Abstract
Background:
HLA class I polymorphism has the greatest impact of human genetic variation on viral load set point. A substantial part of this effect is due to the action of HLA-B and HLA-C alleles. With few exceptions the role of HLA-A molecules in immune control of HIV is unclear.
Methods:
We here study HLA-A*68:02, one of the most highly prevalent HLA-A alleles in C-clade infected sub-Saharan African populations, and one that plays a prominent role in the HIV-specific CD8+ T-cell responses made against the virus.
Results:
We define eight epitopes restricted by this allele and propose the peptide binding motif for HLA-A*68:02. Although one of these epitopes almost exactly overlaps an HLA-B*57-restricted epitope in Gag linked with immune control of HIV, this HLAA*68:02-restricted Gag-TA10 response imposed only weak selection pressure on the virus and was not associated with significantly lower viral setpoint. The only HLAA*68:02-restricted responses imposing strong selection pressure on HIV were in the flanking regions of Pol-EA8 and Pol-EA11 and within the Vpr-EV10 epitope (P=8×10−8). However, targeting of this latter epitope was associated with significantly higher viral loads (P=0.003), suggesting lack of efficacy.
Conclusion:
This study is consistent with previous data showing that HLA-A-restricted Gag-specific responses can impose selection pressure on HIV. In the case of HLAA*68:02 the Gag response is subdominant, and apparently has little impact in natural infection. However, these data suggest the potential for high frequency vaccine-induced Gag responses restricted by this allele to have significant antiviral efficacy in vaccine recipients.
Keywords: HIV-1, CD8+ T cells, HLA-peptide binding
INTRODUCTION
Polymorphism in the HLA region of chromosome 6 has the strongest impact of any human genetic variation on HIV viral set point and therefore on the rate of HIV disease progression [1–3]. In particular this is due to variation at the HLA-B and HLA-C loci [1–4]. The mechanism underlying this effect has been attributed in part to the antiviral activity from CD8+ T cell responses. In particular it has been proposed that Gag-specific responses are important for improved immune control of HIV, because Gag is highly abundant and Gag epitopes are presented early following viral infection of target cells [5–7]; and also because regions of Gag are highly conserved and so escape mutants can have a significant impact on viral replicative capacity [8, 9]. Thus, a broad Gag-specific response is associated with immune control, whereas increasing breadth of responses to Envelope or towards the accessory/regulatory proteins, Tat, Rev, Nef, Vif, Vpr and Vpu, typically is not [10, 11].
A number of additional factors contribute to the effectiveness of the CD8+ T-cell response [12], and these are collectively reflected in the selection of escape mutants within the relevant epitopes by the virus. Thus it has been proposed that Gag responses that also drive selection pressure on HIV are those specifically associated with immune control [10, 13–15]. In contrast, although escape mutants are often selected in epitopes in Env and the accessory/regulatory proteins, these generally do not have significant impact on viral replication capacity or on viral load [9, 10].
It remains open to speculation why variation at the HLA-A locus appears to have less of an impact on immune control than HLA-B or HLA-C [1–3, 16]. Although some HLA-A alleles have been associated with viraemic control [17–21], what distinguishes HLA-A alleles that are somewhat protective from the remainder is unknown.
To begin to address the potential role of HLA-A alleles in immune control of HIV, we here focused on HLA-A*68:02, since this is one of the HLA-A alleles that is most prevalent in sub-Saharan Africa where the global HIV epidemic is substantially focused and in particular on subclade C infection [22]. In addition, a high contribution to the HIV specific response is made by HLA-A*68:02-restricted CTL activity [4], and yet no peptide binding motif for this allele has been established, nor have the HIV epitopes restricted by this allele been systematically defined and characterized. Therefore, the aim of this study was first to systematically define the CD8+ T cell responses restricted by HLA-A*68:02, and then to assess the contribution of each of these responses to immune control.
