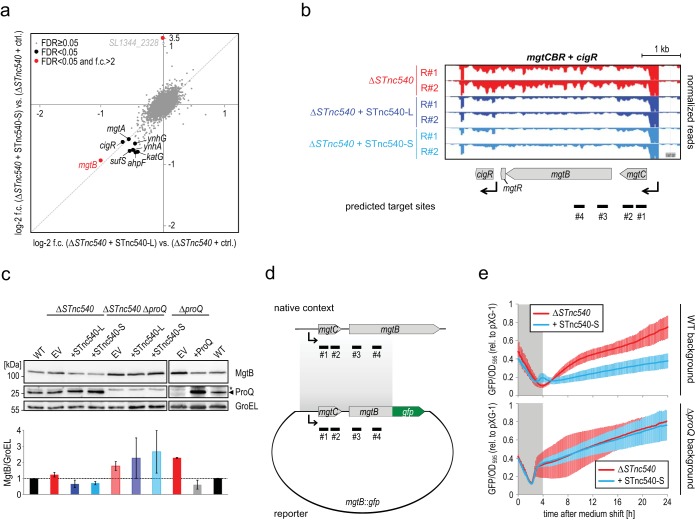
Given that ProQ is an RNA chaperone and that previously characterized ProQ-associated sRNAs all function by base pairing mechanisms with other cellular RNAs (23, 44), we opted for an sRNA pulse expression approach (46, 47) to identify potential STnc540 RNA targets. To perform this in a STnc540 null background, we removed most of the STnc540 sequence from the Salmonella chromosome while maintaining the terminator for ihfA mRNA (ΔSTnc540 strain; Fig. S6a and b). The resulting mutant strain did not show any significant growth difference compared to the parental wild-type strain (Fig. S7). STnc540 was subsequently induced from a plasmid-borne promoter under the condition of maximal expression of the endogenous sRNA, i.e., under SPI-2-inducing conditions (Fig. 4c). RNA samples from the sRNA overexpression strains (the long or short STnc540 version) were collected 10 min after induction, analyzed by RNA-seq, and compared to samples from a control strain in which the empty vector was induced (Fig. 5a; Table S1D). Of the 2,312 mRNAs captured (≥100 aligned reads/gene), eight were downregulated >1.5-fold upon overexpression of STnc540. There was a high correlation between the RNA-seq data for the long and short STnc540 isoforms, arguing that the short sRNA version possesses the full regulatory capacity.
FIG 5.
STnc540 target identification screen. (a) Pulse expression of STnc540 under SPI-2 conditions. Either one of the two STnc540 isoforms was overexpressed for 10 min prior to total RNA isolation and sequencing. The log2 fold changes in expression upon induction of the long (x axis) or short (y axis) STnc540 isoform compared to an isogenic strain carrying the empty control vector are plotted. The results represent the means from two biological replicate experiments. mRNAs whose expression was significantly altered (FDR < 0.05) upon induction of at least one STnc540 isoform compared to the control are labeled. SL1344_2328 is a low-confidence target (as inferred from manual read coverage inspection; not shown). (b) Read coverage plot of the expression of the mgt locus upon pulse expression of the long (dark blue) or short (light blue) STnc540 isoform relative to the empty control vector. The position of predicted interaction sites with STnc540 (Fig. S6d) are denoted. TSS annotations (black arrows) were derived from reference 64. (c) Western blot analysis indicates a slight repression of MgtB protein levels upon constitutive overexpression of STnc540 in the presence of ProQ. Wild-type, ΔSTnc540, ΔproQ, or ΔSTnc540 ΔproQ Salmonella with 3xFLAG-tagged MgtB expressed from its native promoter and harboring the indicated, constitutive sRNA overexpression plasmids or pZE-ProQ, respectively, were grown under SPI-2-inducing conditions to an OD600 of 0.5. Total protein samples were harvested and analyzed by Western blotting using FLAG-specific or endogenous ProQ-detecting antibodies (the position of ProQ is indicated by an arrowhead; the asterisk denotes an unspecific signal). GroEL serves as a loading control. A representative Western blot experiment out of three independent biological replicates is shown, and the quantification of the MgtB signal intensity in the different strains (normalized to GroEL and relative to the wild-type strain) over the three replicates is given in the graph below the Western blot. (d) Schematic representation of the mgtB::gfp reporter construct used in panel f. The predicted interaction sites with STnc540 (#1 to #4; see Fig. S6d) are indicated. (e) GFP reporter assay for STnc540-dependent regulation of mgtB. The reporter construct in panel d was cotransformed with a constitutive STnc540 (short isoform) overexpression plasmid or the respective vector control into a ΔSTnc540 (top) or ΔSTnc540 ΔproQ (bottom) Salmonella background. The resulting strains were grown under SPI-2-inducing conditions in a 96-well plate for 24 h, and the GFP intensity (as a proxy for MgtB levels) was monitored in 10-min intervals. The data show the means ± SD of the GFP intensity normalized to OD595 values and relative to the GFP intensity of the same strains harboring a constitutive GFP expression control plasmid (pXG-1) from three biological replicates, each comprising technical triplicates. In the first 3-4 h, OD, but not GFP intensity, increased (grey windows).
Construction of an STnc540 deletion mutant and in silico prediction of target interactions. (a) To generate the ΔSTnc540 strain, the 5′ portion of the sRNA gene was removed, while its terminator was kept. TSS annotations (black arrows) are according to reference 64. (b) Northern blot validation of the successful deletion of STnc540. Salmonella strains were grown in LB to stationary phase (ON) or under the SPI-2-inducing condition, and total RNA samples were analyzed by Northern blotting using probes against the 3′ end of STnc540 or the body of the ihfA mRNA. 5S rRNA serves as a loading control. (c) qRT-PCR for mgtB mRNA at the indicated time points upon the addition of arabinose to strains harboring the long or short STnc540 isoform on the pBAD plasmid relative to the control strain harboring the empty vector. gfp mRNA was the reference. The data are means ± SD from three biological replicate experiments. Significance (*, P value of <0.05) was evaluated using a two-tailed Student’s t test. (d) In silico prediction of RNA-RNA interactions between STnc540 and its putative target mgtCBR. RNAhybrid (49) was used to predict interaction sites. The numbering of nucleotides is relative to the respective TSS of the mRNA or the annotated 5′ end of STnc540, respectively, based on the annotations from reference 64. Download FIG S6, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth of different mutant strains of Salmonella. The indicated strains were grown in LB (left) or minimal SPI-2-inducing medium (right), and OD600 values were measured in 30-min intervals. The data are means ± SD from two biological replicate experiments. Download FIG S7, PDF file, 1.3 MB (1.4MB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The top candidate target was the mgtB mRNA encoding a magnesium import protein (Fig. 5a and b). The gene is part of the mgtCBR locus that is silent in rich medium but whose transcription is activated by the two-component system PhoP/Q under SPI-2 conditions (48). Interestingly, the downregulation upon STnc540 induction selectively applied to mgtB and not to the other members of this operon. Using the RNAhybrid software (49), we predicted possible RNA-RNA interactions between STnc540 and mgtB (Fig. S6d). Notably, in three of the four proposed interactions (#2 to #4 in Fig. S6d), the 3′ region of STnc540 where ProQ binds (22), was engaged in base pairing. This may explain why the short sRNA isoform is sufficient for target repression.
In the pulse expression experiment above, either isoform of STnc540 reduced the level of the mgtB mRNA to ∼50% (Fig. 5a; Fig. S6c). On the level of the MgtB protein which we rendered detectable via addition of a chromosomal 3xFLAG tag, repression by constitutively expressed STnc540 was slightly less pronounced (Fig. 5c). Importantly, however, removing both STnc540 and ProQ substantially increased the basal levels of MgtB-3xFLAG (compared to the wild type), and abrogated the reduction of MgtB when STnc540 was coexpressed in the ΔproQ background (Fig. 5c). Furthermore, STnc540 repressed a fluorescent reporter of MgtB, harboring the 5′ portion of its open reading frame fused to GFP (Fig. 5d and e; Fig. S8a). In contrast, removing the putative interaction sites #3 and #4 from this construct (mgtC::gfp in Fig. S8a) abrogated repression by STnc540, arguing that one or both of these sites may be the actual base-pairing region(s) with STnc540.
