Abstract
It is estimated that up to 5% of cystic fibrosis transmembrane conductance regulator (CFTR) pathogenic alleles are unidentified. Some of these errors may lie in noncoding regions of the locus and affect gene expression. To identify regulatory element variants in the CFTR locus, SureSelect targeted enrichment of 460 kb encompassing the gene was optimized to deep sequence genomic DNA from 80 CF patients with an unequivocal clinical diagnosis but only one or no CFTR-coding region pathogenic variants. Bioinformatics tools were used to identify sequence variants and predict their impact, which were then assayed in transient reporter gene luciferase assays. The effect of five variants in the CFTR promoter and four in an intestinal enhancer of the gene were assayed in relevant cell lines. The initial analysis of sequence data revealed previously known CF-causing variants, validating the robustness of the SureSelect design, and showed that 85 of 160 CF alleles were undefined. Of a total 1737 variants revealed across the extended 460-kb CFTR locus, 51 map to known CFTR cis-regulatory elements, and many of these are predicted to alter transcription factor occupancy. Four promoter variants and all those in the intestinal enhancer significantly repress reporter gene activity. These data suggest that CFTR regulatory elements may harbor novel CF disease–causing variants that warrant further investigation, both for genetic screening protocols and functional assays.
CME Accreditation Statement: This activity (“JMD 2019 CME Program in Molecular Diagnostics”) has been planned and implemented in accordance with the accreditation requirements and policies of the Accreditation Council for Continuing Medical Education (ACCME) through the joint providership of the American Society for Clinical Pathology (ASCP) and the American Society for Investigative Pathology (ASIP). ASCP is accredited by the ACCME to provide continuing medical education for physicians.
The ASCP designates this journal-based CME activity (“JMD 2019 CME Program in Molecular Diagnostics”) for a maximum of 18.0 AMA PRA Category 1 Credit(s)™. Physicians should claim only credit commensurate with the extent of their participation in the activity.
CME Disclosures: The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
Most disease-causing variants for monogenic disorders fall within the coding region or splice site sequences of genes. Although sequencing efforts to identify causal variants have focused mainly on the exome, it is estimated that 98% of the human genome is composed of noncoding DNA. It is likely that many pathogenic variants lie within these noncoding sequences, especially those that influence chromatin structure/organization or gene expression. Next-generation sequencing protocols have facilitated the discovery and characterization of many cis-regulatory element variants that underlie human disease.1 However, generating and interpreting full genomic sequence data for disease-associated loci in patients are not yet common practice.
Cystic fibrosis, a common life-shortening autosomal recessive disorder, is caused by pathogenic variants in the cystic fibrosis transmembrane conductance regulator (CFTR) gene, which encodes a cyclic AMP-activated chloride channel. Newborn screening for CF is routine in the countries with advanced health care, because most of the estimated 70,000 individuals with CF are of European ancestry.2, 3, 4, 5, 6 Commercial tests for approximately 100 variants detect approximately 90% of CF-causal alleles in North America and northern European countries. There are >2000 documented CFTR variants listed in the CFTR Mutation Database (www.genet.sickkids.on.ca); however, as of December 2017, only 312 of the most frequently observed variants in the CF population have been annotated as disease-causing in the Clinical and Functional Translation of CFTR Database (https://cftr2.org). Notably, the variants listed in these databases largely include those that fall within the exons (coding regions) or at intron-exon boundary segments (encompassing splice sites) of the 189-kb CFTR locus, which spans from 1 kb upstream of the transcription start site to the 3′ untranslated region. However, even with extensive sequencing of these exonic and boundary regions, and identification of novel intronic cryptic splice sites by transcript analysis, it is estimated that 1% to 5% of CF alleles have unidentified causal variants, which are presumed to lie in regulatory noncoding regions of the CFTR locus.7
We previously identified and characterized CFTR structural and cis-regulatory elements, which extend approximately 80 kb upstream of the CFTR transcriptional start site and approximately 90 kb downstream of the CFTR translational stop site, spanning a distance of approximately 350 kb.8 The distal sites at −80.1 kb (from the CFTR start codon) and +48.9 kb (from the CFTR stop codon) function as topologically associating domain boundaries (TADs), which isolate CFTR and its regulatory elements from those of other genes.9, 10 Many important CFTR cis-regulatory elements are found in the intergenic and intronic sequences encompassed by this topologically associating domain.8, 9, 10, 11, 12
Our goal was to develop a robust targeted-enrichment approach to sequence the approximately 460-kb region encompassing the CFTR topologically associating domain, thus including known structural and cis-regulatory elements. This pipeline was then applied to look for variants in the noncoding regions of CFTR in CF patients with an unequivocal diagnosis but incomplete causal variant information. The following were used: an archived cohort of Canadian CF patients with undefined pathogenic variants on one or both alleles after coding region evaluation by earlier multiplex heteroduplex analysis13 and exclusion of large deletions in some patients; and CF patients from Brittany, France, and Japan in whom all exons had been fully sequenced (those from France were also screened for large deletions).14, 15 After deep sequencing and genotyping of 80 CF genomic DNA (gDNA) samples, known disease-causing variants were identified on 75 alleles, whereas 85 CFTR alleles still contained unknown pathogenic CF variants. Taking all alleles together, 1737 variants [1426 substitutions and 311 insertions/deletions (indels)] were identified across the 460-kb region, of which 52 occurred in 20 CFTR structural and cis-regulatory elements. Many of these alterations are predicted to alter the binding of transcription factors (TFs) known to regulate CFTR expression or other TFs with a pivotal role in differentiated epithelia. Functional analysis of five variants found within the CFTR promoter region (2 kb) showed them to impair its activity in airway and intestinal cell lines. Moreover, four variants in an intestinal intronic enhancer of CFTR also significantly reduced enhancer activity in transient assays in an intestinal cell line. Further characterization of these intronic and intergenic variants may reveal novel pathogenic mechanisms causing CF.
Materials and Methods
Genomic DNA Samples
Genomic DNA from 80 deidentified CF patients, with one or both CFTR alleles having unknown pathogenic variants, were obtained from three CF centers in accordance with approved local guidelines. All patients were observed as having CF on the basis of their clinical characteristics, with diagnosis based on elevated sweat chloride levels and/or other CF-presenting features, including failure to thrive, progressive lung disease, and infection histories. gDNA samples were drawn populations in Canada (n = 72), Brittany, France (n = 6), and Japan (n = 2). The integrity of each gDNA sample was determined by Bioanalyzer (Agilent Technologies, Santa Clara, CA) before target enrichment and sequencing.
SureSelect Targeted Enrichment Design
Agilent's SureDesign eArray tool (Agilent Technologies) was used to design approximately 120-bp SureSelect biotinylated complementary RNA probes to span approximately 460 kb that encompasses the extended CFTR locus (GRCh37/hg19 chromosome 7:116,970,000 to 117,429,999), including previously published cis-regulatory elements.8 The first design included 12,776 probes with a total coverage of 491,070 kb (9158_repeat_moderate stringency_maximum_boost_1X set and an additional 3618 least stringency_max_boost_1X, to add coverage for regions omitted from the moderate stringency group). To reduce off-target hybridization, an optimized design of 12,053 probes covering 463,793 kb was generated by removing the 630 most repetitive probes from the least stringency group (9158_repeat_moderate stringency_maximum_boost_1X set and an additional 2895 least stringency_max_boost_1X). SureSelect libraries (containing 24 or 48 samples) were prepared, according to the manufacturer's protocol, and up to 48 libraries were pooled and sequenced on a HiSeq2500 (Illumina Inc., San Diego, CA) for 100-bp paired-end reads.
