Abstract
Several RNA aptamers that bind small molecules and enhance their fluorescence have been successfully used to tag and track RNAs in vivo, but these genetically-encodable tags have not yet achieved single-fluorophore resolution. Recently, Mango-II, an RNA that binds TO1-Biotin with ~1 nM affinity and enhances its fluorescence by >1,500-fold was isolated by fluorescence selection from the pool that yielded the original RNA Mango. We now determined crystal structures of Mango-II in complex with two fluorophores, TO1-Biotin and TO3-Biotin, and found that despite their high affinity, the ligands adopt multiple distinct conformations, indicative of a binding pocket with modest stereoselectivity. Mutational analysis of the binding site led to Mango-II(A22U), which retains high affinity for TO1-Biotin but now discriminates over 5-fold against TO3-biotin. Moreover, fluorescence enhancement of TO1-Biotin increases 18% while that of TO3-Biotin decreases by 25%. Crystallographic, spectroscopic, and analog studies show that the A22U mutation improves conformational homogeneity and shape complementarity of the fluorophore-RNA interface. Our work demonstrates that even after extensive functional selection, aptamer RNAs can be further improved through structure-guided engineering.
Keywords: RNA, aptamer, fluorescence, structure, G-quadruplex, fluorophore, thiazole orange
Several aptamers that induce fluorescence of their cognate small molecules by >1000-fold, and that can be used in vivo as RNA analogues of fluorescent proteins have been described.1-3 Among these, RNA Mango is noteworthy because of its small size (<30 nucleotides) and its high affinity for the thiazole orange-derived fluorophore, TO1-Biotin (Kd = 3.1 nM). Moreover, its complex with the related fluorophore TO3-Biotin (Kd = 8 nM) is one of the most red-shifted fluorogenic tags described to date, having an emission maximum of longer wavelength than the fluorescent protein mPlum.4 The Co-crystal structure of RNA Mango revealed a three-tiered G-quadruplex. TO1-Biotin binds on one of its flat faces, with each of its three heterocylces, benzothiazole (BzT), mehtylquinoline (MQ) and biotin sequestered under an unpaired nucleotide.5,6
Mango-II was recently obtained by subjecting the final pool of the RNA Mango selection to compartmentalization coupled to fluorescence sorting.7 Compared to the original aptamer (hereafter Mango-I), the new RNA is brighter (11,000 M−1cm−1 vs. 17,000 M−1cm−1 for Mango-I and Mango-II, respectively), and binds to both TO1-Biotin and TO3-Biotin (Figure 1A) with higher affinity (Kd = 1.1±0.3 nM and 1.4±0.3 nM, respectively). To elucidate the structural basis for the enhanced properties of Mango-II, and as a starting point for structure-guided engineering, we have now determined its co-crystal structures in complex with TO1-Biotin (Figure 1B,C) and TO3-Biotin (Supplementary Fig. 1) at 2.9 Å and 3.0 Å resolution, respectively (Supplementary Table 1). The two complexes crystallized in similar unit cells with three crystallographically independent RNAs (chains A, B, C) in the asymmetric unit (A.U.). Overall, the three RNAs in each structure are very similar (rmsd ~ 0.7 Å for all non-hydrogen RNA atoms), and so are the RNAs in complex with the two different fluorophores (rmsd ~ 0.2 Å between the best-ordered chains in each structure; the coordinate precisions of the two structures are 0.42Å and 0.40Å, respectively). The folds of Mango-I and Mango-II are overall similar, and this similarity extends to crystal packing, in which adjacent aptamers stack pairwise through their fluorophore-distal quadruplex faces. Nonetheless, and as in the case of Mango-I, analytical ultracentrifugation indicates the aptamer RNA is monomeric in solution (Supplementary Fig. 2).
