Abstract
Multiple sclerosis (MS) is a chronic neurological disorder characterized by inflammation, demyelination, and axonal damage. Increased levels of reactive oxygen species (ROS), produced by macrophages and leading to oxidative stress, have been implicated as mediators of demyelination and axonal injury in both MS and experimental autoimmune encephalomyelitis, the murine model of the disease. On the other hand, reduced ROS levels can increase susceptibility to autoimmunity. In this work, we screened for association with MS 11 single nucleotide polymorphisms (SNPs) and two microsatellite markers in the five genes (NCF1, NCF2, NCF4, CYBA, and CYBB) of the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (NOX2) system, the enzymatic pathway producing ROS in the brain and neural tissues, in 347 Finnish patients with MS and 714 unaffected family members. This analysis showed suggestive association signals for NCF1 and CYBB (lowest p = 0.038 and p = 0.013, respectively). Functional relevance for disease predisposition was further supported for the CYBB gene, by microarray analysis in CD4+/− mononuclear cells of 21 individuals from five Finnish multiplex MS families, as well as by real-time RT-PCRs performed on RNA extracted from peripheral blood mononuclear cells of an Italian replication cohort of 21 MS cases and 21 controls. Our results showed a sex-specific differential expression of CYBB, suggesting that this gene, and more in general the NOX2 system, deserve to be further investigated for their possible role in MS.
Keywords: multiple sclerosis, association study, reactive oxygen species, NADPH oxidase, CYBB
1. Introduction
Multiple sclerosis (MS) (Online Mendelian Inheritance in Men, OMIM #126200) is a chronic disease of the central nervous system (CNS) characterized by multifocal inflammation, plaques of myelin degeneration, axonal damage, and by a high degree of individual variability in the severity and progression of symptoms [1,2,3]. Among neurological disorders of young adults, MS is the most common: in Europe, where a latitude-correlated distribution of prevalence and incidence rates can be observed, prevalence ranges from 70 to 100 per 100,000, whereas incidence varies between 2 and 4 per 100,000 person/year; the highest prevalence and incidence rates have been reported in Finland [4]. Here, incidence and prevalence reach the exceptional rate of 11.6 and 200 per 100,000 in the Southern Ostrobothnian region (western coast of Finland), reflecting several centuries of genetic drift in a small and isolated population [5].
In agreement with the multigenic character of MS, genome-wide linkage scans, genome-wide association studies (GWAS), as well as meta-analyses performed in large cohorts disclosed more than 200 non-HLA (human leukocyte antigen) single nucleotide polymorphisms (SNPs) associated with MS, each having a small effect size on MS predisposition [6,7,8,9,10]. Typically, the identified association signals point to genes belonging to innate and adaptive pathways [9,10]. The deep involvement of a dysregulated immune system in MS is also supported by the efficacy of drugs targeting T- and B-cell functions in the treatment of the disease, as well as by the numerous studies indicating CD4+ and CD8+ T cells as strong contributors to the pathogenic process [11,12,13,14].
Apart from immune-mediated mechanisms, a growing body of evidence indicates that oxidative stress (OS) may play a role in the etiology of MS [15,16,17]. Increased levels of oxygen-free radicals (collectively called reactive oxygen species, ROS), produced by macrophages and leading to OS, have been implicated as mediators of demyelination and axonal injury in both MS and experimental autoimmune encephalomyelitis, the murine model of the disease [18,19,20]. Moreover, ROS: (i) activate specific transcription factors, such as the nuclear transcription factor kappa B, which in turn upregulates the expression of genes associated with MS or its progression (e.g., tumor necrosis factor α) [21,22]; (ii) mediate the activity of matrix metalloproteinases, which are involved in T-cell activation and trafficking into the CNS and hence probably involved in some of the early pro-inflammatory events in MS [23,24,25]; (iii) are produced in increased amount by activated mononuclear cells of patients, resulting in oxygen damage to DNA, lipids, and proteins—these molecules are frequently found in active MS lesions and are associated with apoptotic oligodendrocytes and neurodegeneration in the brains of patients with MS [16,26,27,28].
Besides reactions involving the electron-transport chain of mitochondria or those related to the metabolism of amino acids and neurotransmitters, in the brain and neural tissue ROS are also produced by enzymatic pathways, such as xanthine oxidase, lipoxygenase, and cyclooxygenase, as well as by the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (NOX) systems [29,30]. Specifically, the NOX2 system is a complex that, in resting cells, is compartmentalized between cytosol and plasma membrane: the core enzyme is composed of three cytoplasmic polypeptides (p47phox, p67phox, and p40phox, encoded by NCF1, NCF2, and NCF4 genes, respectively), and a membrane-bound flavo-hemo-cytochrome, b558, constituted by two components, the p22phox and gp91phox subunits (encoded by CYBA and CYBB, respectively) [29,30]. When the resting cell is exposed to stimuli (i.e., chemotactic reactants or particulate stimuli, such as bacteria and fungi), p47phox becomes heavily phosphorylated, inducing all cytoplasmic components to translocate to the membrane-bound cytochrome in order to form the active enzyme complex [29,30].
Given the proposed role of OS in MS, its contribution to the pathogenesis of the disease was here investigated by genetic association and expression analyses of the five genes coding for the main components of the NOX2 complex.
2. Results
2.1. NCF1 and CYBB Are Associated with MS in the Finnish Population
For investigating the potential role of OS genes in the pathogenesis of MS, we performed an association analysis on a Finnish cohort of 63 MS families (547 individuals) with two microsatellite markers and 11 SNPs mapping in or located close to the five genes coding for the main components of the NOX2 complex (the soluble factors NCF1, NCF2, and NCF4, and the membrane-bound redox core proteins of the complex, CYBA and CYBB) (see Table S1 in the Supplementary Materials). No significant deviation from Hardy-Weinberg equilibrium (HWE) and no Mendelian inheritance errors were observed for any of the markers included in this study. The overall average genotype call rate was 96.5%, and the accuracy was >99.5% according to duplicated genotyping (5%) of all samples.
Table 1 shows the results of the allelic association analysis for each of the 11 variants for the whole study cohort. Two polymorphisms, D7S1870 in NCF1 and rs5963310 in CYBB, showed suggestive evidence of association with MS in the transmission/disequilibrium (TDT) test (p = 0.038 and p = 0.027 for NCF1 and CYBB, respectively; not corrected for multiple testing). By using the more powerful approach based on the TDT statistic implemented with the parental discordance test (see the Materials and Methods section), we confirmed the association signal for the rs5963310 polymorphism in CYBB (p = 0.013), which confers a protective effect against MS (the minor allele A is untransmitted in MS cases). No significant allelic association between MS and the other NADPH variants could be detected (Table 1).
Table 1.
