Abstract
One of the most interesting findings from genome-wide expression analysis is that a considerable amount of noncoding RNA (ncRNA) is present in the cell. Recent studies have identified diverse biological functions of ncRNAs, which are expressed in a much wider array of forms than proteins. Certain ncRNAs associated with diseases, in particular, have attracted research attention as novel therapeutic targets and diagnostic markers. BC200 RNA, a 200-nucleotide ncRNA originally identified as a neuron-specific transcript, is abnormally over-expressed in several types of cancer tissue. A number of recent studies have suggested mechanisms by which abnormal expression of BC200 RNA contributes to the development of cancer. In this article, we first provide a brief review of a recent progress in identifying functions of BC200 RNA in cancer cells, and then offer examples of other ncRNAs as new therapeutic targets and diagnostic markers for human cancer. Finally, we discuss future directions of studies on BC200 RNA for new cancer treatments.
Keywords: BC200 RNA, cancer, diagnostic marker, ncRNA (noncoding RNA), therapeutic target
INTRODUCTION
The 2001 Human Genome Project revealed that only ~2% of human genome sequences encode proteins (Lander et al., 2001). This discovery was shocking because protein-based analyses had long been the mainstay of molecular biology. Moreover, the subsequent ENCODE project showed that approximately 60% of genome sequence are transcribed into RNA (Consortium, 2004), revealing the existence of an enormous amount of noncoding RNAs (ncRNAs) in the human cell. Although some early reports argued that these ncRNAs are just nonfunctional byproducts produced during gene expression (Ebisuya et al., 2008; Struhl, 2007), more recent studies have demonstrated that they have intrinsic functions in regulating gene expression, cell differentiation and development, and maintaining genomic integrity (Brosnan and Voinnet, 2009; Johnsson and Morris, 2014; Ko et al., 2017; Wilusz et al., 2009).
Given their multiple roles in diverse biological processes, ncRNAs are closely associated with various diseases (Jain et al., 2017; Wapinski and Chang, 2011). Transcriptome analyses of disease models have confirmed that various ncRNAs exhibit disease-specific altered expression or mutations (Chung et al., 2011; Pasmant et al., 2011; Ward and Kellis, 2012). These changes in ncRNA expression directly or indirectly affect the pathogenesis of human diseases. For example, abnormally overexpressed HOX antisense intergenic RNA (HOTAIR) in some types of cancers promotes metastasis by reprogramming the chromatin state (Gupta et al., 2010). Subsequent studies showed that expressional levels of HOTAIR RNA are highly correlated with poorer prognosis (Luo et al., 2017; Song and Zou, 2016; Zhang et al., 2018), prompting current clinical trials of HOTAIR RNA as a diagnostic marker. ncRNA studies provide a broader understanding of disease and are being used in the development of new therapeutic strategies and diagnostic techniques.
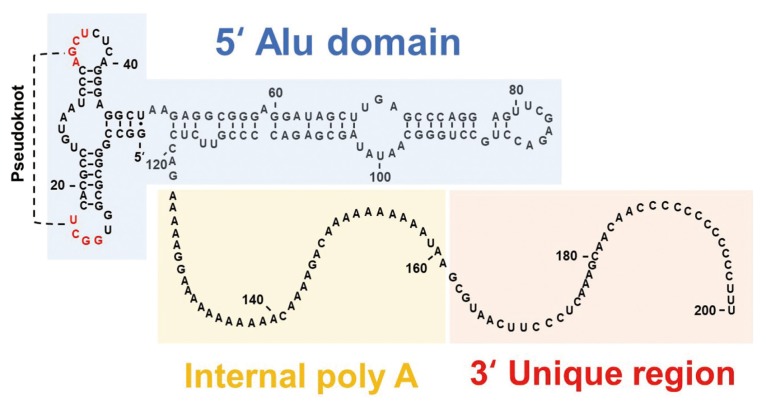
Brain cytoplasmic 200 RNA (BC200 RNA), first identified in human neurons in 1987, is a 200-nucleotide (nt) ncRNA that shows high functional similarity to BC1 RNA found in rodent neurons (Watson and Sutcliffe, 1987). BC200 RNA is composed of a 5′-terminal Alu domain, an internal A-rich domain and a unique 3′ region containing poly C residues (Fig. 1)(Tiedge et al., 1993). In normal cells, BC200 RNA is localized exclusively to neuronal dendrites, where it contributes to formation of neuronal plasticity by controlling postsynaptic translation (Briggs et al., 2015; Muddashetty et al., 2002). However, the fine control of BC200 RNA expression is disrupted in some diseases. In 1997, it was found that BC200 RNA is overexpressed in carcinomas of the breast, cervix, esophagus, lung, ovary, parotid, and tongue (Chen et al., 1997), representing the first confirmation of BC200 RNA expression in tissues of non-neuronal origin. BC200 RNA was subsequently shown to play important roles in the growth and development of cancer cells (Samson et al., 2018), and has thus become an emerging novel therapeutic target and diagnostic marker for human cancers.