PATIENTS, MATERIALS AND METHODS
Study subjects and HIV samples
All study subjects were antiretroviral therapy-naïve in chronic HIV-1 infection. The subclade C cohort used for peptide HLA-A*68:02 associations consisted of n=1,010 subjects and identification of escape mutations comprised of a total of n=1,826 subjects of whom sequences and HLA were available as previously described [13, 23]. Peripheral blood mononuclear cells (PBMCs) for IFNγ elispot peptide titration, intracellular cytokine staining (ICS) and HLA-A*68:02 tetramer staining were derived from treatment naïve subclade C infected individuals recruited from a local UK cohort as previously described [5]. Informed consent was obtained from all participating individuals, and institutional review boards at the University of KwaZulu-Natal and the University of Oxford approved the study (E028/99 and 06/Q1604/12, respectively).
Reagents, IFNγ elipost assays and flow cytometry.
IFNγ elispot assays for optimal peptide mapping were performed as previously described [13]. Peptide-HLA-A*68:02 tetramers were generated using one pot – one mix method where the peptide, β2m and peptide was refolded for 48 hrs in the presence of protease inhibitors [24] and used as previously described [13]. Briefly, cells were stained with PE conjugated tetramers for 30 min at room temperature and subsequently stained for live/dead (Invitrogen), CD8 Qdot605 (Invitrogen) and CD3 Pacific Orange (Invitrogen). Ex vivo ICS was performed as described previously [13]. Briefly, PBMCs from responding subjects were stimulated with the optimal peptide for 6 hours and stained for live/dead cells (Invitrogen) and with surface markers CD3 PacificOrange (Invitrogen), CD8 Pacific Blue (BD) and intracellular with IFNγ PE-Cy7 (BD) according manufacturers protocol using the cytofix/cytoperm kit from BD. All samples were acquired on a LSR II flow cytometer within 12 hours and analysed using FlowJo version 8.8.6 (Treestar).
HLA peptide binding assay.
The measurement of peptide HLA-A*68:02 affinity was undertaken using alphaScreen technology as previously described [25]. Briefly, recombinant, biotinylated MHC-I heavy chains were diluted to a concentration of 2 nM in a mixture of 30 nM β2m and peptide, and allowed to fold for 48 h at 18°C. The pMHC-I complexes were detected using streptavidin donor beads and a conformation-dependent anti-HLA-I antibody, W6/32, conjugated to acceptor beads. The beads were added to a final concentration of 5 μg/mL and incubated over night at 18°C. One hour prior to reading the plates, these were placed next to the reader to equilibrate to reader temperature. Detection was done on an EnVision multilabel reader (Perkin Elmer).
Statistical analysis for HLA-A*68:02 and associations to OLP and HIV sequence polymorphisms.
The statistical method to identify HLA associations to OLP has been described previously [13, 14]. Briefly, we used a decision tree based on Fisher’s exact test to identify associations between the recognition of individual 18-mer peptides and the expression of HLA-A*68:02. For each 18-mer peptide, we computed the Fisher exact test p-value against all 4-digit HLA alleles observed in the cohort and added the most significant HLA allele to the decision tree. We next removed all individuals who expressed that allele and repeated until the most significant HLA allele had p>0.2. The statistical method to identify HLA associations to HIV sequence polymorphism has previously been described [14, 15]. Briefly, our approach corrected for the phylogenetic structure of the sequences and then we computed P and q values for every association with the q value being used to correct for multiple comparisons by calculation of a false detection rate [26].
