Abstract
Paralytic shellfish toxins (PSTs) are a group of potent neurotoxic alkaloids that are produced mainly by marine dinoflagellates. PST biosynthesis in dinoflagellates is a discontinuous process that is coupled to the cell cycle. However, little is known about the molecular mechanism underlying this association. Here, we compared global protein expression profiles of a toxigenic dinoflagellate, Alexandrium catenella, collected at four different stages of toxin biosynthesis during the cell cycle, using an isobaric tags for relative and absolute quantification (iTRAQ)-based quantitative proteomic approach. The results showed that toxin biosynthesis occurred mainly in the G1 phase, especially the late G1 phase. In total, 7232 proteins were confidently identified, and 210 proteins exhibited differential expression among the four stages. Proteins involved in protein translation and photosynthetic pigment biosynthesis were significantly upregulated during toxin biosynthesis, indicating close associations among the three processes. Nine toxin-related proteins were detected, and two core toxin biosynthesis proteins, namely, sxtA and sxtI, were identified for the first time in dinoflagellates. Among these proteins, sxtI and ompR were significantly downregulated when toxin biosynthesis stopped, indicating that they played important roles in the regulation of PST biosynthesis. Our study provides new insights into toxin biosynthesis in marine dinoflagellates: nitrogen balance among different biological processes regulates toxin biosynthesis, and that glutamate might play a key modulatory role.
Keywords: dinoflagellates, Alexandrium catenella, paralytic shellfish toxins, cell cycle, toxin biosynthesis, quantitative proteomics, iTRAQ
1. Introduction
Paralytic shellfish toxins (PSTs) are a group of neurotoxic alkaloids that selectively block voltage-gated sodium channels in excitable cells, resulting in approximately 2000 incidents of paralytic shellfish poisoning (PSP) annually worldwide [1]. In the past few decades, PSP has caused increasing concern due to the global increase in the frequency, intensity and geographic distribution of PST-producing algal blooms [1,2].
Recently, the PST biosynthetic pathway has been unveiled in several species of cyanobacteria, which are another important PST-producing group in addition to dinoflagellates [3,4,5,6]. A group of core genes (sxtA, sxtG, sxtB, sxtD, sxtS, sxtU, sxtH/T and sxtI) that are directly involved in toxin biosynthesis, tailor genes (sxtL, sxtN, sxtX) that participate in toxin transformation and some additional genes that are responsible for toxin transportation and regulation are characterized, and several toxin-related proteins are also identified in cyanobacteria [3,7]. In contrast, the toxin biosynthetic pathway in dinoflagellates remains unclear, although the pathway is believed to be similar to that in cyanobacteria [8]. To date, some putative homologs of cyanobacterial toxin genes and proteins have been identified in dinoflagellates [9,10]; however, only sxtA and sxtG, which participate in the first two biosynthetic steps, have been well characterized [11,12]. The sxtA gene of dinoflagellates has two isoforms: the long isoform contains all the sxtA1–A4 catalytic domains, while the short isoform contains only domains sxtA1–A3, not sxtA4, which is essential for PST biosynthesis [11,13,14,15].
Studies have shown that PST production in toxigenic dinoflagellates is a discontinuous process coupled to a restricted time frame of the cell cycle. PST biosynthesis in Alexandrium fundyense is regulated by light and mainly occurs in the G1 phase [16,17,18]. However, PST biosynthesis in Alexandrium tamarense occurs in the early stage of the S phase [19]. Similarly, toxin synthesis in Alexandrium catenella does not occur in the G1 phase but in the S phase or G2/M phase of the cell cycle [20,21]. Although the toxin-producing periods of different species and strains are not consistent, all these results indicate that the production process in dinoflagellates is regulated by the cell cycle and occurs during a specific period of the cell cycle. Further studies have revealed that three genes related to the biosynthesis of PST precursors are upregulated during toxin production in A. fundyense [21], and the expression of nine proteins that are potentially involved in toxin biosynthesis in A. catenella vary significantly at different stages of toxin biosynthesis [22]. However, the molecular mechanism underlying the association between the cell cycle and toxin synthesis remains unclear.
In our recent studies, several genes and cellular processes potentially involved in PST biosynthesis are identified, and toxin production is found to be regulated translationally or post-translationally [10,15,23,24]. In this study, we compared the global protein expression profiles of A. catenella at four different toxin biosynthesis stages in the cell cycle using an isobaric tags for relative and absolute quantification (iTRAQ)-based quantitative proteomic approach. Our goal was to mine toxin-related proteins and to identify molecular processes associated with PST biosynthesis at the protein level.
2. Results
2.1. Cell Cycle Distribution and Toxin Content Variation
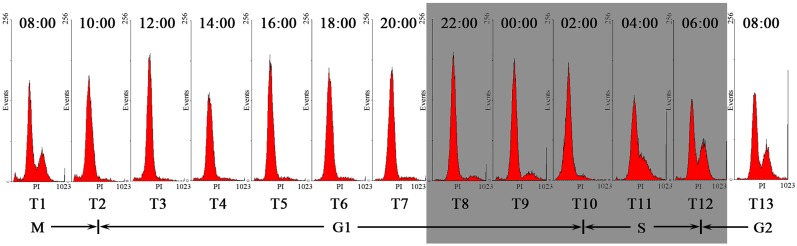
Variations in the DNA histogram indicated that the toxic A. catenella strain, ACHK-T, completed a cell cycle within one day (Figure 1). At 10:00, the cells entered the G1 phase, which lasted for 16 h, until 02:00 the next day. Then, the cells entered the S phase between 02:00 and 06:00, after which, the cells entered the G2/M phase and completed cell division by 10:00.
Figure 1.
Flow cytometric analysis of ACHK-T during the cell cycle. X-axis: Relative DNA content; Y-axis: Cell number. The gray box represents the dark period.