NADPH polymorphisms: Results of family-based association TDT and haplotype relative risk (HRR) tests.
| Gene | Marker | ANALYZE Analyses 1 | PLINK Analyses | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HRR p Value | TDT p Value | Minor Allele 2 | MAF | Transmitted/Untransmitted 3 | TDT p Value | OR [95% CI] | p Par 4 | p Comb 4 | ||
| NCF1 | D7S1870 | 0.0491 | 0.0383 | 20 repeats | 0.4533 | Untransmitted | 0.330 | 0.727 [0.38–1.385] | 0.157 | 0.206 |
| D7S2518 | 0.376 | 0.431 | 7 repeats | 0.3986 | / | 1 | 1 [0.351–2.851] | 0.414 | 0.655 | |
| NCF2 | rs796860 | 0.895 | 0.500 | C | 0.06548 | / | 1 | 1 [0.250–3.998] | 1 | 1 |
| rs2296164 | 0.568 | 0.399 | T | 0.4861 | Untransmitted | 0.297 | 0.643 [0.278–1.485] | 0.564 | 0.239 | |
| rs3818364 | 0.604 | 0.500 | T | 0.4688 | Transmitted | 0.847 | 1.0770 [0.506–2.291] | 0.564 | 0.715 | |
| rs789192 | 0.183 | 0.253 | G | 0.4268 | Transmitted | 0.144 | 1.727 [0.822–3.630] | 0.317 | 0.423 | |
| rs2274065 | 0.293 | 0.500 | C | 0.0974 | Transmitted | 0.180 | 4 [0.447–35.790] | 0.317 | 0.414 | |
| NCF4 | rs1883112 | 0.991 | 0.500 | A | 0.3861 | Transmitted | 0.353 | 1.417 [0.677–2.966] | 0.257 | 0.182 |
| rs741999 | 0.680 | 0.444 | A | 0.4444 | Transmitted | 0.336 | 1.455 [0.675–3.134] | 0.479 | 0.237 | |
| CYBA | rs4673 | 0.319 | 0.199 | T | 0.1795 | Untransmitted | 0.317 | 0.600 [0.218–1.651] | 0.564 | 0.251 |
| rs2306422 | 0.661 | 0.500 | T | 0.494 | Transmitted | 0.739 | 1.118 [0.581–2.150] | 0.655 | 0.876 | |
| CYBB | rs5963310 | 0.239 | 0.0274 | A | 0.09848 | Untransmitted | 0.0578 | 0.250 [0.0531–1.177] | 0.0833 | 0.0126 |
| rs9330580 | 0.663 | 0.206 | A | 0.1203 | Untransmitted | 0.366 | 0.571 [0.167–1.952] | 0.0833 | 0.109 | |
Suggestive p values (p < 0.05) are indicated in bold. Results are presented not corrected for multiple testing (total number of performed analyses for each test: 11; Bonferroni threshold for significance: p < 0.0045). 1 Analyses were run both considering MS frequency of 0.01 and penetrance of 0.05, as well as considering MS frequency of 0.0006 and penetrance of 0.76 (identical results). 2 In the case of multiallelic markers (D7S1870, D7S2518), the minor allele corresponds to the most frequent repeat, which was analyzed against all the others clumped together. 3 Indication referred to the minor allele. 4 p par (p values for parents) and p comb (combined p value, deriving from the asymptotic TDT p value and the p par value). NADPH, nicotinamide adenine dinucleotide phosphate; CI, confidence interval; HRR, haplotype relative risk; MAF, minor allele frequency; OR, odds ratio; TDT, transmission disequilibrium test.
2.2. CYBB Is Differentially Expressed in MS Cases
To verify the possible functional relevance of NADPH genes for MS predisposition, we performed a genome-wide microarray-based gene expression analysis. We extracted total RNA from CD4+ and CD4− samples isolated from peripheral blood mononuclear cells (PBMCs) of 21 individuals from five Finnish multiplex MS families. An adequate amount of high-quality total RNA was obtained from nine MS cases (four males, five females) and ten controls (four males, six females) for the CD4− cells, and from nine cases (five males, four females) and 11 controls (six males, five females) for the CD4+ cells. All patients had a relapsing-remitting (RR) subtype, i.e., the most common form of MS (~80% of cases), which is characterized by periods of acute intensification of symptoms followed by phases of almost complete remission [31].
To screen for differential expression of the genes of interest in MS, we specifically searched for probe sets corresponding to NCF1, NCF2, NCF4, CYBA, and CYBB in the NetAffx annotation site (http://www.affymetrix.com/analysis/index.affx). The 11 identified probes were individually searched in the University of California Santa Cruz (UCSC) Genome Browser (hg19; Blat search) to identify the correct target gene (http://genome.ucsc.edu/cgi-bin/hgGateway). Probe sets recognizing only intronic sequences or mapping to several loci were excluded: in this way expression data were available for seven probes mapping in the five genes.
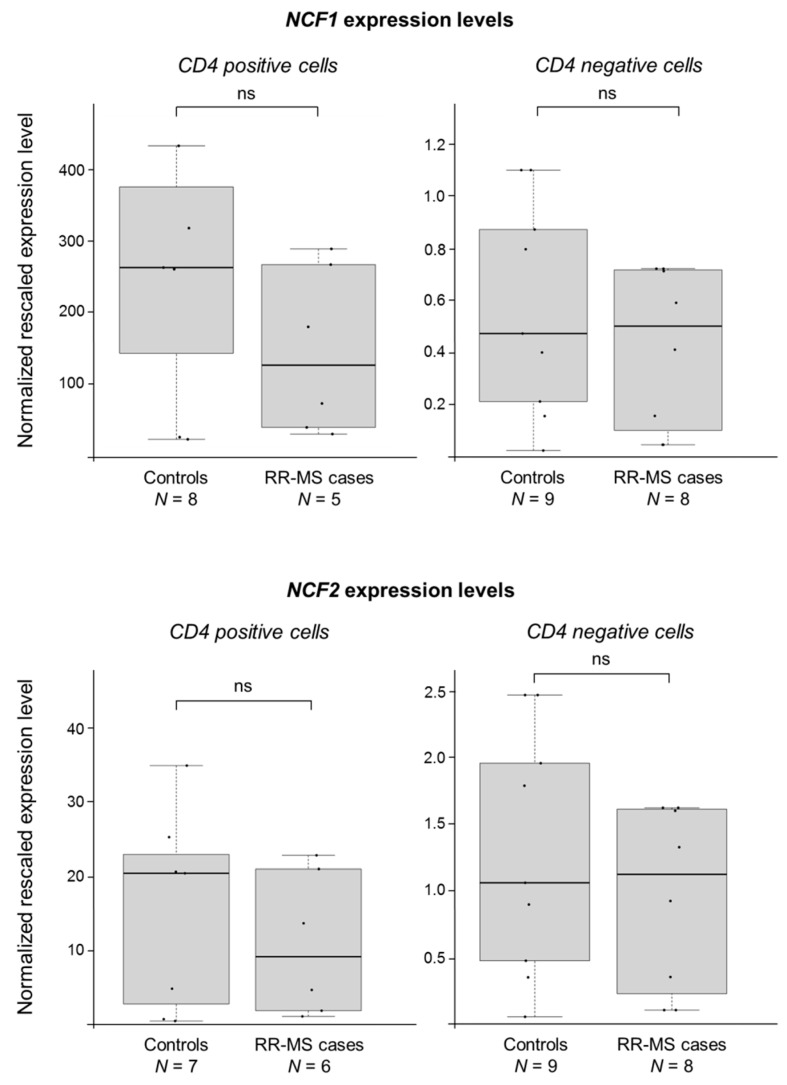
As for the NCF1 gene, none of the probes were mapping correctly/specifically. As for NCF2, it did not show appreciable expression signals in any of the CD4+/CD4− samples. We hence monitored for potential differential expression of these two genes by using the more sensitive semi-quantitative real-time reverse transcription-PCR (RT-PCR) approach. Figure 1 shows the results of RT-PCR assays: no significant difference between cases and controls in the expression levels of either genes was evidenced (no significant difference between cases and controls was seen even stratifying individuals on the basis of the D7S1870 genotype in NCF1; data not shown).
