Abstract
As sessile organisms, plants have evolved to adjust their growth and development to environmental changes. It has been well documented that the crosstalk between different plant hormones plays important roles in the coordination of growth and development of the plant. Here, we describe a novel recessive mutant, mildly insensitive to ethylene (mine), which displayed insensitivity to the ethylene precursor, ACC (1-aminocyclopropane-1-carboxylic acid), in the root under the dark-grown conditions. By contrast, mine roots exhibited a normal growth response to exogenous IAA (indole-3-acetic acid). Thus, it appears that the growth responses of mine to ACC and IAA resemble those of weak ethylene insensitive (wei) mutants. To understand the molecular events underlying the crosstalk between ethylene and auxin in the root, we identified the MINE locus and found that the MINE gene encodes the pyridoxine 5′-phosphate (PNP)/pyridoxamine 5′-phosphate (PMP) oxidase, PDX3. Our results revealed that MINE/PDX3 likely plays a role in the conversion of the auxin precursor tryptophan to indole-3-pyruvic acid in the auxin biosynthesis pathway, in which TAA1 (TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1) and its related genes (TRYPTOPHAN AMINOTRANSFERASE RELATED 1 and 2; TAR1 and TAR2) are involved. Considering that TAA1 and TARs belong to a subgroup of PLP (pyridoxal-5′-phosphate)-dependent enzymes, we propose that PLP produced by MINE/PDX3 acts as a cofactor in TAA1/TAR-dependent auxin biosynthesis induced by ethylene, which in turn influences the crosstalk between ethylene and auxin in the Arabidopsis root.
Keywords: Arabidopsis, auxin biosynthesis, ethylene, PDX3, PLP
INTRODUCTION
Plants, which are sessile in nature, need to adapt their growth and development to a changing environment for survival and reproduction. Plant hormones play important roles in ensuring flexible growth and development by coordinating the interactions between environmental signals and genetic programs (Wolters and Jürgens, 2009). Furthermore, accumulating evidence indicates that the crosstalk between different hormone pathways is crucial for the plasticity of plant growth and development under various environmental conditions (reviewed in Depuydt and Hardtke, 2011; Gazzarrini and McCourt, 2003; Vanstraelen and Benková, 2012). Substantial data have been published on the interactions between auxin and ethylene in the regulation of Arabidopsis (Arabidopsis thaliana) root growth (Alonso et al., 2003; He et al., 2011; Luschnig et al., 1998; Pickett et al., 1990; Robles et al., 2013; Roman et al., 1995; Růžička et al., 2007; Stepanova and Alonso, 2005; Stepanova et al., 2005; 2007; 2008; Swarup et al., 2002; 2007). For example, weak ethylene insensitive 2 (wei2) and wei7 mutants, which have defects in the α and β subunits of anthranilate (Ant) synthase (ASA and ASB), a key biosynthetic enzyme for the auxin precursor tryptophan (Trp), display root-specific insensitivity to ethylene or its precursor 1-aminocyclopropane-1-carboxylic acid (ACC) (Stepanova et al., 2005). In addition, quantitative analysis of auxin in the root under ACC treatment has revealed that ethylene induces auxin biosynthesis (Růžička et al., 2007; Swarup et al., 2007). Interestingly, it has been demonstrated that mutations in WEI8, which encodes TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1), a key enzyme converting Trp to indole-3-pyruvic acid (IPyA) in one of the auxin biosynthesis pathways (Stepanova et al., 2008; 2011; Tao et al., 2008), cause root-specific insensitivity to ethylene. In addition to TAA1, TRYPTOPHAN AMINOTRANSFERASE RELATED 1 (TAR1) and TAR2, two homologs of TAA1, have also been shown to play key roles in ethylene-mediated auxin biosynthesis (He et al., 2011; Stepanova et al., 2008; 2011). Taken together, the previous studies indicate that ethylene-induced inhibition of root growth is primarily due to high auxin production at the root apex, which is substantially induced by ethylene. Likewise, auxin can also induce ethylene biosynthesis by activating transcription of the genes encoding ACC SYNTHASE (ACS), which catalyzes a rate-limiting step in ethylene production (Abel et al., 1995; Stepanova et al., 2008). Thus, the crosstalk between ethylene and auxin is crucial for growth and development, including root-specific growth inhibition via auxin biosynthesis mediated by ethylene (He et al., 2011; Růžička et al., 2007; Stepanova et al., 2005; 2007; 2008; Swarup et al., 2007).
ACSs, TAA1 and TARs all belong to the pyridoxal-5′-phosphate (PLP)-dependent class of enzymes (He et al., 2011; Huai et al., 2001; Stepanova et al., 2008; Tao et al., 2008). PLP, a B6 vitamer, acts as a cofactor in numerous enzymatic reactions, including ethylene and auxin biosynthesis. Vitamin B6 collectively refers to PLP and its vitamers, which comprise pyridoxine (PN), pyridoxamine (PM), pyridoxal (PL), and their 5′ phosphorylated forms: pyridoxine 5′-phosphate (PNP), pyridoxamine 5′-phosphate (PMP), and PLP (Colinas et al., 2016; Fitzpatrick et al., 2007; Rueschhoff et al., 2013). PLP can be produced by both de novo and salvage pathways. In Arabidopsis, previous research has demonstrated that both pyridoxal phosphate synthase 1 (PDX1) and PDX2 play central roles in the de novo pathway (Boycheva et al., 2015; Chen and Xiong, 2005; Denslow et al., 2007; Tambasco-Studart et al., 2005; Titiz et al., 2006; Wagner et al., 2006). In the Arabidopsis genome, there are three PDX1 homologs (referred to as PDX1.1, PDX1.2, and PDX1.3 ), and one PDX2 homolog (Boycheva et al., 2015; Chen and Xiong, 2005; Denslow et al., 2007; González et al., 2007; Tambasco-Studart et al., 2005; Titiz et al., 2006; Wagner et al., 2006). In the salvage pathway, PN, PM, and PL can be phosphorylated by the SALT OVERLY SENSITIVE 4 (SOS4) kinase (González et al., 2007; Rueschhoff et al., 2013; Shi and Zhu, 2002; Shi et al., 2002). In addition to SOS4, the PNP/PMP oxidase, PDX3 (also known as PPOX), catalyzes the conversion of the phosphorylated forms of PN and PM into PLP (Colinas et al., 2016; González et al., 2007; Sang et al., 2007; 2011). Intriguingly, it has been shown that mutations in the PDX1.1 and PDX1.3 genes, which are involved in the de novo pathway, display growth phenotypes linked to impaired ethylene and auxin biosynthesis (Boycheva et al., 2015; Chen and Xiong, 2009a; 2009b). However, it is currently unknown whether PDX3 in the salvage pathway is also involved in the biosynthesis of the two hormones.
To identify additional molecular components involved in ethylene-auxin crosstalk, in this study, we performed a genetic screen for root-specific growth responses to ethylene. By utilizing the GAL4/UAS activation-tagging system previously reported (Waki et al., 2013), we identified a novel Arabidopsis mutant with insensitivity to root growth inhibition in the presence of ACC. Through genetic, physiological, and molecular analyses, we revealed that the PNP/PMP oxidase (PDX3) in the salvage pathway plays a role in root-growth response to ethylene via ethylene-induced auxin biosynthesis.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia-0 (Col) and its derivatives were used in all experiments, except for genetic mapping, in which mine was crossed with Landsberg erecta (Ler) to generate a mapping population. The Col T-DNA insertion mutant, pdx3-3 (SALK_054167C), was identified on the SIGnAL website (http://signal.salk.edu) and obtained from the Arabidopsis Biological Resource Center (ABRC). Other mutant and transgenic lines used in this study have been described previously: ein2-5 (Alonso et al., 1999; Guzmán and Ecker, 1990), sur2 (Barlier et al., 2000; Delarue et al., 1998; Stepanova et al., 2011; SALK_028573), wei2-2 (Stepanova et al., 2005; SALK_017444C), wei7-4 (Stepanova et al., 2005; CS16700), wei8-4 (Stepanova et al., 2008; SALK_022743C), CYCB1::GUS (Donnelly et al., 1999), DR5 (Ulmasov et al., 1997), ProSCR::GFP-SCR (Di Laurenzio et al., 1996; Gallagher et al., 2004), ProSHR::SHR-GFP (Helariutta et al., 2000; Nakajima et al., 2001), and ProWOX5::GFP (Sarkar et al., 2007). Seeds were sterilized with 50% sodium hypochlorite and 0.15% Tween 20 for 2 min, rinsed with sterile water for four times, and imbibed at 4°C in the dark as previously described (Lee et al., 2012). Imbibed seeds were placed onto half-strength (0.5X) Murashige and Skoog (MS) plates containing 1% sucrose and 1% agar supplemented with or without aminocyclopropane-1-carboxylic acid (ACC), indole-3-acetic acid (IAA), and tryptophan (Trp), and grown vertically for 4 d in the dark at 22°C after a 6 h-exposure to light. For multiplication of crossed or transgenic plants, seedlings grown in MS agar plates were transferred to soil and were further incubated to full maturity at 22°C under a long photoperiod (16 h light/ 8 h dark), as described previously (Lee et al., 2012).