RESULTS
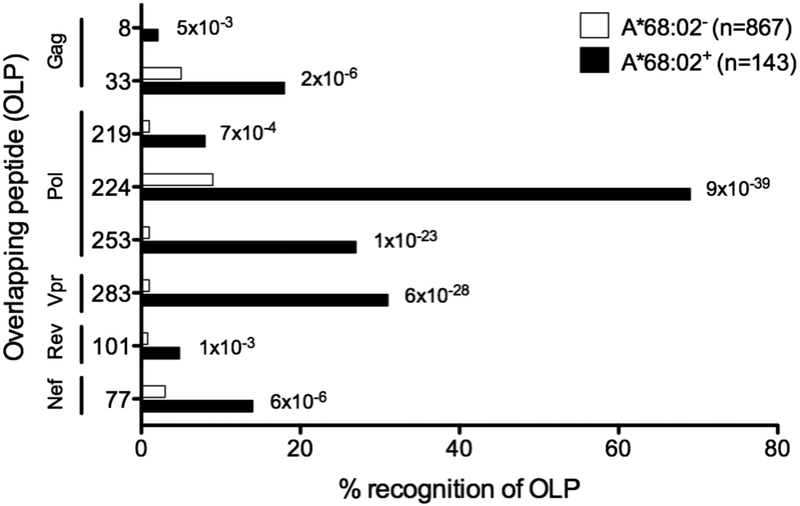
We first sought to determine the HIV-specific CD8+ T-cell responses restricted by HLA-A*68:02 in a C-clade cohort of 1,010 individuals of whom 143 expressed HLA-A*68:02. In each subject we tested in IFNγ elispot assays recognition of a panel of 410 18mer peptides overlapping by 10 amino acids and spanning the entire HIV C clade consensus proteome, as previously described [10]. We identified responses to 8 overlapping 18mer peptides that were statistically associated with HLA-A*68:02 expression (Fig 1) (q<0.1). The dominant responses were against Pol and Vpr epitopes. We then used a previously validated approach to predict the peptide binding motif for HLA-A*68:02 [27]. The HLA residues predicted to form the A and B pockets of the peptide binding groove [28] are identical for HLA-A*68:01 and HLA-A*68:02, and the former has a defined motif with Asp or Glu at position-1 (P1) in the binding peptide, and Thr/Val at P2 [29]. In contrast, the F pocket for HLA-A*68:02 differs substantially from that of HLA-A*68:01 in having Tyr at residue 116 as opposed to Asp; however the HLA residues predicted to form the F pocket for HLA-A*68:02 are almost identical to that of HLA-A*02:01, which has Val as the primary anchor residue at the carboxy-terminal position (PC) of the binding peptide [30]. The only difference at the F pocket is Val at HLA residue 95 in HLA-A*02:01, and Ile in this position in HLA-A*68:02, suggesting a slightly smaller hydrophobic residue than Val, such as Ala, might be the primary anchor at PC for HLA-A*68:02.
Figure 1. HLA-A*68:02 restricts 8 epitopes in which dominant responses are directed towards Pol and Vpr epitopes.
Screening of HIV infected individuals with 410 overlapping peptides from the entire HIV proteome reveals eight 18mer peptides significantly associated with HLA-A*68:02 expression by IFNγ elispot. For OLP-33 associations all subjects expressing HLA-B*57/58:01 were removed from analysis because of HLA-B*57/58:01 TW10 restriction. q-value <0.1 used as cut off for HLA and OLP associations (n=1,010) with P-values determined using Fishers’s exact test [38].
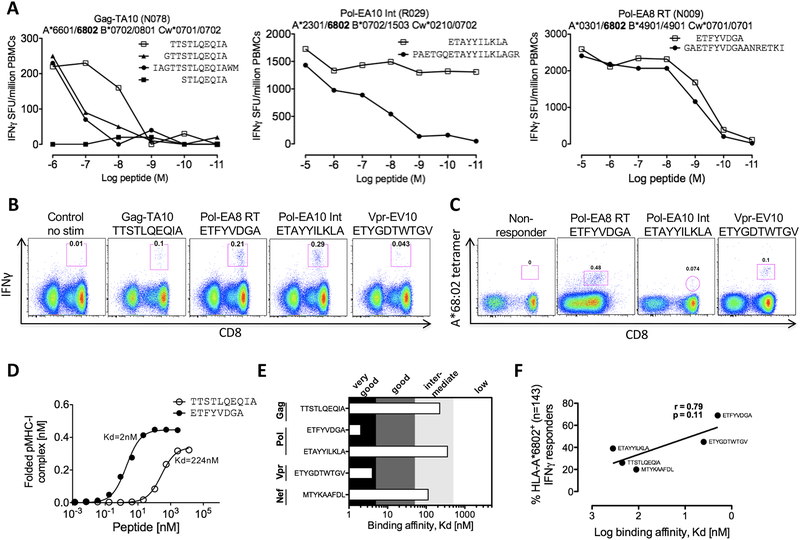
We then used a previously validated tool for predicting the optimal epitopes from longer peptides [31] to define the 8 HLA-A*68:02 optimal epitopes from the 18mer sequences recognised. We next confirmed the optimal epitope for the four most well targeted peptides (Fig 2). The HLA-A*68:02-peptide binding avidity [25] was measured for the optimal epitopes defined in this way (Fig 2D and E) and a strong correlation was observed between frequency of peptide recognition and peptide binding avidity to HLA-A*68:02 (Fig 2F).
