Abstract
Lafora disease is a severe, autosomal recessive, progressive myoclonus epilepsy. The disease usually manifests in previously healthy adolescents, and death commonly occurs within 10 years of symptom onset. Lafora disease is caused by loss-of-function mutations in EPM2A or NHLRC1, which encode laforin and malin, respectively. The absence of either protein results in poorly branched, hyperphosphorylated glycogen, which precipitates, aggregates and accumulates into Lafora bodies. Evidence from Lafora disease genetic mouse models indicates that these intracellular inclusions are a principal driver of neurodegeneration and neurological disease. The integration of current knowledge on the function of laforin-malin as an interacting complex suggests that laforin recruits malin to parts of glycogen molecules where overly long glucose chains are forming, so as to counteract further chain extension. In the absence of either laforin or malin function, long glucose chains in specific glycogen molecules extrude water, form double helices and drive precipitation of those molecules, which over time accumulate into Lafora bodies. In this article, we review the genetic, clinical, pathological and molecular aspects of Lafora disease. We also discuss traditional antiseizure treatments for this condition, as well as exciting therapeutic advances based on the downregulation of brain glycogen synthesis and disease gene replacement.
Lafora disease (Online Mendelian Inheritance in Man (OMIM) #254780) is a rare autosomal recessive and severe form of progressive myoclonus epilepsy. After onset, which usually occurs during late childhood or early adolescence, Lafora disease is invariably fatal, typically within 10 years1,2. The condition was first described by Lafora and Glück over 100 years ago3. A post-mortem study showed profuse accumulation of small inclusion bodies in many tissues, including the brain. These inclusions, subsequently termed Lafora bodies, became the hallmark of the disease. They were shown to be composed primarily of abnormal glycogen4, placing Lafora disease in the context of glycogen metabolism disorders.
The affected genes in Lafora disease — EPM2A, which encodes laforin glucan phosphatase (henceforth termed laforin) and NHLRC1 (also known as EPM2B), which encodes E3 ubiquitin protein ligase 1 (henceforth termed malin) — were discovered only two decades ago5–7 (FIG. 1). The exact roles of laforin and malin in glycogen metabolism are still under investigation. However, recent progress affords an improved understanding of the disease mechanisms, leading to the identification of new therapeutic avenues for what is arguably the severest of the epilepsies.
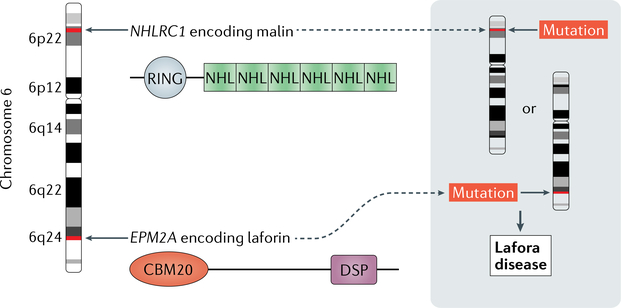
Fig. 1 |. Causative mutations in Lafora disease.
EPM2A and NHLRC1 encode laforin and malin, respectively. Both genes are located on chromosome 6. Laforin contains an amino- terminal family 20 carbohydrate-binding module (CBM20) and a carboxy-terminal dual-specificity phosphatase (DSP) domain. Malin contains RING and NHL domains, which are typical for E3 ubiquitin ligases. Mutations — usually missense, nonsense or frameshift — in either of the two genes cause Lafora disease.
This Review provides a broad overview of Lafora disease, including its clinical features, genetics and existing management strategies. Our current understanding of Lafora disease pathogenesis is outlined, laying the groundwork for discussion of new disease-mechanism-based therapeutic strategies.
Clinical features of Lafora disease
At the time of onset, Lafora disease is difficult to distinguish from idiopathic generalized epilepsies8. Apparently healthy older children or teenagers start having seizures, which can initially be controlled with antiepileptic drugs (AEDs)8,9. Several types of seizure typically occur in patients with Lafora disease, including myoclonic, occipital, generalized tonic-clonic, absence and atonic seizures10,11. In retrospect, parents often recall that their child had experienced isolated febrile or non-febrile seizures earlier in childhood10. Additional symptoms in the first few years comprise behavioural changes, confusion, depression, dysarthria, ataxia and intellectual decline (BOX 1). Over time, the seizures become more frequent and increasingly intractable. Behavioural and cognitive deterioration increases, gradually leading to dementia. Eventually, the patient enters a vegetative state with continuous myoclonus and requires tube feeding and artificial respiration. Death commonly results from status epilepticus or aspiration pneumonia and other complications of chronic neurodegeneration2,11.
Box 1 | Lafora disease symptoms
The first 2–3 years of Lafora disease are characterized by the following symptoms:
Ataxia
confusion
Depression
Grand mal seizures
Staring spells and/or absence seizures
Drop in school performance
Drop attacks
Myoclonus
visual hallucinations
Headaches
Dysarthria
Genetics
Many types of pathogenic variant have been identified in the genes that encode laforin and malin including missense, nonsense and frameshift mutations, as well as some larger deletions, particularly in EPM2A12. Human laforin consists of two functional domains: an amino-terminal family 20 carbohydrate-binding module (CBM20) and a carboxy-terminal dual-specificity phosphatase (DSP) domain. Malin is an E3 ubiquitin ligase containing a RING domain and six NHL repeats, both of which are typical for this family of enzymes (FIG. 1). In general, mutations in EPM2A and NHLRC1 are distributed evenly across both genes, with no particular clustering in the functional domains mentioned above (see The Lafora Progressive Myoclonus Epilepsy Mutation and Polymorphism Database13).
Studies have shown that certain mutations within the DSP domain of laforin compromise its phosphatase activity, which might render the protein pathogenic14,15. However, two independent studies demonstrated that phosphatase activity is not required to rescue laforin- deficient mice from Lafora disease16,17. Evidently, in many cases, other properties of laforin, including glycogen binding, subcellular localization and the interaction with malin or protein phosphatase 1 regulatory subunit 3 C (PP1 subunit R5; also known as PTG), are additionally affected by DSP mutations14,15,18,19. For example, mutations in the DSP, such as Gly279Ser, Gln293Leu, Tyr294Asn and Pro301Leu14, can lead to decreased glycogen binding, although the CBM20 domain is not directly affected. Conversely, mutations within the CBM20, such as Trp32Gly, Phe84Leu and Arg108Cys14, can lead to loss of phosphatase activity. Together, these findings clearly show that the overall effect of a mutation on laforin function cannot be deduced solely from the mutation site. Furthermore, the role of laforin in glycogen metabolism and Lafora disease is not confined to its function as a phosphatase.
In a few patients with Lafora disease, sequence analysis of EPM2A and NHLRC1 coding regions reveals no mutation in either gene20–23. In such cases, the disease could be caused by mutations in non-coding regulatory regions, such as promoter or intronic regions20,24. Mutations in non-coding regions might also explain some cases in which only one heterozygous mutation in the coding region could be identified23. Patients who carried an apparently homozygous mutation in NHLRC1 but had one parent who lacked mutations in this gene were found to have large deletions in one allele, which were undetectable by PCR25. In addition, disease-causing mutations in a third locus have been proposed in cases in which causative involvement of EPM2A or NHLRC1 was excluded24. Further analysis revealed a mutation in the PR domain zinc-finger protein 8 gene (PRDM8) in a single family26. Of note, patients with PRDM8 mutations presented with an atypical form of Lafora disease, characterized by early childhood onset and negative skin (but positive muscle) biopsies26. Finally, some patients with no EPM2A or NHLRC1 mutations might not actually have Lafora disease but were misdiagnosed on the basis of a false-positive skin biopsy (see ‘Diagnostic strategy’ section below). Therefore, it seems likely that essentially all classic cases of Lafora disease can be explained by mutations in EPM2A or NHLRC1.
The clinical features of classic Lafora disease are similar in most patients10,12, although the time of onset and the rate of disease progression can vary substantially25,27. In the past, attempts have been made to establish genotype-phenotype correlations12,18. One example is the early-onset learning disorder phenotype in some patients with Lafora disease who harbour a mutation in exon 1 of EPM2A18,28, although not all patients with such mutations have this atypical subsyndrome21,29. Another example is the observation that patients with NHLRC1 mutations — in particular, the Asp146Asn mutation — tend to live longer than those with EPM2A muta- tions20,21,23,30,31. However, some patients with NHLRC1 mutations have very severe phenotypes29,32–34.
What factors make genotype-phenotype correlation studies so difficult? The fact that Lafora disease is rare and shows considerable mutational heterogeneity, with more than 90 known pathogenic variants in EPM2A and almost 80 in NHLRC1 (REF 13), hampers progress in understanding the genetic epidemiology of the condition. Various combinations of compound heterozygosity for mutations12,20,29,35 further complicate correlation studies. In addition, the quality of available care has a strong impact on the frequency of disease complications and survival and varies between countries and health sys- terns10,12,18. Moreover, evidence indicates that genetic factors other than the disease-causing EPM2A or NHLRC1 mutations, such as modifier genes, can substantially modulate the course of the disease36. For example, of two siblings with homozygous exon 1 mutations in EPM2A, only one presented with the early-onset learning disorder described above, even though both siblings carried the same pathogenic variant35. Another example is the highly variable age of onset among siblings carrying the same mutation25,27. Candidate factors that influence the pathogenesis of Lafora disease include interacting partners of laforin and/or malin. Indeed, a variant of PP1 subunit R5 — a protein that usually interacts with the laforin-malin complex — was suggested to be responsible for a milder course of Lafora disease37. This protein variant exhibits reduced function, and its effects in humans are consistent with the observation that knockout of the gene encoding PP1 subunit R5 leads to rescue of the Lafora disease phenotype in mice38.