Screening for mine and isolation of the MINE locus
For identification of mine, approximately 1,000 M2 activation-tagging lines were screened for insensitivity to growth arrest in the presence of the ethylene precursor ACC. In particular, the GAL4/UAS activation-tagging system was utilized for mutant screening. For this, we first generated a driver line that expressed a synthetic transcriptional activator, GAL4:VP16 (GV), under the control of the quiescent center (QC)-specific WOX5 promoter (ProWOX5::GV ). Subsequently, this host line was transformed with T-DNA containing five copies of an upstream activation sequence (pBIB-UAS) as previously reported (Waki et al., 2013). In the first round of mutant screening, we identified one line that exhibited mild insensitivity to ethylene-induced inhibition of root growth. The newly isolated mutant was back crossed to Col wild-type (WT) plants at least 3 times for further study. To identify the MINE locus, thermal asymmetric interlaced-PCR (TAIL-PCR) was conducted as described previously (Liu et al., 1995). Simultaneously, genetic mapping of F2 progeny of the mutant and Ler was carried out using CAPS (Cleaved Amplified Polymorphic Sequences) markers as previously described (Konieczny and Ausubel, 1993; Lukowitz et al., 2000). The primer sequences are listed in Supplementary Table S1.
Growth assay of hypocotyls and roots
Digital images of seedlings at given times were taken with SP-560UZ digital camera (Olympus, Japan). Lengths of hypocotyls and roots from the digital images were measured using ImageJ software (http://rsbweb.nih.gov/ij). There were three replicates for each experiment and statistical significance was determined by the Student’s t-test using Excel (Microsoft, USA), as previously described (Lee et al., 2012). Error bars represent the standard deviation (SD) of the mean. To measure the length of fully expanded cortex cells, the roots were cleared as previously described, with modifications (Lee et al., 2012). Seedlings were incubated in 70%, 80%, and 90% ethanol (EtOH) for 30 min, incubated in 100% EtOH for 20 min, and finally in 70% EtOH for 30 min. After dehydration, the roots were incubated in clearing solution (5% NaOH and 60% EtOH) for 15 min, incubated in a series of EtOH/glycerol mixtures (10% glycerol/50% EtOH and 30% glycerol/30% EtOH) for 30 min each, and finally in 50% glycerol/0.05% Triton X-100 for 30 min. The cleared roots were mounted in the final solution (50% glycerol/0.05% Triton X-100) and observed with an Axio Imager.A1 microscope (Carl Zeiss, Germany). The lengths of ten randomly picked cells from the elongation zone were measured and seven independent roots were used in this analysis.
RNA isolation and reverse transcription-associated quantitative PCR (RT-qPCR)
Total RNA was extracted from roots of 4-d-old WT and mutant seedlings using the RNeasy Plant Mini Kit (Qiagen, Germany), as previously described (Lee et al., 2012). Approximately 0.5 μg of the isolated RNA samples were used for synthesis of cDNA using TOP script™ RT DRY MIX (d18/dN6 Plus) (Enzynomics, Korea) according to the manufacturer’s instructions. The RT-qPCR assays were performed with an Mx3000P QPCR machine (Agilent Technologies, USA) using RbTaq™ qPCR 2X PreMIX (Enzynomics, Korea). The UBQ10 (AT4G05320) gene was used as an internal reference as previously described (Lee et al., 2016). Each experiment was independently conducted with at least three biological replicates, and the data were analyzed using Excel (Microsoft, USA). Error bars represent the SD of the mean. The PCR primer sequences are listed in Supplementary Table S1.
Plasmid construction and plant transformation
To generate transgenic plants with a ProMINE::MINE-GFP fusion, both the MINE promoter (3,012 bp) and the coding region (1,593 bp) of MINE with no stop codon were amplified using Phusion High-Fidelity DNA polymerase (ThermoFisher, USA) as previously described, but with minor modifications (Lee et al., 2012). The amplified fragment was first cloned into the pENTR/D-TOPO vector (Invitrogen, USA), and then recombined into the pMDC123 destination vector (Curtis and Grossniklaus, 2003). The error-free plasmid was transferred into Agrobacterium tumefaciens (GV3101) and then introduced into the mine plant by using floral dipping method (Clough and Bent, 1998). Transgenic plants were initially selected in the T1 generation by antibiotic- or herbicide-resistance. Homozygous T2 plants were identified through confirmation in the T3 generation, and were used for further analysis (Lee et al., 2012; Yoon et al., 2016). The sequence information of the primers used for plasmid constructs is listed in Supplementary Table S1.
Histochemical GUS detection and confocal microscopy
The histochemical detection of GUS activity was carried out as previously described with minor modifications (Lee et al., 2012; Yoon et al., 2016). Dark-grown seedlings were incubated in GUS staining solution [0.4 mM 5-bromo-4-chloro-3-indoxyl-b-D-glucuronic acid, 2 mM K3Fe(CN)6, 2 mM K4Fe(CN)6, 0.1 M sodium phosphate, 10 mM EDTA, and 0.1% Triton X-100] for 2 h. After GUS staining, samples were rinsed with 70% EtOH for 30 min. Subsequently, samples were observed with differential interference contrast (DIC) optics using an Axio Imager.A1 microscope (Carl Zeiss, Germany). Digital images of the samples were obtained with an AxioCam MRc5 digital camera (Carl Zeiss, Germany). For GFP images, samples were stained with 10 μM propidium iodide (Sigma-Aldrich, USA) and observed with an Olympus FV-1000 confocal laser scanning microscope (Olympus, Japan).
Staining and detection of 5–ethynyl–2′–deoxyuridine (EdU)
EdU staining was performed as described previously, but with minor modifications (Choe et al., 2017; Hong et al., 2015). Four-day-old seedlings were transferred to 0.5X MS media with 0.8% agar containing 5 μM EdU (Invitrogen, USA) for 30 min. The seedlings were fixed in fixative solution (3.7% formaldehyde and 1% Triton x-100 in 1X PBS) for 10 min with vacuum infiltration. After fixation, samples were incubated for 50 min at room temperature. Subsequently, samples were washed twice with 3% bovine serum albumin (BSA) in 1X PBS. Finally, the seedlings were incubated with 250 μL of freshly prepared Click-iT® reaction cocktail for 2 h in the dark at room temperature, according to the manufacturer’s instructions (Invitrogen, USA). The EdU-stained seedlings were washed once with 3% BSA in 1X PBS and stored in 1X PBS in the dark. Stained seedlings were observed with an Olympus FV-1000 confocal laser scanning microscope (Olympus, Japan).
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: EIN2 (AT5G03280), MINE/PDX3 (AT5G49970), PDX1 (AT5G01410), PDX2 (AT5G60540), SCR (AT3G54220), SHR (AT4G37650), SUR2 (AT4G31500), TAR1 (AT1G23320), TAR2 (AT4G24670), UBQ10 (AT4G05320), WEI2/ASA (AT5G05730), WEI7/ASB (AT1G25220), WEI8/TAA1 (AT1G10560), and WOX5 (AT3G11260).