Figure 1.
NCF1 and NCF2 do not show any significant difference between MS cases and controls. Boxplots show the expression levels of the NCF1 and NCF2 genes measured by semi-quantitative real-time RT-PCR in CD4+ and CD4− cells (prepared from peripheral blood mononuclear cells, PBMCs). MS cases and controls belong to five multiplex Finnish families (comprising both male and female individuals). Boxes define the interquartile range; the thick line refers to the median. Results were normalized to expression levels of the HMBS and ACTB housekeeping genes and are presented as rescaled values. The number of subjects belonging to each group is indicated (N). The significance level of t-tests was in all cases not significant (ns).
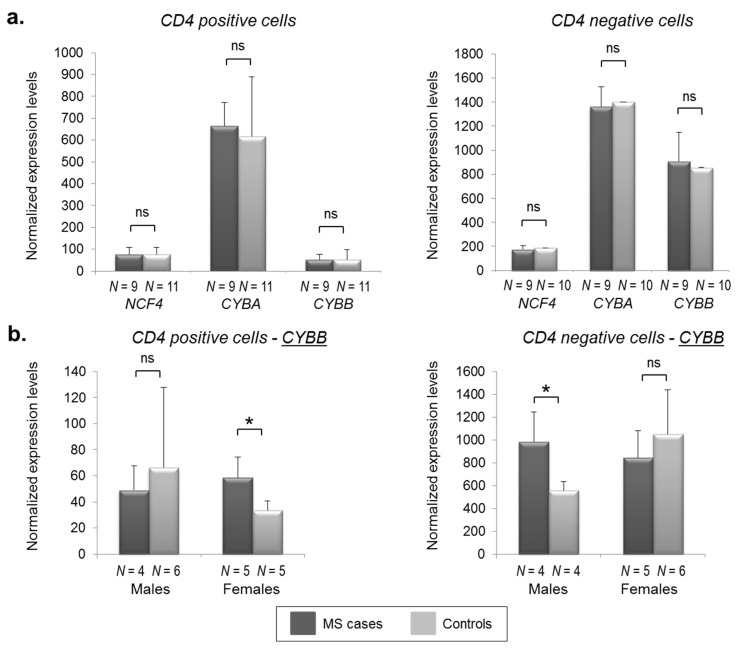
Concerning NCF4, CYBA, and CYBB, probes were correctly mapping on the corresponding transcripts and gave appreciable signals. In Figure 2a, differential expression analysis for MS cases and controls is shown for both CD4− and CD4+ cells, again indicating no differences between the two groups. However, considering that the CYBB gene is located on chromosome X, we stratified patients according to gender. Interestingly, this analysis allowed us to highlight an opposite molecular signature not only between males and females, but also between CD4− and CD4+ cells (Figure 2b). In particular, we found a 1.76-fold significant up-regulation (p = 0.021) of CYBB in RR-MS male patients in CD4− cells (with females showing an opposite, though not significant, trend; 0.80-fold decrease). Conversely, in CD4+ cells, we noticed the opposite situation, with a 1.75-fold significant CYBB increased expression in RR-MS females patients (p = 0.014) and a 0.74-fold decrease (not significant) in MS male cases compared to healthy controls.
Figure 2.
CYBB shows sex-related and cell-specific differential expression in MS cases and controls. NCF4, CYBA, and CYBB expression levels (shown by histograms) were measured by means of microarray-based experiments (see the “Materials and Methods” section). The number of subjects belonging to each group is indicated (N). Error bars represent means + SD (standard deviation). Significance levels of t-tests are shown. *: p < 0.05.
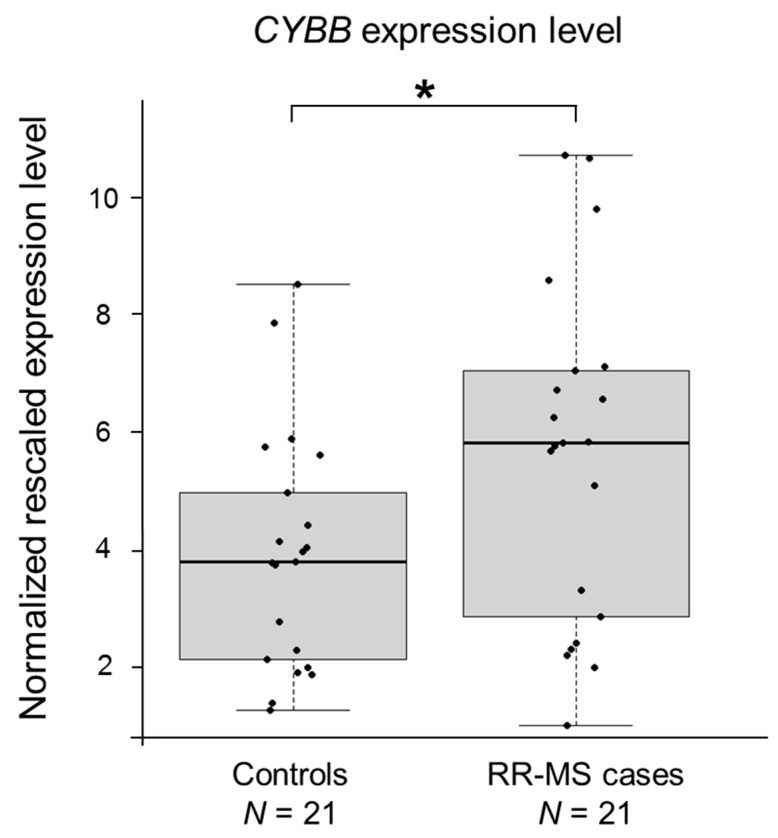
To validate the most interesting result obtained from the microarray profiling, expression of CYBB was quantitated by real-time RT-PCR assays, performed on RNA extracted from PBMCs of an independent cohort of 21 RR-MS Italian cases and an equal number of age-matched controls. Only females were included in this analysis. We found a 1.43-fold significant up-regulation (p = 0.032) of CYBB in RR-MS patients (Figure 3), which is consistent with the high abundance of CD4+ cells in the heterogeneous pool of PBMCs (25–60% of PBMC is composed of CD4+ cells) [32].