Fig. 1. Schematic diagram of BC200 RNA.
BC200 RNA is composed of three domains: a 5′ Alu domain, an internal A-rich domain, and a unique 3′ domain carrying C-rich sequences.
In this review, we summarize recent investigations of BC200 RNA in cancer cells and consider future research directions for its clinical application. To place these observations in a broader context, we also highlight recent results obtained with representative oncogenic ncRNAs, including HOTAIR and micro RNAs (miRNAs), with the hope that these strategies for preventing cancers may provide insight that helps guide future research of BC200 RNA for possible clinical applications.
BC200 RNA IN NEURONS
Identification of BC200 RNA
When the 160-nt BC1 RNA was first identified in rodent neurons, it was detected only in brain tissues (Sutcliffe et al., 1982). Because of the small size of the transcript, it was presumed at the time to be a nuclear byproduct generated during the maturation of neuronal mRNA. However, it was found in the cytoplasm and named brain cytoplasm 1 RNA to emphasize its specific location (Sutcliffe et al., 1984). In 1987, the primate analog BC200 RNA was found in the human brain (Watson and Sutcliffe, 1987). It contained an internal A-rich region and was localized to the cytoplasm of neurons in the brain, characteristics similar to those of BC1 RNA. In addition to the internal A-rich domain, BC200 contains a 5′ Alu domain and a unique 3′ region containing poly C residues (Fig. 1)(Tiedge et al., 1993; Wu et al., 2014). The BC200 RNA gene has an internal promoter consisting of an A box and B box, common to members of the Alu family, and a specific upstream promoter (Kim et al., 2017). The TATA binding protein (TBP) binds to the BC200 RNA promoter and recruits the RNA polymerase III complex, resulting in transcription of BC200 RNA (Schramm and Hernandez, 2002).
Early studies identified neuron-specific expression of BC200 RNA by Northern blotting analysis and in situ hybridization (Tiedge et al., 1993; Watson and Sutcliffe, 1987). Recently, highly sensitive real-time PCR analysis was able to detect BC200 RNA in normal testis and ovary tissues, albeit at much lower levels than in brain tissues (Booy et al., 2017). ncRNAs are expressed in a more tissue-specific manner than coding mRNAs (Amin et al., 2015; Derrien et al., 2012), probably because expression of most ncRNAs is elaborately controlled by epigenetic regulatory mechanisms (Baer et al., 2013; Bandres et al., 2009; Wu et al., 2014). BC200 RNA expression may also be similarly tightly controlled by epigenetic regulation.
Function of BC200 RNA in neurons
In neurons, BC200 RNA regulates translation in postsynaptic regions. Immediately after expression, it migrates to neuronal dendrites and binds to the translational initiation factors, poly A binding protein (PABP), eukaryotic translation initiation factor 4A (eIF4A) and eIF4B, thereby inhibiting local translation (Eom et al., 2011; Kondrashov et al., 2005; Muddashetty et al., 2002; Lin et al., 2008). This may lead to the expression of specific proteins in neurons that are required for neuronal plasticity (Qureshi and Mehler, 2012; Richter and Klann, 2009; Smalheiser, 2014) and whose function could be controlled by cellular regulators, such as synaptotagmin binding cytoplasmic RNA interacting protein (SYNCRIP), heterogeneous nuclear ribonucleoprotein E1 (hnRNPE1) and hnRNPE2 (Duning et al., 2008; Jang et al., 2017).
BC200 RNA IN CANCER
Abnormal expression of BC200 RNA in cancer
At the time BC200 RNA was first identified as a neuron-specific ncRNA in the brains of monkeys and humans, it was also observed to be expressed in the non-neuronal HeLa cancer cell line (Watson and Sutcliffe, 1987). However, the significance of this exceptional expression was not clarified at that time. A later study showed that BC200 RNA is highly expressed in cancer tissues of the breast, lung, esophagus and parotid, but not in normal tissues (Chen et al., 1997). The abnormal expression of BC200 RNA has been further reported in clinical samples from patients with lung, ovarian, esophageal, colorectal, breast or cervical cancers (Hu and Lu, 2015; Iacoangeli et al., 2004; Li et al., 2016; Peng et al., 2018; Wu et al., 2016; Zhao et al., 2016). In colorectal and gastric cancers, BC200 RNA expression is highly correlated with tumor node metastasis (TNM) stage (Wu et al., 2018) and tumor size (Gu et al., 2018; Ren et al., 2018). In addition, upregulation of BC200 RNA is associated with poor prognosis of cancer patients (Wu et al., 2018; Zhao et al., 2016). BC200 RNA was also recently detected in blood of breast cancer patients (Iacoangeli et al., 2018). These data suggest the possibility of using BC200 RNA as a diagnostic and prognostic marker for human cancers.
