Abstract
Aflatoxins are carcinogenic metabolites produced primarily by fungi within Aspergillus section Flavi. These fungi infect a wide range of crops in warm regions. Molecular phylogenetic analyses of fungi with S morphology (average sclerotium size < 400 µm) within section Flavi collected from across the United States (US) resulted in the discovery of a novel aflatoxin-producing species, Aspergillus texensis. Aspergillus texensis was isolated from maize grown in Arkansas, Louisiana, and Texas, and from soils cropped to maize in Texas. Aspergillus texensis produces sparse conidia and abundant sclerotia on various culture media, and on maize. Physiological studies have revealed optimal growth on culture media at 35 °C. All isolates of A. texensis produced B and G aflatoxins, cyclopiazonic acid and aspergillic acid. Aspergillus texensis and A. flavus S strain morphotypes produced similar concentrations of total aflatoxins on maize (p > 0.05). Phylogenetic analyses of aflatoxin-producers based on partial gene sequences of the β-tubulin (0.9 kb), calmodulin (1.2 kb), and nitrate reductase (2.1 kb) genes placed A. texensis in a highly supported monophyletic clade closely related to A. minisclerotigenes and a previously reported unnamed lineage designated Lethal Aflatoxicosis Fungus.
Keywords: Aflatoxins, Aspergillus texensis, S morphology, Molecular phylogenetics
1. Introduction
Aflatoxins (AF) are extremely potent naturally-occurring hepatocarcinogenic mycotoxins produced by several members of the genus Aspergillus on important commodities such as maize, groundnut, tree nuts, spices, and cottonseed [1,2,3,4,5]. Naturally occurring aflatoxin-producers contaminate food and feed with four major aflatoxins, i.e., aflatoxins B1, B2, G1 and G2. Bio-transformation of aflatoxins from consumption of contaminated food results in formation of aflatoxins M1 and M2, which are secreted into milk [6,7]. Although certain species within Aspergillus section Ochraceorosei and Aspergillus section Nidulantes also produce aflatoxins [8,9], the most economically important aflatoxin-producers belong to Aspergillus section Flavi [10,11,12]. Aspergillus flavus, A. parasiticus, A. aflatoxiformans from West Africa, and an unnamed lineage referred to as the Lethal Aflatoxicosis Fungus (LAF) from Kenya are particularly notorious members of Aspergillus section Flavi responsible for contamination of crops with high levels of aflatoxins [3,13,14,15,16,17]. These fungi colonize a wide range of host crops resulting in dangerous concentrations of aflatoxins under conducive environmental conditions (high temperature, i.e., >28 °C, humidity and plant stress) [18].
Aflatoxins are both health and economic threats. Aflatoxin B1 has been categorized as a Group 1 human carcinogen by IARC [19]. In developing nations, aflatoxin regulations largely remain unenforced and crops are consumed without monitoring, resulting in frequent exposure of humans and animals [20,21]. Chronic dietary exposure to sub-lethal concentrations can cause immune suppression [22], impaired growth [23,24], and liver cancer [25,26]. Ingestion of high concentrations of aflatoxins may result in liver necrosis followed by rapid death [20,21]. Acute cases of aflatoxin poisoning have resulted in human deaths in Kenya and Tanzania [27,28,29]. Aflatoxins are a severe economic burden in the developed world where regulations are stringently enforced leading to heavy economic losses incurred by growers [30]. These regulatory enforcements result in rejection of contaminated food/feed followed by loss of markets and expense to the exporter. Since aflatoxins remain a global concern, it becomes crucial to identify and characterize aflatoxin-producing species in order to design management strategies for aflatoxin contamination of crops.
Aspergillus flavus and A. parasiticus are the most commonly implicated causal agents of aflatoxin contamination of crops [31]. The filamentous fungus A. flavus produces only B aflatoxins and has two morphotypes. The L strain morphotype is characterized by the production of copious conidia, few large sclerotia (>400 µm), and variable concentrations of aflatoxins, such that many L strain morphotype fungi are atoxigenic (do not produce aflatoxins); the S strain morphotype produces large quantities of small sclerotia (<400 µm) and sparse conidia [32]. The A. flavus S strain morphotype consistently produces high concentrations of aflatoxins. Aspergillus parasiticus produces both B and G aflatoxins. The A. flavus S strain morphotype produces only B aflatoxins and is the more commonly occurring S morphology fungus in North America [2,32,33]. However, several highly aflatoxigenic S morphology fungi belonging to genetically distinct taxa, i.e., A. minisclerotigenes, A. aflatoxiformans, A. cerealis (previously A. korhogoensis), and LAF, are known from Sub-Saharan Africa [4,14,15,17,34]. During phylogenetic analysis of fungi with S morphology belonging to Aspergillus section Flavi isolated from soils and maize collected from across the US, we discovered fungal isolates that produced both B and G aflatoxins, but were morphologically indistinguishable from the A. flavus S strain morphotype. Phylogenetic reconstruction using multi locus gene sequences with previously described members of Aspergillus section Flavi indicated that these B and G aflatoxin producers represent a novel, undescribed species.
Aspergillus section Flavi contains several phylogenetically distinct species with S morphology characterized by production of small sclerotia (<400 µm) [16,17,32]. Description of species within section Flavi solely based on phenotypic characteristics can be erroneous due to overlapping character states [35,36]. The current study used a polyphasic approach to compare newly discovered species to those previously described. Phenotypic description included macro- and micromorphology, growth at different temperatures, and production of aflatoxins, aspergillic acid, and cyclopiazonic acid. Phylogenetic reconstructions based on multiple unlinked loci were utilized to determine relationships of the novel species to those previously described with S morphology.
2. Results
2.1. Molecular Analyses and Phylogenetics
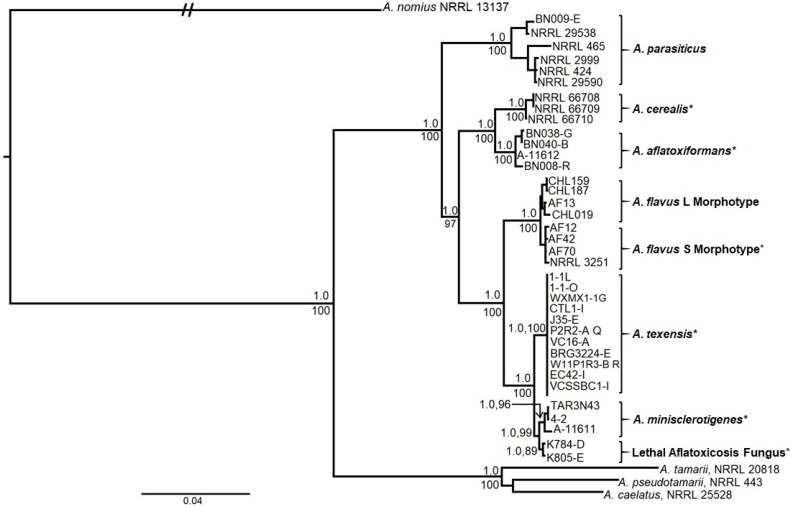
Homologous DNA sequences of reference isolates of described species within section Flavi with S morphology (e.g., A. flavus S strain morphotype, A. minisclerotigenes, A. cerealis, A. aflatoxiformans), representatives of the fungi associated with lethal aflatoxicosis in Kenya (LAF), and other aflatoxin-producers (A. nomius, A. pseudotamarii, A. parasiticus) were aligned with sequences of S morphology fungi recovered from the US (Table 1). Phylogenetic reconstruction using Bayesian Inference (BI) and Maximum Likelihood (ML) analyses from partial gene sequences of β-tubulin (benA, 0.9 kb, chromosome 6), calmodulin (cmdA, 1.2 kb, chromosome 2), and nitrate reductase (niaD, 2.1 kb, chromosome 4) genes for individual and concatenated sequences yielded similar topologies. The new species, named Aspergillus texensis, occupied a highly supported monophyletic clade in concatenated benA, cmdA, and niaD gene phylogenies (Figure 1), and was sister to a clade containing A. minisclerotigenes and LAF fungi. Phylogenetic reconstruction based individually upon cmdA and niaD sequences showed that each of these genes was sufficient to resolve A. texensis into a monophyletic clade with high Bayesian posterior probability and bootstrap support (Figures S1 and S2). The A. flavus S strain morphotype, which is a frequently reported S morphology fungus in the US, is phylogenetically distinct from A. texensis (Figure 1). Additionally, molecular analysis of the norB-cypA region did not detect any deletions in the cypA gene of A. texensis, whereas the A. flavus S strain morphotype isolates contained either a 0.9 or 1.5 kb deletion in this region of the aflatoxin biosynthesis cluster required for G aflatoxin production.
