Abstract
Warfarin is an oral anticoagulant frequently used in the treatment of different cardiovascular diseases. Genetic polymorphisms in the CYP2C9 and VKORC1 genes have produced variants with altered catalytic properties. A total of 212 cardiovascular patients were genotyped for 17 Single Nucleotide Polymorphisms (SNPs) within the CYP2C9 and VKORC1 genes. This study confirmed a genetic association of the CYP2C9*3 and VKORC1 rs10871454, rs8050894, rs9934438, and rs17708472 SNPs with warfarin sensitivity. This study also found an association between CYP2C9 and VKORC1 genetic haplotype blocks and warfarin sensitivity. The initial warfarin dose was significantly related to the CYP2C9*3 polymorphism and the four VKORC1 SNPs (p < 0.001). There were significant associations between rs4086116 SNP and TAT haplotype within CYP2C9 gene and rs17708472 SNP and CCGG haplotype within VKORC1 gene and warfarin responsiveness. However, possessing a VKORC1 variant allele was found to affect the international normalized ratio (INR) outcomes during initiation of warfarin therapy. In contrast, there was a loose association between the CYP2C9 variant and INR measurements. These findings can enhance the current understanding of the great variability in response to warfarin treatment in Arabs.
Keywords: CYP2C9, VKORC1, warfarin, warfarin initiation phase of therapy, INR, pharmacogenetics study
1. Introduction
Warfarin is a commonly prescribed oral anticoagulant that is employed for the treatment of venous and arterial thromboembolic disorders and cardiac valve replacements [1]. However, interindividual genetic variation causes great variability in dosage requirements, making the latter a problematic issue for physicians. A higher or lower dose than needed could lead to bleeding and thrombotic risk, respectively [2,3]. Two-thirds of warfarin dose variation was due to environmental factors like age, body mass index, smoking status, gender, and diet, among others, while the remaining one-third is caused by genetic factors such as the CYP2C9 and VKORC1 genes [4,5,6].
Belonging to the cytochrome P450 superfamily, the CYP2C9 gene is involved in the metabolism and clearance of S-warfarin, the latter of which is a racemic form of warfarin together with R-warfarin [7,8]. CYP2C9 is located on the long arm of chromosome 10, and like other members of CYP2C, CYP2C9 is highly polymorphic [9,10,11]. Although there are over 50 single nucleotide polymorphisms (SNPs) located in the regulatory and coding region of the CYP2C9 sequence, the most studied CYP2C9 polymorphisms are CYP2C9*2 (R144C) and CYP2C9*3 (I359L) [12,13]. It was found that individuals carrying the CYP2C9 *2 *3 alleles reduced the elimination of S-warfarin and, therefore, plasma concentrations of the latter increased significantly compared to the wild-type allele and the individuals with these variant alleles need a lower warfarin maintenance dose [14].
Similarly, the VKORC1 gene encodes the vitamin K epoxide reductase complex subunit 1 and it is a warfarin target [5]. The vitamin K epoxide reductase complex subunit 1 normally catalyzes the carboxylation reaction of the vitamin K-dependent protein glutamic acid residues in order to activate it, the latter of which are responsible for catalyzing the clotting factor pathway [15]. Several genetic studies conducted in different populations suggest that the G3673A (rs9923231), C6484T (rs9934438), and G9041A (rs7294) polymorphisms of the VKORC1 gene are the most common and well-studied [7,16].
Analysis of CYP2C9 and VKORC1 gene polymorphisms revealed that they were responsible for 10% and 40% of warfarin dose requirement variance, respectively [17]. Combined with clinical data, both of the aforementioned genes can explain up to 60% of the warfarin variance [18,19]. More than 50 years ago, the dose of warfarin was determined by trial and error, with an initial dose (2–10 mg/day) dependent on the indication of warfarin and clinical factors, regardless of the effect of the genetic factor [20]. Patients are treated with warfarin in two stages, the first called the initiation phase of treatment, which is considered as the stage in which the initial international normalized ratio (INR) value of the patient is unstable and fluctuates up and down. With the second (maintenance) stage of the therapy, the patient is within the therapeutic INR for at least two consecutive visits [21]. However, pharmacogenetics for specific populations including Jordanian Arabs is necessary. Therefore, the objective of this study was to recognize genetic variations within the CYP2C9 and VKORC1 genes that are involved in warfarin sensitivity and responsiveness in Jordanian cardiovascular patients of Arab descent during initiation of treatment.
2. Materials and Methods
2.1. Patient Population and Study Design
The study population consisted of 212 unrelated warfarin intake patients selected from the Jordanian-Arab population, from the Anticoagulation Clinic at the Queen Alia Heart Institute (QAHI) in Amman, Jordan. Informed consent was obtained from all subjects. The study protocol was approved by the Human Ethics Committee at Jordan University of Science and Technology in Irbid, Jordan, and the Royal Medical Services in Amman, Jordan. Ethical approval code: 13/78/2014.
In this study, patients have cardiovascular diseases and are prescribed warfarin as an anticoagulant therapy. Inclusion criteria involved patients being 18 years or older and having received warfarin for at least three months. Patients who did not provide informed written consent, did not visit the anticoagulation clinic regularly, took CYP2C9 inducers or inhibitors, or did not have a complete data set were excluded.