GFP reporter assays for STnc540/ProQ-mediated control of MgtB. Schematic representation (top) and measurement (bottom) of a transcriptional reporter (Pmgt::gfp) and two transcriptional/posttranscriptional reporters (mgtC::gfp and mgtB::gfp) for which the open reading frame of GFP was fused to the mgt locus at the indicated positions. The predicted interaction sites with STnc540 (#1 to #4; see Fig. S6d) are indicated below the schematic representations. These reporter constructs were cotransformed with a constitutive STnc540 (long or short isoform) overexpression plasmid, the ProQ expression plasmid, or the respective vector control into the ΔSTnc540 (a) or ΔSTnc540 ΔproQ (b) Salmonella background. The resulting strains were grown in SPI-2-inducing medium in a 96-well plate for 24 h, and the GFP intensity (as a proxy for MgtC or MgtB levels) was monitored in 10-min intervals. The data show the means ± SD of the GFP intensity normalized against OD595 and relative to the GFP intensity of the same strains harboring a constitutive GFP expression control plasmid (pXG-1) from three biological replicates, each comprising technical triplicates. Download FIG S8, PDF file, 1.6 MB (1.7MB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Most importantly, the repression of the mgtB::gfp reporter was abrogated in the ΔproQ background (Fig. 5d and e; Fig. S8b), supporting the basic requirement of ProQ for STnc540-mediated repression of the mgtB mRNA. Interestingly, ProQ delayed the induction of the mgtB::gfp reporter in the absence of STnc540 (Fig. S8b), arguing that this RBP, directly or indirectly, counteracts mgtB expression by an additional, STnc540-independent, mechanism. Although more mechanistic analysis is needed to dissect the regulatory interplay between STnc540, ProQ, and mgtB, our results support the idea that ProQ engages directly in the regulation of Salmonella virulence genes.
DISCUSSION
With ProQ biology being in its infancy (19), a fundamental question remains as to whether and how the global RNA-binding activity of this RBP, which in Salmonella includes many interactions with transcripts from major virulence regions (22), impacts pathogenesis. The present study reveals that ProQ is required for optimal virulence gene expression in Salmonella and directly compares the molecular consequences of loss of function of ProQ with those of the primary enterobacterial sRNA-binding chaperone, Hfq.
Molecular consequences of the loss of ProQ for Salmonella pathogenesis.
In Salmonella, ProQ controls motility and chemotaxis gene expression and affects specific SPI-1 transcripts. ProQ further exerts a positive influence on the expression of members of the PhoP regulon and, particularly, SPI-2 genes, implicating ProQ in the cross talk between the two major pathogenicity islands of this bacterium, SPI-1 and SPI-2. Overall, the impact of ProQ on Salmonella expression of coding genes is most likely a combination of the dysregulated expression of direct mRNA ligands (such as cspE mRNA [22]) and indirect derepression of mRNAs normally targeted by ProQ-associated sRNAs (such as hupA mRNA [23]). It is important to note that ProQ binding can have divergent outcomes for different RNA ligands, even if they belong to the same regulon. For example, ProQ targets transcripts of both the SPI-1-controlled operons prgHIJK and sicP-sptP (22). However, we found here that the deletion of proQ reduced the steady-state levels of prgI mRNA, while it enhanced sptP expression. Therefore, more insight into the potentially diverse molecular mechanisms of ProQ is needed to fully correlate ProQ binding with changes in gene expression.
On the side of the infected host, this transcriptome analysis revealed that the ΔproQ mutant elicits less MAPK signaling compared to wild-type Salmonella. Notably, the MAPK signaling cascade, which activates nuclear factor κB (NF-κB) and interferon responses, is one of the prime targets of bacterial pathogens as they manipulate host immunity for their own benefit (50), and several Salmonella effector proteins are known to target MAPKs (51). For example, specific effectors of the SPI-2 regulon—expression of which is reduced in Salmonella ΔproQ bacteria—are known activators of MAPK signaling in the host (52–54), whereas the SPI-1 effector SptP, whose level increases in the absence of ProQ, inhibits the MAPK cascade through ERK (extracellular signal-regulated kinase) (55). Therefore, the ProQ-dependent expression changes in the bacterial transcriptome provide an explanation for the observed differences between the host response to wild-type and ΔproQ Salmonella (Fig. 6a).
FIG 6.
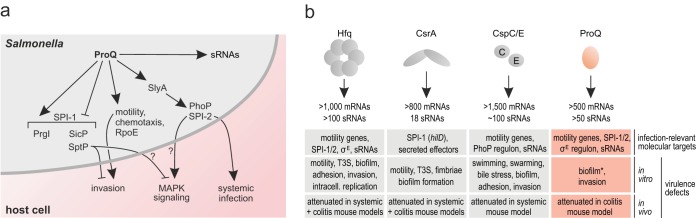
Roles of global RBPs in Salmonella virulence. (a) Model of infection-relevant regulations mediated by ProQ that were uncovered in the present study. (b) Known contributions of the five global RBPs for Salmonella virulence mechanisms and phenotypes associated with their respective deletion. Information about the number of RNA ligands for the indicated proteins is derived from references 5, 6, 13, and 22. E. coli mutants devoid of proQ exhibit defects in biofilm formation (70) (indicated by biofilm*). See main text for further information.
Comparing the ProQ and Hfq regulons under virulence conditions.
Hfq and ProQ are the two major sRNA-binding proteins of Salmonella (13), and each of them targets hundreds of different transcripts with little overlap, at least when assayed in early stationary-phase cells (5, 22). Strikingly, however, our comparative transcriptomics here revealed an unexpected overlap between the ProQ and Hfq regulons under infection-relevant conditions. Key virulence systems, such as the flagellum or the SPI-1 secretion system, were dysregulated in the absence of either one of these RNA chaperones. However, interactome studies suggest that different regulatory cascades are affected by ProQ and Hfq and that these branches converge at the level of flagellin expression or SPI-1 secretion, respectively. For example, Hfq binds flhDC, encoding the master regulator of flagellar expression (5), while ProQ directly interacts with the major flagellin-encoding fliC mRNA (22). Likewise, Hfq (5), but not ProQ (22), binds the mRNA for the SPI-1 master regulator HilD. Instead, ProQ directly binds to individual SPI-1 transcripts for structural components of the SPI-1 needle or secreted effectors (e.g., the prg and sicP-sptP operons) (22). In summary, despite overall similar virulence processes being affected by both global RBPs, the higher-resolution dissection on transcript level reveals that Hfq and ProQ target distinct branches in the corresponding pathways.
Importantly, the positive effect on SPI-2 genes was exclusive to ProQ, whereas the deletion of hfq had no effect on SPI-2 expression. As ProQ interacts with the mRNAs of two key regulators of the SPI-2 pathway, SlyA and SsrB (22), we probed the levels of these proteins in the presence or absence of the RBP. While SsrB levels were minimally affected, SlyA abundance decreased in ΔproQ Salmonella, proposing the latter as the relevant target through which ProQ promotes SPI-2 expression. We note, however, that former RIP-seq (13) and CLIP-seq data (22) were obtained from Salmonella grown to exponential phase in LB, i.e., a condition under which SPI-2 genes are poorly expressed. Therefore, our comparative expression data should motivate future ProQ interactome studies under similar in vitro conditions which better reflect the intracellular lifestyle of this pathogen.
Identification of infection-relevant ProQ-dependent sRNAs.
Our Dual RNA-seq experiments revealed a set of infection-induced sRNAs whose levels depend on ProQ (Fig. 4a and b) and which include STnc540. This sRNA is made from the 3′ UTR of the ihfA mRNA which encodes the α-subunit of the conserved, heterodimeric integration host factor (IHF) complex. The protein made from the mRNA is important for Salmonella pathogenesis: by alleviating H-NS-mediated transcriptional repression, IHF contributes to the activation of virulence genes (56, 57). Interestingly, the experimental target screen for STnc540 suggests that the noncoding region of the ihfA mRNA may also be involved in the regulation of virulence genes. At least when overexpressed, the STnc540 sRNA selectively represses the mRNA of the MgtB magnesium ion transporter.
Regulation of mgt genes could hardly be more complex. Early work showed that mgtCBR expression is transcriptionally activated via PhoQ/P (48). However, subsequent work (58–63) revealed multiple additional regulatory layers that make expression of the mgt operon responsive to disparate external and internal cues. Among those, a PhoP-activated cis-antisense RNA (AmgR) constitutes a negative-feedback loop to counteract mgtCBR expression in an Hfq- and RNase E-dependent manner. Additionally, two short open reading frames (termed mgtM and mgtP) within the long mgtC leader respond to increasing ATP or decreasing proline levels, respectively, and control translation elongation into mgtCBR via an attenuation-like mechanism.
Here, we identified STnc540 as the first trans-acting riboregulator of mgtB. We hypothesize that STnc540 may serve as a molecular device setting an upper limit to the expression of this infection-induced transporter. It should be noted, however, that mgtB mRNA was not identified in previous in vivo mappings of ProQ ligands, the caveat being that mgtB was hardly expressed under those conditions (13, 22, 64). Alternatively, ProQ may interact only with STnc540 to stabilize and/or unwind the sRNA, that is, without making contact with the target mRNA. This would be different from Hfq-dependent regulations which usually involve the formation of Hfq-sRNA-mRNA complexes. However, it would also differ from the previously reported ProQ-dependent regulation of hupA mRNA by the RaiZ sRNA in which a trimeric complex with ProQ is formed (23). Similarly, the ProQ-related RocC protein in Legionella pneumophila is also assumed to bind both the RocR sRNA and its mRNA targets (24).