Pathogenesis
Understanding the pathogenesis of Lafora disease depends on providing a mechanistic link between a deficiency in laforin or malin and the neurodegenerative disease. Studies of the pathological hallmark of the disease, the Lafora bodies, provided insight into the affected metabolic pathways even before the genetic causes were found; Lafora bodies were classified as polyglucosan bodies, as they are largely composed of glucan chains and, hence, are chemically similar to glycogen4 (FIG. 2a). Intriguingly, in contrast to cytosolic glycogen, these polyglucosans are water-insoluble. The exact roles of laforin and malin are still under investigation, but they are likely to be involved in processes that prevent the accumulation of insoluble glycogen-like particles (FIG. 2b). The sections that follow discuss Lafora body accumulation as the cause of Lafora disease, review the proposed roles of laforin and malin in glycogen metabolism (BOX 2) and propose a model of Lafora disease pathogenesis whereby laforin and malin act as part of a glycogen quality control mechanism to prevent glycogen insolubility.
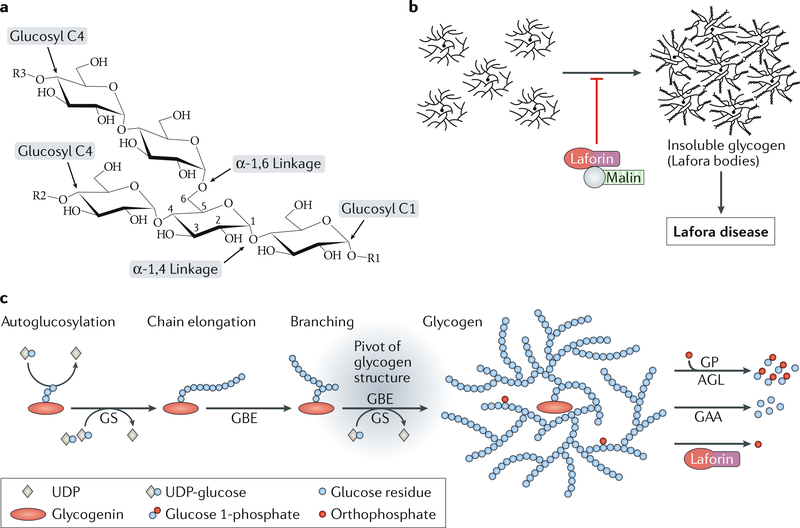
Fig. 2 |. Impaired glycogen metabolism in Lafora disease.
a | The chemical basis of the polyglucans glycogen and starch. Glucan chains are formed by chain-elongating enzymes that incorporate glucosyl residues at terminal glucosyl C4 carbons of pre-existing chains, forming α−1,4 glycosidic linkages. Branching points are introduced by branching enzymes that cleave part of an existing chain, which is then reattached to form an α−1,6 glycosidic linkage. b | Laforin and malin act as a complex to prevent the accumulation of insoluble glycogen. In the absence of functional laforin or malin, normally soluble glycogen contains abnormally long chains and precipitates and aggregates as Lafora bodies, which drive Lafora disease progression. c | Glycogen metabolism. Glycogen is synthesized from glycogenin-containing glycogen primers by the concerted action of glycogen synthase (GS) and glycogen branching enzyme (GBE). This well-balanced reaction determines glycogen chain length and, hence, represents the pivot of glycogen structure. BOX 2 provides a more detailed explanation of glycogen metabolism. AGL, glycogen debranching enzyme; GAA, lysosomal α-giucosidase; GP, glycogen phosphorylase. Part a adapted with permission from REF58, Elsevier. Part c adapted with permission from REF87, CC-BY-4.0.
Box 2 | Glycogen metabolism
Glycogen serves as a cellular store of energy and reduced carbon and is found in many tissues, including the brain. It is a heterogeneous mix of roughly spherical macromolecules, each composed of branched glucan chains with up to 55,000 glucosyl residues per molecule82. De novo synthesis of glycogen requires the formation of small glycogen primers. This process usually involves dimers of the protein glycogenin144, which mediate first autoglucosylation and then initial chain elongation through formation of α−1,4 glycosidic linkages88. Subsequently, chain elongation is continued by glycogen synthase, which, like glycogenin, uses uDP-glucose as a glucosyl donor. Branching points are introduced by glycogen branching enzyme, which detaches the downstream portion of a glucan chain and reattaches it upstream at the same or a neighbouring chain through an α−1,6 glycosidic linkage. In the cytosol, glycogen is degraded by glycogen phosphorylase and glycogen debranching enzyme (AGL). Glycogen phosphorylase removes glucosyl units from the end of the glycogen chains but is stalled by branching points, which are cleaved by AGL. cytosolic glycogen degradation largely yields glucose 1-phosphate, which is available for glycolysis and other cellular pathways82,88 (FiG. 2c).
Lafora bodies
The accumulation of dense cytoplasmic aggregates is a feature of many neurodegenerative diseases, including Parkinson disease and Alzheimer disease39,40, and the accumulation of Lafora bodies seems to be the primary cause of Lafora disease progression. This conclusion has been supported by several studies in mouse models of Lafora disease, which lack either laforin or malin and recapitulate the human disease41,42. Genetic approaches that reduce or abolish glycogen synthesis prevent Lafora body formation and rescue other features of the Lafora disease phenotype in mice, including autophagy impairment, neurodegeneration and seizure susceptibility38,43,44. In addition, overexpression or increased activation of the glycogen-synthesizing enzyme glycogen synthase in the presence of laforin and malin can result in polyglucosan body formation, as well as down-stream effects such as impaired autophagy and neuro- degeneration43,45. Evidence of a link between impaired autophagy and neurodegeneration46–48 completes the current view on the pathogenesis of Lafora disease. The roles of laforin and malin in glycogen metabolism need to be unravelled to enable us to understand the mechanisms that lead to Lafora body formation and accumulation.
The role of laforin as a phosphatase
CBM20s are found in various glucan chain-binding proteins across many species49,50, and the presence of this module in laforin places this protein in the context of glycogen metabolism (BOX 2; FIGS. 1,2c). Laforin is known to bind to glycogen51, but how it prevents glycogen insolubility and protects against Lafora disease is open to question.
The identification of a DSP in laforin and the fact that protein phosphorylation has a substantial role in the regulation of the chain-elongating enzyme glycogen synthase implicated laforin in the regulation of glycogen synthesis. Glycogen synthase kinase 3 (GSK3) is activated by dephosphorylation and itself phosphorylates and inactivates glycogen synthase52. Yeast two- hybrid and cell culture overexpression experiments showed that laforin interacts with and dephosphorylates the muscle isoform of GSK3 (REF53). A defect in laforin, resulting in insufficient GSK3 activation, might therefore lead to abnormally high glycogen synthase activity. However, no increase in glycogen synthase activity could be demonstrated in a laforin-deficient mouse model of Lafora disease42.
Another interesting discovery was the ability of laforin to dephosphorylate glycogen (FIG. 2c). Over 30 years ago, glycogen was discovered to contain small amounts of covalently bound phosphate54. The role of glycogen phosphate gained importance when laforin was shown to act as a glucan phosphatase55,56. Glycogen in Lafora disease mice was found to have elevated levels of covalently bound phosphate and to contain an abnormally high proportion of long glucan chains57,58. On the basis of these observations, it was hypothesized that an excess of glycogen phosphate lies at the root of Lafora disease pathogenesis, and that laforin keeps glycogen phosphate levels low to prevent abnormal glycogen structure, glycogen insolubility and, hence, Lafora disease59. However, more recent studies contradict this hypothesis by showing that the laforin-deficient Lafora disease mouse model can be rescued by overexpression of laforin with a mutated DSP lacking phosphatase activity. In fact, the glycogen in the rescued mice was still hyperphosphorylated, as in the laforin knockout, but the chain length was normal, Lafora bodies were absent, and the behavioural phenotype was nor- malized16,17. These findings largely excluded glycogen hyperphosphorylation as the underlying cause of Lafora disease.
The laforin-malin interaction
Investigations into the role of laforin in cell metabolism revealed several interaction partners for this protein, some of which were related to endoplasmic reticulum stress60, protein clearance61,62, iron homeostasis63 or tumour suppression64. Besides GSK3, additional laforin interaction partners with a close connection to glycogen metabolism were identified, including glycogen synthase55, subunits R4, R5 and R6 of the glycogen synthase activator PP1 (REFS14,55,65) and AMP-activated kinase (AMPK), a protein that is involved in glycogen phosphorylase regulation. The array of putative interaction partners implies a complex role for laforin, involving multiple functions66.