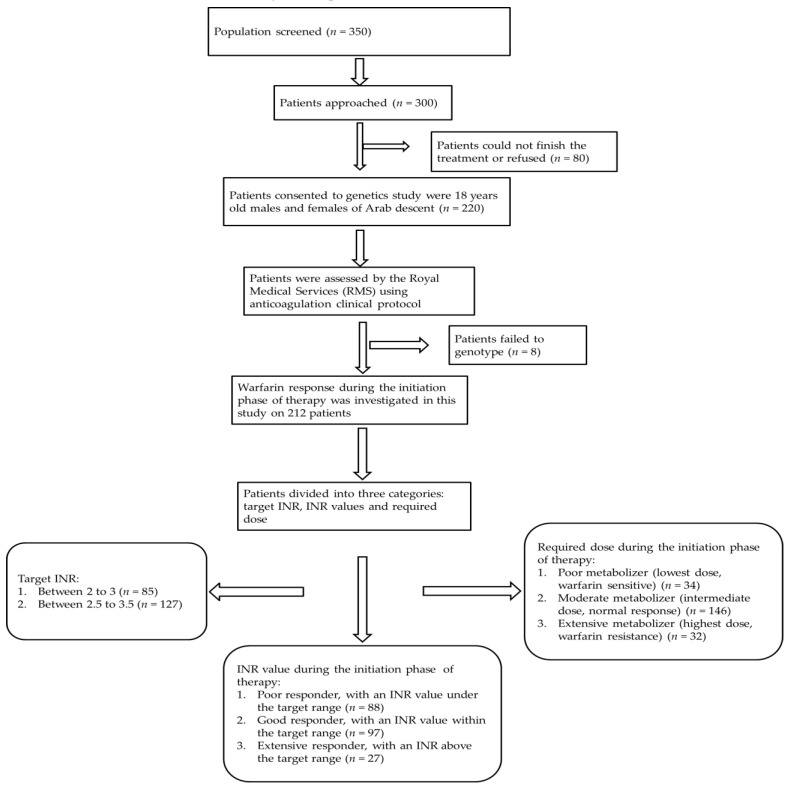
Initially, 350 patients were screened and, based on the aforementioned inclusion and exclusion criteria, 300 patients were approached to participate in this study (Figure 1). 80 patients were subsequently excluded because of inability to complete the treatment program or refusal. Of the remaining patients, 220 accepted to be part of this study, after which an additional eight patients were excluded from the final analysis because of a failure in genotyping. In total, whole sets of data were obtained from 212 patients with cardiovascular disease who were being treated with warfarin. Data was collected on demographic (age, gender, and body mass index) and lifestyle (smoking status and diet) characteristics as well as medical history (diseases and clinical features of warfarin therapy). Clinical features included the target INR, mean weekly warfarin doses required to reach the target INR, and the use of concomitant medication, all of which are required to be known in order to adjust warfarin doses. All data were blinded and obtained through semi-standardized interviews and medical records.
Figure 1.
Flow chart depicting study design. INR: international normalized ratio.
All subjects in this study received warfarin anticoagulant therapy according to RMS anticoagulation protocol, which began with 2.5 to 10 mg nightly doses. INR monitoring is required at least once a week for the first three to four weeks after the initiation of therapy. After three consecutive visits, a patient with stable INR reaches the maintenance dose and is monitored for warfarin treatment administration by clinic protocol.
2.2. Outcome Measurement
Oral anticoagulant therapy was mandated by the prothrombin time (PT) that is, evaluated using an automated method over STAGO coagulometric unit in the QAHI laboratory. To calculate INR, there was a blood coagulation (clotting) test. INR monitoring for dose adjustments was determined by the physician and pharmacist. INR values between January 2014 and November 2015 were obtained from medical records, and these values were then used to divide the patients according to warfarin responsiveness into: (1) good responders, with an INR value within the target range; (2) poor responders, with an INR value under the target range; and (3) extensive responders, with an INR above the target range.
Further, based on their warfarin sensitivity, patients were divided into resistant, normal, or sensitive to warfarin as follows: (1) warfarin resistance (or Poor metabolizer), largest daily doses were required to keep a patient’s INR within a therapeutic range (dose required > 49 mg/week); (2) warfarin normal patient (or Intermediate metabolizer), intermediate doses required (doses between 21–49 mg/week); and (3) warfarin sensitive patient (or Extensive metabolizer) lowest doses required (required dose <21 mg/week) [22].
2.3. SNP Selection, DNA Extraction, and Genotyping
In this study, 17 SNPs in the CYP2C9 and VKORC1 genes were selected from public databases and genotypes. Information about the aforementioned SNPs is shown in Table S1. Genomic DNA was extracted within one week of blood collection using the commercially available Wizard Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA) according to the manufacturer’s instructions. After extraction, the DNA was diluted in 96-well plates using an automated robotic system to achieve concentrations of 20 ng/μL (50–500 μL). Concentrations were confirmed with the Nano-Drop ND-100 (Thermo Scientific, Wilmington, DE, USA). Genotyping was carried out by means of the MassARRAY® system (iPLEX GOLD) (Sequenom, San Diego, CA, USA), which was carried out at the Australian Genome Research Facility (AGRF) (Sequenom).
2.4. Statistical Analysis
Discrepancy and call rates were calculated using Microsoft Excel, and the deviation from Hardy Weinberg Equilibrium (HWE) was assessed using the Pearson X2 test. Minor allele frequencies (MAF) and HWE p-values for genotypic distribution were calculated via the Court lab-HW calculator. To test which of the chosen SNPs is associated with warfarin response, various statistical genetic association analyses were conducted, such as the chi-square, nonparametric correlation tests (Kruskal-Wallis and Tukey Pairwise comparison) and haplotype genetic analysis test. The Statistical Package for the Social Sciences (SPSS) version 21.0 and the SNPStat Web Tool (https://www.snpstats.net/start.htm) were used to perform all analyses.
3. Results
3.1. Study Group
The study group comprised of 212 unrelated Jordanian-Arabs patients treated with warfarin, with a mean age (±SD) of 56.03 (±17.68) years, a median age of 60, and an age range of 18 to 85 years. There were 34 poor responders (16%), 146 moderate responders (68.9%), and 32 extensive responders (15.1%). Table 1 summarizes the demographic, lifestyle, and medical characteristics of each of the three groups.
Table 1.
Descriptive analysis of demographics and clinical characteristics of 212 cardiovascular patients treated with warfarin at the Queen Alia Heart Institute.
