Abstract
A plant-specific B3 domain and AP2 domain-containing transcription factor, RAV1 acts as a negative regulator of growth in many plant species and its transcription was downregulated by BR and ABA. In this study, we found that RAV1-overexpressing transgenic plants showed abnormally developed ovules, resulting in reduced seed size, weight, and number in a silique. Interestingly, the endogenous expression of RAV1 fluctuated during seed development; it remained low during the early stage of seed development and sharply increased in the seed maturation stage. In plants, seed development is a complex process that requires coordinated growth of the embryo, endosperm, and maternal integuments. Among many genes that are associated with endosperm proliferation and embryo development, three genes consisting of SHB1, MINI3, and IKU2 form a small unit positively regulating this process, and their expression was regulated by BR and ABA. Using the floral stage-specific RNAs, we found that the expression of MINI3 and IKU2, the two downstream genes of the SHB1-MINI3-IKU2 cascade in the seed development pathway, were particularly reduced in the RAV1-overexpressing transgenic plants. We further determined that RAV1 directly binds to the promoter of MINI3 and IKU2, resulting in their repression. Direct treatment with brassinolide (BL) improved seed development of RAV1-overexpressing plants, but treatment with ABA severely worsened it. Overall, these results suggest that RAV1 is an additional negative player in the early stages of seed development, during which ABA and BR signaling are coordinated.
Keywords: seed development, SHB1-MINI3-IKU2 pathway, RAV1
INTRODUCTION
Seed size is an important agronomic trait that influences many aspects of regeneration strategies, such as seedling growth, survival rate, and seed dispersal syndrome (Zhang et al., 2015). Seed development begins with double fertilization, followed by differentiation of seed coat derived from the integuments (Lopes and Lakins, 1993). Once double fertilization succeeds in flowering plants, endosperm development proceeds through syncytium formation, followed by cellularization and differentiation (Li and Li, 2015). The embryo then grows from the endosperm, utilizing nutrients and occupying most of the mature seed. During this period, the integument, which begins to form at ovule development, also undergoes cell differentiation and accumulates pigments, mucilage, and starch granules, eventually forming the mature seed coat (Windsor et al., 2000). Therefore, successful seed development and the resulting final size of seeds are determined by the coordinated growth of the embryo, endosperm, and maternal integuments.
In Arabidopsis, many genes associated with seed development have been identified. SHORT HYPOCOTYL UNDER BLUE1 (SHB1 ), HAIKU1 (IKU1 ), HAIKU2 (IKU2 ), and MINISEED3 (MINI3 ) are involved in endosperm size determination and embryo development (Luo et al., 2005; Wang et al., 2010; Zhou et al., 2009). SHB1 is a positive regulator of MINI3 and IKU2 expression, although it cannot directly bind to DNA sequences (Zhou et al., 2009). MINI3 encodes a WRKY family transcription factor and is expressed in both the endosperm and embryo; it promotes the expression of IKU2, which encodes a leucine-rich repeat receptor kinase in the endosperm. Therefore, SHB1, MINI3, and IKU2 form a small signaling unit that successively acts to expand the seed cavity (Luo et al., 2005). In addition to SHB1, IKU1 also interacts and promotes MINI3 expression (Wang et al., 2010). The genes encoding MADS-box transcription factors, such as AGAMOUS-LIKE1 61 (AGL61), AGL62, and AGL80, also regulate endosperm development (Bemer et al., 2008; Kang et al., 2008; Steffen et al., 2008).
Plant hormones, such as brassinosteroids (BRs), cytokinins, auxin, and abscisic acid (ABA), regulate many aspects of seed development (Sun et al., 2010), and their activities seem to be interrelated (Locascio et al., 2014). ABA negatively regulates endosperm development, because the seed size of the ABA-deficient mutant aba2 or ABA-insensitive mutant abi5 was greater than that of the wild-type plants (Cheng et al., 2014). In contrast, BRs promote the enlargement of seed size, because the BR-insensitive mutant bri1-5 or BR-deficient mutant det2-1 showed reduced seed size compared with that of the wild type (Jiang et al., 2013). BR signaling triggered from the cell surface receptor complex BRI1/BAK1 (Nam and Li, 2002; Wang et al., 2005) is transduced to the transcription factors BRASSINAZOLE-RESISTANT1 (BZR1) and BRI1-EMS-SUPPRESSOR1 (BES1) (Wang et al., 2002; Yin et al., 2002) to regulate numerous BR-responsive genes (Kim et al., 2009; Sun et al., 2010). Recently, the expression of the SHB1, IKU2, and MINI3 was shown to be induced by BR. BZR1 directly bound to the promoters of SHB1 and IKU1 and activated them (Jiang et al., 2013). SHB1 activates MINI3 and IKU2 expression, and IKU1 interacts with MINI3 to activate IKU2 (Luo et al., 2005; Zhou et al., 2009).
In Arabidopsis, 118 genes encoding plant-specific B3 transcription factors have been identified (Swaminathan et al., 2008). Among them, a subset related to the ABI3/VP1 (RAV) transcription factor subfamily is encoded by six genes (RAV1, RAV1L, TEM1, TEM2/RAV2, At1g50680, and At1g51120 ) and possesses the AP2-domain (Pérez-Rodríguez et al., 2010). RAV1 was the first transcription factor identified in this family as a novel DNA-binding protein with bipartite sequence motifs (Kagaya et al., 1999). Since then, RAV family transcription factors have been considered negative growth regulators by antisense and overexpression approaches (Castillejo et al., 2008; Hu et al., 2004; Osnato et al., 2012). Previously, the growth promotion in rav1 knockout mutant plants in the early stage of development and growth retardation in transgenic plants overexpressing RAV1 were considered under the control of a native promoter (Fu et al., 2014). The expression of RAV1 was regulated by many plant hormones and down-regulated by ABA (Fu et al., 2014; Hu et al., 2004). BR-induced repression of RAV1 seems to be mediated by the direct binding of BZR1 to the promoter of RAV1 (He et al., 2005; Sun et al., 2010).