RESULTS
Identification of a novel mutant mildly insensitive to ethylene-induced root growth inhibition
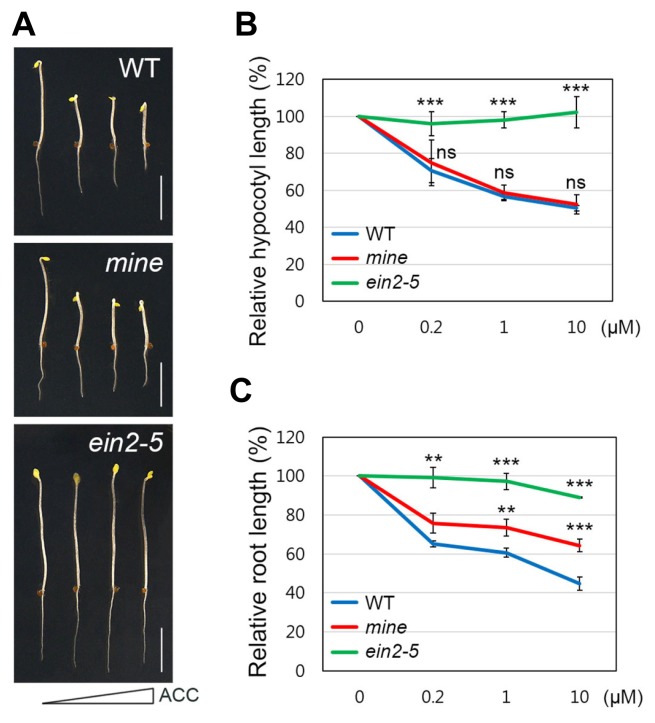
Genetic screening for mutants with root-specific ethylene insensitivity has previously been used to successfully identify molecular components involved in the ethylene-auxin crosstalk (Alonso et al., 2003; Stepanova et al., 2005; 2008). To identify additional root-specific modulators of the ethyleneauxin crosstalk, we utilized the GAL4/UAS activation-tagging system. In particular, we used transgenic plants harboring a driver of the QC-specific WOX5 promoter (ProWOX5::GV) and T-DNA with five tandem UAS sequences (pBIB-UAS), as previously reported (Waki et al., 2013) (see ‘MATERIALS AND METHODS’). Under dark-grown conditions with the ethylene precursor ACC, Columbia wild-type (hereafter WT) seedlings displayed typical triple response phenotypes, including exaggerated curvature of the apical hook, radial swelling of the hypocotyl, and inhibition of root and hypocotyl elongation (Alonso et al., 2003; Bleecker et al., 1988; Guzmán and Ecker, 1990; Roman et al., 1995; Stepanova et al., 2005; 2008). Initially, we screened a population of approximately 1,000 M2 activation-tagged lines for insensitivity to root growth inhibition under ACC (10 μM) treatment. In the genetic screening, we identified one line that exhibited “mild” insensitivity to ethylene-induced inhibition of root growth. In the presence of ACC, root growth of the newly isolated mutant showed less inhibition than that of the WT, but slightly more than that of the ethylene insensitive mutant, ein2-5 (Alonso et al., 1999; Roman et al., 1995; Supplementary Fig. S1). We further analyzed the growth phenotypes of the newly isolated line in comparison with WT and ein2-5 in varying concentrations of ACC (0, 0.2, 1, and 10 μM). The dark-grown shoot phenotype of the new line was nearly indistinguishable from that of WT in the range of concentrations tested (Figs. 1A and 1B). By contrast, we found that the mutant root showed a mild insensitive phenotype to ACC treatment (Figs. 1A and 1C). These findings suggest that the newly isolated line is preferentially insensitive to ACC in the root, in which the ProWOX5::GV driver is presumed to be active. For further genetic analysis, we first backcrossed the line to WT. Because of its insensitivity to ACC in the root, we found that the mutant was reproducibly identified in the F2 progeny of the cross, regardless of the presence of the ProWOX5::GV driver. In the presence of ACC, the line segregated in the typical 3:1 (WT:mutant) Mendelian ratio, indicating inheritance of a single recessive mutation. Therefore, we concluded that the causal root-specific phenotype was not due to overexpression by the ProWOX5::GV driver but instead was likely due to insertional mutation by pBIB-UAS. Furthermore, we generated a mapping population by crossing the mutant line to Landsberg erecta (Ler) plants. In our genetic mapping, we found that the locus responsible for the root-specific insensitivity to ACC mapped to the bottom of chromosome 5, which did not include the known WEI loci (Supplementary Fig. S2).
Fig. 1. The mine mutant shows root-specific insensitivity to ACC.
(A) Growth phenotypes of 4-d-old etiolated WT, mine, and ein2-5 seedlings grown in MS plates supplemented with increasing concentrations of ACC (0, 0.2, 1, and 10 μM). Scale bars= 0.5 cm. (B) The graph illustrates relative hypocotyl responses of 4-d-old etiolated WT, mine, and ein2-5 to ACC, with increasing concentrations. (C) Relative root responses of 4-d-old etiolated WT, mine, and ein2-5 seedlings to ACC, with increasing concentrations. Error bars indicate ±SD from three biological replicates. Asterisks indicate statistically significant differences compared to WT, as determined by Student’s t-test (**P < 0.01 and ***P < 0.001; ns: statistically not significant).
Taken together, our genetic and physiological results suggest that the mild insensitive phenotype is likely due to an insertional mutation, which results in a defect in the root-specific ethylene response. Therefore, we named the newly isolated loss-of-function mutant as mildly insensitive to ethylene (mine).
MINE likely plays a role in ethylene-induced auxin biosynthesis
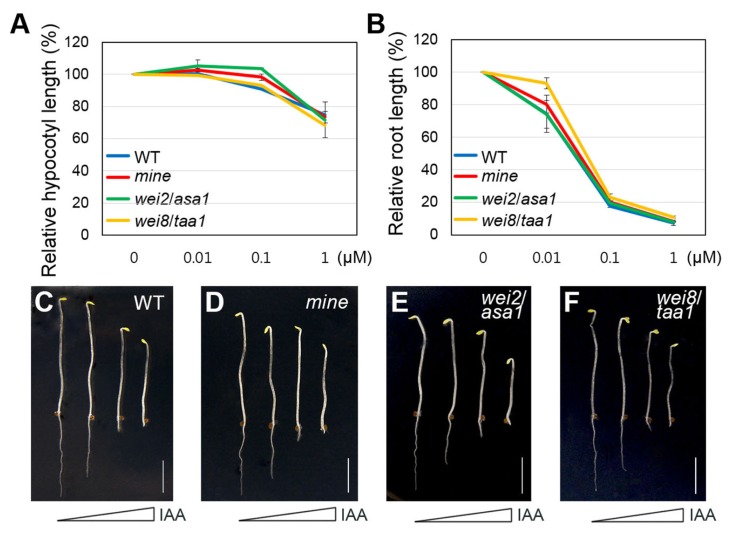
Previous studies have reported that under IAA treatment, wei mutants showed similar growth phenotypes to WT seedlings (Stepanova et al., 2005; 2008). Considering that the ethylene response of dark-grown mine seedlings resembled that of wei mutants, we examined auxin responses of mine seedlings in the presence of IAA. In varying concentrations (0, 0.01, 0.1, and 1 μM), dark-grown mine seedlings displayed nearly identical responses to those of WT and wei (wei2/asa1 and wei8/taa1) seedlings in both hypocotyls and roots (Fig. 2).
Fig. 2. The mine mutant exhibits normal growth responses to IAA.
(A) The graph illustrates relative hypocotyl responses of 4-d-old etiolated WT, mine, wei2/asa1, and wei8/taa1 to IAA, with increasing concentrations. (B) Relative root responses of 4-d-old etiolated WT, mine, wei2/asa1, and wei8/taa1 to IAA with increasing concentrations. Error bars indicate ±SD from three biological replicates. (C–F) Growth phenotypes of 4-d-old etiolated WT, mine, wei2/asa1, and wei8/taa1 seedlings grown in MS plates supplemented with increasing concentrations of IAA (0, 0.01, 0.1, and 1 μM). Scale bars = 0.5 cm.
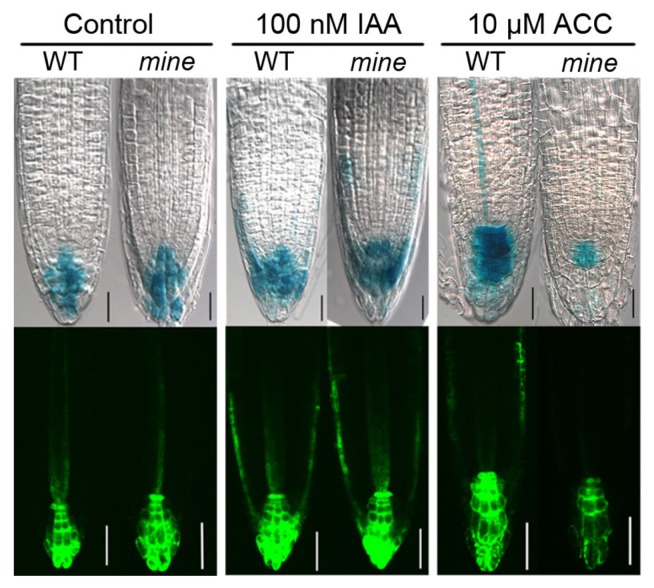
Given that the root growth phenotypes of dark-grown mine seedlings were reminiscent of those of wei mutants under ACC or IAA treatment (Stepanova et al., 2005; 2008; Fig. 2), we hypothesized that MINE might be involved in ethylene-induced auxin biosynthesis. To test this, we first analyzed expression of an auxin maximum marker (DR5), which was previously shown to be induced by ethylene and auxin (He et al., 2011; Stepanova et al., 2005; 2008). In the mine mutant background, the reporter expression of either DR5::GUS or DR5::GFP was similar to that of WT, both in the absence and presence of IAA (100 nM) (Fig. 3). In contrast, the auxin maxima in the mine root tip were discernibly attenuated by exogenous ACC (10 μM) (Fig. 3).