| Category | Subcategory | Extensive Metabolizer |
Good Metabolizers |
Poor Metabolizers |
|---|---|---|---|---|
| Demographics | Patients (N, %) | (32/212) 15.1% | (146/212) 68.9% | (34/212) 16% |
| Age a (years) | 56.0 (17.68) | 55.0 (14.64) | 48.29 (15.09) | |
| BMI a | 27.87 (3.72) | 27.7 (4.85) | 27.42 (3.45) | |
| Smoking (N, %) | 31.25% | 18.6% | 41.2% | |
| Male | 59.4% | 51.4% | 67.6% | |
| Female | 40.6% | 48.6% | 32.4% | |
| Concomitant Disease | Co morbidity | 56.3% | 68.5% | 55.9% |
| Hypertension | 34.4% | 42.5% | 23.5% | |
| Diabetes mellitus | 18.8% | 21.9% | 26.5% | |
| CHD b | 28.1% | 25.3% | 29.4% | |
| Thyroid | 0% | 3.4% | 2.9% | |
| Lipid | 3.1% | 6.8% | 2.9% | |
| Medication | Aspirin | 62.5% | 65.8% | 76.5% |
| Indication of Treatment | MVR c | 18.8% | 10.3% | 20.6% |
| AVR d | 6.3% | 24.0% | 20.6% | |
| AF e | 34.4% | 19.2% | 20.6% | |
| DVR f | 9.4% | 15.8% | 11.8% | |
| Others | 9.4% | 7.5% | 0.0% | |
| Target INR | 2–3 | 43.8% | 39.7% | 38.2% |
| 2.5–3.5 | 56.3% | 60.3% | 61.8% | |
| Mean weakly dose a | 16.699 (2.79) | 35.896 (7.39) | 67.44 (42.48) | |
| Mean INR a | 2.82 (0.72) | 2.38 (0.75) | 2.44 (0.83) |
a Mean Standard deviation in square brackets. b CHD: Chronic heart disease. c MVR:Mitral valve replacement. d AVR: Aortic valve replacement. e AF: Atrial Fibrillation. f DVR: Double valve replacement.
In total, 17 SNPs (100%) passed the quality control measures for throughput genotyping and were analyzed by the MassARRAY® system (iPLEX GOLD) with high accuracy and a 97% average success rate. The genotypic discrepancy average (±SD) rate over the 17 loci was only 0.06% (±0.0004%) out of the entire cohort (212 subjects). Genotypic and allelic frequencies are shown in Table S2.
For the 17 SNPs examined in this study, all were in accordance with the HWE. Ten polymorphisms (rs104894539, rs104894540, rs104894541, rs104894542, and rs61742245 in VKORC1, and rs28371685, rs28371686, rs72558191, rs9332131, and rs9332239 in CYP2C9) were non-polymorphic. In contrast, the seven remaining SNPs (rs10871454, rs8050894, rs9934438, and rs17708472 located in VKORC1, and rs1799853, rs4086116, and rs1057910 located in CYP2C9) were polymorphic and thus included in the study. The minor alleles and their frequencies for the successful genotyped SNPs are shown in Table S3.
3.2. Effect of CYP2C9 and VKORC1 Polymorphisms on Warfarin Sensitivity during Initiation Phase of Therapy
Regarding the association of VKORC1 and CYP2C9 SNPs with warfarin sensitivity among the three inclusion groups, significant differences in proportions among genotypes were observed at all tested VKORC1 SNPs (p < 0.001) (Table 2). Significant differences were also observed between two SNPs of the CYP2C9 gene (rs4086116 (p = 0.012) and rs1057910 (p < 0.001)), as shown in Table 2. Moreover, there was a significant association observed between VKORC1 and CYP2C9 haplotypes and warfarin sensitivity (p < 0.0001) (Table 3).
Table 2.
Association of VKORC1 and CYP2C9 single nucleotide polymorphism (SNPs) with warfarin sensitivity during the initiation phase of therapy of 212 cardiovascular patients.
| Gene | SNP ID | Genotype | Sensitive | Moderate | Resistance | p-Value * |
|---|---|---|---|---|---|---|
| VKORC1 | rs10871454 | CC | 4.3% | 57.4% | 38.3% | <0.001 |
| CT | 10.0% | 76.4% | 13.6% | |||
| TT | 34.5% | 63.6% | 1.8% | |||
| rs8050894 | CC | 2.3% | 60.5% | 37.2% | <0.001 | |
| CG | 10.9% | 74.5% | 14.5% | |||
| GG | 32.2% | 64.4% | 3.4% | |||
| rs9934438 | CC | 4.2% | 58.3% | 37.5% | <0.001 | |
| CT | 9.9% | 76.6% | 13.5% | |||
| TT | 35.8% | 62.3% | 1.9% | |||
| rs17708472 | CC | 18.1% | 68.8% | 13.1% | <0.001 | |
| CT | 6.1% | 73.5% | 20.4% | |||
| TT | 0.0% | 0.0% | 100% | |||
| CYP2C9 | rs1799853 | CC | 14% | 70.1% | 15.9% | 0.744 |
| CT | 19.6% | 63% | 17.4% | |||
| TT | 0.0% | 100% | 0.0% | |||
| rs4086116 | CC | 8.1% | 73.2% | 18.7% | 0.012 | |
| CT | 24.1% | 62.0% | 13.9% | |||
| TT | 30.0% | 70.0% | 0.0% | |||
| rs1057910 | AA | 8.8% | 72.5% | 18.7% | <0.001 | |
| AC | 41.5% | 53.7% | 4.9% |
* Chi-Square Test with p-value < 0.05 is considered significant.
Table 3.
Frequencies of the haplotypes of VKORC1 and CYP2C9 genes among the 212 warfarin sensitive patients.
| Gene | Haplotypes | Frequency * (%) | Odds Ratio (95% CI) | p-Value ** |
|---|---|---|---|---|
| VKORC1 | TGAG | 0.512 | 0.00 | ------ |
| CCGG | 0.324 | 0.32 (0.2–0.43) | <0.0001 | |
| CCGA | 0.129 | 0.38 (0.23–0.54) | <0.0001 | |
| CGGG | 0.028 | 0.34 (0.03–0.66) | 0.034 | |
| TCGG | 0.007 | 0.21 (−0.38–0.8) | 0.48 | |
| CYP2C9 | CAC | 0.767 | 0.00 | ----- |
| TAT | 0.116 | −0.05 (−0.21–0.11) | 0.53 | |
| TCC | 0.094 | −0.45 (−0.64–−0.27) | <0.0001 | |
| TAC | 0.021 | 0.01 (−0.34–0.37) | 0.95 | |
| TCT | 0.002 | −1.11 (−2.14–−0.08) | 0.037 |
* Genetic haplotype frequency of 212 warfarin intake patients, ** p-value < 0.05 is considered significant.
3.3. Effect of CYP2C9 and VKORC1 Polymorphisms on Warfarin Required Dose during Initiation Phase of Therapy
Carriers of CYP2C9 and VKORC1 polymorphisms had a significantly increased required dose compared with wild-type subjects or carriers of only one polymorphism of CYP2C9 or VKORC1 (Table 4).
Table 4.