Table 1.
Origin of fungal isolates examined in the current study.
| Isolate # | Species/Taxon | Source | Citation |
|---|---|---|---|
| NRRL 13137 | A. nomius | Wheat, USA | [37] |
| NRRL 443 | A. pseudotamarii | Argentina | [36] |
| NRRL 25528 | A. caelatus | Soil, USA | [38] |
| NRRL 465 | A. parasiticus | USA | [39] |
| NRRL 29538 | A. parasiticus | Soil, Georgia, USA | Horn B.W., National Peanut Lab, Dawson, GA (in NRRL database) |
| NRRL 29590 | A. parasiticus | Soil, Georgia, USA | Horn B.W., National Peanut Lab, Dawson, GA (in NRRL database) |
| NRRL 424 | A. parasiticus | Soil, Georgia, USA | Scales F.M. (in NRRL database) |
| NRRL 2999 | A. parasiticus | Peanut, Uganda | [40] |
| BN009-E | A. parasiticus | Soil, Benin | None |
| NRRL A-11611 | A. minisclerotigenes | Soil, Nigeria | [41,42] |
| 4-2 | A. minisclerotigenes | Soil, Australia | [43] |
| TAR3N43 | A. minisclerotigenes | Peanut, Argentina | [16] |
| AF70/ATCC MYA-384 | A. flavus S Morphotype | Soil, Arizona, USA | [32] |
| AF42/ATCC MYA-383 | A. flavus S Morphotype | Soil, Arizona, USA | [32] |
| AF12/ATCC MYA-382 | A. flavus S Morphotype | Soil, Arizona, USA | [32] |
| NRRL 3251 | A. flavus S Morphotype | Walnut, California, USA | [41] |
| AF13/ATCC 96044 | A. flavus L Morphotype | Soil, Arizona, USA | [32] |
| CHL019 | A. flavus L Morphotype | Chili | [12] |
| CHL159 | A. flavus L Morphotype | Chili | [12] |
| CHL187 | A. flavus L Morphotype | Chili, Pakistan | [12] |
| W11P1R3-B R/NRRL 66855/A2292 | A. texensis | Soil, Texas, USA | Current Study |
| EC42-I/NRRL 66856/A2295 | A. texensis | Maize, Texas, USA | Current Study |
| BRG3224-E/NRRL 66857/A2296 | A. texensis | Maize, Louisiana, USA | Current Study |
| WXMX1-1G/NRRL 66858/A2294 | A. texensis | Soil, Texas, USA | Current Study |
| VCSSBC1-I/NRRL 66859/A2293 | A. texensis | Maize, Arkansas, USA | Current Study |
| J35-E | A. texensis | Soil, Texas, USA | Current Study |
| VC16-A | A. texensis | Maize, Texas, USA | Current Study |
| CTL-1I | A. texensis | Soil, Texas, USA | Current Study |
| P2R2-A Q | A. texensis | Soil, Texas, USA | Current Study |
| 1-1-O | A. texensis | Soil, Texas, USA | Current Study |
| 1-1L | A. texensis | Soil, Texas, USA | Current Study |
| K805-E/A1170 | Lethal Aflatoxicosis Fungus | Maize, Kenya | [44] |
| K784-D/A1168 | Lethal Aflatoxicosis Fungus | Maize, Kenya | [44] |
| NRRL A-11612 | A. aflatoxiformans | Groundnut, Nigeria | [17,41] |
| BN040-B/ATCC MYA-381 | A. aflatoxiformans | Soil, Benin | [17,45] |
| BN038-G/ATCC MYA-380 | A. aflatoxiformans | Soil, Benin | [17,45] |
| BN008-R/ATCC MYA-379 | A. aflatoxiformans | Soil, Benin | [17,45] |
# Isolates were obtained from the ARS Culture Collection, Peoria, IL, USA (NRRL), the American Type Culture Collection, Manassas, USA (ATCC), Fungal Genetics Stock Center, Manhattan, KS (A) or were present at the USDA-ARS, Tucson Laboratory Culture Collection.
Figure 1.
Mid-point rooted Bayesian phylogeny of A. texensis and closely related S morphology fungi with several additional species within section Flavi for reference. Phylogeny is based on concatenated benA (0.9 kb), cmdA (1.2 kb), and niaD (2.1 kb) genes of Aspergillus section Flavi. Values above nodes or before commas are Bayesian posterior probabilities and values below nodes or after commas are bootstrap values from 500 replicates. Aspergillus nomius NRRL 13137 was used as the outgroup. * Aflatoxin producers with S morphology.
All isolates of A. texensis, regardless of state of origin, contained only the MAT1-1 idiomorph at the mating-type locus, suggesting that A. texensis is heterothallic. Each isolate amplified only a single amplicon of approximately 390 bp in the MAT locus PCR assay. This is characteristic of the MAT1-1 idiomorph. The MAT1-2 idiomorph that is highly conserved in Aspergillus section Flavi should produce a 270 bp amplicon, but was not detected in any A. texensis isolate.
2.2. Aspergillic Acid
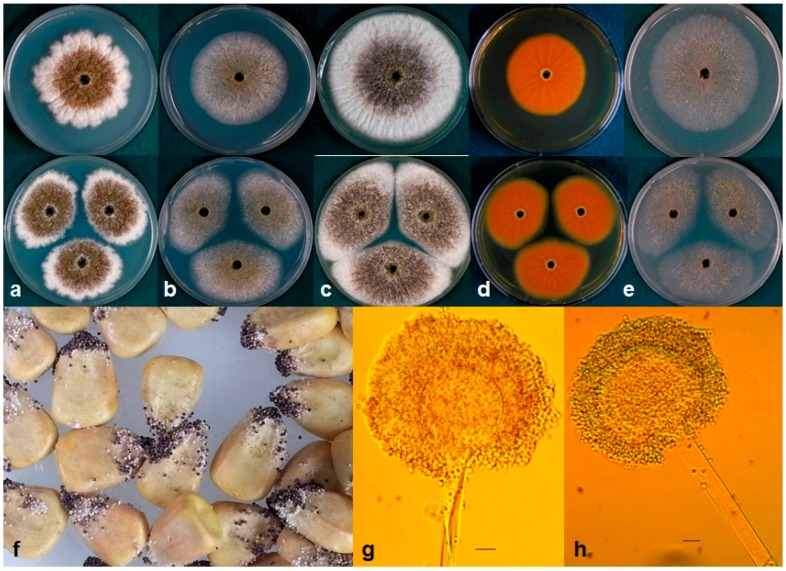
All isolates of A. texensis produced aspergillic acid, which was detected by the bright-orange reaction on the reverse side of AFPA medium (Figure 2; Table 2), similar to A. flavus and A. minisclerotigenes. Colony texture of A. texensis on AFPA after 7 d at 25, 30 and 35 °C was floccose with abundant white mycelia and immature white sclerotia.
Figure 2.
(a–e) Colonies of A. texensis grown at 25 °C for 7 d on (a) CZ, (b) CYA, (c) Malt agar, (d) reverse on AFPA, and (e) V8 agar. (f) A. texensis on maize grown at 31 °C for 7 d; (g–h) Conidiophores. Bars—10 µm.
Table 2.