Fig. 3. The mine mutant shows aberrant expression of auxin maxima monitored by DR5 under ACC treatment.
DR5::GUS and DR5::GFP markers examined in 4-d-old etiolated WT and mine roots in the absence or presence of IAA (100 nM) or ACC (10 μM). The mine roots specifically display attenuated expression of both DR5::GUS and DR5::GFP compared with WT roots in the presence of ACC. Scale bars = 50 μm. mine to respond to growth inhibition under ACC treatment. Next, we crossed mine with the wei8/taa1 mutant, which is defective in an auxin biosynthesis gene encoding a PLP-dependent tryptophan aminotransferase (Stepanova et al., 2008). Unlike in mine sur2, the root growth response of mine wei8 double mutants was marginally but significantly more insensitive to ACC than mine and wei8/taa1 single
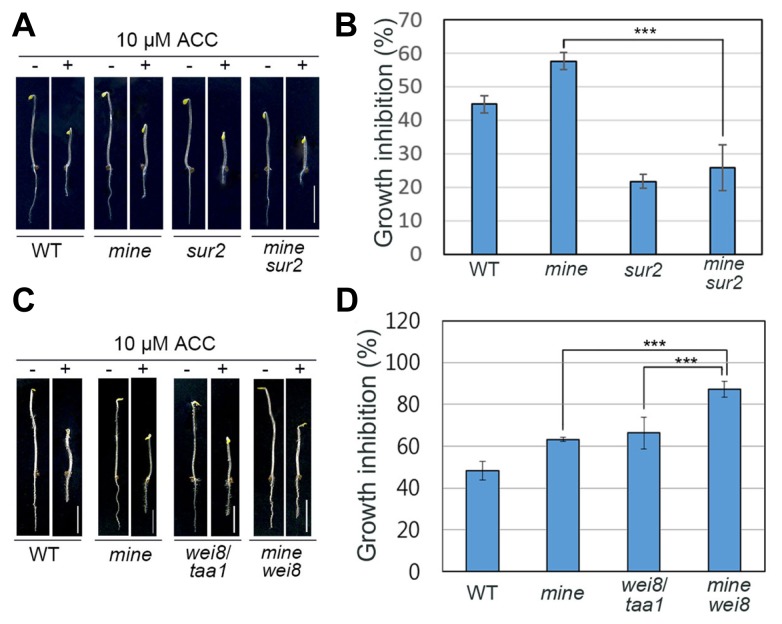
To further evaluate the role of mine, we performed genetic analyses by constructing double mutants, with lines known to have defects in auxin biosynthesis. First, we crossed mine with an auxin-overproducing superroot 2 (sur2 ) mutant, which is flawed in the enzyme P450 CYP83B1 involved in the auxin biosynthesis pathway (Barlier et al., 2000; Boerjan et al., 1995; Delarue et al., 1998; Pacurar et al., 2014; Stepanova et al., 2005; 2011). In the presence of ACC, growth inhibition of mine sur2 roots was indistinguishable from that of sur2 (Figs. 4A and 4B). This finding suggests that sur2-mediated auxin overproduction caused mutants (Figs. 4C and 4D), implying that MINE might play a role in a TAA1-dependent auxin biosynthesis pathway.
Fig. 4. Genetic analysis of ethylene-induced auxin biosynthesis in mine roots.
(A) Growth phenotypes of 4-d-old etiolated WT, mine, sur2, and mine sur2 seedlings in the presence of ACC (10 μM). (B) The graph illustrates relative growth inhibition of WT, mine, sur2, and mine sur2 roots under 10 μM ACC treatment. (C) Growth phenotypes of 4-d-old WT, mine, wei8/taa1, and mine wei8 seedlings in the presence of 10 μM ACC. (D) Relative growth inhibition of WT, mine, wei8/taa1, and mine wei8 roots under 10 μM ACC treatment. Error bars indicate ±SD from three biological replicates. Statistically significant differences were determined by Student’s t-test (***P < 0.001). Scale bars = 0.5 cm.
Collectively, our results strongly support the hypothesis that MINE is involved in ethylene-induced auxin biosynthesis in the root.
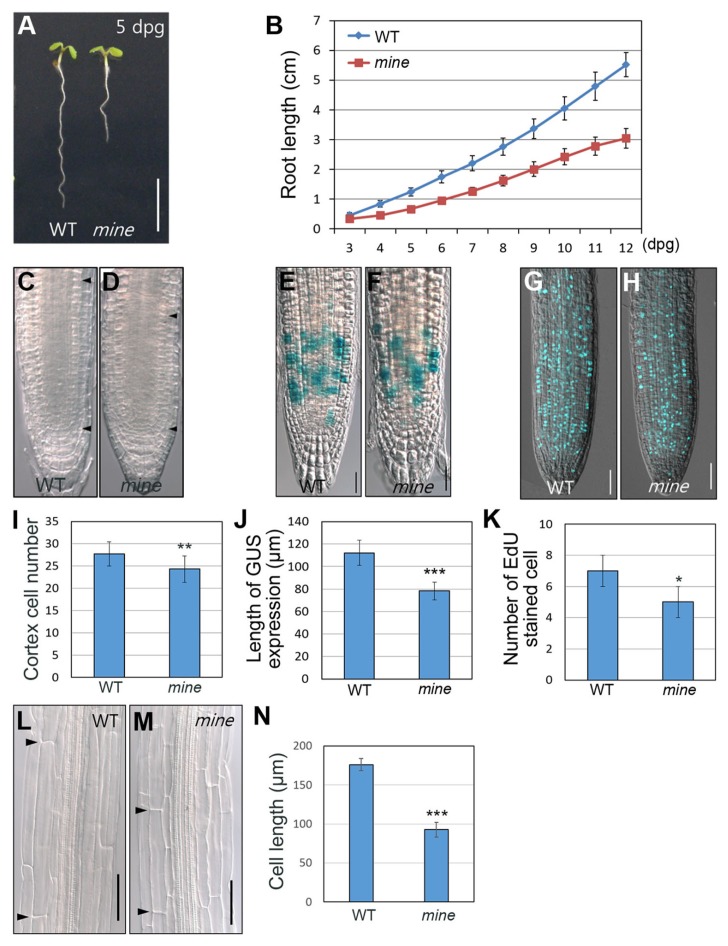
The mine mutant exhibits growth retardation under normal growth conditions
In a root growth assay, besides its insensitive root growth to ethylene under dark-grown conditions, we found that mine showed a short-root phenotype under normal growth conditions (Fig. 5A). As seedlings grew, the root growth rate of mine gradually decreased in comparison with that of WT seedlings (Fig. 5B). To investigate whether mine has developmental defects in the root, we analyzed the expression of SCARECROW (SCR), SHORT-ROOT (SHR), and WOX5 (Di Laurenzio et al., 1996; Gallagher et al., 2004; Helariutta et al., 2000; Nakajima et al., 2001; Sabatini et al., 2003; Sarkar et al., 2007). Under normal growth conditions, the stem cell niche of mine, monitored by expression of the marker genes, was nearly indistinguishable from that of WT seedlings, suggesting that the retarded root growth of mine is not due to defects in the stem cell niche (Supplementary Fig. S3).
Fig. 5. The mine mutant exhibits retarded root growth under normal growth conditions.