Association of VKORC1 and CYP2C9 SNPs with variability on warfarin required doses and with INR treatment outcome.
| SNP ID | Initiation Dose | p-Value * | Initiation INR | p-Value * |
|---|---|---|---|---|
| rs10871454 | 38.1 (23.02) | <0.001 | 2.46 (0.77) | 0.006 |
| rs8050894 | <0.001 | 0.008 | ||
| rs9934438 | <0.001 | 0.009 | ||
| rs17708472 | <0.001 | 0.511 | ||
| rs1799853 | 0.118 | 0.184 | ||
| rs4086116 | 0.001 | 0.08 | ||
| rs1057910 | 0.001 | 0.572 |
* Kurskal Wallis test with p-value < 0.05 is considered significant, Mean Standard deviation in square brackets.
3.4. Effect of CYP2C9 and VKORC1 Polymorphisms on Warfarin Responsiveness during Initiation of Therapy
There were no significant differences in patient responder groups regarding the VKORC1 and CYP2C9 SNPs except for the VKORC1 rs17708472 (p = 0.042) and the CYP2C9 rs4086116 (p = 0.005) SNPs (Table 5). However, significant associations were found between genetic haplotypes of CCGG VKORC1 and TAT CYP2C9 and warfarin sensitivity, with p = 0.02 and p = 0.018, respectively (Table 6).
Table 5.
Association of VKORC1 and CYP2C9 SNPs with response to warfarin during the initiation phase of therapy of 212 cardiovascular patients.
| Gene | SNP ID | Genotype | Poor Responder |
Good Responder |
Extensive Responder |
p-Value * |
|---|---|---|---|---|---|---|
| VKORC1 | rs10871454 | CC | 55.3% | 36.2% | 8.5% | 0.171 |
| CT | 40.9% | 45.5% | 13.6% | |||
| TT | 30.9% | 54.5% | 14.5% | |||
| rs8050894 | CC | 53.5% | 39.5% | 7% | 0.235 | |
| CG | 41.8% | 43.6% | 14.5% | |||
| GG | 32.2% | 54.2% | 13.6% | |||
| rs9934438 | CC | 54.2% | 37.5% | 8.3% | 0.226 | |
| CT | 40.5% | 45% | 14.4% | |||
| TT | 32.1% | 54.7% | 13.2% | |||
| rs17708472 | CC | 38.1% | 50.6% | 11.3% | 0.042 | |
| CT | 55.1% | 28.6% | 16.3% | |||
| TT | 0.0% | 66.7% | 33.3% | |||
| CYP2C9 | rs1799853 | CC | 45.1% | 44.5% | 10.4% | 0.076 |
| CT | 28.3% | 52.5% | 19.6% | |||
| TT | 50.0% | 0.0% | 50.0% | |||
| rs4086116 | CC | 45.5% | 43.9% | 10.6% | 0.005 | |
| CT | 39.2% | 49.4% | 11.4% | |||
| TT | 10.0% | 40.0% | 50.0% | |||
| rs1057910 | AA | 42.1% | 45.0% | 12.9% | 0.910 | |
| AC | 39% | 48.8% | 12.2% |
* Chi-Square Test with p-value < 0.05 is considered significant.
Table 6.
Frequencies of the haplotypes of VKORC1 and CYP2C9 genes among the 212 warfarin responsiveness patients.
| Gene | Haplotypes | Frequency * (%) | Odds Ratio (95% CI) | p-Value ** |
|---|---|---|---|---|
| VKORC1 | TGAG | 0.512 | 0.00 | ------ |
| CCGG | 0.326 | −0.18 (−0.33–−0.03) | 0.02 | |
| CCGA | 0.129 | −0.08 (−0.28–0.12) | 0.46 | |
| CGGG | 0.026 | −0.05 (−0.47–0.36) | 0.8 | |
| TCGG | 0.007 | 0.55 (−0.22–1.32) | 0.16 | |
| CYP2C9 | CAC | 0.767 | 0.00 | ------ |
| TAT | 0.115 | 0.25 (0.04–0.45) | 0.018 | |
| TCC | 0.094 | 0.06 (−0.17–0.3) | 0.59 | |
| TAC | 0.021 | 0.44 (−0.01–0.89) | 0.059 | |
| TCT | 0.003 | 0.4 (−0.9–1.71) | 0.55 |
* Genetic haplotype frequency of 212 warfarin intake patients, ** p-value < 0.05 is considered significant.
3.5. Effect of CYP2C9 and VKORC1 Polymorphisms on INR Treatment Outcome
There were no significant differences observed between the CYP2C9 SNP genotypes and INR values measured at start of treatment for 212 cardiovascular patients treated with warfarin. In contrast, significant differences were observed between INR values measured at the initiation phase of therapy and certain VKORC1 SNPs, namely rs10871454 (p = 0.006), rs8050894 (p = 0.007), and rs9934438 (p = 0.009), as shown in Table 4.
3.6. Correlation Between Warfarin Dose and Clinical Data
Finally, there was no significant correlation between warfarin dose and body mass index, age, gender, co-morbidities, or the treatment indication (p = 0.505).
4. Discussion
Earlier studies on warfarin pharmacogenetics provide evidence that common VKORC1 and CYP2C9 polymorphisms with clinical and environmental factors are responsible for over half of the variability in warfarin required dose [23,24]. Genotyping patients who are carriers of VKORC1 and CYP2C9 variant alleles has been proven to reduce the risk of over-anticoagulation compared to the traditional initial dose approach [25,26].
In this study, our goal was to identify genetic factors associated with sensitivity and responsiveness to warfarin treatment during the initiation phase of treatment in Jordanian-Arab patients with cardiovascular disease. The results of the current pharmacogenetic study strongly suggest that there is a significant association of the VKORC1 rs8050894, rs10871454, rs9934438, and rs17708472 SNPs and the CYP2C9 rs4086116 and rs1057910 SNPs and their haplotypes with the required warfarin dosage and warfarin sensitivity. This study also reported that there is a genetic association between VKORC1 rs17708472 SNP and CCGG genetic haplotype block and CYP2C9 rs4086116 SNP and TAT genetic haplotype block with warfarin responsiveness during the initiation phase of therapy.