It also remains to be fully established whether mgtB is a physiologically relevant target of STnc540. Admittedly, the observed ∼2-fold decrease in mgtB mRNA and MgtB protein levels upon ectopic STnc540 expression is moderate. However, the STnc540/mgtB pair has value because it increases the currently small number of target transcripts for the study of the molecular mechanisms of ProQ-mediated regulation. Importantly, our preliminary results suggest that STnc540 binds mgtB mRNA in the coding sequence where sRNAs have to compete with elongating ribosomes and for which few cases of sRNA-mediated regulation are currently known (65). This mechanism would also be different from the ProQ/RocC-mediated regulations by the RaiZ/RocR sRNAs which similarly to Hfq-associated sRNAs sequester the ribosome binding sites of their target mRNAs (23, 24). In addition to the intergenic and UTR-derived sRNAs that were inspected in the present study, recent observations imply that stable RNAs may also be carved out from coding regions (87) and some of these “decay-generated noncoding RNAs” in Salmonella bind ProQ. What relevance these RNAs might have for physiology and virulence remains an open question for future efforts.
What is the role of ProQ in vivo?
The observed infection defects notwithstanding, the general robustness of Salmonella virulence in the face of the lack of the central RNA modulator ProQ is astonishing. A concept referred to as “phase transition” was recently proposed for eukaryotic RBPs (66). According to this hypothesis, surplus cellular transcripts may localize in distinct speck-like complexes together with RBPs, and thus be sequestered in a nonfunctional state. Upon detection of a specific stimulus, these specks can rapidly dissolve, giving rise to an immediate pool of ready-to-use transcripts. If bacterial RBPs may work in a similar fashion, one would expect rather mild phenotypic effects upon their deletion, as reported here for ΔproQ Salmonella. The major consequence of ProQ removal in this case would be that more or less RNA is stored in the corresponding complexes, whereas the steady-state levels of free (i.e., functional) RNA would hardly be affected. Future efforts, e.g., subcellular localization studies, should address whether ProQ might fulfill such an RNA-buffering function during Salmonella infection.
Alternatively, simplistic cell culture models reflect host complexity only to a certain degree. It is not unheard of that bacterial mutant strains with a weak macroscopic virulence defect in vitro still produce a robust phenotype in whole-animal models (67). In a first attempt to assess the impact of ProQ on Salmonella virulence in an animal model, we compared the colonization ability of the ΔproQ strain to that of wild-type bacteria after oral infection of streptomycin-pretreated C57BL/6 mice (see Fig. S9 in the supplemental material). This revealed an attenuation in survival/growth of ProQ-deficient Salmonella at systemic sites, although not reaching the level of a functional SPI-2 knockout (ΔssaV). Future efforts should aim at the characterization of proQ mutant Salmonella in further in vivo models of infection and also consider the role of this RBP in breaching the barrier functions imposed by the intestinal microbiota.
In vivo phenotype of ΔproQ Salmonella. Streptomycin-pretreated C57BL/6 mice were infected (5 × 107 CFU in total by gavage) with a 1:1 mixture of wild-type Salmonella (Strr) and the indicated isogenic mutant strains (Strr Kanr). The ΔssaV strain has a functional knockout of the T3SS of SPI-2 and was included as a positive control of an attenuated strain. Three days postinfection, the indicated organs were isolated and homogenized, and competitive indices were determined by selective plating. The data stem from five animals per group from two independently performed experiments (single data points are shown, and the geometric mean for a group is indicated by a black bar). Statistical significance was assessed by one-tailed Wilcoxon matched pair rank test, comparing the proQ-deficient strain to its coinfecting wild-type control strain. Statistically significant differences are indicated by asterisks as follows: *, P = 0.0313; **, P values of ≤0.01. Despite the slight induction in basal prc expression in the ΔproQ background (Fig. S1a), we confidently assign the in vivo attenuation of the proQ mutant strain to the absence of this RBP, since prc overexpression would be expected to result in hyperreplication (86). Download FIG S9, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Perspective.
We now know that deletion of any of the five global RBPs in Salmonella is associated with diverse virulence defects (Fig. 6b). In Salmonella, Hfq binds many virulence-related RNA transcripts (5, 7, 68) and hence, hfq deletion mutants show severe virulence defects, including dysregulated virulence factor expression and secretion, chemotaxis defects, and reduced motility (9, 10, 25). The translational control protein CsrA tunes expression of the master regulator of Salmonella invasion genes, HilD (8), and interacts with mRNAs for effectors of both major SPIs (5). Consequently, Salmonella mutants lacking CsrA are avirulent (69). Most recently, the cold shock proteins CspC and CspE were shown to interact with transcripts of PhoP regulon members in Salmonella, and a ΔcspC ΔcspE double deletion mutant is strongly attenuated during infection of both cell lines and mice (6). With respect to ProQ, the only pathogenesis-related process previously known to be affected was biofilm formation which was shown to be strongly reduced in E. coli ΔproQ (70). The results presented here for Salmonella ProQ further highlight the importance of RBP functions for bacterial virulence and should encourage more efforts to address the molecular mechanisms employed by global RBPs under infection settings.
MATERIALS AND METHODS
Salmonella strain construction and growth conditions.
Salmonella enterica serovar Typhimurium strain SL1344 (strain JVS-1574 [43]) and a constitutively GFP-expressing derivative thereof (JVS-3858 [28]) are considered wild-type. Chromosomal mutations were generated as previously described (71), the mutated alleles were subsequently transduced into one of the wild-type backgrounds (GFP negative or positive, respectively) using P22 phage (72), and the respective resistance cassettes were eliminated using the FLP helper plasmid pCP20 (71) at 42°C. For plasmid transformation, the respective Salmonella strains were electroporated with ∼10 ng of DNA. The complete lists of bacterial strains and plasmids used in this study are provided in Tables S1E and F in the supplemental material. Routinely, Salmonella strains were grown in liquid Lennox broth (LB) medium or on solid LB agar medium at 37°C. When appropriate, the liquid or solid medium was supplemented with 30 μg/ml chloramphenicol (Cm), 100 μg/ml ampicillin (Amp), 50 μg/ml kanamycin (Kan), or 0.02% (wt/vol) L-arabinose (final concentrations).
For in vitro assays reflecting defined virulence conditions, Salmonella overnight cultures were diluted 1:100 in 10 ml of LB and grown at 37°C with shaking at 220 rpm to an OD600 of 2.0 (ESP, i.e., a SPI-1-inducing condition [9, 64]). To reflect the conditions of the Salmonella-containing vacuole, 1 ml of an LB culture at an OD600 of 2.0 was pelleted, and the bacterial cells were washed two times with PBS and once with SPI-2-inducing minimal medium (42) and diluted 1:50 in 10 ml of this minimal medium. If not mentioned otherwise, cultures were grown at 37°C and 220 rpm until they reached an OD600 of 0.3.
Mammalian cell culture techniques.
Human cervix carcinoma HeLa cells (ATCC CCL-2; for Fig. 2f) or HeLa-S3 cells (ATCC CCL-2.2; if not mentioned otherwise), and human monocytic THP-1 cells (ATCC TIB-202) were cultured as previously described (30). HeLa cells were passaged in DMEM (Gibco), and THP-1 cells were passaged in RPMI (Gibco), with each medium supplemented with 10% fetal calf serum (FCS) (Biochrom), 2 mM L-glutamine (Gibco), and 1 mM sodium pyruvate (Gibco) in T-75 flasks (Corning) in a 5% CO2, humidified atmosphere at 37°C, and routinely tested for mycoplasma contamination using the MycoAlert Mycoplasma Detection kit (Lonza). Two days prior to infection, 2 × 105 cells were seeded in 2 ml of antibiotic-free medium (six-well format), resulting in a cell density of 1 × 106 cells/well at the time of infection. To differentiate THP-1 cells into macrophages, monocytic THP-1 were seeded at 1 × 106 cells/well (six-well format) 3 days prior to infection in medium supplemented with 50 ng/ml (final concentration) of phorbol 12-myristate 13-acetate (PMA) (Sigma), and the medium was replenished after 2 days.
Infection assays.
Infection assays were conducted as previously described (30). To avoid loss of the proQ overexpression plasmid (and the respective empty control vector) during intracellular bacterial growth, we performed HeLa infection assays in the presence or absence of the respective selection marker (ampicillin) in the host cell medium (Fig. S1c). During the first 24 h of infection, no significant plasmid loss was observed, even in the absence of the antibiotic. Nevertheless, all future infection experiments were conducted in the presence of 1× ampicillin (=100 µg/ml final concentration) in the cell medium.
