Abstract
N6-methyladenosine (m6A) is known to occur in plant and animal messenger RNAs (mRNAs) since the 1970s. However, the scope and function of this modification remained un-explored till very recently. Since the beginning of this decade, owing to major technological breakthroughs, the interest in m6A has peaked again. Similar to animal mRNAs, plant mRNAs are also m6A methylated, within a specific sequence motif which is conserved across these kingdoms. m6A has been found to be pivotal for plant development and necessary for processes ranging from seed germination to floral development. A wide range of proteins involved in methylation of adenosine have been identified alongside proteins that remove or identify m6A. This review aims to put together the current knowledge regarding m6A in Arabidopsis thaliana.
Keywords: N6-methyladenosine; mRNA m6A methyltransferase (MTA), MTB; FIP37; ALKBH; YTDHF; Arabidopsis
1. Introduction
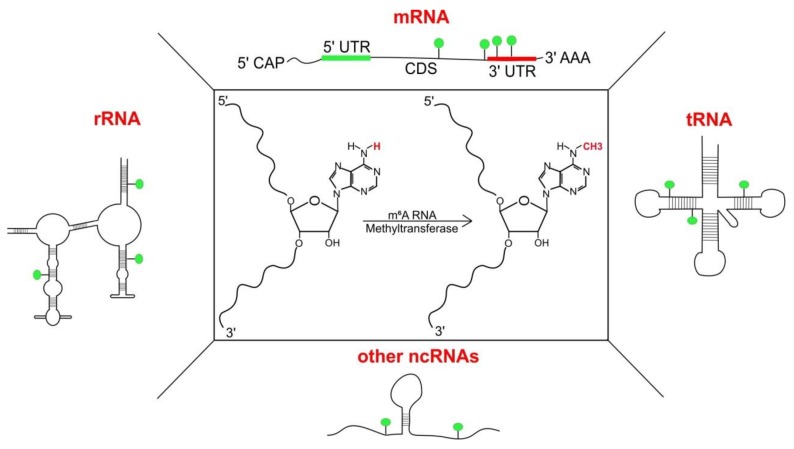
Methylation of internal adenosine at nitrogen-6 position (m6A) is a RNA modification abundant across Eukaryota, and is found in messenger RNAs (mRNAs), transfer RNAs (tRNAs) [1], ribosomal RNAs (rRNAs) [2] and other non-coding RNAs (ncRNAs) [3,4] (Figure 1). m6A was first found in mammalian mRNAs in the 1970s [5,6,7] followed by plants [8,9] and viruses [10,11]; later in yeast [12] and much more recently in bacteria [13]. With its discovery, m6A was added to a pool of other RNA modifications [14,15] but its role remained largely obscured till the late 1990s. m6A was challenging to study, as it does not cause any changes in Watson–Crick base pairing and thus could not be detected by traditional reverse transcription methods [16]. With the technological advances surrounding scientific techniques and new methods being developed for functional analysis of such RNA modifications, m6A became relevant again, surprisingly, with the discovery that this modification is reversible [17] and has an influence on a variety of essential processes in cells [18].
Figure 1.
Structure and presence of adenosine methylation at nitrogen-6 position (m6A): Substitution of ‘H’ at N6 with ‘CH3’ (central panel, marked in red) in adenosine results in the formation of N6-methyladenosine. m6A has been detected indifferent classes of RNA including messenger RNAs (mRNAs), transfer RNAs (tRNAs) [1], ribosomal RNAs (rRNAs) [2] and other non-coding RNAs (ncRNAs) like small nucleolar RNAs (snoRNAs), long non-coding RNAs (lncRNAs) and primary micro-RNAs (pri-miRNAs).
First reports of m6A in plant mRNAs date back to 1979, when it was first found in wheat (Triticum aestivum) and maize (Zea mays) [8,9]. Soon after, m6A mark in mRNA was reported in oat (Avena sativa) [19]. Interestingly, shortly after the first discovery of m6A in mRNAs of plants, scientists were able to identify the sequence motif RRACH (where R = G/A, H = A/C/U, bold letter stands for adenosine being modified to m6A) which was in consensus with the animal sequence motif [20]. After these initial discoveries, the interest in plant m6A gradually decreased and was regained after its discovery in Arabidopsis thaliana which was reported much later in 2008 by Zhong and colleagues [21]. It was also found, that although the sequence motif in animals and plants is conserved, the sites of m6A enrichment show some differences between them: While m6A in animals is highly enriched near the 3′UTRs and stop codons [3,22], Arabidopsis m6A mRNA methylome also shows enrichment near the start codon [23] besides 3′UTRs and stop codons.