In this study, we focused on how RAV1 affects seed development, because overexpression of the RAV1 adversely affected all aspects, such as seed size and mass, seed numbers in a silique, and seed shape. The endogenous expression of RAV1 remained low during the seed developmental stage. We examined the expression of the genes associated with ovule development or embryo and endosperm development in the RAV1-overexpressing transgenic plants. We found that the expressions of MINI3 and IKU2, the two downstream genes of the SHB1-MINI3-IKU2 cascade in the seed development pathway, were particularly reduced, because RAV1 directly binds to the promoter of MINI3 and IKU2, resulting in their repression.
MATERIALS AND METHODS
Plant growth and conditions
Arabidopsis thaliana Columbia (Col-0) was used as wild type. RAV1-overexpressing transgenic plants in a Col-0 background were previously described (Fu et al., 2014). For growth in soil, seeds were planted directly onto Sunshine #5 top-layered with fine granules of vermiculite. All plants were grown at 22°C under long-day conditions (16 of light and 8 of darkness).
Assessment of seed germination
Seeds were sterilized in 75% ethanol containing 0.05% Tween-20 for 15 min, washed twice with 95% ethanol, and placed onto 1/2 Murashige & Skoog (MS) (Duchefa) media containing 0.8% phytoagar. The plate was sequentially exposed to far-red light (3.2 μmol/m2/s) and red light (20 μmol/m2/s) as previously described (Oh et al., 2006), and then placed in the growth room set to long day conditions. Each day, germinating seeds were counted based on radicle protrusion and the germination rate was calculated.
Morphological analysis of seeds
Seed morphology was analyzed for the seeds in green siliques of the RAV1-overexpressing transgenic plants and wild-type plants using a dissecting microscope (Leica MZ12.5 stereomicroscope). To examine the effect of direct application of BL and ABA, 2 μl of BL (1 μM) or ABA (10 μM) solution was absorbed onto the floral bud at stage 12–13. After 7 days, siliques were collected and analyzed. The number of seeds showing various defects per silique counted. Seed weight was determined by weighing 500 dry seeds at a time. Seed length and width were measured using the images that were taken using the dissecting microscope equipped with a CCD camera, and were analyzed using Image J software.
RNA expression analysis
Total RNA was extracted from various floral stages from 12 to 20 using TRI Reagent® (SIGMA). First-strand cDNAs were synthesized using RNA treated with RNase-free RQ1 DNases (Promega) and adding SuperScriptIII-MMLV reverse transcriptase (Invitrogen) and the Oligo(dT)-15 primer. An aliquot of the cDNA samples was used as template for quantitative RT-PCR analyses using SYBR Green PCR Master Mix in the Step-One Plus Real Time PCR system (Applied Biosystems). Expression of the Ubiquitin gene (At3g62250 ) was used to normalize the data.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed following the procedure previously described (Gendrel et al., 2005) with some modifications. In brief, 0.8 g of floral tissue at stages 15 to 16 from the RAV1-overexpressing transgenic plants were cross-linked with 1% formaldehyde for 15 min under a vacuum; the reaction was stopped by adding 0.125 M glycine. Nuclei were isolated in an extraction buffer (1.7 M sucrose, 10 mM Tris-HCl, pH 8.0, 2 mM MgCl2, 0.15% Trion X-100, 5 mM β-mercaptoethanol, protease inhibitor cocktail (Roche)), and were re-suspended in a sonication buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS, protease inhibitor cocktail (Roche)). The chromatin was released by sonication and DNA fragments with an average size of 0.2–1.5 kb only were recovered. The extracted DNA fragments were diluted with an equal volume of ChIP dilution buffer (16.7 mM Trs-HCl, pH 8.0, 1.2 mM EDTA, 167 mM NaCl, 1.1% Triton X-100, protease inhibitor cocktail (Roche)) and incubated with polyclonal anti-RAV1 antibody (Ab Frontier) at 4°C overnight. The chromatin-antibody complex was precipitated with protein-A beads (GE Healthcare) at 4°C for 1 h. Reverse crosslinked DNA was captured and cleaned using the PCR cleanup kit (Favorgen Biotech Corp). PCR was performed using the specific primer sets indicated, and the fold change was normalized by the data from an experiment done using the same procedure without the antibody.
Primer sequences
Primer sequences for the genes used in this study are listed in Supplementary Table S1.