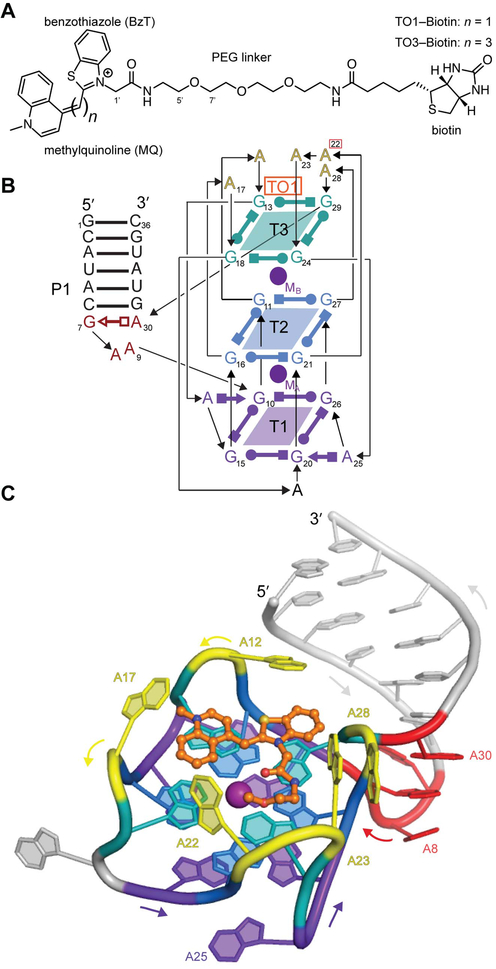
Figure 1 ∣. Structure of Mango-II in complex with TO1-Biotin.
(A) Chemical structures of TO1-Biotin and TO3-Biotin. The latter contains two additional methine carbons. (B) Secondary structure of the Mango-II-TO1-Biotin complex. Thin lines with arrows denote connectivity. Base pairs are represented with Leontis-Westhof symbols.11 Location of fluorophore and two potassium ions (TO1, MA, and MB) is indicated. This color scheme is used throughout the manuscript except where noted. (C) Cartoon representation of the Mango-II-TO1-Biotin complex. Arrows adjacent to the cartoon denote chain direction.
Mango-II has an almost identical melting profile in both the presence and absence of TO1-Biotin and is more thermostable than Mango-I.7 In addition, Mango-II is remarkably formaldehyde resistant; Mango-II reacted with formaldehyde, can still bind and induce fluorescence of TO1-Biotin. Together, these data suggest that the Mango-II aptamer has a pre-organized binding pocket.
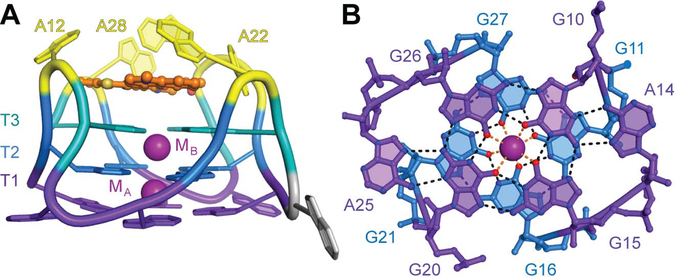
Three structural differences between Mango-I and Mango-II are consistent with a higher stability of the latter RNA and its improved fluorophore affinity. First, each guanine in the T3 G-quartet is isolated from the T2 G-quartet by one of four adenine containing propeller loops (Figure 2B). This approximate four-fold rotational symmetry is lacking in Mango-I where one T3 guanine buckles 30˚ out of plane of the other guanines.5 Second, the GAA^A junction between the P1 duplex and the quadruplex of Mango-II appears to have more hydrogen bonding interactions than that of Mango-I (Supplementary Fig. 3). Consistent with a higher stability of the Mango-II junction, all P1 helices in the A.U. are crystallographically resolved. In contrast, only one of the two P1s in the Mango-I A.U. exhibited electron density. Third, the T1 G-quartet (distal from the fluorophore) of Mango-II is augmented into a hexad by two adenines, A14 and A25 (Figure 2A). The corresponding T1 in Mango-I is augmented by one adenine into only a pentad. 5 The higher stability of Mango-II may reflect the higher temperature at which it was selected (45 °C vs. 37 °C for Mango-I).
Figure 2 ∣. The Mango-II G-quadruplex.