2. m6A: The Methyltransferase Complex
The fact that mRNA m6A methyltransferase complex in animals is a multi-protein complex [24] was known for a long time. Using nuclear extracts from HeLa cells, Bokar and colleagues pulled down the m6A methyltransferase complex (which was named as m6A-MT) using a synthetic RNA with the known binding site as a bait. They identified three major components of the complex and designated them as MT-A1 (30 KDa), MT-A2 (200 KDa) and MT-B (875 KDa). It was also noted that all three components had to be present to observe any m6A methylation activity. Later, Bokar and colleagues also identified a 70 kDa protein from MT-A2 subcomplex as containing the S-adenosylmethionine (AdoMet) binding domain and it was named as MT-A70/METTL3 [25].
The characterization of Arabidopsis mRNA m6A methyltransferase complex was initiated by the identification of a homolog of human mRNA m6A methyltransferase (MTA-70/METTL3): mRNA adenosine methylase (MTA) [21]. The functional importance of MTA was also demonstrated by Zhong and colleagues by loss of function studies, where disruption of the MTA gene led to embryo lethality. This observation is in consensus with results from studies on animals, as disruption of MTA/METTL3 homologs has been shown to be embryo lethal in mice [26], causes apoptosis in mammalian cells [27] and leads to increased female lethality in Drosophila melanogaster [28,29]. To overcome the obvious difficulties in studying the role of m6A or lack thereof in Arabidopsis, Bodi and colleagues complemented the KO mutant of MTA with ABI3:MTA. The ABI3 promoter drives the expression of MTA in the embryonic stages of development, enough to rescue the embryo-lethal phenotype, and at a vastly reduced level at later stages [30]. Expression analysis of MTA also showed that it is mostly expressed in dividing tissue (apical meristems, reproductive organs and seeds) while the expression levels are quite low in other tissues; m6A methylation levels also vary in the tissues accordingly [21].
While the characterization of Arabidopsis MTA has been performed and shown in detail, its protein partner, Arabidopsis MTB, has been characterized less intensively. MTB is the homolog of human METTL14, which has also been shown to be a part of m6A methyltransferase complex [31]. On the lines of homology with the human METTL14, MTB was considered to be a possible m6A methyltransferase complex partner [32], but surprisingly no study focusing solely on MTB and its role on m6A methylation can be found. MTB has been characterized as embryo defective and complete lack of MTB is also embryo lethal. A 2017 study, focusing on newer components of m6A methyltransferase complex (discussed below) also takes MTB into account [33]. In their study, Růžička and colleagues, found that MTB associated with MTA and also formed homodimers. MTB was also shown to interact with the newly discovered components of m6A methyltransferase complex as identified in the same study. Experiments on RNA interference RNAi lines with inducible knockdown of MTB show that knockdown of MTB leads to nearly a 50% reduction in m6A levels [33].
The first characterization of an MTA protein partner in the mRNA m6A methyltransferase complex was reported from Arabidopsis, in the form of FKBP12 interacting protein 37 kDa (AtFIP37) [21,34]. FIP37 is a homolog of human WTAP (Wilms’ Tumour1-Associating Protein) and Drosophila FL(2)D (Female Lethal2) [21]; disruption of AtFIP37 is also embryo lethal [35]. Using an approach identical to Bodi et al., transgenic lines with LEC1:FIP37 (embryo specific) and also ABI3:FIP37 constructs were used by researchers to show that low levels of FIP37 cause developmental defects in the shoot apical meristem (SAM), without proper leaf production and eventual death of plants. FIP37 expression patterns are similar to that of MTA’s, and as one would expect, m6A levels in plants with low levels of FIP37 were found to be much lower than in wild type (WT) plants [34].
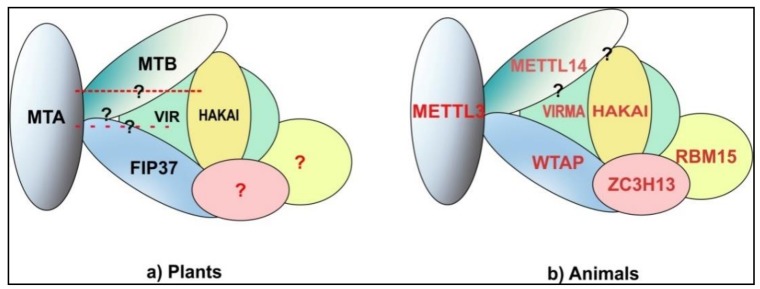
While MTA, MTB and FIP37 are largely considered to form the core of m6A methyltransferase complex, they definitely are not the only proteins involved in m6A methylation. Růžička and colleagues showed that a homolog of human VIRMA (KIAA1429) (Drosophila—Virilizer (VIR)) [36,37] is also a part of Arabidopsis m6A methyltransferase complex, along with the homolog of human HAKAI (an E3 Ubiquitin ligase) [33,38]. VIR is also essential for embryo development and null alleles are embryo lethal. Furthermore, the authors also show significant reductions of m6A levels in vir and hakai mutants (where both proteins are expressed inefficiently). Interestingly, while the lack of all other proteins discussed above result in similar sever phenotypes, HAKAI null mutants are not only viable but also show phenotypes similar to WT plants while some phenotypic defects are still present [33]. While the association of these new members with the m6A methyltransferase complex and their necessity for m6A methylation has been proven, they have not yet been attributed to any specific role within the complex. Figure 2 shows the comparison of m6A methyltransferase complexes in plants and animals.