The interaction of laforin with malin is of particular interest and has been supported by a large number of studies14,53,65,67,68. The existence of a patient mutation that causes Lafora disease merely through the loss of the laforin-malin interaction69 emphasizes the relevance of this interaction in vivo. Moreover, the similar clinical progression in patients with EPM2A and NHLRC1 mutations is consistent with a crucial role for both proteins in a single functional complex. The laforin- malin complex was demonstrated to incorporate lysine 63 (K63)-linked polyubiquitin chains19,70,71, which principally promote autophagic inclusion and degradation of ubiquitylated targets72–74. In agreement with evidence for proteasomal degradation of laforin-malin targets in the cytosol65,69,75, this finding implies that targets of the laforin-malin complex are generally subjected to degradation. Interestingly, the targets of malin-mediated ubiquitylation that have been demonstrated ex vivo include glycogen synthase69 and PP1 subunit R5 (REF65). Glycogen synthase drives glycogen chain elongation, and PP1 subunit R5 indirectly activates glycogen synthase by targeting PP1 to glycogen76. Therefore, both proteins help to determine a pivotal aspect of glycogen structure — the balance between chain elongation and branching (FiG. 2c).
The importance of glycogen structure
Elongation and branching of glucan chains, as well as chain degradation and debranching, are enzymatic features that are not restricted to glycogen metabolism but are also involved in starch metabolism in plants77. Glycogen and amylopectin, the main component of plant starch, are both polyglucans78,79 (FiG. 2a). Unlike plant starch, however, glycogen is normally water-soluble. Glycogen and amylopectin both consist of glucan chains with a wide distribution of chain lengths17,80, but the average chain length is generally higher in amylopectin than in glycogen81. In glycogen, branching points are essentially distributed evenly, whereas in amylopectin, they are arranged in clus- ters82,83, giving rise to regions with fewer branching points where neighbouring glucan chains form double helices. In turn, these structures form semi-crystalline layers that render the entire starch granule water-insoluble81,83,84. Interestingly, genetically engineered plants with impaired clustering of branching points produce a soluble form of polyglucan, termed phytoglycogen85,86. Thus, the frequency of branching points, which is controlled by the concerted action of glycogen synthase and glycogen branching enzyme (GBE) in the case of glycogen metabolism, seems to determine the water solubility of a polyglucan87 (FiG. 2c).
Glycogen synthase is regulated post-translationally through complex mechanisms involving phosphorylation and allosteric effectors88, which allow a fine-tuned balance of chain elongation and branching. In mice with laforin or malin deficiency, glycogen shows structural abnormalities consisting of substantially increased proportions of long glucan chains58, suggesting a role for the laforin-malin complex in the regulation of glycogen chain elongation.
Glycogen quality control
The laforin-malin complex is thought to downregulate glycogen chain elongation, probably by targeting both glycogen synthase and PP1 subunit R5, which is involved in glycogen synthase activation, to degradation65,69. Accordingly, in Lafora disease, insufficient downregulation of this process would lead to an imbalance of elongation and branching reactions, resulting in overly long glucan chains, double-helix formation and, hence, glycogen insolubility and deposition as Lafora bodies. However, studies showing that levels of glycogen synthase and PP1 subunit R5 were not substantially altered in tissue lysates from Lafora disease mice cast doubt on whether the two proteins are substrates of the laforin- malin complex in vivo42. This apparent inconsistency might be resolved by assuming a locally focused action of the laforin-malin complex, targeting glycogen synthase and PP1 subunit R5 only on a small subgroup of glycogen molecules that are especially prone to precipitation87. Such subcellular changes would not be detected if protein levels and activity are quantified in whole-tissue lysates42.
This theory is corroborated by the fact that normal glycogen is a heterogeneous mixture of macromolecules. The range of molecule sizes within one preparation spans at least one order of magnitude, and evidence exists that some molecules are composed of longer chains than others58,89. As long chains promote polyglucan insolubility, glycogen molecules might vary in their propensity to undergo precipitation (FiG. 3a,b). The differences between glycogen molecules are likely to be inherent to normal glycogen metabolism and could be caused by subcellular inhomogeneity of constituents that affect local glycogen synthase activity, including the enzyme itself, the allosteric activator glucose 6-phosphate and the substrate UDP-glucose. Subcellular inhomogeneity has been demonstrated for a number of metabolites, including ATP, reduced NADH and glucose90–92.
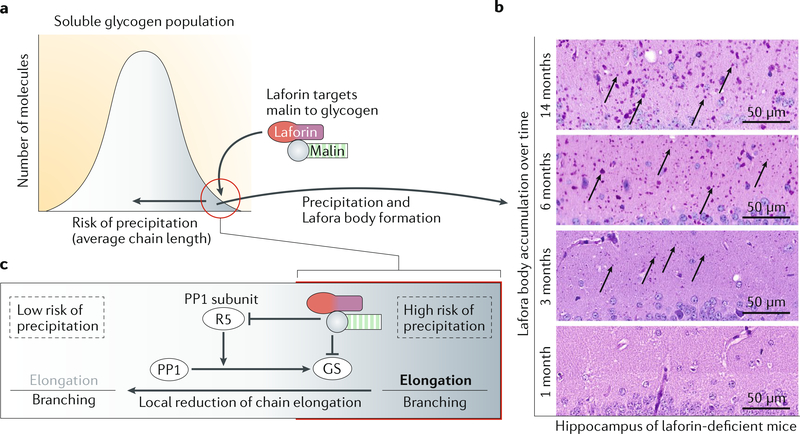
Fig. 3 |. Mechanistic model for Lafora body formation and accumulation.
a | Glycogen is a heterogeneous mixture of molecules that differ in their long glucan chain content and possess different risks of precipitation. To prevent precipitation, a small proportion of molecules that are precipitation-prone (circled in red) might be modified by local reduction of chain elongation, which is achieved by the mechanism explained in part c. This modification leads to an increased relative branching frequency and a decreased risk of precipitation (horizontal arrow). b | If glycogen precipitation cannot be entirely prevented, precipitated glycogen accumulates over time, as observed in Lafora disease mice of different ages. Arrows indicate some typical Lafora bodies in diastase-resistant periodic acid-Schiff-stained sections of mouse hippocampus. c | A functional iaforin-maiin complex mediates a reduction in glycogen synthase (GS) activity by targeting GS and protein phosphatase 1 (PP1) subunit R5 to degradation, resulting in an increased branching frequency. Rather than affecting the entire GS protein population, this process might occur locally, that is, predominantly on the small proportion of glycogen molecules with a high risk of precipitation (red circle in part a). Parts a and c adapted with permission from REF87, CC-BY-4.0.
Through its CBM20, laforin preferentially binds polyglucans composed of longer glucan chains. This fact is illustrated by the predilection of laforin for solubilized potato starch over glycogen with respect to substrate binding51 and is consistent with the strong enrichment of laforin in Lafora bodies in malin-deficient Lafora disease mice93. Via its interaction with laforin, malin is likely to be sequestered preferentially to glycogen molecules with a larger proportion of long chains (FiG. 3a), where it ubiquitylates glycogen synthase and PP1 subunit R5 for subsequent degradation. Accordingly, chain elongation would be specifically decreased at glycogen molecules with an increased probability of precipitation, leading to a localized relative increase in branching frequency. This phenomenon would result in shorter chains, higher solubility and, through avoidance of precipitation, availability of these glycogen molecules for normal glycogen degradation87 (FiG. 3c). In addition to this putative role of the laforin-malin complex in avoiding glycogen insolubility, laforin removes glycogen phosphate to allow complete glycogen degradation by glycogen phosphorylase and glycogen debranching enzyme80.
In addition to the strategy of preventing the formation of glycogen with a high risk of precipitation, the laforin-malin complex could be involved in an autophagic process to remove abnormal or already insoluble glycogen. Starch-binding domain-containing protein 1 (STBD1) anchors glycogen to intracellular membranes and also interacts with autophagic proteins94. STBD1 could have a role in targeting malstructured glycogen to lysosomal degradation, although its function does not seem to be impaired in Lafora disease. Cell culture experiments have shown that the laforin-malin complex interacts with p62 (also known as sequestosome 1), an autophagy-related adaptor protein that binds ubiquitylated proteins that are targeted to autophagy68. Lafora body-containing tissue does not always exhibit a general defect in autophagy, and if present, this defect seems to be secondary to glycogen accumula- tion17,43,45,80,95. However, it is conceivable that in mammalian cells, several layers of glycogen quality control are established to avoid deposition and accumulation of abnormal glycogen. The first layer is avoidance of glycogen precipitation through tight local regulation of glycogen synthase, with an evident role for the laforin-malin complex. A putative second layer would be the removal of abnormal and precipitated glycogen, also possibly involving the laforin-malin complex. In the absence of a functional laforin-malin complex, cellular glycogen quality control is impaired, with fatal consequences, as observed in Lafora disease.