(A) Augmented Tier 1 (T1, purple) and Tier 2 G-quartets (T2, blue) of the Mango-II-TO1-Biotin complex. T1 is augmented into a hexad. (B) Side view.
The five adenines in the four propeller loops and the planar T3 quadruplex they create together define the fluorophore binding pocket of Mango-II. In this complex, the MQ and BzT of TO1-Biotin are coplanar and, in chains A and B, stack on G13 and G29 of T3, respectively (Figure 3A). The binding modes of the headgroup of the fluorophore in the Mango-I and Mango-II complexes are near-mirror images. When seen from above the plane of T3, the BzT and MQ heterocycles are arranged clockwise and counter-clockwise in the two structures, respectively. As a result, the long axes of the BzT and MQ heterocycles are parallel and perpendicular to those of the T3 guanines on which they stack in the Mango-I and Mango-II complexes, respectively (ref. 5, Supplementary Fig. 4). The perpendicular stacking arrangement is also present in chain C of the Mango-II structure but the heterocycles of the fluorophore are rotated by 90° around the 4-fold symmetry axis of the G-quartet with respect to that found in the A and B chains (Figure 3B). Concomitant with this rotation, four of the flap adenines of chain C of the A.U. (A17, A22, A23, and A28) adopt different conformations from those of chains A and B (Figure 3B). A17 reorients from anti to syn; A22 and A28 become disordered; and A23 reorients from syn to anti. In additional contrast to the Mango-I structure, electron density for atoms 1’ through 7’ of the PEG linker is coplanar with the T3 guanine layer, with the biotin moiety being disordered in all three complexes. The increased coplanarity of the TO1 heterocycles is consistent with the higher quantum yield of Mango-II-TO1-Biotin complex (0.21 vs. 0.14 for the corresponding Mango-I complex).
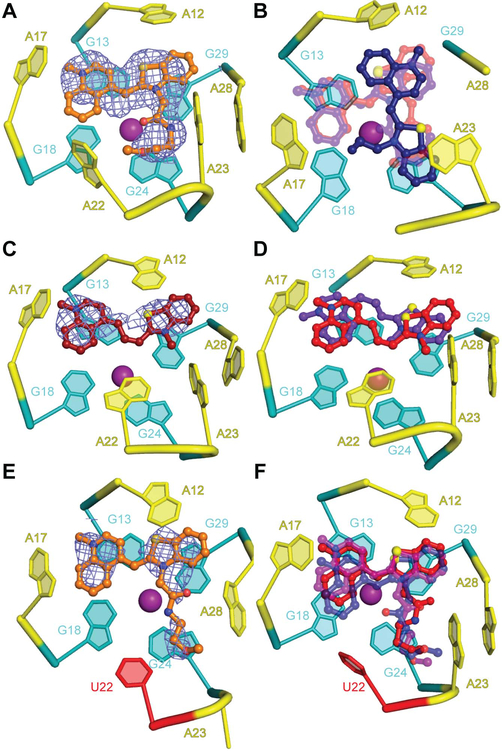
Figure 3 ∣. Plasticity of the Mango-II ligand binding pocket.
(A) Cartoon representation of the ligand binding pocket of chain A of the Mango-II-TO1-Biotin complex structure, with fluorophore electron density (2∣Fo∣-∣Fc∣ prior to building the ligand, contoured at 1.2 σ, blue mesh) shown in mesh. (B) Overlay of the fluorophores from the three complexes in the asymmetric unit aligned on chain C. TO1-Biotin from chains A, B, and C are shown in transparent red, transparent purple and blue, respectively. In chain C, A22 and A28 are disordered. (C) Fluorophore electron density (2∣Fo∣-∣Fc∣) prior to building the ligand, contoured at 1.2 σ, blue mesh) for chain A of the Mango-II-TO3-Biotin complex structure. (D) Overlay of the ligands in chain A (purple) and chain B (red). The chain A RNA is shown. (E) Cartoon representation of the ligand binding pocket of chain B of the Mango-II(A22U)-TO1-Biotin complex Fluorophore electron density (2∣Fo∣-∣Fc∣ prior to building the ligand, contoured at 1.2 σ) blue mesh. (F) Overlay of the ligands in chains A, B and C (purple,blue and red, respectively). Chain A RNA shown.