Briefly, overnight cultures of Salmonella were diluted 1:100 in fresh LB medium and grown aerobically to an OD600 of 2.0. Bacterial cells were harvested by centrifugation (2 min at 12,000 rpm, room temperature) and resuspended in host cell medium. Alternatively, to reduce THP-1 cytotoxicity (Fig. S2), overnight cultures were opsonized in mouse serum (Sigma) for 20 min at room temperature and used for infection. Either way, infection was carried out by adding the respective bacterial suspension directly to each well of seeded host cells. HeLa cells were infected at a multiplicity of infection (MOI) of 5, unless otherwise indicated, and THP-1 cells (both monocytes and macrophages) were infected at an MOI of 10. Immediately after addition of the bacteria, the plates were centrifuged for 10 min at 250 × g at room temperature, followed by 30-min incubation in 5% CO2, humidified atmosphere at 37°C. Thereafter, gentamicin and ampicillin were added to the medium at a final concentration of 50 µg/ml or 100 µg/ml, respectively. When infecting HeLa cells or differentiated THP-1 cells (i.e., adherent cells), the medium was replaced after a further 30-min incubation for host medium containing 10 μg/ml of gentamicin and 100 µg/ml of ampicillin and incubated for the remainder of the experiment. For the infection of nondifferentiated THP-1 cells (that grow in suspension), the medium was not replaced, and cells were kept in 50 µg/ml gentamicin and 100 µg/ml ampicillin until harvest. In each case, time zero was defined as the time when gentamicin was first added to the infected cells.
Flow cytometry and fluorescence-activated cell sorting (FACS).
For flow cytometry-based analyses, infected cell cultures were washed twice with PBS, if necessary cells were detached from the bottom of the plate by trypsinization (HeLa cells) or scraping (differentiated THP-1 cells) and resuspended in host cell medium. Upon pelleting the cells (5 min at 250 × g, room temperature), they were resuspended in 4% (wt/vol) paraformaldehyde (PFA) and stored at 4°C until the analysis. To prepare the PFA-fixed cells for flow cytometry, the samples were centrifuged as described above, the pelleted cells were washed with PBS, centrifuged again, and resuspended in 250 µl of PBS per 1 × 106 cells (i.e., one well of the six-well format). The cells were measured by flow cytometry using a BD Accuri C6 instrument (BD Biosciences), gating for GFP-positive cells in the FITC channel versus the autofluorescence in the PE channel as described elsewhere (29), and the data were analyzed using FlowJo software (Tree Star Inc.).
For the FACS-based enrichment of invaded (GFP-positive) HeLa-S3 cells, the infected cultures were washed once with PBS and trypsinized, and each three wells were pooled and transferred into 15-ml tubes and pelleted (5 min at 250 × g, 4°C). The supernatant was removed, the cell pellet was resuspended in 500 µl of RNAlater (Qiagen), and stored at 4°C until sorting. Immediately prior to FACS, RNAlater-fixed cell samples were diluted by adding each 10 ml of ice-cold PBS and pelleted (5 min at 500 × g, 4°C). The RNAlater was removed by aspiration. The cell pellet was washed once in ice-cold PBS, resuspended in 250 µl of ice-cold PBS/1 × 106 cells, and sorted using the FACSAria III device (BD Biosciences) at 4°C (cooling both the input tube holder and the collection tube rack) and at a medium flow rate as described previously (29). Typically ∼2 × 105 cells of each fraction were collected and used for RNA isolation.
CFU assays.
To assess plasmid loss during intracellular replication, infected HeLa cultures were solubilized with PBS containing 0.1% (vol/vol) Triton X-100 (Gibco) at 24 h p.i. The resulting cell lysates were serially diluted in PBS, plated onto LB or LB-Amp plates and incubated at 37°C overnight. The number of colony-forming units (CFU) recovered on the next day was compared to that obtained from the bacterial input solution used for infection. Biological triplicates were collected, each comprised of two technical replicates.
RNA extraction and gDNA removal.
For Dual RNA-seq analysis, total bacterial and host RNA from infected cells was isolated using the mirVana kit (Ambion). To this end, the fixed and sorted cells were pelleted (5 min, 1,000 × g, 4°C), lysed in 600 µl of the Lysis/Binding buffer of the mirVana kit (Ambion), and the samples were further processed following the manufacturer’s instructions for total RNA isolation. For bacterial RNA-seq analyses, qRT-PCR experiments or Northern blots, total RNA was isolated using the TRIzol LS reagent (Invitrogen) according to the manufacturer’s recommendations. To remove contaminating genomic DNA, samples for (Dual) RNA-seq or qRT-PCR were further treated with 0.25 U of DNase I (Fermentas) per 1 µg of RNA for 45 min at 37°C.
cDNA library preparation for (Dual) RNA-seq analyses.
To deplete ribosomal transcripts, RNA samples were treated with the Ribo-Zero “Epidemiology” (Dual RNA-seq) or “Bacteria” (Salmonella-only RNA-seq) kits (Illumina). Following the manufacturer’s instructions, ∼500 ng of total, DNase I-treated RNA was used as an input to the ribo-depletion procedure. rRNA-depleted RNA was precipitated in ethanol for 3 h at −20°C.
cDNA libraries for Illumina sequencing were generated by Vertis Biotechnologie AG, Freising-Weihenstephan, Germany. rRNA-free RNA samples were first sheared via ultrasound sonication (four 30-s pulses at 4°C) to generate on average 200- to 400-nt fragments. Fragments of <20 nt were removed using the Agencourt RNAClean XP kit (Beckman Coulter Genomics) and the Illumina TruSeq adapter was ligated to the 3′ ends of the remaining fragments. First-strand cDNA synthesis was performed using M-MLV reverse transcriptase (NEB) wherein the 3′ adapter served as a primer. The first-strand cDNA was purified, and the 5′ Illumina TruSeq sequencing adapter was ligated to the 3′ end of the antisense cDNA. The resulting cDNA was PCR amplified to about 10 to 20 ng/μl using a high fidelity DNA polymerase. The TruSeq barcode sequences were part of the 5′ and 3′ TruSeq sequencing adapters. The cDNA library was purified using the Agencourt AMPure XP kit (Beckman Coulter Genomics) and analyzed by capillary electrophoresis (Shimadzu MultiNA microchip).
Illumina sequencing.
Generally, for sequencing, cDNA samples were pooled in approximately equimolar amounts. The cDNA pool was size fractionated in the size range of 200 to 600 bp using a differential cleanup with the Agencourt AMPure kit (Beckman Coulter Genomics). Aliquots of the cDNA pools were analyzed by capillary electrophoresis (Shimadzu MultiNA microchip). Sequencing was performed on a NextSeq 500 platform (Illumina) at Vertis Biotechnologie AG, Freising-Weihenstephan, Germany (single-end mode; 75 cycles).
Computational methods to interpret RNA-seq data sets.
The adapter sequences as well as low-quality ends of the raw sequences were removed using cutadapt version 1.13 (73). Reads were mapped to the Salmonella reference sequence (NC_016810.1, NC_017718.1, NC_017719.1, and NC_017720.1) and for the Dual RNA-Seq samples additionally to the human reference genome (GRCh38.p10 from GENCODE) using READemption’s subcommand align (READemption version 0.4.3) (74) and segemehl 0.2.0. Coverage plots were generated with READemption’s subcommand coverage, and reads overlapping genomic features were quantified with the subcommand gene_quanti followed by expression level comparison with edgeR (version 3.22.1) (75). Sequencing coverages were visualized using the Integrated Genome Browser (IGB) (76) and are based on uniquely mapped reads normalized by the total number of aligned reads per organism. Host-pathogen pathway enrichment analyses in the Dual RNA-seq data set (Fig. 1a and 2a) were performed using gene set enrichment analysis (GSEA; version 2.1.0) (77) fed with the log2 fold changes reported by edgeR. Salmonella pathway enrichment analyses in the in vitro RNA-seq data set (Fig. 3e) were performed using the Gene Ontology (GO) knowledgebase and resources (78, 79).
qRT-PCR.
qRT-PCR was performed with the Power SYBR Green RNA-to-CT1-Step kit (Applied Biosystems) according to the manufacturer’s instructions and a CFX96 Touch real-time PCR detection system (Bio-Rad). gfp mRNA (where applicable) or 5S rRNA served as Salmonella reference transcripts. Fold changes in expression were determined using the 2−ΔΔCt method (80). Primer sequences are given in Table S1G, and their specificity had been confirmed using Primer-BLAST (NCBI).
PCR to detect Salmonella flagellar phase variation.
To extract genomic DNA from wild-type Salmonella, 100 µl of an LB overnight culture of strain JVS-1574 was pelleted, and the bacteria were resuspended in 100 µl of water. The suspension was boiled for 5 min at 100°C, vortexed briefly, and centrifuged (5 min at 13,000 rpm, room temperature). To the supernatant, one volume of chloroform was added, the mixture was vortexed for 30 s and centrifuged for 10 min at 13,000 rpm and 4°C. One microliter of the aqueous phase served as the template for PCR amplification with Taq polymerase (NEB). Flanking PCR primers (JVO-14315 and JVO-14316) were designed to anneal outside of—and the internal primer (JVO-14313) to bind to—the invertible chromosomal region in front of the fljB promoter (according to reference 81). Denaturation was at 95°C for 30 s, annealing was at 60°C for 30 s, and elongation was for 1.5 min at 72°C. After 35 cycles, the PCR products were resolved on a 1.5% (wt/vol) agarose/TAE gel, stained with ethidium bromide, and visualized using a UV transilluminator (Intas Science Imaging).
