Abstract
OST1 (open stomata 1) protein kinase plays a central role in regulating freezing tolerance in Arabidopsis; however, the mechanism underlying cold activation of OST1 remains unknown. Here, we report that a plasma membrane‐localized clade‐E growth‐regulating 2 (EGR2) phosphatase interacts with OST1 and inhibits OST1 activity under normal conditions. EGR2 is N‐myristoylated by N‐myristoyltransferase NMT1 at 22°C, which is important for its interaction with OST1. Moreover, myristoylation of EGR2 is required for its function in plant freezing tolerance. Under cold stress, the interaction of EGR2 and NMT1 is attenuated, leading to the suppression of EGR2 myristoylation in plants. Plant newly synthesized unmyristoylated EGR2 has decreased binding ability to OST1 and also interferes with the EGR2‐OST1 interaction under cold stress. Consequently, the EGR2‐mediated inhibition of OST1 activity is released. Consistently, mutations of EGRs cause plant tolerance to freezing, whereas overexpression of EGR2 exhibits decreased freezing tolerance. This study thus unravels a molecular mechanism underlying cold activation of OST1 by membrane‐localized EGR2 and suggests that a myristoyl switch on EGR2 helps plants to adapt to cold stress.
Keywords: Arabidopsis, EGR2 phosphatase, freezing tolerance, myristoylation, OST1 kinase
Subject Categories: Plant Biology; Post-translational Modifications, Proteolysis & Proteomics; Signal Transduction
Introduction
Temperature is a key environmental cue along with sunlight and water that regulates plant growth and development. Extreme temperatures (high or low) adversely affect plant growth and survival, and these conditions are becoming more common due to climate change. Plants have evolved sophisticated mechanisms to respond and adapt to extreme temperatures (Guy, 1990). Cold acclimation is a process by which plants increase their tolerance to frost damage (< 0°C) after exposure to chilling temperatures (> 0°C; Guy, 1990; Thomashow, 1999). C‐repeat (CRT)‐binding factor (CBF)/dehydration‐responsive element (DRE)‐binding protein 1 (DREB1)‐dependent signaling has extensive roles in cold acclimation (Stockinger et al, 1997; Liu et al, 1998; Thomashow, 1999). Cold stress rapidly induces the expression of CBF genes, and their encoding proteins directly activate a set of cold‐regulated (COR) genes, leading to enhanced freezing tolerance (Stockinger et al, 1997; Liu et al, 1998; Thomashow, 1999). Cold‐induced CBF expression is positively regulated by several transcription factors, including ICE1 (inducer of CBF expression 1), CAMTA3 (calmodulin‐binding transcription activator 3), and BZR1 (brassinazole‐resistant 1; Chinnusamy et al, 2003; Doherty et al, 2009; Li et al, 2017b). ICE1 functions as a master regulator of CBF expression (Chinnusamy et al, 2003). ICE1 is targeted for degradation after ubiquitination by HOS1 (high expression of osmotically responsive gene 1; Dong et al, 2006), whereas it is stabilized after sumoylation by SIZ1 (SAP and Miz; Miura et al, 2007). Recent studies showed that ICE1 protein is phosphorylated by protein kinases, including OST1 (open stomata 1) and MPK3/6 in Arabidopsis, and OsMAPK3 in rice (Ding et al, 2015; Li et al, 2017a; Zhang et al, 2017; Zhao et al, 2017; Shi et al, 2018). Under cold conditions, MPK3/6 phosphorylate ICE1 and promote ICE1 degradation in Arabidopsis, while OsMAPK3 phosphorylates and stabilizes OsICE1 in rice (Li et al, 2017a; Zhang et al, 2017; Zhao et al, 2017; Guo et al, 2018). Importantly, ICE1 is stabilized under cold stress after phosphorylation by cold‐activated OST1, thereby promoting plant freezing tolerance (Ding et al, 2015). More recently, OST1 was found to interact with and phosphorylate BTF3 proteins, β‐subunits of a nascent polypeptide‐associated complex (NAC), and enhances their interaction with CBF proteins, thereby stabilizing CBF proteins under cold stress (Ding et al, 2018; Liu et al, 2018). Nevertheless, the molecular mechanism underlying cold activation of OST1 remains unclear.
Protein phosphatases that dephosphorylate phosphorylated proteins cooperate with sensor proteins to regulate stress responses in both eukaryotes and prokaryotes (Fuchs et al, 2013). Type 2C protein phosphatases (PP2Cs) are Ser‐Thr protein phosphatases and are the largest protein phosphatase family in plants. In Bacillus subtilis, energy and nutritional deficiency responses are controlled by the energy sensor RsbQ together with the PP2C RsbP (Martinez et al, 2010). In Arabidopsis thaliana, ABA signaling is negatively regulated by the ABA receptors PYR/PYL/RCAR interacting with PP2Cs of clade A, such as ABI1 and ABI2 (Ma et al, 2009; Park et al, 2009; Yin et al, 2009). In B. subtilis, cold signals are perceived by plasma membrane‐localized histidine kinase DesK, which has a dual role as protein kinase and protein phosphatase (Aguilar et al, 2001; Albanesi et al, 2004, 2009). Under normal temperatures, DesK acts as a phosphatase that removes the phosphoryl group from DesR. Cold temperatures induce conformational changes in the DesK tertiary structure that stimulate its histidine kinase activity, resulting in phosphorylation of the downstream regulator DesR, activation of the target gene Des, and maintenance of membrane fluidity (Albanesi et al, 2009; Cybulski et al, 2015). There is no evidence for a protein with both kinase and phosphatase activities under different stress conditions in plants, so it is possible that phosphatase(s) might cooperatively function with protein kinase(s) for cold signal sensing and signaling.
In this study, we identified a plasma membrane‐localized clade‐E growth‐regulating 2 (EGR2) phosphatase as a negative regulator of plant freezing tolerance by inhibiting OST1 kinase activity. At normal temperature, EGR2 is associated with and myristoylated at the N‐terminus by N‐myristoyltransferase NMT1, which is important for EGR2 to interact with and inhibit OST1. Cold stress induces the accumulation of newly synthesized unmyristoylated EGR2 by compromising the EGR2‐NMT1 interaction, leading to the decreased interaction of EGR2 and OST1, which consequently activates OST1 under cold stress. These results provide novel insights into the mechanism of cold activation of OST1 by membrane‐localized EGR2.
Results
Cold‐induced OST1 activity is independent of ABA receptors
We previously reported that OST1 protein kinase, a core component in ABA signaling, is activated by cold and positively regulate plant freezing tolerance (Ding et al, 2015). However, the mechanism underlying cold‐mediated OST1 activation is hitherto unknown. It has been reported that the PYR/PYL (pyrabactin resistance/pyrabactin resistance like) ABA receptors are prominent regulators of OST1 activation in ABA signaling (Ma et al, 2009; Park et al, 2009). This prompted us to investigate whether ABA receptors are involved in cold‐mediated OST1 activation by performing in‐gel kinase assays using total proteins of wild‐type plants and ABA receptor mutants. OST1 kinase activity was induced within 30 min of cold treatment and peaked after 2 h in wild‐type plants (Figs 1A and B, and EV1A). These results are consistent with our previous results using OST1‐overexpressing transgenic plants and Arabidopsis protoplasts (Ding et al, 2015). Moreover, we observed that OST1 was obviously activated by cold stress in ABA receptor mutants, including pyr1 pyl1,4, pyr1 pyl1,2,4, pyr1 pyl4,5,8, and pyr1 pyl1,4,5,8 (Figs 1A and B, and EV1A). As a control, ABA‐induced OST1 activity was drastically decreased in the pyr1 pyl1,4 triple mutant after ABA treatment (Fig EV1B), in agreement with previous reports (Ma et al, 2009; Park et al, 2009). These combined results suggest that cold activation of OST1 is independent of ABA receptors. Considering that OST1 kinase activity was significantly activated at 2 h after cold treatment, we selected 2 h as a time point for further studies.
Figure 1. Cold‐induced OST1 kinase activity is independent of ABA and ABA receptors.

-
A, BIn‐gel kinase assay of OST1 activity in ABA receptor mutants under cold stress. Ten‐day‐old wild‐type, ost1‐3, pyr1 pyl1,4, pyr1 pyl1,2,4, pyr1 pyl4,5,8 and pyr1 pyl1,4,5,8 mutants were treated at 4°C for 2 h. Total protein extracts were prepared and separated on SDS–PAGE gel containing 0.1 mg/ml GST‐∆ABF2 as a substrate, and incubated with 60 μCi of [γ‐32P]ATP. Representative pictures are shown in (A), and relative kinase activity is shown in (B).
-
C, DIn‐gel kinase assay of OST1 activity in ABI1‐OE14 and abi1 abi2hab1 mutants under cold stress. Representative pictures are shown in (C), and relative kinase activity is shown in (D).
Figure EV1. Cold‐induced OST1 kinase activity is independent of ABA receptors.
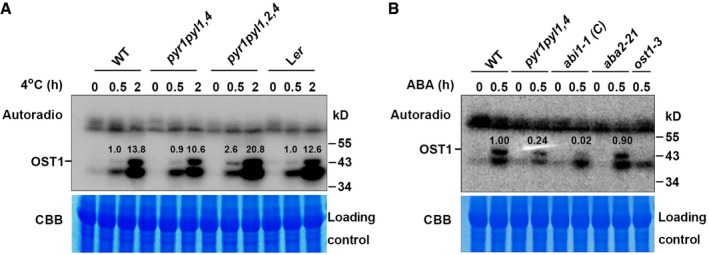
- In‐gel kinase assay of OST1 in wild‐type (Col and Ler), pyr1 pyl1,4, and pyr1 pyl1,2,4 mutants under cold stress. Total proteins prepared from 10‐day‐old seedlings were treated with 4°C for 0, 0.5, and 2 h, separated on a 10% SDS–PAGE gel containing 0.1 mg/ml GST‐∆ABF2 as substrate, and incubated with 60 μCi [γ‐32P]ATP. Top, gel autoradiograph; bottom, Coomassie Brilliant Blue (CBB) staining of RuBisCO large subunit was used as a loading control. The ratio of band intensity of OST1 to RuBisCO in wild type with cold treatment for 0.5 h was set to 1.0.
- In‐gel kinase assay of OST1 in the presence of ABA. Total proteins prepared from 10‐day‐old wild‐type, pyr1 pyl1,4, abi1‐1 (C), aba2‐21 and ost1‐3 mutants with or without 50 μM ABA treatment for 30 min and subjected to in‐gel kinase assay using GST‐∆ABF2 as substrate. Top, gel autoradiograph; bottom, CBB staining. The ratio of band intensity of OST1 to RuBisCO in wild type with ABA treatment for 0.5 h was set to 1.00.
Source data are available online for this figure.
Next, we determined OST1 kinase activity in ABI1‐overexpressing (ABI1‐OE14) transgenic plants (Ding et al, 2015), and abi1 abi2 hab1 triple mutant after cold treatment using in‐gel kinase assay. The OST1 kinase activity was decreased approximately 30% in ABI1‐OE14, whereas enhanced twofold in abi1 abi2 hab1 triple mutant (Fig 1C and D), suggesting that clade‐A PP2Cs contribute to inhibiting OST1 kinase activity in response to cold stress, which is consistent with our previous study (Ding et al, 2015). However, it is worthy to note that OST1 kinase activity was not fully inhibited in ABI1‐overexpressing plants (Fig 1C and D), implying that unknown protein phosphatase(s) might be also responsible for modulating cold‐mediated OST1 activity.