Expression of BC200 RNA is associated with estrogen receptor-α (ERα) in breast cancers (Singh et al., 2016) and tends to be high in ERα-positive cell lines. Its expression is induced by estrogen, which promotes BC200 RNA transcription through binding of ERα to the −585 bp region relative to the transcription start site. In lung cancers, the oncogenic transcription factor c-MYC promotes BC200 RNA expression (Hu and Lu, 2015). Modulation of c-Myc expression, including inhibition of c-MYC, c-MYC knockdown and c-MYC overexpression, affects cellular levels of BC200 RNA, which are also proportional to those of c-MYC protein in lung cancer.
Functions of BC200 RNA in cancer
BC200 RNA is more highly expressed in invasive cancer cells than in benign tumors (Iacoangeli et al., 2004), giving rise to the hypothesis that BC200 RNA plays a role in cell metastasis. This hypothesis was first experimentally demonstrated by showing that cell migration and invasion are altered according to the BC200 RNA level in the lung cancer cell line A594 (Hu and Lu, 2015). In A594 cells, BC200 RNA promotes cell migration and invasion by upregulating the matrix metalloproteases, MMP9 and MMP13. BC200 RNA also modulates the expression of MMP9 in colon cancers (Wu et al., 2018). In these cancers, BC200 RNA not only regulates MMP9 expression, it also induces epithelial-mesenchymal transition through activation of signal transducer and activator of transcription 3 (STAT3) (Wu et al., 2018). In gastric carcinoma cells, BC200 RNA upregulates expression of epithelial cell adhesion molecule (EpCAM) (Ren et al., 2018). Using a systematic ribosome profiling approach to identify downstream targets of BC200 RNA, Lee and colleagues (Shin et al., 2017) sought to elucidate additional functions of BC200 RNA in cervical cancer cells, revealing 29 genes whose ribosomal occupation was affected by more than 2-fold by BC200 RNA knockdown. They further showed that BC200 RNA acts through stabilization of mRNA for S100A11, one of the genes downregulated by BC200 RNA knockdown, to promote cell migration and invasion. In addition, BC200 RNA is associated with another ncRNA miR138, which is known to act as a tumor-suppressive miRNA (Long et al., 2013; Mitomo et al., 2008; Zhang et al., 2013) to promote cell metastasis of cervical cancer (Peng et al., 2018). Overexpression of BC200 RNA downregulates miR138, promoting cell migration.
BC200 RNA also contributes to cancer development by suppressing apoptosis and activating proliferation. In colorectal cancers, BC200 RNA knockdown leads to apoptosis and decreased proliferation via down-regulating natriuretic peptide receptor C (NPR3) expression (Gu et al., 2018). Furthermore, BC200 RNA blocks splicing of the Bcl gene to the pro-apoptotic Bcl-xS isoform in breast cancer cells, thereby inhibiting apoptosis (Singh et al., 2016). Interestingly, apoptosis induced by BC200 knockdown is limited to proliferating cancer cells (Booy et al., 2017), making targeting BC200 RNA a rationale cancer therapy strategy for reducing side effects on normal cells.
REPRESENTATIVE CANCER-RELATED NCRNAS
HOTAIR
HOTAIR (Hox transcript antisense intergenic RNA) is the best-known oncogenic ncRNA (Gupta et al., 2010; Kogo et al., 2011). HOTAIR contributes to the genesis and development of breast, gastric, pancreatic, liver, hepatocellular, colorectal, lung, and ovarian cancer (Hajjari and Salavaty, 2015). Mechanistic studies have identified HOTAIR as an epigenetic regulator (Gupta et al., 2010; Kogo et al., 2011), showing that it plays a pivotal role in guiding histone-modifying complexes to target loci. Overexpression of HOTAIR in cancer cells changes methylation status and expression patterns of hundreds of genes, including the metastasis-suppressor genes homeobox D10 (HOXD10), junction adhesion molecule 2 (JAM2), progesterone receptor (PGR), protocadherin 10 (PCDH10) and protocadherin beta 5 (PCDHB5); HOTAIR contributes to cancer formation by inhibiting the expression of these genes (Balas and Johnson, 2018).