(A) Growth phenotypes of WT and mine seedlings grown in MS plates at 5 dpg (days postgermination). (B) Root growth assay of WT and mine seedlings under normal growth conditions. (C, D, I) Meristem size was determined by counting the cortex cell numbers from the QC to the point at which cell size starts to increase (arrowheads demarcate the meristem zone). (E, F, J) CYCB1::GUS expression in 5-d-old WT and mine roots. (G, H, K) EdU assays in WT and mine roots at 5 dpg. (L, M, N) Analysis of cortex cell elongation in the elongation zone (arrowheads indicate the boundaries of a cortex cell). Error bars indicate ±SD from three biological replicates. Asterisks indicate statistically significant differences compared to WT, as determined by Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001). Scale bars = 0.5 cm in A and 50 μm in E, F, G, H, L, M.
When measured as described previously (Achard et al., 2009; Dello Ioio et al., 2007; Heo et al., 2011; Lee et al., 2012; Ubeda-Tomás et al., 2009), the root meristem size of mine was smaller than that of WT (Figs. 5C, 5D, and 5I), suggesting a reduction of cell division in the meristem zone. To further characterize this defect, we monitored cell division with EdU, a thymidine analog (S-phase marker) (Choe et al., 2017; Hong et al., 2015; Kotogány et al., 2010) and CYCB1::GUS (G2/M-phase marker) (Donnelly et al., 1999). Cell division potential in mine roots was slightly decreased compared with that in WT (Figs. 5E–5H, 5J, and 5K). Next, in addition to cell division, we analyzed cell elongation in the mine root as root growth also depends on the extent of elongation of cells exiting the meristem zone (Beemster and Baskin, 1998). We found that under normal growth conditions, the length of mine root cells was considerably reduced compared with that of WT (Figs. 5L–5N). Taken together, these findings strongly suggest that the retarded root growth observed in mine mutants is attributable to reduction in both cell division and cell elongation.
Molecular cloning of the MINE gene
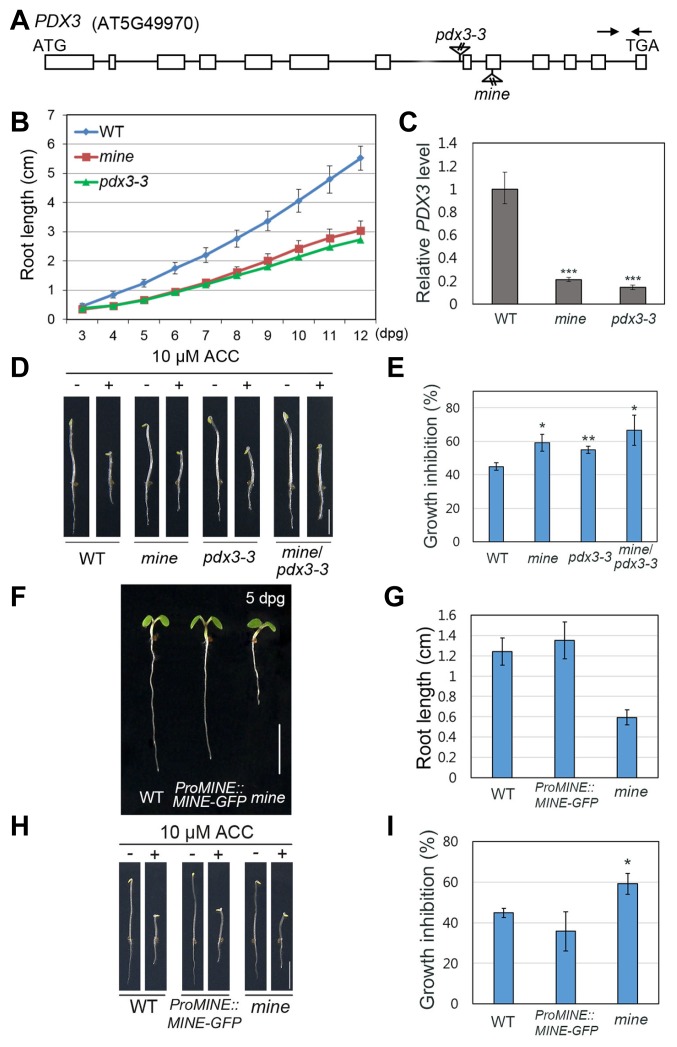
To identify the locus responsible for the mine phenotype, we used a TAIL-PCR method (Liu et al., 1995) as the mutant contained a pBIB-UAS tag. Through TAIL-PCR, we found a T-DNA insertion in the ninth exon of the locus (AT5G49970) encoding the PNP/PMP oxidase, PDX3 (also known as PPOX) (Colinas et al., 2016; González et al., 2007; Sang et al., 2007; Fig. 6A), which is known to convert PMP and PNP into PLP through a salvage pathway (Colinas et al., 2016; González et al., 2007; Sang et al., 2007). In the SALK database (http://signal.salk.edu), we identified another T-DNA insertion allele of PDX3, pdx3-3 (SALK_054167C). Under normal growth conditions, the root growth of pdx3-3 was also reduced as in the mine mutant (Fig. 6B). Next, the levels of PDX3 transcripts were analyzed in WT, mine, and pdx3-3 roots by RT-qPCR. Consistent with our genetic analysis of mine, the level of PDX3 mRNA was substantially reduced in mine roots, confirming that mine is indeed a loss-of-function mutant (Fig. 6C). As in mine, we found that PDX3 expression was also attenuated in pdx3-3 roots when compared with WT (Fig. 6C). Subsequently, we reciprocally crossed these two mutants for a complementation test. In the presence of ACC, the F1 progeny (mine/pdx3-3 ) of the cross between mine and pdx3-3 was also insensitive to ethylene-induced inhibition of root growth compared to each parental line under dark-grown conditions (Figs. 6D and 6E), corroborating their allelic relationship.
Fig. 6. The MINE gene encodes the PNP/PMP oxidase, PDX3.
(A) The MINE locus (AT5G49970) with locations of T-DNA insertion. The boxes depict the coding regions, whereas the lines represent the noncoding regions. The triangles indicate T-DNA insertions in mine isolated from our activation-tagging population and in pdx3-3 identified from the SALK T-DNA database. The arrows show the location of PCR primers used for RT-qPCR. (B) Growth assay of WT, mine, and pdx3-3 roots under normal growth conditions. (C) Expression levels of PDX3 in WT, mine, and pdx3-3 roots. (D, E) Allelism test of mine and pdx3-3 in the absence or presence of 10 μM ACC treatment. From left to right, WT, mine, pdx3-3, and F1 progeny (mine/pdx3-3 ) of crosses between mine and pdx3-3. The F1 progeny show insensitivity to root growth inhibition under ACC treatment. (F, G) Molecular complementation of mine. Under normal growth conditions, the reduced root growth phenotype of mine is rescued by the MINE transgene (ProMINE::MINE-GFP ). (H, I) Root responses of mine are restored to WT levels by the MINE transgene (ProMINE::MINE-GFP ) in the presence of 10 μM ACC. Error bars indicate ±SD from three biological replicates. Statistically significant differences were determined by Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001). Scale bars = 0.5 cm in D, F, H.
To further verify whether mutations in the PDX3 locus cause the root growth insensitivity to ethylene in mine mutants, we introduced a translational fusion, which includes the MINE/PDX3 promoter, coding region, and GFP (ProMINE::MINE-GFP ), into the mine mutant by the floral dipping method (Clough and Bent, 1998). Under normal growth conditions, Arabidopsis transgenic seedlings with the translational fusion were also practically indistinguishable from WT (Figs. 6F and 6G), and the transgenic plants exhibited a similar phenotype to that of WT under ACC treatment (Figs. 6H and 6I). Consequently, our results strongly suggest that the mine phenotypes, in the presence of ACC under dark-grown conditions and in the absence of ACC under light-grown conditions, are indeed due to an insertional mutation in the PDX3 locus, which encodes the PNP/PMP oxidase involved in the PLP salvage pathway (Colinas et al., 2016; González et al., 2007; Sang et al., 2007).