Bioinformatic Variant Analysis
Base calls were determined by the the Illumina CASAVA pipeline. After filtering for base quality and adapter sequences, the sequencing reads were aligned to the human reference genome (hg19) using bowtie2 version 2.2.2.16 The aligned reads were formatted for input into the Genome Analysis Toolkit version 3.317; the pipeline was modeled on the Broad Institute’s Best Practices Guideline for variant calling,18, 19 including local realignment, removal of PCR duplicates, and base quality recalibration. Single-nucleotide variants and small indel variants were called by the Genome Analysis Toolkit HaplotypeCaller, and variants for each sample were consolidated with GenotypeGVCFs. The combined variant call format file was then annotated with ANNOVAR version 2015June1720 and filtered for quality with VCFtools version 0.1.12.b.21 Large deletions/duplications were identified using custom Perl scripts and manually called using Integrative Genomics Viewer (IGV) version 2.3.92 (http://www.broadinstitute.org/igv) (Supplemental Code S1).22, 23
CFTR and Variant Nomenclature
CFTR introns and exons are numbered using legacy nomenclature,24 for consistency with our previous work.8 All variants are numbered with respect to the A (+1) of the ATG start codon of the major CFTR transcript [LRG_663; NG_016465.4 (NM_000492.3)], following the recommendations of the Human Genome Variation Society.25
In Silico Predictions
MatInspector version 8.4 (Genomatix, Munich, Germany) was used to predict TF binding sites (Matrix Library 10.0) in reference genome versus variant sequences for CFTR cis-regulatory elements surrounding the variants of interest using default matrix search parameters (core similarity, 0.75; matrix similarity, optimized).
Plasmid Construction
Site-directed mutagenesis on pGL3B.1963,26 which contains an approximately 2-kb CFTR promoter fragment (hg19 chromosome 7:117,118,152 to 117,120,148), was performed using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent Technologies). Approximately 1700 bp encompassing the CFTR intron 11 enhancer element27 (hg19 chromosome 7: 117,227,831 to 117,229,503) was PCR amplified and cloned into pSCB using the StrataClone Blunt PCR Cloning Kit (Agilent Technologies). The fragment was sub-cloned using SalI into the enhancer site of pGL3B.245, containing the 787-bp minimal CFTR promoter,28, 29 and site-directed mutagenesis was performed. Primers are listed in Table 1, and all plasmids were verified using Sanger sequencing.
Table 1.
Oligonucleotides
| Oligonucleotide | Sequence | Description |
|---|---|---|
| oJLK021 | 5′-GTAATTACGCAAAGCATTATCTCTTCTTACCTCCTTGCAGATTTTTT-3′ | CFTR promoter -887C>T |
| oJLK022 | 5′-CTCCTCTTACCTCCTTGCAGATTTTTTTTCTCTTTCAGTACG-3′ | CFTR promoter -869delT |
| oJLK023 | 5′-CCACCCTTGGAGTTCACGCACCTAAACCTGAAACT-3′ | CFTR promoter -812T>G |
| oJLK025 | 5′-GGATGGGCCTGCTGCTGGGCGGT-3′ | CFTR promoter -410G>C |
| oJLK026 | 5′-CCCCAGCGCCCCAGAGACCA-3′ | CFTR promoter -8G>C |
| SalI DHS11F | 5′-CGTCGACTGGAGAAGGTGGAATCACACTG-3′ | DHS11-long forward cloning primer |
| oJLK048 | 5′-CGTCGACTTCTCTGTTTATACATGTAATTGTTGG-3′ | DHS11-long reverse cloning primer |
| oJLK044 | 5′-GTCCAAGCATTTTAAAGCTGTCAAAGATATGTAAATATAGATAATGTATGTCAAG-3′ | c.1679+566G>T (DHS11) |
| oJLK045 | 5′-ACTTTGAGGAACTAAAAATAATTGTCTATTCTTATTCTGATCAGAATGTGTAATG-3′ | c.1679+1280G>A (DHS11) |
| oJLK046 | 5′-GATCCATTATGTAGCTCTTGCATGCTGTCTTCAAAAATAAGTTACA-3′ | c.1679+1449A>G (DHS11) |
| oJLK047 | 5′-CCATTGGTTTTTAAAAAAATTTTTAAATTGGCTTCAAAAATTTCTTAATTGTGTGCTGAATACAATTTT-3′ | c.1679+1539T>C (DHS11) |
Cell Culture and Transient Luciferase Assays
Human colorectal carcinoma Caco230 and bronchial epithelial 16HBE14o-31 cell lines were cultured in Dulbecco's modified Eagle's medium, low glucose supplemented with 10% fetal bovine serum. Using standard methods,27 cells were cotransfected with pGL3B luciferase reporter constructs and a modified pRL Renilla luciferase control vector using Lipofectin (Thermo Fisher Scientific, Waltham, MA). Cells were lysed after 48 hours and assayed on a GloMax Navigator (Promega Corp., Madison, WI) for firefly and Renilla luciferase activities using the Dual-Luciferase Reporter Assay Kit (Promega Corp.). Transfections were performed three times, in triplicate, using two different plasmid preparations.
Results
SureSelect Targeted Sequencing Study Design
Using a targeted enrichment approach (chromosome 7:116,970,000 to 117,429,999) and 12,053 SureSelect biotinylated complementary RNA probes, 463 kb encompassing the extended CFTR locus were deep sequenced. This region extends beyond the −80.1 kb and +48.9 kb topologically associating domain boundaries that demarcate the limits of the CFTR locus, irrespective of cell type or CFTR expression,9, 10 and includes at least 20 other defined CFTR regulatory elements (Figure 1).8 Genomic DNA isolated from 80 deidentified CF patients from Toronto, ON, Canada (n = 72), Brittany, France (n = 6), and Japan (n = 2) was analyzed. At the time of clinical CF diagnosis and initial genotype analysis, all patients had at least one undefined CF allele.
Figure 1.
SureSelect targeted enrichment experimental design. A total of 12,053 SureSelect biotinylated probes span the 463-kb CFTR locus and encompass 20 previously identified CFTR cis-regulatory elements. Gaps in probe coverage correspond to highly repetitive regions that were excluded. Repeats shown correspond to RepeatMasker track from University of California, Santa Cruz, Genome Browser (hg19).
Analysis of CF Alleles
To first evaluate the sensitivity of SureDesign, reliable detection of the known pathogenic alleles among the 80 CF patient gDNA samples was confirmed. Next, currently understood CFTR variants, which were not identified by earlier screening methods, were considered.13, 14, 15 These variants were classified as disease-causing, varying clinical significance, or unknown significance, according to the Clinical and Functional Translation of CFTR Database (which includes 374 variants; https://www.cftr2.org). Eight pathogenic variants, S489X, C524X, W1282X, L558S, CFTRdele2,3, CFTRdele4-7, CFTRdup6b-10, and CFTRdele14b-17b, were newly identified in this patient cohort after SureDesign enrichment and deep sequencing compared with original genotyping efforts (Table 2 and Supplemental Table S132, 33). The large deletions and duplication were detected as approximately twofold reduction or twofold increase in sequence read depth across the affected regions, and all were reported in the literature previously.34, 35, 36, 37, 38 Also, a previously identified approximately 7.2-kb deletion in one CF patient was confirmed.33 The only common CFTR variant among the patient gDNAs that was not detected by the SureDesign was the T(n) polymorphic tract 5T [c.1210-1212(5)] allele. However, this was likely because of sequence alignment issues that will be discussed later. Of 160 CF alleles in our sample set, 75 contained known CF-causing variants that affect the CFTR coding sequence or mRNA splicing (Table 2). Thus, 85 CF alleles in our analysis had unknown pathogenic CF variants.
Table 2.