EGR protein phosphatases interact with OST1
To identify protein phosphatases modulating OST1 activity under cold stress, we analyzed RNA‐Seq data from our previous study (Jia et al, 2016). Nine genes encoding PP2C‐type phosphatases were up‐ or down‐regulated by cold treatment (Appendix Fig S1A).
Yeast two‐hybrid analyses were performed to test whether these phosphatases interacted with OST1 and EGR2 (clade‐E growth‐regulating 2; At5g27930) was found to be an OST1‐interacting protein (Fig 2A). In vitro pull‐down assays showed that MBP‐His‐EGR2, but not MBP‐His, interacted with GST‐OST1 (Fig 2B). The interaction of OST1 and EGR2 was further verified in N. benthamiana leaves by co‐immunoprecipitation (co‐IP) assay (Fig 2C). EGR2 protein was previously shown to be localized at the plasma membrane (Bhaskara et al, 2017). We performed bimolecular fluorescence complementation (BiFC) assay and found that the interaction between OST1 and EGR2 occurred at the plasma membrane in protoplast cells (Fig 2D). In addition, we created Super:EGR2‐Myc construct and generated EGR2‐Myc‐overexpressing transgenic plants. Mass spectrometry analysis with these transgenic plants identified OST1 as an EGR2‐interacting partner (Appendix Table S1), further supporting that EGR2 interacts with OST1 in planta.
Figure 2. OST1 interacts with EGR2 and inhibits OST1 activity under cold stress.
-
AInteraction of OST1 and EGR2 in yeast. Yeast cells harboring different vector combinations were grown on SC/−Leu/−Trp medium for 3 days or SC/−Leu/−Trp/−His/−Ade medium for 6 days. OST1‐BD/AD and BD/EGR2‐AD were used as negative controls.
-
BOST1 interacts with EGR2 in vitro. Purified recombinant MBP‐His‐EGR2 or MBP‐His proteins from E. coli were immunoprecipitated with MBP beads and then incubated with purified recombinant GST‐OST1. Precipitated proteins were detected with anti‐His and anti‐GST antibodies.
-
CCo‐IP assay of OST1 with EGR2 in vivo. 35S:HF‐OST1/Super:EGR2‐Myc or 35S:HF‐OST1/Super:Myc was expressed in N. benthamiana leaves. Total proteins were immunoprecipitated with anti‐Myc agarose beads, and the co‐immunoprecipitation products were subjected to immunoblot analysis. EGR2‐Myc and HF‐OST1 were detected with anti‐Myc and anti‐HA antibodies, respectively.
-
DBimolecular fluorescence complementation (BiFC) assay. Wild‐type Arabidopsis protoplasts transformed with OST1‐YFP C and EGR2‐YFP N were incubated for 18 h. The interaction signal was detected by confocal microscopy. The combinations of OST1‐YFPC/GUS‐YFPN and GUS‐YFPC/EGR2‐YFPN were used as negative controls. Scale bars: 100 μm.
-
EEGR2 inhibits OST1 kinase activity in vitro. Proteins were incubated with 1 μCi [γ‐32P]ATP in kinase reaction buffer for 30 min at 30°C and then separated by SDS–PAGE. Top, autoradiograph; bottom, CBB staining.
-
F, GIn‐gel assay of OST1 activity in 10‐day‐old wild‐type, egr1 and egr2 single mutants, and the egr1 egr2 double mutant under cold stress.
-
H, IIn‐gel assay of OST1 activity in 10‐day‐old wild‐type and EGR2‐overexpressing plants under cold stress.
EGR2 was reported as a negative regulator of plant growth during drought stress, and it has two close homologs, EGR1 (At3g05640) and EGR3 (At3g16800; Bhaskara et al, 2017). We found that the expression of EGR1 and EGR3, like EGR2, was up‐regulated by cold stress (Appendix Fig S1B). Furthermore, EGR1 and EGR3 also interacted with OST1 in vivo (Appendix Fig S1C and D).
EGRs inhibit OST1 kinase activity
We next examined whether EGR2 was involved in regulating OST1 activity using in vitro phosphorylation assay. OST1 has auto‐phosphorylation activity and phosphorylates its substrate ABF2 protein (Fig 2E). Interestingly, the auto‐phosphorylation and kinase activities of OST1 were largely abolished when it was incubated with EGR2 (Fig 2E). A previous study showed that a conserved amino acid Gly180 of ABI1 or Gly168 of ABI2 located closely to Mg2+ coordination center that is important for their phosphatase activity (Vlad et al, 2009). This Gly was found in a conserved motif “DGHG*” in type 2C phosphatases in Arabidopsis and other plant species (Robert et al, 2006). We analyzed EGR2 amino acid sequence and found the conserved Gly100 in EGR2 which might be involved in its dephosphorylation activity. We therefore mutated Gly100 to Asp100 (EGR2G100D) to mimic its inactive form and performed in vitro phosphorylation assay. As expected, EGR2G100D failed to repress OST1 auto‐phosphorylation or kinase activity (Fig 2E). These results indicate that EGR2 dephosphorylates OST1 and thus represses OST1 kinase activity in vitro.
A previous study showed that PYL4 inhibits activities of clade‐A PP2C phosphatases in ABA‐dependent and ABA‐independent manners (Hao et al, 2011). Therefore, we examined whether the phosphatase activity of clade‐E phosphatase EGR2 was also regulated by ABA‐PYL4 in an in vitro phosphorylation assay. Consistent with the previous study (Hao et al, 2011), ABA‐PYL4 repressed ABI1‐mediated OST1 dephosphorylation in vitro (Fig EV2A). However, ABA‐PYL4 could not repress EGR2‐mediated dephosphorylation of OST1 (Fig EV2B). These results suggest that EGR2‐mediated inhibition of OST1 activity is independent of the ABA receptor PYL4.
Figure EV2. The inhibition of OST1 activity by EGR2 is independent of ABA‐PYL4.
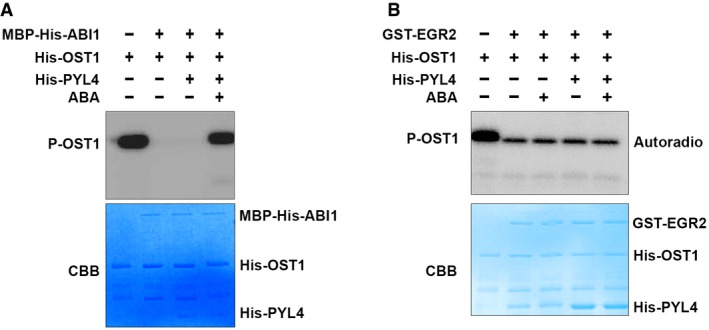
- The effect of ABA‐PYL4 on ABI1‐mediated inhibition of OST1 in an in vitro kinase assay.
- The effect of ABA‐PYL4 on EGR2‐mediated inhibition of OST1 in an in vitro kinase assay.The combination proteins were incubated with 1 μCi [γ‐32P]ATP in kinase reaction buffer for 30 min at 30°C and then separated by 10% SDS–PAGE. Top, autoradiograph; bottom, CBB staining.
Source data are available online for this figure.
To determine whether OST1 activity was regulated by EGRs in planta, we obtained egr1‐1, egr1‐2, egr2‐1, egr2‐3, egr3‐1, and egr3‐2 single mutants and subsequently generated the egr1‐2 egr2‐3 (egr1 egr2) double mutant. Semi‐qPCR analysis showed that egr1‐1 was a knockdown mutant, whereas the others were knockout mutants (Appendix Fig S2A and B). Next, we performed in‐gel kinase assays using total proteins prepared from wild‐type, egr1, egr2, and egr3 single mutants, and egr1 egr2 double mutant subjected to 4°C treatment for 2 h. The OST1 activity was consistently enhanced in egr1, egr2, and egr3 single mutants compared with the wild‐type plants under cold stress (Figs 2G and H, and Appendix Fig S2C and D). Strikingly, cold‐induced OST1 activity was much higher in the egr1 egr2 double mutant than in the single mutants (Fig 2F and G), suggesting that EGR1 and EGR2 function redundantly in regulating OST1 activity. We also examined the OST1 activity in EGR2‐overexpressing transgenic plants (Appendix Fig S2E) and observed that cold‐induced OST1 activity was obviously decreased compared to the wild type (Fig 2H and I). These results suggest that EGRs negatively regulate OST1 activity under cold stress.
SnRK2.2 and SnRK2.3 are homologs of OST1 (Fujii et al, 2011). Thus, we tested whether EGR2 interacted with SnRK2.2 and SnRK2.3 and inhibited their kinase activity. We found that EGR2 also interacted with SnRK2.2 and SnRK2.3 proteins (Appendix Fig S2F and G). In vitro kinase assay proved that SnRK2.2 or SnRK2.3 kinase activity was also inhibited by EGR2 (Appendix Fig S2H).
OST1 was reported to stabilize ICE1 by phosphorylating ICE1 under cold conditions (Ding et al, 2015), which tempts us to ask whether ICE1 stability was affected by EGR2. HF‐ICE1 proteins were expressed in wild‐type, egr2‐3, and egr2‐3 egr1‐2 protoplasts and treated with or without 4°C for 3 h. We found that ICE1 protein was much more stable in the egr2‐3 and egr1 egr2 mutants than that in the wild type after cold treatment (Appendix Fig S3A–D). Therefore, EGR1/2 promotes ICE1 degradation by repressing OST1 activity under cold stress.
Next, we dissected whether EGR2 affected the subcellular localization of OST1. Cell fractionation assays were performed, and the results showed that OST1 was localized at the cytosol and nucleus at both 22 and 4°C in wild‐type and egr2‐3 mutant plant (Appendix Fig S4A and B). Interestingly, we found that a very little amount of OST1 protein was localized at the plasma membrane in wild‐type and egr2‐3 plants (Appendix Fig S4C and D). Because no transmembrane domain was found in OST1, it is possible that OST1 is bound to the PM by binding with PM‐localized proteins, including SLAC1, KAT1, RbohF (Geiger et al, 2009; Sato et al, 2009; Sirichandra et al, 2009), and EGR2. These combined results suggest that the activity of OST1 around PM is inhibited by EGR2 under normal temperature.