Diagnostic strategy
Several factors, including the rare nature of the disease, cultural consanguinity, founder effects and limited treatment options, pose considerable challenges for clinicians and caregivers when working with patients with Lafora disease and their families. Diagnosis is usually based on clinical findings (described above), EEG abnormali- ties2,8,96 with normal brain MRI97, and diastase-resistant periodic acid-Schiff (PAS-D)-positive skin biopsies, revealing the presence of Lafora bodies29,98 (FiG. 4). As virtually all cases of Lafora disease can be attributed to pathogenic variants of either EPM2A or NHLRC1 (REFS20,22,23,99), targeted genetic testing is the gold standard to confirm the diagnosis. Genetic testing is not only less invasive than skin biopsy but is also more readily available and less expensive. Furthermore, skin biopsy testing is fraught with false positivity. The sweat glands where Lafora bodies form contain equally PAS-D-positive and visually impressive normal secretory contents that are frequently falsely identified as Lafora bodies owing to a lack of experience with the disease in most pathology laboratories98. False negativity of skin biopsy in patients with genetically confirmed Lafora disease has also been reported29. Therefore, clinicians should await genetic confirmation before disclosing the diagnosis to caregivers and — if appropriate and with the approval of the family — to the young patients.
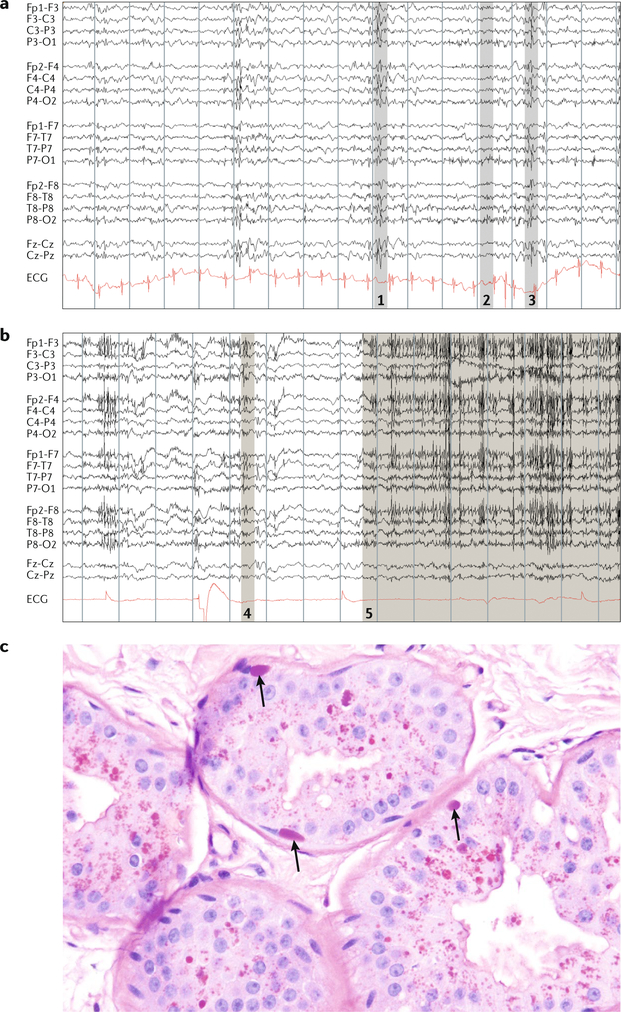
Fig. 4 |. Diagnosis of Lafora disease.
a | A 16 s EEG trace in an awake patient with Lafora disease (compound heterozygous for EPM2A: Arg241X and Pro211Leu) at age 20 years, 5 years into the disease. Note the slow background rhythms and generalized irregular spike-wave discharges associated with myoclonus. At this age, the patient was still conversant, but each thought and sentence was interrupted by myoclonic absences. b | EEG in the same patient at age 28 years in an unresponsive wakeful state. Grey shaded areas in parts a and b denote patient movements: 1, arm jerk; 2, head jerk; 3, head and arm jerk; 4, twitch; and 5, eyes upward and blinking. c | Skin biopsy in the same patient at age 20 years. Arrows indicate Lafora bodies in the myoepithelium of apocrine glands.The equally stained structures near the lumina of the glands are not Lafora bodies but the normal secretory materials of these cells. Owing to a lack of experience with the disease, many laboratories mistake these materials for Lafora bodies, leading to false-positive diagnoses. ECG, electrocardiogram
A diagnosis of Lafora disease can cause severe psychological trauma to patients and their families, including but not limited to feelings of guilt and resentment, fear of younger siblings also becoming affected, and financial costs and concern regarding the availability of treatment options. Consequently, the diagnosis should be followed up with sustained, attentive genetic and psychological counselling and support.
Current management strategies
Currently, AEDs are the only available treatments that control the severity and frequency of seizures and myoclonus to some degree in patients with Lafora disease. Among these drugs, valproic acid is the mainstay. Other effective medications include topiramate, etho- suximide, phenobarbital, zonisamide, felbamate and benzodiazepines. Most recently, perampanel, a new α-amino- 3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptor antagonist AED, was shown to be effective in two single-case studies and a group of ten patients100–102.
Besides AEDs, vagal nerve stimulation resulted in temporary cessation of generalized tonic-clonic seizures and status epilepticus in two single-case studies103,104. The ketogenic diet was also tried in a group of patients with relatively advanced disease but was shown to be ineffective99. This finding was surprising given that the diet converts brain energy usage from glucose to fatty acids, thus presumably reducing the neuronal glucose availability for glycogen (and Lafora body) synthesis. Unpublished work from our laboratory did show the effectiveness of this diet in a Lafora disease mouse model, and the possibility remains that the failure in the clinical setting was attributable to the overly advanced disease in the treated patients rather than to actual ineffectiveness.
In 2016, the European Commission granted orphan designation and permission to use metformin for the treatment of Lafora disease (European Medicines Agency orphan decision number EU/3/16/1803). Metformin is an activator of AMPK and is widely used to treat type 2 diabetes105. AMPK activation is associated with the inhibition of several ATP-consuming pathways, including glycogen synthesis106,107. In mice and rats, metformin was shown to have positive effects on neuronal survival and seizure termination108–110. Studies in a mouse model of Lafora disease showed that metformin ameliorated neuropathological symptoms, reduced seizure susceptibility and slightly reduced the numbers of Lafora bodies111,112. No clinical data are yet available regarding the efficacy of metformin as a treatment for Lafora disease.
The dietary supplement sodium selenate has been shown to reduce neurodegeneration, gliosis, seizure susceptibility and memory loss in a mouse model of Lafora disease113. However, a gradual decline in overall motor conditioning following an initial improvement in the treated mice raised doubts about the efficacy of the drug as a potential treatment for Lafora disease.
Aminoglycoside antibiotics, such as gentamicin, can suppress translation termination at premature termination codons114,115 and could be repositioned for potential use in patients with Lafora disease who have nonsense mutations. However, clinical studies involving patients with cystic fibrosis, Duchenne muscular dystrophy or McArdle disease resulted in various outcomes115. At best, only subpopulations of patients benefited from the treatment116–119, and the use of aminoglycosides is also limited by adverse effects120,121. No preclinical data are available for the use of gentamicin to treat Lafora disease.
The road to new therapies
Virus-mediated gene replacement
As discussed above, Lafora disease is primarily a neurodegenerative disease, and the symptoms are caused by Lafora body accumulation in the brain due to deficiency in either laforin or malin87. Gene therapy to deliver a functional copy of the defective gene would thus be an obvious option for Lafora disease treatment (FiG. 5). Undoubtedly, gene therapy has considerably advanced the development of treatments for hereditary diseases. Several strategies to introduce a transgene have been developed, including the use of viral vectors, such as adeno-associated virus (AAV)122 and lentivirus123. Other delivery methods, using nanoparticles, liposomes and exosomes, have also been established124,125.
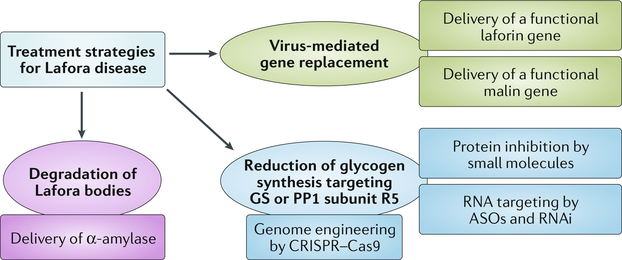
Fig. 5 |. Overview of therapeutic strategies in Lafora disease.
In addition to the viral delivery of functional laforin or malin, the therapeutic options for Lafora disease include intracellular degradation of Lafora bodies by glucan-degrading enzymes such as α-amylase, as well as the downregulation of glycogen synthesis by targeting glycogen synthase (GS) or protein phosphatase 1 (PP1) subunit R5 at the gene, RNA or protein level. ASOs, antisense oligonucleotides; RNAi, RNA interference.
AAV has become the vector of choice for gene replacement, owing to its non-pathogenic nature, fairly widespread transduction, long-lasting and high transgene expression, and very low frequency of integration into the host genome122,123. Although packing capacity is a limiting factor for AAV-mediated gene delivery122, cDNA sequences of EPM2A and NHLRC1 are below the size limit and can, therefore, be delivered using this vector. The greatest hurdle to AAV-mediated delivery of these genes is the transduction efficiency. As Lafora bodies form throughout the brain, widespread CNS transduction is paramount. The transduction efficiency has been improved through the use of serotypes with high neuronal transduction efficiency, such as AAV9123, or modified serotypes, such as AAV-PHP.eB or AAV-PHPS126.