Several recent meta-analyses have evaluated the prognostic value of HOTAIR in cancers. In an analysis of data from 1,297 gastrointestinal cancer patients published in 15 articles, Zhang et al. (Zhang et al., 2018) showed that HOTAIR is strongly positively correlated with lymph node metastasis, distant metastasis and poor clinical stage, and negatively correlated with overall survival. A similar tendency has also been found in studies of esophageal cancer and ovarian cancer, suggesting that HOTAIR RNA has potential value as a prognostic marker (Luo et al., 2017; Song and Zou, 2016).
miRNAs
miRNAs are small ncRNAs that regulate gene expression at the post-transcriptional level. They exert their regulatory influence by interacting with the untranslated region of the target mRNA, resulting in mRNA degradation or translational arrest (Bushati and Cohen, 2007). Abnormal expression of miRNAs in cancer immediately leads to abnormal expression of their target mRNAs. Through this mechanism, a large number of miRNAs are involved in the genesis and development of cancer (Iorio and Croce, 2012; Ma et al., 2018). For example, let-7, a representative miRNA associated with cancers (Barh et al., 2010), inhibits expression of the oncogenes, RAS and HMGA2 (Johnson et al., 2005; Lee and Dutta, 2007); thus, downregulation of let-7 could promote tumorigenesis.
miRNA profiling analysis can be a practical cancer diagnostic method. There are many miRNA expression signatures in cancers as well as characteristic patterns of miRNA expression for each cancer stage (Lu et al., 2005; Volinia et al., 2006). The ability to classify cancer histology by miRNA profiling has been successfully demonstrated for renal and lung cancers (Benjamin et al., 2010; Spector et al., 2013). Some miRNAs are detected in body fluids (Chen et al., 2008). Such circulating miRNAs are remarkably stable and have characteristic patterns determined by their origins, characteristics that are ideal for use as biomarkers. Various studies and clinical trials utilizing circulating miRNAs for cancer diagnosis in carcinomas of the prostate and cervix are currently underway (Brase et al., 2011; Hasanzadeh et al., 2018; Qu et al., 2011). miRNAs themselves are also utilized as drug candidates. MRX34, a liposomal miR-34a mimic, is the first case of a miRNA mimic entering clinical trials (Beg et al., 2017). In this phase I study, MRX34 has shown evidence of antitumor activity in a subset of patients with refractory, advanced solid tumors.
DIRECTION OF FUTURE INVESTIGATIONS OF BC200 RNA
HOTAIR and miRNAs are potentially good models for clinical applications of BC200 RNA. A number of considerations need to be addressed before BC200 RNA can be applied clinically. First, BC200 RNA expression needs to be systematically analyzed in a wider variety of samples. To date, expression patterns of BC200 RNA have been analyzed in a total of 478 patients with breast, lung, ovarian, esophageal, colorectal, cervical, or gastric cancers (Table 1). Studies have been biased towards specific areas and, in many cases, are small-scale, and are retrospective in nature. Thus, reducing experimental and regional biases will require analysis of BC200 RNA expression in a wider range of patient samples. Second, existing data need to be integrated through meta-analyses. For example, a meta-analysis of 1297 patient samples from 15 individual studies validated the prognostic value of HOTAIR in gastrointestinal cancer (Zhang et al., 2018). Such comprehensive analyses are needed to verify the diagnostic value of ncRNAs determined based on data accumulated through individual experiments, as in the case of HOTAIR. Third, a technology that can reliably reduce BC200 RNA levels in patients’ cancer cells is needed. To date, BC200 RNA knockdown has been achieved by transfection of oligonucleotides such as siRNAs or Gapmers using commercial reagents (Booy et al., 2017; Shin et al., 2017). This approach works well in cell lines, but such transfection reagents are not suitable for use in clinical applications, and oligonucleotides lack cell permeability on their own. Applying the results of recent studies designed to increase the cell permeability of oligonucleotides to the BC200 RNA-knockdown process might ultimately solve this problem (Chen et al., 2018).
Table 1.
Summary of clinical studies of BC200 RNA in cancer
| Cancer type | Sample number | Detection method | BC200 RNA expression | Characteristics correlated with BC200 RNA expression | References |
|---|---|---|---|---|---|
| Breast | 42 | ISH | Up | (Iacoangeli et al., 2004) | |
| Lung | 20 | qRT-PCR | Up | (Hu and Lu, 2015) | |
| Ovary | 22 | qRT-PCR | Down | (Wu et al., 2016) | |
| Esophagus | 70 | qRT-PCR | Up | Lower survival rate | (Zhao et al., 2016) |
| Colon | 36 | qRT-PCR | Up | (Li et al., 2016) | |
| Colon | 82 | qRT-PCR | Up | Higher TNM stage, lower survival rate | (Wu et al., 2018) |
| Colon | 96 | qRT-PCR | Up | Higher TNM stage, larger tumor size | (Gu et al., 2018) |
| Stomach | 85 | qRT-PCR | Up | Higher TNM stage, larger tumor size | (Ren et al., 2018) |
| Cervix | 25 | qRT-PCR, NB | Up | (Peng et al., 2018) |
*ISH, in situ hybridization; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; NB, Northern blotting; TNM, tumor node metastasis.