Northern blotting.
Each 5 μg of total Salmonella RNA prepared with TRIzol LS reagent (Invitrogen) was loaded per lane and separated in 6% (vol/vol) polyacrylamide–7 M urea gels. Blotting was performed as previously described (30). After the transfer onto Hybond XL membranes (Amersham), RNA was cross-linked with UV light and hybridized at 42°C with gene-specific 32P-end-labeled DNA oligonucleotides (see Table S1G for probe sequences) in Hybri-Quick buffer (Carl Roth AG). After exposure, the screens were read out on a Typhoon FLA 7000 phosphorimager (GE Healthcare).
Western blotting.
Immunoblotting was performed as previously described (30). Briefly, samples from Salmonella in vitro cultures were taken corresponding to an OD600 of 0.4 and centrifuged for 4 min at 16,100 × g at 4°C, and pellets were resuspended in sample loading buffer to a final concentration of 0.01 OD per μl. After denaturation for 5 min at 95°C, 0.05-OD equivalents of the sample were separated via SDS-PAGE (10% [wt/vol] PAA). Gel-fractionated proteins were blotted for 90 min (0.2 mA per cm2; 4°C) in a semidry blotter (Peqlab) onto a PVDF membrane (Perkin Elmer) in transfer buffer (25 mM Tris base, 190 mM glycine, 20% [vol/vol] methanol). Blocking was for 1 h at room temperature in 10% (wt/vol) dry milk/TBST20. Appropriate primary antibodies (Table S1H) were hybridized at 4°C overnight, and after three 10-min washes with TBST20, secondary antibodies (Table S1H) were hybridized for 1 h at room temperature. After three additional washing steps for each 10 min in TBST20, blots were developed using Western lightning solution (Perkin Elmer) in a Fuji LAS-4000 imager (GE Healthcare). When needed, quantification of the signal intensities of protein bands was performed using ImageJ (82).
For Western blotting of human proteins, infected HeLa cultures were trypsinized and sorted, and each ∼2 × 105 GFP-positive or -negative cells were collected, pelleted, and lysed in 50 µl of sample loading buffer. Alternatively, ∼2 × 106 cells (i.e., one well; six-well format) of an infected culture were directly lysed in 500 µl of sample loading buffer. In each case, the lysates were boiled for 5 min at 95°C, and each 20 -μl lysate was loaded per lane of a 10% (wt/vol) PAA gel for SDS-PAGE as described above. After blotting and blocking (as described above), the membrane was probed with the respective primary antibody at 4°C overnight, and after washing (as described above), the membrane was probed with the secondary antibody for 1 h at room temperature (see Table S1H for a list of antibodies). Detection of tubulin was as described above. To detect cFOS, the SuperSignal West Femto maximum sensitivity substrate (Thermo Scientific) was used.
sRNA pulse expression.
The plasmids pBAD-ctrl. (pKP-8), pBAD-STnc540-L (pAW-16), or pBAD-STnc540-S (pAW-17) (Table S1F) were transformed into the ΔSTnc540 background. Pulse expression of the respective isoform of STnc540 was induced from the pBAD promoter by the addition of 0.02% (wt/vol) L-arabinose (Sigma) to the respective cultures in minimal SPI-2-inducing medium at an OD600 of 0.3. Ten minutes after the induction, total RNA was extracted using TRIzol LS reagent (Invitrogen), treated with DNase I, and analyzed by RNA-seq.
GFP reporter assays.
To monitor ssaG or mgtC promoter induction and translational MgtC and MgtB activity in real time, GFP reporters were reused (PssaG::gfp [30]) or newly constructed by fusing truncations of the 5′ part of the mgt operon to the GFP open reading frame as shown in Fig. S8a. To this end, the respective regions were PCR amplified from Salmonella genomic DNA and inserted into the plasmid pAS-93 via AatII/NheI sites as previously described (83). The resulting reporter plasmids (pYC-104, pYC-101, pAW-37, and pAW-38) or a constitutive GFP expression plasmid (pXG-1) were cotransformed with either pZE-ctrl. (pJV-300), pZE-STnc540-L (pAW-24), pZE-STnc540-S (pAW-25), or pZE12-ProQ into the ΔSTnc540, ΔproQ, or ΔSTnc540 ΔproQ strain background. The resulting strains were grown overnight in LB-Amp/Cm, then diluted 1:100, and further grown in the same medium to an OD600 of 2.0. A volume of 1 ml of the cultures was pelleted and the collected cells were shifted to SPI-2-inducing medium (defined as time point zero). For the PssaG reporter assay (Fig. 2g), the respective strains were grown to defined optical densities in 96-well plates (Nunc Microwell 96F; Thermo Scientific) at 37°C (with shaking), and the GFP signal intensity was recorded using the Infinite F200 PRO plate reader (Tecan). For the temporal measurement of the activity of mgt reporters (Fig. 5e and Fig. S8), the strains were grown under the same conditions for 24 h, and GFP levels were monitored in 10-min intervals (Infinite F200 PRO plate reader; Tecan). In parallel, the OD595 was recorded, and the respective values were used to normalize the GFP signals to cell density.
In silico prediction of sRNA-mRNA interactions.
RNA duplex formation between STnc540 and the mgt operon (Fig. S6d) was predicted using the RNAhybrid program (49) with default settings.
Animal infection experiments.
Animal housing and experimentation were approved by the Kantonales Veterinäramt Zürich, Switzerland (licenses 223/2010 and 222/2013). Wild-type C57BL/6 mice were kept under specific-pathogen-free (SPF) barrier conditions in individually ventilated cages at the EPIC facility, ETH Zürich. Salmonella infections were performed according to previously published protocols (84). Briefly, 8- to 12-week-old mice were pretreated with 25 mg of streptomycin by gavage 24 h before the infection. The indicated Salmonella strains were grown in LB/0.3 M NaCl with appropriate antibiotics at 37°C for 12 h and subcultured to late log phase for 4 h in the same medium lacking antibiotics. The bacteria were washed in PBS and mixed in a 1:1 ratio, and a total of 5 × 107 CFU were inoculated by gavage. Salmonella loads in the indicated organs were monitored by plating serial dilutions of homogenized cecal content or tissue on MacConkey agar supplemented with appropriate antibiotics. The competitive index (CI) was calculated as the ratio of competitor population sizes in the infected organ corrected for the initial ratio in the inoculum. A CI of <1 means that the indicated strain loses against the wild-type strain (JVS-1574).
Data availability.
All RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE117256. The workflows of the different analyses represented as Shell scripts are deposited at Zenodo (https://doi.org/10.5281/zenodo.1311214).
ACKNOWLEDGMENTS
We thank Barbara Plaschke, Tatjana Achmedov, Manja Barthel, and Sarah Reichardt for excellent technical support, Alexandre Smirnov, Erik Holmqvist, and Yanjie Chao for the construction of bacterial strains and plasmids and the design of oligonucleotides, and Stan Gorski for editing the manuscript.
This work was supported by a DFG Leibniz Award to J.V. (grant DFG Vo875-18), a grant from the Swiss National Science Foundation to W.-D.H. (310030B-173338), and a grant from the Swedish Research Council to M.E.S. (2015-00635).
Footnotes
Citation Westermann AJ, Venturini E, Sellin ME, Förstner KU, Hardt W-D, Vogel J. 2019. The major RNA-binding protein ProQ impacts virulence gene expression in Salmonella enterica serovar Typhimurium. mBio 10:e02504-18. https://doi.org/10.1128/mBio.02504-18.
Contributor Information
Julian Parkhill, The Sanger Institute.
Xavier Charpentier, Centre International de Recherche en Infectiologie.
Jay Hinton, University of Liverpool.