Figure 2. Optimal and unequivocal characterization of HLA-A*68:02 restricted epitopes with peptide binding affinity to HLA-A*68:02 confirming optimal epitopes and correlation to targeting frequency.
A. Three of the 18mer HLA-A*68:02 restricted peptides were optimised by peptide titration with PBMCs from responding HLA-A*68:02 positive individuals (N078, R029 and N009) using IFNγ elispot with their respective HLA genotyping shown.
B. Optimal defined epitopes were used to stimulate CD8+ T cell responses to Gag-TA10, Pol-EA8RT, Pol-EA10 and Vpr-EV10 from PBMCs derived from HLA-A*68:02 positive individuals using intracellular cytokine staining to confirm these optimal epitopes.
C. HLA-A*68:02 tetramers were loaded with optimal peptides Pol-EA8 (ETFVVDGA), Pol-EA10 (ETAYYILKLA) and Vpr-EV10 (ETYGDTWTGV) and used to stain epitope specific CD8+ T cells from subject R029 (Vpr-EV10) and N078 (Pol-EA10 and controlled by a non-responding HLA-A*68:02 positive subject.
D. Peptide binding affinity to the HLA-A*68:02 molecule for Gag-TA10 and Pol-EA8-RT.
E. HLA-A*68:02 peptide binding compared to previous standard definitions of peptide-MHC affinity.
F. Correlation of immunodominant targeting for five epitopes (located within 18mer peptides) in n=143 HLA-A*68:02 positive individuals. Spearman rank correlation, r and P-values shown in F.
There are five peptides listed in the Los Alamos HIV Immunology Database ‘A’ list [32] as HLA-A*68:02 restricted epitopes. We did not observe significant responses to three of these C-clade consensus peptides (ITLWQRPLV, IAARAVELL and DTVLEEWNL) in the study cohort. These three peptides also proved to be weak binders to HLA-A*68:02 (Kd>500nM in each case; data not shown). In addition, we optimised the ‘A’ list 10mer Pol epitope GAETFYVDGA to the 8mer ETFYVDGA epitope, which was also a significant better binder than the 10mer to HLA-A*68:02 (Kd=1246 and Kd=2, respectively). Thus, in total we identified 8 epitopes restricted by HLA-A*68:02, which support a peptide binding motif comprising Glu at P1 and primary anchors of Thr at P2 and Ala in the C-terminal position of the binding peptide (Table 1).
Table 1. HLA-A*68:02 associations to 18mer overlapping peptide targeting.
q-values correcting for multiple comparison of <0.1 used as cut off.
| OLP number | HXB2 Location | OLP sequence | Optimal sequence | Epitope name | p-value | q-value |
|---|---|---|---|---|---|---|
| 8 | pl7 52–60 | GLLETSEGCKQIMKQL | ETSEGCKQ I | Gag-EI9 | 4.9E-03 | 6.7E-02 |
| 33 | p24 239–247 | SDlAGTTSTLQEQIAWM | TTSTLQEQIA | Gag-TA10 | 1.5E-06 | 3.9E-05 |
| 219 | RT 399–408 | TWETWWTDYWQATWIPEW | ETWWTDYWQA | Pol-EAI 0 RT | 6.9E-04 | 5.7E-03 |
| 224 | RT 430–437 | GAETFYVDGAANRETKI | ETFYVDG A | P0I-EA8 RT | 8.7E-39 | 3.9E-05 |