Genotype Summary
| Variant class | CFTR variants | Alleles, n (total n = 160) |
|---|---|---|
| CF-causing | 75 | |
| Coding variants | G85E, S489X, I507del, F508del, C524X, G551D, c.3744delA,∗W1282X, Q1313X | 59/75 |
| Splice site variants | c.489+1G>T, c.1116+1G>A, c.1393-1G>A, c.1585-1G>A, c.1679+1634A>G, c.1680-877G>T, c.3718-2477C>T, c.3718-3T>G | 10/75 |
| Large deletions/duplications† | CFTRdele2,3, CFTRdele4-7, CFTRdup6b-10, CFTRdele14b-17b, CFTRdele16-17b | 6/75 |
| Varying clinical significance | R117H, 5T | 3‡ |
| Unknown significance | L558S | 1 |
Newly detected variants in this study are shown in bold.
Alias p.Lys1250ArgfsX9.
CFTR legacy nomenclature, see Supplemental Table S1 for Clinical and Functional Translation of CFTR Database full nomenclature.
No patients with R117H, 5T genotype.
Variation Analysis within the Extended CFTR Locus
To use the full depth of the data set, variant analysis was performed on all 160 CF alleles, irrespective of whether they carried defined or undefined causal variants. Substitution and small indel variants, identified by SureDesign–deep sequencing in the extended CFTR locus, were cross referenced against The Database of Single Nucleotide Polymporhisms (dbSNP Build ID: 138; National Center for Biotechnology Information, National Library of Medicine, Bethesda, MD; http://www.ncbi.nlm.nih.gov/SNP) (Table 3). Of the 1737 variants identified, 19% of substitutions and 67% of indels were not annotated in dbSNP138. The high percentage of unannotated indels is likely because many of them occur as more than two alleles. Next, the 20 regions of CFTR that were previously defined as regulatory were studied (Figure 1).8, 27, 39 These regulatory elements cover approximately 20 kb of genomic sequence within which 51 total variants (37 substitutions and 11 indels) were observed in 17 elements (Table 3). Although four of these variants were seen in patients with two CF disease-causing variants (Supplemental Table S1), they were included in further analyses because they may nonetheless be functional in regulating CFTR expression. Many of the variants alter the predicted binding of TFs, either disrupting sites or generating novel ones. Table 4 summarizes these changes for selected known CFTR enhancer elements, including airway (−44 kb and −35 kb) and intestinal (introns 1 and 11) selective enhancers, and an enhancer that is common to both cell lineages (intron 23).39, 40, 41, 42, 43, 44 Changes in TF binding sites in the variant sequences compared with the wild-type CFTR sequence were predicted using MatInspector version 8.4 (Table 4). This approach successfully identified key factors driving CFTR cis-regulatory elements in our earlier work.41, 42, 43, 44, 45
Table 3.
Variant Summary
| Regulatory region | Size, bp | Total variants | Total variants in dbSNP138 | Substitutions | Substitutions in dbSNP138 | Insertions/deletions | Insertions/deletions in dbSNP138 |
|---|---|---|---|---|---|---|---|
| 460-kb CFTR locus | 1737 | 1253 | 1426 | 1152 | 311 | 101 | |
| −80.1 kb | 382 | 1 | 1 | 1 | 1 | 0 | NA |
| −44 kb | 1200 | 1 | 1 | 1 | 1 | 0 | NA |
| −35 kb | 1600 | 4 | 2 | 3 | 2 | 1 | 0 |
| −20.9 kb | 395 | 0 | NA | 0 | NA | 0 | NA |
| −3.4 kb | 1214 | 3 | 3 | 3 | 3 | 0 | NA |
| Promoter | 1999 | 6 | 5 | 5 | 4 | 1 | 1 |
| 185 + 10 kb (Intron 1) | 1100 | 4 | 3 | 2 | 2 | 2 | 1 |
| 1716 + 13.2/13.7 kb (Intron 10a, b) | 1300 | 6 | 3 | 1 | 1 | 5 | 2 |
| 1716 + 23 kb (Intron 10c) | 700 | 1 | 0 | 1 | 0 | 0 | NA |
| 1811 + 0.8 kb (Intron 11) | 1400 | 4 | 2 | 4 | 2 | 0 | NA |
| 3600 + 1.6 kb (Intron 18a) | 1000 | 6 | 5 | 4 | 4 | 2 | 1 |
| 3600 + 10 kb (Intron 18b) | 500 | 2 | 1 | 2 | 1 | 0 | NA |
| 3849 + 12.5 kb (Intron 19) | 900 | 3 | 3 | 3 | 3 | 0 | NA |
| 4374 + 1.3 kb (Intron 23) | 1400 | 1 | 1 | 1 | 1 | 0 | NA |
| +6.8 kb/+7.0 kb | 399 | 0 | NA | 0 | NA | 0 | NA |
| +15.6 kb | 1500 | 3 | 3 | 3 | 3 | 0 | NA |
| +21.5 kb | 1000 | 2 | 2 | 2 | 2 | 0 | NA |
| +36.6 kb | 1200 | 1 | 0 | 1 | 0 | 0 | NA |
| +48.9 kb | 250 | 0 | NA | 0 | NA | 0 | NA |
| +83.7 kb | 459 | 3 | 2 | 3 | 2 | 0 | NA |
| Regulatory regions totals | 19,898 | 51 | 37 | 40 | 32 | 11 | 5 |
NA, not applicable.
Table 4.
Predicted TFBS Losses and Gains in CFTR Promoter and Enhancers
| Regulatory region | Variant | dbSNP138 | Predicted TFBS loss | Predicted TFBS gain | MAF (1000 Genomes Project)40 | MAF (this study) |
|---|---|---|---|---|---|---|
| −44 kb DHS39, 41 | c.-43626 G>A | rs185018312 | V$HMX2.02 | V$HOXC12.01 | 0.0004 | 0.0125 |
| V$MEIS1A_HOXA9.01 | V$CPHX.01 | |||||
| V$VMYB.05 | V$OSNT.01 | |||||
| V$TST.01 | V$SMARCA3.01 | |||||
| −35 kb DHS39, 42 | c.-35564T>G | rs6972168 | None | None | 0.4295 | 0.4063 |
| c.-35147T>G | rs6972819 | V$HHEX.01 | None | 0.1458 | 0.0063 | |
| V$HBP1.01 | ||||||
| V$TEAD.01 | ||||||
| V$NF1.01 | ||||||
| V$NF1.02 | ||||||
| c.-34893G>C | -35 novel.1 | V$GZF1.01 | V$EOMES.02 | NA | 0.0063 | |
| V$ZBED1.01 | ||||||
| V$ZBED1.02 | ||||||
| V$PAX5.02 | ||||||
| 2 kb Promoter26 | c.-966T>G | rs4148682 | V$STAT.01 | V$PRDM4.01 | 0.2238 | 0.0875 |
| V$BRN2.01 | ||||||
| c.-887C>T | rs34465975 | V$RAR_RXR.01 | V$ZFP652.01 V$IR2_NGRE.01 | 0.0164 | 0.0063 | |
| V$TR4.02 | ||||||
| V$PPARG.02 | ||||||
| c.-869delT | rs4148683 | V$IRF4.01 | V$IRF3.01 | 0.0048 | 0.0063 | |
| c.-812T>G | rs181008242 | V$NKX25.05 | V$AHRARNT.03 V$TAXCREB.01 | 0.0012 | 0.0188 | |
| V$AP1.02 | ||||||
| V$FXRE.01 | ||||||
| V$FXRE.01 | ||||||
| V$DELTAEF1.01 | ||||||
| V$VDR_RXR.06 | ||||||
| V$PSE.01 | ||||||
| c.-410G>C | promoter novel.1 | V$ZFX.01 | None | NA | 0.0063 | |
| V$NRSF.02 | ||||||
| c.-8G>C | rs1800501 | None | V$ZNF300.01 | 0.0274 | 0.0250 | |
| Intron 1 DHS28, 43∗ | c.53+9941A>C | rs35714998 | V$PLZF.01 | V$SOX9.06 | 0.0160 | 0.0063 |
| c.53+10442G>C | rs1557630 | V$HNF1.03 | V$ZEC.01 | 0.3073 | 0.4063 | |
| V$PLZF.01 | ||||||
| V$PAX8.01 | ||||||
| Intron 11 DHS27, 44∗ | c.1679+566G>T | 11 novel.1 | V$GATA3.02 V$BRACH.01 | V$PBX1_MEIS1.03 | NA | 0.0125 |
| V$SRY.04 | ||||||
| V$E4BP4.01 | ||||||
| c.1679+1280G>A | rs213963 | V$PRE.01 | V$FAST1.01 | 0.4265 | 0.5938 | |
| c.1679+1449A>G | rs213964 | V$POU3F3.01 | None | 0.4263 | 0.5938 | |
| c.1679+1539T>C | 11 novel.2 | V$CDX2.01 | V$HMGIY.01 | NA | 0.0125 | |
| Intron 23 DHS39∗ | c.4242+198T>C | rs1429568 | None | V$SOX9.09 | 0.1879 | 0.1750 |
DHS, DNaseI hypersensitive site; MAF, minor allele frequency; NA, not applicable; TFBS, transcription factor binding site.