EGRs negatively regulate plant freezing tolerance
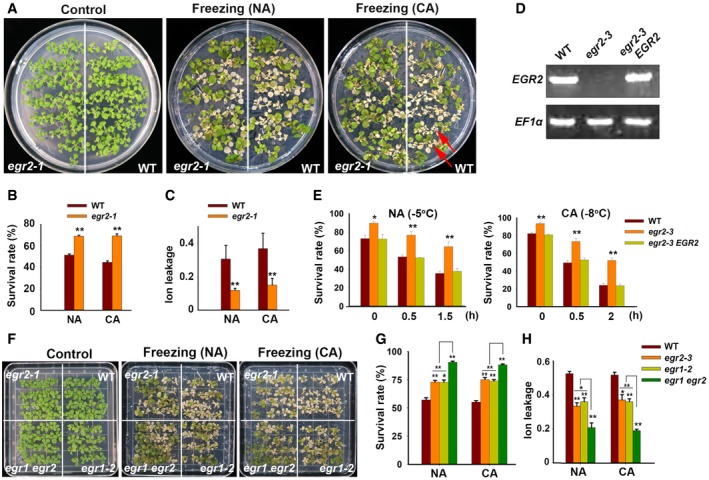
We next assessed whether EGRs were involved in regulating plant freezing tolerance. Under non‐acclimated (NA) and cold‐acclimated (CA) conditions, the egr2‐1 and egr2‐3 mutants displayed enhanced freezing tolerance with higher survival rates than the wild‐type plants (Figs 3A and B, and EV3A and B). Ion leakage, an indicator of stress‐induced PM injury, was dramatically lower in egr2 mutants than in wild‐type plants after freezing treatment (Figs 3C and EV3C). The freezing tolerance of egr2‐3 was fully complemented by Myc‐tagged EGR2 genomic fragments driven by the native promoter (EGR2:EGR2‐Myc; Figs 3A–C, and EV3D and E). In addition, the egr1‐1, egr1‐2, egr3‐1, and egr3‐2 mutants also displayed increased freezing tolerance compared with the wild type (Appendix Fig S5A–L). We also analyzed the freezing phenotypes of egr1 egr2 double mutants and found that egr1 egr2 double‐mutant plants showed much more freezing tolerance than egr1‐2 and egr2‐3 single mutants under NA and CA conditions, indicating that EGR2 and EGR1 function redundantly in regulating plant freezing tolerance (Fig EV3F–H). Next, we analyzed the freezing tolerance of EGR2‐overexpressing transgenic plants, which had lower freezing tolerance than the wild type with or without cold acclimation (Fig 3D). The survival rates of these transgenic plants were lower, whereas ion leakage was higher than the wild type after freezing treatment (Fig 3E and F). These results indicate that EGRs negatively regulate plant freezing tolerance.
Figure 3. Freezing phenotypes of egr2‐3 mutant and EGR2‐overexpressing plants.
-
A–CFreezing phenotype (A), survival rate (B), and ion leakage (C) of the egr2‐3 mutant and egr2/EGR2:EGR2‐Myc complementation lines under non‐acclimated (NA) and acclimated (CA; 4 days at 4°C) conditions. Two‐week‐old seedlings grown on MS medium containing 0.8% agar were treated at −5°C for 0.5 h (NA) or −8°C for 0.5 h (CA).
-
D–FFreezing phenotype (D), survival rate (E), and ion leakage (F) of EGR2‐overexpressing plants. Two‐week‐old Super:EGR2‐Myc plants were treated as described in (A–C).
-
G–JExpression of CBFs (G, I) and their target genes (H, J) in egr2‐3 mutant and EGR2‐overexpressing plants. Two‐week‐old seedlings were treated at 4°C for the indicated period and subjected to qRT–PCR analysis.
Figure EV3. Mutation of EGR2 results in enhanced freezing tolerance.
-
A–CFreezing phenotype (A), survival rate (B), and ion leakage (C) of the egr2‐1 mutant. Two‐week‐old seedlings grown on MS medium containing 0.8% agar were treated at −5°C for 0.5 h (NA) or −8°C for 0.5 h (CA). Red arrows show dead plants.
-
DExpression of EGR2 in 2‐week‐old wild‐type, egr2‐3, and egr2/EGR2:EGR2‐Myc complementation plants. EF1α was used as a loading control.
-
ESurvival rates of wild‐type, egr2‐3, and EGR2 egr2‐3.
-
F–HFreezing phenotype (F), survival rate (G) and ion leakage (H) of wild‐type, egr2‐3, egr1‐2, and egr1 egr2 mutants under NA and CA conditions. Two‐week‐old seedlings grown on MS medium containing 0.8% agar were treated at −5°C for 0.5 h (NA) or −8°C for 0.5 h (CA).
We previously showed that OST1‐mediated plant freezing tolerance is dependent on the CBF signaling pathway (Ding et al, 2015). Therefore, we examined whether EGRs regulate the expression of CBFs and their target COR genes by qRT–PCR. The basal and cold‐induced CBF expression was much higher in egr1 and egr2 mutants than that in the wild type (Figs 3G and Appendix Fig S5M). Consistently, expression of the CBF target genes, such as COR15A, KIN1, and COR47, was significantly higher in egr1 and egr2 mutants than that in the wild type before and after cold treatment (Figs 3H and Appendix Fig S5N). By contrast, basal expression levels and cold‐induced CBF and COR expression in Super:EGR2‐Myc transgenic plants were much lower than those in the wild type (Fig 3I and J). These combined results demonstrate that EGRs inhibit OST1 activity and negatively regulate the CBF pathway and freezing tolerance.
EGR2 is myristoylated in plants
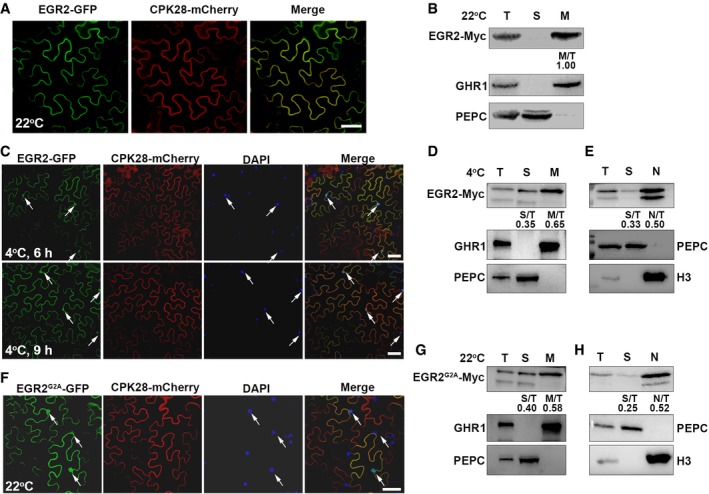
To gain insight into the mechanism underlying EGR2 regulation in plant freezing tolerance, we first examined whether the subcellular localization of EGR2 is affected by cold stress. EGR2 was co‐localized with a plasma membrane‐localized protein CPK28 (Monaghan et al, 2014) at the plasma membrane of N. benthamiana leaf pavement cells, and it was detected only in the PM fraction (Fig EV4A and B), which is consistent with a previous study (Bhaskara et al, 2017). However, confocal images and cell fractionation assays showed that EGR2 was not only localized at the PM, but also in the cytosol and nucleus under cold stress (Fig EV4C–E). It is worthy to note that EGR2 harbors a predicated N‐terminal myristoylation motif (MGXXXS/T; Fig 4A), which is usually important for the PM localization of proteins (Hannoush, 2015). To determine whether possible myristoylation of EGR2 is responsible for its PM localization, we mutated the second amino acid Gly of EGR2 to Ala (G2A) that inhibits N‐myristoylation (Ishitani et al, 2000), and found that EGR2G2A was localized at the PM, cytosol as well as nucleus (Fig EV4F–H), which is similar to the localization of EGR2 at 4°C (Fig EV4C and E). These results indicate that the PM localization of EGR2 is affected by low temperature.
Figure EV4. Characterization of EGR2.
-
A–F(A, C, F) Subcellular localization of EGR2 at 22°C (A) and 4°C (C), and EGR2G2A at 22°C (F). Super:EGR2‐GFP/CPK28‐mCherry or Super:EGR2G2A‐GFP/CPK28‐mCherry proteins were co‐expressed in N. benthamiana leaves for 48 h and then treated at 22 or 4°C for additional 6 or 9 h. GFP fluorescence was detected by confocal microscopy. N. benthamiana leaves were stained with DAPI (C, F). Scale bars: 20 μm (A) and 75 μm (C, F). In (C, F), the arrows indicate nuclei of N. benthamiana leaf pavement cells. (B, D, E) Cell fractionation assays of EGR2 under normal and cold conditions. Proteins were prepared from 2‐week‐old EGR2:EGR2‐Myc transgenic plants treated at 4°C for 0 h (B) and 6 h (D, E). EGR2 was detected with anti‐Myc antibody.
-
G, HCell fractionation assays of EGR2G2A. Proteins were prepared from 2‐week‐old EGR2:EGR2 G2A ‐Myc transgenic plants. EGR2G2A was detected with anti‐Myc antibody.
Figure 4. N‐myristoylated modification of EGR2 is important for its function under cold stress.

-
APossible conserved N‐myristoylated motif in EGR2.
-
BN‐myristoylation of EGR2 and EGR2G2A in N. benthamiana leaves. Protein extracts prepared from N. benthamiana leaves expressing EGR2‐Myc or EGR2G2A‐Myc were immunoprecipitated by anti‐Myc agarose beads. Precipitated proteins were detected by anti‐Myc and anti‐myristic acid antibodies.
-
CDiagram of the protocol for detecting EGR2 N‐myristoylation in planta.
-
DImmunoblot analysis showing that the total level of myristoylated proteins in wild‐type plants. Total proteins were prepared from 2‐week‐old wild‐type seedlings treated with or without 80 μM myristic acid alkyne at 22°C for 72 h and then subjected to click reaction with the biotin azide. The reaction was protected from light at room temperature for 3 h. The products were detected by anti‐streptavidin–HRP antibody.
-
EN‐myristoylation of EGR2 in plants. Total proteins were prepared from 2‐week‐old EGR2‐Myc seedlings treated as described in (D). The products were immunoprecipitated by anti‐Myc agarose beads and detected by anti‐streptavidin–HRP antibody. EGR2‐Myc detected by anti‐Myc antibody was used as a loading control.
-
FImmunoblot assay of myristoylated EGR2 under cold stress. Total proteins were prepared from EGR2‐Myc plants treated with 4°C for the indicated period and immunoprecipitated by anti‐Myc agarose beads. Precipitated proteins were detected by anti‐Myc and anti‐myristic acid antibodies. The ratio of band intensity detected by anti‐myristic acid antibody to anti‐Myc antibody without cold treatment was set to 1.00. Representative pictures are shown (left), and quantitative analysis is shown (right). Each bar represents the mean ± SEM of three biological repeats. **P < 0.01 (two‐tailed t‐test).
-
GExpression of EGR2 in egr2‐3 EGR2:EGR2 G2A ‐Myc complementation transgenic plants. Total RNAs were extracted from 2‐week‐old seedlings and subjected to RT–PCR analysis. EF1α was used as a loading control.
-
H, IFreezing tolerance assays of the egr2‐3 mutant and EGR2:EGR2 G2A ‐Myc transgenic plants in egr2‐3 mutant. Two‐week‐old seedlings grown on MS medium containing 0.8% agar at 22°C were treated at −5°C for 0–1.5 h (NA) or −8°C for 0–1.5 h (CA). Representative photographs (−5°C, 0.5 h; −8°C, 0.5 h) are shown in (H), and survival rate is shown in (I). Data shown in (I) are mean values ± SEM of three biological replicates, each of which has three technical replicates. Asterisks indicate significant differences compared with wild type for the same treatment (*P < 0.05, **P < 0.01, two‐tailed t‐test); n. s., not significant.
Source data are available online for this figure.