Production of major mycotoxins a by certain species within Aspergillus section Flavi.
| Species | Aflatoxin B1 | Aflatoxin B2 | Aflatoxin G1 | Aflatoxin G2 | CPA | Aspergillic Acid |
|---|---|---|---|---|---|---|
| A. texensis | + | + | + | + | + | + |
| A. flavus | + | + | − | − | ± | + |
| A. minisclerotigenes | + | + | + | + | + | + |
| A. parasiticus | + | + | + | + | − | + |
2.3. Cyclopiazonic Acid
Aspergillus texensis produced cyclopiazonic acid (CPA) (Table 2); CPA concentrations ranged from 7.5–26.6 µg/g with a mean of 17.4 µg/g. Similarly, all S strain morphotype isolates of A. flavus (n = 3) tested in the current study produced CPA (Range: 16.0–30.6 µg/g; Mean = 25.0 µg/g).
2.4. Aflatoxins
Aspergillus texensis produced B1, B2, G1, and G2 aflatoxins in maize. Aflatoxin concentrations ranged from 11–71 µg/g AFB1, 0.6–2.6 µg/g AFB2, 66–225 µg/g AFG1, and 2.0–7.7 µg/g AFG2. The S strain morphotype of A. flavus, which is the most commonly reported S morphology fungus in the US, produces only B aflatoxins. In an experiment comparing total aflatoxin production by genetically distinct S morphology fungi, mean aflatoxin concentrations produced by A. texensis and A. flavus S morphotype fungi were similar at 31 °C (Table 3; p > 0.05). Aspergillus texensis produced at least two-fold more G aflatoxins than B aflatoxins (Table 3). Fungi with S morphology from the African continent, including isolates of A. aflatoxiformans and LAF, produced the highest concentrations of total aflatoxins in maize, while A. minisclerotigenes produced the lowest quantities. However, all fungi with S morphology evaluated in the current study overall produced lethal concentrations of total aflatoxins in maize at 31 °C (Range: 33–568 µg/g).
Table 3.
Aflatoxin production by fungi with S morphology within Aspergillus section Flavi.
| Species | Isolate | Source/Location | Aflatoxin (µg/g) | ||
|---|---|---|---|---|---|
| AFB | AFG | Total AF | |||
| A. texensis | BRG3224 E | Maize/USA | 42 | 111 | 153 |
| EC42-I | Maize/USA | 40 | 99 | 139 | |
| W11P1R3-B R | Soil/USA | 47 | 111 | 158 | |
| Average | 43C | 107B | 150B | ||
| A. flavus S Morphotype | AF12 | Soil/USA | 190 | NA | 190 |
| AF42 | Soil/USA | 181 | NA | 181 | |
| AF70 | Soil/USA | 191 | NA | 191 | |
| Average | 187AB | NA* | 187B | ||
| A. minisclerotigenes | A-11611 | Groundnut/Nigeria | 31x | 72x | 103x |
| 4-2 | Soil/Australia | 9y | 24y | 33y | |
| TAR3N43 | Soil/Argentina | 9y | 25y | 34y | |
| Average | 17D | 40C | 57C | ||
| A. aflatoxiformans | A-11612 | Groundnut/Nigeria | 92y | 202y | 294y |
| BN038-G | Soil/Benin | 105y | 129z | 234y | |
| BN040-B | Soil/Benin | 207x | 361x | 568x | |
| Average | 134B | 231A | 365A | ||
| Lethal Aflatoxicosis Fungus | K805-E | Maize/Kenya | 304 | NA | 304xy |
| K784-D | Maize/Kenya | 361 | NA | 361x | |
| K849-B | Maize/Kenya | 215 | NA | 215y | |
| Average | 294A | NA* | 294A | ||
B aflatoxin, G aflatoxin and total aflatoxin concentrations were compared between and within species. Each toxin concentration is a mean of four replicates. Means followed by different upper case letters in bold (A,B,C) or lower case letters (x,y) within a column differ significantly according to Tukey’s HSD (p < 0.01). Values within a column lacking a letter do not differ (ANOVA, p > 0.05). * Fungi which do not produce G aflatoxins were excluded when comparing G aflatoxin production between and within species.
2.5. Taxonomy
Aspergillus texensis P.Singh, M.J.Orbach, and P.J.Cotty sp. nov. MycoBank MB828668. Figure 2.
Etymology: The species epithet texensis is Latin for “from Texas”, where the first isolates of the new species were collected.
Diagnosis: Aspergillus texensis is closely related to A. minisclerotigenes and the unnamed lineage LAF. Aspergillus minisclerotigenes grows faster than A. texensis on Czapek agar at 20 °C and 37 °C, and on V8 agar at 37 °C; however, A. texensis grows faster on V8 agar at 40 °C (Table 4). The unnamed lineage LAF lacks the ability to produce G aflatoxins unlike A. texensis, which produces both B and G aflatoxins (Table 3).
Table 4.
Influence of temperature on radial growth of three S morphology species in Aspergillus section Flavi on agar media.
| Medium Φ | Species | Colony Diameter (mm) ρ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 15 °C | 20 °C | 25 °C | 30 °C | 35 °C | 37 °C | 40 °C | 42 °C | ||
| CZ | A. texensis | 3b | 31b | 53ab | 67 | 73 | 55b | 46 | 7 |
| A. flavus | 7a | 35b | 50b | 66 | 65 | 54b | 37 | 8 | |
| A. minisclerotigenes | 3b | 41a | 62a | 75 | 75 | 63a | 39 | 8 | |
| CZ with NaCl | A. texensis | 11 | 44 | 60 | 72 | 77 | 72 | 68 | 13 |
| A. flavus | 12 | 44 | 57 | 69 | 71 | 65 | 69 | 12 | |
| A. minisclerotigenes | 10 | 44 | 66 | 73 | 77 | 72 | 73 | 15 | |
| CYA | A. texensis | 9 | 46 | 71 | 77 | 78 | 65 | 54 | 9 |
| A. flavus | 11 | 42 | 64 | 72 | 71 | 57 | 56 | 11 | |
| A. minisclerotigenes | 9 | 44 | 68 | 77 | 78 | 64 | 49 | 10 | |
| Malt | A. texensis | 7a | 30 | 46ab | 68 | 69 | 55 | 41 | 11 |
| A. flavus | 5ab | 34 | 44b | 57 | 67 | 55 | 30 | 7 | |
| A. minisclerotigenes | 3b | 43 | 69a | 67 | 68 | 58 | 32 | 8 | |
| V8 | A. texensis | 4a | 33 | 60 | 68 | 72 | 52b | 42a | 7 |
| A. flavus | 3ab | 32 | 53 | 66 | 68 | 61a | 28b | 7 | |
| A. minisclerotigenes | 2b | 31 | 59 | 74 | 73 | 67a | 31b | 9 | |
Φ Media compositions in Materials and Methods; CZ-Czapek solution agar, CZ with NaCl-Czapek solution agar with 2% NaCl, CYA-Czapek solution with yeast extract agar, Malt-Malt solution agar, V8-V8 juice agar. ρ Colony diameter 7 d post inoculation. Statistical comparisons among species were performed independently for each medium/temperature combination. Values are means of four replicates; means followed by the same letter do not differ significantly (Tukey’s HSD, p > 0.05).
Typus: United States of America, Texas, Waxahachi, soil cropped to maize (Zea mays), collected by P.J.Cotty (holotype NRRL 66855, ex-type: WXMXP1R3-B R = A2292).
Colony characteristics: Aspergillus texensis colonies attained an average diameter of 53 mm (51–58 mm) at 25 °C, 55 mm (53–59 mm) at 37 °C, and 7 mm (4–9 mm) at 42 °C on Czapek agar after 7 d. Colony diameters were greater on Czapek agar with 2% NaCl and Czapek agar with yeast extract (CYA) media at these temperatures (Table 4). Maximum radial growth of A. texensis occurred at 35 °C on all media. Aspergillus texensis did not germinate at 5 °C or 10 °C. Colony surface on Malt agar was floccose with dominant white mycelia, velvety on CYA and V8 (Figure 2). Colony reverse buff on CZ, CYA and Malt.