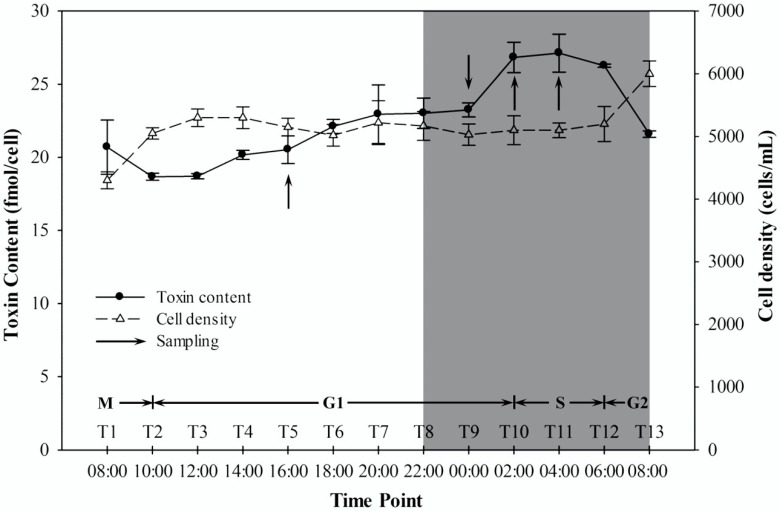
The cell density remained stable during the cell cycle except for a sharp increase in the G2/M phase (Figure 2). The toxin content increased slowly during the light period (the early and middle G1 phases) but increased sharply between 00:00 to 02:00, the last 2 h of the G1 phase. Subsequently, the toxin content remained at the highest level for approximately 4 h and decreased sharply when cell division started at 06:00 (Figure 2).
Figure 2.
Variations of cell density and intracellular toxin content during the cell cycle. Both values are reported as the means of biological triplicates with standard deviations. The gray box represents the dark period.
2.2. Proteome Overview and Protein Annotation
Overall, 71,413 of the total 345,182 output spectra were matched to 22,299 peptides with an approximately 20.7% spectrum utilization rate. Using the Mascot search engine (version 2.3.02; Matrix Science, London, United Kingdom), 7232 proteins were identified from 19,227 unique peptides that collectively matched 51,904 unique spectra. All of the proteins identified were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and National Center for Biotechnology Information nonredundant protein sequences (NCBInr) databases (Supplementary Table S1).
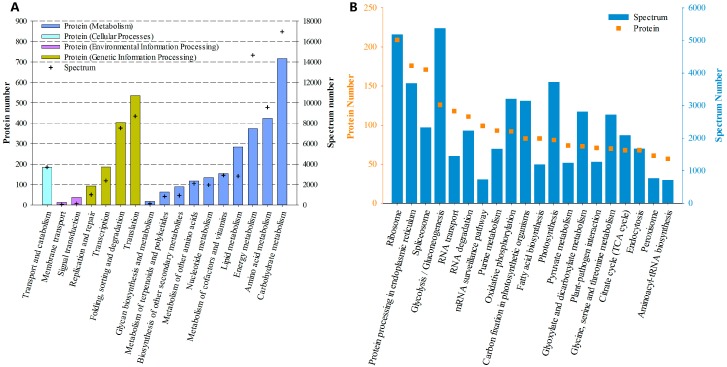
Based on the KEGG categories, proteins were annotated into 17 groups. “Carbohydrate metabolism”, “Amino acid metabolism” and “Energy metabolism” were the top three categories under “Metabolism”, while “Translation” and “Folding, sorting and degradation” were the dominant categories under “Genetic Information Processing” (Figure 3A). Further tertiary hierarchical classification showed that “Ribosome”, “Protein processing in endoplasmic reticulum” and “Spliceosome” were the most frequently detected pathways (Figure 3B).
Figure 3.
Functional distribution of proteins based on Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation. (A) Distribution based on the secondary hierarchy. (B) Top 20 abundant pathways (according to protein number) based on the tertiary hierarchy.
To further annotate the protein functions, all the proteins were subjected to NCBInr database analysis. With a criterion of e-value ≤ 1 × 10−5, a total of 4924 proteins were successfully assigned with diverse functional annotations (Supplementary Table S1). The most abundant proteins (according to spectrum number) were associated with photosynthesis, such as the peridinin-chl a protein, chlorophyll a/c-binding protein and ribulose 1,5-bisphosphate carboxylase oxygenase. Moreover, the luciferin-binding protein, which is involved in bioluminescence, was also present in the list of the 10 most abundant proteins.
2.3. Differentially Expressed Proteins
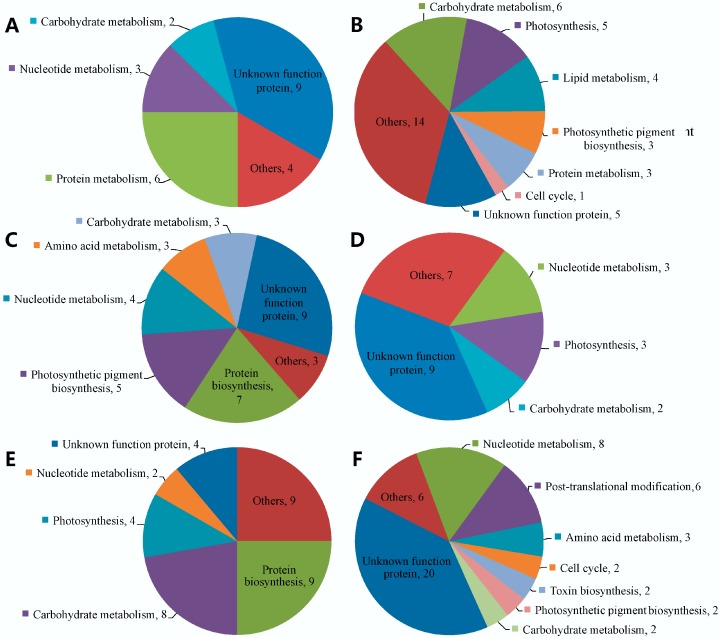
Of the 7232 proteins identified, 65 proteins in T5 vs. T9, 58 proteins in T9 vs. T10 and 87 proteins in T10 vs. T11 exhibited differential expression. In T5 vs. T9, 24 proteins were upregulated and 41 proteins were downregulated; in T9 vs. T10, 34 proteins were upregulated and 24 proteins were downregulated; and in T10 vs. T11, 36 proteins were upregulated and 51 proteins were downregulated. Functional classification of these proteins based on KEGG and NCBInr annotation is presented in Figure 4 and Supplementary Table S2.