Figure 2.
m6A methyltransferase complex: (a) PlantsMTA-MTB-FIP37 form the core components of the Arabidopsis m6A methyltransferase complexof which VIR and HAKAI are also a part.VIR and HAKAI interact with MTB and FIP37 but no interaction with MTA has been shown yet. Similarly, direct interactions between MTB and FIP37 have not been shown yet. Whether homologs of animal ZC3H13 and RBM15 are a part of plant methyltransferase complex still needs to be shown. The components of methyltransferase complex here are shown on the basis of data from studies on Arabidopsis. (b) Animal interactions between all three core components METTL3-WTAP-METTL14 has been shown. VIRMA (KIAA1429) (VIR homolog) and HAKAI have been shown to be part of the complex via interactions with WTAP and their interaction with METTL14 is yet to be elucidated. Most recent data suggests that WTAP along with RBM15/ZC3H13/HAKAI/VIRMA provides a scaffold for METTL3/METTL14 for methylation [39,40,41,42]. The animal model is based on data from mammals and Drosophila. Question marks depict possible but not yet identified interactions.
3. m6A: Readers and Erasers
It is known that methylation of adenosine does not alter nucleotide base pairing, but it alters the secondary structure of RNA and plays a role in RNA-protein interactions [43,44,45,46,47]. The change in RNA secondary structure is mainly brought about by lowered thermodynamic stability of RNA duplexes containing the m6A mark [43]. It was also shown that the presence of m6A in a hairpin loop leaves the double stranded region more accessible to various proteins, a phenomenon that is termed as “m6A switch” [44,45,46]. It was also shown that the presence of m6A in hairpin loop destabilizes the double stranded structure, while single stranded RNA (ssRNA) is stabilized by m6A and presence of m6A causes the adjacent area to resemble single stranded RNA, thus leaving it exposed to RNA binding proteins [47]. Lastly, proteins that directly identify the m6A mark have also been identified. YT521-B homology (YTH) domain family proteins (YTHDF) is an example of such a family of proteins that has been shown to recognize m6A and bind RNA [48,49,50,51,52]. Thus, it is not hard to imagine that a majority of biological effects of reduced m6A levels are a result of RNA secondary structure distortions and/or lack of interactions between certain proteins identifying m6A marks in mRNAs and facilitating further processes in plants. These proteins, that can identify m6A, are called “reader” proteins. On the other hand, proteins (demethylases) that identify m6A and affect RNA metabolism by removing m6A are called “erasers.” Recently, researchers working on plants have focused on readers and erasers to explain the physiological effects that accompany the lack of m6A.
In plants, 13 YTH family proteins are known to occur and out of those 11 have been identified as Evolutionarily Conserved C-Terminal Region proteins 1–11 (ECT 1–11). The first report of two ECT proteins (ECT1/2) in 2005 by Ok and colleagues associated them with calcium signaling and showed that the conserved C-terminal domain (YTH domain) is necessary for their nuclear localization. ECT2 has since been shown to bind m6A in Arabidopsis [48,50,51,52]. Scutenaire and colleagues show that ECT2 binds to m6A via a tri-tryptophan pocket, and if these amino acids are mutated, ECT2 loses its m6A binding ability. They also show that ect mutants share phenotypes (defective trichomes) with mta mutants and FIP37 overexpressing transgenic lines. They also show that the altered trichome morphology is a result of higher cell ploidy caused by endoreduplication [50]. On the other side, Wei and colleagues demonstrate that ECT2 binds to m6A at a motif, URUAY (R=G>A, Y=U>A), that is not similar to the MTA methylation motif of RRACH. They present URUAY as a new plant specific motif that plant m6A methyltransferase can recognize and methylate [51] and this data is supported by recently published data from another group who also found enrichment of U rich motif in their analysis regarding global mapping of uncapped and cleaved transcripts [53]. Providing a more molecular mechanism for the altered trichome morphology by ECT2, Wei and colleagues show that ECT2 improves the stability of m6A methylated RNAs transcribed from genes involved in trichome morphogenesis. This observation is opposite to the reported decrease in stability of RNAs caused by YTHDF proteins’ binding this mark in animal system [54]. While animal YTH domain protein (YTHDF2) causes accelerated deadenylation of transcripts carrying the m6A mark, Anderson and group show that, in plants, m6A prevents ribonucleolytic cleavage of such transcripts (as observed by Wei and colleagues too) [51,53,54]. In a study focused more on the morphological aspects of ECT proteins, including ECT2/3 and4, Arribas-Hernández and colleagues show that these proteins are intrinsically important for proper leaf morphogenesis including trichome branching [52]. These results provide answers to the observations made in the very beginning of MTA characterization regarding trichome branching [30].