CONCLUSIONS
As long-unappreciated functions of ncRNAs have begun to emerge, it has become clear that ncRNAs play key roles in multiple cellular functions that are essential for various physiological or pathological processes. Therefore, it is critical to understand functions of disease-related ncRNAs and utilize them for the development of new strategies for cancer therapy.
Studies conducted during the last four years have identified abnormal expression mechanisms and functions of BC200 RNA in lung, colorectal, breast, and cervical cancer (Fig. 2). These studies have shown that there is a reliable correlation between BC200 RNA expression and tumor stage, and that BC200 RNA promotes cancer cell development by promoting cell proliferation and metastasis, and averting apoptosis. These basic studies highlight the promise that BC200 RNA holds as a potential cancer diagnostic marker and therapeutic target. Future systematic analyses and experimental studies will test these possibilities.
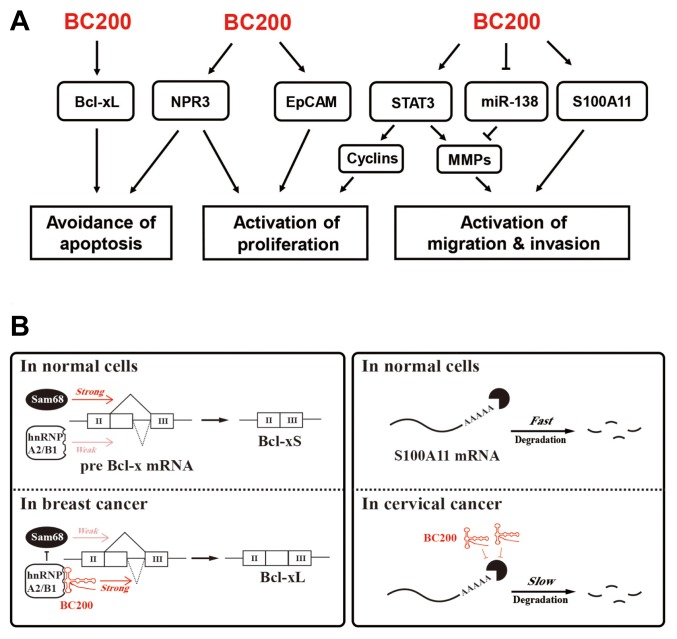
Fig. 2. Involvement of BC200 RNA in the genesis and development of cancers.
(A) Diagram depicting BC200 RNA signaling involved in induction of cancer-related phenotypes. BC200 RNA shifts Bcl splicing toward production of the Bcl-xL isoform, thereby preventing apoptosis; this effect is further achieved by upregulation of NPR3. BC200 RNA promotes cell proliferation by upregulating NPR3, EpCAM, and STAT3. Activation of STAT3 or upregulation of S100A11 by BC200 RNA causes stimulation of migration and invasion. BC200 RNA inhibits synthesis of the tumor-suppressive miRNA, miR-138, resulting in activation of migration and invasion. (B) Functional mechanisms of BC200 RNA. Left: Altering the splicing pattern of Bcl-x mRNA. In the absence of BC200 RNA, Sam68 regulates the splicing of Bcl-x by interacting with its pre-mRNA, leading to expression of Bcl-xS. BC200 RNA interacts with Bcl-x pre-mRNA and facilitates recruitment of hnRNP A2/B1 to the pre-mRNA. This recruitment interferes with binding of Sam68 to the pre-mRNA and consequently changes the fate of Bcl-x splicing to generation of Bcl-xL. Right: Stabilization of S100A11 mRNA. BC200 RNA stabilizes S100A11 mRNA, possibly by binding to its mRNA.
ACKNOWLEDGMENTS
This study was supported by grants from the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (MSIT) of the Korean government (NRF-2017R1A2B4010713 and 2017R1A6A3A11031308) and the Intelligent Synthetic Biology Center of Global Frontier Project, funded by the MSIT (2013M3A6A8073557).