Figure 4.
Functional distribution of differentially expressed proteins. (A) Upregulated proteins in T5 vs. T9. (B) Downregulated proteins in T5 vs. T9. (C) Upregulated proteins in T9 vs. T10. (D) Downregulated proteins in T9 vs. T10. (E) Upregulated proteins in T10 vs. T11. (F) Downregulated proteins in T10 vs. T11.
Quantitative analysis showed that proteins involved in protein translation were upregulated in T9 vs. T10, which was the toxin biosynthesis period. These proteins included ribosomal protein subunits, translation initiation factors, elongation factors and some aminoacyl-tRNA synthetases. Proteins participating in photosynthetic pigment biosynthesis were downregulated in T5 vs. T9 and T10 vs. T11 but upregulated significantly in T9 vs. T10. Moreover, the cell cycle-related protein E3 SUMO-protein ligase RanBP2 was downregulated in T5 vs. T9, and two other proteins, namely, kinesin family member 11 and cell division control protein 47, were downregulated in T10 vs. T11. However, notably, a large number of differentially expressed proteins were annotated with unknown functions, especially in T9 vs. T10, in which the toxin content increased sharply. These proteins might be involved in toxin biosynthesis or might participate in other biological processes associated with toxin production.
2.4. Toxin-Related Proteins
With a cutoff e-value ≤ 1 × 10−5, a total of 43 homologs of nine toxin-related proteins were identified in A. catenella, including sxtA, sxtG, sxtH, sxtI, sxtO, sxtT, sxtU, sxtZ and ompR (Table 1). Among these proteins, sxtA and sxtI were detected in dinoflagellates for the first time. Quantitative analysis showed that the expression levels of sxtI and ompR were significantly downregulated in T10 vs. T11, while those of other toxin-related proteins remained relatively stable.
Table 1.
Identification and quantitation of toxin-related proteins in Alexandrium catenella.
| Protein | Top Hit Protein | T5 vs. T9 | T9 vs. T10 | T10 vs. T11 | |||
|---|---|---|---|---|---|---|---|
| Ratio 1 | Ratio 2 | Ratio 1 | Ratio 2 | Ratio 1 | Ratio 2 | ||
| sxtA | CL2951.Contig2_All | - | - | - | - | - | - |
| sxtG | CL1611.Contig1_All | 0.86 | 1.09 | 1.12 | 0.87 | 0.95 | 0.86 |
| sxtH | comp55174_c0_orf1 | 0.76 | 0.90 | 1.14 | 0.80 | 0.88 | 1.23 |
| sxtI | comp65949_c0_orf1 | 0.92 | 1.49 * | 1.57 | 0.92 | 0.71 * | 0.81 * |
| sxtO | CL4284.Contig1_All | 1.02 | 1.07 | 0.92 | 1.00 | 1.07 | 0.93 |
| sxtT | comp55174_c0_orf1 | 0.76 | 0.90 | 1.14 | 0.80 | 0.88 | 1.23 |
| sxtU | Unigene83583_All | 1.22 | 1.53 | 1.13 | 0.83 | 0.94 | 1.23 |
| sxtZ | comp61883_c0_orf1 | 0.87 | 1.14 * | 1.19 * | 1.04 | 1.06 | 1.02 |
| ompR | comp17794_c0_orf1 | 1.02 | 0.97 | 1.10 | 1.29 * | 0.81 * | 0.77 * |
-: unquantifiable; *: p-value ≤ 0.05.
3. Discussion
In this study, we compared the global protein expression profiles of ACHK-T at four different toxin biosynthesis stages using an iTRAQ-based proteomic approach. A total of 210 proteins exhibited differential expression in T5 vs. T9, T9 vs. T10 and T10 vs. T11. These proteins are involved in various biological processes, such as PST biosynthesis, protein translation and photosynthetic pigment biosynthesis, and some of these proteins exhibited different variations in these three pairwise comparisons, which might be associated with toxin production.
3.1. Toxin Biosynthesis
High-performance liquid chromatography (HPLC) analysis showed that the toxin content exhibited periodic variation: toxin content increased rapidly in the late G1 phase and decreased sharply when the cells entered the G2/M phase. This variation was similar to that observed in our previous study [24] but differed from the results of previous studies on Alexandrium [16,18,20,22], which might be caused by species [16,18] or strain specific differences [20,22]. In addition, different culture conditions such as temperature or irradiance might be other possibilities resulting in the difference [16,18,20].
Nine toxin-related proteins were identified, and sxtA and sxtI were first detected in dinoflagellates. Among these proteins, sxtA is the key protein involved in the first step of the PST biosynthetic pathway [3,11]. In toxigenic dinoflagellates, the gene sxtA is transcribed into two types of transcripts: a long transcript containing all four domains (sxtA1–sxtA4), and a short transcript containing the first three domains but not sxtA4 [11]. Several studies have revealed that the sxtA4 domain performs a Claisen condensation reaction and is essential for PST production in dinoflagellates [9,15,25]. In our study, sxtA was identified as the long isoform (see Protein ID 6131 in Supplementary Table S1); therefore, the one unique peptide identified for this protein should belong to the sxtA4 domain. However, this protein was not quantified among the four different toxin production stages because of the presence of only one unique peptide. This finding indicated that the abundance of some proteins involved in PST production might be very low.
SxtI is another core protein that is directly involved in toxin production. This protein catalyzes the transfer of a carbamoyl group from carbamoyl phosphate to an intermediate product of PSTs [3]. Quantitative analysis showed that sxtI was significantly downregulated in T10 vs. T11 but varied nonsignificantly in T5 vs. T9 and T9 vs. T10 (Table 1). A similar expression pattern was also observed for ompR, a transcriptional regulator of the toxin biosynthetic pathway [3] (Table 1). Downregulation of these two proteins in T10 vs. T11 was consistent with the toxin biosynthesis pattern because the cells stopped toxin production during this period (Figure 2), and the variation in ompR level suggested that the regulation of toxin production in dinoflagellates might be similar to that in cyanobacteria. However, the expression of these two proteins varied nonsignificantly in T5 vs. T9 and T9 vs. T10, especially in T9 vs. T10, in which the toxin content increased sharply (Figure 2). Moreover, the expression of other toxin-related proteins varied nonsignificantly among the three pairwise comparisons. Previous studies reported that some important biological processes including toxin production are regulated at translational or post-translational level in dinoflagellates [25,26,27,28]. Therefore, our results indicated that the toxin biosynthetic process in dinoflagellates was more complex than that in cyanobacteria, and the PST biosynthesis might be regulated post-translationally rather than translationally.