Similar to the “reader” proteins, “erasers” can also modulate various biological processes that result in certain distinct phenotypes and hence help in identification of newer pathways where m6A plays a role. AlkB family of non-heme Fe(II)/α-ketoglutarate (α-KG)-dependent dioxygenases family proteins and their homologs (ALKBH), are known to act as m6A demethylases in mammalian systems [17,55]. ALKBH family proteins are also found in Arabidopsis and two of them, ALKBH9B and ALKBH10B, have been shown to be active m6A demethylases concerning plant systems [56,57]. ALKBH9B was the first m6A demethylase reported from the Arabidopsis system, although it is shown to affect viral ssRNA methylation status. With the spotlight on alfalfa mosaic virus (AMV), researchers demonstrate that ALKBH9B positively affects viral abundance in plant cells [57]. Working on this further, the group also found that ALKBH9B can demethylate m6A from ssRNA in vitro. They also show that the viral RNA gets m6A methylated upon infection. Depletion of ALKBH9B led to the viral RNA being hypermethylated and eventually degraded by Non-sense Mediated Decay (NMD). These observations, when combined with the fact that lack of ALKBH9B led to lower infection levels, allowed the authors to conclude that methylation status plays a vital role in modulating viral infections in Arabidopsis [57].
Interestingly, a study done in Nicotiana tobaccum also co-relates m6A with viral infections [58]. Li and colleagues show that upon infection with tobacco mosaic virus (TMV), methylation levels were decreased in Nicotiana plants while simultaneously the expression level of a protein homologous to human ALKBH5 demethylase went up [58]. This indicates that a mechanism similar to Arabidopsis exists in other plants as well.
ALKBH9B, while interesting as m6A demethylase, was not demonstrated to work on any RNA substrate endogenous to Arabidopsis. In order to focus on demethylase that would be more involved in plant metabolism, Duan and colleagues targeted another ALKBH family protein, ALKBH10B. ALKBH10B binds to and demethylates m6A RNA both in vitro and in vivo. Scientists observed that disruption of ALKBH10B leads to global upregulation of mRNA methylation. This upregulation could be seen in genes that are involved in several developmental pathways and organ development among others. One of the phenotypic effects of ALKBH10B over/under expression observed by the authors was altered flowering time. alkbh10b null mutants flower later than WT plants and ALKBH10B overexpressing plants flower earlier when compared to wild type plants. This phenomenon was attributed to the methylation status of mRNAs corresponding to several genes involved in proper flowering, especially but not limited to, Flowering Locus T (FT) [56]. Researchers were able to show that ALKBH10B directly binds to FT mRNA (which contains m6A residues) and demethylates it. In alkbh10b background, FT transcripts are degraded faster as compared to WT plants, linking m6A levels to the degradation of mRNA.
4. m6A: Physiological Roles
The fact that the lack of m6A methylation (or any of the core mRNA m6A methyltransferase complex proteins) is lethal for plants highlights the importance of this modification in primary metabolism of plants. m6A methylation in Arabidopsis could be associated with a variety of stress and/or stimuli responses, as was indicated by the analysis of gene expression comparing WT and mta mutants [23,30]. m6A marks have been found in mRNAs of photosynthesis related proteins, notably Serine/Threonine Protein Kinase (STN8), indicating towards some role of m6A in this vital process [23]. Upon investigation of m6A levels in different organs, it was observed that m6A methylation levels, varied in different organs indicating towards some role of m6A in organ development [21,59]. Zhong and colleagues show that m6A abundance is the highest in young seedlings and flower buds followed by leaves and roots. Contrary to this, Wan and colleagues reported that methylation levels were the highest in leaves, followed by flowers and roots. They also report that transcripts with higher levels of methylation in each organ are different and somehow co-related. For example: transcripts related to photosynthesis were methylated at a higher level in leaves as compared to flowers or roots; transcripts related to alkaloid biosynthesis had higher methylation levels in roots as compared to the other two organs etc. In the same study, Wan and colleagues, also report presence of the m6A mark in mRNA of various transporter and signal transduction proteins. They also observed that methylation levels were generally higher in transcripts that are normally present at lower levels in the cell. The authors speculate that m6A methylation may play a role in providing stability to such low expressed transcripts [59]. m6A methylation patterns were also found to be strain specific, as Luo et al. found that two accessions (Can-0 and Hen-16) had strain specific enrichment of the m6A mark. The enrichment of m6A in these two strains also co-related to the different gene expression levels pointing towards the differences in m6A patterns to different geographical habitat of these strains [23]. Recently, studies to understand the role m6A in plants other than Arabidopsis like tobacco (discussed earlier) and rice have also been undertaken [60]. Genome wide identification of rice m6A transcripts in two different tissue types (differentiated callus and leaves) revealed an organ specific m6A pattern in transcripts (as observed previously in Arabidopsis); and a negative correlation between m6A methylation enrichment and gene expression [60].