AAV9 is known to pass through the blood-brain barrier (BBB)127, which makes intravenous delivery possible. However, vector loss through high off-target transduction (mainly accumulation in the liver) is a concern with systemic delivery123. Direct CNS delivery — for example, intrathecal administration — would circumvent this problem and enable relatively high transduction throughout the CNS. Intrathecal delivery is considered fairly safe and is routinely used to deliver various drugs to the CNS128,129. Delivery via the cerebrospinal fluid might also circumvent loss of the virus by neutralizing antibodies130. As AAV infects humans naturally, the pre-existence of neutralizing antibodies leads to reduced transgene expression when the vector is delivered systemically131,132. In addition, despite the low immunogenicity of AAV, efficient therapy can only be accomplished once133.
A clinical trial using intrathecal AAV administration is ongoing for Batten disease, a late infantile neuronal ceroid lipofuscinosis, and is likely to establish a proof of concept for this delivery method in AAV therapy for CNS disorders. Several other clinical trials are ongoing in CNS disorders123. In Lafora disease, early intervention is likely to be the best option; in metachromatic leukodystrophy134 and adrenoleukodystrophy135, gene therapy has been shown to arrest neurodegeneration if administered before extensive CNS damage has occurred.
Degradation of Lafora bodies
Another therapeutic option for Lafora disease is the degradation of accumulated Lafora bodies (FIG. 5). This goal could be achieved by the delivery of a polyglucan-degrading enzyme to the brain. For instance, α-amylase is known to break the α−1,4 glycosidic linkages in glycogen and starch and thus could mediate Lafora body-degradation. Efficient delivery strategies are essential to effectively introduce the Lafora-body-de- grading protein into the cells that harbour these inclusions. Fused to a non-toxic form of diphtheria toxin, α-amylase can be translocated into cells, is active and can degrade intracellular glycogen136. Among the vast variety of α-amylases that are present in different species, an enzyme that both effectively degrades insoluble polyglucans (such as Lafora bodies) under cytosolic conditions and provokes a minimal immune reaction should be selected.
Reducing brain glycogen synthesis
One of the most promising therapeutic avenues for Lafora disease is the reduction of brain glycogen synthesis (FIG. 5). Partial or full removal of glycogen [starch] synthase, muscle (GYS1) — the glycogen synthase isoform that is expressed in the brain — in Lafora disease mouse models prevented Lafora body formation and led to rescue of the neurological phenotype. These findings imply that Lafora disease could be prevented by inhibiting glycogen synthesis, and that inhibition by ~50% might be sufficient to halt the progression of the disease43,137,138. Patients with loss- of-function GYS1 mutations, including those with complete loss of GYS1 (GSDOb, OMIM #611556), experience cardiac problems, epilepsy and exercise intolerance139–141. However, no health problems were reported in parents of these individuals in whom GYS1 levels were reduced to 50%140,141. Gysl-null mice have only 10% survival, with pups dying perinatally owing to cardiac dysfunction. However, the mice that do survive have apparently normal heart function, and heterozygous mice, with ~50% GYS1 levels, have no reported health issues142.
Another more indirect way to prevent glycogen synthesis is to decrease glycogen synthase activity by reducing levels of PP1 subunit R5, an indirect activator of this enzyme. Removal of PP1 subunit R5 in Lafora disease mice almost fully prevented Lafora body formation, and rescued neurodegeneration and the subsequent myoclonic epilepsy phenotype38,44. However, PP1 subunit R5 is only one of several PP1 subunits that promote glycogen activation55,65, and removal of this subunit led to a substantial reduction in but not a complete absence of Lafora bodies. In addition, PP1 subunit R5 promotes inactivation of glycogen phosphorylase, which mediates glycogen degradation76; therefore, the absence of this subunit might increase glycogen degradation. Targeting of other PP1 subunits in addition to the R5 subunit could be beneficial.
In principle, glycogenin 1 (encoded by the GYG1 gene) could also be considered as a therapeutic target for Lafora disease owing to its function as the primer for glycogen synthesis143. However, data concerning the Gygl - null mouse model and patients with loss-of-function mutations in GYG1 (GSDXV, OMIM #613507) suggest that glycogenin 1 depletion leads to glycogen accumulation, cardiomyopathy and muscle weakness. Therefore, this protein might not be the best therapeutic target for Lafora disease144–146.
Antisense oligonucleotides.
Antisense oligonucleotides (ASOs) are emerging as an excellent therapy platform, and though not a new concept147, ASO therapies have made substantial progress in recent years148,149. ASOs are short synthetic nucleic acids that are chemically modified to increase their stability in biological fluids and potency in binding to the target mRNA147. ASOs function in several different ways, one of which entails Watson-Crick base pairing with the target mRNA, leading to RNase H-mediated target degradation148.
The use of ASO therapy to target the mRNA encoding GYS1, PP1 subunit R5 and/or other PP1 subunits in the brain is a viable option for the treatment of Lafora disease (FIG. 5). As ASOs do not cross the BBB, direct CNS delivery is required in Lafora disease, as for other neurological diseases150. Intrathecally delivered ASOs spread widely throughout the brain and spinal cord in mice, rats and nonhuman primates150–154. However, ASOs require repeated administration to maintain therapeutic levels because they gradually degrade over time155.
ASO therapy targeting the splicing of the SMN2 gene is already prescribed to patients with spinal muscular atrophy156,157, and ASO drugs have also been developed for non-neurological diseases158. Clinical trials are ongoing to develop ASO therapies in amyotrophic lateral sclerosis (targeting SOD1 and C9orf72), Huntington disease (targeting HTT), tauopathies (targeting MAPT) and Alzheimer disease (targeting APP)149.
RNA interference.
RNA interference (RNAi) is another potential option for post-transcriptional suppression of Lafora disease-related therapeutic targets (FIG. 5). RNAi is achieved by the delivery of artificial short RNAs, such as microRNAs (miRNAs) and short hairpin RNAs (shRNAs), which are designed to pair with their target mRNA, leading to target degradation159. Artificial miRNAs and shRNAs are recognized by the short-RNA-processing machinery of the cell and are processed as endogenous small RNAs160. RNAi cell toxicity is a concern, as the endogenous RNA-processing machinery might become saturated159. In addition, an expression vector (AAV) and direct CNS delivery are required for wide CNS distribution. Regarding neurodegenerative disease, good results have been achieved with RNAi-mediated gene knockdown in Huntington disease161–163.
Genome engineering.
Another way to reduce the levels of glycogen synthase and/or glycogen-targeting subunits of PP1 is to modify the genes that encode these proteins (FIG. 5). The recent development of CRISPR-Cas9 as a biotechnological tool has substantially facilitated genome engineering, and this system is now being harnessed to edit the mammalian genome164,165.
A commonly used application of CRISPR-Cas9 is a target gene knockout whereby Cas9 introduces doublestranded DNA breaks (DSBs) adjacent to a guide RNA (sgRNA) recognition sequence, known as a protospacer adjacent motif. DSBs are repaired by non-homologous end-joining (NHEJ), leading to permanent indel formation and a non-functional protein164–166. This strategy is applicable to postmitotic cells, such as neurons, in which NHEJ is the preferred DNA repair pathway167,168, and the system has been successfully applied for target gene knockout in the mouse brain169–172.
CRISPR-Cas9-mediated therapy requires AAV- mediated delivery of Cas9 and sgRNA to the CNS. AAV transduction efficiency again determines which cells express Cas9. In addition, only a proportion of indels lead to biallelic mutation and a non-functional protein, which further decreases the overall efficiency. Other possible downsides include off-target effects, as partial sgRNA sequence similarity might lead to alterations in non-targeted genes, and an immune response provoked by permanently expressed bacterial Cas9 protein.
Technically, correction of laforin or malin at the gene level is possible. A study has shown successful alteration of single DNA bases using the CRISPR-Cas9 system173. However, in conditions such as Lafora disease where several different mutations in the associated gene(s) have been identified, base correction therapy would have to be individualized for each patient and would, therefore, be extremely expensive.
Small-molecule therapies.
A small-molecule therapy for Lafora disease would provide the least invasive option for the patient, as administration would be in the form of a digestible pill. Small molecules cannot replace the function of laforin or malin at the gene level but could affect the metabolic pathway that leads to the development of Lafora bodies. For instance, inhibitors of glycogen synthase or the PP1 subunits could prove to be effective therapeutics (FIG. 5).
Identification of small molecules requires high- throughput screening of thousands of molecules and development of in vitro assays to measure the target response. A high-throughput screening assay has been developed to identify small molecules that inhibit glycogen synthase activity to treat adult polyglucosan body disease, a form of GBE deficiency174. Inhibitory small molecules were successfully identified using an ex vivo cell-based assay and could potentially provide a therapy for glycogen storage diseases, including Lafora disease. Glycogen synthase inhibition has already been shown to be effective in glycogen storage diseases. Rapamycin, a tuberculosis drug, was found to lower the levels of pol- yglucosans and glycogen by interfering with the mechanistic target of rapamycin (mTOR) pathway, which is known to be disrupted in other epilepsies175,176.
To treat neurological disorders, the BBB usually needs to be crossed, which presents a tremendous challenge. To address this issue, repeated direct and invasive CNS delivery might be required. The target specificity of small molecules is also a potential challenge, and adverse events caused by off-target effects could limit the long-term utility of these therapies.