3.2. Protein Translation
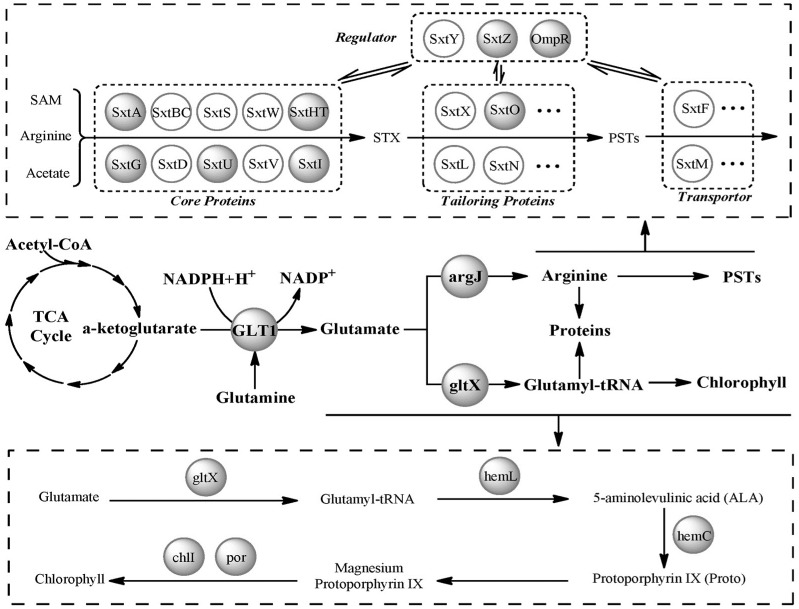
In our study, a number of proteins involved in protein translation were significantly upregulated in T9 vs. T10, including one ribosomal protein, three translation factors that directly participate in the translation process, four aminoacyl-tRNA synthetases and three amino acid metabolism proteins that provide substrates for protein translation (Supplementary Table S2). The upregulation of these proteins suggested that the protein translation process was enhanced, which was coincident with PST production. A similar correlation between these two biological processes was also observed in A. fundyense [17]. It is postulated that PST production of dinoflagellates is a specific pathway integrated in the cellular nitrogen metabolism [17]. PST biosynthesis is indirectly modulated by protein biosynthesis and amino acid metabolism, and, as a common substrate, arginine acts as the link between these processes (Figure 5) [17,29,30]. Among the three amino acid metabolism proteins mentioned above, glutamate N-acetyltransferase/amino acid N-acetyltransferase is a key bifunctional protein that is involved in arginine biosynthesis, while glutamate synthase catalyzes the biosynthesis of glutamate, which is the substrate of arginine and glutamine—another nitrogen-enriched intermediate for the biosynthesis of many macromolecules [31]. Upregulation of these two proteins suggested that glutamate and arginine biosynthesis were enhanced, which provided enough substrate and intermediate for protein translation and toxin production during this period. Overall, our results supported the previous opinion that PST production might be a mechanism by which the balance of intracellular nitrogen metabolism is maintained in dinoflagellates.
Figure 5.
Relationships among paralytic shellfish toxin (PST) production, protein translation and chlorophyll biosynthesis. Dark circles represent the proteins identified in this study. The dashed box on top shows the putative PST biosynthesis pathway in dinoflagellates (modified from previous reports [4,9,32]), and the dashed box on the bottom summarizes the route of chlorophyll biosynthesis (modified from previous reports [33,34]). The abbreviations are: TCA: tricarboxylic acid cycle; gltX: glutamyl-tRNA synthetase; chlI: magnesium chelatase subunit I; por: protochlorophyllide reductase; hemL: oxygen-dependent protoporphyrinogen oxidase; argJ: glutamate N-acetyltransferase/amino acid N-acetyltransferase; GLT1: glutamate synthase (NADH).
3.3. Photosynthetic Pigment Biosynthesis
In marine dinoflagellates, chlorophyll and carotenoids are two major groups of photosynthetic pigments and play essential roles in photosynthetic light harvesting and energy transduction [35]. The biosynthetic process of chlorophyll can be divided into three steps (Figure 5): (i) the formation of 5-aminolevulinic acid (ALA), which is the rate limited and committed step of the whole pathway; (ii) the formation of protoporphyrin IX (Proto) from eight molecules of ALA; and (iii) the formation of chlorophyll in the magnesium branch [33,34]. Previous studies have revealed that the accumulation of photosynthetic pigments and the proteins involved in this process are adapted to the circadian clock or light–dark cycle [36,37]. Chlorophyll biosynthesis is restricted to the light period, and the proteins involved in this process are coordinately expressed [36,38,39]. In our study, the T9, T10 and T11 sampling points were in the dark period, and the T5 sampling point was in the light period. Among the differentially expressed proteins, glutamate-1-semialdehyde 2,1-aminomutase (hemL), hydroxymethylbilane synthase (hemC) and protochlorophyllide reductase (por), which participate in the three steps of chlorophyll biosynthesis were significantly downregulated in T5 vs. T9. In addition, glutamyl-tRNA synthetase (gltX) and oxygen-dependent protoporphyrinogen oxidase (hemL), which are involved in the first step of the process, were downregulated in T10 vs. T11. The downregulation of these proteins was consistent with the circadian biosynthesis of chlorophyll. However, in T9 vs. T10, five proteins—gltX, hemL, magnesium chelatase subunit I (chlI), por and zeaxanthin epoxidase—were significantly upregulated, indicating that photosynthetic pigment biosynthesis was enhanced during this period. Given the previous finding that chlorophyll biosynthesis is a light-induced process, the enhancement of photosynthetic pigment biosynthesis during the dark period is abnormal. Considering the sharp increase in toxin content during this period, there may exist an association between PST production and photosynthetic pigment biosynthesis.