The above results imply that the possible myristoylation of EGR2 might be regulated by low temperatures. To examine whether EGR2 has myristoylated modification in planta, we expressed EGR2‐Myc and EGR2 G2A ‐Myc in N. benthamiana leaves and performed an immunoblot analysis with anti‐myristic acid antibody, which specifically conjugates myristic acid (Hannoush, 2015). After immunoprecipitated EGR2‐Myc or EGR2G2A‐Myc with anti‐Myc beads, myristoylated EGR2 was detected with the anti‐myristic acid antibody in N. benthamiana leaves expressing EGR2‐Myc, but not in leaves expressing EGR2 G2A ‐Myc (Fig 4B). This result indicates that EGR2 is myristoylated in planta. To further deeply dissect myristoylated modification of EGR2, we performed a click reaction assay which has been used in mammals and Caenorhabditis elegans (Fig 4C; Tang & Han, 2017). Two‐week‐old wild‐type seedlings grown on MS medium were shifted on MS medium containing 0 or 80 μM myristic acid alkyne for additional 72 h at 22°C, and total proteins were extracted, followed by subjected to click reaction with the biotin azide at 22°C for 3 h under dark conditions. The reaction products were then separated on SDS–PAGE gel and detected with anti‐streptavidin–HRP antibody. The myristoylated proteins were detected in plant materials treated with myristic acid alkyne (Fig 4D), suggesting that this method is suitable for detecting myristoylated proteins in plants. Using this approach, we prepared proteins from 2‐week‐old EGR2‐Myc transgenic plants treated with the above conditions, immunoprecipitated EGR2 using anti‐Myc agarose beads, and performed immunoblot analysis with anti‐streptavidin–HRP antibody. The myristoylated EGR2 was detected in EGR2‐Myc transgenic plants treated with myristic acid alkyne (Fig 4E). Next, we tested whether N‐myristoylated EGR2 was affected by low temperatures. To this end, total proteins were prepared from 2‐week‐old EGR2‐Myc seedlings treated with 4°C for 0, 3, and 6 h and immunoprecipitated by anti‐Myc agarose beads. The precipitants were immunoblotted with anti‐myristic acid antibody. The myristoylated form of EGR2 was detected before cold treatment, and this modification form was gradually decreased after cold treatment (Fig 4F). Together, these in vivo data demonstrate that EGR2 is N‐myristoylated in planta, which is suppressed by cold stress.
To further explore the function of EGR2 N‐myristoylation in plant freezing tolerance, we generated transgenic plants expressing EGR2:EGR2 G2A ‐Myc in egr2‐3 mutant (Fig 4G). EGR2 fully rescued the freezing tolerance phenotype of egr2‐3 (Fig 3A–C). However, EGR2 G2A ‐Myc failed to complement the enhanced freezing tolerance of egr2‐3 (Fig 4H and I), suggesting that N‐myristoylation of EGR2 is required for its function in regulating plant freezing tolerance.
Cold stress disrupts the interaction between NMT1 and EGR2
Next, we explored why N‐myristoylated EGR2 was decreased under cold treatment. To this end, we determined whether EGR2 interacted with NMT1 (At5g57020), a major N‐myristoyltransferase responsible for N‐myristoylation in plants (Pierre et al, 2007). In a pull‐down assay, MBP‐His‐EGR2, but not MBP‐His, pulled down GST‐NMT1 (Fig 5A), indicating that NMT1 physically interacts with EGR2 in vitro. Moreover, NMT1 co‐localized with EGR2 at the PM of leaf pavement cells in N. benthamiana (Fig 5B). The interaction between NMT1 and EGR2 was further verified by a co‐IP assay (Fig 5C). Furthermore, we found that the EGR2‐NMT1 interaction was severely disrupted after cold treatment (Fig 5C and D). To further assess the interaction of EGR2 and NMT1 under cold stress, we immunoprecipitated EGR2 from 2‐week‐old EGR2‐Myc transgenic plants treated at 4°C for 6 h and performed an in vitro pull‐down assay. The amount of GST‐NMT1 pulled down by EGR2 (4°C) was much lower than that by EGR2 (22°C; Fig 5E). All these results indicate that low temperature attenuates the formation of NMT1‐EGR2 protein complex.
Figure 5. Cold stress disrupts the interaction of NMT1 and EGR2.

-
AIn vitro pull‐down assay showing the interaction of NMT1 and EGR2. Recombinant GST‐NMT1 was incubated with immunoprecipitated MBP‐His‐EGR2 or MBP‐His at 4°C for 2 h. Anti‐GST and anti‐MBP were used to detect NMT1, EGR2, and MBP‐His, respectively.
-
BCo‐localization of NMT1 and EGR2 in N. benthamiana leaves. Scale bar: 50 μm.
-
C, DEffect of cold stress on the interaction of EGR2 and NMT1 determined by co‐IP assay. Arabidopsis protoplasts expressing HF/NMT1‐Myc and HF‐EGR2/NMT1‐Myc were treated with or without 4°C for 2 h. Total proteins were immunoprecipitated by anti‐HA agarose beads. Anti‐Myc and anti‐HA were used to detect NMT1‐Myc and HF‐EGR2, respectively. In (C), relative band intensity is shown below the co‐IP blot. In (D), each bar represents the mean ± SEM of three biological experiments (**P < 0.01, two‐tailed t‐test).
-
EIn vitro pull‐down assay showing decreased interaction intensity of EGR2 (4°C) and NMT1 compared with that of EGR2 (22°C) and NMT1. Total proteins prepared from 2‐week‐old EGR2‐Myc plants grown at 4°C for 0 and 6 h were immunoprecipitated by anti‐Myc agarose beads and incubated with GST‐NMT1 in pull‐down buffer for 2 h at 4°C. Anti‐Myc and anti‐GST antibodies were used to detect EGR2 and NMT1 proteins. Relative band intensity is shown below the output blot.
-
FExpression of NMT1 in NMT1‐Myc‐overexpressing plants. EF1α was used as a loading control.
-
G, HFreezing phenotype (G) and survival rate (H) of NMT1‐Myc‐overexpressing transgenic plants. Two‐week‐old Super:NMT1‐Myc plants were treated at −5°C (NA) or −8°C for 0.5 h (CA). In (H), each bar represents the mean ± SEM of three biological experiments, and asterisks indicate significant differences compared with the wild type for the same treatment (*P < 0.05, two‐tailed t‐test).
-
I, JOST1 kinase activity in NMT1‐Myc‐overexpressing plants under cold stress. Top, autoradiograph; bottom, CBB staining. Representative pictures are shown in (I), and the relative kinase activity is shown in (J). In (I), the ratio of band intensity of OST1 to RuBisCO in wild type with cold treatment for 2 h was set to 1.00. In (J), each bar represents the mean ± SEM of three independent experiments (**P < 0.01, two‐tailed t‐test).
Source data are available online for this figure.
Mutation of NMT1 causes late‐embryo abortion (Pierre et al, 2007), which is impossible for freezing phenotype and related biochemical analyses. Thus, we generated transgenic plants overexpressing NMT1 (Super:NMT1‐Myc) to assess the function of NMT1 in plant freezing tolerance. However, the T1 transgenic seedlings highly expressing NMT1 were seedling lethal. Therefore, we selected two independent lines expressing NMT1 at relatively low levels, which grew normally on soil and did not show obvious morphological defects under normal growth conditions (Fig 5F), for further study. They consistently displayed reduced freezing tolerance compared to the wild type (Fig 5G and H). These plants also displayed reduced OST1 activity after cold treatment (Fig 5I and J). These combined data demonstrate that the EGR2‐NMT1 interaction is impaired under low temperatures, which consequently results in the OST1 activation.
Cold stress attenuates the interaction of OST1 and EGR2
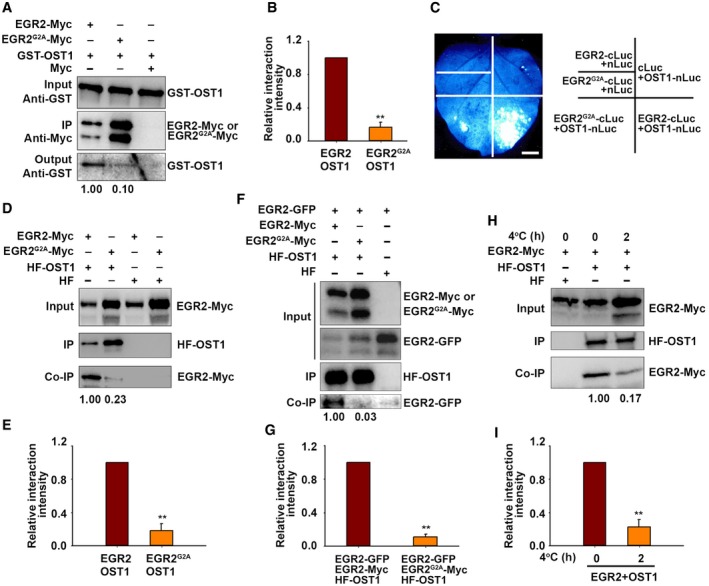
N‐myristoylation is not only important for protein–membrane interaction, but also important for protein–protein interaction (Mclaughlin & Aderem, 1995; Wright et al, 2010). To test whether N‐myristoylation of EGR2 is important for its interaction with OST1, we performed a pull‐down assay using EGR2‐Myc and ERG2G2A‐Myc proteins extracted from EGR2:EGR2‐Myc and EGR2:ERG2 G2A ‐Myc stable transgenic plants. Immunoprecipitated EGR2‐Myc or ERG2G2A‐Myc was incubated with GST‐OST1 recombinant protein and then subjected to the immunoblot analysis with anti‐Myc and anti‐GST antibodies, respectively. Interestingly, the interaction of EGR2G2A‐OST1 was much weaker than that of EGR2‐OST1 (Fig 6A and B). The split luciferase complementation assay in N. benthamiana leaves also showed that the interaction of EGR2G2A‐OST1 is weak compared to that of EGR2‐OST1 (Fig 6C). A co‐IP assay further verified this result (Fig 6D and E). These results suggest that N‐myristoylation of EGR2 is important for its interaction with OST1. Interestingly, we also found that the interaction between EGR2 and OST1 was disrupted by EGR2G2A in a co‐IP assay (Figure 6F and G). Low temperature inhibited EGR2 myristoylation in planta (Fig 4F); therefore, we performed a co‐IP assay to examine whether the OST1‐EGR2 interaction was affected by low temperature. OST1 was associated with EGR2 in planta at 22°C, and this association was significantly disrupted by cold treatment (Fig 6H and I). Next, we performed a pull‐down assay to detect the interaction of EGR2 and OST1 under normal and cold conditions. EGR2‐Myc proteins were extracted from 2‐week‐old EGR2‐Myc stable transgenic plants treated at 4°C for 0 and 6 h and then incubated with GST‐OST1. We found that the interaction of EGR2 (4°C)‐OST1 was obviously reduced compared to that of EGR2 (22°C)‐OST1 (Fig EV5A).
Figure 6. Cold stress attenuates the interaction between EGR2 and OST1.
-
A, BPull‐down assay. Total proteins prepared from EGR2‐Myc or EGR2 G2A ‐Myc transgenic plants were immunoprecipitated with anti‐Myc agarose beads and then incubated with GST‐OST1. Anti‐Myc and anti‐GST antibody were used to detect GST‐OST1 and EGE2‐Myc. Representative pictures are shown in (A). Relative interaction is shown in (B).