Micromorphology: Abundant production of dark black sclerotia (150–300 µm) on the agar surface was observed on Czapek, CYA, Malt, and V8 agar (Figure 2). Fungal isolates produced sparse yellow-green conidia on all media tested; conidia were circular and smooth walled (3-6 µm diameter). Vesicle globose, 30–60 µm in diameter. Conidiophores with stippled stipes, hyaline, 400–800 × 10–14 µm, phialides 6–11 × 3–5. Aspergillus texensis produced copious quantities of sclerotia but fewer conidia on maize after incubation at 31 °C for 7 d (Figure 2).
The type and other representative isolates of A. texensis have been submitted to the ARS Culture Collection (NRRL) (Peoria, IL, USA) and the Fungal Genetics Stock Center, Manhattan, KS (Table 1).
3. Discussion
Most aflatoxin producing fungi belong to Aspergillus section Flavi with several members recognized as economically important agents of aflatoxin contamination of crops [13]. Although many species within section Flavi produce aflatoxins, fungi that have repeatedly been recovered at high frequencies from crops and agricultural soils, such as A. flavus L and S strain morphotypes, A. parasiticus, A. minisclerotigenes, A. aflatoxiformans from West Africa, and the fungi associated with lethal aflatoxicoses in Kenya (LAF), are the primary concern as causal agents of aflatoxin contamination of crops [1,12,14,15,16]. Morphology (sclerotia, conidia, colony characteristics) and physiology (growth rates at different temperatures), and mycotoxin production have been conventional tools in identification and characterization of species within section Flavi [41,46]. However, in the past decade, fungi within section Flavi with overlapping morphological and physiological characteristics have been assigned to genetically distinct taxa based on DNA sequences [16,17,34,42]. For instance, the A. flavus S Strain morphotype and LAF fungi produce small sclerotia (<400 µm) and only B aflatoxins; however, multi locus gene genealogies, and differences in deletions in the norB-cypA region of the aflatoxin biosynthesis gene cluster, clearly show that A. flavus and LAF are distinct species [16,44]. Likewise, A. minisclerotigenes, A. aflatoxiformans, and A. cerealis show high morphological similarity (numerous small sclerotia), and produce B and G aflatoxins. DNA sequence data from multiple unlinked loci strongly support placement of each of these into a distinct monophyletic taxon [17,34,42]. The utilization of a polyphasic approach, which combines morphology, physiology, and DNA-based analyses for recognition of novel species, provides a powerful tool for researchers to approach cryptic diversity within Aspergillus section Flavi. However, multi-locus DNA sequence-based phylogenetics are frequently sufficient for delineation of new taxa [17,47,48].
Aspergillus texensis forms a highly supported distinct terminal group in phylogenies of sequence data from three unlinked loci (benA, cmdA, niaD). The branch containing A. texensis is congruent, and the same isolates occur as a terminal group in trees constructed from individual or concatenated genes with strong statistical support, both by Bayesian posterior probability and bootstrap analyses (Figure 1; Figures S1 and S2). In light of the aforementioned results, A. texensis fulfills requirements of the phylogenetic species concept, which limits species boundaries to a monophyletic group within which a pattern of ancestry and descent exists [36,47,49]. This, along with physiological data and types of secondary metabolites produced, distinguishes A. texensis as a new species.
DNA sequences resulting from the portion of the β-tubulin gene amplified using primers from [50] were highly conserved among closely-related aflatoxin producing species. The current study therefore incorporated partial 5′ untranslated region (UTR) of β-tubulin using primer pair Bt3a-3b (Table 5) to include more variable characters to seek better resolution. However, 0.9 kb of individual β-tubulin sequences (112 5′ UTR positions, 320 exon positions and 453 intron positions) still did not contain enough variable characters to resolve multiple aflatoxin producing species. Nevertheless, individually, calmodulin and nitrate reductase sequences could clearly separate previously reported aflatoxin-producers and A. texensis in both Bayesian and Maximum Likelihood topologies with high Bayesian posterior probabilities and Bootstrap support (Figures S1 and S2). The coding sequence of β-tubulin encodes for proteins, which along with alpha tubulin, polymerize into microtubules, cellular structures that are crucial in multiple cellular processes. Owing to this highly conserved function, DNA sequences of β-tubulin may not be sufficient to reveal the cryptic diversity among more closely related species within Aspergillus section Flavi, while calmodulin and nitrate reductase could be superior choices to identify novel species within section Flavi. This is in agreement with previous studies describing novel species within section Flavi that have often reported phylogenies based on concatenated gene sequences including β-tubulin, calmodulin, RNA polymerase, internal transcribed spacers (ITS), etc. with high support for the clade representing a new species [34,36,48]. However, gene trees based on individual sequences of RNA polymerase, ITS, and β-tubulin either did not resolve members of section Flavi or did so with low branch support [36,48]. On the other hand, the calmodulin gene provides a useful tool to identify and resolve closely-related aflatoxin producers of section Flavi [17,35,36,48]. The nitrate reductase gene, which encodes for an enzyme essential for nitrate assimilation in fungi, is also a powerful tool to identify and characterize species within section Flavi, as demonstrated in the current study, and by [4,12].
Table 5.
Primers and locus specific annealing temperatures (Ta) for PCR amplifications.
| Primer Pair | Target Gene | Sequence | Ta (°C) | Reference |
|---|---|---|---|---|
| Bt2a-2b | β-tubulin | F-GGTAACCAAATCCGTGCTGCTTTC | 51 | [50] |
| R-ACCCTCAGTGTAGTGACCCTTGGC | ||||
| Bt3a-3b | F-CGTCGTTCATTCGAGGTGTA | 56 | Current study | |
| R-CCGCTCAACTTCAAGTCCAT | ||||
| cmd42-637 | Calmodulin | F-GGCCTTCTCCCTATTCGTAA | 56 | [16] |
| R-CTCGCGGATCATCTCATC | ||||
| cmd2F-2R | F-GGCTGGATGTGTGTAAATC | 48 | [16] | |
| R-ATTGGTCGCATTTGAAGGG | ||||
| niaDF-AR | Nitrate reductase | F-CGGACGATAAGCAACAACAC | 52 | [16] |
| R-GGATGAACACCCGTTAATCTGA | ||||
| niaDBF-BR | F-ACGGCCGACAGAAGTGCTGA | 57 | [16] | |
| R-TGGGCGAAGAGACTCCCCGT | ||||
| niaDCF-CR | F-GCAGCCCAATGGTCACTACGGC | 55 | [12] | |
| R-GGCTGCACGCCCAATGCTTC |
The fungal isolates representing A. texensis sp. nov. produce abundant quantities of small sclerotia (<400 µm diameter) on several growth media, and on maize kernels (Figure 2). Conidia are smooth walled and appear light green to yellowish green, similar to the S strain morphotype of A. flavus. Although microscopic characters and growth at different temperatures in various media largely overlap (Table 4), A. texensis is characterized by production of both B and G aflatoxins, unlike the A. flavus S strain morphotype, which produces only B aflatoxins. This is due to either a 0.9 or 1.5 kb deletion in the norB-cypA region of the aflatoxin biosynthesis gene cluster in A. flavus, which has resulted in loss of G aflatoxin production by this species [51]; however, A. texensis has an intact norB-cypA region. Total aflatoxin production on maize by A. flavus S strain morphotype did not differ from that of A. texensis (Table 3; p > 0.05); however, A. texensis produced significantly lower concentrations of B aflatoxins (Table 3; p < 0.05). Additionally, phylogenetic reconstruction based on 4.1 kb sequence data from three unlinked genes (benA, cmdA and niaD) clearly demonstrate that A. texensis is a genetically distinct, distant relative of A. flavus.