The presence or lack of m6A marks in transcripts is not a definitive sign that it plays some significant role in plant metabolism. A more physiologically relevant proof of such role comes from experiments that show the physiological effects of reduced m6A levels. When MTA level is lower, plants show developmental defects like reduced inflorescence length, increased trichome branching with crinkled leaves and differences in flower morphology with reduced seed production [30]. Increased trichome branching is also observed in Arabidopsis line over-expressing FIP37 protein [35]. A pioneering study that related m6A levels directly to morphological changes, focused on FIP37 [34]. Shen and colleagues found that the deficiency of FIP37 caused overproliferation of SAM tissue. After showing that FIP37 is otherwise highly expressed in dividing tissue and it is necessary for m6A methylation, the authors found two genes (WUSCHEL (WUS) and SHOOTMERISTEMLESS (STM)) involved in SAM proliferation that have an m6A mark in their mRNAs. Upon depletion of FIP37, the depletion of m6A was accompanied by over accumulation of these two mRNAs. This overaccumulation was shown to be a result of slower degradation of these mRNAs because of the lack of m6A marks. The authors also demonstrated the overaccumulation of these transcripts and their lack of methylation when MTA levels were reduced, using artificial microRNA (miRNA) based RNAi technology [34]. Through their experiments Shen and colleagues clearly demonstrated a pathway where the m6A mark enhances degradation of certain mRNAs and plays a significant role in their homeostasis which is reflected in proper plant growth and development.
Recent work by Anderson and colleagues, shows that m6A stabilizes transcripts as it protects them from ribonucleolytic cleavage [53]. An increase in cleavage of transcripts, 4–5 nt upstream of adenosine, in the absence of m6A proved that m6A indeed protects these transcripts from cleavage. Moreover, under stress, m6A also stabilizes stress responsive gene transcripts [53]. As discussed in earlier sections, m6A is also associated with viral infections (biotic stress) and its presence in a variety of transcripts could link this modification to many more vital processes.
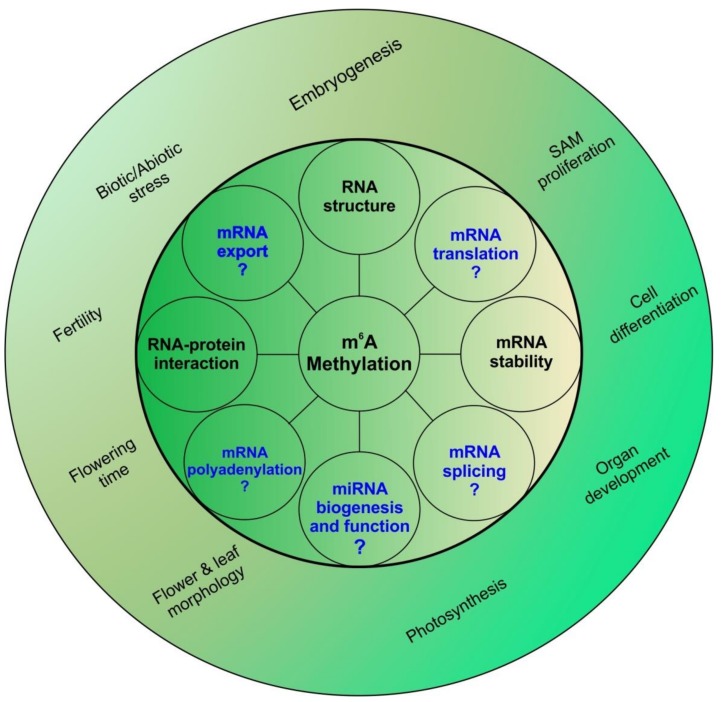
Apart from affecting mRNA metabolism, some other roles of m6A have already been elucidated in animal systems; one such role is in miRNA biogenesis. In 2015, researchers showed that m6A methylation acts as a mark for further processing of mammalian miRNAs [61]. Developing on this further, it was also shown that Heterogenous Nuclear Ribonuclear Protein A2B1 (HNRNPA2B1) identifies the m6A mark and facilitates further processing of pri-miRNAs [62]. To our knowledge, no information regarding the role of m6A marks in plant miRNA biogenesis is available. Figure 3 summarizes physiological and molecular roles of m6A in plants.
Figure 3.
m6A and its many roles: m6A methylation has been known to alter mRNA stability and structure along with altering RNA-protein interactions at the molecular level. These changes in molecular levels are translated to a large array of physiological changes ranging from photosynthesis to stress response. In animal systems, m6A has also been shown to affect mRNA splicing, export, polyadenylation and translation; whether these also play a role in plants remains to be understood. m6A has been shown to be vital for proper shoot apical meristem (SAM) proliferation and organ development. Flower growth, morphology and fertility are also affected by m6A. In animals m6A has also been associated with microRNA (miRNA) biogenesis; whether this is true for plants is still unknown.