Precise genomic locations defined in Table 3.
CFTR Promoter Variants Repress Promoter Activity
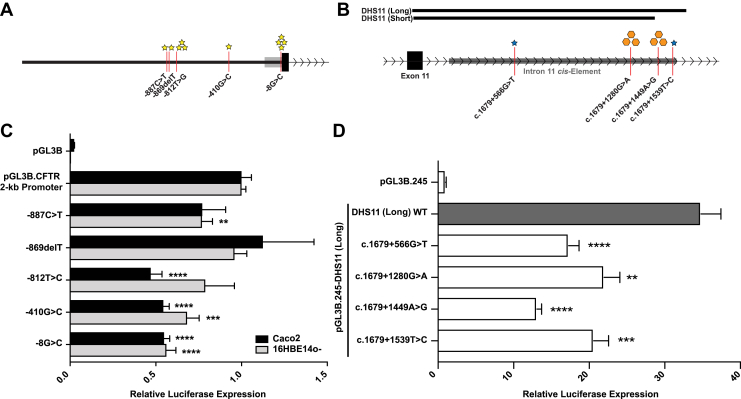
We previously used transient luciferase reporter gene assays to determine the extent of the CFTR promoter sequence required to drive the most robust gene expression.26 An approximately 2-kb fragment (1963 bp) was defined previously that has strong promoter activity in airway and intestinal cell lines.26 Herein, the effect of five promoter variants identified in our SureSelect screen was investigated on the activity of the 2-kb CFTR promoter. All five variants (four substitutions and one indel) in the 2-kb CFTR promoter (Figure 2A) are predicted to alter TF binding sites, and all, except c.-410G>C, were previously annotated in dbSNP138 (Table 4). Site-directed mutagenesis was used to independently introduce variants into the pGL3B.2kb CFTR promoter-luciferase construct. CFTR promoter constructs were transfected into airway (16HBE14o-) and intestinal (Caco2) cell lines that express high levels of endogenous CFTR transcript,27, 39 and luciferase expression was compared with the wild-type promoter (Figure 2C). An additional variant detected (c.-966T>G) was not examined independently, because the 2-kb wild-type promoter fragment contained the minor allele (c.-966G). This variant has a high minor allele frequency (0.2238) in the general population (Table 4). Among the five variants tested, four significantly repressed CFTR promoter activity, with c.-812T>G being the strongest repressor of promoter activity (53% in Caco2) and c.-869delT not significantly affecting promoter activity in either cell type (Figure 2C). Consistent with the CFTR promoter lacking tissue-specific control elements,46 the effect of the promoter variants examined herein is cell type independent, with little differences observed between 16HBE14o- and Caco2 cells for most variants.
Figure 2.
Promoter variants reduce activity of 2-kb CFTR promoter in a cell type–independent manner. A and B: Schematic of five variants identified within the 2-kb CFTR promoter (A) and four substitutions identified within the CFTR intron 11 cis-element (B). Stars represent individual patients with observed variant (blue, homozygous variant; yellow, heterozygous variant), and orange hexagons represent 10 patients heterozygous for the observed variant. C: Luciferase expression vectors containing the 2-kb CFTR promoter [wild type (WT) or with variants] were transiently cotransfected into 16HBE14o- or Caco2 cells. Data are shown relative to the CFTR 2-kb promoter parental vector. pGL3B, lacking a promoter, is shown for control. D: Luciferase expression vectors containing the 787-bp minimal CFTR promoter and the DHS11 long (WT or with variants) cis-element were transiently cotransfected into Caco2 cells. Data are shown relative to the CFTR minimal promoter parental vector (pGL3B.245). C and D: Luciferase expression levels were compared against pGL3B.2 kb (C) and pGL3B.245-DHS11(long) (D) using unpaired t-tests. Data are expressed as means ± SEM (C and D). n = 12 (C); n = 9 (D). ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 versus pGL3B.2 kb (C; unpaired t-test) and versus pGL3B.245-DHS11(long) (D; unpaired t-test). DHS, DNaseI hypersensitive site.
Variants in the CFTR Intron 11 Intestinal Enhancer Repress Enhancer Activity
To determine whether sequence variants identified in cis-regulatory elements affect their function, four variants found in a robust intestine-selective enhancer element within intron 11 of CFTR were first evaluated.9, 27, 44, 45 These four substitutions within the 1400 bp encompassing the element were predicted to alter TF binding sites (Table 4). Two of the four single-nucleotide polymorphisms (SNPs) were novel (not annotated in dbSNP138), whereas the annotated SNPs both had high minor allele frequencies. The intron 11 enhancer/CFTR promoter luciferase construct (DHS11 short), used previously,27 lacked the terminal 200 bp of DHS11, which included the location of two of the four variants (Figure 2B). Hence, a new construct (pGL3B.245-DHS11 long) was designed to assay their function. Site-directed mutagenesis was used to independently introduce variants into this construct, and plasmids were transfected into Caco2 cells. The four variants tested all reduce intron 11 enhancer activity in Caco2 cells by 37% to 63% (Figure 2D).
Discussion
Among disease-causing variants currently annotated in the CFTR gene, only those in the promoter or those that disrupt or generate splice sites occur in noncoding regions. However, because 1% to 5% of CF patients have unknown molecular lesions, additional noncoding variants within the CFTR locus likely contribute to the pathogenicity of CF. Advances in next-generation sequencing technologies have enabled whole CFTR locus sequencing to search for novel and/or noncoding variants.47, 48 Herein, we describe a targeted-enrichment method to deep sequence 460 kb of the CFTR locus in 80 CF patients with at least one unknown CF allele (Figure 1). This method was robust in identifying known CF alleles, but the disease-associated variant on 85 alleles in this cohort remains unknown (Table 2). We predicted that, among the 85 alleles, there are uncharacterized variants in CFTR regulatory elements, including the promoter, enhancers, or other structural regulatory elements, that could reduce or abolish CFTR transcription (potentially class I or V CFTR pathogenic variants, resulting in no or low transcript synthesis49). Some variants may also cause disease by generating cryptic splice sites or altering splicing efficiency, which, although potentially detectable by in silico prediction programs, would need to be validated using relevant RNA samples and so are not considered herein.
Overall, the SureDesign targeted enrichment and deep-sequencing method identified nearly all previously known CF variants present in our cohort, except for the T(n) polymorphic tract 5T [c.1210-1212(5)] variant. Previous genotyping efforts indicated that at least two individuals contained a 5T allele (Supplemental Table S1); however, the inability of our bioinformatic analysis to identify this variant is likely because of low confidence of mapping and alignment of the T(n) tract. Manual inspection of sequence reads that mapped to the three probes that spanned the T(n) tract revealed the 5T genotype in only the two expected patients, and no others. 5T contributes to complex CF alleles and is pathogenic, for example, when found in cis with R117H.50 Although the phase of variants cannot be distinguished with this analysis, the only patient with an R117H allele was negative for the 5T allele (Supplemental Table S1). Therefore, the 5T alleles in isolation were not considered as CF-causing in this analysis.