The allelic frequencies of CYP2C9 and VKORC1 SNPs in our population were similar to those found in other ethnic groups, as in the case of CYP2C9*2, with 10% in our population, 6% in American and European populations, and 1% in Africans [27]. However, allelic frequencies of VKORC1 SNPs were found to differ drastically from other populations. For example, rs10871454 was 52% in our population compared to 41% in Americans, 39% in Europeans, and 6% in African populations [27]. With regard to the association of CYP2C9 polymorphisms with warfarin sensitivity, our results are consistent with the study by Takahashi et al. (2001) which shows that CYP2C9 *2 and *3 polymorphisms reduce warfarin clearance [28] as CYP2C9 is the major metabolizing enzyme of warfarin, therefore, reduction of activity results in lower required doses needed to achieve the therapeutic INR. We found a strong association of CYP2C9*3 (rs1057910 A>C) and CYP2C9 (rs4086116 C>T) genotypes with warfarin sensitivity during the initiation stage of treatment with p < 0.001 and p = 0.012, respectively (Table 2). This study reported that individuals with one variant allele were associated with an increased risk of warfarin sensitivity. For example, 41.5% of the patients who carried the rs1057910 A>C variant allele were sensitive to warfarin, compared to 8.8% of the wild-type patients were sensitive. Moreover, carrying a CYP2C9 TCC genetic haplotype block was significantly associated with warfarin sensitivity with p < 0.0001 (Table 3).
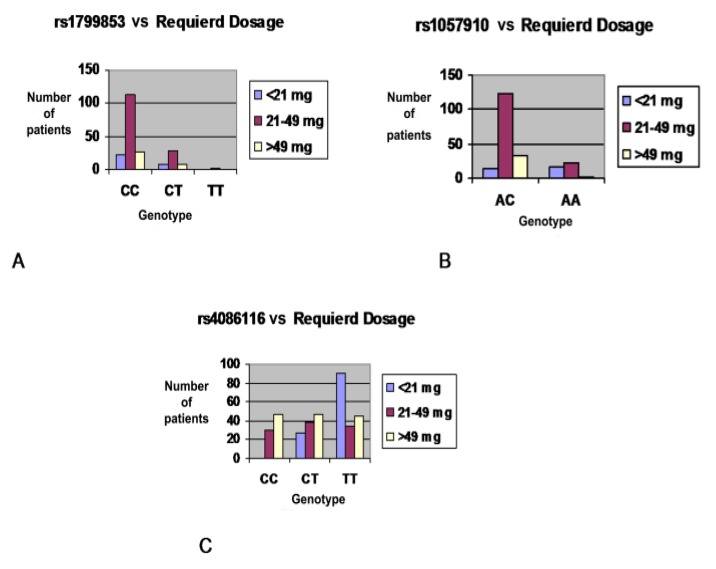
In disagreement with other studies, the CYP2C9*2 variant did not show a significant association with warfarin sensitivity (p = 0.744). This can possibly be explained by the genotypic frequency having an impact on the association; in our population only two patients were homozygous for the variant allele (TT), 32 were heterozygous patients (CT), and 105 patients were homozygous for the wild-type allele (CC) (Table 2). Moreover, it has been proposed that patients who carry one CYP2C9* 2 allele results in a dose reduction compared with the wild-type dose [14,29,30]. Although the allelic frequency of CYP2C9*2 (rs1799853) in our samples was in agreement with the American and European populations [27], this study did not find significant differences between this SNP and variability in required doses in the initiation phase of therapy (p = 0.366) as shown in Table 4 and Figure 2.
Figure 2.
The distribution of warfarin dose by CYP2C9 genotypes during the initiation phase of therapy for 212 Jordanian cardiovascular patients: X axis represents different CYP2C9 genotypes, Y axis represents the proportion of patients across each genotype, Blue column represents a sensitive group who required the lowest warfarin dose (<21 mg/week), Purple column represents the intermediate group who required moderate warfarin dose ((21–49) mg/week), Yellow column represents a resistant group who required the highest warfarin dose (>49 mg/week). (A) Distribution of warfarin dose by rs1799853 variant. (B) Distribution of warfarin dose by rs1057910 variant. (C) Distribution of warfarin dose by rs4086116 variant.
Clinical pharmacogenetic studies suggested that patients who carry the CYP2C9*3 (rs1057910) C allele leads to a dose reduction of 28–41% [12,29,30] in the Caucasian American population and from 12–38% of the Asian population compared to the wild-type [31,32,33,34]. In alignment with the aforementioned studies, we revealed that there is a 34.3% reduction in warfarin dose. Patients carrying one variant allele (C) required 41.75 mg/week, in comparison with the wild-type allele which required 27.43 mg/week (p < 0.001), as shown in Table 4 and Figure 2. Therefore, in our study the CYP2C9*3 allele has a greater effect on variation in warfarin dose during the initiation phase of therapy compared with CYP2C9*2. In the case of individuals carrying rs4086116 C>T variant allele, this resulted in 23.9% and 37.9% reduction on warfarin dose compared to wild-type with p = 0.016, as shown in Table 4 and Figure 2.
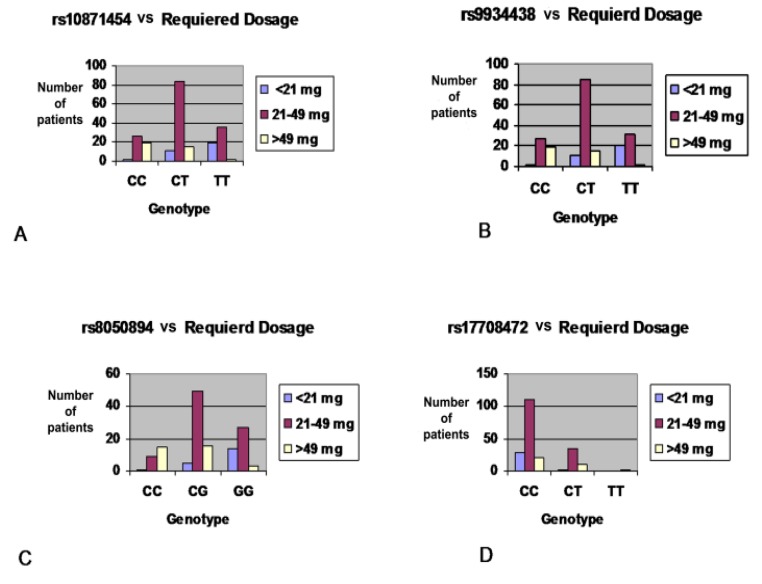
For the four studied VKORC1 SNPs, we observed a strong association of VKORC1 SNPs with warfarin sensitivity (p < 0.001). For example, patients with the T allele for rs10871454 C>T showed a high risk of warfarin sensitivity with 25.5% (CT) and 48.3% (TT) reduction of the required dose, respectively. In this case, the drug target enzyme could be expressed in smaller amounts and, therefore, low doses of the drug can obtain a therapeutic INR in an initial phase of therapy (Table 2). Furthermore, VKORC1 genetic haplotype analysis showed a significant association between three VKORC1 genetic haplotype blocks and sensitivity to warfarin with p < 0.0001 (Table 3). Moreover, Schelleman et al. (2007) and Wadelius et al. (2005) reported that a correlation exists between VKORC1 SNP 1173 C> T (rs9934438), and the variation in warfarin dose patients carrying the variant allele of this SNP is related to a reduction in the required dose compared to the wild-type [5,35]. In alignment with these results, we found that patients who carry one variant allele required an average dose of 39.47mg/week and two variant allele carriers needed an average of 26.82 mg/week, while patients carrying the wild type CC needed an average dose of 53.02 mg/week with p < 0.001 as shown in Figure 3. Limdi et al. (2007) and Shrif et al. (2011) showed that VKORC1 rs8050894 (1542G>C) were associated with lower warfarin doses in European Americans and Sudanese patients, respectively [36,37]. Accordingly, our results found a significant association between lower required warfarin dose and this SNP at an initial phase of therapy with p < 0.001 as shown in Figure 3.