REFERENCES
- 1.Holmqvist E, Vogel J. 2018. RNA-binding proteins in bacteria. Nat Rev Microbiol 16:601–615. doi: 10.1038/s41579-018-0049-5. [DOI] [PubMed] [Google Scholar]
- 2.Patel JC, Galan JE. 2005. Manipulation of the host actin cytoskeleton by Salmonella–all in the name of entry. Curr Opin Microbiol 8:10–15. doi: 10.1016/j.mib.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Jennings E, Thurston TLM, Holden DW. 2017. Salmonella SPI-2 type III secretion system effectors: molecular mechanisms and physiological consequences. Cell Host Microbe 22:217–231. doi: 10.1016/j.chom.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Matilla MA, Krell T. 2018. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol Rev 42:fux052. doi: 10.1093/femsre/fux052. [DOI] [PubMed] [Google Scholar]
- 5.Holmqvist E, Wright PR, Li L, Bischler T, Barquist L, Reinhardt R, Backofen R, Vogel J. 2016. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J 35:991–1011. doi: 10.15252/embj.201593360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michaux C, Holmqvist E, Vasicek E, Sharan M, Barquist L, Westermann AJ, Gunn JS, Vogel J. 2017. RNA target profiles direct the discovery of virulence functions for the cold-shock proteins CspC and CspE. Proc Natl Acad Sci U S A 114:6824–6829. doi: 10.1073/pnas.1620772114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. 2012. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J 31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez LC. 2011. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol 80:1637–1656. doi: 10.1111/j.1365-2958.2011.07674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sittka A, Pfeiffer V, Tedin K, Vogel J. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol 63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ansong C, Yoon H, Porwollik S, Mottaz-Brewer H, Petritis BO, Jaitly N, Adkins JN, McClelland M, Heffron F, Smith RD. 2009. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation. PLoS One 4:e4809. doi: 10.1371/journal.pone.0004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogel J, Luisi BF. 2011. Hfq and its constellation of RNA. Nat Rev Microbiol 9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santiago-Frangos A, Woodson SA. 2018. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip Rev RNA 9:e1475. doi: 10.1002/wrna.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smirnov A, Förstner KU, Holmqvist E, Otto A, Günster R, Becher D, Reinhardt R, Vogel J. 2016. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci U S A 113:11591–11596. doi: 10.1073/pnas.1609981113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunte HJ, Crane RA, Culham DE, Richmond D, Wood JM. 1999. Protein ProQ influences osmotic activation of compatible solute transporter ProP in Escherichia coli K-12. J Bacteriol 181:1537–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milner JL, Wood JM. 1989. Insertion proQ220::Tn5 alters regulation of proline porter II, a transporter of proline and glycine betaine in Escherichia coli. J Bacteriol 171:947–951. doi: 10.1128/jb.171.2.947-951.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith MN, Crane RA, Keates RA, Wood JM. 2004. Overexpression, purification, and characterization of ProQ, a posttranslational regulator for osmoregulatory transporter ProP of Escherichia coli. Biochemistry 43:12979–12989. doi: 10.1021/bi048561g. [DOI] [PubMed] [Google Scholar]
- 17.Attaiech L, Glover JN, Charpentier X. 2017. RNA chaperones step out of Hfq's shadow. Trends Microbiol 25:247–249. doi: 10.1016/j.tim.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 18. Glover JNM, Chaulk SG, Edwards RA, Arthur D, Lu J, Frost LS. 2015. The FinO family of bacterial RNA chaperones. Plasmid 78:79–87. doi: 10.1016/j.plasmid.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Olejniczak M, Storz G. 2017. ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers? Mol Microbiol 104:905–915. doi: 10.1111/mmi.13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaulk SG, Smith-Frieday MN, Arthur DC, Culham DE, Edwards RA, Soo P, Frost LS, Keates RAB, Glover JNM, Wood JM. 2011. ProQ is an RNA chaperone that controls ProP levels in Escherichia coli. Biochemistry 50:3095–3106. doi: 10.1021/bi101683a. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez GM, Hardwick SW, Maslen SL, Skehel JM, Holmqvist E, Vogel J, Bateman A, Luisi BF, Broadhurst RW. 2017. Structure of the Escherichia coli ProQ RNA-binding protein. RNA 23:696–711. doi: 10.1261/rna.060343.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmqvist E, Li L, Bischler T, Barquist L, Vogel L. 2018. Global maps of ProQ binding in vivo reveal target recognition via RNA structure and stability control at mRNA 3′ ends. Mol Cell 70:971–982.e6. doi: 10.1016/j.molcel.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 23.Smirnov A, Wang C, Drewry LL, Vogel J. 2017. Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J 36:1029–1045. doi: 10.15252/embj.201696127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Attaiech L, Boughammoura A, Brochier-Armanet C, Allatif O, Peillard-Fiorente F, Edwards RA, Omar AR, MacMillan AM, Glover M, Charpentier X. 2016. Silencing of natural transformation by an RNA chaperone and a multitarget small RNA. Proc Natl Acad Sci U S A 113:8813–8818. doi: 10.1073/pnas.1601626113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chao Y, Vogel J. 2010. The role of Hfq in bacterial pathogens. Curr Opin Microbiol 13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Santiviago CA, Reynolds MM, Porwollik S, Choi S-H, Long F, Andrews-Polymenis HL, McClelland M. 2009. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog 5:e1000477. doi: 10.1371/journal.ppat.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhuri RR, Morgan E, Peters SE, Pleasance SJ, Hudson DL, Davies HM, Wang J, van Diemen PM, Buckley AM, Bowen AJ, Pullinger GD, Turner DJ, Langridge GC, Turner AK, Parkhill J, Charles IG, Maskell DJ, Stevens MP. 2013. Comprehensive assignment of roles for Salmonella typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet 9:e1003456. doi: 10.1371/journal.pgen.1003456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papenfort K, Said N, Welsink T, Lucchini S, Hinton JCD, Vogel J. 2009. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol 74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- 29.Westermann AJ, Vogel J. 2018. Host-pathogen transcriptomics by Dual RNA-Seq. Methods Mol Biol 1737:59–75. doi: 10.1007/978-1-4939-7634-8_4. [DOI] [PubMed] [Google Scholar]
- 30.Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Müller L, Reinhardt R, Stadler PF, Vogel J. 2016. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 529:496–501. doi: 10.1038/nature16547. [DOI] [PubMed] [Google Scholar]
- 31.Bruno VM, Hannemann S, Lara-Tejero M, Flavell RA, Kleinstein SH, Galán JE. 2009. Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog 5:e1000538. doi: 10.1371/journal.ppat.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hannemann S, Gao B, Galan JE. 2013. Salmonella modulation of host cell gene expression promotes its intracellular growth. PLoS Pathog 9:e1003668. doi: 10.1371/journal.ppat.1003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. 2007. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5:2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou D, Chen LM, Hernandez L, Shears SB, Galan JE. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol Microbiol 39:248–259. doi: 10.1046/j.1365-2958.2001.02230.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhou D, Galan J. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect 3:1293–1298. doi: 10.1016/S1286-4579(01)01489-7. [DOI] [PubMed] [Google Scholar]
- 37.Mirold S, Ehrbar K, Weissmüller A, Prager R, Tschäpe H, Rüssmann H, Hardt WD. 2001. Salmonella host cell invasion emerged by acquisition of a mosaic of separate genetic elements, including Salmonella pathogenicity island 1 (SPI1), SPI5, and sopE2. J Bacteriol 183:2348–2358. doi: 10.1128/JB.183.7.2348-2358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu Y, Galan JE. 1999. A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 401:293–297. doi: 10.1038/45829. [DOI] [PubMed] [Google Scholar]
- 39.Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, Danino V, Bongaerts RJM, Ahmad N, Rhen M, Hinton JCD. 2008. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol 10:958–984. doi: 10.1111/j.1462-5822.2007.01099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srikumar S, Kröger C, Hébrard M, Colgan A, Owen SV, Sivasankaran SK, Cameron ADS, Hokamp K, Hinton JCD. 2015. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog 11:e1005262. doi: 10.1371/journal.ppat.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortega ÁD, Gonzalo-Asensio J, García-del Portillo F. 2012. Dynamics of Salmonella small RNA expression in non-growing bacteria located inside eukaryotic cells. RNA Biol 9:469–488. doi: 10.4161/rna.19317. [DOI] [PubMed] [Google Scholar]
- 42.Lober S, Jackel D, Kaiser N, Hensel M. 2006. Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int J Med Microbiol 296:435–447. doi: 10.1016/j.ijmm.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Stocker BA, Hoiseth SK, Smith BP. 1983. Aromatic-dependent “Salmonella sp.” as live vaccine in mice and calves. Dev Biol Stand 53:47–54. [PubMed] [Google Scholar]