RESULTS
RAV1-overexpressing transgenic plants showed defective seed development
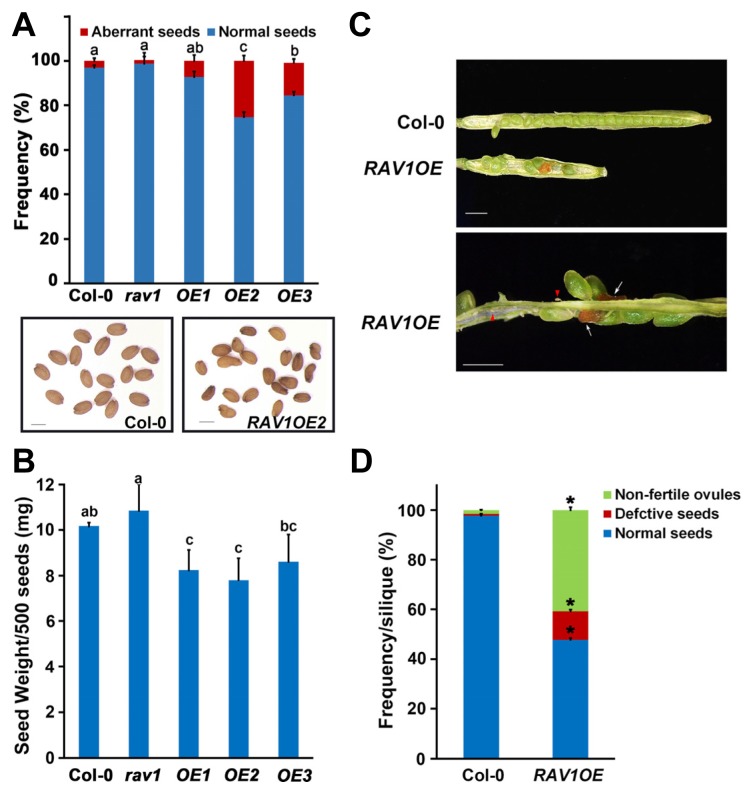
Previously, we reported that RAV1 is a negative regulator of growth (Fu et al., 2014), because overall growth retardation occurred in the RAV1-overexpressing transgenic plants compared with wild-type plants. The growth pattern of the rav1 mutant appeared similar to that of wild-type plant showing a slight promotion only in the early stage of growth (Supplementary Fig. S1). To further explore the specific functions of RAV1, we observed detailed morphological alterations of the RAV1-overexpressing transgenic plants. We frequently observed wrinkled and shrunken seeds in the RAV1 -overexpressing plants, compared with the regularly ellipsoidal seed shape in wild-type plants. The frequency of the appearance of this aberrant seed shape was considerably higher in RAV1-overexpressing plants than in wild-type plants (Fig. 1A). We further found that even normal shaped seeds from the RAV1-overexpressors were lighter and smaller than that of the wild-type plants (Fig. 1B). These phenotypic alterations of seeds were not observed in the rav1 mutant. Based on these results, we decided to further explore the effects of RAV1 overexpression on the seed development and selected the RAV1OE2 transgenic plant as a representative (hereafter RAV1OE ). In addition to the abnormal seeds that were already harvested, we found that the silique shape of the transgenic plants was different from that of wild-type plants. Unlike the long and flat cylindrical siliques in wild-type plants, short and sunken siliques with empty space were observed in the RAV1-overexpressors. Green siliques of the RAV1-overexpressing plants contained several brown-colored ovules and aborted ovules (Fig. 1C). The brown-colored ovules are un-fertilized ovules, and the withering aborted whitish ovules results from ovule development failure (Kirioukhova et al., 2011). To analyze the results quantitatively, we separately counted the numbers of normal-looking seeds, unfertilized ovules, and aborted ovules in each silique. Less than 1% of seeds showed defective development of approximately 40 seeds in the wild-type silique. In contrast, more than 45% of developing seeds of the total of approximately 20 seeds in a silique of RAV1-overexpressing plants were defective, and degenerating seed development was detected (Fig. 1D). This phenomenon was likely to be specific to RAV1’s action, because similar defective phenotypes did not occur in the rav1l or rav2 mutant and in the transgenic plants overexpressing RAV1L or RAV2 (Supplementary Fig. S2).
Fig. 1. Defective seed development in the RAV1-overexpressing transgenic plants.
(A) Frequency of aberrant seeds in rav1 mutant and the RAV1-overexpressing plants compared with those in the wild-type plant (n = 300 seeds per each plant). Seed shape of the RAV1OE2 transgenic plant is shown compared with that of the wild-type plant. Scale bars indicate 1 mm. (B) Seed weight of rav1 mutant and the RAV1-overxpressing plants compared with that of wild-type plants. Values labeled with different letters in (A) and (B) indicate statistical differences across each plant line by one-way ANOVA (P < 0.05). (C) Shape of a silique and the seeds within it of the RAV1-overexpressing plants compared with those of wild-type plants. Un-fertilized ovules and aborted whitish ovules are marked with white arrow and red arrowhead, respectively. Scale bars indicate 1 mm. (D) Frequency of the various seeds in a silique of the RAV1-overexpressing plants compared with those in wild-type plants (n = 15 siliques in each plant, *P < 0.001 for all kinds of seeds).
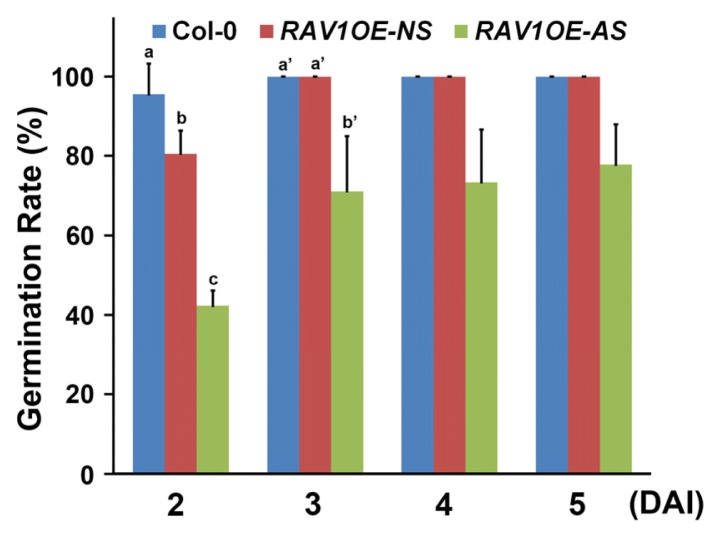
Next, we investigated whether the seeds from RAV1-overexpressing plants had any defects in germination after maturation. Because considerable proportions (approximately 5%) of the final seed shape was aberrant in the RAV1-overexpressing plants (Fig. 1A), we performed the germination test separately for the normal-looking seeds (NS) and aberrant-looking seeds (AS). Under normal conditions, wild-type seeds germinated almost completely at 2 days after imbibition (DAI). Although NS from the RAV1-overexpressing plants showed a germination rate similar to that of the wild type, it was slightly lower at 2 DAI than that of wild-type seeds. In contrast, AS from the RAV1 -overexpressing plants showed a significantly reduced germination rate; only 40% of seeds had germinated at 2 DAI, and more than 20% of the seeds did not germinate even after 5 DAI (Fig. 2). These results suggest that seed germination was delayed, and seed viability was reduced in the AS of the RAV1-overexpressing plants.