In previous studies, PST biosynthesis has been proposed to be closely associated with chloroplast biosynthesis. PSTs and chlorophyll are thought to share the same polyketide precursor [40]. Chlorophyll formation starts from the synthesis of ALA, where gltX and hemL are the two enzymes that catalyze the conversion of glutamate to ALA [33,34]. Therefore, glutamate provides a carbon skeleton for chlorophyll, and upregulation of gltX and hemL in T9 vs. T10 suggested that utilization of carbon from glutamate to synthesize chlorophyll was enhanced during the toxin production period. For PST production, the carbon skeleton of PSTs originates from arginine, which is synthesized from glutamate [3,31]. Therefore, glutamate, instead of some polyketides, was the common substrate for both chlorophyll biosynthesis and PST production (Figure 5), and the enhanced biosynthesis of glutamate provided sufficient substrate and intermediate for photosynthetic pigment biosynthesis, PST production and protein translation. This close association between photosynthetic pigment biosynthesis and PST production was also observed in our previous study [24]. Because photosynthetic pigments, PSTs and proteins are all nitrogen-enriched macromolecules in toxigenic dinoflagellates and because alterations in these macromolecules always occur simultaneously [17,41], an ecological role of PSTs as intracellular nitrogen stores is plausible in dinoflagellates [42,43].
4. Materials and Methods
4.1. Culture Conditions and Sample Collection
Unialgal cultures of ACHK-T were provided by the Collection Center of Marine Algae, Xiamen University, China. Cultures of ACHK-T were grown in K-medium [44] at 20 °C. Irradiance of approximately 100 μE·m−2·s−1 was provided using cool white fluorescent bulbs under a 14:10 h light: dark photoperiod.
Synchronization and sampling of ACHK-T cells were conducted using a previously reported method [24,45]. Briefly, the algal cells floating in the upper layer of the culture flask were inoculated into fresh medium with algal cell suspension:medium = 1:3 every 4 days for a month. Then, the synchronized cells were finally transferred to 3 5-L flasks filled with 4 L of K-medium and grown for 2 days. Sample collection was started at 08:00 (T1) and carried out every 2 h for 24 h. One milliliter of each culture was collected and fixed with Lugol’s solution for cell counting; 50 mL of culture was collected and the cell pellets were resuspended in 1 mL of 70% ethanol for flow cytometric (FCM) analysis; and 50 mL of culture was collected by centrifugation and stored at −20 °C for toxin analysis. For proteomic analysis, cells were harvested at 16:00 (T5), 00:00 (T9), 02:00 (T10) and 04:00 (T11) based on the toxin analysis result. T5 was the middle time point, at which the toxin content increased slowly; T9 was the time point at which the toxin content started to increase rapidly; and T10 and T11 were the transition and termination time points of this rapid increase. For each time point, 500 mL of each culture (approximately 106 cells) was collected and the cell pellets were resuspended in 1 mL of TRIzol reagent (Invitrogen, Carlsbad, CA, USA), frozen in liquid nitrogen and stored at −80 °C.
4.2. Toxin Analysis
PST analysis was conducted by following our previous study [46]. Cell pellets were suspended in 0.5 mL of 50 mM acetic acid and homogenized with sonication. Then, the supernatant was collected by centrifugation at 10,000 ×g for 30 min and filtered with 0.22-μm-pore filter membranes (Millipore). The toxin analysis was carried out using HPLC with post column derivatization by an Intersil C8-5 column. Toxin standards were purchased from the National Research Council, Canada and the concentration of PSTs was determined by comparing the peak area of the sample with that of the toxin standards.
4.3. Flow Cytometric Analysis
FCM analysis was conducted by following the previous study [22]. The FCM sample was centrifuged at 10,000× g for 5 min and washed twice with phosphate-buffered saline (1× PBS, pH 8.0). Then the sample was suspended in 100 μL of RNase A, incubated at room temperature for 10 min. After that, the sample was suspended in 1 mL of 1× PBS containing 2.5 mg·mL−1 propidium iodide and stained for 1 h at 37 °C. The DNA content of stained cells was analyzed using an Epics XL flow cytometer (Beckman Coulter, Miami, FL, USA), and the cell cycle was determined based on the histograms of the relative DNA content, which was analyzed using MultiCycle software (Version 4.0, Phoenix Flow Systems, San Diego, CA, USA).
4.4. Protein Preparation
The protein samples were homogenized with sonication, and protein was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), chloroform and ethanol, and precipitated using isopropanol. Then, the protein pellets were washed with 95% ethanol and air-dried [46]. After that, 100 μL of rehydration buffer was added to dissolve the pellets. Then, the proteins were reduced with 10 mM DTT at 56 °C for 1 h, alkylated with 55 mM iodacetamide and precipitated with 4 volumes of acetone. After air-drying, 500 μL of 0.5 M tetraethyl-ammonium bromide (TEAB) (Applied Biosystems, Milan, Italy) was added to dissolve the pellets and the protein concentration was quantified with the Bradford assay, using bovine serum albumin (BSA) as a standard.
4.5. Peptide Labeling
Protein samples from 4 time points were compared using 2 biological replicates each. A total of 100 μg protein from each sample was digested using Trypsin Gold (Promega, Madison, WI, USA) with protein:trypsin = 30:1 at 37 °C for 16 h. After drying by vacuum centrifugation, the trypsin-digested samples were reconstituted in 0.5 M TEAB and labeled with 8-plex iTRAQ reagent (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Samples were labeled with the iTRAQ tags as follows: T5-1 (119 tag), T5-2 (121 tag), T9-1 (117 tag), T9-2 (118 tag), T10-1 (115 tag), T10-2 (116 tag), T11-1 (113 tag) and T11-2 (114 tag). After 2 h of incubation at room temperature, the labeled samples were pooled and dried using vacuum centrifugation.