Figure 3.
CYBB is upregulated in MS patients. Boxplots show the expression levels of the CYBB gene measured by semi-quantitative real-time RT-PCR in PBMCs of an Italian case-control cohort (only female individuals). Boxes define the interquartile range; the thick line refers to the median. Results were normalized to expression levels of the HMBS and ACTB housekeeping genes and are presented as rescaled values. The number of subjects belonging to each group is indicated (N). Significance levels of t-tests are shown. * p < 0.05.
3. Discussion
Apart from the well-known implication of mutations affecting the NOX2 complex in the pathogenesis of chronic granulomatous disease (CGD) [33], some information is currently available in the literature on the possible predisposing role of polymorphisms in NCF1, NCF2, NCF4, CYBA, and CYBB genes in other immune disorders. Among others, missense variants in the NCF1 gene have been associated with systemic lupus erythematosus (SLE), Sjögren’s syndrome, and rheumatoid arthritis (RA) [34]; two missense polymorphisms in NCF2 were described as predisposing to SLE [35]; and common polymorphisms in NCF4 were associated both with RA and Crohn’s disease [36,37]. In our exploratory study, we aimed at investigating the possible genetic association between MS and all of the five genes coding for the NADPH oxidase complex. We found suggestive association signals for the NCF1 and CYBB genes (lowest p = 0.038 and p = 0.013, respectively), which were paralleled, in the case of CYBB, by a gender-specific differential gene expression between MS cases and controls.
However, we acknowledge the limited size of our study material, and a lack of a replication step confirming the association of CYBB with MS. No significant association between the CYBB locus and MS was reported in the previous GWAS analyses, which, however, excluded the X chromosome harboring the CYBB gene from the genome-wide analysis. The role of the X chromosome in MS—a pathology characterized by higher susceptibility in females than males—remains still largely unknown. For instance, in the largest MS-related meta-analysis so far published, genome-wide, a total of 233 loci were found to be significantly associated with the disease, and only one maps to chromosome X [10]. The case-control analysis did not take gender into account, while the study presented here was utilizing family-based transmission analysis. Overall, the lack of association signals on chromosome X is indeed a common feature of complex traits: the X chromosome accounts for 5% of the nuclear genome and underlies almost 10% of Mendelian disorders [38]. Nonetheless, only 114 associations (0.8%) at p ≤ 5 × 10−8 have been reported on the X chromosome, on a total of more than 14,700 signals identified by GWAS for ~300 traits [39]. These data could potentially be explained by the fact that the vast majority of GWAS have not included chromosome X in their analyses and have not considered gender. There is also a lack of specific bioinformatics pipelines to adopt in the analytic steps [40].
To overcome the limitation of the lack of a replication cohort in the association study, we focused on the investigation of an “intermediate phenotype”, i.e., the gene expression profile of NOX2 components in CD4+ and CD4− cells. We used a two-tiered approach, based on both microarray and real-time RT-PCR experiments. The most striking result was the differential gene expression for the CYBB gene in MS cases, with opposite molecular signatures not only between males and females, but also between CD4− and CD4+ cells. Indeed, sex-differential expression has recently emerged as a common feature of many genes. For instance, by comprehensively analyzing the Genotype-Tissue Expression (GTEx) data across 53 tissues (publicly available at https://www.gtexportal.org/home/), Gershoni and Pietrokovski were able to demonstrate that more than 6500 protein-coding genes showed significant sex-differential expression in one or more tissues [41]. In most of the cases, these genes are differentially expressed according to gender in just one or a few tissues. The CYBB gene seems to conform to this rule, showing a certain degree of sex-differential expression in kidney cortex, bladder, and thyroid (see Figure S1 in the Supplementary Materials); similar observations can be made for NCF1, NCF2, NCF4, and CYBA. Of note, data relative to blood cell sub-populations are missing in the GTEx portal.
The differential gene expression evidenced by stratifying patients and controls on the basis of the gender and, above all, according to the cell type, could contribute to explaining—if confirmed in other immune disorders—some of the puzzling observations that have accumulated over the years on CGD, SLE, or RA. For instance, RA patients show in blood a marked increase in ROS formation, protein oxidation, as well as lipid peroxidation [42]. In addition, they also present a synovial tissue that is invaded by inflammatory cells, a large proportion of which are activated CD4+ T cells [43]. Both these phenomena can collectively account for tissue damage and for the chronicity of the disease. However, the development of RA has been associated with a lower copy number of the NCF1 gene [44], and this association was supported by ncf1 mutant rodents [45,46]. In this regard, it could be speculated that lower expression levels of the NOX2 complex could be protective against the chronic inflammation in RA relevant tissues; on the other hand, a lower oxidative burst response in antigen-presenting cells, especially in the thymus during priming, could change the antigen-presentation capacity of the cells, thus inducing autoimmunity [46]. Besides RA and SLE, previously shown examples of immune pathology characterized by inappropriate over-activation or down-regulation of ROS also include psoriasis, Hashimoto thyroiditis, vitiligo, and inflammatory bowel disease [47,48].
Concerning specifically MS, our results well fit with the observations of Fischer and colleagues [16], who described a global NOX2 over-expression in microglia and infiltrating macrophages of MS patients’ autopsy brain tissues. These data also suggest that an inflammation-associated oxidative burst could play a fundamental role in the demyelination process typical of MS.
In conclusion, our work adds another piece of information on the possible involvement of NOX2 in the pathogenesis of MS, suggesting that this particular topic deserves to be further investigated, especially in the light of potential therapies based on decreasing/enhancing the oxidative burst.
4. Materials and Methods
4.1. Materials
All oligonucleotides for SNP genotyping and quantitative real-time RT-PCRs were purchased from Proligo (Paris, France). Primer couples for microsatellite analysis were from Applied Biosystems (Foster City, CA, USA). All sequences can be provided on request. The AmpliTaq Gold, DynaZyme, and HotStar Taq DNA polymerases were from Applied Biosystems, Finnzymes (Espoo, Finland), and Qiagen (Hilden, Germany), respectively.