-
CLCI of EGR2 and EGR2G2A with OST1 in N. benthamiana leaves. EGR2‐cLuc/OST1‐nLuc and EGR2 G2A ‐cLuc/OST1‐nLuc were expressed in N. benthamiana leaves for 48 h. EGR2‐cLuc/nLuc, EGR2 G2A ‐cLuc/nLuc, and cLuc/OST1‐nLuc were used as negative controls. Scale bar: 2 cm.
-
D, ECo‐IP assay. EGR2‐Myc/HF‐OST1, EGR2 G2A ‐Myc/HF‐OST1, HF/EGR2‐Myc, and HF/EGR2 G2A ‐Myc were expressed in N. benthamiana leaves. Total proteins were immunoprecipitated by anti‐HA agarose beads. Anti‐HA and anti‐Myc antibodies were used to detect HF‐OST1, EGR2‐Myc, and EGR2G2A‐Myc, respectively. Representative pictures are shown in (D), and relative interaction is shown in (E).
-
F, GEffect of EGR2G2A on the interaction of OST1 and EGR2 in Arabidopsis protoplasts expressing HF‐OST1/EGR2‐GFP/EGR2‐Myc, HF‐OST1/EGR2‐GFP/EGR2 G2A ‐Myc, or HF‐OST1/EGR2‐GFP. Total proteins were immunoprecipitated by anti‐HA agarose beads. Anti‐Myc, anti‐GFP, and anti‐HA antibodies were used to detect EGR2‐Myc, EGR2‐GFP, and HF‐OST1, respectively. Representative pictures are shown in (F), and relative interaction is shown in (G).
-
H, IEffect of cold stress on the interaction of OST1 and EGR2. Arabidopsis protoplasts expressing EGR2‐Myc/HF‐OST1 and EGR2‐Myc/HF were treated at 4°C for 0 and 2 h. Total proteins were immunoprecipitated by anti‐HA agarose beads. The co‐immunoprecipitated products were detected by anti‐HA and anti‐Myc antibodies, respectively. Representative pictures are shown in (H), and relative interaction is shown in (I).
Figure EV5. Cold stress activates OST1 kinase activity by attenuating the interaction between EGR2 and OST1.

-
AIn vitro pull‐down assay showing decreased interaction intensity of EGR2 (4°C) and OST1 compared to that of EGR2 (22°C) and OST1. Total proteins prepared from 2‐week‐old EGR2‐Myc plants treated at 4°C for 0 and 6 h were immunoprecipitated with anti‐Myc agarose beads and incubated with GST‐OST1. EGR2 and OST1 proteins were detected by anti‐Myc and anti‐GST antibodies. Relative band intensity is shown below the output blot.
-
B, CIn vitro kinase assay showing decreased activity of EGR2 (4°C; B) and EGR2G2A (C) on OST1 activation. Total proteins extracted from 2‐week‐old EGR2‐Myc seedlings treated at 4°C for 6 h (B) or from EGR2 G2A ‐Myc (C) were immunoprecipitated with anti‐Myc agarose beads and incubated with His‐OST1 in the presence of 1 μCi [γ‐32P]ATP. MBP (myelin basic protein) was used a substrate. Relative OST1 kinase activity was calculated and shown below the relative bands.
-
DIn‐gel kinase assay of OST1 activity in EGR2 G2A ‐Myc‐overexpressing transgenic lines. Total proteins prepared from 10‐day‐old seedlings were treated at 4°C for 2 h and subjected to in‐gel kinase assay using GST‐∆ABF2 as substrate. Top, autoradiograph; bottom, CBB staining. The ratio of band intensity of OST1 to RuBisCO in wild type with cold treatment for 2 h was set to 1.00. Each bar represents the mean ± SEM of three independent experiments (*P < 0.05, **P < 0.01, two‐tailed t‐test).
-
ESurvival rate of the transgenic plants overexpressing EGR2 G2A ‐Myc. Two‐week‐old seedlings grown on MS medium containing 0.8% agar were treated at −5°C for 0.5 h (NA) or −8°C (CA). Each bar represents the mean ± SEM of three biological repeats. Asterisks indicate significant differences compared with the wild type for the same treatment (*P < 0.05, two‐tailed t‐test).
-
FExpression of EGR2 G2A in EGR2 G2A ‐Myc‐overexpressing plants. Actin2 was used as a loading control.
Source data are available online for this figure.
Above results indicate that cold‐treated EGR2 and EGR2G2A have decreased interaction ability to OST1, which prompted us to ask whether these forms of EGR2 has less activity on repressing OST1 kinase activity. To this end, we extracted EGR2‐Myc (22°C) or EGR2‐Myc (4°C) protein from 2‐week‐old EGR2‐Myc transgenic plants treated at 4°C for 0 and 6 h and performed in vitro kinase assays. The results showed that OST1 kinase activity was repressed by both EGR2 (22°C) and EGR2 (4°C); however, EGR2 (4°C) had less capacity on repressing OST1 activity than EGR2 (22°C; Fig EV5B). Next, we examined the effect of EGR2G2A on OST1 activity in vitro using immunoprecipitated EGR2G2A‐Myc from EGR2 G2A ‐Myc transgenic plants. Similar to the effect of EGR2 (4°C) on OST1, the inhibition of EGR2G2A on OST1 activity was much lower than EGR2 (Fig EV5C). Intriguingly, we also found that transgenic plants overexpressing EGR2 G2A ‐Myc showed enhanced freezing tolerance and OST1 activity compared to the wild type (Fig EV5D–F), indicating that EGR2G2A could prevent the inhibition of OST1 by myristoylated EGR2 in vivo. These combined results suggest that plants mainly synthesize unmyristoylated EGR2 under cold stress, which disrupts the formation of EGR2‐OST1 protein complex, thereby activating OST1 kinase activity.
Genetic interaction of OST1 and EGR2
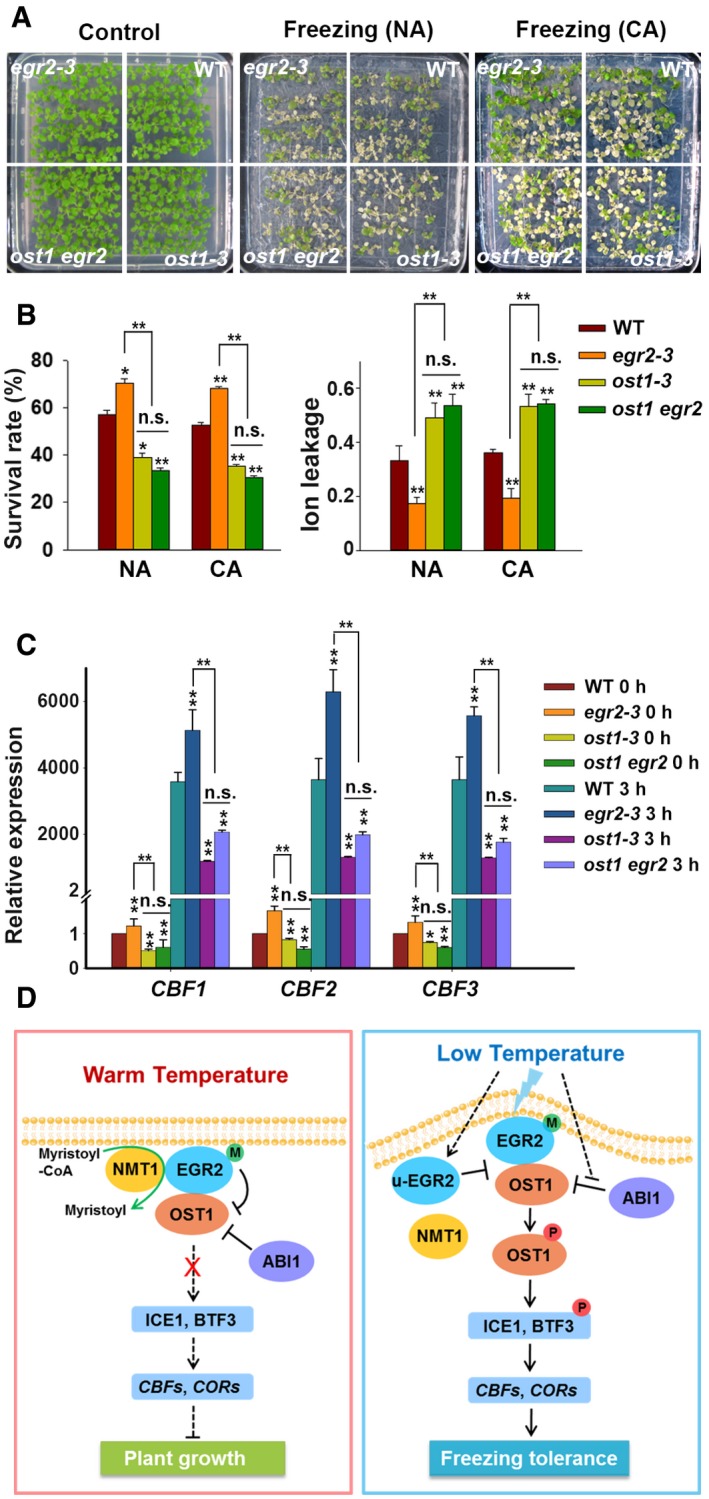
To further determine the genetic interaction between OST1 and EGR2, we crossed ost1‐3 with egr2‐3 to generate the ost1 egr2 double mutant. The ost1‐3 mutant displayed reduced freezing tolerance, whereas the egr2 mutant displayed enhanced freezing tolerance (Fig 7A). The ost1 egr2 double mutant phenocopied the ost1 single mutant in terms of survival rate and ion leakage (Fig 7B).
Figure 7. Genetic Interaction of OST1 and EGR2 .
-
A, BFreezing phenotype (A), survival rate and ion leakage (B) of wild‐type, ost1‐3, egr2‐3, and ost1 egr2 mutants. Two‐week‐old seedlings grown on MS medium were treated at −5°C (NA) or −8°C for 0.5 h (CA).
-
CExpression of CBFs in wild‐type, ost1‐3, egr2‐3, and ost1 egr2‐3 plants under cold stress. Two‐week‐old seedlings grown at 22°C were treated at 4°C for 0 and 3 h.
-
DModel of EGR2‐OST1 modulation of cold stress responses in Arabidopsis. At warm temperature (22°C), EGR2 can be modified by myristoylation, which allows it to recognize OST1 and inhibit its activity to ensure plant growth. Under cold stress (4°C), the interaction of NMT1 and EGR2 is decreased, thereby causing the accumulation of unmyristoylated EGR2, which shows reduced interaction with OST1 and disrupts the interaction of EGR2‐OST1 as well. This ultimately leads to activating OST1 protein kinases and enhancing plant freezing tolerance. Clade‐A PP2Cs, such as ABI1, also partially contributes to regulating OST1 activity in response to cold stress. EGR2‐M and u‐EGR2 represent myristoylated and unmyristoylated EGR2, respectively.
We also evaluated the expression levels of CBFs in wild‐type, ost1‐3, egr2‐3, and ost1 egr2 plants. The expression of CBFs in ost1 egr2 resembled ost1, both much lower than the wild type before and after cold treatment (Fig 7C). These combined results indicate that EGR2 acts upstream of OST1 to negatively regulate CBF expression and freezing tolerance.