Fig. 2. Defective seed development continues to adversely affect seed germination.
Comparison of seed germination of the normal-looking (NS) and aberrant-looking (AS) seeds from the RAV1-overexpressing plants with those from wild-type plants. Values labeled with different letters in 2 or 3 DAI indicate a statistical difference across each day by one-way ANOVA (P < 0.05).
Natural expression of RAV1 kept low in the early stage of seed development
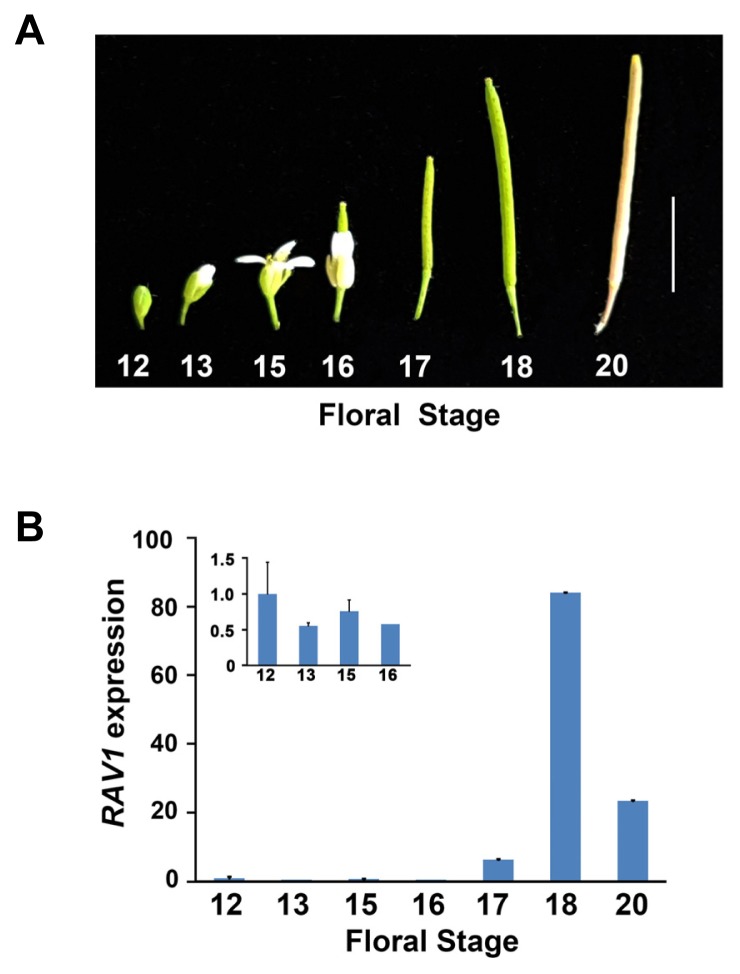
Because normal seed development was perturbed in the RAV1-overexpressing transgenic plants as shown above, we thought that it would be important to first know how natural expression of the endogenous RAV1 changes during seed development. To address this issue, we isolated RNA from different stages of developing flowers and siliques from fertilization to seed maturation stages, which were determined based on flower morphology (Smyth et al., 1990). Flower organ development was almost complete at stage 12. Then, complete anthesis and fertilization occurred successively at stage 13, and developing siliques elongated through stage 15, and petals and sepals were withered at stage 16. All floral organs fell off from green siliques, and siliques became more elongated and yellow during stages 17–20 (Fig. 3A). Using the RNAs of each stage, we performed quantitative real time-polymerase chain reaction (qRT-PCR) analyses for RAV1 expression. Whereas RAV1 expression remarkably increased up to 60-fold at stage 18 when the green siliques became distinct, RAV1 expression did not considerably change during stage 12 to 16, during which the early stage of seed development occurred (Fig. 3B). This result suggests that RAV1 expression at low levels during the early stage of seed development is required for proper seed development. Excessive expression of RAV1 caused by ectopic overexpression, such as in the RAV1-overexpressing plants, inhibits normal seed development.
Fig. 3. Expression analyses of RAV1 during seed development.
(A) Picture of developing flower and siliques of a wild-type plant from which RNA was isolated. Scale bar indicates 10 mm. (B) Relative expression of RAV1 determined using qRT-PCR analysis during indicated flower stages. Inset denotes the expression of RAV1 from stages 12 to 16. qRT-PCR was conducted in triplicate in each experiment and data were normalized to the expression of Ubiquitin. Experiments were independently repeated twice. Error bars indicate standard error.