- 44.Silva IJ, Ortega AD, Viegas SC, Garcia-del Portillo F, Arraiano CM. 2013. An RpoS-dependent sRNA regulates the expression of a chaperone involved in protein folding. RNA 19:1253–1265. doi: 10.1261/rna.039537.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol 11:941–950. doi: 10.1016/S0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 46.Masse E, Vanderpool CK, Gottesman S. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JCD, Vogel J. 2006. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol 62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soncini FC, Garcia Vescovi E, Solomon F, Groisman EA. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol 178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kruger J, Rehmsmeier M. 2006. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arthur JS, Ley SC. 2013. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol 13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 51.Shan L, He P, Sheen J. 2007. Intercepting host MAPK signaling cascades by bacterial type III effectors. Cell Host Microbe 1:167–174. doi: 10.1016/j.chom.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Odendall C, Rolhion N, Förster A, Poh J, Lamont DJ, Liu M, Freemont PS, Catling AD, Holden DW. 2012. The Salmonella kinase SteC targets the MAP kinase MEK to regulate the host actin cytoskeleton. Cell Host Microbe 12:657–668. doi: 10.1016/j.chom.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou J-M, Shao F. 2007. The phosphothreonine lyase activity of a bacterial type III effector family. Science 315:1000–1003. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- 54.Mazurkiewicz P, Thomas J, Thompson JA, Liu M, Arbibe L, Sansonetti P, Holden DW. 2008. SpvC is a Salmonella effector with phosphothreonine lyase activity on host mitogen-activated protein kinases. Mol Microbiol 67:1371–1383. doi: 10.1111/j.1365-2958.2008.06134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin SL, Le TX, Cowen DS. 2003. SptP, a Salmonella typhimurium type III-secreted protein, inhibits the mitogen-activated protein kinase pathway by inhibiting Raf activation. Cell Microbiol 5:267–275. doi: 10.1046/j.1462-5822.2003.t01-1-00274.x. [DOI] [PubMed] [Google Scholar]
- 56.Queiroz MH, Madrid C, Paytubi S, Balsalobre C, Juarez A. 2011. Integration host factor alleviates H-NS silencing of the Salmonella enterica serovar Typhimurium master regulator of SPI1, hilA. Microbiology 157:2504–2514. doi: 10.1099/mic.0.049197-0. [DOI] [PubMed] [Google Scholar]
- 57.Mangan MW, Lucchini S, Danino V, Croinin TO, Hinton JCD, Dorman CJ. 2006. The integration host factor (IHF) integrates stationary-phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Mol Microbiol 59:1831–1847. doi: 10.1111/j.1365-2958.2006.05062.x. [DOI] [PubMed] [Google Scholar]
- 58.Nam D, Choi E, Shin D, Lee EJ. 2016. tRNA(Pro)-mediated downregulation of elongation factor P is required for mgtCBR expression during Salmonella infection. Mol Microbiol 102:221–232. doi: 10.1111/mmi.13454. [DOI] [PubMed] [Google Scholar]
- 59.Sevostyanova A, Groisman EA. 2015. An RNA motif advances transcription by preventing Rho-dependent termination. Proc Natl Acad Sci U S A 112:E6835–E6843. doi: 10.1073/pnas.1515383112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee EJ, Choi J, Groisman EA. 2014. Control of a Salmonella virulence operon by proline-charged tRNA(Pro). Proc Natl Acad Sci U S A 111:3140–3145. doi: 10.1073/pnas.1316209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee EJ, Groisman EA. 2012. Tandem attenuators control expression of the Salmonella mgtCBR virulence operon. Mol Microbiol 86:212–224. doi: 10.1111/j.1365-2958.2012.08188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park SY, Cromie MJ, Lee EJ, Groisman EA. 2010. A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell 142:737–748. doi: 10.1016/j.cell.2010.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee EJ, Groisman EA. 2010. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol Microbiol 76:1020–1033. doi: 10.1111/j.1365-2958.2010.07161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JC. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 65.Wagner EG, Romby P. 2015. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv Genet 90:133–208. doi: 10.1016/bs.adgen.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 66.Nielsen FC, Hansen HT, Christiansen J. 2016. RNA assemblages orchestrate complex cellular processes. Bioessays 38:674–681. doi: 10.1002/bies.201500175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Figueira R, Watson KG, Holden DW, Helaine S. 2013. Identification of Salmonella pathogenicity island-2 type III secretion system effectors involved in intramacrophage replication of S. enterica serovar Typhimurium: implications for rational vaccine design. mBio 4:e00065. doi: 10.1128/mBio.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JCD, Vogel J. 2008. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet 4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vakulskas CA, Potts AH, Babitzke P, Ahmer BM, Romeo T. 2015. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev 79:193–224. doi: 10.1128/MMBR.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sheidy DT, Zielke RA. 2013. Analysis and expansion of the role of the Escherichia coli protein ProQ. PLoS One 8:e79656. doi: 10.1371/journal.pone.0079656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sternberg NL, Maurer R. 1991. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol 204:18–43. doi: 10.1016/0076-6879(91)04004-8. [DOI] [PubMed] [Google Scholar]
- 73.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 74.Forstner KU, Vogel J, Sharma CM. 2014. READemption—a tool for the computational analysis of deep-sequencing-based transcriptome data. Bioinformatics 30:3421–3423. doi: 10.1093/bioinformatics/btu533. [DOI] [PubMed] [Google Scholar]
- 75.McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nicol JW, Helt GA, Blanchard SG, Raja A, Loraine AE. 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gene Ontology Consortium. 2017. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene Ontology: tool for the unification of biology. Nat Genet 25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 81.Zieg J, Simon M. 1980. Analysis of the nucleotide sequence of an invertible controlling element. Proc Natl Acad Sci U S A 77:4196–4200. doi: 10.1073/pnas.77.7.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J. 2007. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol Microbiol 66:1174–1191. doi: 10.1111/j.1365-2958.2007.05991.x. [DOI] [PubMed] [Google Scholar]
- 84.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt W-D. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hara H, Yamamoto Y, Higashitani A, Suzuki H, Nishimura Y. 1991. Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J Bacteriol 173:4799–4813. doi: 10.1128/jb.173.15.4799-4813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baumler AJ, Kusters JG, Stojiljkovic I, Heffron F. 1994. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun 62:1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dar D, Sorek R. 2018. Bacterial noncoding RNAs excised from within protein-coding transcripts. mBio 9:e01730-18. doi: 10.1128/mBio.01730-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Virulence parameters of Salmonella proQ mutants in HeLa cell culture assays. (a) proQ deletion strategy. The 5′ portion of proQ (including its own promoter; red arrow) is deleted from the Salmonella genome, while the 3′ portion is kept as it contains the promoter of the downstream gene prc (as deduced from the respective site in the E. coli genome [85]; blue arrow with the asterisk). The remaining TSS annotations were obtained from reference 64 (black arrows). The read coverage plots show RNA-seq data for two biological replicates of wild-type or ΔproQ Salmonella and confirm the deletion of the respective genomic region. Note that the downstream prc gene is slightly induced in the proQ deletion background. (b) Wild-type Salmonella harboring an empty control vector, a ΔproQ strain with the same empty vector, or a ΔproQ strain overexpressing proQ from that plasmid (strain proQ++) in either the wild-type background (GFP-negative; black) or Ptet::gfp background (green) were grown under SPI-1- or SPI-2-inducing conditions (see Materials and Methods), and the levels of ProQ protein were determined by Western blotting using an antibody against the endogenous protein. GroEL served as a loading control. (c) Assessment of the risk of plasmid loss during infection. HeLa cells were infected with wild-type Salmonella containing an ampicillin resistance-mediating plasmid (MOI of 10) in the presence or absence of ampicillin in the infection medium, and after 24 h, host cells were lysed, and intracellular bacteria were recovered and plated in parallel on LB only or LB-Amp plates. (d) proQ deletion causes a ∼2-fold reduction in invasion of Salmonella into HeLa cells. Salmonella input suspensions were derived from bacteria in late exponential phase (OD600 of 2.0). The fraction of infected cells at 0 h and 24 h p.i. is shown. Significance was evaluated using a two-tailed Student’s t test, and P values of <0.05 were considered statistically significant and are denoted by an asterisk (*). P values of <0.01 were considered very significant (**), and P values of <0.001 were considered extremely significant (***). n.s., not significant. (e) Intracellular replication inside HeLa cells is not affected by proQ deletion or overexpression. In panels d and e, HeLa cells were infected with the indicated Salmonella strains (GFP-expressing background) at an MOI of 5. The proportion of invaded (GFP-positive) cells and the rate of intracellular replication (as deduced from the increase in GFP signal intensity per infected HeLa cell over time) were determined by flow cytometry. Data in panels c to e represent the means ± SD from at least three biological replicate experiments. Download FIG S1, PDF file, 0.1 MB (125.6KB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Assessment of an effect of ProQ on Salmonella virulence in monocyte and macrophage models. (a) Rate of infected (GFP-positive) THP-1 cells at 30 min and 24 h p.i. (MOI of 10) for the indicated Salmonella strains. THP-1 cells were either nondifferentiated (monocytes) or differentiated in vitro by adding phorbol 12-myristate 13-acetate (PMA) into the cell medium 3 days prior to infection (macrophages). Salmonella input suspensions were either derived from exponentially growing cultures (OD600 of 2.0) (exponential) or from overnight cultures opsonized for 20 min in mouse serum (stationary). Significance was evaluated using a two-tailed Student’s t test, and P values of <0.05 were considered statistically significant and are denoted by an asterisk. n.s., not significant. In differentiated THP-1 cells, none of the ProQ-dependent changes were statistically significant. (b) Fold