Discussion
We previously identified OST1 as a key protein kinase that positively regulates plant freezing tolerance (Ding et al, 2015). In the present study, we found that EGR2 interacts with OST1 and inhibits OST1 kinase activity in vitro and in vivo at 22°C. EGR2 has N‐myristoylated modification catalyzed by Arabidopsis N‐myristoyltransferase NMT1 under normal conditions, which promotes EGR2‐OST1 protein complex formation. Under cold stress, the interaction of NMT1 and EGR2 is compromised, causing the accumulation of newly synthesized unmyristoylated EGR2, which dissociates from OST1 and competes for the interaction of myristoylated EGR2 and OST1. As a result, OST1 is activated, thereby promoting CBF expression and freezing tolerance in Arabidopsis (Fig 7D).
In both eukaryotes and prokaryotes, PP2Cs usually cooperate with sensor proteins to respond to changes in environmental cues throughout the life cycle (Fuchs et al, 2013). Energy sensor RsbQ and the PP2C RsbP regulate energy and nutritional response in B. subtilis (Martinez et al, 2010). Dynamic regulation of clade‐A PP2Cs with ABA receptors PYR/PYL/RCAR and ABA is required for downstream protein kinase activation to confer plant ABA response (Ma et al, 2009; Park et al, 2009; Yin et al, 2009). In the present study, we found that PP2C‐type clade‐E phosphatase EGR2 negatively regulates plant freezing tolerance by repressing OST1 kinase activity. Although this negative regulation is repressed by low temperature, expression of EGR2 gene is up‐regulated by low temperature (Appendix Fig S1A). This phenomenon is reminiscent of ABA signaling, in which group A PP2Cs is induced by ABA, but their ability for repressing OST1 activity is inhibited by ABA (Fujii et al, 2009; Nakashima & Yamaguchi‐Shinozaki, 2013). It appears a feedback regulation in plant responses to adverse conditions. Despite the induction of EGR2 gene, cold‐induced unmyristoylated EGR2 is mainly synthesized, which shows low binding ability to OST1 and thus releases the inhibition of EGR2 on OST1 under cold conditions.
Clade‐A PP2Cs, such as ABI1 and ABI2, are also responsible for repressing cold‐induced OST1 activity (Ding et al, 2015; Fig 1C and D). In the ABA signaling pathway, the function of ABI1 on inhibiting OST1 kinase activity is repressed by ABA and ABA receptors (Fujii et al, 2009). However, we showed that cold‐activated OST1 activity is not dependent on ABA and ABA receptors (Ding et al, 2015; Fig 1A and B). Thus, it is unlikely that the release of OST1 by ABI1 is coupled with ABA or ABA receptors under cold stress. The mechanism of OST1 activation mediated by ABI1 under cold stress needs further investigation. It is noteworthy that cold‐induction of OST1 activity is approximately ninefold in egr1 egr2 double mutant, whereas it is reduced to half level in EGR2‐overexpressing lines when compared to the WT in in‐gel kinase assay (Fig 2F–I). In contrast, OST1 activity is 30% less in ABI1‐overexpressing line than the WT but twofold in abi1 abi2 hab1 mutant compared to WT (Fig 1C and D). These results suggest that clade‐E PP2C EGRs and clade‐A PP2Cs in parallel or coordinately regulate OST1 activity at/around the PM, and in the cytosol and nucleus, respectively. Compared to ABI1/2, EGRs play a predominant role to modulate OST1 activity in response to cold stress.
Intriguingly, we found that the EGR2 phosphatase is localized at the PM under warm temperatures, but at the PM, cytosol, and nucleus under cold stress (Fig EV4). Lipid modification of proteins, such as myristoylation and palmitoylation, is important for PM localization in plants (Hannoush, 2015). We found that EGR2 is N‐myristoylated in planta, and it is partially responsible for the PM localization of EGR2 (Figs 4 and EV4). Moreover, N‐myristoylation is also important for protein–protein interactions (Mclaughlin & Aderem, 1995; Wright et al, 2010). It is possible that N‐myristoylation on EGR2 may be involved in its interaction with other proteins. Interestingly, N‐myristoylation of EGR2 is important for its interaction with OST1 in plants. It is likely that protein conformation of myristoylated EGR2 is suitable for recognizing its target proteins. This possibility was hypothesized from some examples in animals. For instance, activation of ADP ribosylation factor (ARF) GTPase in animals requires its membrane localization conferred by N‐myristoylation, and the protein conformation of ARFs allows ARF‐GTP to be an active form that has increased affinity for multiple effectors, thereby initiating some cellular and physiological processes (Goldberg, 1998; Wright et al, 2010). More importantly, EGR2‐NMT1 interaction is weakened under cold stress, which consequently leads to production of more unmyristoylated EGR2 in plants (Fig 5). The possible conformation change from myristoylated to unmyristoylated form renders attenuated association of EGR2 and OST1, thereby releasing OST1 inhibition under cold stress. It is worthy to note that EGR2 purified from E. coli (in which myristoylated modification does not exist) also interacts with OST1. Given that the interaction of EGR2G2A (purified from plants) and OST1 is not fully dissociated, the myristoylated modification on EGR2 is likely sufficient but unnecessary for its interaction with OST1. However, we cannot rule out the other possibility: Protein conformation of EGR2 for recognizing its substrates in E. coli and plants is different.
Our result showed that N‐myristoylated EGR2 is gradually decreased after 3 h and 6 h of cold treatment (Fig 4), which activates OST1 followed by stabilizing ICE1. Therefore, it is expected that CBF expression still keeps high level at 6 h after cold treatment. However, the expression of CBF genes peaks at 3 h after cold treatment and gradually reduces later. This phenomenon supports the notion that in addition to OST1‐ICE1 module as positive regulators, some other negative regulators are involved in regulating CBF gene expression, such as PIFs (Lee & Thomashow, 2012; Jiang et al, 2017), MYB15 (Agarwal et al, 2006), and EIN3 (Shi et al, 2012), so that plants adjust an appropriate cold response to balance plant growth and cold tolerance.
PM‐localized proteins are involved in sensing and transducing signals into the cell, thereby activating subsequent cellular responses (de Mendoza, 2014). In Oryza sativa (rice), the COLD1 protein located at the PM and endoplasmic reticulum membrane may sense cold signal and activate chilling tolerance responses (Ma et al, 2015). In B. subtilis, the PM‐localized protein histidine kinase DesK functions as a cold sensor with characteristic bifunctional enzyme activities (containing both kinase and phosphatase activities; Albanesi et al, 2009; Cybulski et al, 2015). DesK protein structures differ under different temperature conditions; these structures correspond with its bifunctional enzyme activities (Cybulski et al, 2015). We hypothesized that higher plants might have evolved a specialized protein or protein complex that regulates responses to extreme environmental temperatures, in a manner similar to DesK in B. subtilis. N‐myristoylation is an evolutionarily conserved mechanism of protein post‐translational modification (Farazi et al, 2001). The myristoyl group of protein fosters protein–protein and protein–membrane interactions and stabilizes protein conformation (Mclaughlin & Aderem, 1995; Wright et al, 2010). This so‐called myristoyl switch makes it an essential feature of many cellular processes and signal transduction pathways, such as proliferation, differentiation, and disease resistance in human (Wright et al, 2010). Thus, we postulate that myristoyl switch on the PM‐localized EGR2 may be involved in cold perception and transduction of plants. Under normal temperatures, myristoylated EGR2 interacts with and dephosphorylates OST1, which keeps the stress response at “off” state, ensuring plant growth and development. Under cold conditions, N‐myristoylation of EGR2 is at “off” state, which fosters activation of OST1‐mediated cold stress response, leading to enhanced freezing tolerance in plants (Fig 7D).
Materials and Methods
Plants materials and growth conditions
The Arabidopsis thaliana Col‐0 ecotype was used in this study, and the plants were grown on soil or Murashige and Skoog (MS) medium (Sigma‐Aldrich) containing 0.8% agar and 2% sugar under long‐day conditions (16‐h light/8‐h dark) at 22°C. The mutants ost1‐3/snrk2.6 (Salk_008068; Ding et al, 2015), egr1‐1 (Salk_011589), egr1‐2 (GK‐013B05), egr2‐1 (Salk_048861), egr2‐3 (Salk_0424824), egr3‐1 (Salk_061302), and egr3‐2 (Salk_132380) were obtained from the Arabidopsis Biological Resource Center.
Plasmid construction and plant transformation
EGR2 cDNA was amplified and cloned into pSuper1300 (Ni et al, 1995; Yang et al, 2010), pGEX4T‐1, pMalC2, and pGADT7 vectors to obtain Super:EGR2‐Myc, Super:EGR2‐GFP, GST/MBP‐His‐EGR2, EGR2‐pGADT constructs, respectively.
EGR2 promoter together with genomic DNA amplified by PCR was fused with GFP or Myc in the pCAMBIA1300 vector to generate EGR2:EGR2‐Myc vector.
Mutated forms of EGR2 were obtained by site‐directed mutagenesis using EGR2 genomic DNA as templates, and then amplified by PCR. They were cloned into pSuper1300 or pCAMBIA1300 vectors to generate Super:EGR2 G2A ‐Myc, Super:EGR2 G2A ‐GFP and EGR2:EGR2 G2A ‐Myc vectors.
EGR1 and EGR3 cDNAs were amplified and cloned into pSuper1300 vector to generate Super:EGR1‐Myc and Super:EGR3‐Myc.
NMT1 cDNA was amplified and cloned into pSuper1300 and pGEX4T‐1 vectors to generate Super:NMT1‐Myc and GST‐NMT1 constructs, respectively.
SnRK2.2 and SnRK2.3 cDNAs were amplified and cloned into pCM1307 and pGEX4T‐1 vectors to generate HF‐SnRK2.2/HF‐SnRK2.3 and GST‐SnRK2.2/GST‐SnRK2.3 constructs, respectively.
OST1 cDNA was amplified and cloned into pCM1307 vector to generate HF‐OST1 construct, respectively. All primers used in vector construction are listed in Appendix Table S2.
All transgenic plants were generated through Agrobacterium‐mediated transformation by floral dip (Clough & Bent, 1998), and selected by hygromycin B. T4 homozygous transgenic plants were used in this study.
Freezing tolerance and ion leakage assays
The freezing tolerance was performed as described (Ding et al, 2015). Briefly, plants were grown on MS plates containing 0.8% agar and 2% sugar at 22°C for 2 weeks and transferred to 4°C chamber for 4 days as cold acclimation, or directly subjected to freezing treatment in a freezer chamber (RUMED4501). The freezing program was set at 0°C and dropped 1°C/h to desired temperatures. After freezing treatment, the seedlings were transferred to 4°C for 12 h at dark condition and then put into normal conditions for recovery for 3 days and the number of seedlings that generated new leaves was counted for survival rate. After freezing treatment, the injured seedlings were subjected to the ion leakage assay as described (Ding et al, 2015).
Gene expression assays
Total RNA was extracted from 14‐day‐old seedlings with or without cold treatment using TRI Reagent (Thermo Fisher Scientific) and reverse‐transcribed using the M‐MLV Reverse Transcriptase (Promega). Quantitative real‐time PCR was performed using Applied Biosystems 7500 Real‐Time PCR system with SYBR Premix Ex Taq (Takara). The expression of various genes was calculated as previously described (Shi et al, 2012). The expression of actin2 was used as an internal control. The primers used for gene expression were listed in Appendix Table S2.