| 253 | Int 96–105 | PAETGQETAYYILKLAGR | ETAYYILKLA | Pol-EAI 0 Int | 1.1E-23 | 3.9E-05 |
| 283 | Vpr48–57 | SLGQYIYETYGDTWTGV | ETYGDTWTGV | Vpr-EVl 0 | 5.7E-28 | 3.9E-05 |
| 101 | Rev 61–68 | QIHSISERILSTCLGRPA | STCLGRP A | Rev-SA8 | 1.4E-03 | 1.7E-02 |
| 77 | Nef 79–87 | QVPLRPMTYKAAFDLSFF | MTYKAAFD L | Nef-ML9 | 6.2E-06 | 3.9E-05 |
| HLA-A*68:02 peptide binding motif | ETxxxxxxxA | |||||
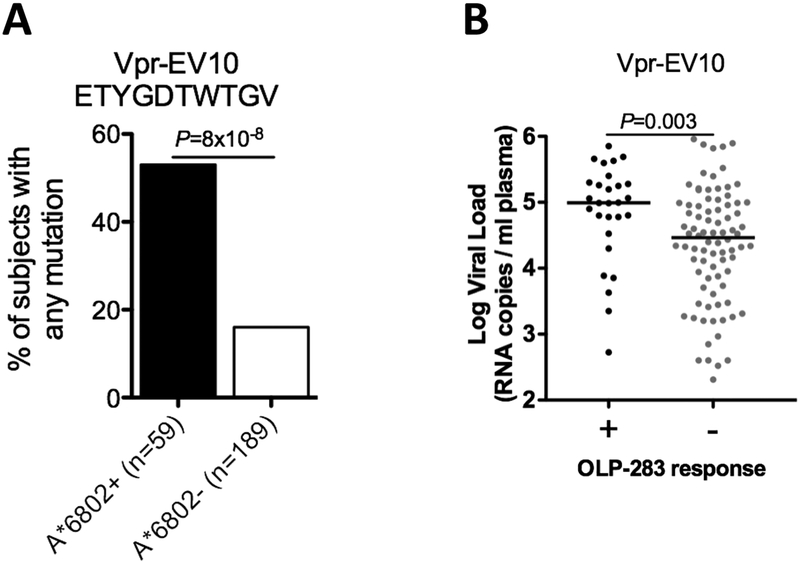
As described above, it is suggested that effective CD8+ T-cell responses are more likely both to be Gag-specific and to drive selection pressure on the virus. We therefore first sought evidence of selection pressure on HIV imposed by these responses. For Vpr EV10, 53% of HLA-A*68:02 individuals carried mutations within the epitope compared to 16% in A*68:02-negative persons, (P=8×10−8) (Fig 3A), in most cases selecting for the E48D polymorphism at P1 (P=0.0016) (Table 2), a mutation also selected in association with HLA-A*68:02 in B-clade infection [33]. However, the viral set points in HLAA*68:02-positive responders to this Vpr-EV10 epitope were higher than in the HLAA*68:02 positive non-responders (P=0.003) (Fig 3B), suggesting lack of efficacy. Two Pol responses, Pol-EA8 and Pol-EA11, selected mutations in their flanking regions (Table 2); however no response to Pol-EA11 was identified in elispot assays. The GagEI9, Gag-TA10 and Pol-EA8 responses showed non-significant trends towards lower viral setpoint in HLA-A*68:02-positive responders, but, of note, significant selection pressure was imposed by these responses (Table 2). The I247V mutant was observed in association with HLA-A*68:02 at P9 in the epitope TA10 in 4 out of 278 HLA-A*68:02 expressing individuals.
Figure 3. Vpr-EV10 specific CD8+ T cells are associated with higher viral loads and select escape mutations in HLA-A*68:02 positive individuals.
A. Vpr sequencing of proviral HIV DNA from 248 subjects were used to identify escape mutations within the Vpr-EV10 epitope and expressed as % of subjects not having the ETYGDTWTGV wildtype sequence (‘any mutation’).
B. Viral loads in n=112 HLA-A*68:02 positive individuals for the Vpr-EV10 specific response located within OLP-283 comparing IFNγ elispot responders (‘+’) to HLA matched non-responders (‘-’). P-values by Fisher’s Exact test in A and Mann-Whitney U test in B.
Table 2. HLA-A*68:02 associated sequence polymorphisms within or in flanking regions of known or newly defined epitopes.
q-value <0.05 within known epitopes based on the defined HLA-A*68:02 motif was used as cut off for HLA and HIV sequence residue association in C, with one exception being the Vpr-E48D polymorphism due to the lower n-values for statistical power.