Figure 3.
The distribution of warfarin dose by VKORC1 genotypes during the initiation phase of therapy for 212 Jordanian cardiovascular patients: X axis represents different VKORC1 genotypes, Y axis represents the proportion of patients across each genotype, Blue column represents a sensitive group who required the lowest warfarin dose (<21 mg/week), Purple column represents the intermediate group who required moderate warfarin dose ((21–49) mg/week), Yellow column represents a resistant group who required the highest warfarin dose (>49 mg/week). (A) Distribution of warfarin dose by rs10871454 variant. (B) Distribution of warfarin dose by rs9934438 variant. (C) Distribution of warfarin dose by rs8050894 variant. (D) Distribution of warfarin dose by rs17708472 variant.
For the last VKORC1 SNP VKORC1*4 C<T (rs17708472), our study is similar to the study by Haug et al. (2008), which reported that this SNP was associated with higher dose requirements [38]. Our results showed that this variant was associated with significant differences in initial warfarin required dose; patients who were homozygous for the variant allele (TT) genotype required an average dose of 57.8 mg/week, heterozygous (CT) patients required an average dose of 41.63 mg/week, while wild-type (CC) patients required an average dose of 33.65 with p < 0.001 (Table 4).
With regard to the correlation of CYP2C9 and VKORC1 SNPs and warfarin responsiveness, we compared SNP genotypes with the warfarin responder groups (poor, good, and extensive responders). Significant differences were found between VKORC1 rs17708472 (C>T) genotypes and the three different responder groups (Table 5); 33.3% of the patients carrying the variant allele (TT) were within the extensive responder group (meaning this variant allele was associated with increased risk of over-anticoagulation), compared to 11.3% of the wild-type (CC) patients who were extensive responders (p = 0.042). Accordingly, Kringen et al. (2011) have also shown that patients who carry this SNP are associated with an increased risk of the existence of therapeutic INR (over-anticoagulation) [39]. Otherwise, VKORC1 rs10871454, rs8050894, and rs9934438 alleles show no significant differences between SNP genotypes within the three responder groups in our population, with p = 0.171, 0.235, and 0.226, respectively (Table 5). In contrast, VKORC1 CCGG genetic haplotype block showed a significant association with warfarin responsiveness with p = 0.02 (Table 6).
Moreover, significant differences were observed between VKORC1 rs10871454, rs8050894, and rs9934438 SNPs and INR value during the initiation phase of therapy (p = 0.006, 0.007, and 0.009, respectively), while rs17708472 SNP showed no significant differences (p = 0.493). Therefore, in our population the VKORC1 SNP genotypes are associated with the generation of a high or low INR during the initiation phase of therapy (Table 4).
Taube et al. (2000) reported that an individual carrying an allelic variant of CYP2C9 was not associated with an increased incidence of severe over-coagulation during long-term treatment [40]. Correspondingly, our results show no significant differences between the CYP2C9*2 and *3 genotypes within the three responder groups with p = 0.076 and 0.910, respectively (Table 5). Conversely, our study reported significant differences between the proportion of CYP2C9 rs4086116 (C>T) genotype and the three different responder groups; 50% of TT carriers were within the extensive responder group compared with 10.6% of wild type CC carriers (p = 0.005), which means that TT carriers are associated with increased risk of over-anticoagulation (Table 5). Moreover, significant association was observed between CYP2C9 TAT genetic haplotype block and warfarin responsiveness with p = 0.018 (Table 6).
In addition, we did not observe significant differences in the three studied SNPs (CYP2C9*2, *3, and CYP2C9 (C> T) rs4086116) and the INR value during the initiation phase of therapy. Therefore, in our population CYP2C9 is not associated with the generation of a high or low INR as shown in Table 4.
Confirmation of our results and ongoing research including additional factors will be accomplished in a larger patient cohort, including genetic factors such as OATP transporters (mediates the uptake of warfarin into hepatocytes), CYP3A4, CYP1A1, and CYP1A2 enzymes (metabolizing of R-warfarin), or GGCX encoded gamma-glutamyl carboxylase (the reduced vitamin K–form to activate coagulation factors) [5,41]. Application for individualized warfarin treatment will be both beneficial and efficient for cardiovascular patients in the future. Finally, the majority of the population included in this study is elderly, with only 15% of the subjects under 40 years of age. Therefore, additional study is needed in children and young adults.
Acknowledgments
We are grateful to Pharmacist Nadia Al-Omary, and engineer of Nutrition Miss Hanna from anticoagulation clinic for cooperation and providing excellent samples for this study and for coordinating the Anticoagulant Service, and Miss Diana, laboratory technician, for the blood collection and INR measurements.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/12/578/s1. Table S1: SNPs ID, their position and genotyping data based on whole cohort (N = 220). Table S2: List of SNPs, their minor allele frequencies, and HWE p values for genotypic distribution at each locus based on 220 patients. Table S3: The frequency of the allele and genotype for all polymorphisms in cardiovascular patients treated with warfarin.