Protein expression and purification from E. coli
To obtain purified EGR2 and OST1 proteins, His‐OST1, MBP‐His‐EGR2, GST‐OST1, GST‐SnRK2.2/GST‐SnRK2.3, and GST‐EGR2 plasmids were transformed into E. coli (BL21), and then induced by 0.5 mM IPTG at 18°C for 12 h. The expressed recombinant proteins were affinity‐purified using nickel nitriloacetic acid and glutathione‐linked resins according to the protocols (GE). The purified proteins were used in pull‐down and in vitro protein kinase assays.
Yeast two‐hybrid assays
The yeast two‐hybrid assay was performed as described (Ding et al, 2015). In brief, OST1‐pGBKT7 and EGR2‐pGADT7, OST1‐pGBKT7 and pGADT7 or pGBKT7 and EGR2‐pGADT7 were transformed into yeast strain AH109. The yeast cells were grown on SC/–Leu/−Trp (Clontech, #630417; 3 days) or SC/−Leu/−Trp/−His/−Ade medium (Clontech, #630428) for 6 days.
In vitro pull‐down assays
For pull‐down assay, 5 μg purified MBP‐His‐EGR2 or MBP‐His proteins was incubated with MBP agarose beads (GE Healthcare, #71502076‐EG) in PBS buffer containing 0.1% NP‐40 (pull‐down buffer) for 2 h at 4°C and then washed by PBS buffer for three times. The immunoprecipitated MBP‐His‐EGR2 or MBP‐His was incubated with 0.5 μg GST‐OST1 for 2 h at 4°C in pull‐down buffer. The proteins were washed by PBS buffer for five times and separated by SDS–PAGE. Anti‐His (Beijing Protein Innovation, #AbM59012‐18‐PU) and anti‐GST antibodies (Beijing Protein Innovation, #AbM59001‐2H5‐PU) were used to detect MBP‐His‐EGR2, MBP‐His, and GST‐OST1, respectively.
For detecting the interaction of NMT1 and EGR2, 0.5 μg GST‐NMT1 incubated with the immunoprecipitated MBP‐His‐EGR2 or MBP‐His (5 μg) by MBP agarose beads (GE Healthcare, #71502076‐EG). The proteins were washed by PBS buffer for five times and separated by SDS–PAGE. Anti‐His (Beijing Protein Innovation, #AbM59012‐18‐PU) and anti‐GST antibodies (Beijing Protein Innovation, #AbM59001‐2H5‐PU) were used to detect MBP‐His‐EGR2, MBP‐His, and GST‐NMT1, respectively.
To dissect the interaction of SnRK2.2 and EGR2 or SnRK2.3 and EGR2, the immunoprecipitated MBP‐His‐EGR2 or MBP‐His (5 μg) by MBP agarose beads (GE Healthcare, #71502076‐EG) was incubated with 0.5 μg GST‐SnRK2.2 or GST‐SnRK2.3 in pull‐down buffer for 2 h. The precipitants were washed with PBS buffer for five times and then separated by SDS–PAGE. EGR2, SnRK2.2, and SnRK2.3 were detected with anti‐His (Beijing Protein Innovation, #AbM59012‐18‐PU) and anti‐GST antibodies (Beijing Protein Innovation, #AbM59001‐2H5‐PU).
To analyze the interaction of EGR2G2A and OST1, immunoprecipitated EGR2G2A protein prepared from 2‐week‐old EGR2G2A‐Myc transgenic plants was incubated with 1 μg GST‐OST1 in pull‐down buffer for 2 h. After five times washing with PBS buffer, the proteins were separated by SDS–PAGE and detected by anti‐Myc (Sigma‐Aldrich, #M4439) and anti‐GST (Beijing Protein Innovation, #AbM59001‐2H5‐PU) antibodies.
To detect the interaction of EGR2 (4°C) and OST1, EGR2‐Myc (4°C) protein extracted from 2‐week‐old EGR2‐Myc plants treated at 4°C for 6 h was immunoprecipitated by anti‐Myc agarose beads for 2 h at 4°C and then incubated with 1 μg GST‐OST1 in pull‐down buffer for additional 2 h at 4°C. Immunoprecipitated proteins were washed five times with PBS buffer and then separated by SDS–PAGE. Anti‐Myc (Sigma‐Aldrich, #M4439) and anti‐GST (Beijing Protein Innovation, #AbM59001‐2H5‐PU) antibodies were used to detect EGR2 and OST1 proteins. The same method was used to examine the interaction of EGR2 (4°C) and NMT1.
Co‐IP assays
Co‐IP assay was performed as previously described (Ding et al, 2015). In brief, the total proteins were extracted from N. benthamiana leaves expressing 35S:HF‐OST1/Super:EGR2‐Myc, or 35S:HF‐OST1/Super:Myc constructs with protein extraction buffer (2 mM EDTA pH 8.0, 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% NP‐40, and 0.1% Triton X‐100) and then incubated with anti‐Myc agarose (Sigma‐Aldrich, #A7470) for 2 h at 4°C. After washing by extraction buffer for five times, the co‐immunoprecipitated products were separated by SDS–PAGE and detected with anti‐Myc (Sigma‐Aldrich, #M4439) and anti‐HA (Sigma‐Aldrich, #H3663) antibodies. The same method was used to examine the interaction of EGR1/OST1 and EGR3/OST1.
For the NMT1 and EGR2 interaction assay, proteins were prepared from Arabidopsis protoplasts expressing Super:NMT1‐Myc/35S:HF‐EGR2 and Super:NMT1‐Myc/35S:HF with or without cold treatment for 2 h. The proteins were extracted and incubated by anti‐HA agarose at 4°C for 2 h and detected with anti‐Myc (Sigma‐Aldrich, #M4439) and anti‐HA antibodies (Sigma‐Aldrich, #H3663).
For the interaction of OST1 and EGR2 under cold stress, 35S:HF‐OST1/Super:EGR2‐Myc or 35S:HF/SuperEGR2‐Myc plasmids were transformed into Arabidopsis protoplasts and expressed for 16 h and then treated at 4°C for 2 h. Proteins were extracted and incubated by anti‐HA agarose (Sigma‐Aldrich, #A2095) at 4°C for 2 h and detected with anti‐HA (Sigma‐Aldrich, #H3663) and anti‐Myc antibodies (Sigma‐Aldrich, #M4439).
For the interaction of SnRK2.2 and EGR2, HF‐SnRK2.2/EGR2‐Myc or HF/EGR2‐Myc proteins were expressed in N. benthamiana leaves for 48 h. Total proteins were extracted with protein extraction buffer and incubated with anti‐HA agarose (Sigma‐Aldrich, #A2095) at 4°C for 2 h. The immunoprecipitated products were separated by SDS–PAGE and detected by anti‐HA (Sigma‐Aldrich, #H3663) and anti‐Myc (Sigma‐Aldrich, #M4439) antibodies. The same method was used to examine the interaction between SnRK2.3 and EGR2.
Protein kinase assays
In vitro kinase assay was performed as previously described (Ding et al, 2015). Purified recombinant protein combinations (His‐OST1/MBP‐His‐EGR2, His‐OST1/MBP‐His) were incubated with kinase reaction buffer containing 20 mM MgCl2, 20 mM Tris–HCl pH 7.5, 1 mM DTT, 50 μM ATP, and 1 μCi [γ‐32P]ATP at 30°C for 30 min and then heated at 100°C for 5 min with 5× loading buffer. The proteins were separated by SDS–PAGE and detected by Typhoon 9410 imager.
For in‐gel kinase assay, total proteins were extracted from 10‐day‐old seedlings grown at 22°C at long‐day conditions with or without cold treatment with protein extraction buffer containing 5 mM EDTA, 5 mM EGTA, 5 mM DTT, 25 mM NaF, 1 mM Na3VO4, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail (Roche), and 50 mM HEPES‐KOH, pH 7.5. Proteins were separated on a SDS–PAGE gel containing 0.1 mg/ml GST‐∆ABF2 substrate. After washing by washing buffer (1 mM DTT, 5 mM NaF, 0.1 mM Na3VO4, 0.5 mg/ml BSA, 0.1% Triton X‐100, and 25 mM Tris–HCl, pH 7.5) for three times, 20 min each, the gel was renatured with buffer containing 2 mM DTT, 5 mM NaF, 0.1 mM Na3VO4, and 25 mM Tris–HCl, pH 7.5, for 1, 12, and 1 h at 4°C. Then, the gel was incubated with kinase reaction buffer (2 mM EGTA, 12 mM MgCl2, 1 mM DTT, 0.1 mM Na3VO4, and 25 mM HEPES‐KOH, pH 7.5) for 30 min, and then incubated with reaction buffer supplemented with 60 μCi [γ‐32P]ATP and 9 μl 1 mM cold ATP at room temperature for 1.5 h. The gel was washed by 5% TCA and 1% sodium pyrophosphate five times for 30 min each. The signal was detected by Typhoon 9410 imager.
Cell fractionation assays
The details for plasma membrane fractionation were performed as described (Liu et al, 2017) with some modifications. Briefly, total proteins were prepared from 2‐week‐old seedlings of EGR2:EGR2‐Myc or EGR2:EGR2 G2A ‐Myc treated at 4°C for 0 and 6 h with 200 μl protein extraction buffer and filtered through Miracloth (Calbiochem) and 20 μl solution was saved as total protein fraction. The remainder was centrifuged at 5,000 g for 5 min to remove organelles and debris. Supernatants were centrifuged at 100,000 g for 1.5 h at 4°C to separate soluble and membrane fractions. Supernatants were used for the soluble sample. Membrane pellets were resolved in 180 μl protein extraction buffer. Anti‐Myc (Sigma‐Aldrich, #M4439) was used to detect EGR2 and EGR2G2A proteins. Anti‐PEPC (Agrisera, #AS09 458; RRID: AB_1312) and anti‐GHR1 (produced from Shanghai Youke Biotech Co., LTD) antibodies were used to detect the cytosolic and plasma membrane proteins.
The procedure for nucleus/cytosolic proteins fractionation assays was followed according to the protocol of CelLytic PN Isolation/Extraction kit (Sigma‐Aldrich, #CELLYTPN1). Briefly, total proteins were prepared from 2‐week‐old seedlings of EGR2:EGR2‐Myc or EGR2:EGR2 G2A ‐Myc treated with or without low temperature 400 μl protein extraction buffer and incubated on ice for 20 min and filtered through Miracloth (Calbiochem), and 40 μl solution was saved as total protein. The protein was centrifuged at 12,000 g for 10 min, and supernatant was transferred into other 1.5‐ml tube as soluble fraction. The sediment was washed by protein extraction buffer for five times, five min each, and then resolved by 40 μl nuclear protein extraction buffer. Anti‐OST1 (produced from Shanghai Youke Biotech Co., LTD) was used to detect OST1 protein. Anti‐PEPC (Agrisera, #AS09 458; RRID: AB_1312) and anti‐H3 (Millipore, #05‐499; RRID: AB_2787688) antibodies were used to detect the cytosolic and nucleus proteins, respectively.