4.6. Cation Exchange Fractionation
The cation exchange fractionation was performed using an LC-20AB HPLC pump system (Shimadzu, Kyoto, Japan). The labeled peptide mixtures were reconstituted with 4 mL of buffer A (25 mM NaH2PO4 in 25% ACN, pH 2.7) and loaded onto a column containing 5-μm particles (Phenomenex). The peptides were eluted at a flow rate of 1 mL·min−1 with the following gradient: buffer A for 10 min, 5–60% buffer B (25 mM NaH2PO4, 1 M KCl in 25% ACN, pH 2.7) for 27 min, and 60–100% buffer B for 1 min. The system was then maintained at 100% buffer B for 1 min before equilibrating with buffer A for 10 min prior to the next injection. Elution was monitored by measuring the absorbance at 214 nm, and fractions were collected every 1 min. The eluted peptides were pooled into 20 fractions, desalted with a Strata X C18 column (Phenomenex, Torrance, CA, USA) and vacuum-dried.
4.7. LC-MS/MS Analysis
All peptide samples were separated on a LC-20AD nanoHPLC system (Shimadzu, Kyoto, Japan) and analyzed on a Q-EXACTIVE system (Thermo Fisher Scientific, San Jose, CA, USA). Each fraction was resuspended in buffer C (2% ACN, 0.1% formic acid) and the final concentration of peptide was about 0.5 μg·μL−1 on average. The samples were loaded at 8 μL·min−1 for 4 min, and then, a 44-min gradient was run at 300 nL·min−1 starting from 2% to 35% buffer D (98% ACN, 0.1% formic acid), followed by a 2-min linear gradient to 80% and maintenance at 80% for 4 min, finally returning to 5% in 1 min.
Peptides were selected for MS/MS using the high-energy collision dissociation (HCD) operating mode with a normalized collision energy setting of 27.0. Survey scans were acquired at a resolution of 70,000 and resolution for HCD spectra was set to 17,500. A data-dependent procedure that alternated between 1 MS scan followed by 15 MS/MS scans was applied for the 15 most abundant precursor ions that were above a threshold ion count of 20,000 in the MS survey scan with a subsequent dynamic exclusion duration of 15 s. The electrospray voltage applied was 1.6 kV. Automatic gain control was used to optimize the generated spectra. For MS scans, the m/z scan range was 350 to 2000 Da. For MS2 scans, the m/z scan range was 100–1800.
4.8. Bioinformatics Analysis
MASCOT genetic format files that were obtained by using Proteome Discoverer 1.2 (PD 1.2, Thermo Fisher Scientific, San Jose, CA, USA) to covert raw data files were searched, and protein identification was performed using the MASCOT search engine (Matrix Science, version 2.3.02) against database containing amino acid sequences translated from unigenes in two previous projects [15,23].
For protein identification, a mass tolerance of 0.1 Da was permitted for fragmented ions and 0.05 Da for peptide masses, with allowance for one missed cleavage in the trypsin digests. The charge states of peptides were set to 2+ and 3+. iTRAQ labeling and carbamidomethylation were defined as fixed modifications, and Gln->pyro-Glu (N-term Q), oxidation (M), and dehydration (NQ) were the potential variable modifications. Specifically, an automatic decoy database search was performed in MASCOT by choosing the decoy checkbox in which a random sequence of database is generated and tested for raw spectra as well as the real database. Peptides with significance scores (≥20) at the 99% confidence interval were counted as being identified, and each identified protein contained at least one unique peptide.
For protein quantitation, it was required that a protein contained at least two unique peptides. The quantitative ratios were weighted and normalized using the median ratio in MASCOT. Proteins were subjected to three pairwise comparisons: T5 vs. T9, T9 vs. T10 and T10 vs. T11. A protein was considered to be differentially expressed if it contained at least two unique peptides, had similar and significant ratios ≥1.2 (upregulated) or ≤0.83 (downregulated) in two replicates [7].
Functional annotation of the proteins was conducted using the Blast2GO program against the KEGG and NCBInr databases.
4.9. Identification of Toxin-Related Proteins
To identify proteins associated with PST biosynthesis, a BLAST comparison was performed of the toxin-related protein sequences from the cyanobacterium Cylindrospermopsis raciborskii T3 [3] against the proteins identified. All hits with e-value ≤ 1 × 10−5 were retrieved as confident identifications.
5. Conclusions
This study investigated PST production in toxigenic A. catenella during the cell cycle and compared the global protein expression profiles at four different stages of toxin biosynthesis using an iTRAQ-based quantitative proteomic approach. Toxin content increased rapidly in the late G1 phase and decreased sharply when the cells entered the G2/M phase. SxtA and sxtI were identified in dinoflagellates for the first time, and the expression of sxtI and ompR was consistent with the toxin biosynthesis pattern. The regulation of toxin biosynthesis in dinoflagellates was similar to that in cyanobacteria, and some steps of the PST biosynthesis process might be regulated post-translationally. Moreover, PST production was modulated by protein translation and photosynthetic pigment biosynthesis via the common substrate glutamate. Overall, our study provided new insights into PST biosynthesis of dinoflagellates and clarified the complex relationship between the cell cycle and PST production.
Acknowledgments
D.-Z.W. was also supported by the ‘Ten Thousand Talents Program’ for leading talents in science and technological innovation.
Supplementary Materials
The following are available online at http://www.mdpi.com/1660-3397/16/12/491/s1. Supplementary Table S1: The identification, quantitation and annotation of the 7232 proteins. Supplementary Table S2: Functional classifications of the differentially expressed proteins.
Author Contributions
Conceptualization, D.-Z.W. and S.-F.Z.; investigation, S.-F.Z., Y.Z. and L.L.; writing—original draft preparation, S.-F.Z.; writing—review and editing, D.-Z.W.