The CFTR promoter is the most extensively studied region of the gene,46 with approximately 20 variants identified in the 2-kb CFTR promoter in patients with CF or CF-related disorders.51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61 Indeed, four of the six variants identified in this study, c.-966T>G, c.-887C>T, c.-812T>G, and c.-8G>C (Table 4), were reported previously,51, 52, 56, 60, 61 although none are listed in the Clinical and Functional Translation of CFTR Database because their disease-causing status has not been assayed. Herein, it was shown that the c.-887T variant reduces CFTR promoter activity by 33% in intestinal and airway cell lines (Figure 2C). However, a previous study found that c.-887T had no effect on promoter activity in A549 (lung adenocarcinoma, CFTR mRNA+), Panc1 (pancreatic adenocarcinoma, CFTR mRNA+), or HepG2 (hepatocellular carcinoma, CFTR mRNA−) cells.60 This disparity could be a consequence of the promoter fragments assayed (2-kb versus extended 6-kb), or might suggest that the functional consequence of the c.-887T allele is cell type dependent. The 16HBE14o- and Caco2 cells used in our experiments likely express substantially more CFTR transcript than A549 and Panc1 cells, implicating higher levels of activating TFs. The c.-887T allele is predicted to generate binding motifs for zinc finger protein 652 (ZNF652) and an inverted repeat 2 (IR2) negative glucocorticoid response element (nGRE). A C2H2-type zinc finger protein, ZNF652, functions as a transcriptional repressor,62, 63 like glucocorticoid receptor when bound to negative glucocorticoid response elements.64 Aberrant recruitment of either of these factors to the CFTR promoter may have negative consequences on expression.
This study also shows that the c.-812G variant decreased CFTR promoter activity 53% in Caco2 cells, although it did not significantly decrease CFTR promoter activity in 16HBE14o- cells (Figure 2C). Two recent studies also investigated the functional consequence of this variant. In the first, c.-812G increased the extended 6-kb promoter activity by approximately 1.5-fold in HepG2 cells, which do not express endogenous CFTR transcript.60 In the second, this variant decreased promoter activity in another CFTR mRNA+ bronchial epithelial cell line, Beas2B, possibly because of the generation of a potential binding motif for an E2F TF family member.61 E2F TFs serve as activators and repressors during development, differentiation, and the cell cycle.65 Although MatInspector analysis did not predict the generation of an E2F motif in the c.-812G sequence, it predicts loss of a δEF1 motif (Table 4), now known as zinc finger E-box binding homeobox 1 (ZEB1). ZEB1 is an important regulator of epithelial-to-mesenchymal transition in development and cancer, and like E2F family members, ZEB1 functions as both a transcriptional activator and a repressor.66 Further work is required to determine how the c.-812G>T transversion can both activate and repress CFTR promoter activity.
The novel promoter variant observed in one patient, c.-410G>C, also reduces the activity of the CFTR promoter (Figure 2C). This substitution abolishes a putative X-linked zinc finger protein (ZFX) motif (Table 4), a transcriptional activator that plays an important role in maintaining stem cell pluripotency.67 Conversely, the c.-8G>C variant is predicted to gain a zinc finger protein 300 (ZNF300) consensus site (Table 4). ZNF300, a broadly expressed C2H2/Krüppel-associated box (KRAB) zinc finger factor, helps mediate the NF-κB immune response and functions as a transcriptional repressor.68, 69
Disruption of TF recruitment to CFTR structural and cis-regulatory elements can have a dramatic effect on CFTR expression and chromatin organization of the locus8, 9, 11, 42, 44, 45; however, until recently, it has been challenging to identify and investigate CF disease–associated variants in these regions. These data indicate that multiple TF binding sites are predicted to be lost or gained by the variants in CFTR enhancers identified herein (Table 4). These enhancers are experimentally validated27, 28, 39, 41, 42, 43, 44 and are known to regulate CFTR expression. Moreover, the altered TF binding sites recruit factors that are relevant to lung and intestinal biology. For example, a nuclear factor I (NFI) motif is destroyed by the c.-35147G variant in the −35 kb airway-selective enhancer. The four nuclear factor I family members bind the same consensus motif and play critical roles during development through activation and repression of target genes.70, 71 Notably, Nfib-null mice die shortly after birth because of severe lung hypoplasia, which is a direct result of developmental defects in mesenchymal and epithelial lung cells in Nfib−/− embryos.72 Also, the c.53+10442G>C variant in the intron 1 intestine-selective enhancer is predicted to destroy a binding motif for hepatocyte nuclear factor 1 homeobox (HNF1) in intron 1, which was previously shown to be an important regulator of CFTR expression in human and mouse intestinal cells, in part through its interaction with the intron 1 enhancer element.43, 44, 73 Loss of binding sites for these factors could reduce CFTR expression to contribute to CF disease pathogenicity in patients carrying these alleles. The observations that variants in the intron 11 intestinal enhancer reduce luciferase reporter gene expression by 37% to 63% in Caco2 cells clearly demonstrate this point (Figure 2D), although in vitro assays are not always indicative of significance in vivo. The novel c.1679+566G>T substitution is predicted to destroy a putative site of occupancy by GATA binding protein 3 (GATA3). GATA3 acts as a pioneer factor to facilitate the opening of previously inaccessible chromatin74 and is known to precede forkhead box A (FOXA) binding at some loci in breast cancer cells.75 Notably, FOXA factors, which also function as pioneer TFs, are crucial for expression of CFTR in intestinal cells, in part through binding to the intron 11 enhancer.44, 45 Furthermore, loss of a predicted caudal-type homeobox 2 (CDX2) binding site may contribute to the reduced enhancer activity of the c.1679+1539T>C novel variant (Table 4 and Figure 2D). More important, in intestinal cells, CDX2 binds at multiple sites within the CFTR locus, including in intron 11, and siRNA-mediated depletion of CDX2 reduces CFTR mRNA abundance.44
One aspect of this study that warrants further discussion is whether novel rSNPs occur in cis or in trans with each other, and with known pathogenic variants. Because our study cohort includes gDNA only from index cases and not their parents, phase cannot be readily established. Moreover, unequivocal phasing may require long-read sequencing of CFTR alleles to establish reference haplotypes and confirm imputations. However, this limitation does not detract from the utility of this study in defining rSNPs, because they may have an important impact irrespective of the haplotype. For example, where a functional rSNP impairs a CFTR tissue-specific enhancer, this on its own could reduce transcript abundance below a threshold required for normal CFTR channel activity. If that same rSNP was in cis with a known coding region variant that causes partial loss of function of CFTR, it might lead to a more severe phenotype. Although there is controversy about how much functional CFTR is required to prevent disease, it is possible that 5% of mean wild-type levels of CFTR transcript results in less-severe CF phenotypes, whereas 10% may protect against CF disease.76 It may also be important to know the impact of rSNPs in cis with another pathogenic variant when designing novel personalized therapeutics.
Identification of noncoding CFTR variants in the CF population will rapidly increase with the application of next-generation sequencing screening protocols. The challenge will lie in determining the functional consequence of these variants, especially those that fall within tissue-specific regulatory elements. Few molecular diagnostic laboratories are currently equipped to perform these tests. However, the robust cellular assays for coding region pathogenic variants and splice-site errors developed through the Clinical and Functional Translation of CFTR Database,77, 78 together with cell-specific enhancer assays in CF-relevant epithelial cell lines that express endogenous CFTR mRNA, as described herein, provide a toolbox and reagents that can be used for testing many novel variants. For future studies, it will be important to use patient-derived or CRISPR-generated variant induced pluripotent stem cells (iPSCs) to test the effect of these polymorphisms on endogenous CFTR expression in the appropriate differentiated cell types.