MINE/PDX3 likely plays a role in the TAA1/TAR-dependent auxin biosynthesis pathway induced by ethylene
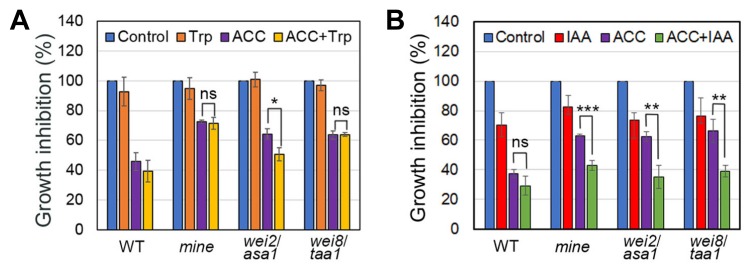
Previous studies have shown that by adding Trp or IAA, wei mutants can be correctly placed in the Trp-dependent auxin biosynthesis pathway (Stepanova et al., 2005; 2008; 2011). For example, both Ant and Trp could rescue the ethylene sensitivity of wei2 and wei7 roots to WT levels (Stepanova et al., 2005). On the other hand, defects in the ethylene-induced root responses of wei8/taa1 could be restored by exogenous IAA, but not by Trp, suggesting that TAA1 and its related genes (TARs) play key roles in the IPyA-dependent route of the conversion of Trp to IAA (Stepanova et al., 2008). Therefore, we examined growth responses of mine roots under ACC (10 μM) only, Trp (10 μM) only, IAA (10 nM) only, ACC plus Trp (ACC+Trp), and ACC plus IAA (ACC+IAA) conditions. With ACC+Trp, mine seedlings exhibited a similar insensitive phenotype in the roots, as compared to those with only ACC, which is reminiscent of that seen in wei8/taa1 seedlings (Fig. 7A). Interestingly, the root responses of mine seedlings were rescued in the presence of both ACC and IAA (ACC+IAA condition) (Fig. 7B). These findings strongly suggest that MINE/PDX3 is likely involved in ethylene-induced auxin biosynthesis downstream of Trp, in which TAA1/WEI8 and its related genes (TARs) participate (Stepanova et al., 2008; 2011).
Fig. 7. Ethylene-insensitive responses of mine roots are restored by IAA, but not by Trp.
(A) Growth inhibition of 4-d-old etiolated WT, mine, wei2/asa1, and wei8/taa1 roots grown in MS plates only (control; blue), or MS plates supplemented with Trp (10 μM; orange) only, ACC (10 μM; purple) only, or ACC+Trp (10 μM; yellow). (B) Root growth inhibition of WT, mine, wei2/asa1, and wei8/taa1 seedlings grown in MS plates only (control; blue), or MS plates with IAA (10 nM; red) only, ACC (10 μM; purple) only, or ACC+IAA (10 nM; green). Error bars indicate ±SD from three biological replicates. Asterisks indicate statistically significant differences compared to WT, as determined by Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001; ns: statistically not significant).
DISCUSSION
We had initially attempted to tissue-specifically overexpress T-DNA with tandem UAS sequences (pBIB-UAS) by using the QC-specific driver (ProWOX5::GV ) (Waki et al., 2013). Unexpectedly, we identified a novel recessive mutant, named mildly insensitive to ethylene (mine), which showed insensitivity to growth inhibition in the presence of the ethylene precursor, ACC. Under dark-grown conditions supplemented with ACC, mine roots were insensitive to ethylene-induced inhibition. By contrast, mine hypocotyls displayed typical triple responses in a similar manner to those of WT. The root-specific insensitivity of mine seedlings to ethylene-induced growth inhibition is reminiscent of that seen in wei mutants (Alonso et al., 2003; Stepanova et al., 2005; 2008). Therefore, we also investigated the growth response of mine seedlings to auxin, and found that mine seedlings exhibited normal response phenotypes to exogenous IAA, as did wei (wei2 and wei8) mutants (Stepanova et al., 2005; 2008; 2011). Considering the mine phenotypes under ACC and IAA treatments, we hypothesized that MINE might play a role in ethylene-induced auxin biosynthesis. The auxin maxima in the root, monitored by DR5::GUS and DR5::GFP, were indeed attenuated in mine, when compared with WT under ACC treatment. In our genetic analysis, we found that root growth of mine seedlings in the auxin-overproducing sur2 background (Barlier et al., 2000; Boerjan et al., 1995; Delarue et al., 1998; Pacurar et al., 2014; Stepanova et al., 2005; 2011) was substantially inhibited in the presence of ACC. This finding suggests that in the sur2 background, auxin overproduction likely causes mine roots to respond to growth inhibition by ethylene. Moreover, the growth response of mine wei8 double mutants was discernibly insensitive to exogenous ACC compared with that of mine and wei8/taa1 single mutants (Stepanova et al., 2008). Previous work has shown that WEI8 encodes a PLP-dependent tryptophan aminotransferase, TAA1, which is involved in the conversion of Trp to IPyA in the Trp-dependent auxin biosynthesis pathway (Stepanova et al., 2008; Fig. 8). Thus, this result suggests that loss of both MINE and TAA1 has compound effects on the root growth response to ethylene. Taken together, our results lend strong support for the hypothesis that MINE likely plays a role in the ethylene-induced auxin biosynthesis pathway.
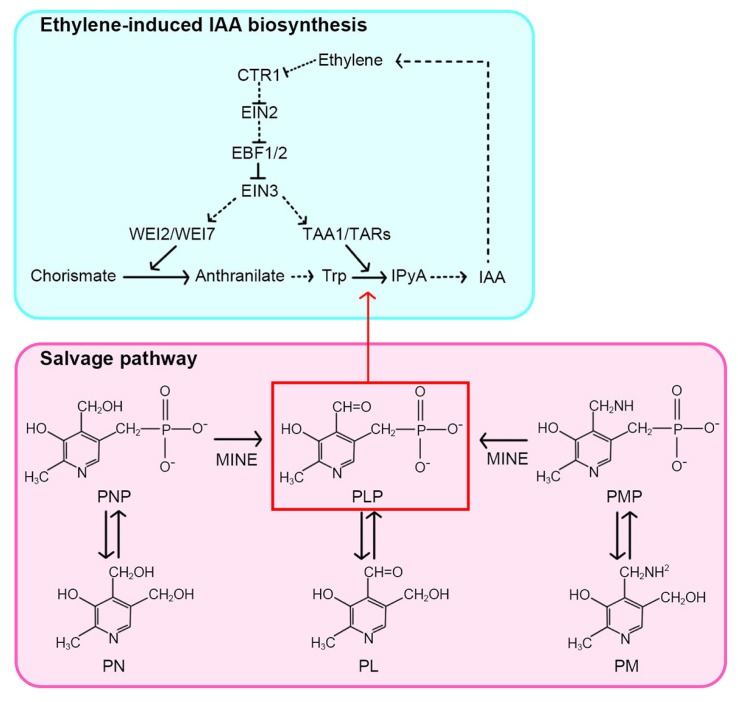
Fig. 8. A schematic model for the involvement of MINE/PDX3 in ethylene-induced auxin biosynthesis.
Anthranilate synthase (ASA and ASB), encoded by WEI2 and WEI7, is a key biosynthesis enzyme for the auxin precursor, Trp. By contrast, TAA1 and its related genes (TARs) are involved in the conversion of Trp to IPyA in the auxin biosynthesis pathway. The MINE gene, which encodes the PNP/PMP oxidase (PDX3) in the PLP salvage pathway, likely plays a role in ethylene-induced auxin biosynthesis.
Ethylene is known to inhibit root growth primarily by affecting cell elongation (Alarcón et al., 2014; Bleecker and Kende, 2000; Le et al., 2001; Růžička et al., 2007; Swarup et al., 2007). We also found that cell elongation in the mine root was substantially reduced under normal growth conditions. Previous studies have demonstrated that auxin redistribution into the root elongation zone is attributable to ethylene-induced inhibition of root cell elongation (Růžička et al., 2007; Swarup et al., 2007). Given that the auxin maxima were discernibly attenuated in the mine root by exogenous ACC, it is tempting to speculate that auxin redistribution from the auxin maxima in the root tip is likely affected in the mine seedling as well.
Vitamin B6 plays crucial roles in plant development and hormone homeostasis (Boycheva et al., 2015; Chen and Xiong, 2005; Percudani and Peracchi, 2003; Titiz et al., 2006). Therefore, a complete loss of de novo vitamin B6 biosynthesis results in embryonic lethal (Colinas et al., 2016; Tambasco-Studart et al., 2005; Titiz et al., 2006). Previous studies have reported that mutations in the PDX1.1 and PDX1.3 genes, which are involved in the de novo PLP biosynthesis pathways, promote growth phenotypes linked to impaired ethylene and auxin biosynthesis that require PLP as a cofactor (Boycheva et al., 2015; Chen and Xiong, 2009a; 2009b; Colinas et al., 2016). However, it remains unclear whether PDX3 in the salvage pathway is also involved in the biosynthesis of the two hormones.