The expressions of MINI3 and IKU2, which are involved in endosperm and embryo development, were reduced in the RAV1-overexpressing plants
As endogenous expression of the RAV1 was low at stages 12–16 during seed development (as shown above (Fig. 3B), it is reasonable to assume that the overexpression of RAV1 can interfere with timely expression of the genes involved in proper seed development. Therefore, we next focused on identifying the genes affected by RAV1 overexpression during these stages.
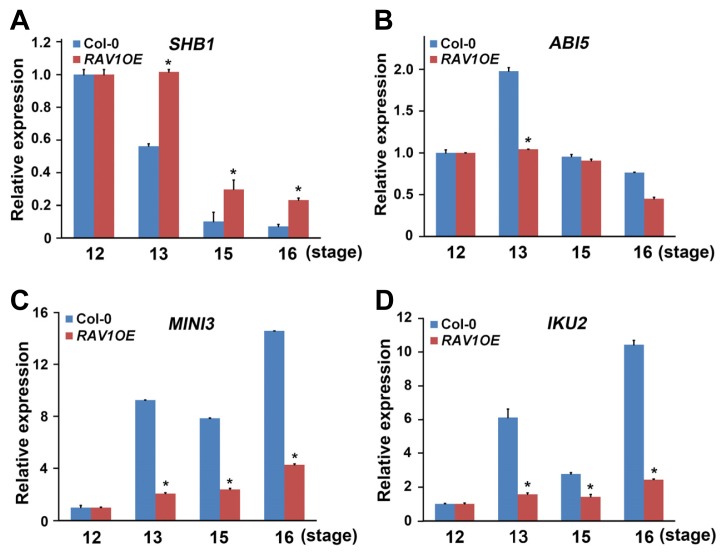
Among many genes, the SHB1, MINI3, and IKU2 were good candidates to test, because the SHB1-MINI3-IKU2 pathway is a well-known cascade for the positive regulation of endosperm proliferation and embryo development. SHB1 activates MINI3 and IKU2 expression and MINI3 activates IKU2 (Luo et al., 2005). In addition, the ABA signaling component ABI5 negatively acts upstream of this pathway by directly binding to the promoter of SHB1, leading to its repression (Cheng et al., 2014). Therefore, using the RNAs from stage 12 to 16, we compared the expressions of these genes in the wild type and RAV1-overexpressing plants. In wild-type plants, SHB1 expression gradually decreased from stages 12 to 16 (Fig. 4A), and ABI5 expression changed with the opposite pattern of that of SHB1, especially at stages 12 and 13 (Fig. 4B). Compared to these wild-type patterns, SHB1 accumulation levels in RAV1-overexpressing plants were higher at all monitored stages, with the gradual reduction in expression delayed (Fig. 4A). ABI5 expression did not increase in RAV1-overexpressing plants at 13 stage (Fig. 4B). These results suggest that ABI5 expression was repressed by the overexpression of RAV1, resulting in the increased expression of SHB1 in the RAV1-overexpressing plants. Next, we compared expression of the MINI3 and IKU2. In wild-type plants, both MINI3 and IKU2 expression increased from stage 13 and remained continuously high until the stage 16, implying that SHB1 that was encoded by the previous activation of the SHB1 transcript at stage 12, which caused the subsequent activation of these genes (Figs. 4C and 4D). However, in the RAV1-overexpressing plants, despite the higher expression of SHB1 at stage 12 and 13, the accumulations of MINI3 and IKU2 after stage 13 were much lower compared with those of wild-type plants (Figs. 4C and 4D).
Fig. 4. Expression analyses of the genes associated with embryo and endosperm development.
Expressions level of SHB1 (A), ABI5 (B), MINI3 (C), and IKU2 (D) were analyzed during the period indicated as the flower stage of the RAV1-overexpressing plants compared with those of wild-type plants. In each experiment, RNAs were examined in triplicate in each experiment and data were normalized to the expression of Ubiquitin. Experiments were independently repeated three times. Error bars indicate standard error. (*P < 0.01 compared with wild-type plant at the same floral stage)
RAV1 directly binds to the promoter of MINI3 and IKU2
Because the expressions of MINI3 and IKU2 were not coupled with that of the expression of SHB1 in the RAV1-overexpressing plants, we hypothesized that these genes could be the direct targets of RAV1. Indeed, the repression of ABI5 by RAV1 was reported during seed germination and post-germination seedling growth. In addition to ABI5, RAV1 also directly binds to the promoters of ABI3 and ABI4, leading to the perturbation of ABA signaling (Feng et al., 2014). Therefore, it is plausible that repression of ABI5 by the RAV1 would be effective in seed development. To validate this hypothesis, we performed a chromatin immunoprecipitation (ChIP) assay. Immunoprecipitated chromatin samples against anti-RAV1 antibodies were prepared from the stage 15 and 16 flowers of RAV1-overexpressing plants. Then, the promoter regions of ABI5 that were already reported to be the binding site for RAV1 in seedlings were targeted to be amplified. Compared with that of amplification results without antibody, we found that RI and RII fragments of the ABI5 were significantly amplified in the samples containing the RAV1 antibody (Supplementary Fig. S3).