Aspergillus texensis is closely related to A. minisclerotigenes (first described from Argentinian groundnuts) [42] and LAF (the lineage with S morphology responsible for severe aflatoxin contamination that led to hundreds of deaths in Kenya) [16,44] (Figure 1). Both A. minisclerotigenes and LAF have been reported in very low frequencies in the US (one isolate of A. minisclerotigenes and three isolates of LAF) [16,42]. However, higher incidences of A. minisclerotigenes are known from groundnuts in Argentina [42], almonds and maize in Portugal [48], maize in Eastern and Central Africa [4], and chilies in Nigeria [12]. Similarly, LAF has been recovered in high frequencies from maize in Kenya [16]. Production of G aflatoxins by A. texensis differentiates it from LAF, which produces only B aflatoxins due to a characteristic 2.2 kb deletion in the norB-cypA region of the aflatoxin biosynthesis gene cluster [16]. Aspergillus texensis is distinguished in growth from A. minisclerotigenes on Czapek agar at 20 °C and 37 °C, and on V8 agar at 37 °C and 40 °C (Table 4). Furthermore, A. texensis produces higher concentrations of aflatoxins than A. minisclerotigenes on maize (Table 3; p < 0.05). Also, phylogenetic analyses of DNA sequence data strongly support the delineation of these two species (Figure 1, Figures S1 and S2).
Fungal isolates of most species within section Flavi contain either MAT1-1 or MAT1-2 idiomorphs at the mating type locus, and are therefore heterothallic [34]. All isolates of A. texensis examined to date have the MAT1-1 idiomorph suggesting clonal evolution in the absence of meiotic recombination, which within heterothallic species requires the presence of MAT1-2 idiomorph strains for sexual reproduction. However, further sampling is needed to determine whether isolates of A. texensis with the MAT1-2 idiomorph are present, as only 11 isolates have been collected to date.
Aflatoxigenic fungi frequently reported across the US include A. flavus L strain morphotype (isolates can be toxigenic or atoxigenic), A. flavus S strain morphotype, and A. parasiticus [52,53,54,55]. Accurate identification and characterization of aflatoxin producing fungi allows clarification of the etiology of crop contamination and development of procedures for aflatoxin mitigation. Aspergillus texensis is known from 11 isolates currently, and was recovered from soils cropped to maize, and maize grain produced in Arkansas, Louisiana, and Texas. Aflatoxin contamination of crops is severe in these regions [56,57,58,59]. The majority of A. texensis isolates (82%) discovered in the current study are from Texas, where contamination is a perennial issue. The highly toxic A. flavus S strain morphotype has been reported in high frequencies from hot and dry regions of Texas, and increased proportions of these fungi have been associated with increased soil temperature [33,60]. The morphology of Aspergillus texensis is highly similar to that of the S strain morphotype of A. flavus with numerous small (<400 µm) sclerotia. Furthermore, both A. texensis and the A. flavus S strain morphotype produce high and comparable concentrations of aflatoxins in maize. These data, along with the aforementioned observations, suggest that co-occurrence of these S morphology aflatoxin producers in the US may lead to crop contamination with high aflatoxin concentrations. The current study also indicates that G aflatoxins in maize are not solely attributable to A. parasiticus.
4. Materials and Methods
4.1. Fungal Isolates and Morphology
Eleven fungal isolates were chosen from a survey of S morphology fungi, collected from across the US, based on their ability to produce both B and G aflatoxins. These isolates were recovered from soil and maize samples at the USDA-ARS laboratory in Tucson, Arizona. Fungal isolates with known phylogenetic placement were obtained from the ARS Culture Collection, Peoria, IL, USA (NRRL in Table 1), the American Type Culture Collection, Manassas, USA (ATCC in Table 1), the Fungal Genetics Stock Center, Manhattan, KS, USA (A in Table 1), or the USDA-ARS Laboratory Culture Collection in Tucson, Arizona (Table 1).
In order to compare morphological features and growth, fungal isolates were grown for 7 d in the dark at 5 °C, 10 °C, 15 °C, 20 °C, 25 °C, 30 °C, 35 °C, 37 °C, 40 °C, and 42 °C using center point inoculation on Czapek agar (20 g Bacto agar, 30 g sucrose, 3 g NaNO3, 0.5 g KH2PO4, 0.5 g K2HPO4, 0.5 g MgSO4 * 7H2O, 0.5 g KCl, and 1 mL micronutrients per liter of deionized distilled water, pH = 6.0), Czapek with 2% NaCl, Czapek yeast extract agar (CYA) (Czapek agar with 0.5% yeast extract), Malt agar and V8 agar (5% V8 juice and 2% agar, pH = 6.0). Micronutrients were prepared according to [61]. Colony diameters were recorded (four replicates per isolate) at each temperature. For micromorphological observations, 3 d old cultures grown on Czapek agar were viewed and captured with a differential interference contrast microscope (Model BH-2, Olympus, Shinjuku, Tokyo, Japan) equipped with an OMAX 5.0MP USB2.0 digital camera and the software package ToupView (v 3.7, ToupTek Photonics, Hangzhou, China, 2014).
4.2. Production of Aspergillic Acid, Aflatoxins, and Cyclopiazonic Acid
Aspergillic acid production was analyzed by inoculating fungal isolates onto Aspergillus flavus and parasiticus agar (AFPA) [62], which was incubated for 7 d in the dark at 25 °C, 30 °C, and 35 °C. Isolates were replicated four times at each temperature.
Aflatoxins were analyzed and quantified for fungi with S morphology belonging to five phylogenetically distinct taxa. Each taxon was represented by three isolates. Conidial suspensions were inoculated onto 10 g autoclaved maize (Pioneer hybrid N82VGT), as previously described [63]. Briefly, single spored isolates were seeded at the center of V8 agar plates and incubated for 7 d at 31°C. Conidia were swabbed from plates with sterile cotton swabs into sterile distilled water (10 mL). The quantity of conidia was measured using a turbidity meter (Turbidimeter, Orbeco Analytical Systems) and maize was inoculated with 106 conidia/mL of each fungal isolate. Water content of maize was adjusted to 30%. Maize cultures were incubated at 31 °C for 7 d in the dark to allow fungal growth and aflatoxin formation. The experiment was terminated by addition of 50 mL 85% acetone and crop cultures were ground to homogeneity in a laboratory grade Waring Blender (seven-speed laboratory blender, Waring Laboratory, Torrington, CT, USA) at full speed for 30 s. The culture filtrate was spotted directly onto thin-layer chromatography (TLC) plates (Silica gel 60, EMD, Darmstadt, Germany) adjacent to aflatoxin standards (Aflatoxin Mix Kit-M, Supelco, Bellefonte, PA, USA) containing a mixture of known concentrations of aflatoxins B1, B2, G1, and G2. Plates were developed in ethyl ether-methanol-water (96:3:1), air-dried, and aflatoxins were visualized under 365-nm UV light. Total aflatoxins were quantified directly on TLC plates with a scanning densitometer (TLC Scanner 3, Camag Scientific Inc., Wilmington, NC, USA). Treatments were replicated four times and each experiment was performed twice.
Cyclopiazonic acid analyses were performed according to [64]. Conidial suspensions were inoculated onto 20 g of autoclaved maize using the method described above. For CPA analyses, inoculated maize was incubated for 7 d at 31 °C in the dark, and the experiment was terminated by addition of 100 mL of extraction solvent (20% methanol, 80% chloroform and 0.2% of 85% phosphoric acid). Maize-fungal cultures were ground at full speed to homogeneity in a laboratory grade Waring Blender. The slurry was transferred to polypropylene containers and shaken for 60 min at 200 rpm on a rotary shaker (HS501digital, IKA Works Inc., Wilmington, NC, USA). Extracts were filtered through Whatman No. 4 filter paper into 100 mL cylinders and the volume of extract was recorded. Each extract was transferred into a separatory funnel followed by addition of 100 mL of 0.5 N NaHCO3 with 3% (w/v) NaCl and gently shaken. The mixture was allowed to settle for 30 min and the bottom layer was discarded. Seven mL of concentrated HCl was added dropwise to each sample and gently shaken for 30 s. Once the bubbling subsided, samples were extracted twice with 25 mL of chloroform. Extracts were combined, dried, and re-suspended in 4 mL of chloroform. Extracts were spotted directly on TLC plates alongside a CPA standard of known concentration. Plates were dried at 50 °C for 15 min and then cooled for 1 min. Each plate was then dipped in Ehrlich’s reagent, quickly removed and air dried. Appearance of purple bands alongside the CPA standard indicated that samples were positive for CPA. Aspergillus flavus L strain isolate AF36, which is a registered biological control in the US, was used as the positive control [65]. CPA was quantified on TLC plates by scanning fluorescence densitometry at 546 nm (TLC Scanner 3, Camag Scientific Inc., Wilmington, NC).