REFERENCES
- Amin V., Harris R.A., Onuchic V., Jackson A.R., Charnecki T., Paithankar S., Lakshmi Subramanian S., Riehle K., Coarfa C., Milosavljevic A. Epigenomic footprints across 111 reference epigenomes reveal tissue-specific epigenetic regulation of lincRNAs. Nat Commun. 2015;6:6370. doi: 10.1038/ncomms7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer C., Claus R., Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73:473–477. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- Balas M.M., Johnson A.M. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res. 2018;3:108–117. doi: 10.1016/j.ncrna.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandres E., Agirre X., Bitarte N., Ramirez N., Zarate R., Roman-Gomez J., Prosper F., Garcia-Foncillas J. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- Barh D., Malhotra R., Ravi B., Sindhurani P. MicroRNA let-7: an emerging next-generation cancer therapeutic. Curr Oncol. 2010;17:70–80. doi: 10.3747/co.v17i1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg M.S., Brenner A.J., Sachdev J., Borad M., Kang Y.K., Stoudemire J., Smith S., Bader A.G., Kim S., Hong D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35:180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin H., Lebanony D., Rosenwald S., Cohen L., Gibori H., Barabash N., Ashkenazi K., Goren E., Meiri E., Morgenstern S., et al. A diagnostic assay based on microRNA expression accurately identifies malignant pleural mesothelioma. J Mol Diagn. 2010;12:771–779. doi: 10.2353/jmoldx.2010.090169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy E.P., McRae E.K., Koul A., Lin F., McKenna S.A. The long non-coding RNA BC200 (BCYRN1) is critical for cancer cell survival and proliferation. Mol Cancer. 2017;16:109. doi: 10.1186/s12943-017-0679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brase J.C., Johannes M., Schlomm T., Falth M., Haese A., Steuber T., Beissbarth T., Kuner R., Sultmann H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128:608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- Briggs J.A., Wolvetang E.J., Mattick J.S., Rinn J.L., Barry G. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. 2015;88:861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- Brosnan C.A., Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009;21:416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Bushati N., Cohen S.M. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Chen W., Bocker W., Brosius J., Tiedge H. Expression of neural BC200 RNA in human tumours. J Pathol. 1997;183:345–351. doi: 10.1002/(SICI)1096-9896(199711)183:3<345::AID-PATH930>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X., et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Chen X., Mangala L.S., Rodriguez-Aguayo C., Kong X., Lopez-Berestein G., Sood A.K. RNA interference-based therapy and its delivery systems. Cancer Metastasis Rev. 2018;37:107–124. doi: 10.1007/s10555-017-9717-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Nakagawa H., Uemura M., Piao L., Ashikawa K., Hosono N., Takata R., Akamatsu S., Kawaguchi T., Morizono T., et al. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 2011;102:245–252. doi: 10.1111/j.1349-7006.2010.01737.x. [DOI] [PubMed] [Google Scholar]
- Consortium E.P. The ENCODE (ENCyclopedia Of DNA Elements) Project. Sci. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duning K., Buck F., Barnekow A., Kremerskothen J. SYNCRIP, a component of dendritically localized mRNPs, binds to the translation regulator BC200 RNA. J Neurochem. 2008;105:351–359. doi: 10.1111/j.1471-4159.2007.05138.x. [DOI] [PubMed] [Google Scholar]
- Ebisuya M., Yamamoto T., Nakajima M., Nishida E. Ripples from neighbouring transcription. Nat Cell Biol. 2008;10:1106–1113. doi: 10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- Eom T., Berardi V., Zhong J., Risuleo G., Tiedge H. Dual nature of translational control by regulatory BC RNAs. Mol Cell Biol. 2011;31:4538–4549. doi: 10.1128/MCB.05885-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Lu L., Zhou D., Liu Z. Long noncoding RNA BCYRN1 promotes the proliferation of colorectal cancer cells via upregulating NPR3 expression. Cell Physiol Biochem. 2018;48:2337–2349. doi: 10.1159/000492649. [DOI] [PubMed] [Google Scholar]
- Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjari M., Salavaty A. HOTAIR: an oncogenic long noncoding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasanzadeh M., Movahedi M., Rejali M., Maleki F., Moetamani-Ahmadi M., Seifi S., Hosseini Z., Khazaei M., Amerizadeh F., Ferns G.A., et al. The potential prognostic and therapeutic application of tissue and circulating microRNAs in cervical cancer. J Cell Physiol. 2019;234:1289–1294. doi: 10.1002/jcp.27160. [DOI] [PubMed] [Google Scholar]