Conclusions and future prospects
Common genetic epilepsies tend to be comparatively mild and genetically complex, whereas rare genetic epilepsies are generally severe and genetically simple. The severity of these conditions usually prevents procreation and generational spread, which partially accounts for their rarity. However, until general prenatal or preconceptual screening becomes available, Lafora disease and similar neurodegenerative conditions are likely to persist.
One positive aspect of Lafora disease is its monogenicity, which could permit curative gene replacement in the coming years. Hyperelongation of glycogen chains lies at the root of the disease, and as reducing an activity is generally simpler than replacing a function, therapies aimed at reducing brain glycogen synthesis to mitigate and perhaps reverse the disease are relatively close at hand. Interventions could act at the DNA, RNA or protein level to target a number of enzymes that contribute to glycogen synthesis.
Owing to the considerable progress in Lafora disease research over the past few years, this condition is likely to become treatable before many of the other severe epilepsies. In addition, the insights into laforin and malin function that are emerging from this research are uncovering previously unsuspected roles for these proteins in glycogen metabolism. Further genetic studies in patients with Lafora disease genes are expected to teach us more about the body’s main energy store and the bioenergetics of brain function.
Key points
Lafora disease, a lethal, autosomal recessive, progressive myoclonus epilepsy, is caused by loss-of-function mutations in EPM2A or NHLRC1, which encode laforin and malin, respectively.
A large variety of pathogenic variants in EPM2A and NHLRC1 exists, with even distribution across both genes; variants include missense, nonsense and frameshift mutations, as well as larger deletions.
laforin and malin are implicated in glycogen metabolism and are presented as part of a glycogen quality control mechanism, which decreases the risk of precipitation of individual glycogen molecules.
In the absence of a functional laforin-malin complex, structurally abnormal glycogen becomes insoluble and accumulates as Lafora bodies, which drive disease progression in the brain.
The lack of curative treatments and the current understanding of pathogenesis are driving investigations into a variety of therapeutic strategies for Lafora disease, including reduction in brain glycogen synthesis and replacement of the non-functional gene.
Acknowledgements
The authors’ research work, some of which is described in this Review, is funded by families and friends of the Chelsea’s Hope Lafora Disease Research Fund, Associazione Italiana Lafora (AILA), France-Lafora, the Milana and Tatjana Gajic Lafora Disease Foundation, Genome Canada, the Ontario Brain Institute and the National Institute of Neurological Disorders and Stroke of the NIH under award number P01 NS097197. B.A.M. holds the University of Texas Southwestern Jimmy Elizabeth Westcott Chair in Pediatric Neurology. The authors thank S. Arnold (University of Texas Southwestern) for providing images of EEG and skin biopsy material from a patient with Lafora disease. This Review is dedicated to the memory of Adela Richer, a patient with Lafora disease who inspired this work and sadly passed away as the article was being completed.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Delgado-Escueta AV, Ganesh S & Yamakawa K Advances in the genetics of progressive myoclonus epilepsy. Am. J. Med. Genet. 106, 129–138 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Girard JM, Turnbull J, Ramachandran N & Minassian BA Progressive myoclonus epilepsy. Handb. Clin. Neurol. 113, 1731–1736 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Lafora GR & Glueck B Beitrag zur Histopathologie der myoklonischen Epilepsie [German]. Z. Gesamte Neurol. Psychiatr. 6, 1–14 (1911). [Google Scholar]
- 4.Sakai M, Austin J, Witmer F & Trueb L Studies in myoclonus epilepsy (Lafora body form). II. Polyglucosans in systemic deposits of myoclonus epilepsy and in corpora amylacea. Neurology 20, 160–176 (1970). [DOI] [PubMed] [Google Scholar]
- 5.Minassian BA et al. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat. Genet. 20, 171–174 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Serratosa JM et al. A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2). Hum. Mol. Genet. 8, 345–352 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Chan EM et al. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat. Genet. 35, 125–127 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Shahwan A, Farrell M & Delanty N Progressive myoclonic epilepsies: a review of genetic and therapeutic aspects. Lancet Neurol. 4, 239–248 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Madhavan D & Kuzniecky RI Lafora disease. Rev. Neurol. Dis. 3, 131–135 (2006). [PubMed] [Google Scholar]
- 10.Minassian BA et al. Mutation spectrum and predicted function of laforin in Lafora’s progressive myoclonus epilepsy. Neurology 55, 341–346 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Minassian BA Lafora’s disease: towards a clinical, pathologic, and molecular synthesis. Pediatr. Neurol. 25, 21–29 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Singh S & Ganesh S Lafora progressive myoclonus epilepsy: a meta-analysis of reported mutations in the first decade following the discovery of the EPM2A and NHLRC1 genes. Hum. Mutat. 30, 715–723 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Ianzano L et al. Lafora progressive myoclonus epilepsy mutation database — EPM2A and NHLRC1 (EPM2B) genes. Hum. Mutat. 26, 397 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Fernández-Sánchez ME et al. Laforin, the dual-phosphatase responsible for Lafora disease, interacts with R5 (PTG), a regulatory subunit of protein phosphatase-1 that enhances glycogen accumulation. Hum. Mol. Genet. 12, 3161–3171 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Raththagala M et al. Structural mechanism of laforin function in glycogen dephosphorylation and lafora disease. Mol. Cell 57, 261–272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gayarre J et al. The phosphatase activity of laforin is dispensable to rescue Epm2a−/− mice from Lafora disease. Brain 137, 806–818 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Nitschke F et al. Abnormal glycogen chain length pattern, not hyperphosphorylation, is critical in Lafora disease. EMBO Mol. Med. 9, 906–917 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganesh S et al. Genotype-phenotype correlations for EPM2A mutations in Lafora’s progressive myoclonus epilepsy: exon 1 mutations associate with an early-onset cognitive deficit subphenotype. Hum. Mol. Genet. 11, 1263–1271 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Roma-Mateo C et al. Lafora disease E3-ubiquitin ligase malin is related to TRIM32 at both the phylogenetic and functional level. BMC Evol. Biol. 11, 225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gómez-Abad C et al. Lafora disease due to EPM2B mutations: a clinical and genetic study. Neurology 64, 982–986 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Franceschetti S et al. Clinical and genetic findings in 26 Italian patients with Lafora disease. Epilepsia 47, 640–643 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Lohi H, Chan EM, Scherer SW & Minassian BA On the road to tractability: the current biochemical understanding of progressive myoclonus epilepsies. Adv. Neurol 97, 399–415 (2006). [PubMed] [Google Scholar]
- 23.Singh S et al. Novel NHLRC1 mutations and genotype-phenotype correlations in patients with Lafora’s progressive myoclonic epilepsy. J. Med. Genet. 43, e48 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan EM et al. Progressive myoclonus epilepsy with polyglucosans (Lafora disease): evidence for a third locus. Neurology 63, 565–567 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Lohi H et al. Genetic diagnosis in Lafora disease: genotype-phenotype correlations and diagnostic pitfalls. Neurology 68, 996–1001 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Turnbull J et al. Early-onset Lafora body disease. Brain 135, 2684–2698 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gómez-Abad C et al. Founder effect with variable age at onset in Arab families with Lafora disease and EPM2A mutation. Epilepsia 48, 1011–1014 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Annesi G et al. A novel exon 1 mutation in a patient with atypical lafora progressive myoclonus epilepsy seen as childhood-onset cognitive deficit. Epilepsia 45, 294–295 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Lesca G et al. Novel mutations in EPM2A and NHLRC1 widen the spectrum of Lafora disease. Epilepsia 51, 1691–1698 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Baykan B et al. Late-onset and slow-progressing Lafora disease in four siblings with EPM2B mutation. Epilepsia 46, 1695–1697 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Ferlazzo E et al. Mild Lafora disease: clinical, neurophysiologic, and genetic findings. Epilepsia 55, e129–e133 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Traoré M et al. Novel mutation in the NHLRC1 gene in a Malian family with a severe phenotype of Lafora disease. Neurogenetics 10, 319–323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brackmann FA, Kiefer A, Agaimy A, Gencik M & Trollmann R Rapidly progressive phenotype of Lafora disease associated with a novel NHLRC1 mutation. Pediatr. Neurol. 44, 475–477 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Kecmanovic M et al. Lafora disease: severe phenotype associated with homozygous deletion of the NHLRC1 gene. J. Neurol. Sci. 325, 170–173 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Jara-Prado A et al. Late onset Lafora disease and novel EPM2A mutations: breaking paradigms. Epilepsy Res. 108, 1501–1510 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Singh S & Ganesh S Phenotype variations in Lafora progressive myoclonus epilepsy: possible involvement of genetic modifiers? J. Hum. Genet. 57, 283–285 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Guerrero R et al. A PTG variant contributes to a milder phenotype in Lafora disease. PLoS ONE 6, e21294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turnbull J et al. PTG depletion removes Lafora bodies and rescues the fatal epilepsy of Lafora disease. PLoS Genet. 7, e1002037 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spillantini MG et al. α-Synuclein in Lewy bodies. Nature 388, 839–840 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Nuovo G et al. Increased expression of importin-β, exportin-5 and nuclear transportable proteins in Alzheimer’s disease aids anatomic pathologists in its diagnosis. Ann. Diagn. Pathol. 32, 10–16 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Ganesh S et al. Targeted disruption of the Epm2a gene causes formation of Lafora inclusion bodies, neurodegeneration, ataxia, myoclonus epilepsy and impaired behavioral response in mice. Hum. Mol. Genet. 11, 1251–1262 (2002). [DOI] [PubMed] [Google Scholar]