In the structure of Mango-II bound to TO3-Biotin, electron density corresponding to the fluorophore was only observed for the headgroup (BzT, MQ, and the trimethine linker), and only in RNA chains A and B (Figure 3C,D). Notably, in the two chains with bound TO3-Biotin, the nucleobase of A22 stacks directly on G24 of T3. In chain B, A22 also is in van der Waals contact with the methine linker of TO3-Biotin. These interactions may further stabilize the complex, and the packing of A22 observed in chain B may directly promote fluorescence of TO3-Biotin by stabilizing a coplanar conformation BzT, MQ, and the extended methine linker of this fluorophore.
Highly selective fluorogenic RNAs would facilitate development of orthogonal fluorescent tags. The structural heterogeneity revealed by our Mango-II TO1-Biotin and TO3-Biotin co-crystal structures, as well as the comparable affinity of the aptamer for the two fluorophores, suggests that the fluorophore binding pocket of this aptamer is not highly stereoselective. To increase selectivity of Mango-II, we constructed mutants focusing on residue A22, which in our structures is conformationally plastic and appeared to have the most potential to influence TO3-Biotin discrimination. Mutants were analyzed for affinity and fluorescence enhancement of TO1-Biotin and TO3-Biotin (Supplementary Table 3). Mutation to guanine reduced the binding affinity and fluorescence enhancement of both TO1-Biotin and TO3-Biotin (Supplementary Table 3). Such an outcome is not unexpected, as G-quadruplex nucleic acids can undergo alternative folding when additional guanine bases are present.8 However, mutation to uridine or cytosine maintained the binding affinity for TO1-Biotin (0.9 nM) while decreasing the affinity of TO3-Biotin from ~1.4 nM to ~5.0 nM (Supplementary Table 3). Unexpectedly, these mutations also increased the fluorescence enhancement of TO1-Biotin by 18% while decreasing the fluorescence enhancement of TO3-Biotin by 25% (Supplementary Table 3).
To understand how the Mango-II A22U mutation results in increased TO1-Biotin fluorescence enhancement, we determined the co-crystal structure of this complex at 2.8 Å resolution (Supplementary Table 2). The RNA structure is generally unchanged from that of the wild-type. The nucleobase of U22 is ordered but projects into solvent rather than interacting with T3. TO1-Biotin adopts the same orientation in all three chains in the A.U. with Bzt and MQ stacking on G29 and G13, respectively (Figure 3 E,F). Among our crystal structures, the binding pocket of the mutant is better ordered than that of either wild-type complex as judged by the mean real-space correlation coefficient (Supplementary Table 5). Ligand-RNA shape complementarity (SC) analysis9 of each co-crystal structure indicates that the A22U binding pocket conforms to TO1-Biotin better than wildtype, with SC statistics of 0.796 (rms=0.004) and 0.750(rms=0.009), respectively. The SC statistic of Mango-II-TO3-Biotin complex, 0.800 (rms=0.019), was similar to that of Mango-II(A22U)-TO1-Biotin.