4.2. MS Pedigrees
The study was approved by the ethical committee of the Helsinki and Uusimaa hospital district (Decision 46/2002, #192/E9/02), and all individuals included in the study gave their informed consent. The study material consisted of a total of 63 families: 22 of them were multiplex MS families, having two to six affected cases per pedigree, whereas the remaining 41 families were composed of MS patients with their parents and unaffected siblings. These families were previously described [49] and they all originate from the high-risk region for MS in Southern Ostrobothnia, Finland. In total, the cohort consisted of 547 individuals, 116 being MS cases. Patient selection was hospital-based; only definite cases were included (clinically or laboratory-supported definite). Diagnosis of MS in affected individuals strictly followed Poser’s diagnostic criteria [50].
4.3. Genotyping
Genomic DNA was extracted from peripheral blood cells following standard procedures.
Two non-chimeric microsatellite markers (D7S1870 and D7S2518) were selected from databases for the association analysis of the NCF1 gene, avoiding the highly duplicated regions characterizing this chromosomal locus (7q11.23). Microsatellite markers were PCR amplified in two-plex format in 10-μL reaction mixtures under standard conditions, using VIC (D7S1870) or PET (D7S2518) labeled primers, the AmpliTaq Gold DNA polymerase, a MJ Tetrad thermal cycler (MJ Research INC, Waltham, MA, USA), and a touch-down thermal profile. PCR products were run on an ABI-3730 Genetic Analyzer (Applied Biosystems) and analyzed using the Genotyper software (Applied Biosystems).
For the association analysis of the other genes, we selected 21 SNPs, having heterozygosity >10%. Primers flanking the putative SNPs were designed for multiplex PCR amplification using the web-based program MPprimer (https://omictools.com/mpprimer-tool?t=tab-tool-variant-0). SNPs were first tested for polymorphic content by direct sequencing the relevant PCR-amplified fragment in 32 healthy Finnish individuals. Nine SNPs were monomorphic in the Finnish population, whereas one was tri-allelic; those ten SNPs were not further investigated. The remaining 11 SNPs were typed in MS samples using an in-house developed microarray, based on allele-specific primer extension [51]. In particular, two allele-specific detection oligonucleotides were designed for each SNP: these primers were characterized by the presence of a 5’ aminolinker and a stretch of 9T (poly-T), followed by the SNP-specific sequence. The 5’ aminolinker and the poly-T served to spot and anchor detection oligonucleotides onto silane/isothiocyanate-coated chrome mirror microscope slides (Evaporated Coatings, Willow Grove, PA, USA). Spotting was performed in duplicate using an OmniGrid 100 arrayer (Discovery Scientific, Vancouver, BC, Canada). Regions containing the SNPs were PCR amplified with a touch-down protocol from genomic DNA in two multiplex reactions, using SNP-specific primers with a T3 (forward) or a T7 (reverse) tail. Multiplex reactions were in-vitro transcribed using the AmpliScribe T7 or T3 High Yield Transcription Kit (Epicentre Technologies, Madison, WI, USA). The DNAseI-treated T3 or T7 RNA pools were hybridized to the arrays in a hybridization buffer (1.67 M NaCl) at 42 °C for 20 min. Arrays were washed twice in a washing buffer (0.3 M NaCl, 0.5× TE, 0.1% TritonX-100) and subsequently rinsed in ice-cold water. The allele-specific extension was performed using the MMLV Reverse Transcriptase (Epicentre Technologies) in a 20-μL reaction containing 50 mM Tris-HCl (pH 8.3), 10 mM MgCl2, 75 mM KCl, 10 mM DTT, 0.5 μM dATP, 0.5 μM dGTP, 0.5 μM ddATP, 0.5 μM ddGTP, 1 μM Cy5-labeled dCTP, 1 μM Cy5-labeled dUTP (Cy5-labeled nucleotides were from Amersham Pharmacia Biotech, Uppsala, Sweden), 15% glycerol, and 0.24 M trehalose. Reverse transcription was carried out at 52 °C for 20 min. Finally, arrays were put in washing buffer under mild agitation for 15 min, dipped quickly in 50 mM NaOH, washed again in washing buffer, rinsed in ice-cold water, air dried, and scanned using a ScanArray 4000 instrument (Packard Biochip Technologies, Perkin Elmer Life Sciences, Boston, MA, USA). The image was analyzed using the ScanArray software (Packard Biochip Technologies, Perkin Elmer Life Sciences). Genotypes were called using an in-house developed software (W. Wong and C. Li, unpublished data).
The list of selected polymorphisms is presented in the Supplementary Materials (Table S1).
Important note: the entire analysis was performed a while ago, i.e., before the genomic databases became available with all the associated information on allele frequencies in different ethnicities.
4.4. Statistical Analyses
All genotypes were checked for Mendelian errors using PedCheck [52]. The check for the HWE was performed using the Genepop program (option 1), which also admits multiallelic markers [53]; only genotypes of women were considered for X-linked SNPs.
TDT [54] and HRR (a modified TDT test that uses siblings as “pseudo-controls”) [55] analyses, both implemented in the ANALYZE package [55], were used in the association study. Since the inheritance pattern of MS is unknown, analyses were performed using two different modes of inheritance, i.e., dominant with reduced penetrance of f = 0.05 (with disease allele-frequency estimate of 0.01) or f = 0.76 (with disease allele-frequency estimate of 0.0006) [56].
Analyses were repeated using the PLINK software v.1.07 [57] by applying a more powerful method based on the TDT statistic implemented with the parental discordance test. This test is based on counting the number of alleles in affected versus unaffected parents (using each nuclear-family parental pair as a matched pair). These counts are then combined with the transmitted and untransmitted counts of the basic TDT to give a combined test statistic [58]. In this analysis, multiallelic markers were analyzed as biallelic, considering from one hand the most common allele, and from the other all the remaining ones clumped together as they were a unique alternative allele. In the text, p values, ORs, and 95% CIs always referred to the minor allele.
Considering the exploratory hypothesis-generating nature of the present study and the limited sample size as compared to the number of investigated genetic variants, we did not adjust for multiple testing, and p values < 0.05 were considered to be statistically significant. However, the total number of performed tests as well as the threshold of significance based on the over-conservative Bonferroni correction has been indicated in the footnote of the relevant table.
4.5. RNA Samples
Fresh blood samples from 25 members (both male and female subjects) belonging to five of the above-mentioned multiplex MS families were collected. PBMCs were separated using a Ficoll-Paque (Amersham Pharmacia Biotech) density gradient centrifugation, and hence stratified to CD4+ and CD4− cell populations by negative selection using the CD4+ T-cell isolation kit (Miltenyi Biotec, Auburn, CA, USA) and an autoMACS instrument (Miltenyi Biotec). Total RNA was extracted from CD4+ and CD4− samples using the Trizol reagent (Ambion, Austin, TX, USA), treated with DNAse-I, and further purified with RNeasy Mini Kit columns (Qiagen, Valencia, CA, USA) according to the manufacturers’ instruction. RNA quality was assessed using the RNA 6000 Nano assay in a Bioanalyzer (Agilent Technologies, Foster City, CA, USA).