| HXB2 Location | Sequencea | Optimal epitope name | Polymorphism | p-value | q-value | number of sequences (n) |
|---|---|---|---|---|---|---|
| Gag 54 | ELERFALNPGLLETSEGCKQIIKQLQPAL | Gag-EI9b | A | 3.1E-07 | 3.1E-04 | 1753 |
| Gag 247 | GSDIAGTTSTLQEQIAWMTSNPPIPVGDI | Gag-TA10 | V | 4.2E-06 | 3.2E-03 | 1826 |
| Pol 592 | VKLWYQLEKEPIAGAETFYVDGAANRETK | P0I-EA8 RT | V | 6.5E-05 | 3.3E-02 | 1040 |
| Pol 601 | EPIAGAETFYVDGAANRETKIGKAGYVTD | P0I-EA8 RT | S | 7.1E-06 | 4.7E-03 | 1046 |
| Pol 607 | ETFYVDGAANRETKIGKAGYVTDRGRQKI | P0I-EA8 RT | K | 6.5E-10 | 2.7E-07 | 1044 |
| Pol 622 | GKAGYVTDRGRQKIVSLTETTNQKTELQA | Pol-EA11b | I | 6.5E-05 | 3.3E-02 | 558 |
| Vpr 48 | FPRPWHGSLGQYIYETYGDTWTGVEALIR | Vpr-EV10 | D | 1.6E-03 | 5.8E-01 | 255 |
optimal epitope underlined
epitope not optimized
DISCUSSION
In this study we have undertaken a comprehensive analysis of the HIV epitopes restricted by HLA-A*68:02 that are significantly targeted in a large cohort of study subjects in sub Saharan Africa infected with C-clade virus. We have defined 8 HLA-A*68:02-restricted epitopes, and propose a peptide-binding motif for HLA-A*68:02. Although five HIV epitopes are currently listed as HLA-A*68:02-restricted in the Los Alamos HIV Immunology database (www.HIV.lanl.gov), these are not supported by published experimental data, and only two of these epitopes (Vpr-EV10 and Pol-EA8) are in common with those described here. As described above, the remaining three epitopes included in the Los Alamos immunology database list were not targeted to a significant degree in this cohort and also proved to be weak binders to HLA-A*68:02 (data not shown).
The inability of HLA-A*68:02 to have a significant impact on viral load set-point [34] may be reviewed in the light of these data. There is one HLA-A*68:02-restricted response in Gag-TA10 (Gag 239–248), which shows evidence of selection pressure on the virus, albeit a weak selection pressure. In contrast, the TW10 epitope (Gag 240–249) restricted by the protective HLA-B*57/58:01 alleles, that overlaps the HLA-A*68:02 TA10 epitope, is under strong selection pressure in HLA-B*57/58:01 expressing individuals [8, 15, 35, 36]. The fact that the most frequently selected HLA-B*57/58:01 escape mutant T242N has a significant fitness cost on the virus [36] may significantly contribute to immune control mediated by these alleles [12].
The dominant HLA-A*68:02-restricted responses in Pol and Vpr were also associated with no beneficial impact on viral load. In fact, we observed an overall trend towards higher viral load with increasing breadth of HLA-A*68:02 restricted responses (P=0.1, data not shown). Although the Vpr-EV10 response was capable of driving selection pressure on the virus, consistent with B-clade infection [33], this did not appear to be a protective response, in keeping with previous studies suggesting in general a lack of immune control in subjects targeting epitopes in the accessory/regulatory HIV proteins [10, 14].
These studies are therefore consistent with the hypothesis that an HIV-specific CD8+ Tcell response comprised of multiple Gag specificities that are capable of imposing selection pressure on the virus [13] is associated with improved immune control of viraemia. Although the HLA-A*68:02 makes a significant contribution to HIV-specific CD8+ T-cell activity, the Gag-specific response was subdominant and perhaps for that reason did not have a significant protective effect. Previous studies of protective HLA-A alleles such as HLA-A*74:01, however, have shown dominant Gag-specific responses and the selection of escape mutants within Gag [21]. These data therefore do not suggest an intrinsic difference between HLA-A and HLA-B molecules to explain the lack of contribution of HLA-A to immune control of HIV. Indeed, where there are HLAA*68:02-restricted Gag responses they tend to be associated with lower viral load, and in some instances there is evidence of selection pressure, albeit in neither case are these effects strong. However this may simply reflect a lack of dominant Gag responses restricted by HLA-A molecules in comparison to HLA-B, in which the protective HLAB*57/58:01-, HLA-B*42:01, HLA-B*81:01, HLA-B*13:02-restricted molecules present dominant responses directed towards p24 Gag and also impose strong selection pressure within or in flanking regions of these epitopes [12]. However, accumulation of escape mutations may represent epitopes being driven towards extinction [37] and thereby affect immunogenicity. These data are therefore consistent with the concept that a vaccine inducing Gag specific CD8+ T-cell responses might be effective not only for individuals expressing protective HLA alleles such as HLA-B*27 and HLA-B*57, for whom the dominant responses are Gag-specific in natural infection [14, 38], but also for individuals expressing alleles such as HLA-A*68:02 that are not protective in natural infection, and in whom Gag responses exist but are subdominant.