Acknowledgments
We thank Dr. Pieter Faber and staff (University of Chicago Genomics Core) for sequencing, Dr. Ricky Chan (Case Western Reserve University Institute for Computational Biology) for bioinformatics variant analysis, the SureDesign team (Agilent Technologies) for technical advice, and the late Dr. Julian Zielenski for efforts toward the initial CFTR analysis of the Canadian samples.
J.L.K., S.G., and A.H. designed the experiments; J.L.K. and A.H. wrote the manuscript; J.L.K., A.P., S.G., and W.R.C. performed experiments; J.L.K. and S.G. analyzed the data; C.F., M.N., H.I., and J.R. shared genomic DNA samples.
Footnotes
Supported by Cystic Fibrosis Foundation grants Harris 14P0 and Harris 16G0, NIH grant R01HL094585 (principal investigator: A.H.), Genome Canada through the Ontario Genomics Institute grant 2004-OGI-3-05, and the Canadian Cystic Fibrosis Foundation (also known as Cystic Fibrosis Canada).
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.jmoldx.2018.08.011.
Supplemental Data
References
- 1.Scacheri C.A., Scacheri P.C. Mutations in the noncoding genome. Curr Opin Pediatr. 2015;27:659–664. doi: 10.1097/MOP.0000000000000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Programme WHG . 2004. The Molecular Genetic Epidemiology of Cystic Fibrosis: Report of a Joint Meeting of WHO/IECFTN/ICF(M)A/ECFS, Genoa, Italy, 19 June 2002. [Google Scholar]
- 3.Massie J., Clements B., Australian Paediatric Respiratory Group Diagnosis of cystic fibrosis after newborn screening: the Australasian experience--twenty years and five million babies later: a consensus statement from the Australasian Paediatric Respiratory Group. Pediatr Pulmonol. 2005;39:440–446. doi: 10.1002/ppul.20191. [DOI] [PubMed] [Google Scholar]
- 4.Southern K.W., Munck A., Pollitt R., Travert G., Zanolla L., Dankert-Roelse J., Castellani C., ECFS CF Neonatal Screening Working Group A survey of newborn screening for cystic fibrosis in Europe. J Cyst Fibros. 2007;6:57–65. doi: 10.1016/j.jcf.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Ross L.F. Newborn screening for cystic fibrosis: a lesson in public health disparities. J Pediatr. 2008;153:308–313. doi: 10.1016/j.jpeds.2008.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Massie R.J., Curnow L., Glazner J., Armstrong D.S., Francis I. Lessons learned from 20 years of newborn screening for cystic fibrosis. Med J Aust. 2012;196:67–70. doi: 10.5694/mja11.10686. [DOI] [PubMed] [Google Scholar]
- 7.Castellani C., Cuppens H., Macek M., Jr., Cassiman J.J., Kerem E., Durie P., Tullis E., Assael B.M., Bombieri C., Brown A., Casals T., Claustres M., Cutting G.R., Dequeker E., Dodge J., Doull I., Farrell P., Ferec C., Girodon E., Johannesson M., Kerem B., Knowles M., Munck A., Pignatti P.F., Radojkovic D., Rizzotti P., Schwarz M., Stuhrmann M., Tzetis M., Zielenski J., Elborn J.S. Consensus on the use and interpretation of cystic fibrosis mutation analysis in clinical practice. J Cyst Fibros. 2008;7:179–196. doi: 10.1016/j.jcf.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosalia N., Harris A. Chromatin dynamics in the regulation of CFTR expression. Genes (Basel) 2015;6:543–558. doi: 10.3390/genes6030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang R., Kerschner J.L., Gosalia N., Neems D., Gorsic L.K., Safi A., Crawford G.E., Kosak S.T., Leir S.H., Harris A. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res. 2016;44:3082–3094. doi: 10.1093/nar/gkv1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith E.M., Lajoie B.R., Jain G., Dekker J. Invariant TAD boundaries constrain cell-type-specific looping interactions between promoters and distal elements around the CFTR locus. Am J Hum Genet. 2016;98:185–201. doi: 10.1016/j.ajhg.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gosalia N., Neems D., Kerschner J.L., Kosak S.T., Harris A. Architectural proteins CTCF and cohesin have distinct roles in modulating the higher order structure and expression of the CFTR locus. Nucleic Acids Res. 2014;42:9612–9622. doi: 10.1093/nar/gku648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moisan S., Berlivet S., Ka C., Le Gac G., Dostie J., Ferec C. Analysis of long-range interactions in primary human cells identifies cooperative CFTR regulatory elements. Nucleic Acids Res. 2016;44:2564–2576. doi: 10.1093/nar/gkv1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zielenski J., Aznarez I., Onay T., Tzounzouris J., Markiewicz D., Tsui L.C. CFTR mutation detection by multiplex heteroduplex (mHET) analysis on MDE gel. Methods Mol Med. 2002;70:3–19. doi: 10.1385/1-59259-187-6:03. [DOI] [PubMed] [Google Scholar]
- 14.Audrezet M.P., Chen J.M., Raguenes O., Chuzhanova N., Giteau K., Le Marechal C., Quere I., Cooper D.N., Ferec C. Genomic rearrangements in the CFTR gene: extensive allelic heterogeneity and diverse mutational mechanisms. Hum Mutat. 2004;23:343–357. doi: 10.1002/humu.20009. [DOI] [PubMed] [Google Scholar]
- 15.Ferec C., Casals T., Chuzhanova N., Macek M., Jr., Bienvenu T., Holubova A., King C., McDevitt T., Castellani C., Farrell P.M., Sheridan M., Pantaleo S.J., Loumi O., Messaoud T., Cuppens H., Torricelli F., Cutting G.R., Williamson R., Ramos M.J., Pignatti P.F., Raguenes O., Cooper D.N., Audrezet M.P., Chen J.M. Gross genomic rearrangements involving deletions in the CFTR gene: characterization of six new events from a large cohort of hitherto unidentified cystic fibrosis chromosomes and meta-analysis of the underlying mechanisms. Eur J Hum Genet. 2006;14:567–576. doi: 10.1038/sj.ejhg.5201590. [DOI] [PubMed] [Google Scholar]
- 16.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., McKenna A., Fennell T.J., Kernytsky A.M., Sivachenko A.Y., Cibulskis K., Gabriel S.B., Altshuler D., Daly M.J. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., Banks E., Garimella K.V., Altshuler D., Gabriel S., DePristo M.A. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., McVean G., Durbin R., Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsui L.C., Dorfman R. The cystic fibrosis gene: a molecular genetic perspective. Cold Spring Harb Perspect Med. 2013;3:a009472. doi: 10.1101/cshperspect.a009472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.den Dunnen J.T., Dalgleish R., Maglott D.R., Hart R.K., Greenblatt M.S., McGowan-Jordan J., Roux A.F., Smith T., Antonarakis S.E., Taschner P.E. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37:564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 26.Lewandowska M.A., Costa F.F., Bischof J.M., Williams S.H., Soares M.B., Harris A. Multiple mechanisms influence regulation of the cystic fibrosis transmembrane conductance regulator gene promoter. Am J Respir Cell Mol Biol. 2010;43:334–341. doi: 10.1165/rcmb.2009-0149OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ott C.J., Blackledge N.P., Kerschner J.L., Leir S.H., Crawford G.E., Cotton C.U., Harris A. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc Natl Acad Sci U S A. 2009;106:19934–19939. doi: 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith A.N., Barth M.L., McDowell T.L., Moulin D.S., Nuthall H.N., Hollingsworth M.A., Harris A. A regulatory element in intron 1 of the cystic fibrosis transmembrane conductance regulator gene. J Biol Chem. 1996;271:9947–9954. doi: 10.1074/jbc.271.17.9947. [DOI] [PubMed] [Google Scholar]