replication of the same strains in the same host cell types between 30 min and 24 h p.i. (MOI of 10). Panels a and b report flow cytometry data (mean ± SD) from at least three biological replicate experiments. Note that for fully differentiated THP-1 cells (but not monocytic THP-1), the rate of Salmonella-containing cells increased over time, because macrophages but not monocytic cells engulf bacteria released from neighboring cells. (c) THP-1 cell death after infection with the indicated Salmonella strains (MOI of 10). Cell death was determined by measuring the rate of lactate dehydrogenase (LDH) released into the culture supernatant at 24 h p.i. The data indicate the means ± SD from at least three biological replicates. Download FIG S2, PDF file, 0.04 MB (46.3KB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Experimental overview of the Dual RNA-seq screen for ProQ-dependent expression changes during Salmonella infection of HeLa cells. (a) Dual RNA-seq pipeline. HeLa cells were infected with wild-type or ΔproQ Salmonella (GFP-expressing strains), and invaded (GFP-positive) cells were enriched by FACS. Total RNA was extracted, and ribosomal transcripts of host and pathogen were depleted prior to sequencing. (b) Comparative Dual RNA-seq outline. RNA samples were taken before infection (0 h; lysate from Salmonella in exponential growth phase after the medium shift from LB to host cell media, artificially mixed with HeLa lysate prior to joint RNA extraction) or at 8 and 16 h after infection (mixed human-Salmonella infection samples). Each two biological replicates were analyzed. (c) Dual RNA-seq mapping statistics. Of the ∼20 million reads per sample that could be aligned to the reference annotations, between 1% (0 h and 8 h) and 2% (16 h) mapped to Salmonella and the remaining 98 or 99% mapped to the human genome. (d) RNA class distribution of reads mapped to the Salmonella genome. (e) RNA class distribution of reads mapped to the human genome. Abbreviations in panels d and e: rRNA, ribosomal RNA; tRNA, transfer RNA; mRNA, messenger RNA; sRNA, bacterial small regulatory RNA; IGRs, intergenic regions; lncRNA, long noncoding RNA; miRNA, microRNA; sn(o)RNA, small nucle(ol)ar RNA; miscRNA, miscellaneous RNA. The percentage values denote the range of the fraction of reads mapping to the corresponding RNA class (relative to the total fraction of reads assigned to the respective organism) for the two replicate experiments. Download FIG S3, PDF file, 0.1 MB (98.5KB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Compiled data sets of the present study. Access table for detailed descriptions. Download Table S1, XLSX file, 23.0 MB (23.6MB, xlsx) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA-seq coverage plots and validation data for ProQ-dependent effects on Salmonella motility, chemotaxis, and the σE response. (a) Read coverage over the flagellar locus of Salmonella and zoom-in on the fliC gene. At the time of infection (0 h), fliC mRNA is downregulated in ΔproQ Salmonella (red) compared to wild-type Salmonella (black). (b) Orientation-specific PCR of the fliC locus of the indicated Salmonella strains using genomic DNA as the template. Information about the invertible region comes from reference 81. While all three strains contain populations of both fliC-ON and fliC-OFF bacteria, the majority express fliC (ON state). Importantly, the ratio (fliC-ON/fliC-OFF) is unchanged in the three strains. (c) Read coverage over the chemotaxis regulon and close-up view of the cheW locus. (d) qRT-PCR data for cheW mRNA. The indicated Salmonella strains were grown in LB to an OD600 of 2.0, total RNA was extracted and served as the template for qRT-PCR measurements. gfp mRNA was the reference transcript. Bars and error bars indicate the means ± SD from four biological replicate experiments. Significance was evaluated using a two-tailed Student’s t test (**, P < 0.01; ***, P < 0.001). (e) Read coverage over the rpoE operon and close-up view of the rpoE gene. (f) qRT-PCR data for rpoE mRNA. Data shown as described above for panel d. (g) Northern blot shows the expression of the σE-dependent sRNAs RybB and MicA upon growth in LB to an OD600 of 2.0 (ESP) or in the minimal SPI-2-inducing medium to an OD600 of 0.3 (in SPI-2), respectively. Since both sRNAs were expressed at low levels in LB at an OD600 of 2.0, the respective membrane was reexposed for an extended time period (shown to the right). 5S rRNA serves as a loading control. Download FIG S4, PDF file, 0.3 MB (315.5KB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA-seq coverage plots and validation data for ProQ-dependent expression changes of Salmonella pathogenicity islands. (a) Read coverage over the SPI-1 locus of Salmonella and zoom-in onto the TSS of the sicP-sptP transcript. TSS annotation (black arrow) obtained from reference 64. (b) qRT-PCR data on the expression of selected Salmonella SPI-1 virulence genes (the sicP-sptP operon and the mRNA of the SPI-1 master regulator, hilD) in proQ-expressing versus non-proQ-expressing strains during the infection of HeLa cells (MOI of 50). gfp mRNA was used as a reference. The plot displays the mean ± SD from three biological replicate experiments. (c) Read coverage over the SPI-2 locus. (d) qRT-PCR data on the expression of selected SPI-2 genes (a gene for a secreted effector, pipB2; the gene for the SPI-2 master regulator, ssrB) in proQ-expressing versus non-proQ-expressing strains during the infection of HeLa cells (MOI of 50). gfp mRNA was the reference. The data are the means ± SD from three biological replicate experiments. Download FIG S5, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of an STnc540 deletion mutant and in silico prediction of target interactions. (a) To generate the ΔSTnc540 strain, the 5′ portion of the sRNA gene was removed, while its terminator was kept. TSS annotations (black arrows) are according to reference 64. (b) Northern blot validation of the successful deletion of STnc540. Salmonella strains were grown in LB to stationary phase (ON) or under the SPI-2-inducing condition, and total RNA samples were analyzed by Northern blotting using probes against the 3′ end of STnc540 or the body of the ihfA mRNA. 5S rRNA serves as a loading control. (c) qRT-PCR for mgtB mRNA at the indicated time points upon the addition of arabinose to strains harboring the long or short STnc540 isoform on the pBAD plasmid relative to the control strain harboring the empty vector. gfp mRNA was the reference. The data are means ± SD from three biological replicate experiments. Significance (*, P value of <0.05) was evaluated using a two-tailed Student’s t test. (d) In silico prediction of RNA-RNA interactions between STnc540 and its putative target mgtCBR. RNAhybrid (49) was used to predict interaction sites. The numbering of nucleotides is relative to the respective TSS of the mRNA or the annotated 5′ end of STnc540, respectively, based on the annotations from reference 64. Download FIG S6, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth of different mutant strains of Salmonella. The indicated strains were grown in LB (left) or minimal SPI-2-inducing medium (right), and OD600 values were measured in 30-min intervals. The data are means ± SD from two biological replicate experiments. Download FIG S7, PDF file, 1.3 MB (1.4MB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
GFP reporter assays for STnc540/ProQ-mediated control of MgtB. Schematic representation (top) and measurement (bottom) of a transcriptional reporter (Pmgt::gfp) and two transcriptional/posttranscriptional reporters (mgtC::gfp and mgtB::gfp) for which the open reading frame of GFP was fused to the mgt locus at the indicated positions. The predicted interaction sites with STnc540 (#1 to #4; see Fig. S6d) are indicated below the schematic representations. These reporter constructs were cotransformed with a constitutive STnc540 (long or short isoform) overexpression plasmid, the ProQ expression plasmid, or the respective vector control into the ΔSTnc540 (a) or ΔSTnc540 ΔproQ (b) Salmonella background. The resulting strains were grown in SPI-2-inducing medium in a 96-well plate for 24 h, and the GFP intensity (as a proxy for MgtC or MgtB levels) was monitored in 10-min intervals. The data show the means ± SD of the GFP intensity normalized against OD595 and relative to the GFP intensity of the same strains harboring a constitutive GFP expression control plasmid (pXG-1) from three biological replicates, each comprising technical triplicates. Download FIG S8, PDF file, 1.6 MB (1.7MB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In vivo phenotype of ΔproQ Salmonella. Streptomycin-pretreated C57BL/6 mice were infected (5 × 107 CFU in total by gavage) with a 1:1 mixture of wild-type Salmonella (Strr) and the indicated isogenic mutant strains (Strr Kanr). The ΔssaV strain has a functional knockout of the T3SS of SPI-2 and was included as a positive control of an attenuated strain. Three days postinfection, the indicated organs were isolated and homogenized, and competitive indices were determined by selective plating. The data stem from five animals per group from two independently performed experiments (single data points are shown, and the geometric mean for a group is indicated by a black bar). Statistical significance was assessed by one-tailed Wilcoxon matched pair rank test, comparing the proQ-deficient strain to its coinfecting wild-type control strain. Statistically significant differences are indicated by asterisks as follows: *, P = 0.0313; **, P values of ≤0.01. Despite the slight induction in basal prc expression in the ΔproQ background (Fig. S1a), we confidently assign the in vivo attenuation of the proQ mutant strain to the absence of this RBP, since prc overexpression would be expected to result in hyperreplication (86). Download FIG S9, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2019 Westermann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE117256. The workflows of the different analyses represented as Shell scripts are deposited at Zenodo (https://doi.org/10.5281/zenodo.1311214).