To extend our crystallographic observations into solution conditions, we examined the fluorescence lifetimes of the wildtype and A22U mutant Mango-II RNAs in complex with TO1-Biotin. The A22U mutation increases the average lifetime of the emission from 1.51 (±0.06) ns to 1.92 (±0.03) ns. The lifetime of the Mango-II-TO1-Biotin complex has two components, and both are increased in the mutant compared to the wild-type: the first from 0.52 (±0.02) ns to 0.72 (±0.01) ns, and the second from 2.33 (±0.12) ns to 2.904 (±0.001) ns (Supplementary Figure 5, Supplementary Table 6). Our X-ray structures show that the wild-type Mango-II has two different modes of TO1-Biotin binding, while the A22U mutant is more homogeneous (Figure 3). The longer lifetimes of the mutant are consistent with this, and in particular suggest reduced torsional freedom in the atoms of the methine linker, as has been shown previously for other complexes of thiazole orange with nucleic acids.10
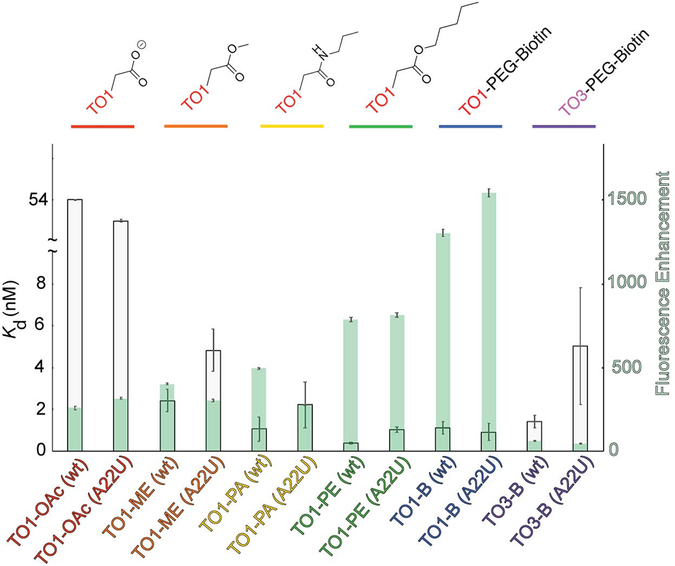
Our mutant co-crystal structure shows no direct contacts between U22 and TO1-Biotin. To gain further insight into how the U22 mutation improves Mango-II properties, we examined the effect of varying the length of the linker between the thiazole orange and biotin moieties of TO1-Biotin. We determined the affinity and fluorescence enhancement of four compounds: TO1-OAc (TO1-acetate), TO1-ME (TO1-methylester), TO1-PA (TO1-propanylamide), TO1-PE (TO1-pentenoylester) (Figure 4, Supplementary Table 3) Binding affinity and fluorescence enhancement of TO1-OAc is drastically diminished, and nearly identical between the wild-type and A22U mutant. However, the TO1-ME and TO1-PA exhibit greater fluorescence enhancement and binding affinity with the wild-type than with A22U. This suggests that in the absence of an extended linker, the A22 stabilizes the binding pocket, presumably by stacking on G24 as observed in chain B of the Mango-I-TO3-Biotin co-crystal structure (Figure 3 C,D). Further extension of the linker stabilizes the binding pocket by stacking on T3 but becomes sterically hindered by a bulky purine in position 22. The A22U mutation would limit steric clashes while preventing misfolding of the G-quadruplex core. Thus, the improved properties of the Mango-II A22U mutant may arise from improved packing of the linker of the fluorophore with an RNA with a homogeneous or predominant conformation.
Figure 4 ∣. Mango-II binding and fluorescence enhancement of thiazole orange derivatives.
Dissociation constants, Kd, and fluorescence enhancement, (gray and green bars, respectively) for various TO1-Biotin derivatives. Length of the linker between TO1 and biotin increases from left to right. Error bars report standard error, see Supplementary Table 3 for values.
We demonstrate that Mango-II is a versatile platform for fluorescence enhancement of thiazole-orange derived fluorophores, and have shown that variants of this RNA tag, designed with the aid of crystal structures, have improved properties. Analysis of the fluorophore-RNA shape complementarity of each of our structures reveals that fluorescence enhancement correlates directly with the SC statistic. The Mango-II-TO3-Biotin and Mango-II(A22U)-TO1-Biotin complexes both yield similarly high SC statistics, possibly indicating an upper limit to the shape complementarity attainable from this RNA scaffold. The high SC statistic observed for the A22U mutant-TO1-Biotin complex also correlates with improved binding affinity, fluorescence enhancement and fluorescence lifetime. Fluorescence binding studies on TO1-Biotin variants independently suggest that these improvements are due to removal of steric constraints on the ligand by the transversion mutation to a pyrimidine, consistent with our crystallographic analysis. By achieving enhanced fluorescence properties through improved ligand-RNA packing and structural homogeneity, this work demonstrates that even functionally selected fluorogenic RNAs can potentially be improved through detailed structural and mechanistic analysis.