ACKNOWLEGEMENTS
We are grateful to Eric Hunter for providing additional HIV sequence data to this study. This work was supported by the Welcome Trust (P.G.) and the National Institutes of Health, grant R01 AI46995. H.N.K holds a grant from the Danish Agency for Science, Technology and Innovation #12–132295.
Footnotes
The authors have declared that no competing interests exist.
REFERENCES
- 1.Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet 2009,5:e1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, et al. A whole-genome association study of major determinants for host control of HIV-1. Science 2007,317:944–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 2010,330:1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 2004,432:769–775. [DOI] [PubMed] [Google Scholar]
- 5.Payne RP, Kloverpris H, Sacha JB, Brumme Z, Brumme C, Buus S, et al. Efficacious Early Antiviral Activity of HIV Gag- and Pol-Specific HLA-B*2705-Restricted CD8+ T Cells. J Virol 2010,84:10543–10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sacha JB, Chung C, Rakasz EG, Spencer SP, Jonas AK, Bean AT, et al. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J Immunol 2007,178:2746–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kloverpris HN, Payne RP, Sacha JB, Rasaiyaah JT, Chen F, Takiguchi M, et al. Early Antigen Presentation of Protective HIV-1 KF11Gag and KK10Gag Epitopes from Incoming Viral Particles Facilitates Rapid Recognition of Infected Cells by Specific CD8+ T Cells. J Virol 2013,87:2628–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crawford H, Lumm W, Leslie A, Schaefer M, Boeras D, Prado JG, et al. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J Exp Med 2009,206:909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Troyer RM, McNevin J, Liu Y, Zhang SC, Krizan RW, Abraha A, et al. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoSPathog 2009,5:e1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med 2007,13:46–53. [DOI] [PubMed] [Google Scholar]
- 11.Masemola A, Mashishi T, Khoury G, Mohube P, Mokgotho P, Vardas E, et al. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J Virol 2004,78:3233–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulder PJ, Walker BD. HIV and HLA Class I: An Evolving Relationship. Immunity 2012,37:426–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kloverpris HN, Stryhn A, Harndahl M, van der Stok M, Payne RP, Matthews PC, et al. HLA-B*57 micropolymorphism shapes HLA allele-specific epitope immunogenicity, selection pressure and HIV immune control. J Virol 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matthews PC, Prendergast A, Leslie A, Crawford H, Payne R, Rousseau C, et al. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J Virol 2008,82:8548–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlson JM, Listgarten J, Pfeifer N, Tan V, Kadie C, Walker BD, et al. Widespread impact of HLA restriction on immune control and escape pathways of HIV-1. J Virol 2012,86:5230–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas R, Apps R, Qi Y, Gao X, Male V, O’HUigin C, et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet 2009,41:1290–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawashima Y, Satoh M, Oka S, Shirasaka T, Takiguchi M. Different immunodominance of HIV-1-specific CTL epitopes among three subtypes of HLA-A*26 associated with slow progression to AIDS. Biochem Biophys Res Commun 2008,366:612–616. [DOI] [PubMed] [Google Scholar]
- 18.Koehler RN, Walsh AM, Saathoff E, Tovanabutra S, Arroyo MA, Currier JR, et al. Class I HLA-A*7401 is associated with protection from HIV-1 acquisition and disease progression in Mbeya, Tanzania. J Infect Dis 2010,202:1562–1566. [DOI] [PubMed] [Google Scholar]