4.3. DNA Extraction and PCR Amplifications
Fungal isolates were grown and DNA extraction was performed as described previously [66]. DNA concentration was adjusted to 5 ng/µl for PCR reactions. Partial gene fragments of β-tubulin (benA) (0.9 kb), calmodulin (cmdA) (1.2 kb), and nitrate reductase (niaD) (2.1 kb) genes were amplified (Table 5). The primer pair Bt3a-3b was designed in the current study to include the 5′ untranslated region of β-tubulin. Primers were designed based on genome sequence of A. nomius NRRL 13137 (GenBank accession no. JNOM01000216) using Primer3 version 0.4.0 [67,68]. PCR reactions were performed in 20 µl using 5 ng genomic DNA with a PCR premix (AccuPower® HotStart, Bioneer, Alameda, CA, USA) on a MyCycler thermocycler (Bio-Rad Laboratories, Richmond, CA, USA) under the following conditions: for β-tubulin, 5 min at 94 °C followed by 35 cycles of 96 °C for 30 s, locus-specific annealing temperature for 1 min (Table 5), 72 °C for 1 min, and 5 min at 72 °C; for cmdA and niaD genes, 5 min at 94 °C followed by 38 cycles of 94 °C for 20 s, locus-specific annealing temperature for 30 s (Table 5), 72 °C for 1 min, and 5 min at 72 °C. Amplicons were visualized with SYBR Gold after 1.0% agarose gel electrophoresis and sequenced at the University of Arizona Genetics Core facility (UAGC, Tucson, AZ, USA) using primers mentioned in Table 5. For mating type analyses, portions of MAT1-1 and MAT1-2 were amplified and characterized from all 11 A. texensis isolates following [69] with slight modifications: 2 min at 95 °C followed by 30 cycles of 94 °C for 30 s, 54 °C for 30 s, 72 °C for 45 s, and 2 min at 72 °C. Amplicons were visualized using a 1.5% agarose gel. DNA sequences of representative isolates are deposited at Genbank (Table S1).
4.4. Molecular Analysis and Phylogenetics
Bidirectional sequences were used to create a consensus sequence of each amplicon. Gene segments were assembled with either two (benA and cmdA) or six (niaD) amplicons per gene, corrected manually and gene alignments were generated using the MUSCLE algorithm within Geneious Pro Version 7.1.9 (Biomatters Ltd., Auckland, New Zealand). Phylogenetic trees were generated for concatenated and individual gene sequences following Bayesian inference using MrBayes version 3.2.6 [70] and maximum likelihood (ML) analysis with PhyML at Phylogeny.fr [71,72] to confirm tree topologies. Bayesian inference was conducted by running Markov Chain Monte Carlo analysis for up to 10 million generations. For ML analysis, datasets were bootstrapped with 500 replicates. Trees were mid-point rooted using FigTree v.1.4.3 [73]. The presence of a complete norB-cypA sequence in the A. texensis aflatoxin biosynthesis gene cluster was confirmed with previously described primer sets [51].
4.5. Data Analysis
Total aflatoxin was measured in µg/g. Aflatoxin concentrations produced by each isolate and each taxon were analyzed using Analysis of Variance as implemented in JMP 11.1.1 (SAS Institute, Cary, NC, USA, 2013). Means were separated using Tukey’s HSD test (p = 0.05). Fungal colony diameters were measured in millimeters (mm). Diameters for each medium and temperature were compared among species with Tukey’s HSD (p = 0.05). Data were tested for normality prior to statistical analyses and, if required, log-transformed. True means are presented for clarity.
Acknowledgments
This research was supported by the Agricultural Research Service, US Department of Agriculture [CRIS project 2020-42000-020-00D]. We thank Eve Beauchemin for excellent technical assistance.
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6651/10/12/513/s1, Figure S1: Mid-point rooted Bayesian phylogeny of A. texensis and closely related S morphology fungi with several additional species for reference based on partial calmodulin gene, cmdA (1.2 kb). Values above nodes are Bayesian posterior probabilities and values below nodes are bootstrap values from 500 replicates. Figure S2: Mid-point rooted Bayesian phylogeny of A. texensis and closely related S morphology fungi with several additional species for reference based on a portion of nitrate reductase gene, niaD (2.1 kb). Values above nodes or before commas are Bayesian posterior probabilities and values below nodes or after commas are bootstrap values from 500 replicates. Table S1: Isolates used in the current study with GenBank accession numbers. Sequences recovered from GenBank are indicated in bold.
Author Contributions
Conceptualization, P.S. and P.J.C.; Methodology, P.S. and P.J.C.; Validation, M.J.O. and P.J.C.; Formal Analysis, P.S.; Investigation, P.S.; Resources, P.J.C.; Data Curation, P.S.; Writing-Original Draft Preparation, P.S.; Writing-Review & Editing, M.J.O. and P.J.C.; Visualization, P.S., M.J.O. and P.J.C.; Supervision, M.J.O. and P.J.C.; Project Administration, P.J.C.; Funding Acquisition, P.J.C.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Key Contribution
A novel aflatoxin producing species from the United States is reported. Fungal isolates have an S morphology and produce B and G aflatoxins.