- 29.Phylactides M., Rowntree R., Nuthall H., Ussery D., Wheeler A., Harris A. Evaluation of potential regulatory elements identified as DNase I hypersensitive sites in the CFTR gene. Eur J Biochem. 2002;269:553–559. doi: 10.1046/j.0014-2956.2001.02679.x. [DOI] [PubMed] [Google Scholar]
- 30.Fogh J., Wright W.C., Loveless J.D. Absence of HeLa cell contamination in 169 cell lines derived from human tumors. J Natl Cancer Inst. 1977;58:209–214. doi: 10.1093/jnci/58.2.209. [DOI] [PubMed] [Google Scholar]
- 31.Cozens A.L., Yezzi M.J., Kunzelmann K., Ohrui T., Chin L., Eng K., Finkbeiner W.E., Widdicombe J.H., Gruenert D.C. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am J Respir Cell Mol Biol. 1994;10:38–47. doi: 10.1165/ajrcmb.10.1.7507342. [DOI] [PubMed] [Google Scholar]
- 32.Bonini J., Varilh J., Raynal C., Theze C., Beyne E., Audrezet M.P., Ferec C., Bienvenu T., Girodon E., Tuffery-Giraud S., Des Georges M., Claustres M., Taulan-Cadars M. Small-scale high-throughput sequencing-based identification of new therapeutic tools in cystic fibrosis. Genet Med. 2015;17:796–806. doi: 10.1038/gim.2014.194. [DOI] [PubMed] [Google Scholar]
- 33.Nakakuki M., Fujiki K., Yamamoto A., Ko S.B., Yi L., Ishiguro M., Yamaguchi M., Kondo S., Maruyama S., Yanagimoto K., Naruse S., Ishiguro H. Detection of a large heterozygous deletion and a splicing defect in the CFTR transcripts from nasal swab of a Japanese case of cystic fibrosis. J Hum Genet. 2012;57:427–433. doi: 10.1038/jhg.2012.46. [DOI] [PubMed] [Google Scholar]
- 34.Dork T., Macek M., Jr., Mekus F., Tummler B., Tzountzouris J., Casals T., Krebsova A., Koudova M., Sakmaryova I., Macek M., Sr., Vavrova V., Zemkova D., Ginter E., Petrova N.V., Ivaschenko T., Baranov V., Witt M., Pogorzelski A., Bal J., Zekanowsky C., Wagner K., Stuhrmann M., Bauer I., Seydewitz H.H., Neumann T., Jakubiczka S. Characterization of a novel 21-kb deletion, CFTRdele2,3(21 kb), in the CFTR gene: a cystic fibrosis mutation of Slavic origin common in Central and East Europe. Hum Genet. 2000;106:259–268. doi: 10.1007/s004390000246. [DOI] [PubMed] [Google Scholar]
- 35.Morral N., Nunes V., Casals T., Cobos N., Asensio O., Dapena J., Estivill X. Uniparental inheritance of microsatellite alleles of the cystic fibrosis gene (CFTR): identification of a 50 kilobase deletion. Hum Mol Genet. 1993;2:677–681. doi: 10.1093/hmg/2.6.677. [DOI] [PubMed] [Google Scholar]
- 36.Quemener S., Chen J.M., Chuzhanova N., Benech C., Casals T., Macek M., Jr., Bienvenu T., McDevitt T., Farrell P.M., Loumi O., Messaoud T., Cuppens H., Cutting G.R., Stenson P.D., Giteau K., Audrezet M.P., Cooper D.N., Ferec C. Complete ascertainment of intragenic copy number mutations (CNMs) in the CFTR gene and its implications for CNM formation at other autosomal loci. Hum Mutat. 2010;31:421–428. doi: 10.1002/humu.21196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niel F., Martin J., Dastot-Le Moal F., Costes B., Boissier B., Delattre V., Goossens M., Girodon E. Rapid detection of CFTR gene rearrangements impacts on genetic counselling in cystic fibrosis. J Med Genet. 2004;41:e118. doi: 10.1136/jmg.2004.022400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Girardet A., Guittard C., Altieri J.P., Templin C., Stremler N., Beroud C., des Georges M., Claustres M. Negative genetic neonatal screening for cystic fibrosis caused by compound heterozygosity for two large CFTR rearrangements. Clin Genet. 2007;72:374–377. doi: 10.1111/j.1399-0004.2007.00850.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z., Ott C.J., Lewandowska M.A., Leir S.H., Harris A. Molecular mechanisms controlling CFTR gene expression in the airway. J Cell Mol Med. 2012;16:1321–1330. doi: 10.1111/j.1582-4934.2011.01439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z., Leir S.H., Harris A. Oxidative stress regulates CFTR gene expression in human airway epithelial cells through a distal antioxidant response element. Am J Respir Cell Mol Biol. 2015;52:387–396. doi: 10.1165/rcmb.2014-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z., Leir S.H., Harris A. Immune mediators regulate CFTR expression through a bifunctional airway-selective enhancer. Mol Cell Biol. 2013;33:2843–2853. doi: 10.1128/MCB.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ott C.J., Suszko M., Blackledge N.P., Wright J.E., Crawford G.E., Harris A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J Cell Mol Med. 2009;13:680–692. doi: 10.1111/j.1582-4934.2008.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerschner J.L., Harris A. Transcriptional networks driving enhancer function in the CFTR gene. Biochem J. 2012;446:203–212. doi: 10.1042/BJ20120693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kerschner J.L., Gosalia N., Leir S.H., Harris A. Chromatin remodeling mediated by the FOXA1/A2 transcription factors activates CFTR expression in intestinal epithelial cells. Epigenetics. 2014;9:557–565. doi: 10.4161/epi.27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCarthy V.A., Harris A. The CFTR gene and regulation of its expression. Pediatr Pulmonol. 2005;40:1–8. doi: 10.1002/ppul.20199. [DOI] [PubMed] [Google Scholar]
- 47.Vecchio-Pagan B., Blackman S.M., Lee M., Atalar M., Pellicore M.J., Pace R.G., Franca A.L., Raraigh K.S., Sharma N., Knowles M.R., Cutting G.R. Deep resequencing of CFTR in 762 F508del homozygotes reveals clusters of non-coding variants associated with cystic fibrosis disease traits. Hum Genome Var. 2016;3:16038. doi: 10.1038/hgv.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Straniero L., Solda G., Costantino L., Seia M., Melotti P., Colombo C., Asselta R., Duga S. Whole-gene CFTR sequencing combined with digital RT-PCR improves genetic diagnosis of cystic fibrosis. J Hum Genet. 2016;61:977–984. doi: 10.1038/jhg.2016.101. [DOI] [PubMed] [Google Scholar]
- 49.Rowe S.M., Miller S., Sorscher E.J. Cystic fibrosis. N Engl J Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- 50.Kiesewetter S., Macek M., Jr., Davis C., Curristin S.M., Chu C.S., Graham C., Shrimpton A.E., Cashman S.M., Tsui L.C., Mickle J., Amos J., Highsmith W.E., Shuber A., Witt D.R., Crystal R.G., Cutting G.R. A mutation in CFTR produces different phenotypes depending on chromosomal background. Nat Genet. 1993;5:274–278. doi: 10.1038/ng1193-274. [DOI] [PubMed] [Google Scholar]
- 51.Bienvenu T., Lacronique V., Raymondjean M., Cazeneuve C., Hubert D., Kaplan J.C., Beldjord C. Three novel sequence variations in the 5′ upstream region of the cystic fibrosis transmembrane conductance regulator (CFTR) gene: two polymorphisms and one putative molecular defect. Hum Genet. 1995;95:698–702. doi: 10.1007/BF00209490. [DOI] [PubMed] [Google Scholar]
- 52.Verlingue C., Vuillaumier S., Mercier B., Le Gac M., Elion J., Ferec C., Denamur E. Absence of mutations in the interspecies conserved regions of the CFTR promoter region in cystic fibrosis (CF) and CF related patients. J Med Genet. 1998;35:137–140. doi: 10.1136/jmg.35.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romey M.C., Guittard C., Carles S., Demaille J., Claustres M., Ramsay M. First putative sequence alterations in the minimal CFTR promoter region. J Med Genet. 1999;36:263–264. [PMC free article] [PubMed] [Google Scholar]