Supplementary Material
ACKNOWLEDGMENT
We thank the staff of beamlines 501 and 502 of ALS and 24-ID-C of APS for crystallographic support, G. Piszczek of the Biophysics Core of NHLBI for AUC support, D.-Y. Lee of the Biochemistry Core of NHLBI and R. Levine for HPLC support, and for discussions, and members of the laboratories of A. Ferré-D’Amaré, P. Unrau, and M. Ryckelynck for helpful discussions.
Funding Sources
This work was partly conducted at the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory, on the Berkeley Center for Structural Biology beamlines, and at the Advanced Photon Source (APS), Argonne National Laboratory, on the NE-CAT beamlines which are supported by the U.S. National Institutes of Health (NIH). Use of ALS and APS was supported by the US Department of Energy. AUC was performed at the Biophysics Core Facility of the National Heart, Lung and Blood Institute (NHLBI). PJU was supported by an NSERC (Canada) operating grant. MR was supported by the LabEx NetRNA (ANR-10-LABX-0036 NETRNA). This work was supported in part by the intramural program of the NHLBI, NIH.
ABBREVIATIONS
- BzT
benzothiazole
- MQ
methylquinoline
Footnotes
Supporting Information
Experimental methods, electron density maps, fluorescence binding data, fluorescence lifetime data. This material can be found free of charge via the Internet at http://pubs.acs.org. Coordinates and structure factors have been deposited with the Protein Data Bank under accession codes 6C63, 6C64, and 6C65 for the Mango-II-TO1-Biotin, Mango-II-TO3-Biotin and Mango-II(A22U)-TO1-Biotin co-crystal structures, respectively.
The authors declare no competing financial interest.
REFERENCES
- 1.Paige JS; Wu KY; Jaffrey SR (2011) RNA mimics of green fluorescent protein. Science, 333 (6042), 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dolgosheina EV; Jeng SCY; Panchapakesan SSS; Cojocaru R; Chen PSK; Wilson PD; Hawkins N; Wiggins PA; Unrau PJ (2014) RNA Mango aptamer-fluorophore: A bright, high-affinity complex for RNA labeling and tracking. ACS Chem. Biol, 9 (10), 2412–2420. [DOI] [PubMed] [Google Scholar]
- 3.Tan XH; Constantin TP; Sloane KL; Waggoner AS; Bruchez MP; Armitage BA (2017) Fluoromodules consisting of a Promiscuous RNA aptamer and red or blue fluorogenic cyanine dyes: selection, characterization, and bioimaging. J. Am. Chem. Soc. 139 (26), 9001–9009. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez EA; Campbell RE; Lin JY; Lin MZ; Miyawaki A; Palmer AE; Shu XK; Zhang J; Tsien RY (2017) The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem. Sci, 42 (2), 111–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trachman RJ; Demeshkina NA; Lau MWL; Panchapakesan SSS; Jeng SCY; Unrau PJ; Ferré-D’Amaré AR (2017) Structural basis for high-affinity fluorophore binding and activation by RNA Mango. Nat Chem Biol, 13 (7), 807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trachman RJ; Truong L; Ferre-D’Amare AR (2017) Structural principles of fluorescent RNA aptamers. Trends Pharmacol. Sci, 38 (10), 928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Autour A; Jeng SCY; Cawte AD; Abdolahzahdeh A; Galli A; Panchapakesan SSS; Rueda D; Ryckelynck M; Unrau PJ (2018) Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat. Comm, 9(1):656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phan AT; Kuryavyi V; Gaw HY; Patel DJ (2005) Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol, 1 (3), 167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence MC; Colman PM Shape complementarity at protein/protein interfaces. (1993) J. Mol. Biol, 234, 946–950. [DOI] [PubMed] [Google Scholar]
- 10.Jarikote DV; Krebs N; Tannert S; Roder B; Seitz O (2007) Exploring base-pair-specific optical properties of the DNA stain thiazole orange. Chem. Eur. J, 13, 300–310. [DOI] [PubMed] [Google Scholar]
- 11.Leontis NB; Westhof E (2001) Geometric nomenclature and classification of RNA base pairs. RNA, 7 (4), 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.