Author Contributions
L.N.A.-E. designed the study. L.N.A.-E., A.Y.A. and R.H.K. were responsible for clinical data and blood samples collection. L.N.A.-E. and A.Y.A. analyzed and interpreted the data. L.N.A.-E. and A.Y.A. prepared the manuscript. All authors helped in reviewing the manuscript.
Funding
This work was supported by the Deanship of Research at Jordan University of Science and Technology under grant number 203/2014.
Conflicts of Interest
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Kim Y., Smith A., Wu A. C3435T polymorphism of MDR1 gene with warfarin resistance. Clin. Chim. Acta. 2013;425:34–36. doi: 10.1016/j.cca.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Motulsky A.G., Qi M. Pharmacogenetics, pharmacogenomics and ecogenetics. J. Zhejiang Univ. Sci. B. 2006;7:169–170. doi: 10.1631/jzus.2006.B0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eriksson N., Wadelius M. Prediction of warfarin dose: Why, when and how? Pharmacogenomics. 2012;13:429–440. doi: 10.2217/pgs.11.184. [DOI] [PubMed] [Google Scholar]
- 4.Sconce E.A., Khan T.I., Wynne H.A., Avery P., Monkhouse L., King B.P., Wood P., Kesteven P., Daly A.K., Kamali F. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: Proposal for a new dosing regimen. Blood. 2005;106:2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 5.Wadelius M., Chen L.Y., Downes K., Ghori J., Hunt S., Eriksson N., Wallerman O., Melhus H., Wadelius C., Bentley D. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogen. J. 2005;5:262–270. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 6.Eichelbaum M., Ingelman-Sundberg M., Evans W. Pharmacogenomics and individualized drug therapy. Annu. Rev. Med. 2006;57:119–137. doi: 10.1146/annurev.med.56.082103.104724. [DOI] [PubMed] [Google Scholar]
- 7.D’Andrea G., D’Ambrosio R.L., Di Perna P., Chetta M., Santacroce R., Brancaccio V., Grandone E., Margaglione M. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005;105:645–649. doi: 10.1182/blood-2004-06-2111. [DOI] [PubMed] [Google Scholar]
- 8.Muszkat M., Blotnik S., Elami A., Krasilnikov I., Caraco Y. Warfarin metabolism and anticoagulant effect: A prospective, observational study of the impact of CYP2C9 genetic polymorphism in the presence of drug-disease and drug-drug interactions. Clin. Ther. 2007;29:427–437. doi: 10.1016/S0149-2918(07)80081-6. [DOI] [PubMed] [Google Scholar]
- 9.Rendic S. Summary of information on human CYP enzymes: Human P450 metabolism data. Drug Metab. Rev. 2002;34:83–448. doi: 10.1081/DMR-120001392. [DOI] [PubMed] [Google Scholar]
- 10.Dean L. Warfarin Therapy and VKORC1 and CYP Genotype. National Center for Biotechnology Information; Bethesda, MD, USA: 2012. [Google Scholar]
- 11.Zhou S., Zhou Z., Yang L., Cai J. Substrates, inducers, inhibitors and structure-activity relationships of human cytochrome P450 2C9 and implications in drug development. Curr. Med. Chem. 2009;16:3480–3675. doi: 10.2174/092986709789057635. [DOI] [PubMed] [Google Scholar]
- 12.Kirchheiner J., Brockmoller J. Clinical consequences of cytochrome P450 2C9 polymorphisms. Clin. Pharmacol. Ther. 2005;77:1–16. doi: 10.1016/j.clpt.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y., Ingelman-Sundberg M., Lauschke V.M. Worldwide distribution of cytochrome P450 alleles: A meta-analysis of population-scale sequencing projects. Clin. Pharmacol. Ther. 2017;102:688–700. doi: 10.1002/cpt.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linder M.W., Homme M.B., Reynolds K.K., Gage B.F., Eby C., Silvestrov N., Valdes R. Interactive modeling for ongoing utility of pharmacogenetic diagnostic testing: Application for warfarin therapy. Clin. Chem. 2009;55:1861–1868. doi: 10.1373/clinchem.2009.125898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tie J.K., Stafford D.W. Structural and functional insights into enzymes of the vitamin K cycle. J. Thromb. Haemost. 2016;14:236–247. doi: 10.1111/jth.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loebstein R., Dvoskin I., Halkin H., Vecsler M., Lubetsky A., Rechavi G., Amariglio N., Cohen Y., Ken-Dror G., Almog S., et al. A coding VKORC1 Asp36Tyr polymorphism predisposes to warfarin resistance. Blood. 2007;109:2477–2480. doi: 10.1182/blood-2006-08-038984. [DOI] [PubMed] [Google Scholar]
- 17.Schalekamp T., De Boer A. Pharmacogenetics of oral anticoagulant therapy. Curr. Pharm. Des. 2010;16:187–203. doi: 10.2174/138161210790112737. [DOI] [PubMed] [Google Scholar]
- 18.Wadelius M., Chen L.Y., Lindh J.D., Eriksson N., Ghori M.J., Bumpstead S., Holm L., McGinnis R., Rane A., Deloukas P. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113:784–792. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenzini P., Wadelius M., Kimmel S., Anderson J.L., Jorgensen A.L., Pirmohamed M., Caldwell M.D., Limdi N., Burmester J.K., Dowd M.B., et al. Integration of genetic, clinical, and INR data to refine warfarin dosing. Clin. Pharmacol. Ther. 2010;87:572–578. doi: 10.1038/clpt.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gage B., Lesko L. Pharmacogenetics of warfarin: Regulatory, scientific, and clinical issues. J. Thromb. Thrombolysis. 2007;25:45–51. doi: 10.1007/s11239-007-0104-y. [DOI] [PubMed] [Google Scholar]
- 21.Kuruvilla M., Gurk-Turner C. A review of warfarin dosing and monitoring. Proc. Bayl. Univ. Med. Cent. 2001;14:305–306. doi: 10.1080/08998280.2001.11927781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hylek E., D’Antonio J., Evans-Molina C., Shea C., Henault L., Regan S. Translating the results of randomized trials into clinical practice: The challenge of warfarin candidacy among hospitalized elderly patients with atrial fibrillation. Stroke. 2006;37:1075–1080. doi: 10.1161/01.STR.0000209239.71702.ce. [DOI] [PubMed] [Google Scholar]