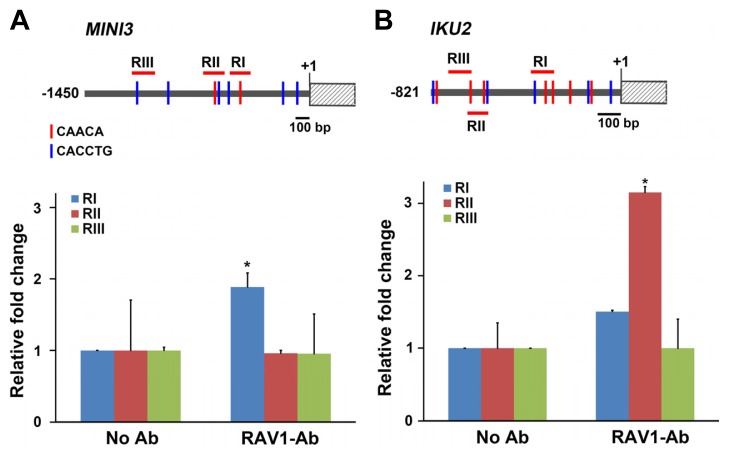
In this study, we further focused on how RAV1 affects MINI3 and IKU2 expression. First, we analyzed the promoter sequences of MINI3 and IKU2 to determine whether any RAV1-binding motifs were present in these regions. RAV1 bound to two distinct DNA binding motifs, 5′-CAACA-3′ and 5′-CACCTG-3′ (Kagaya et al., 1999). The latter is similar to the E-box (CANNTG) motif. We found two 5′-CAACA-3′ motifs and six E-box-like 5′-CACCTG-3′ motifs within the 1 Kb region of the MINI3 promoter, and seven 5′-CAACA-3′ motifs and five 5′-CACCTG-3′ motifs in the 1 Kb region of the IKU2 promoter (Figs. 5A and 5B). To confirm whether these motifs could be the actual target sites for RAV1, we performed a ChIP assay with the same immunoprecipitated chromatin samples against anti-RAV1 antibodies that were used for ABI5. We determined that several fragments containing the RAV1-binding motifs in the promoters of MINI3 and IKU2 were amplified. We found that the RI fragment contained one 5′-CAACA-3′ motif, spanning from −368 to −521 bases in the promoter of MINI3, and it was approximately two-fold more amplified than that of the negative control (Fig. 5A). In the promoter of IKU2, the RII fragment spanning from −543 to −646 bases was significantly amplified (Fig. 5B). Taken together, our findings suggest that the reduced expression of MINI3 and IKU2 in addition to ABI5 in the RAV1-overexpressing plants was caused by direct binding of the RAV1 to their promoters, resulting in their repression.
Fig. 5. RAV1 directly binds to the promoter of MIN3 and IKU2.
A and B. Two different RAV1-binding sites are marked with red and blue bars in a schematic representation of the MINI3 and IKU2 promoters, respectively (upper panels). Graph showing the relative amplification of the fragments containing RAV1-binding sites of MINI3 or IKU2 using ChIP analyses (lower panels). (*P < 0.05 compared with the same amplified region without antibody treatment)
Defective seed development of RAV1-overexpressing plants was partially rescued by BL treatment
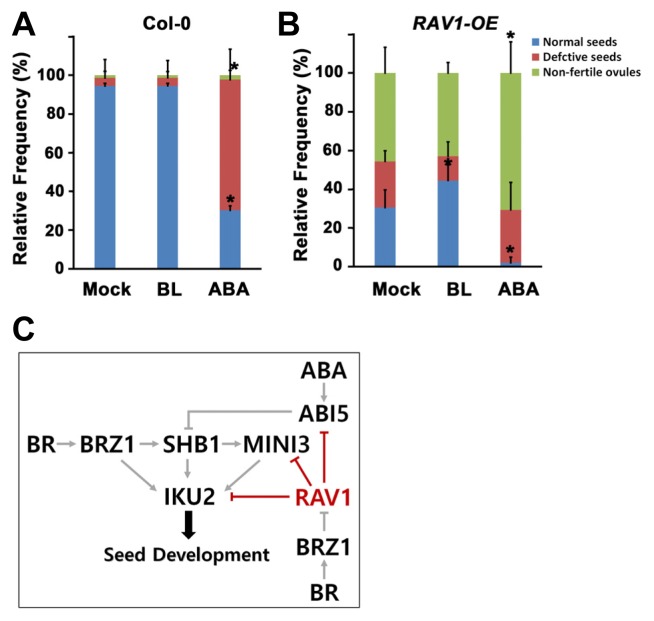
BRs promote seed development (Jiang et al., 2013) and repress RAV1 expression (He et al., 2005). RAV1 expression decreased naturally during seed development (Fig. 2). If the overexpression of RAV1 conferred inhibitory effects on seed development, direct application of BR may promote seed development of the RAV1-overexpressing plants. To prove this hypothesis, we treated brassinolide (BL), the most bioactive BR, to the floral buds at stage 12–13, just before seed development commenced. We also examined the effect of ABA on seed development for comparison. After 7 days, we collected elongated siliques and observed the seeds inside them. In the wild-type silique, most of the seeds were normal. We found that ABA treatment in the wild-type flower severely inhibited seed development thereafter, whereas BL treatment did not much alter the proportion of normally developed seeds compared with that of the mock-treatment (Fig. 6A). In comparison, more than half of the seeds were defective or non-fertile ovules in the siliques of the RAV1-overexpressing plants. Interestingly, we observed that BL treatment improved normal seed development approximately 15% (Fig. 6B). ABA significantly inhibited normal seed development in RAV1-overexpressing plants, as well as in wild-type plants.
Fig. 6. RAV1 interacts with BR and ABA signaling.
Treatment with BL or ABA of the floral bud of wild-type (A) and RAV1-overexpressing transgenic plant (B) affects seed development. After hormone treatment, frequency of the various seeds in a silique from each plant were counted and compared with those from mock-treated plants (n = 15 siliques from each plant, *p < 0.001 compared with mock-treated sample). C. Model for RAV1 action in seed development. The SHB1-MINI3-IKU2 pathway is positively regulated by BR via the action of BZR1 and is negatively regulated by ABA via the action of ABI5. In this known pathway, RAV1, whose transcriptional expression is down-regulated by BZR1, represses the expression of ABI5, MINI3, and IKU2 as a transcriptional repressor, which provides information for the fine regulation of seed development.