- 42.DePaoli-Roach AA et al. Genetic depletion of the malin E3 ubiquitin ligase in mice leads to Lafora Bodies and the accumulation of insoluble laforin. J. Biol. Chem 285, 25372–25381 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duran J, Gruart A, Garcia-Rocha M, Delgado-Garcia JM & Guinovart JJ Glycogen accumulation underlies neurodegeneration and autophagy impairment in Lafora disease. Hum. Mol. Genet. 23, 3147–3156 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Turnbull J et al. PTG protein depletion rescues malin-deficient Lafora disease in mouse. Ann. Neurol. 75, 442–446 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Duran J et al. Deleterious effects of neuronal accumulation of glycogen in flies and mice. EMBO Mol. Med. 4, 719–729 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Komatsu M et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Hara T et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006). [DOI] [PubMed] [Google Scholar]
- 48.McMahon J et al. Impaired autophagy in neurons after disinhibition of mammalian target of rapamycin and its contribution to epileptogenesis. J. Neurosci. 32, 15704–15714 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dias DM et al. Biophysical characterization of laforin-carbohydrate interaction. Biochem. J. 473, 335–345 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Christiansen C et al. The carbohydrate-binding module family 20 — diversity, structure, and function. FEBS J. 276, 5006–5029 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Chan EM et al. Laforin preferentially binds the neurotoxic starch-like polyglucosans, which form in its absence in progressive myoclonus epilepsy. Hum. Mol. Genet. 13, 1117–1129 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Zhang W, DePaoli-Roach AA & Roach PJ Mechanisms of multisite phosphorylation and inactivation of rabbit muscle glycogen synthase. Arch. Biochem. Biophys. 304, 219–225 (1993). [DOI] [PubMed] [Google Scholar]
- 53.Lohi H et al. Novel glycogen synthase kinase 3 and ubiquitination pathways in progressive myoclonus epilepsy. Hum. Mol. Genet. 14, 2727–2736 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Fontana JD The presence of phosphate in glycogen. FEBS Lett. 109, 85–92 (1980). [DOI] [PubMed] [Google Scholar]
- 55.Worby CA, Gentry MS & Dixon JE Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J. Biol. Chem. 281, 30412–30418 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tagliabracci VS et al. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc. Natl Acad. Sci. USA 104, 19262–19266 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tagliabracci VS et al. Abnormal metabolism of glycogen phosphate as a cause for Lafora disease. J. Biol. Chem. 283, 33816–33825 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nitschke F et al. Hyperphosphorylation of glucosyl C6 carbons and altered structure of glycogen in the neurodegenerative epilepsy Lafora disease. Cell Metab. 17, 756–767 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Roach PJ Glycogen phosphorylation and Lafora disease. Mol. Aspects Med. 46, 78–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vernia S, Rubio T, Heredia M, Rodriguez de Cordoba S & Sanz P Increased endoplasmic reticulum stress and decreased proteasomal function in lafora disease models lacking the phosphatase laforin. PLoS ONE 4, e5907 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rao SN et al. Sequestration of chaperones and proteasome into Lafora bodies and proteasomal dysfunction induced by Lafora disease-associated mutations of malin. Hum. Mol. Genet. 19, 4726–4734 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Mittal S, Dubey D, Yamakawa K & Ganesh S Lafora disease proteins malin and laforin are recruited to aggresomes in response to proteasomal impairment. Hum. Mol. Genet. 16, 753–762 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Ganesh S et al. The Lafora disease gene product laforin interacts with HIRIP5, a phylogenetically conserved protein containing a NIfU-like domain. Hum. Mol. Genet. 12, 2359–2368 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Wang Y et al. Epm2a suppresses tumor growth in an immunocompromised host by inhibiting Wnt signaling. Cancer Cell 10, 179–190 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Worby CA, Gentry MS & Dixon JE Malin decreases glycogen accumulation by promoting the degradation of protein targeting to glycogen (PTG). J. Biol. Chem. 283, 4069–4076 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gentry MS, Roma-Mateo C & Sanz P Laforin, a protein with many faces: glucan phosphatase, adapter proteinet et alii. FEBS J. 280, 525–537 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Solaz-Fuster MC et al. Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway. Hum. Mol. Genet. 17, 667–678 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Sanchez-Martin P, Roma-Mateo C, Viana R & Sanz P Ubiquitin conjugating enzyme E2-N and sequestosome-1 (p62) are components of the ubiquitination process mediated by the malin-laforin E3-ubiquitin ligase complex. Int. J. Biochem. Cell Biol. 69, 204–214 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Vilchez D et al. Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy. Nat. Neurosci. 10, 1407–1413 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Moreno D, Towler MC, Hardie DG, Knecht E & Sanz P The laforin-malin complex, involved in Lafora disease, promotes the incorporation of K63-linked ubiquitin chains into AMP-activated protein kinase β subunits. Mol. Biol. Cell 21, 2578–2588 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rubio-Villena C, Garcia-Gimeno MA & Sanz P Glycogenic activity of R6, a protein phosphatase 1 regulatory subunit, is modulated by the laforin-malin complex. Int. J. Biochem. Cell Biol. 45, 1479–1488 (2013). [DOI] [PubMed] [Google Scholar]
- 72.Deshaies RJ & Joazeiro CA RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Tan JM et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum. Mol. Genet. 17, 431–439 (2008). [DOI] [PubMed] [Google Scholar]
- 74.Nathan JA, Kim HT, Ting L, Gygi SP & Goldberg AL Why do cell proteins linked to K63- polyubiquitin chains not associate with proteasomes? EMBO J. 32, 552–565 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sharma J, Mulherkar S, Mukherjee D & Jana NR Malin regulates Wnt signaling pathway through degradation of dishevelled2. J. Biol. Chem. 287, 6830–6839 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fong NM et al. Identification of binding sites on protein targeting to glycogen for enzymes of glycogen metabolism. J. Biol. Chem. 275, 35034–35039 (2000). [DOI] [PubMed] [Google Scholar]
- 77.Zeeman SC, Kossmann J & Smith AM in In Annual Review of Plant Biology Vol. 61 (eds Merchant S et al. ) 209–234 (2010). [DOI] [PubMed] [Google Scholar]
- 78.Gilbert RG et al. Improving human health through understanding the complex structure of glucose polymers. Anal. Bioanal. Chem. 405, 8969–8980 (2013). [DOI] [PubMed] [Google Scholar]
- 79.Powell PO et al. Extraction, isolation and characterisation of phytoglycogen from su-1 maize leaves and grain. Carbohydr. Polym. 101, 423–431 (2014). [DOI] [PubMed] [Google Scholar]
- 80.Irimia JM et al. Muscle glycogen remodeling and glycogen phosphate metabolism following exhaustive exercise of wild type and laforin knockout mice. J. Biol. Chem. 290, 22686–22698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu AC & Gilbert RG Molecular weight distributions of starch branches reveal genetic constraints on biosynthesis. Biomacromolecules 11, 3539–3547 (2010). [DOI] [PubMed] [Google Scholar]
- 82.Manners DJ Recent developments in our understanding of glycogen structure. Carbohydr. Polym. 16, 37–82 (1991). [Google Scholar]
- 83.Buleon A, Colonna P, Planchot V & Ball S Starch granules: structure and biosynthesis. Int. J. Biol. Macromol. 23, 85–112 (1998). [DOI] [PubMed] [Google Scholar]
- 84.Umeki K & Kainuma K Fine structure of Naegeli amylodextrin obtained by acid treatment of defatted waxy-maize starch — structural evidence to support the double-helix hypothesis. Carbohydr. Res. 96, 143–159 (1981). [Google Scholar]