Funding
This research was funded by the National Key R & D Program of China, grant number 2017YFC1404300, and the National Natural Science Foundation of China, grant numbers 41230961 and 41425021.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hallegraeff G.M. A review of harmful algal blooms and their apparent global increase. Phycologia. 1993;32:79–99. doi: 10.2216/i0031-8884-32-2-79.1. [DOI] [Google Scholar]
- 2.Anderson D.M., Cembella A.D., Hallegraeff G.M. Progress in understanding harmful algal blooms: Paradigm shifts and new technologies for research, monitoring, and management. Ann. Rev. Mar. Sci. 2012;4:143–176. doi: 10.1146/annurev-marine-120308-081121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kellmann R., Mihali T.K., Jeon Y.J., Pickford R., Pomati F., Neilan B.A. Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria. Appl. Environ. Microbiol. 2008;74:4044–4053. doi: 10.1128/AEM.00353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mihali T.K., Kellmann R., Neilan B.A. Characterisation of the paralytic shellfish toxin biosynthesis gene clusters in Anabaena circinalis AWQC131C and Aphanizomenon sp. NH-5. BMC Biochem. 2009;10:8. doi: 10.1186/1471-2091-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stucken K., John U., Cembella A., Murillo A.A., Soto-Liebe K., Fuentes-Valdés J.J., Friedel M., Plominsky A.M., Vásquez M., Glöckner G. The smallest known genomes of multicellular and toxic cyanobacteria: Comparison, minimal gene sets for linked traits and the evolutionary implications. PLoS ONE. 2010;5:e9235. doi: 10.1371/journal.pone.0009235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mihali T.K., Carmichael W.W., Neilan B.A. A putative gene cluster from a Lyngbya wollei bloom that encodes paralytic shellfish toxin biosynthesis. PLoS ONE. 2011;6:e14657. doi: 10.1371/journal.pone.0014657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Agostino P.M., Song X.M., Neilan B.A., Moffitt M.C. Comparative proteomics reveals that a saxitoxin-producing and a nontoxic strain of Anabaena circinalis are two different ecotypes. J. Proteome Res. 2014;13:1474–1484. doi: 10.1021/pr401007k. [DOI] [PubMed] [Google Scholar]
- 8.Shimizu Y. Microalgal metabolites. Chem. Rev. 1993;93:1685–1698. doi: 10.1021/cr00021a002. [DOI] [Google Scholar]
- 9.Hackett J.D., Wisecaver J.H., Brosnahan M.L., Kulis D.M., Anderson D.M., Bhattacharya D., Plumley F.G., Erdner D.L. Evolution of saxitoxin synthesis in cyanobacteria and dinoflagellates. Mol. Biol. Evol. 2013;30:70–78. doi: 10.1093/molbev/mss142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang S.F., Zhang Y., Xie Z.X., Zhang H., Lin L., Wang D.Z. iTRAQ-based quantitative proteomic analysis of a toxigenic dinoflagellate Alexandrium catenella and its non-toxic mutant. Proteomics. 2015;15:4041–4050. doi: 10.1002/pmic.201500156. [DOI] [PubMed] [Google Scholar]
- 11.Stüken A., Orr R.J.S., Kellmann R., Murray S.A., Neilan B.A., Jakobsen K.S. Discovery of nuclear-encoded genes for the neurotoxin saxitoxin in dinoflagellates. PLoS ONE. 2011;6:e20096. doi: 10.1371/journal.pone.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orr R.J.S., Stüken A., Murray S.A., Jakobsen K.S. Evolutionary acquisition and loss of saxitoxin biosynthesis in dinoflagellates: The second “core” gene, sxtG. Appl. Environ. Microbiol. 2013;79:2128–2136. doi: 10.1128/AEM.03279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray S.A., Wiese M., Stüken A., Brett S., Kellmann R., Hallegraeff G., Neilan B.A. SxtA-based quantitative molecular assay to identify saxitoxin-producing harmful algal blooms in marine waters. Appl. Environ. Microbiol. 2011;77:7050–7057. doi: 10.1128/AEM.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray S.A., Diwan R., Orr R.J.S., Kohli G.S., John U. Gene duplication, loss and selection in the evolution of saxitoxin biosynthesis in alveolates. Mol. Phylogenet. Evol. 2015;92:165–180. doi: 10.1016/j.ympev.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Zhang S.F., Lin L., Wang D.Z. Comparative transcriptome analysis of a toxin-producing dinoflagellate Alexandrium catenella and its non-toxic mutant. Mar. Drugs. 2014;12:5698–5718. doi: 10.3390/md12115698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taroncher-Oldenburg G., Kulis D.M., Anderson D.M. Toxin variability during the cell cycle of the dinoflagellate Alexandrium fundyense. Limnol. Oceanogr. 1997;42:1178–1188. doi: 10.4319/lo.1997.42.5_part_2.1178. [DOI] [Google Scholar]
- 17.Taroncher-Oldenburg G., Kulis D.M., Anderson D.M. Coupling of saxitoxin biosythesis to the G1 phase of the cell cycle in the dinoflagellate Alexandrium fundyense: Temperature and nutrient effects. Nat. Toxins. 1999;7:207–219. doi: 10.1002/1522-7189(200009/10)7:5<207::AID-NT61>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 18.Taroncher-Oldenburg G., Anderson D.M. Identification and characterization of three differentially expressed genes, encoding S-adenosylhomocysteine hydrolase, methionine aminopeptidase, and a histone-like protein, in the toxic dinoflagellate Alexandrium fundyense. Appl. Environ. Microbiol. 2000;66:2105–2112. doi: 10.1128/AEM.66.5.2105-2112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho Y., Ogawa M., Hirota M., Oshima Y. Effects of mitomycin C and colchicine on toxin production and cell cycle regulation in the dinoflagellate Alexandrium tamarense. Harmful Algae. 2011;10:235–244. doi: 10.1016/j.hal.2010.10.003. [DOI] [Google Scholar]
- 20.Siu G.K.Y., Young M.L.C., Chan D.K.O. Environmental and nutritional factors which regulate population dynamics and toxin production in the dinoflagellate Alexandrium catenella. Hydrobiologia. 1997;352:117–140. doi: 10.1023/A:1003042431985. [DOI] [Google Scholar]
- 21.Harlow L.D., Negri A., Hallegraeff G.M., Koutoulis A. Sam, Sahh and Map gene expression during cell division and paralytic shellfish toxin production of Alexandrium catenella (Dinophyceae) Phycologia. 2007;46:666–674. doi: 10.2216/07-25.1. [DOI] [Google Scholar]