- 54.Romey M.C., Guittard C., Chazalette J.P., Frossard P., Dawson K.P., Patton M.A., Casals T., Bazarbachi T., Girodon E., Rault G., Bozon D., Seguret F., Demaille J., Claustres M. Complex allele [-102T>A+S549R(T>G)] is associated with milder forms of cystic fibrosis than allele S549R(T>G) alone. Hum Genet. 1999;105:145–150. doi: 10.1007/s004399900066. [DOI] [PubMed] [Google Scholar]
- 55.Romey M.C., Pallares-Ruiz N., Mange A., Mettling C., Peytavi R., Demaille J., Claustres M. A naturally occurring sequence variation that creates a YY1 element is associated with increased cystic fibrosis transmembrane conductance regulator gene expression. J Biol Chem. 2000;275:3561–3567. doi: 10.1074/jbc.275.5.3561. [DOI] [PubMed] [Google Scholar]
- 56.Wu C.C., Alper O.M., Lu J.F., Wang S.P., Guo L., Chiang H.S., Wong L.J. Mutation spectrum of the CFTR gene in Taiwanese patients with congenital bilateral absence of the vas deferens. Hum Reprod. 2005;20:2470–2475. doi: 10.1093/humrep/dei077. [DOI] [PubMed] [Google Scholar]
- 57.Taulan M., Lopez E., Guittard C., Rene C., Baux D., Altierl J.P., DesGeorges M., ClaustreS A., Romey M.C. First functional polymorphism in CFTR promoter that results in decreased transcriptional activity and Sp1/USF binding. Biochem Biophys Res Commun. 2007;361:775–781. doi: 10.1016/j.bbrc.2007.07.091. [DOI] [PubMed] [Google Scholar]
- 58.Lopez E., Viart V., Guittard C., Templin C., Rene C., Mechin D., Des Georges M., Claustres M., Romey-Chatelain M.C., Taulan M. Variants in CFTR untranslated regions are associated with congenital bilateral absence of the vas deferens. J Med Genet. 2011;48:152–159. doi: 10.1136/jmg.2010.081851. [DOI] [PubMed] [Google Scholar]
- 59.Viart V., Des Georges M., Claustres M., Taulan M. Functional analysis of a promoter variant identified in the CFTR gene in cis of a frameshift mutation. Eur J Hum Genet. 2012;20:180–184. doi: 10.1038/ejhg.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giordano S., Amato F., Elce A., Monti M., Iannone C., Pucci P., Seia M., Angioni A., Zarrilli F., Castaldo G., Tomaiuolo R. Molecular and functional analysis of the large 5′ promoter region of CFTR gene revealed pathogenic mutations in CF and CFTR-related disorders. J Mol Diagn. 2013;15:331–340. doi: 10.1016/j.jmoldx.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 61.Bergougnoux A., Viart V., Miro J., Bommart S., Molinari N., des Georges M., Claustres M., Chiron R., Taulan-Cadars M. Should diffuse bronchiectasis still be considered a CFTR-related disorder? J Cyst Fibros. 2015;14:646–653. doi: 10.1016/j.jcf.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 62.Kumar R., Cheney K.M., McKirdy R., Neilsen P.M., Schulz R.B., Lee J., Cohen J., Booker G.W., Callen D.F. CBFA2T3-ZNF652 corepressor complex regulates transcription of the E-box gene HEB. J Biol Chem. 2008;283:19026–19038. doi: 10.1074/jbc.M709136200. [DOI] [PubMed] [Google Scholar]
- 63.Kumar R., Manning J., Spendlove H.E., Kremmidiotis G., McKirdy R., Lee J., Millband D.N., Cheney K.M., Stampfer M.R., Dwivedi P.P., Morris H.A., Callen D.F. ZNF652, a novel zinc finger protein, interacts with the putative breast tumor suppressor CBFA2T3 to repress transcription. Mol Cancer Res. 2006;4:655–665. doi: 10.1158/1541-7786.MCR-05-0249. [DOI] [PubMed] [Google Scholar]
- 64.Surjit M., Ganti K.P., Mukherji A., Ye T., Hua G., Metzger D., Li M., Chambon P. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–241. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 65.Dimova D.K., Dyson N.J. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 66.Zhang P., Sun Y., Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14:481–487. doi: 10.1080/15384101.2015.1006048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Galan-Caridad J.M., Harel S., Arenzana T.L., Hou Z.E., Doetsch F.K., Mirny L.A., Reizis B. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129:345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gou D., Wang J., Gao L., Sun Y., Peng X., Huang J., Li W. Identification and functional analysis of a novel human KRAB/C2H2 zinc finger gene ZNF300. Biochim Biophys Acta. 2004;1676:203–209. doi: 10.1016/j.bbaexp.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 69.Wang T., Wang X.G., Xu J.H., Wu X.P., Qiu H.L., Yi H., Li W.X. Overexpression of the human ZNF300 gene enhances growth and metastasis of cancer cells through activating NF-kB pathway. J Cell Mol Med. 2012;16:1134–1145. doi: 10.1111/j.1582-4934.2011.01388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gronostajski R.M. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 71.Harris L., Genovesi L.A., Gronostajski R.M., Wainwright B.J., Piper M. Nuclear factor one transcription factors: divergent functions in developmental versus adult stem cell populations. Dev Dyn. 2015;244:227–238. doi: 10.1002/dvdy.24182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hsu Y.C., Osinski J., Campbell C.E., Litwack E.D., Wang D., Liu S., Bachurski C.J., Gronostajski R.M. Mesenchymal nuclear factor I B regulates cell proliferation and epithelial differentiation during lung maturation. Dev Biol. 2011;354:242–252. doi: 10.1016/j.ydbio.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mouchel N., Henstra S.A., McCarthy V.A., Williams S.H., Phylactides M., Harris A. HNF1alpha is involved in tissue-specific regulation of CFTR gene expression. Biochem J. 2004;378:909–918. doi: 10.1042/BJ20031157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takaku M., Grimm S.A., Shimbo T., Perera L., Menafra R., Stunnenberg H.G., Archer T.K., Machida S., Kurumizaka H., Wade P.A. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. 2016;17:36. doi: 10.1186/s13059-016-0897-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Theodorou V., Stark R., Menon S., Carroll J.S. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res. 2013;23:12–22. doi: 10.1101/gr.139469.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amaral M.D. Processing of CFTR: traversing the cellular maze--how much CFTR needs to go through to avoid cystic fibrosis? Pediatr Pulmonol. 2005;39:479–491. doi: 10.1002/ppul.20168. [DOI] [PubMed] [Google Scholar]
- 77.Sosnay P.R., Siklosi K.R., Van Goor F., Kaniecki K., Yu H., Sharma N., Ramalho A.S., Amaral M.D., Dorfman R., Zielenski J., Masica D.L., Karchin R., Millen L., Thomas P.J., Patrinos G.P., Corey M., Lewis M.H., Rommens J.M., Castellani C., Penland C.M., Cutting G.R. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat Genet. 2013;45:1160–1167. doi: 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gottschalk L.B., Vecchio-Pagan B., Sharma N., Han S.T., Franca A., Wohler E.S., Batista D.A., Goff L.A., Cutting G.R. Creation and characterization of an airway epithelial cell line for stable expression of CFTR variants. J Cyst Fibros. 2016;15:285–294. doi: 10.1016/j.jcf.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.