- Hu T., Lu Y.R. BCYRN1, a c-MYC-activated long noncoding RNA, regulates cell metastasis of non-small-cell lung cancer. Cancer Cell Int. 2015;15:36. doi: 10.1186/s12935-015-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacoangeli A., Adzovic L., Chen E.Q., Latif Cattie R., Soff G.A., Tiedge H. Regulatory BC200 RNA in peripheral blood of patients with invasive breast cancer. J Investig Med. 2018;66:1055–1063. doi: 10.1136/jim-2018-000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacoangeli A., Lin Y., Morley E.J., Muslimov I.A., Bianchi R., Reilly J., Weedon J., Diallo R., Bocker W., Tiedge H. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis. 2004;25:2125–2133. doi: 10.1093/carcin/bgh228. [DOI] [PubMed] [Google Scholar]
- Iorio M.V., Croce C.M. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Thakkar N., Chhatai J., Pal Bhadra M., Bhadra U. Long non-coding RNA: Functional agent for disease traits. RNA Biol. 2017;14:522–535. doi: 10.1080/15476286.2016.1172756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Shin H., Lee J., Kim Y., Bak G., Lee Y. Regulation of BC200 RNA-mediated translation inhibition by hnRNP E1 and E2. FEBS Lett. 2017;591:393–405. doi: 10.1002/1873-3468.12544. [DOI] [PubMed] [Google Scholar]
- Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Johnsson P., Morris K.V. Expanding the functional role of long noncoding RNAs. Cell Res. 2014;24:1284–1285. doi: 10.1038/cr.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lee J., Shin H., Jang S., Kim S.C., Lee Y. Biosynthesis of brain cytoplasmic 200 RNA. Sci Rep. 2017;7:6884. doi: 10.1038/s41598-017-05097-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.Y., Oh S., Yoo K.H. Functional enhancers as master regulators of tissue-specific gene regulation and cancer Development. Mol Cells. 2017;40:169–177. doi: 10.14348/molcells.2017.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T., Tanaka F., Shibata K., Suzuki A., Komune S., et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- Kondrashov A.V., Kiefmann M., Ebnet K., Khanam T., Muddashetty R.S., Brosius J. Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP) J Mol Biol. 2005;353:88–103. doi: 10.1016/j.jmb.2005.07.049. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Yang B., Xia S., Chen L.-h., Ning N., Ma B., Liu Q., Yang H., Zhang D.S.-z., Du X.N. BC200 RNA is over-expressed in colorectal cancer and promotes migration and invasion of HCT116 cells. Int J Clin Exp Patho. 2016;9:1481–1486. [Google Scholar]
- Lin D., Pestova T.V., Hellen C.U., Tiedge H. Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol Cell Biol. 2008;28:3008–3019. doi: 10.1128/MCB.01800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long L., Huang G., Zhu H., Guo Y., Liu Y., Huo J. Down-regulation of miR-138 promotes colorectal cancer metastasis via directly targeting TWIST2. J Transl Med. 2013;11:275. doi: 10.1186/1479-5876-11-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Luo P., Liu X.F., Wang Y.C., Li N.D., Liao S.J., Yu M.X., Liang C.Z., Tu J.C. Prognostic value of abnormally expressed lncRNAs in ovarian carcinoma: a systematic review and meta-analysis. Oncotarget. 2017;8:23927–23936. doi: 10.18632/oncotarget.14760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Zhang J., Weng Y.C., Wang J.C. EZH2-Mediated microRNA-139-5p regulates epithelial-mesenchymal transition and lymph node metastasis of pancreatic cancer. Mol Cells. 2018;41:868–880. doi: 10.14348/molcells.2018.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitomo S., Maesawa C., Ogasawara S., Iwaya T., Shibazaki M., Yashima-Abo A., Kotani K., Oikawa H., Sakurai E., Izutsu N., et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008;99:280–286. doi: 10.1111/j.1349-7006.2007.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty R., Khanam T., Kondrashov A., Bundman M., Iacoangeli A., Kremerskothen J., Duning K., Barnekow A., Huttenhofer A., Tiedge H., et al. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J Mol Biol. 2002;321:433–445. doi: 10.1016/s0022-2836(02)00655-1. [DOI] [PubMed] [Google Scholar]
- Pasmant E., Sabbagh A., Vidaud M., Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- Peng J., Hou F., Feng J., Xu S.X., Meng X.Y. Long non-coding RNA BCYRN1 promotes the proliferation and metastasis of cervical cancer via targeting microRNA-138 in vitro and in vivo. Oncol Lett. 2018;15:5809–5818. doi: 10.3892/ol.2018.8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu H., Xu W., Huang Y., Yang S. Circulating miRNAs: promising biomarkers of human cancer. Asian Pacific Journal of cancer prevention: APJCP. 2011;12:1117–1125. [PubMed] [Google Scholar]