References
- 1.Cotty P.J. Aflatoxin-producing potential of communities of Aspergillus section Flavi from cotton producing areas in the United States. Mycol. Res. 1997;101:698–704. doi: 10.1017/S0953756296003139. [DOI] [Google Scholar]
- 2.Donner M., Lichtemberg P.S., Doster M., Picot A., Cotty P.J., Puckett R.D., Michailides T.J. Community structure of Aspergillus flavus and A. parasiticus in major almond-producing areas of California, United States. Plant Dis. 2015;99:1161–1169. doi: 10.1094/PDIS-05-14-0450-RE. [DOI] [PubMed] [Google Scholar]
- 3.Kachapulula P.W., Akello J., Bandyopadhyay R., Cotty P.J. Aspergillus section Flavi community structure in Zambia influences aflatoxin contamination of maize and groundnut. Int. J. Food Microbiol. 2017;261:49–56. doi: 10.1016/j.ijfoodmicro.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Probst C., Bandyopadhyay R., Cotty P.J. Diversity of aflatoxin-producing fungi and their impact on food safety in sub-Saharan Africa. Int. J. Food Microbiol. 2014;174:113–122. doi: 10.1016/j.ijfoodmicro.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 5.Singh P., Cotty P.J. Aflatoxin contamination of dried red chilies: Contrasts between the United States and Nigeria, two markets differing in regulation enforcement. Food Control. 2017;80:374–379. doi: 10.1016/j.foodcont.2017.05.014. [DOI] [Google Scholar]
- 6.De Iongh H., Vles R.O., Van Pelt J.G. Milk of mammals fed an aflatoxin-containing diet. Nature. 1964;202:466–467. doi: 10.1038/202466a0. [DOI] [PubMed] [Google Scholar]
- 7.Holzapfel C.W., Steyn P.S., Purchase I.F.H. Isolation and structure of aflatoxins M1 and M2. Tetrahedron Lett. 1966;7:2799–2803. doi: 10.1016/S0040-4039(01)99863-6. [DOI] [PubMed] [Google Scholar]
- 8.Frisvad J.C., Samson R.A., Smedsgaard J. Emericella astellata, a new producer of aflatoxin B1, B2 and sterigmatocystin. Lett. Appl. Microbiol. 2004;38:440–445. doi: 10.1111/j.1472-765X.2004.01520.x. [DOI] [PubMed] [Google Scholar]
- 9.Klich M.A., Mullaney E.J., Daly C.B., Cary J.W. Molecular and physiological aspects of aflatoxin and sterigmatocystin biosynthesis by Aspergillus tamarii and A. ochraceoroseus. Appl. Microbiol. Biotechnol. 2000;53:605–609. doi: 10.1007/s002530051664. [DOI] [PubMed] [Google Scholar]
- 10.Cotty P.J. Aflatoxin contamination of commercial cottonseed caused by the S strain of Aspergillus flavus. Phytopathology. 1996;86:S71. [Google Scholar]
- 11.Ehrlich K.C., Kobbeman K., Montalbano B.G., Cotty P.J. Aflatoxin-producing Aspergillus species from Thailand. Int. J. Food Microbiol. 2007;114:153–159. doi: 10.1016/j.ijfoodmicro.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Singh P., Cotty P.J. Characterization of Aspergilli from dried red chilies (Capsicum spp.): Insights into the etiology of aflatoxin contamination. Int. J. Food Microbiol. 2019;289:145–153. doi: 10.1016/j.ijfoodmicro.2018.08.025. [DOI] [PubMed] [Google Scholar]
- 13.Cotty P.J., Probst C., Jaime-Garcia R. Etiology and management of aflatoxin contamination. In: Leslie J.F., Bandyopadhyay R., Visconti A., editors. Mycotoxins: Detection Methods, Management, Public Health and Agricultural Trade. CAB International; Oxfordshire, UK: 2008. pp. 287–299. [Google Scholar]
- 14.Cardwell K.F., Cotty P.J. Distribution of Aspergillus section Flavi among field soils from the four agroecological zones of the Republic of Benin, West Africa. Plant Dis. 2002;86:434–439. doi: 10.1094/PDIS.2002.86.4.434. [DOI] [PubMed] [Google Scholar]
- 15.Donner M., Atehnkeng J., Sikora R.A., Bandyopadhyay R., Cotty P.J. Distribution of Aspergillus section Flavi in soils of maize fields in three agroecological zones of Nigeria. Soil Biol. Biochem. 2009;41:37–44. doi: 10.1016/j.soilbio.2008.09.013. [DOI] [Google Scholar]
- 16.Probst C., Callicott K.A., Cotty P.J. Deadly strains of Kenyan Aspergillus are distinct from other aflatoxin producers. Eur. J. Plant Pathol. 2012;132:419–429. doi: 10.1007/s10658-011-9887-y. [DOI] [Google Scholar]
- 17.Frisvad J.C., Hubka V., Ezekiel C.N., Hong S.B., Nováková A., Chen A.J., Arzanlou M., Larsen T.O., Sklenář F., Mahakarnchanakul W., et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Studies Mycol. 2019;93:1–63. doi: 10.1016/j.simyco.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotty P.J., Jaime-Garcia R. Influences of climate on aflatoxin producing fungi and aflatoxin contamination. Int. J. Food Microbiol. 2007;119:109–115. doi: 10.1016/j.ijfoodmicro.2007.07.060. [DOI] [PubMed] [Google Scholar]
- 19.International Agency for Research on Cancer (IARC) Traditional Herbal Medicines, Some Mycotoxins, Naphthalene and Styrene. IARC Monogr. Eval. Carcinog. Risks Hum. 2002;82:1–556. [PMC free article] [PubMed] [Google Scholar]
- 20.Shephard G.S. Aflatoxin and food safety: Recent African perspectives. J. Toxicol. Toxin Rev. 2003;22:267–286. doi: 10.1081/TXR-120024094. [DOI] [Google Scholar]
- 21.Williams J.H., Phillips T.D., Jolly P.E., Stiles J.K., Jolly C.M., Aggarwal D. Human aflatoxicosis in developing countries: A review of toxicology, exposure, potential health consequences, and interventions. Am. J. Clin. Nutr. 2004;80:1106–1122. doi: 10.1093/ajcn/80.5.1106. [DOI] [PubMed] [Google Scholar]
- 22.Turner P.C., Moore S.E., Hall A.J., Prentice A.M., Wild C.P. Modification of immune function through exposure to dietary aflatoxin in Gambian children. Environ. Health Perspec. 2003;111:217–220. doi: 10.1289/ehp.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong Y.Y., Hounsa A., Egal S., Turner P.C., Sutcliffe A.E., Hall A.J., Cardwell K., Wild C.P. Postweaning exposure to aflatoxin results in impaired child growth: A longitudinal study in Benin, West Africa. Environ. Health Perspect. 2004;112:1334–1338. doi: 10.1289/ehp.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khlangwiset P., Shephard G.S., Wu F. Aflatoxins and growth impairment: A review. Crit. Rev. Toxicol. 2011;41:740–755. doi: 10.3109/10408444.2011.575766. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y., Wu F. Global burden of aflatoxin-induced hepatocellular carcinoma: A risk assessment. Environ. Health Perspect. 2010;118:818–824. doi: 10.1289/ehp.0901388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J.S., Tang L. Epidemiology of aflatoxin exposure and human liver cancer. Toxin Rev. 2004;23:249–271. doi: 10.1081/TXR-200027834. [DOI] [Google Scholar]
- 27.Centers for Disease Control and Prevention Outbreak of aflatoxin poisoning—Eastern and central provinces, Kenya, January–July 2004. Morb. Mortal. Wkly. Rep. 2004 Jul 20;53:790–793. [PubMed] [Google Scholar]
- 28.Tanzania: Food Poisoning Linked to 14 Deaths in Two Regions. [(accessed on 6 June 2018)]; Available online: http://allafrica.com/stories/201607290685.html.
- 29.Aflatoxin Kills 4 Children in Tanzania. [(accessed on 6 June 2018)]; Available online: https://www.ippmedia.com/en/news/aflatoxin-kills-4-children-tanzania.