Our genetic, physiological, and molecular analyses provide important information on the role of MINE/PDX3 in ethylene-induced auxin biosynthesis and plant growth, with a focus on the root. Subsequent molecular cloning led us to the conclusion that the insensitivity of mine roots to ethylene-induced growth inhibition resulted from the loss of PDX3 activity, which catalyzes the conversion of the phosphorylated forms of PN and PM into PLP (Colinas et al., 2016; González et al., 2007; Sang et al., 2007; 2011). When complemented with the MINE/PDX3 genomic fragment or supplemented with exogenous IAA, defects in the ethylene-induced growth inhibition of mine/pdx3 roots were rescued to levels indistinguishable from WT seedlings. It has previously been shown that the level of PLP was reduced, and the levels of PMP and PNP increased in pdx3 mutants when compared with WT (Colinas et al., 2016). Moreover, Soeno et al. (2010) demonstrated that PLP is required in IPyA formation in both Arabidopsis and wheat in vitro. By adding Trp or IAA along with ACC to mine/pdx3 seedlings, we were able to speculate in which step in ethylene-induced auxin biosynthesis MINE/PDX3 is likely involved (Fig. 8). For instance, under ACC+Trp conditions, mine/pdx3 seedlings still displayed insensitivity in roots like wei8/taa1 seedlings. By contrast, root growth responses of mine/pdx3 seedlings were restored under ACC+IAA conditions. Therefore, MINE/PDX3 likely plays a role in the IPyA-dependent route of the conversion of Trp to IAA, like TAA1 and TARs (Stepanova et al., 2008; 2011). Considering that TAA1 and TARs belong to a subgroup of PLP-dependent enzymes (He et al., 2011; Huai et al., 2001; Stepanova et al., 2008; Tao et al., 2008), it is tempting to speculate that PLP produced by MINE/PDX3 acts as a cofactor in TAA1/TAR-dependent auxin biosynthesis induced by ethylene.
In summary, this study provides lines of evidence that MINE/PDX3-mediated production of PLP in the salvage pathway participates in TAA1/TAR-dependent auxin biosynthesis induced by ethylene, which in turn influences the crosstalk between ethylene and auxin in the Arabidopsis root.
Supplementary data
ACKNOWLEDGMENTS
We thank Keiji Nakajima, the Arabidopsis Biological Resource Center (ABRC), and Nottingham Arabidopsis Stock Center (NASC) for sharing materials. This work was supported by grants from the National Research Foundation (NRF-2017R1A2B4005088) and Next-Generation BioGreen 21 (SSAC-PJ01316101).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Abel S., Nguyen M.D., Chow W., Theologis A. ACS4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana Structural characterization, expression in Escherichia coli, and expression characteristics in response to auxin [corrected] J Biol Chem. 1995;270:19093–19099. doi: 10.1074/jbc.270.32.19093. Erratum. J. Biol. Chem: 270, 26020. [DOI] [PubMed] [Google Scholar]
- Achard P., Gusti A., Cheminant S., Alioua M., Dhondt S., Coppens F., Beemster G.T., Genschik P. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol. 2009;19:1188–1193. doi: 10.1016/j.cub.2009.05.059. [DOI] [PubMed] [Google Scholar]
- Alarcón M.V., Lloret P.G., Salguero J. Synergistic action of auxin and ethylene on root elongation inhibition is caused by a reduction of epidermal cell length. Plant Signal Behav. 2014;9:e28361. doi: 10.4161/psb.28361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova A.N., Solano R., Wisman E., Ferrari S., Ausubel F.M., Ecker J.R. Five components of the ethylene-response pathway identified in a screen for weak ethylene insensitive mutants in Arabidopsis. Proc Natl Acad Sci USA. 2003;100:2992–2997. doi: 10.1073/pnas.0438070100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier I., Kowalczyk M., Marchant A., Ljung K., Bhalerao R., Bennett M., Sandberg G., Bellini C. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci USA. 2000;97:14819–14824. doi: 10.1073/pnas.260502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster G.T.S., Baskin T.I. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 1998;116:1515–1526. doi: 10.1104/pp.116.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker A.B., Estelle M.A., Somerville C., Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1988;241:1086–1090. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Bleecker A.B., Kende H. Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- Boerjan W., Cervera M.T., Delarue M., Beeckman T., Dewitte W., Bellini C., Caboche M., Onckelen H.V., Montagu M.V., Inzé D. superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycheva S., Dominguez A., Rolcik J., Boller T., Fitzpatrick T.B. Consequences of a deficit in vitamin B6 biosynthesis de novo for hormone homeostasis and root development in Arabidopsis. Plant Physiol. 2015;167:102–117. doi: 10.1104/pp.114.247767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Xiong L. Pyridoxine is required for postembryonic root development and tolerance to osmotic and oxidative stresses. Plant J. 2005;44:396–408. doi: 10.1111/j.1365-313X.2005.02538.x. [DOI] [PubMed] [Google Scholar]
- Chen H., Xiong L. Localized auxin biosynthesis and postembryonic root development in Arabidopsis. Plant Signal Behav. 2009a;4:752–754. doi: 10.4161/psb.4.8.9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Xiong L. The short-rooted vitamin B6-deficient mutant pdx1 has impaired local auxin biosynthesis. Planta. 2009b;229:1303–1310. doi: 10.1007/s00425-009-0912-8. [DOI] [PubMed] [Google Scholar]
- Choe J.E., Kim B., Yoon E.K., Jang S., Kim G., Dhar S., Lee S.A., Lim J. Characterization of the GRAS transcription factor SCARECROW-LIKE 28’s role in Arabidopsis root growth. J Plant Biol. 2017;60:462–471. [Google Scholar]
- Clough S., Bent A. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Colinas M., Eisenhut M., Tohge T., Pesquera M., Fernie A.R., Weber A.P., Fitzpatrick T.B. Balancing of B6 vitamers is essential for plant development and metabolism in Arabidopsis. Plant Cell. 2016;28:439–453. doi: 10.1105/tpc.15.01033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M., Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M., Prinsen E., Onckelen H.V., Caboche M., Bellini C. Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 1998;14:603–611. doi: 10.1046/j.1365-313x.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F.S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., Sabatini S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol. 2007;17:678–682. doi: 10.1016/j.cub.2007.02.047. [DOI] [PubMed] [Google Scholar]
- Denslow S.A., Reuschhoff E.E., Daub M.E. Regulation of the Arabidopsis thaliana vitamin B6 biosynthesis genes by abiotic stress. Plant Physiol Biochem. 2007;45:152–161. doi: 10.1016/j.plaphy.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Depuydt S., Hardtke C.S. Hormone signalling crosstalk in plant growth regulation. Curr Biol. 2011;21:R365–R373. doi: 10.1016/j.cub.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Donnelly P.M., Bonetta D., Tsukaya H., Dengler R.E., Dengler N.G. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol. 1999;215:407–419. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick T.B., Amrhein N., Kappes B., Macheroux P., Tews I., Raschle T. Two independent routes of de novo vitamin B6 biosynthesis: not that different after all. Biochem J. 2007;407:1–13. doi: 10.1042/BJ20070765. [DOI] [PubMed] [Google Scholar]
- Gallagher K.L., Paquette A.J., Nakajima K., Benfey P.N. Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol. 2004;14:1847–1851. doi: 10.1016/j.cub.2004.09.081. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S., McCourt P. Cross-talk in plant hormone signalling: what Arabidopsis mutants are telling us. Ann Bot. 2003;91:605–612. doi: 10.1093/aob/mcg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E., Danehower D., Daub M.E. Vitamer levels, stress response, enzyme activity, and gene regulation of Arabidopsis lines mutant in the pyridoxine/pyridoxamine 5′-phosphate oxidase (PDX3) and the pyridoxal kinase (SOS4) genes involved in the vitamin B6 salvage pathway. Plant Physiol. 2007;145:985–996. doi: 10.1104/pp.107.105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán P., Ecker J.R. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Brumos J., Li H., Ji Y., Ke M., Gong X., Zeng Q., Li W., Zhang X., An F., et al. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell. 2011;23:3944–3960. doi: 10.1105/tpc.111.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- Heo J.O., Chang K.S., Kim I.A., Lee M-H, Lee S.A., Song S.K., Lee M.M., Lim J. Funneling of gibberellin signaling by the GRAS transcription regulator SCARECROW-LIKE 3 in the Arabidopsis root. Proc Natl Acad Sci USA. 2011;108:2166–2171. doi: 10.1073/pnas.1012215108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J.H., Chu H., Zhang C., Ghosh D., Gong X., Xu J. A quantitative analysis of stem cell homeostasis in the Arabidopsis columella root cap. Front Plant Sci. 2015;6:206. doi: 10.3389/fpls.2015.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huai Q., Xia Y., Chen Y., Callahan B., Li N., Ke H. Crystal structures of 1-aminocyclopropane-1-carboxylate (ACC) synthase in complex with aminoethoxyvinylglycine and pyridoxal-5′-phosphate provide new insight into catalytic mechanisms. J Biol Chem. 2001;276:38210–38216. doi: 10.1074/jbc.M103840200. [DOI] [PubMed] [Google Scholar]