5. Conclusions and Future Perspectives
While the field of m6A in plants is still evolving, it is clear that m6A methylation plays a definitive role in plant development. In animal systems, m6A has been shown to play a role in processes ranging from mRNA stability, splicing, maturation to cell differentiation and stress response (for a review: Reference [14]). In close agreement, m6A in plant systems also plays roles similar to animal counterparts, as discussed in this review. While we know that m6A alters mRNA stability in Arabidopsis, it appears that these changes are not universal. As discussed earlier, Anderson and colleagues [53] prove that m6A methylation provides stability to mRNAs while Shen and colleagues [34] demonstrate that it promotes transcript degradation. Contradictory results were also reported regarding the extent of m6A methylation between organs, case in point: Two studies by Zhong and colleagues [21], and Wan and colleagues [59]. Zhong and colleagues used TLC to detect m6A and report that flower buds contain the most amount of m6A, while based on m6A sequencing, Wan and colleagues report that m6A methylation is highest in leaves. While highest methylation levels in flowers would be rational, considering that MTA is highly expressed in actively dividing tissue, higher m6A levels in leaves was explained by Wan and colleagues as a consequence of higher photosynthetic and metabolic importance of leaves. Accordingly, we should keep in mind that this could be a matter of different sensitivities of the two different techniques used in the two studies and as the field is still incorporating new tools for analysis, such discrepancies might arise from time to time. On the other hand, the data generated so far clearly suggests that m6A might influence cell metabolism in very complex patterns. Recent data also points to some possible differences between the animal and plant sequence motif within which m6A is deposited. As discussed earlier, UGAU has been reported as a possible m6A methylation motif in Arabidopsis by two different groups [51,53]. Furthermore, data shows that m6A methylation is enriched in some organs of plants than others, and apart from the information that the methylation of transcripts correspond to their function and importance in the organs, a lot needs to be known about the underlying mechanisms regarding such regulation. This is also the case of difference in m6A enrichment across Arabidopsis strains and could point that m6A is also affected by the environmental cues that are not necessarily stress related. Lastly, a possibility that m6A levels in a particular organ in one plant could also vary with time (day or night) cannot be overlooked. As mentioned earlier, m6A also plays a role in miRNA biogenesis in mammals. miRNAs are known to be important players in plant stress responses [63,64,65] and many stress responsive gene transcripts have been found to be m6A methylated. Keeping that in mind, it would be interesting to see whether miRNA biogenesis influenced by m6A works in conjunction with m6A methylation of mRNAs to modulate stress responses. Characterization of more downstream proteins i.e., the “readers” and “erasers”, will further our understanding of the numerous ways m6A may affect plant development.
For an epigenetic modification that has been known since the 1970s, relevant techniques and instruments necessary to study m6A did not develop at the same pace, thus m6A remained relatively unexplored by researchers. Knowing that m6A modification is essential for life and thanks to the rapid advancement of technology and gradual increments in our understanding of this abundant and reversible modification, the future for m6A is filled with many more important questions that need to be answered.
Funding
This review was funded by KNOW Poznan RNA Centre (grant no. 01/KNOW2/2014) and Polish National Science Centre (grants no. UMO-2017/27/N/NZ1/00202, UMO-2016/23/B/NZ9/00862, UMO-2013/10/A/NZ1/00557)
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Saneyoshi M., Harada F., Nishimura S. Isolation and characterization of N6-methyladenosine from Escherichia coli valine transfer RNA. Biochim. Biophys. Acta Nucleic Acids Protein Synth. 1969;190:264–273. doi: 10.1016/0005-2787(69)90078-1. [DOI] [PubMed] [Google Scholar]
- 2.Iwanami Y., Brown G.M. Methylated bases of ribosomal ribonucleic acid from HeLa cells. Arch. Biochem. Biophys. 1968;126:8–15. doi: 10.1016/0003-9861(68)90553-5. [DOI] [PubMed] [Google Scholar]
- 3.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 4.Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R. Single-nucleotide resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei C.M., Gershowitz A., Moss B., Raj N.B., Hennings D. Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell. 1975;4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 7.Schibler U., Kelley D.E., Perry R.P. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J. Mol. Biol. 1977;115:695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- 8.Nichols J.L. N6-methyladenosine in maize poly(A)-containing RNA. Plant Sci. Lett. 1979;15:357–361. doi: 10.1016/0304-4211(79)90141-X. [DOI] [Google Scholar]
- 9.Kennedy T.D., Lane B.G. Wheat embryo ribonucleates. XIII. Methyl-Substituted nucleoside constituents and 5’-terminal dinucleotide sequences in bulk poly(AR)-rich RNA from imbibing wheat embryos. Can. J. Biochem. 1979;57:927–931. doi: 10.1139/o79-112. [DOI] [PubMed] [Google Scholar]