- 23.Wiedermann C.J., Stockner I. Warfarin-induced bleeding complications—Clinical presentation and therapeutic options. Thromb. Res. 2008;122:S13–S18. doi: 10.1016/S0049-3848(08)70004-5. [DOI] [PubMed] [Google Scholar]
- 24.Borgiani P., Ciccacci C., Forte V., Romano S., Federici G., Novelli G. Allelic variants in the CYP2C9 and VKORC1 loci and interindividual variability in the anticoagulant dose effect of warfarin in Italians. Pharmacogenomics. 2007;8:1545–1550. doi: 10.2217/14622416.8.11.1545. [DOI] [PubMed] [Google Scholar]
- 25.Hamberg A., Dahl M.L., Barban M., Scordo M.G., Wadelius M., Pengo V., Padrini R., Jonsson E.N. A PK-PD model for predicting the impact of age, CYP2C9, and VKORC1 genotype on individualization of warfarin therapy. Clin. Pharmacol. Ther. 2007;81:529–538. doi: 10.1038/sj.clpt.6100084. [DOI] [PubMed] [Google Scholar]
- 26.Teichert M., Van Schaik R.H.N., Hofman A., Uitterlinden A.G., de Smet P.A.G.M., Stricker B., Visser L.E. Genotypes associated with reduced activity of VKORC1 and CYP2C9 and their modification of acenocoumarol anticoagulation during the initial treatment period. Clin. Pharmacol. Ther. 2009;85:379–386. doi: 10.1038/clpt.2008.294. [DOI] [PubMed] [Google Scholar]
- 27.1000 Genomes Project Consortium An integrated map of genetic variation from 1092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi H., Echizen H. Pharmacogenetics of warfarin elimination and its clinical implications. Clin. Pharmacokinet. 2001;40:587–603. doi: 10.2165/00003088-200140080-00003. [DOI] [PubMed] [Google Scholar]
- 29.Kealey C., Chen Z., Christie J., Thorn C.F., Whitehead A.S., Price M., Samaha F.F., Kimmel S.E. Warfarin and cytochrome P450 2C9 genotype: Possible ethnic variation in warfarin sensitivity. Pharmacogenomics. 2008;8:217–225. doi: 10.2217/14622416.8.3.217. [DOI] [PubMed] [Google Scholar]
- 30.Limdi N., McGwin G., Goldstein J., Beasley T., Arnett D., Adler B., Baird M., Acton R. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin. Pharmacol. Ther. 2007;83:312–321. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohno M., Yamamoto A., Ono A., Miura G., Funamoto M., Takemoto Y., Otsu K., Kouno Y., Tanabe T., Masunaga Y., et al. Influence of clinical and genetic factors on warfarin dose requirements among Japanese patients. Eur. J. Clin. Pharmacol. 2009;65:1097–1103. doi: 10.1007/s00228-009-0685-9. [DOI] [PubMed] [Google Scholar]
- 32.Huang S.W., Chen H.S., Wang X.Q., Huang L., Xu D.L., Hu X.J., Huang Z.H., He Y., Chen K.M., Xiang D.K., et al. Validation of VKORC1 and CYP2C9 genotypes on interindividual warfarin maintenance dose: A prospective study in Chinese patients. Pharmacogenet. Genomics. 2009;19:226–234. doi: 10.1097/FPC.0b013e328326e0c7. [DOI] [PubMed] [Google Scholar]
- 33.Gan G., Phipps M., Ku C., Teh A., Sangkar V. Genetic polymorphism of the CYP2C9 subfamily of 3 different races in warfarin maintenance dose. Int. J. Hematol. 2004;80:295–296. doi: 10.1532/IJH97.A20401. [DOI] [PubMed] [Google Scholar]
- 34.Tanira M., Al-Mukhaini M., Al-Hinai A., Al Balushi K., Ahmed I. Frequency of CYP2C9 genotypes among Omani patients receiving warfarin and its correlation with warfarin dose. Community Genet. 2007;10:32–37. doi: 10.1159/000096279. [DOI] [PubMed] [Google Scholar]
- 35.Schelleman H., Chen Z., Kealey C., Whitehead A.S., Christie J., Price M., Brensinger C.M., Newcomb C.W., Thorn C.F., Samaha F.F., et al. Warfarin response and vitamin K epoxide reductase complex 1 in African Americans and Caucasians. Clin. Pharmacol. Ther. 2007;81:742–747. doi: 10.1038/sj.clpt.6100144. [DOI] [PubMed] [Google Scholar]
- 36.Limdi N.A., Arnett D.K., Goldstein J.A., Beasley T.M., McGwin G., Adler B.K., Acton R.T. Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European Americans and African Americans. Pharmacogenomics. 2008;9:511–526. doi: 10.2217/14622416.9.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shrif N., Won H.H., Lee S.T., Park J.H., Kim K.K., Kim M.J., Kim S., Lee S.Y., Ki C.S., Osman I.M., et al. Evaluation of the effects of VKORC1 polymorphisms and haplotypes, CYP2C9 genotypes, and clinical factors on warfarin response in Sudanese patients. Eur. J. Clin. Pharmacol. 2011;67:1119–1130. doi: 10.1007/s00228-011-1060-1. [DOI] [PubMed] [Google Scholar]
- 38.Haug K.B., Sharikabad M.N., Kringen M.K., Narum S., Sjaatil S.T., Johansen P.W., Kierulf P., Seljeflot I., Arnesen H., Brørs O. Warfarin dose and INR related to genotypes of CYP2C9 and VKORC1 in patients with myocardial infarction. Thromb. J. 2008;6:7. doi: 10.1186/1477-9560-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kringen M.K., Haug K.B.F., Grimholt R.M., Stormo C., Narum S., Opdal M.S., Fosen J.T., Piehler A.P., Johansen P.W., Seljeflot I., et al. Genetic variation of VKORC1 and CYP4F2 genes related to warfarin maintenance dose in patients with myocardial infarction. J. Biomed. Biotechnol. 2011;2011:739–751. doi: 10.1155/2011/739751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taube J., Halsall D., Baglin T. Influence of cytochrome P-450 CYP2C9 polymorphisms on warfarin sensitivity and risk of over-anticoagulation in patients on long-term treatment. Blood. 2000;96:1816–1819. [PubMed] [Google Scholar]
- 41.Frymoyer A. Effect of single-dose rifampin on the pharmacokinetics of warfarin in healthy volunteers. Clin. Pharmacol. Ther. 2010;88:540–547. doi: 10.1038/clpt.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.