- 22.Wang D.Z., Gao Y., Lin L., Hong H.S. Comparative proteomic analysis reveals proteins putatively involved in toxin biosynthesis in the marine dinoflagellate Alexandrium catenella. Mar. Drugs. 2013;11:213–232. doi: 10.3390/md11010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Zhang S.F., Lin L., Wang D.Z. Whole transcriptomic analysis provides insights into molecular mechanisms for toxin biosynthesis in a toxic dinoflagellate Alexandrium catenella (ACHK-T) Toxins (Basel) 2017;9:213. doi: 10.3390/toxins9070213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S.F., Zhang Y., Lin L., Wang D.Z. iTRAQ-based quantitative proteomic analysis of a toxigenic dinoflagellate Alexandrium catenella and its non-toxigenic mutant exposed to a cell cycle inhibitor colchicine. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiese M., Murray S.A., Alvin A., Neilan B.A. Gene expression and molecular evolution of sxtA4 in a saxitoxin producing dinoflagellate Alexandrium catenella. Toxicon. 2014;95:102–112. doi: 10.1016/j.toxicon.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Van de Waal D.B., Eberlein T., John U., Wohlrab S., Rost B. Impact of elevated pCO2 on paralytic shellfish poisoning toxin content and composition in Alexandrium tamarense. Toxicon. 2014;78:58–67. doi: 10.1016/j.toxicon.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 27.Kellmann R., Neilan B.A. Biochemical characterization of paralytic shellfish toxin biosynthesis in vitro. J. Phycol. 2007;43:497–508. doi: 10.1111/j.1529-8817.2007.00351.x. [DOI] [Google Scholar]
- 28.Perini F., Galluzzi L., Dell’Aversano C., Iacovo E.D., Tartaglione L.U., Ricci F., Forino M., Ciminiello P., Penna A. SxtA and sxtG gene expression and toxin production in the mediterranean Alexandrium minutum (Dinophyceae) Mar. Drugs. 2014;12:5258–5276. doi: 10.3390/md12105258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson D.M., Kulis D.M., Sullivan J.J., Hall S., Lee C. Dynamics and physiology of saxitoxin production by the dinoflagellates Alexandrium spp. Mar. Biol. 1990;104:511–524. doi: 10.1007/BF01314358. [DOI] [Google Scholar]
- 30.Shimizu Y. Toxigenesis and biosynthesis of saxitoxin analogues. Pure Appl. Chem. 1986;58:257–262. doi: 10.1351/pac198658020257. [DOI] [Google Scholar]
- 31.Umbarger H.E. Amino acid biosynthesis and its regulation. Annu. Rev. Biochem. 1978;47:533–606. doi: 10.1146/annurev.bi.47.070178.002533. [DOI] [PubMed] [Google Scholar]
- 32.Wang D.Z., Zhang S.F., Zhang Y., Lin L. Paralytic shellfish toxin biosynthesis in cyanobacteria and dinoflagellates: A molecular overview. J. Proteomics. 2015;135:132–140. doi: 10.1016/j.jprot.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Eckhardt U., Grimm B., Hörtensteiner S. Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol. Biol. 2004;56:1–14. doi: 10.1007/s11103-004-2331-3. [DOI] [PubMed] [Google Scholar]
- 34.Porra R.J. Recent progress in porphyrin and chlorophyll biosynthesis. Photochem. Photobiol. 1997;65:492–516. doi: 10.1111/j.1751-1097.1997.tb08596.x. [DOI] [Google Scholar]
- 35.Jeffrey S.W., Sielicki M., Haxo F.T. Chloroplast pigment patterns in dinoflagellates. J. Phycol. 1975;11:374–384. doi: 10.1111/j.1529-8817.1975.tb02799.x. [DOI] [Google Scholar]
- 36.Braun R., Farré E.M., Schurr U., Matsubara S. Effects of light and circadian clock on growth and chlorophyll accumulation of Nannochloropsis gaditana. J. Phycol. 2014;50:515–525. doi: 10.1111/jpy.12177. [DOI] [PubMed] [Google Scholar]
- 37.Harmer S.L., Hogenesch J.B., Straume M., Chang H.S., Han B., Zhu T., Wang X., Kreps J.A., Kay S.A. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 38.Beator J., Kloppstech K. The circadian oscillator coordinates the synthesis of apoproteins and their pigments during chloroplast development. Plant Physiol. 1993;103:191–196. doi: 10.1104/pp.103.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujita Y. Protochlorophyllide reduction: A key step in the greening of plants. Plant Cell Physiol. 1996;37:411–421. doi: 10.1093/oxfordjournals.pcp.a028962. [DOI] [PubMed] [Google Scholar]
- 40.Jiang X.W., Wang J., Gao Y., Chan L.L., Lam P.K.S., Gu J.D. Relationship of proteomic variation and toxin synthesis in the dinoflagellate Alexandrium tamarense CI01 under phosphorus and inorganic nitrogen limitation. Ecotoxicology. 2015;24:1744–1753. doi: 10.1007/s10646-015-1513-x. [DOI] [PubMed] [Google Scholar]
- 41.Yentsch C.S., Vaccaro R.F. Phytoplankton nitrogen in the oceans. Limnol. Oceanogr. 1958;3:443–448. doi: 10.4319/lo.1958.3.4.0443. [DOI] [Google Scholar]
- 42.Cusick K.D., Sayler G.S. An overview on the marine neurotoxin, saxitoxin: Genetics, molecular targets, methods of detection and ecological functions. Mar. Drugs. 2013;11:991–1018. doi: 10.3390/md11040991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krüger T. Biosynthesis of paralytic shellfish poisoning (PSP) toxins in cyanobacteria and dinoflagellates. J. Endocyt. Cell Res. 2010;68:80. [Google Scholar]
- 44.Keller M.D., Selvin R.C., Claus W., Guillard R.R.L. Media for the culture of oceanic ultraphytoplankton. J. Phycol. 1987;23:633–638. doi: 10.1111/j.1529-8817.1987.tb04217.x. [DOI] [Google Scholar]
- 45.Shi X.G., Zhang H., Lin S.J. Tandem repeats, high copy number and remarkable diel expression rhythm of form II RuBisCO in Prorocentrum donghaiense (dinophyceae) PLoS ONE. 2013;8:e71232. doi: 10.1371/journal.pone.0071232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang D.Z., Li C., Zhang Y., Wang Y.Y., He Z.P., Lin L., Hong H.S. Quantitative proteomic analysis of differentially expressed proteins in the toxicity-lost mutant of Alexandrium catenella (Dinophyceae) in the exponential phase. J. Proteomics. 2012;75:5564–5577. doi: 10.1016/j.jprot.2012.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.