As for the replication cohort, we collected PBMCs from 21 unrelated RR-MS patients and 21 healthy subjects, all females and Caucasians (coming from Northern Italy). Controls were age-matched with cases. To avoid possible confounding effects, we focused only on RR-MS cases in remitting phase, who had not received any immune-modulatory therapy within the month prior to blood withdrawal. PBMCs were isolated from heparinized blood immediately after phlebotomy by centrifugation on a Lympholyte Cell separation media (Cederlane Laboratories Limited, Hornby, ON, Canada) gradient. Total RNA was isolated using the Eurozol kit (Euroclone, Wetherby, UK).
4.6. Sample Preparation, Microarray Processing, and Data Analysis
RNA samples were prepared for hybridization on the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA, USA) according to the manufacturer’s recommendations. In brief, 1 μg of total RNA was converted to biotin-labelled cRNA using the Affymetrix HT One-Cycle cDNA Synthesis Kit and the HT IVT Labelling Kit. Fifteen μg of cRNA were then fragmented and hybridized for 16 hours at 45 °C, washed in Affymetrix Fluidics Station 450, and then scanned with Affymetrix GeneChip Scanner 3000. Hybridization, washing, staining, and scanning were conducted using the same instruments for all samples.
Raw intensity data files were imported to GeneSpring 7.3 (Agilent Technologies) and GC Robust Multi-array Average (GC-RMA) normalized. We then excluded probe sets with low signal intensity (GC-RMA normalized signal < 50). Probe sets with a p ≤ 0.05 were considered to be differentially expressed.
4.7. Semi-Quantitative Real-Time RT-PCRs
Random hexamers and the TaqMan Gold RT-PCR kit (Applied Biosystems) were used to perform first-strand cDNA synthesis starting from 1 μg of total RNA from CD4+/− cells, according to the manufacturer’s instructions. From a total of 20 µL of the RT reaction, 4 µL were used as template for semi-quantitative real-time RT-PCRs for the quantitation of the NCF1 and NCF2 mRNAs. Assays were carried out using the SYBR-Green Kit (Applied Biosystems) and a standard PCR thermal protocol on the ABI Prism 7900 HT Sequence Detection System. Data were analyzed with the Sequence Detector version 2.0 software (Applied Biosystems).
As for CYBB evaluation, random hexamers and the Superscript-III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) were used in the RT step starting from 1 µg of RNA extracted from PBMCs of MS patients and controls. From a total of 20 µL of the RT reaction, 1 µL was used as a template for amplifications, using the FastStart SYBR Green Master Mix (Roche, Basel, Switzerland) on a LightCycler 480 (Roche), following a touch-down thermal protocol. Data were analyzed by the GeNorm software [59].
In all cases, RT-PCRs were performed at least in triplicate. Expression levels were normalized using HMBS (hydroxymethylbilane synthase gene) and ACTB (β-actin) as housekeeping genes. Primer sequences can be provided on request.
Acknowledgments
The European Science foundation is acknowledged for the support to R.A. (Grant program GENOMICS/2003/19).
Abbreviations
| ACTB | β-actin |
| CD4+ | Cluster of differentiation 4 positive |
| CD4− | Cluster of differentiation 4 negative |
| CGD | Chronic granulomatous disease |
| CI | Confidence interval |
| CNS | Central nervous system |
| CYBA | Cytochrome B-245 alpha chain, coding for the p22phox subunit of the NOX2 system |
| CYBB | Cytochrome B-245 beta chain, coding for the gp91phox subunit of the NOX2 system |
| GC-RMA | GC Robust Multi-array Average |
| GWAS | Genome-wide association study |
| HLA | Human leukocyte antigen |
| HMBS | Hydroxymethylbilane synthase gene |
| HRR | Haplotype relative risk |
| HWE | Hardy-Weinberg equilibrium |
| MMLV | Moloney Murine Leukemia Virus |
| MS | Multiple sclerosis |
| NADPH | nicotinamide adenine dinucleotide phosphate |
| NCF1 | Neutrophil cytosolic factor 1, coding for the p47phox subunit of the NOX2 system |
| NCF2 | Neutrophil cytosolic factor 2, coding for the p67phox subunit of the NOX2 system |
| NCF4 | Neutrophil cytosolic factor 4, coding for the p40phox subunit of the NOX2 system |
| NOX | NADPH oxidase system |
| NOX2 | NADPH oxidase system 2 (isoform 2 of the complex) |
| OMIM | Online Mendelian Inheritance in Men |
| OR | Odds ratio |
| OS | Oxidative stress |
| PBMCs | Peripheral blood mononuclear cells |
| RA | Rheumatoid arthritis |
| ROS | Reactive oxygen species |
| RR | Relapsing-remitting |
| RT-PCR | Reverse-transcription polymerase chain reactions |
| SD | Standard deviation |
| SLE | Systemic lupus erythematosus |
| SNP | Single nucleotide polymorphism |
| TDT | Transmission/disequilibrium test |
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/2227-9059/6/4/117/s1.