Analysis of EGR2 myristoylation
The protocol for detecting myristoylated EGR2 in plant was modified from previous study (Tang & Han, 2017). Briefly, 2‐week‐old seedlings grown on MS medium were treated with or without 80 μM myristic acid alkyne (Cayman Chemical, #13267) at 22°C for additional 72 h. Total proteins were prepared with lysis buffer and subjected to click reaction with click reaction kit (Click Chemistry Tools, #1001) and biotin azide (PEG4 carboxamide‐6‐azidohexanyl biotin, Invitrogen, #B10184). The reaction products were immunoprecipitated with anti‐Myc agarose beads (Sigma‐Aldrich, #A7470) and separated on SDS–PAGE. Streptavidin–HRP (CST, #3999) was used for detecting biotin‐labeled proteins.
LC‐MS/MS analysis
The details of LC‐MS/MS for searching interacting proteins were performed as described (Liu et al, 2017) with some modifications. Total proteins (50 mg) were prepared from EGR2‐Myc transgenic plants with protein extraction buffer (50 mM Tris–HCl pH 7.5, 2 mM EDTA pH 8.0, 150 mM NaCl, 20% glycerol, 0.5% Triton X‐100, 0.1% NP‐40) and immunoprecipitated with anti‐Myc agarose (Sigma‐Aldrich, #A7470). The precipitants were washed by 1 ml protein extraction buffer for five times, four min each, and then eluted by 0.1 M glycine‐HCl (pH 2.8) and neutralized by 1 M Tris buffer. The elution products were dissolved in NH4HCO3 solution (30 ml, 50 mM) and reduced with DTT and alkylated with IAM. The reaction products were digested by trypsin (pH 8.5) at 37°C for 12 h and then diluted with 0.1% formic acid, and centrifuged at 15,294 g for 20 min. The supernatant was collected for nanoLC‐MS analysis with a Thermo Q‐Exactive high‐resolution mass spectrometer (Thermo Scientific). The putative interacting proteins of EGR2 and search parameters are listed in Appendix Table S1.
Author contributions
SY directed the project. YD and SY designed the experiments. JL and YD performed localization analysis. YD performed all other experiments. YS, JG, JH, CS, ZG, and SY discussed and interpreted the data. YD, YS, and SY wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We thank Dr. Pedro L Rodriguez for pyr mutant seeds and Dr. Jianru Zuo for helpful discussion and comments on the manuscript. We thank Dr. Zhen Li for the help of LC‐MS/MS analysis. This work was supported by the National Natural Science Foundation of China (31730011 and 31330006), the National Key Scientific Research Project (2015CB910203), and China Postdoctoral Science Foundation (21015051).
The EMBO Journal (2019) 38: e99819
References
- Agarwal M, Hao YJ, Kapoor A, Dong CH, Fujii H, Zheng XW, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281: 37636–37645 [DOI] [PubMed] [Google Scholar]
- Aguilar PS, Hernandez‐Arriaga AM, Cybulski LE, Erazo AC, de Mendoza D (2001) Molecular basis of thermosensing: a two‐component signal transduction thermometer in Bacillus subtilis . EMBO J 20: 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanesi D, Mansilla MC, de Mendoza D (2004) The membrane fluidity sensor DesK of Bacillus subtilis controls the signal decay of its cognate response regulator. J Bacteriol 186: 2655–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanesi D, Martin M, Trajtenberg F, Mansilla MC, Haouz A, Alzari PM, de Mendoza D, Buschiazzo A (2009) Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc Natl Acad Sci USA 106: 16185–16190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara GB, Wen TN, Nguyen TT, Verslues PE (2017) Protein phosphatase 2Cs and microtubule‐associated stress protein 1 control microtubule stability, plant growth, and drought response. Plant Cell 29: 169–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold‐induced transcriptome and freezing tolerance in Arabidopsis . Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cybulski LE, Ballering J, Moussatova A, Inda ME, Vazquez DB, Wassenaar TA, de Mendoza D, Tieleman DP, Killian JA (2015) Activation of the bacterial thermosensor DesK involves a serine zipper dimerization motif that is modulated by bilayer thickness. Proc Natl Acad Sci USA 112: 6353–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S (2015) OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis . Dev Cell 32: 278–289 [DOI] [PubMed] [Google Scholar]
- Ding Y, Jia Y, Shi Y, Zhang X, Song C, Gong Z, Yang S (2018) OST1‐mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO J 37: e98228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF (2009) Roles for Arabidopsis CAMTA transcription factors in cold‐regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Waksman G, Gordon JI (2001) The biology and enzymology of protein N‐myristoylation. J Biol Chem 276: 39501–39504 [DOI] [PubMed] [Google Scholar]
- Fuchs S, Grill E, Meskiene I, Schweighofer A (2013) Type 2C protein phosphatases in plants. FEBS J 280: 681–693 [DOI] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK (2011) Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci USA 108: 1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park S‐Y, Cutler SR, Sheen J, Rodriguez PL, Zhu J‐K (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al‐Rasheid KA, Romeis T, Hedrich R (2009) Activity of guard cell anion channel SLAC1 is controlled by drought‐stress signaling kinase‐phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J (1998) Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP‐myristoyl switching. Cell 95: 237–248 [DOI] [PubMed] [Google Scholar]
- Guo XY, Liu DF, Chong K (2018) Cold signaling in plants: insights into mechanisms and regulation. J Integr Plant Biol 60: 745–756 [DOI] [PubMed] [Google Scholar]
- Guy CL (1990) Cold acclimation and freezing stress tolerance: role of protein metabolism. Annu Rev Plant Physiol Plant Mol Biol 41: 187–223 [Google Scholar]
- Hannoush RN (2015) Synthetic protein lipidation. Curr Opin Chem Biol 28: 39–46 [DOI] [PubMed] [Google Scholar]
- Hao Q, Yin P, Li W, Wang L, Yan C, Lin Z, Wu JZ, Wang J, Yan SF, Yan N (2011) The molecular basis of ABA‐independent inhibition of PP2Cs by a subclass of PYL proteins. Mol Cell 42: 662–672 [DOI] [PubMed] [Google Scholar]
- Ishitani M, Liu JP, Halfter U, Kim CS, Shi WM, Zhu JK (2000) SOS3 function in plant salt tolerance requires N‐myristoylation and calcium binding. Plant Cell 12: 1667–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia YX, Ding YL, Shi YT, Zhang XY, Gong ZZ, Yang SH (2016) The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis . New Phytol 212: 345–353 [DOI] [PubMed] [Google Scholar]
- Jiang BC, Shi YT, Zhang XY, Xin XY, Qi LJ, Guo HW, Li JG, Yang SH (2017) PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis . Proc Natl Acad Sci USA 114: E6695–E6702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Thomashow MF (2012) Photoperiodic regulation of the C‐repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana . Proc Natl Acad Sci USA 109: 15054–15059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ding YL, Shi YT, Zhang XY, Zhang SQ, Gong ZZ, Yang SH (2017a) MPK3‐and MPK6‐mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis . Dev Cell 43: 630–642 [DOI] [PubMed] [Google Scholar]
- Li H, Ye K, Shi Y, Cheng J, Zhang X, Yang S (2017b) BZR1 positively regulates freezing tolerance via CBF‐dependent and CBF‐independent pathways in Arabidopsis . Mol Plant 10: 545–559 [DOI] [PubMed] [Google Scholar]
- Liu JY, Shi YT, Yang SH (2018) Insights into the regulation of C‐repeat binding factors in plant cold signaling. J Integr Plant Biol 60: 780–795 [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi‐Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought‐ and low‐temperature‐responsive gene expression, respectively, in Arabidopsis . Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Jia Y, Ding Y, Shi Y, Li Z, Guo Y, Gong Z, Yang S (2017) Plasma membrane CRPK1‐mediated phosphorylation of 14‐3‐3 proteins induces their nuclear import to fine‐tune CBF signaling during cold response. Mol Cell 66: 117–128 e115 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D, Chong K (2015) COLD1 confers chilling tolerance in Rice. Cell 160: 1209–1221 [DOI] [PubMed] [Google Scholar]
- Martinez L, Reeves A, Haldenwang W (2010) Stressosomes formed in Bacillus subtilis from the RsbR protein of listeria monocytogenes allow sigma(B) activation following exposure to either physical or nutritional stress. J Bacteriol 192: 6279–6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclaughlin S, Aderem A (1995) The myristoyl‐electrostatic switch ‐ a modulator of reversible protein‐membrane interactions. Trends Biochem Sci 20: 272–276 [DOI] [PubMed] [Google Scholar]
- de Mendoza D (2014) Temperature sensing by membranes. Annu Rev Microbiol 68: 101–116 [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007) SIZ1‐mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis . Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J, Matschi S, Shorinola O, Rovenich H, Matei A, Segonzac C, Malinovsky FG, Rathjen JP, MacLean D, Romeis T, Zipfel C (2014) The calcium‐dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16: 605–615 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi‐Shinozaki K (2013) ABA signaling in stress‐response and seed development. Plant Cell Rep 32: 959–970 [DOI] [PubMed] [Google Scholar]
- Ni M, Cui D, Einstein J, Narasimhulu S, Vergara CE, Gelvin SB (1995) Strength and tissue‐specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J 7: 661–676 [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF et al (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre M, Traverso JA, Boisson B, Domenichini S, Bouchez D, Giglione C, Meinnel T (2007) N‐myristoylation regulates the SnRK1 pathway in Arabidopsis . Plant Cell 19: 2804–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert N, Merlot S, N'Guyen V, Boisson‐Dernier A, Schroeder JI (2006) A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis . FEBS Lett 580: 4691–4696 [DOI] [PubMed] [Google Scholar]
- Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, Hibi T, Taniguchi M, Miyake H, Goto DB, Uozumi N (2009) Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA‐activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J 424: 439–448 [DOI] [PubMed] [Google Scholar]
- Shi Y, Ding Y, Yang S (2018) Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci 23: 623–637 [DOI] [PubMed] [Google Scholar]
- Shi Y, Tian S, Hou L, Huang X, Zhang X, Guo H, Yang S (2012) Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type‐A ARR genes in Arabidopsis . Plant Cell 24: 2578–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, Kwak JM (2009) Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett 583: 2982–2986 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain‐containing transcriptional activator that binds to the C‐repeat/DRE, a cis‐acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HY, Han M (2017) Fatty acids regulate germline sex determination through ACS‐4‐dependent myristoylation. Cell 169: 457–469 [DOI] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Lauriere C, Merlot S (2009) Protein phosphatases 2C regulate the activation of the Snf1‐related kinase OST1 by abscisic acid in Arabidopsis . Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MH, Heal WP, Mann DJ, Tate EW (2010) Protein myristoylation in health and disease. J Chem Biol 3: 19–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Shi Y, Liu J, Guo L, Zhang X, Yang S (2010) A mutant CHS3 protein with TIR‐NB‐LRR‐LIM domains modulates growth, cell death and freezing tolerance in a temperature‐dependent manner in Arabidopsis . Plant J 63: 283–296 [DOI] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan XQ, Wu D, Pang YX, Yan CY, Li WQ, Wang JW, Yan N (2009) Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol 16: 1230–U1242 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li J, Li F, Liu H, Yang W, Chong K, Xu Y (2017) OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination to activate OsTPP1 and enhances rice chilling tolerance. Dev Cell 43: 731–743 e735 [DOI] [PubMed] [Google Scholar]
- Zhao C, Wang P, Si T, Hsu CC, Wang L, Zayed O, Yu Z, Zhu Y, Dong J, Tao WA, Zhu JK (2017) MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev Cell 43: 618–629 e615 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6