- Qureshi I.A., Mehler M.F. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nature reviews Neuroscience. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Yang X., Yang Y., Zhang X., Zhao R., Wei R., Zhang X., Zhang Y. Upregulation of LncRNA BCYRN1 promotes tumor progression and enhances EpCAM expression in gastric carcinoma. Oncotarget. 2018;9:4851–4861. doi: 10.18632/oncotarget.23585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J.D., Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 2009;23:1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- Samson J., Cronin S., Dean K. BC200 (BCYRN1) - The shortest, long, non-coding RNA associated with cancer. Noncoding RNA Res. 2018;3:131–143. doi: 10.1016/j.ncrna.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm L., Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- Shin H., Lee J., Kim Y., Jang S., Lee Y., Kim S., Lee Y. Knockdown of BC200 RNA expression reduces cell migration and invasion by destabilizing mRNA for calcium-binding protein S100A11. RNA Biol. 2017;14:1418–1430. doi: 10.1080/15476286.2017.1297913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Gupta S.C., Peng W.X., Zhou N., Pochampally R., Atfi A., Watabe K., Lu Z., Mo Y.Y. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death Dis. 2016;7:e2262. doi: 10.1038/cddis.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser N.R. The RNA-centred view of the synapse: noncoding RNAs and synaptic plasticity. Philos Trans R Soc Lond B Biol Sci. 2014:369. doi: 10.1098/rstb.2013.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Zou S.B. Prognostic role of lncRNA HOTAIR in esophageal squamous cell carcinoma. Clin Chim Acta. 2016;463:169–173. doi: 10.1016/j.cca.2016.10.035. [DOI] [PubMed] [Google Scholar]
- Spector Y., Fridman E., Rosenwald S., Zilber S., Huang Y., Barshack I., Zion O., Mitchell H., Sanden M., Meiri E. Development and validation of a microRNA-based diagnostic assay for classification of renal cell carcinomas. Mol Oncol. 2013;7:732–738. doi: 10.1016/j.molonc.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J.G., Milner R.J., Bloom F.E., Lerner R.A. Common 82-nucleotide sequence unique to brain RNA. Proc Natl Acad Sci USA. 1982;79:4942–4946. doi: 10.1073/pnas.79.16.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J.G., Milner R.J., Gottesfeld J.M., Lerner R.A. Identifier sequences are transcribed specifically in brain. Nature. 1984;308:237–241. doi: 10.1038/308237a0. [DOI] [PubMed] [Google Scholar]
- Tiedge H., Chen W., Brosius J. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J Neurosci. 1993;13:2382–2390. doi: 10.1523/JNEUROSCI.13-06-02382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Ward L.D., Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol. 2012;30:1095–1106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J.B., Sutcliffe J.G. Primate brain-specific cytoplasmic transcript of the Alu repeat family. Mol Cell Biol. 1987;7:3324–3327. doi: 10.1128/mcb.7.9.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D.I., Wang T., Ren C., Liu L., Kong D., Jin X., Li X., Zhang G. Downregulation of BC200 in ovarian cancer contributes to cancer cell proliferation and chemoresistance to carboplatin. Oncol Lett. 2016;11:1189–1194. doi: 10.3892/ol.2015.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Xu K., Liu K., Huang J., Chen J., Zhang J., Zhang N. Long noncoding RNA BC200 regulates cell growth and invasion in colon cancer. Int J Biochem Cell Biol. 2018;99:219–225. doi: 10.1016/j.biocel.2018.04.001. [DOI] [PubMed] [Google Scholar]
- Wu Z., Liu X., Liu L., Deng H., Zhang J., Xu Q., Cen B., Ji A. Regulation of lncRNA expression. Cell Mol Biol Lett. 2014;19:561–575. doi: 10.2478/s11658-014-0212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhang H., Zhao M., Lv Z., Zhang X., Qin X., Wang H., Wang S., Su J., Lv X., et al. MiR-138 inhibits tumor growth through repression of EZH2 in non-small cell lung cancer. Cell Physiol Biochem. 2013;31:56–65. doi: 10.1159/000343349. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang L.J., Li W.F., Zhang X., Yang X.J. The prognostic value of HOTAIR for predicting long-term prognosis of patients with gastrointestinal cancers. Medicine (Baltimore) 2018;97:e11139. doi: 10.1097/MD.0000000000011139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R.H., Zhu C.H., Li X.K., Cao W., Zong H., Cao X.G., Hu H.Y. BC200 LncRNA a potential predictive marker of poor prognosis in esophageal squamous cell carcinoma patients. Onco Targets Ther. 2016;9:2221–2226. doi: 10.2147/OTT.S99401. [DOI] [PMC free article] [PubMed] [Google Scholar]