Author Contributions
R.A. and J.S. conceived and designed the experiments; R.A., G.C., and E.M.P. performed the experiments; R.A., G.S., S.D., and J.S. analyzed the data; R.A. wrote the paper; J.S. and R.A. supervised the entire study.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Trapp B.D., Peterson J., Ransohoff R.M., Rudick R., Mörk S., Bö L. Axonal transection in the lesions of multiple sclerosis. N. Engl. J. Med. 1998;338:278–285. doi: 10.1056/NEJM199801293380502. [DOI] [PubMed] [Google Scholar]
- 2.Hauser S.L., Oksenberg J.R. The neurobiology of multiple sclerosis: Genes, inflammation, and neurodegeneration. Neuron. 2006;52:61–76. doi: 10.1016/j.neuron.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Thompson A.J. Multiple sclerosis—A global disorder and still poorly managed. Lancet Neurol. 2008;7:1078–1079. doi: 10.1016/S1474-4422(08)70246-1. [DOI] [PubMed] [Google Scholar]
- 4.Kingwell E., Marriott J.J., Jetté N., Pringsheim T., Makhani N., Morrow S.A., Fisk J.D., Evans C., Béland S.G., Kulaga S., et al. Incidence and prevalence of multiple sclerosis in Europe: A systematic review. BMC Neurol. 2013;13:128. doi: 10.1186/1471-2377-13-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sumelahti M.L., Tienari P.J., Wikström J., Palo J., Hakama M. Regional and temporal variation in the incidence of multiple sclerosis in Finland 1979–1993. Neuroepidemiology. 2000;19:67–75. doi: 10.1159/000026241. [DOI] [PubMed] [Google Scholar]
- 6.International Multiple Sclerosis Genetics Consortium. Hafler D.A., Compston A., Sawcer S., Lander E.S., Daly M.J., De Jager P.L., de Bakker P.I., Gabriel S.B., Mirel D.B., et al. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 7.Sawcer S., Hellenthal G., Pirinen M., Spencer C.C., Patsopoulos N.A., Moutsianas L., Dilthey A., Su Z., Freeman C., Hunt S.E., et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.International Multiple Sclerosis Genetics Consortium (IMSGC) Beecham A.H., Patsopoulos N.A., Xifara D.K., Davis M.F., Kemppinen A., Cotsapas C., Shah T.S., Spencer C., Booth D., et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bos S.D., Berge T., Celius E.G., Harbo H.F. From genetic associations to functional studies in multiple sclerosis. Eur. J. Neurol. 2016;23:847–853. doi: 10.1111/ene.12981. [DOI] [PubMed] [Google Scholar]
- 10.Patsopoulos N., Baranzini S.E., Santaniello A., Shoostari P., Cotsapas C., Wong G., Beecham A.H., James T., Replogle J., Vlachos I., et al. The Multiple Sclerosis Genomic Map: Role of peripheral immune cells and resident microglia in susceptibility. bioRxiv. 2017 doi: 10.1101/143933. [DOI] [Google Scholar]
- 11.Linker R.A., Kieseier B.C., Gold R. Identification and development of new therapeutics for multiple sclerosis. Trends Pharm. Sci. 2008;29:558–565. doi: 10.1016/j.tips.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Broux B., Stinissen P., Hellings N. Which immune cells matter? The immunopathogenesis of multiple sclerosis. Crit. Rev. Immunol. 2013;33:283–306. doi: 10.1615/CritRevImmunol.2013007453. [DOI] [PubMed] [Google Scholar]
- 13.Salou M., Nicol B., Garcia A., Laplaud D.A. Involvement of CD8(+) T Cells in Multiple Sclerosis. Front. Immunol. 2015;6:604. doi: 10.3389/fimmu.2015.00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones A.P., Kermode A.G., Lucas R.M., Carroll W.M., Nolan D., Hart P.H. Circulating immune cells in multiple sclerosis. Clin. Exp. Immunol. 2017;187:193–203. doi: 10.1111/cei.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilgun-Sherki Y., Melamed E., Offen D. The role of oxidative stress in the pathogenesis of multiple sclerosis: The need for effective antioxidant therapy. J. Neurol. 2004;251:261–268. doi: 10.1007/s00415-004-0348-9. [DOI] [PubMed] [Google Scholar]
- 16.Fischer M.T., Sharma R., Lim J.L., Haider L., Frischer J.M., Drexhage J., Mahad D., Bradl M., van Horssen J., Lassmann H. NADPH oxidase expression in active multiple sclerosis lesions in relation to oxidative tissue damage and mitochondrial injury. Brain. 2012;135:886–899. doi: 10.1093/brain/aws012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adamczyk B., Adamczyk-Sowa M. New Insights into the Role of Oxidative Stress Mechanisms in the Pathophysiology and Treatment of Multiple Sclerosis. Oxidative Med. Cell. Longev. 2016;2016:1973834. doi: 10.1155/2016/1973834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van der Goes A., Brouwer J., Hoekstra K., Roos D., van den Berg T.K., Dijkstra C.D. Reactive oxygen species are required for the phagocytosis of myelin by macrophages. J. Neuroimmunol. 1998;92:67–75. doi: 10.1016/S0165-5728(98)00175-1. [DOI] [PubMed] [Google Scholar]
- 19.Nikić I., Merkler D., Sorbara C., Brinkoetter M., Kreutzfeldt M., Bareyre F.M., Brück W., Bishop D., Misgeld T., Kerschensteiner M. A reversible form of axon damage in experimental autoimmune encephalomyelitis and multiple sclerosis. Nat. Med. 2011;17:495–499. doi: 10.1038/nm.2324. [DOI] [PubMed] [Google Scholar]
- 20.Friese M.A., Schattling B., Fugger L. Mechanisms of neurodegeneration and axonal dysfunction in multiple sclerosis. Nat. Rev. Neurol. 2014;10:225–238. doi: 10.1038/nrneurol.2014.37. [DOI] [PubMed] [Google Scholar]
- 21.Barnes P.J., Karin M. Nuclear factor-kappaB: A pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 22.Srinivasan M., Lahiri D.K. Significance of NF-κB as a pivotal therapeutic target in the neurodegenerative pathologies of Alzheimer’s disease and multiple sclerosis. Expert Opin. Ther. Targets. 2015;19:471–487. doi: 10.1517/14728222.2014.989834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romanic A.M., Madri J.A. Extracellular matrix-degrading proteinases in the nervous system. Brain Pathol. 1994;4:145–156. doi: 10.1111/j.1750-3639.1994.tb00825.x. [DOI] [PubMed] [Google Scholar]
- 24.Merrill J.E., Murphy S.P. Inflammatory events at the blood brain barrier: Regulation of adhesion molecules, cytokines, and chemokines by reactive nitrogen and oxygen species. Brain Behav. Immun. 1997;11:245–263. doi: 10.1006/brbi.1997.0496. [DOI] [PubMed] [Google Scholar]
- 25.Chiurchiù V., Orlacchio A., Maccarrone M. Is Modulation of Oxidative Stress an Answer? The State of the Art of Redox Therapeutic Actions in Neurodegenerative Diseases. Oxidative Med. Cell. Longev. 2016;2016:7909380. doi: 10.1155/2016/7909380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vladimirova O., Lu F.M., Shawver L., Kalman B. The activation of protein kinase C induces higher production of reactive oxygen species by mononuclear cells in patients with multiple sclerosis than in controls. Inflamm. Res. 1999;48:412–416. doi: 10.1007/s000110050480. [DOI] [PubMed] [Google Scholar]
- 27.Lu F., Selak M., O’Connor J., Croul S., Lorenzana C., Butunoi C., Kalman B. Oxidative damage to mitochondrial DNA and activity of mitochondrial enzymes in chronic active lesions of multiple sclerosis. J. Neurol. Sci. 2000;177:95–103. doi: 10.1016/S0022-510X(00)00343-9. [DOI] [PubMed] [Google Scholar]