- 19.Schaubert KL, Price DA, Frahm N, Li J, Ng HL, Joseph A, et al. Availability of a diversely avid CD8+ T cell repertoire specific for the subdominant HLA-A2-restricted HIV-1 Gag p2419–27 epitope. J Immunol 2007,178:7756–7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang J, Shao W, Yoo YJ, Brill I, Mulenga J, Allen S, et al. Human leukocyte antigen class I genotypes in relation to heterosexual HIV type 1 transmission within discordant couples. J Immunol 2008,181:2626–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews PC, Adland E, Listgarten J, Leslie A, Mkhwanazi N, Carlson JM, et al. HLA-A*7401-Mediated Control of HIV Viremia Is Independent of Its Linkage Disequilibrium with HLA-B*5703. J Immunol 2011,186:5675–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novitsky V, Flores-Villanueva PO, Chigwedere P, Gaolekwe S, Bussman H, Sebetso G, et al. Identification of most frequent HLA class I antigen specificities in Botswana: relevance for HIV vaccine design. Hum Immunol 2001,62:146–156. [DOI] [PubMed] [Google Scholar]
- 23.Matthews PC, Koyanagi M, Kloverpris HN, Harndahl M, Stryhn A, Akahoshi T, et al. Differential Clade-Specific HLA-B*3501 Association with HIV-1 Disease Outcome Is Linked to Immunogenicity of a Single Gag Epitope. J Virol 2012,86:12643–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leisner C, Loeth N, Lamberth K, Justesen S, Sylvester-Hvid C, Schmidt EG, et al. One-pot, mix-and-read peptide-MHC tetramers. PLoS One 2008,3:e1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harndahl M, Justesen S, Lamberth K, Roder G, Nielsen M, Buus S. Peptide binding to HLA class I molecules: homogenous, high-throughput screening, and affinity assays. J BiomolScreen 2009,14:173–180. [DOI] [PubMed] [Google Scholar]
- 26.Carlson J, Heckerman D, Shani G. Estimating false discovery rates for contingency tables. Microsoft 2009. [Google Scholar]
- 27.Honeyborne I, Rathod A, Buchli R, Ramduth D, Moodley E, Rathnavalu P, et al. Motif inference reveals optimal CTL epitopes presented by HLA class I alleles highly prevalent in southern Africa. J Immunol 2006,176:4699–4705. [DOI] [PubMed] [Google Scholar]
- 28.Marsh SGE, Parham P, Barber LD. The HLA Facts Book: Academic Press, London: NW1 7DX; 2000. [Google Scholar]
- 29.Guo HC, Jardetzky TS, Garrett TP, Lane WS, Strominger JL, Wiley DC. Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle. Nature 1992,360:364–366. [DOI] [PubMed] [Google Scholar]
- 30.Falk K, Rotzschke O, Takiguchi M, Gnau V, Stevanovic S, Jung G, Rammensee HG. Peptide motifs of HLA-B58, B60, B61, and B62 molecules. Immunogenetics 1995,41:165–168. [DOI] [PubMed] [Google Scholar]
- 31.Erup Larsen M, Kloverpris H, Stryhn A, Koofhethile CK, Sims S, Ndung’u T, et al. HLArestrictor--a tool for patient-specific predictions of HLA restriction elements and optimal epitopes within peptides. Immunogenetics 2011,63:43–55. [DOI] [PubMed] [Google Scholar]
- 32.Llano A, Frahm N, Brander C. How to Optimally Define Optimal Cytotoxic T Lymphocyte Epitopes in HIV Infection? Theoretical Biology and Biophysics, Los Alamos, New Mexico 2009:3–24. [Google Scholar]
- 33.Carlson JM, Brumme CJ, Martin E, Listgarten J, Brockman MA, Le AQ, et al. Correlates of protective cellular immunity revealed by analysis of population-level immune escape pathways in HIV-1. J Virol 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leslie A, Matthews PC, Listgarten J, Carlson JM, Kadie C, Ndung’u T, et al. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J Virol 2010,84:9879–9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med 2004,10:282–289. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol 2006,80:3617–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leslie A, Kavanagh D, Honeyborne I, Pfafferott K, Edwards C, Pillay T, et al. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J Exp Med 2005,201:891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kloverpris HN, Stryhn A, Harndahl M, van der Stok M, Payne RP, Matthews PC, et al. HLA-B*57 Micropolymorphism shapes HLA allele-specific epitope immunogenicity, selection pressure, and HIV immune control. J Virol 2012,86:919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]