DISCUSSION
Successful seed development depends on several criteria, such as seed size/mass, seed number per silique, and proper seed shape (Jiang et al., 2013). In this study, we showed that overexpression of RAV1 adversely affected all aspects of seed development: reduced seed size and mass, decreased seed numbers in a silique, and aberrant seed shape (Fig. 1). Previously, RAV1 was reported to be one of the representative genes that was down-regulated by BR (He et al., 2005). Recently, RAV1 was also shown to be down-regulated by ABA (Fu et al., 2014).
During seed development, two peaks in ABA accumulation have been noted. First, ABA accumulation originates from the maternal tissues and promotes endosperm development. Later, another ABA peak occurs in the embryo during the late maturation stage up to desiccation (Finkelstein et al., 2010), which is the more profound function of ABA during seed development. ABA-deficient mutants, such as aba2-1, aba1, and nced6 exhibit increased seed size because of delayed endosperm cellularization, although their silique length was shorter than that of the wild type (Cheng et al., 2014). The timing of endosperm cellularization is important to the determination of the final seed size (Kang et al., 2008; Sundaresan et al., 2005). Indeed, precocious cellularization of the endosperm causes reduced nuclear proliferation, resulting in smaller seed size; in contrast, delayed cellularization leads to increased seed size (Garcia et al., 2003; 2005; Luo et al., 2005). In aba2-1 mutants, the upregulation of SHB1 expression was repressed by the application of ABA (Cheng et al., 2014). ABI5 directly binds to the promoter region of SHB1, leading to its repression. This might trigger the subsequent embryo development. This notion is consistent with our observation that endogenous expression of ABI5 and SHB1 was oppositely regulated in flowers at stages 12 and 13, and SHB1 expression gradually decreased in flowers from stage 13 to 16 (Figs. 4A and 4B). ABI5 was shown to be a direct target of RAV1 during seedling development (Feng et al., 2014) and seed maturation (Supplementary Fig. S3). In the presence of ABA, RAV1 was phosphorylated by SnRK2 kinases (Feng et al., 2014), which are the positive regulators of ABA signaling (Luo et al., 2005; Mustilli et al., 2002; Yoshida et al., 2002). Phosphorylated RAV1 lost its ability to repress ABI5 expression (Feng et al., 2014). Previously, we and another group reported that RAV1-overexpressing plants exhibited ABA insensitivity (Feng et al., 2014; Fu et al., 2014). Therefore, the timely expression of ABI5 and SHB1 was deregulated in the RAV1-overexpressing plants (Figs. 4A and 4B). In addition, the higher SHB1 expression in these plants did not couple with increased expression of MINI3 and IKU2 (Figs. 4C and 4D).
Recently, reduced seed cavity and endosperm volume in the BR-deficient mutant det2 was shown to result in delayed embryo development and decreased embryo size and number (Jiang et al., 2013). The fact that BR treatment rescued the defects of det2 implied that the activation of BR signaling is required for normal seed development. SHB1, IKU1, MINI3, and IKU2 are genes induced by BR via BZR1 activity (Jiang et al., 2013). BR-induced repression of RAV1 seems to be mediated by the direct binding of BZR1 to the promoter of RAV1 (He et al., 2005; Sun et al., 2010). These findings correlate well with our result that endogenous RAV1 expression was maintained at a low level during early seed development stages (Fig. 3B). Furthermore, we showed that RAV1 overexpression decreased the expression of MINI3 and IKU2 (Figs. 4C and 4D) by directly binding to their promoters (Figs. 5A and 5B). IKU2 encodes a leucine-rich repeat kinase and probably transduces important signaling for endosperm and embryo development (Luo et al., 2005). These results suggest that the overexpression of RAV1 counteracts BZR1 in the activation of MINI3 and then IKU2 expression (Fig. 6C). The results showing that direct treatment with BL improved normal seed development in the RAV1-overexpressing plants also supported this model (Fig. 6B). Under natural conditions, gradients of plant hormones accumulate in different seed compartments during seed development (Locascio et al., 2014). Therefore, the accumulation of plant hormones and transcriptional activity of RAV1 might coordinately regulate the expression of the core components for endosperm and embryo development.