- Konieczny A., Ausubel F.M. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Kotogány E., Dudits D., Horváth G.V., Ayaydin F. A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods. 2010;6:5. doi: 10.1186/1746-4811-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J., Vandenbussche F., Van Der Straeten D., Verbelen J.P. In the early response of Arabidopsis roots to ethylene, cell elongation is up and down regulated and uncoupled from differentiation. Plant Physiol. 2001;125:519–522. doi: 10.1104/pp.125.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.A., Jang S., Yoon E.K., Heo J.O., Chang K.S., Choi J.W., Dhar S., Kim G., Choe J-e, Heo J.B., et al. Interplay between ABA and GA modulates the timing of asymmetric cell divisions in the Arabidopsis root ground tissue. Mol Plant. 2016;9:870–884. doi: 10.1016/j.molp.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Lee S.A., Yoon E.K., Heo J.O., Lee M.H., Hwang I., Cheong H., Lee W.S., Hwang Y.S., Lim J. Analysis of Arabidopsis glucose insensitive growth mutants reveals the involvement of the plastidial copper transporter PAA1 in glucose-induced intracellular signaling. Plant Physiol. 2012;159:1001–1012. doi: 10.1104/pp.111.191726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.G., Mitsukawa N., Oosumi T., Whittier R.F. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Gillmor C.S., Scheible W.R. Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 2000;123:795–805. doi: 10.1104/pp.123.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C., Gaxiola R., Grisafi P., Fink G. EIR1, a root specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 1998;12:2175–2187. doi: 10.1101/gad.12.14.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P.N. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- Pacurar D.I., Pacurar M.L., Bussell J.D., Schwambach J., Pop T.I., Kowalczyk M., Gutierrez L., Cavel E., Chaabouni S., Ljung K., et al. Identification of new adventitious rooting mutants amongst suppressors of the Arabidopsis thaliana superroot2 mutation. J Exp Bot. 2014;65:1605–18. doi: 10.1093/jxb/eru026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani R., Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003;4:850–854. doi: 10.1038/sj.embor.embor914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett F.B., Wilson A.K., Estelle M. The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiol. 1990;94:1462–1466. doi: 10.1104/pp.94.3.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles L., Stepanova A.N., Alonso J.M. Molecular mechanisms of ethylene-auxin interaction. Mol Plant. 2013;6:1734–1737. doi: 10.1093/mp/sst113. [DOI] [PubMed] [Google Scholar]
- Roman G., Lubarsky B., Kieber J.J., Rothenberg M., Ecker J.R. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueschhoff E.E., Gillikin J.W., Sederoff H.W., Daub M.E. The SOS4 pyridoxal kinase is required for maintenance of vitamin B6-mediated processes in chloroplasts. Plant Physiol Biochem. 2013;63:281–291. doi: 10.1016/j.plaphy.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Růžička K., Ljung K., Vanneste S., Podhorská R., Beeckman T., Friml J., Benková E. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell. 2007;19:2197–2212. doi: 10.1105/tpc.107.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Heidstra R., Wildwater M., Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y., Barbosa J.M., Wu H., Locy R.D., Singh N.K. Identification of a pyridoxine (pyridoxamine) 5′-phosphate oxidase from Arabidopsis thaliana. FEBS Lett. 2007;581:344–348. doi: 10.1016/j.febslet.2006.12.028. [DOI] [PubMed] [Google Scholar]
- Sang Y., Locy R.D., Goertzen L.R., Rashotte A.M., Si Y., Kang K., Singh N.K. Expression, in vivo localization and phylogenetic analysis of a pyridoxine 5′-phosphate oxidase in Arabidopsis thaliana. Plant Physiol Biochem. 2011;49:88–95. doi: 10.1016/j.plaphy.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Sarkar A.K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- Shi H., Xiong L., Stevenson B., Lu T., Zhu J.K. The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell. 2002;14:575–588. doi: 10.1105/tpc.010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Zhu J.K. SOS4, a pyridoxal kinase gene, is required for root hair development in Arabidopsis. Plant Physiol. 2002;129:585–593. doi: 10.1104/pp.001982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeno K., Goda H., Ishii T., Ogura T., Tachikawa T., Sasaki E., Yoshida S., Fujioka S., Asami T., Shimada Y. Auxin biosynthesis inhibitors, identified by a genomics-based approach, provide insights into auxin biosynthesis. Plant Cell Physiol. 2010;51:524–536. doi: 10.1093/pcp/pcq032. [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Alonso J.M. Ethylene signaling and response pathway: a unique signaling cascade with a multitude of inputs and outputs. Physiol Plantarum. 2005;123:195–206. [Google Scholar]
- Stepanova A.N., Hoyt J.M., Hamilton A.A., Alonso J.M. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell. 2005;17:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Dolezal K., Schlereth A., Jürgens G., Alonso J.M. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Likhacheva A.V., Alonso J.M. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell. 2007;19:2169–2185. doi: 10.1105/tpc.107.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Robles L.M., Novak O., He W., Guo H., Ljung K., Alonso J.M. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell. 2011;23:3961–3973. doi: 10.1105/tpc.111.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Parry G., Graham N., Allen T., Bennett M. Auxin cross-talk: integration of signalling pathways to control plant development. Plant Mol Biol. 2002;49:411–426. doi: 10.1007/978-94-010-0377-3_12. [DOI] [PubMed] [Google Scholar]
- Swarup R., Perry P., Hagenbeek D., Van Der Straeten D., Beemster G.T., Sandberg G., Bhalerao R., Ljung K., Bennett M.J. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell. 2007;19:2186–2196. doi: 10.1105/tpc.107.052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambasco-Studart M., Titiz O., Raschle T., Forster G., Amrhein N., Fitzpatrick T.B. Vitamin B6 biosynthesis in higher plants. Proc Natl Acad Sci USA. 2005;102:13687–13692. doi: 10.1073/pnas.0506228102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Ferrer J.L., Ljung K., Pojer F., Hong F., Long J.A., Li L., Moreno J.E., Bowman M.E., Ivans L.J., et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titiz O., Tambasco-Studart M., Warzych E., Apel K., Amrhein N., Laloi C., Fitzpatrick T.B. PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis. Plant J. 2006;48:933–946. doi: 10.1111/j.1365-313X.2006.02928.x. [DOI] [PubMed] [Google Scholar]
- Ubeda-Tomás S., Federici F., Casimiro I., Beemster G.T., Bhalerao R., Swarup R., Doerner P., Haseloff J., Bennett M.J. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr Biol. 2009;19:1194–1199. doi: 10.1016/j.cub.2009.06.023. [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S., Bernhardt A., Leuendorf J.E., Drewke C., Lytovchenko A., Mujahed N., Gurgui C., Frommer W.B., Leistner E., Fernie A.R., et al. Analysis of the Arabidopsis rsr4-1/pdx1-3 mutant reveals the critical function of the PDX1 protein family in metabolism, development, and vitamin B6 biosynthesis. Plant Cell. 2006;18:1722–1735. doi: 10.1105/tpc.105.036269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waki T., Miyashima S., Nakanishi M., Ikeda Y., Hashimoto T., Nakajima K. A GAL4-based targeted activation tagging system in Arabidopsis thaliana. Plant J. 2013;73:357–367. doi: 10.1111/tpj.12049. [DOI] [PubMed] [Google Scholar]
- Wolters H., Jürgens G. Survival of the flexible: hormonal growth control and adaptation in plant development. Nat Rev Genet. 2009;10:305–317. doi: 10.1038/nrg2558. [DOI] [PubMed] [Google Scholar]
- Yoon E.K., Dhar S., Lee M.H., Song J.H., Lee S.A., Kim G., Jang S., Choi J.W., Choe J.E., Kim J.H., et al. Conservation and diversification of the SHR-SCR-SCL23 regulatory network in the development of the functional endodermis in Arabidopsis shoots. Mol Plant. 2016;9:1197–1209. doi: 10.1016/j.molp.2016.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.