- 30.Robens J., Cardwell K. The costs of mycotoxin management to the USA: Management of aflatoxins in the United States. J. Toxicol. Toxin Rev. 2003;22:139–152. doi: 10.1081/TXR-120024089. [DOI] [Google Scholar]
- 31.Cotty P.J., Bayman P., Egel D.S., Elias K.S. Agriculture, aflatoxins and Aspergillus. In: Powell K.A., Renwick A., Perberdy J.F., editors. The Genus Aspergillus: From Taxonomy and Genetics to Industrial Application. Plenum Press; New York, NY, USA: 1994. pp. 1–27. [Google Scholar]
- 32.Cotty P.J. Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology. 1989;79:808–881. doi: 10.1094/Phyto-79-808. [DOI] [Google Scholar]
- 33.Jaime-Garcia R., Cotty P.J. Crop rotation and soil temperature influence the community structure of Aspergillus flavus in soil. Soil Biol. Biochem. 2010;42:1842–1847. doi: 10.1016/j.soilbio.2010.06.025. [DOI] [Google Scholar]
- 34.Carvajal-Campos A., Manizan A.L., Tadrist S., Akaki D.K., Koffi-Nevry R., Moore G.G., Fapohunda S.O., Bailly S., Montet D., Oswald I.P., et al. Aspergillus korhogoensis, a Novel Aflatoxin Producing Species from the Côte d’Ivoire. Toxins. 2017;9:353. doi: 10.3390/toxins9110353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson S.W., Ito Y., Horn B.W., Goto T. Aspergillus bombycis, a new aflatoxigenic species and genetic variation in its sibling species, A. nomius. Mycologia. 2001;93:689–703. doi: 10.2307/3761823. [DOI] [Google Scholar]
- 36.Ito Y., Peterson S.W., Wicklow D.T., Goto T. Aspergillus pseudotamarii, a new aflatoxin producing species in Aspergillus section Flavi. Mycol. Res. 2001;105:233–239. doi: 10.1017/S0953756200003385. [DOI] [Google Scholar]
- 37.Kurtzman C.P., Horn B.W., Hesseltine C.W. Aspergillus nomius, a new aflatoxin-producing species related to Aspergillus flavus and Aspergillus tamarii. Antonie van leeuwenhoek. 1987;53:147–158. doi: 10.1007/BF00393843. [DOI] [PubMed] [Google Scholar]
- 38.Horn B.W. Aspergillus caelatus, a new species in section Flavi. Mycotaxon. 1997;61:185–191. [Google Scholar]
- 39.Wei D.L., Jong S.C. Production of aflatoxins by strains of the Aspergillus flavus group maintained in ATCC. Mycopathologia. 1986;93:19–24. doi: 10.1007/BF00437010. [DOI] [PubMed] [Google Scholar]
- 40.Rambo G.W., Tuite J., Crane P. Preharvest inoculation and infection of dent corn ears with Aspergillus flavus and Aspergillus parasiticus. Phytopathology. 1974;64:797–800. doi: 10.1094/Phyto-64-797. [DOI] [Google Scholar]
- 41.Hesseltine C.W., Shotwell O.L., Smith M., Ellis J.J., Vandegraft E., Shannon G. Production of Various Aflatoxins by Strains of the Aspergillus flavus Series. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1970. pp. 202–210. [Google Scholar]
- 42.Pildain M.B., Frisvad J.C., Vaamonde G., Cabral D., Varga J., Samson R.A. Two novel aflatoxin-producing Aspergillus species from Argentinean peanuts. Int. J. Syst. Evol. Microbiol. 2008;58:725–735. doi: 10.1099/ijs.0.65123-0. [DOI] [PubMed] [Google Scholar]
- 43.Geiser D.M., Pitt J.I., Taylor J.W. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc. Natl. Acad. Sci. USA. 1998;95:388–393. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Probst C., Njapau H., Cotty P.J. Outbreak of an acute aflatoxicosis in Kenya in 2004: Identification of the causal agent. Appl. Environ. Microbiol. 2007;73:2762–2764. doi: 10.1128/AEM.02370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cotty P.J., Cardwell K.F. Divergence of West African and North American Communities of Aspergillus section Flavi. Appl. Environ. Microbiol. 1999;65:2264–2266. doi: 10.1128/aem.65.5.2264-2266.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito M., Tsuruta O. A new variety of Aspergillus flavus from tropical soil in Thailand and its aflatoxin productivity. Proc. Jpn. Assoc. Mycotoxicol. 1993;1993:31–36. doi: 10.2520/myco1975.1993.31. [DOI] [Google Scholar]
- 47.Samson R.A., Varga J. What is a species in Aspergillus? Med. Mycol. 2009;47:S13–S20. doi: 10.1080/13693780802354011. [DOI] [PubMed] [Google Scholar]
- 48.Soares C., Rodrigues P., Peterson S.W., Lima N., Venâncio A. Three new species of Aspergillus section Flavi isolated from almonds and maize in Portugal. Mycologia. 2012;104:682–697. doi: 10.3852/11-088. [DOI] [PubMed] [Google Scholar]
- 49.Peterson S.W. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia. 2008;100:205–226. doi: 10.1080/15572536.2008.11832477. [DOI] [PubMed] [Google Scholar]
- 50.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ehrlich K.C., Chang P.K., Yu J., Cotty P.J. Aflatoxin biosynthesis cluster gene cypA is required for G aflatoxin formation. Appl. Environ. Microbiol. 2004;70:6518–6524. doi: 10.1128/AEM.70.11.6518-6524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bayman P., Cotty P.J. Vegetative compatibility and genetic diversity in the Aspergillus flavus population of a single field. Can. J. Bot. 1991;69:1707–1711. doi: 10.1139/b91-216. [DOI] [Google Scholar]
- 53.Boyd M.L., Cotty P.J. Aspergillus flavus and aflatoxin contamination of leguminous trees of the Sonoran Desert in Arizona. Phytopathology. 2001;91:913–919. doi: 10.1094/PHYTO.2001.91.9.913. [DOI] [PubMed] [Google Scholar]
- 54.Jaime-Garcia R., Cotty P.J. Aspergillus flavus in soils and corncobs in south Texas: Implications for management of aflatoxins in corn-cotton rotations. Plant Dis. 2004;88:1366–1371. doi: 10.1094/PDIS.2004.88.12.1366. [DOI] [PubMed] [Google Scholar]
- 55.Horn B.W., Greene R.L., Dorner J.W. Effect of corn and peanut cultivation on soil populations of Aspergillus flavus and A. parasiticus in southwestern Georgia. Appl. Environ. Microbiol. 1995;61:2472–2475. doi: 10.1128/aem.61.7.2472-2475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tubajika K.M., Mascagni H.J., Damann K.E., Russin J.S. Nitrogen fertilizer influence on aflatoxin contamination of corn in Louisiana. J. Agric. Food Chem. 1999;47:5257–5260. doi: 10.1021/jf990535f. [DOI] [PubMed] [Google Scholar]
- 57.King J.M., Walker T., Njapau H., Park D.L., Damann K.E. Managing aflatoxin contamination in corn: Scientists use integrated approach to solution. Louisiana Agric. 2000;43:20–21. [Google Scholar]
- 58.Aflatoxin Prevention Arkansas Corn and Grain Sorghum Board. [(accessed on 14 June 2018)]; Available online: http://www.corn-sorghum.org/aflatoxin-prevention-0.html.
- 59.Jaime-Garcia R., Cotty P.J. Aflatoxin contamination of commercial cottonseed in south Texas. Phytopathology. 2003;93:1190–1200. doi: 10.1094/PHYTO.2003.93.9.1190. [DOI] [PubMed] [Google Scholar]
- 60.Jaime-Garcia R., Cotty P.J. Spatial relationships of soil texture and crop rotation to Aspergillus flavus community structure in South Texas. Phytopathology. 2006;96:599–607. doi: 10.1094/PHYTO-96-0599. [DOI] [PubMed] [Google Scholar]
- 61.Adye J.A.R.I.M., Mateles R.I. Incorporation of labelled compounds into aflatoxins. Biochim. Biophys. Acta Gen. Subj. 1964;86:418–420. doi: 10.1016/0304-4165(64)90077-7. [DOI] [PubMed] [Google Scholar]
- 62.Pitt J.I., Hocking A.D., Glenn D.R. An improved medium for the detection of Aspergillus flavus and A. parasiticus. J. Appl. Microbiol. 1983;54:109–114. doi: 10.1111/j.1365-2672.1983.tb01307.x. [DOI] [PubMed] [Google Scholar]
- 63.Probst C., Cotty P.J. Relationships between in vivo and in vitro aflatoxin production: Reliable prediction of fungal ability to contaminate maize with aflatoxins. Fungal Biol. 2012;116:503–510. doi: 10.1016/j.funbio.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 64.Lansden J.A. Determination of cyclopiazonic acid in peanuts and corn by thin layer chromatography. J. Assoc. Off. Anal. Chem. 1986;69:964–966. [PubMed] [Google Scholar]
- 65.Chang P.K., Horn B.W., Dorner J.W. Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet. Biol. 2005;42:914–923. doi: 10.1016/j.fgb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 66.Callicott K.A., Cotty P.J. Method for monitoring deletions in the aflatoxin biosynthesis gene cluster of Aspergillus flavus with multiplex PCR. Lett. Appl. Microbiol. 2015;60:60–65. doi: 10.1111/lam.12337. [DOI] [PubMed] [Google Scholar]
- 67.Koressaar T., Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 68.Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramirez-Prado J.H., Moore G.G., Horn B.W., Carbone I. Characterization and population analysis of the mating-type genes in Aspergillus flavus and Aspergillus parasiticus. Fungal Genet. Biol. 2008;45:1292–1299. doi: 10.1016/j.fgb.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 70.Huelsenbeck J.P., Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 71.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., et al. Phylogeny. fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dereeper A., Audic S., Claverie J.M., Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rambaut A. FigTree v1. 4. [(accessed on 30 March 2018)];2012 Available online: http://tree.bio.ed.ac.uk/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.