Supplementary data
ACKNOWLEDGMENTS
The authors would like to thank the former laboratory member, Hyun Kyung Kang, for technical assistance to generate transgenic plants. This work was supported by grants from the Next-Generation BioGreen 21 Program (SSAC, PJ01321701 to K.H.N), Rural Development Administration, Republic of Korea, and from the Basic Science Research Program (grant No. 2017R1A2B4002656 to K.H.N, and 2017R1A6A3A11031952 to H.Y.S) through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning Republic of Korea.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Bemer M., Heijmans K., Airoldi C., Davies B., Angenent G.C. The MADS domain protein DIANA acts together with AGAMOUS-LIKE80 to specify the central cell in Arabidopsis ovules. Plant Cell. 2008;20:2088–2101. doi: 10.1105/tpc.108.058958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C., Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol. 2008;18:1338–1343. doi: 10.1016/j.cub.2008.07.075. [DOI] [PubMed] [Google Scholar]
- Cheng Z., Zhao X.Y., Shao X.X., Wang F., Zhou C., Liu Y.G., Zhang Y., Zhang X.S. Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell. 2014;26:1053–1068. doi: 10.1105/tpc.113.121566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C.Z., Chen Y., Wang C., Kong Y.H., Wu W.H., Chen Y.F. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 2014;80:654–668. doi: 10.1111/tpj.12670. [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R. The Role of Hormones during Seed Development and Germination. Plant Hormones. 2010;E4:549–573. [Google Scholar]
- Fu M., Kang H.Y., Son H-Y, Kim S-K, Nam K.H. A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiol. 2014;55:1892–1904. doi: 10.1093/pcp/pcu118. [DOI] [PubMed] [Google Scholar]
- Garcia D., Fitz Gerald J.N., Berger F. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell. 2005;17:52–60. doi: 10.1105/tpc.104.027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D., Saingery V., Chambrier P., Mayer U., Jürgens G., Berge F. Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 2003;131:1661–1670. doi: 10.1104/pp.102.018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.X., Wang Y.X., Liu X.F., Li J.Y. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 2004;4:8–15. doi: 10.1038/sj.cr.7290197. [DOI] [PubMed] [Google Scholar]
- Jiang W.B., Huang H.Y., Hu Y.W., Zhu S.W., Wang Z.Y., Lin W.H. Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol. 2013;162:1965–1977. doi: 10.1104/pp.113.217703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y., Ohmiya K., Hattori T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 1999;27:470–478. doi: 10.1093/nar/27.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang I.H., Steffen J.G., Portereiko M.F., Lloyd A., Drews G.N. The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell. 2008;20:635–647. doi: 10.1105/tpc.107.055137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.X., Sun Y. Burlingame A.L., Wang Z.Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nature Cell Biol. 2009. pp. 1254–1260. [DOI] [PMC free article] [PubMed]
- Kirioukhova O., Johnston A.J., Kleen D., Kägi C., Baskar R., Moore J.M., Bäumlein H., Gross-Hardt R., Grossniklaus U. Female gametophytic cell specification and seed development require the function of the putative Arabidopsis INCENP ortholog WYRD. Development. 2011;138:3409–3420. doi: 10.1242/dev.060384. [DOI] [PubMed] [Google Scholar]
- Li N., Li Y. Maternal control of seed size in plants. J Exp Bot. 2015;66:1087–1097. doi: 10.1093/jxb/eru549. [DOI] [PubMed] [Google Scholar]
- Lopes M., Lakins B.A. Endosperm origin, development, and function. Plant Cell. 1993;5:1383–1399. doi: 10.1105/tpc.5.10.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A., Roig-Villanova I., Bernardi J., Varotto S. Current perspectives on the hormonal control of seed development in Arabidopsis and maize: a focus on auxin. Front Plant Sci. 2014;5:412–422. doi: 10.3389/fpls.2014.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Dennis E.S., Berger F., Peacock W.J., Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli A.C., Merlot S., Vavasseur A., Fenzi F., Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K.H., Li J. BRI1/BAK1, a Receptor Kinase Pair Mediating Brassinosteroid Signaling. Cell. 2002;26:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W.I., Choi G. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006;47:124–139. doi: 10.1111/j.1365-313X.2006.02773.x. [DOI] [PubMed] [Google Scholar]
- Osnato M., Castillejo C., Matías-Hernández L., Pelaz S. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun. 2012;3:808. doi: 10.1038/ncomms1810. [DOI] [PubMed] [Google Scholar]
- Pérez-Rodríguez P., Riaño-Pachón D.M., Corrêa L.G.G., Rensing S.A., Kersten B., Mueller-Roeber B. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010;38:D822–827. doi: 10.1093/nar/gkp805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. Early flower development in Arabídopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen J.G., Kang I.H., Portereiko M.F., Lloyd A., Drews G.N. AGL61 interacts with AGL80 and is required for central cell development in Arabidopsis. Plant Physiol. 2008;148:259–268. doi: 10.1104/pp.108.119404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V. Control of seed size in plants. Proc Natl Acad Sci USA. 2005;102:17887–17888. doi: 10.1073/pnas.0509021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Fan X.Y., Cao D.M., Tang W., He K., Zhu J.Y., He J.X., Bai M.Y., Zhu S., Oh E., Patil S., Kim T.W., Ji H., Wong W.H., Rhee S.Y., Wang Z.Y. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan K., Peterson K., Jack T. The plant B3 superfamily. Trends Plant Sci. 2008;13:647–655. doi: 10.1016/j.tplants.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Wang A., Garcia D., Zhang H., Feng K., Chaudhury A., Berger F., Peacock W.J., Dennis E.S., Luo M. The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 2010;63:670–679. doi: 10.1111/j.1365-313X.2010.04271.x. [DOI] [PubMed] [Google Scholar]
- Wang X., Li X., Meisenhelder J., Hunter T., Yoshida S., Asami T., Chory J. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell. 2005;8:855–865. doi: 10.1016/j.devcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Choty J. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- Windsor J.B., Symonds V.V., Mendenhall J., Lloyd A.M. Arabidopsis seed coat development: morphological differentiation of the outer integument. Plant J. 2000;22:483–493. doi: 10.1046/j.1365-313x.2000.00756.x. [DOI] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Yoshida R., Hobo T., Ichimura K., Mizoguchi T., Takahashi F., Aronso J., Ecker J.R., Shinozaki K. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Du L., Xu R., Cui X.R., Hao J., Lia Y. Transcription Factors SOD7/NGAL2 and DPA4/NGAL3 act redundantly to regulate seed size by directly repressing KLU expression in Arabidopsis thaliana. Plant Cell. 2015;27:620–632. doi: 10.1105/tpc.114.135368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhang X., Kang X., Zhao X., Zhang X., Ni M. SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell. 2009;21:106–117. doi: 10.1105/tpc.108.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.