- 28.Van Horssen J., Witte M.E., Schreibelt G., de Vries H.E. Radical changes in multiple sclerosis pathogenesis. Biochim. Biophys. Acta. 2011;1812:141–150. doi: 10.1016/j.bbadis.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Babior B.M. NADPH Oxidase: An update. Blood. 1999;93:1464–1476. [PubMed] [Google Scholar]
- 30.Vignais P.V. The superoxide-generating NADPH oxidase: Structural aspects and activation mechanism. Cell. Mol. Life Sci. 2002;59:1428–1459. doi: 10.1007/s00018-002-8520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trapp B.D., Nave K.A. Multiple sclerosis: An immune or neurodegenerative disorder? Annu. Rev. Neurosci. 2008;31:247–269. doi: 10.1146/annurev.neuro.30.051606.094313. [DOI] [PubMed] [Google Scholar]
- 32.Kleiveland C.R. Peripheral Blood Mononuclear Cells. In: Verhoeckx K., Cotter P., López-Expósito I., Kleiveland C., Lea T., Mackie A., Requena T., Swiateck D., Wicher H., editors. The Impact of Food Bioactives on Health. Springer; Cham, Switzerland: 2015. [DOI] [PubMed] [Google Scholar]
- 33.Wu J., Wang W.F., Zhang Y.D., Chen T.X. Clinical Features and Genetic Analysis of 48 Patients with Chronic Granulomatous Disease in a Single Center Study from Shanghai, China (2005–2015): New Studies and a Literature Review. J. Immunol. Res. 2017;2017:8745254. doi: 10.1155/2017/8745254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J., Ma J., Deng Y., Kelly J.A., Kim K., Bang S.Y., Lee H.S., Li Q.Z., Wakeland E.K., Qiu R., et al. A missense variant in NCF1 is associated with susceptibility to multiple autoimmune diseases. Nat. Genet. 2017;49:433–437. doi: 10.1038/ng.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Armstrong D.L., Eisenstein M., Zidovetzki R., Jacob C.O. Systemic lupus erythematosus-associated neutrophil cytosolic factor 2 mutation affects the structure of NADPH oxidase complex. J. Biol. Chem. 2015;290:12595–12602. doi: 10.1074/jbc.M115.639021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsson L.M., Lindqvist A.K., Källberg H., Padyukov L., Burkhardt H., Alfredsson L., Klareskog L., Holmdahl R. A case-control study of rheumatoid arthritis identifies an associated single nucleotide polymorphism in the NCF4 gene, supporting a role for the NADPH-oxidase complex in autoimmunity. Arthritis Res. Ther. 2007;9:R98. doi: 10.1186/ar2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rioux J.D., Xavier R.J., Taylor K.D., Silverberg M.S., Goyette P., Huett A., Green T., Kuballa P., Barmada M.M., Datta L.W., et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amberger J., Bocchini C.A., Scott A.F., Hamosh A. McKusick’s Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J., et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wise A.L., Gyi L., Manolio T.A. eXclusion: Toward integrating the X chromosome in genome-wide association analyses. Am. J. Hum. Genet. 2013;92:643–647. doi: 10.1016/j.ajhg.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gershoni M., Pietrokovski S. The landscape of sex-differential transcriptome and its consequent selection in human adults. BMC Biol. 2017;15:7. doi: 10.1186/s12915-017-0352-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mateen S., Moin S., Khan A.Q., Zafar A., Fatima N. Increased Reactive Oxygen Species Formation and Oxidative Stress in Rheumatoid Arthritis. PLoS ONE. 2016;11:e0152925. doi: 10.1371/journal.pone.0152925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kondo Y., Yokosawa M., Kaneko S., Furuyama K., Segawa S., Tsuboi H., Matsumoto I., Sumida T. Review: Transcriptional Regulation of CD4+ T Cell Differentiation in Experimentally Induced Arthritis and Rheumatoid Arthritis. Arthritis Rheumatol. 2018;70:653–661. doi: 10.1002/art.40398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olsson L.M., Nerstedt A., Lindqvist A.K., Johansson S.C., Medstrand P., Olofsson P., Holmdahl R. Copy number variation of the gene NCF1 is associated with rheumatoid arthritis. Antioxid Redox Signal. 2012;16:71–78. doi: 10.1089/ars.2011.4013. [DOI] [PubMed] [Google Scholar]
- 45.Olofsson P., Holmberg J., Tordsson J., Lu S., Akerström B., Holmdahl R. Positional identification of Ncf1 as a gene that regulates arthritis severity in rats. Nat. Genet. 2003;33:25–32. doi: 10.1038/ng1058. [DOI] [PubMed] [Google Scholar]
- 46.Hultqvist M., Olofsson P., Holmberg J., Bäckström B.T., Tordsson J., Holmdahl R. Enhanced autoimmunity, arthritis, and encephalomyelitis in mice with a reduced oxidative burst due to a mutation in the Ncf1 gene. Proc. Natl. Acad. Sci. USA. 2004;101:12646–12651. doi: 10.1073/pnas.0403831101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kienhöfer D., Boeltz S., Hoffmann M.H. Reactive oxygen homeostasis—The balance for preventing autoimmunity. Lupus. 2016;25:943–954. doi: 10.1177/0961203316640919. [DOI] [PubMed] [Google Scholar]
- 48.Di Dalmazi G., Hirshberg J., Lyle D., Freij J.B., Caturegli P. Reactive oxygen species in organ-specific autoimmunity. Autoimmun. Highlights. 2016;7:11. doi: 10.1007/s13317-016-0083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saarela J., Kallio S.P., Chen D., Montpetit A., Jokiaho A., Choi E., Asselta R., Bronnikov D., Lincoln M.R., Sadovnick A.D., et al. PRKCA and multiple sclerosis: Association in two independent populations. PLoS Genet. 2006;2:e42. doi: 10.1371/journal.pgen.0020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poser C.M., Paty D.W., Scheinberg L., McDonald W.I., Davis F.A., Ebers G.C., Johnson K.P., Sibley W.A., Silberberg D.H., Tourtellotte W.W. New diagnostic criteria for multiple sclerosis: Guidelines for research protocols. Ann. Neurol. 1983;13:227–231. doi: 10.1002/ana.410130302. [DOI] [PubMed] [Google Scholar]
- 51.Pastinen T., Raitio M., Lindroos K., Tainola P., Peltonen L., Syvänen A.C. A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res. 2000;10:1031–1042. doi: 10.1101/gr.10.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Connell J.R., Weeks D.E. PedCheck: A program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raymond M., Rousset F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995;86:248–249. doi: 10.1093/oxfordjournals.jhered.a111573. [DOI] [Google Scholar]
- 54.Spielman R.S., McGinnis R.E., Ewens W.J. Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am. J. Hum. Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 55.Terwilliger J.D., Ott J. A haplotype-based ‘haplotype relative risk’ approach to detecting allelic associations. Hum. Hered. 1992;42:337–346. doi: 10.1159/000154096. [DOI] [PubMed] [Google Scholar]
- 56.Kuokkanen S., Sundvall M., Terwilliger J.D., Tienari P.J., Wikström J., Holmdahl R., Pettersson U., Peltonen L. A putative vulnerability locus to multiple sclerosis maps to 5p14-p12 in a region syntenic to the murine locus Eae2. Nat. Genet. 1996;13:477–480. doi: 10.1038/ng0896-477. [DOI] [PubMed] [Google Scholar]
- 57.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;8:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Purcell S., Sham P., Daly M.J. Parental phenotypes in family-based association analysis. Am. J. Hum. Genet. 2005;76:249–259. doi: 10.1086/427886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.