- 10.Krug R.M., Morgan M.A., Shatkin A.J. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. J. Virol. 1976;20:45–53. doi: 10.1128/jvi.20.1.45-53.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kane S.E., Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: Implications for RNA processing. Mol. Cell. Biol. 1985;5:2298–2306. doi: 10.1128/MCB.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clancy M.J., Shambaugh M.E., Timpte C.S., Bokar J.A. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: A potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002;30:4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng X., Chen K., Luo G.-Z., Weng X., Ji Q., Zhou T., He C. Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 2015;43:6557–6567. doi: 10.1093/nar/gkv596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grosjean H., Szweykowska-Kulinska Z., Motorin Y., Fasiolo F., Simos G. Intron-dependent enzymatic formation of modified nucleosides in eukaryotic tRNAs: A review. Biochimie. 1997;79:293–302. doi: 10.1016/S0300-9084(97)83517-1. [DOI] [PubMed] [Google Scholar]
- 16.Dai Q., Fong R., Saikia M., Stephenson D., Yu Y., Pan T., Piccirilli J.A. Identification of recognition residues for ligation-based detection and quantitation of pseudouridine and N6-methyladenosine. Nucleic Acids Res. 2007;35:6322–6329. doi: 10.1093/nar/gkm657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.-G., et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao G., Li H.-B., Yin Z., Flavell R.A. Recent advances in dynamic m6A RNA modification. Open Biol. 2016;6:160003. doi: 10.1098/rsob.160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haugland R.A., Cline M.G. Post-transcriptional modifications of oat coleoptile ribonucleic acids. Eur. J. Biochem. 2018;104:271–277. doi: 10.1111/j.1432-1033.1980.tb04425.x. [DOI] [PubMed] [Google Scholar]
- 20.Nichols J.L., Welder L. Nucleotides adjacent to N6-methyladenosine in maize poly(A)-containing RNA. Plant Sci. Lett. 1981;21:75–81. doi: 10.1016/0304-4211(81)90071-7. [DOI] [Google Scholar]
- 21.Zhong S., Li H., Bodi Z., Button J., Vespa L., Herzog M., Fray R.G. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo G., Macqueen A., Zheng G., Duan H., Dore L.C., Lu Z., Liu J., Chen K., Jia G., Bergelson J., et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 2014;5:1–8. doi: 10.1038/ncomms6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bokar J.A., Rath-Shambaugh M.E., Ludwiczak R., Narayan P., Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei: Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994;269:17697–17704. [PubMed] [Google Scholar]
- 25.Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., Rottman F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 26.Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y.S., et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 27.Bokar J.A. Fine-Tuning of RNA Functions by Modification and Editing. Volume 12. Springer; Berlin/Heidelberg, Germany: 2005. The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA; pp. 141–177. [Google Scholar]
- 28.Kan L., Grozhik A.V., Vedanayagam J., Patil D.P., Pang N., Lim K.-S., Huang Y.-C., Joseph B., Lin C.-J., Despic V., et al. The m6A pathway facilitates sex determination in Drosophila. Nat. Commun. 2017;8:15737. doi: 10.1038/ncomms15737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haussmann I.U., Bodi Z., Sanchez-Moran E., Mongan N.P., Archer N., Fray R.G., Soller M. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 30.Bodi Z., Zhong S., Mehra S., Song J., Graham N., Li H., May S., Fray R.G. Adenosine methylation in Arabidopsis mRNA is associated with the 3’ End and reduced levels cause developmental defects. Front. Plant Sci. 2012;3:48. doi: 10.3389/fpls.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X., et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2013;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bujnicki J.M., Feder M., Radlinska M., Blumenthal R.M. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m6A methyltransferase. J. Mol. Evol. 2002;55:431–444. doi: 10.1007/s00239-002-2339-8. [DOI] [PubMed] [Google Scholar]
- 33.Růžička K., Zhang M., Campilho A., Bodi Z., Kashif M., Saleh M., Eeckhout D., El-Showk S., Li H., Zhong S., et al. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017;215:157–172. doi: 10.1111/nph.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen L., Liang Z., Gu X., Chen Y., Teo Z.W.N., Hou X., Cai W.M., Dedon P.C., Liu L., Yu H. N6-methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev. Cell. 2016:1–15. doi: 10.1016/j.mod.2017.04.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vespa L. The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol. 2004;134:1283–1292. doi: 10.1104/pp.103.028050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., Mertins P., Ter-Ovanesyan D., Habib N., Cacchiarelli D., et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niessen M., Schneiter R., Nothiger R. Molecular identification of virilizer, a gene required for the expression of the sex-determining gene Sex-lethal in Drosophila melanogaster. Genetics. 2001;157:679–688. doi: 10.1093/genetics/157.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horiuchi K., Kawamura T., Iwanari H., Ohashi R., Naito M., Kodama T., Hamakubo T. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 2013;288:33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J., Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., Cheng T., Gao M., Shu X., et al. VIRMA mediates preferential m6A mRNA methylation in 3′ UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4 doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wen J., Lv R., Ma H., Shen H., He C., Wang J., Jiao F., Liu H., Yang P., Tan L., et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 2018;69:1028–1038. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knuckles P., Lence T., Haussmann I.U., Jacob D., Kreim N., Carl S.H., Masiello I., Hares T., Villasenor R., Hess D., et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415–429. doi: 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo J., Tang H.-W., Li J., Perrimon N., Yan D. Xio is a component of the Drosophila sex determination pathway and RNA N6-methyladenosine methyltransferase complex. Proc. Natl. Acad. Sci. USA. 2018;115:3674–3679. doi: 10.1073/pnas.1720945115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kierzek E., Kierzek R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 2003;31:4472–4480. doi: 10.1093/nar/gkg633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou K.I., Parisien M., Dai Q., Liu N., Diatchenko L., Joseph R., Pan T., Program S.T., Division B.S. Hairpin predisposes its conformation to protein binding. J. Mol. Biol. 2017;428:822–833. doi: 10.1016/j.jmb.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu N., Zhou K.I., Parisien M., Dai Q., Diatchenko L., Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45:6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roost C., Lynch S.R., Batista P.J., Qu K., Chang H.Y., Kool E.T. Structure and thermodynamics of N6-methyladenosine in RNA: A spring-loaded base modification. J. Am. Chem. Soc. 2015;137:2107–2115. doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ok S.H., Jeong H.J., Bae J.M., Shin J.-S., Luan S., Kim K.-N. Novel CIPK1-associated proteins in Arabidopsis contain an evolutionarily conserved C-terminal region that mediates nuclear localization. Plant Physiol. 2005;139:138–150. doi: 10.1104/pp.105.065649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z., Theler D., Kaminska K.H., Hiller M., de la Grange P., Pudimat R., Rafalska I., Heinrich B., Bujnicki J.M., Allain F.H.-T., et al. The YTH domain is a novel RNA binding domain. J. Biol. Chem. 2010;285:14701–14710. doi: 10.1074/jbc.M110.104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scutenaire J., Deragon J.-M., Jean V., Benhamed M., Raynaud C., Favory J.-J., Merret R., Bousquet-Antonelli C. The YTH domain protein ECT2 is an m6A reader required for normal trichome branching in Arabidopsis. Plant Cell. 2018;30 doi: 10.1105/tpc.17.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei L.-H., Song P., Wang Y., Lu Z., Tang Q., Yu Q., Xiao Y., Zhang X., Duan H.-C., Jia G. The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell. 2018;30:968–985. doi: 10.1105/tpc.17.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arribas-Hernández L., Bressendorff S., Hansen M.H., Poulsen C., Erdmann S., Brodersen P. An m6A-YTH module controls developmental timing and morphogenesis in Arabidopsis. Plant Cell. 2018;30:952–967. doi: 10.1105/tpc.17.00833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson S.J., Kramer M.C., Gosai S.J., Yu X., Vandivier L.E., Nelson A.D.L., Anderson Z.D., Beilstein M.A., Fray R.G., Lyons E., et al. N6-methyladenosine inhibits local ribonucleolytic cleavage to stabilize mRNAs in Arabidopsis. Cell Rep. 2018;25:1146–1157.e3. doi: 10.1016/j.celrep.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 54.Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., Ma J., Wu L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.-M., Li C.J., Vågbø C.B., Shi Y., Wang W.-L., Song S.-H., et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duan H.-C., Wei L.-H., Zhang C., Wang Y., Chen L., Lu Z., Chen P.R., He C., Jia G. ALKBH10B is an RNA N6-methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell. 2017;tpc.00912.2016 doi: 10.1105/tpc.16.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martínez-Pérez M., Aparicio F., López-Gresa M.P., Bellés J.M., Sánchez-Navarro J.A., Pallás V. Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA. 2017;114:10755–10760. doi: 10.1073/pnas.1703139114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Z., Shi J., Yu L., Zhao X., Ran L., Hu D., Song B. N6-methyl-adenosine level in Nicotiana tabacum is associated with tobacco mosaic virus. Virol. J. 2018;15:1–10. doi: 10.1186/s12985-018-0997-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wan Y., Tang K., Zhang D., Xie S., Zhu X., Wang Z., Lang Z. Transcriptome-wide high-throughput deep m6A-seq reveals unique differential m6A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol. 2015;16:1–26. doi: 10.1186/s13059-015-0839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y., Wang X., Li C., Hu S., Yu J., Song S. Transcriptome-wide N6-methyladenosine profiling of rice callus and leaf reveals the presence of tissue-specific competitors involved in selective mRNA modification. RNA Biol. 2014;11:1180–1188. doi: 10.4161/rna.36281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alarcón C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alarcón C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kruszka K., Pieczynski M., Windels D., Bielewicz D., Jarmolowski A., Szweykowska-Kulinska Z., Vazquez F. Role of microRNAs and other sRNAs of plants in their changing environments. J. Plant Physiol. 2012;169:1664–1672. doi: 10.1016/j.jplph.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 64.Sunkar R., Li Y.-F., Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 65.Barciszewska-Pacak M., Milanowska K., Knop K., Bielewicz D., Nuc P., Plewka P., Pacak A.M., Vazquez F., Karlowski W., Jarmolowski A., et al. Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front. Plant Sci. 2015;6:1–14. doi: 10.3389/fpls.2015.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]