- 85.Bischof S et al. Cecropia peltata accumulates starch or soluble glycogen by differentially regulating starch biosynthetic genes. Plant Cell 25, 1400–1415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fujita N et al. Elongated phytoglycogen chain length in transgenic rice endosperm expressing active starch synthase IIa affects the altered solubility and crystallinity of the storage α-glucan. J. Exp. Bot. 63, 5859–5872 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sullivan M, Nitschke S, Steup M, Minassian B & Nitschke F Pathogenesis of Lafora disease: transition of soluble glycogen to insoluble polyglucosan. Int. J. Mol. Sci. 18, E1743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roach PJ, Depaoli-Roach AA, Hurley TD & Tagliabracci VS Glycogen and its metabolism: some new developments and old themes. Biochem. J. 441, 763–787 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Palmer TN, Macaskie LE & Grewal KK Spatial-distribution of unit chains in glycogen. Carbohydr. Res. 115, 139–150 (1983). [Google Scholar]
- 90.Petty HR, Worth RG & Kindzelskii AL Imaging sustained dissipative patterns in the metabolism of individual living cells. Phys. Rev. Lett. 84, 2754–2757 (2000). [DOI] [PubMed] [Google Scholar]
- 91.Rickey Welch G & Easterby JS Metabolic channeling versus free diffusion: transition-time analysis. Trends Biochem. Sci. 19, 193–197 (1994). [DOI] [PubMed] [Google Scholar]
- 92.Aw TY Intracellular compartmentation of organelles and gradients of low molecular weight species. Int. Rev. Cytol. 192, 223–253 (1999). [DOI] [PubMed] [Google Scholar]
- 93.Tiberia E et al. Increased laforin and laforin binding to glycogen underlie Lafora body formation in malin-deficient Lafora disease. J. Biol. Chem. 287, 25650–25659 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jiang S et al. Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J. Biol. Chem. 285, 34960–34971 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang P et al. SGK1 (glucose transport), dishevelled2 (wnt signaling), LC3/p62 (autophagy) and p53 (apoptosis) proteins are unaltered in Lafora disease. All Results J. Biol 7, 28–33 (2016). [PMC free article] [PubMed] [Google Scholar]
- 96.Khiari HM, Lesca G, Malafosse A & Mrabet A A novel exon 3 mutation in a Tunisian patient with Lafora’s disease. J. Neurol. Sci. 304, 136–137 (2011). [DOI] [PubMed] [Google Scholar]
- 97.Villanueva V, Alvarez-Linera J, Gomez-Garre P, Gutierrez J & Serratosa JM MRI volumetry and proton MR spectroscopy of the brain in Lafora disease. Epilepsia 47, 788–792 (2006). [DOI] [PubMed] [Google Scholar]
- 98.Andrade DM et al. Skin biopsy in Lafora disease: genotype-phenotype correlations and diagnostic pitfalls. Neurology 61, 1611–1614 (2003). [DOI] [PubMed] [Google Scholar]
- 99.Cardinali S et al. A pilot study of a ketogenic diet in patients with Lafora body disease. Epilepsy Res. 69, 129–134 (2006). [DOI] [PubMed] [Google Scholar]
- 100.Schorlemmer K et al. Sustained seizure remission on perampanel in progressive myoclonic epilepsy (Lafora disease). Epilepsy Behav. Case Rep. 1, 118–121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dirani M, Nasreddine W, Abdulla F & Beydoun A Seizure control and improvement of neurological dysfunction in Lafora disease with perampanel. Epilepsy Behav. Case Rep. 2, 164–166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goldsmith D & Minassian BA Efficacy and tolerability of perampanel in ten patients with Lafora disease. Epilepsy Behav. 62, 132–135 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hajnsek S et al. Vagus nerve stimulation in Lafora body disease. Epilepsy Behav. Case Rep. 1, 150–152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mikati MA & Tabbara F Managing Lafora body disease with vagal nerve stimulation. Epilept. Disord 19, 82–86 (2017). [DOI] [PubMed] [Google Scholar]
- 105.Zhou G et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 108, 1167–1174 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ronnett GV, Ramamurthy S, Kleman AM, Landree LE & Aja S AMPK in the brain: its roles in energy balance and neuroprotection. J. Neurochem. 109, 17–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hardie DG AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell. Biol. 8, 774–785 (2007). [DOI] [PubMed] [Google Scholar]
- 108.Yang Y et al. Chronic metformin treatment facilitates seizure termination. Biochem. Biophys. Res. Commun. 484, 450–455 (2017). [DOI] [PubMed] [Google Scholar]
- 109.Dulovic M et al. The protective role of AMP-activated protein kinase in alpha-synuclein neurotoxicity in vitro. Neurobiol. Dis 63, 1–11 (2014). [DOI] [PubMed] [Google Scholar]
- 110.Ashabi G, Khodagholi F, Khalaj L, Goudarzvand M & Nasiri M Activation of AMP-activated protein kinase by metformin protects against global cerebral ischemia in male rats: interference of AMPK/PGC-1α pathway. Metab. Brain Dis. 29, 47–58 (2014). [DOI] [PubMed] [Google Scholar]
- 111.Berthier A et al. Pharmacological interventions to ameliorate neuropathological symptoms in a mouse model of Lafora disease. Mol. Neurobiol. 53, 1296–1309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sanchez-Elexpuru G, Serratosa JM, Sanz P & Sanchez MP 4-PBA and metformin decrease sensitivity to PTZ-induced seizures in a malin knockout model of Lafora disease. Neuroreport 28, 268–271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sanchez-Elexpuru G, Serratosa JM & Sanchez MP Sodium selenate treatment improves symptoms and seizure susceptibility in a malin-deficient mouse model of Lafora disease. Epilepsia 58, 467–475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Burke JF & Mogg AE Suppression of a nonsense mutation in mammalian cells in vivo by the aminoglycoside antibiotics G-418 and paromomycin. Nucleic Acids Res. 13, 6265–6272 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bolze F, Mocek S, Zimmermann A & Klingenspor M Aminoglycosides, but not PTC124 (Ataluren), rescue nonsense mutations in the leptin receptor and in luciferase reporter genes. Sci. Rep. 7, 1020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Clancy JP et al. Evidence that systemic gentamicin suppresses premature stop mutations in patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 163, 1683–1692 (2001). [DOI] [PubMed] [Google Scholar]
- 117.Wilschanski M et al. Gentamicin-induced correction of CFTR function in patients with cystic fibrosis and CFTR stop mutations. N. Engl. J. Med. 349, 1433–1441 (2003). [DOI] [PubMed] [Google Scholar]
- 118.Politano L et al. Gentamicin administration in Duchenne patients with premature stop codon. Preliminary results. Acta Myol. 22, 15–21 (2003). [PubMed] [Google Scholar]
- 119.Schroers A et al. Gentamicin treatment in McArdle disease: failure to correct myophosphorylase deficiency. Neurology 66, 285–286 (2006). [DOI] [PubMed] [Google Scholar]
- 120.Wargo KA & Edwards JD Aminoglycoside-induced nephrotoxicity. J. Pharm. Pract. 27, 573–577 (2014). [DOI] [PubMed] [Google Scholar]
- 121.Guthrie OW Aminoglycoside induced ototoxicity. Toxicology 249, 91–96 (2008). [DOI] [PubMed] [Google Scholar]
- 122.Saraiva J, Nobre RJ & de Almeida LP Gene therapy for the CNS using AAVs: the impact of systemic delivery by AAV9. J. Control. Release 241, 94–109 (2016). [DOI] [PubMed] [Google Scholar]
- 123.Choudhury SR et al. Viral vectors for therapy of neurologic diseases. Neuropharmacology 120, 63–80 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jan AT et al. Perspective insights of exosomes in neurodegenerative diseases: a critical appraisal. Front. Aging Neurosci. 9, 317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Duskey JT et al. in International Review of Neurobiology 1–28 (Elsevier, 2017). [DOI] [PubMed] [Google Scholar]
- 126.Chan KY et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Foust KD et al. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 27, 59–65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bottros MM & Christo P J. Current perspectives on intrathecal drug delivery. J. Pain Res. 7, 615–626 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Haché M et al. Intrathecal injections in children with spinal muscular atrophy: nusinersen clinical trial experience. J. Child Neurol. 31,899–906 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gray SJ, Kalburgi SN, McCown TJ & Samulski RJ Global CNS gene delivery and evasion of anti-AAV-neutralizing antibodies by intrathecal AAV administration in non-human primates. Gene Ther. 20, 450–459 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Samaranch L et al. Adeno-associated virus serotype 9 transduction in the central nervous system of nonhuman primates. Hum. Gene Ther. 23, 382–389 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gray SJ et al. Preclinical differences of intravascular AAV9 delivery to neurons and glia: a comparative study of adult mice and nonhuman primates. Mol. Ther. 19, 1058–1069 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bessis N, GarciaCozar FJ & Boissier MC Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 11 (Suppl. 1), S10–S17 (2004). [DOI] [PubMed] [Google Scholar]
- 134.Biffi A et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 341, 1233158 (2013). [DOI] [PubMed] [Google Scholar]
- 135.Cartier N & Aubourg P Hematopoietic stem cell transplantation and hematopoietic stem cell gene therapy in X-linked adrenoleukodystrophy. Brain Pathol. 20, 857–862 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Auger A et al. Efficient delivery of structurally diverse protein cargo into mammalian cells by a bacterial toxin. Mol. Pharm. 12, 2962–2971 (2015). [DOI] [PubMed] [Google Scholar]
- 137.Pederson BA et al. Inhibiting glycogen synthesis prevents Lafora disease in a mouse model. Ann. Neurol. 74, 297–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Duran J & Guinovart JJ Brain glycogen in health and disease. Mol. Aspects Med. 46, 70–77 (2015). [DOI] [PubMed] [Google Scholar]
- 139.Cameron JM et al. Identification of a novel mutation in GYS1 (muscle-specific glycogen synthase) resulting in sudden cardiac death, that is diagnosable from skin fibroblasts. Mol. Genet. Metab. 98, 378–382 (2009). [DOI] [PubMed] [Google Scholar]
- 140.Kollberg G et al. Cardiomyopathy and exercise intolerance in muscle glycogen storage disease 0. N. Engl. J. Med. 357, 1507–1514 (2007). [DOI] [PubMed] [Google Scholar]
- 141.Sukigara S et al. Muscle glycogen storage disease 0 presenting recurrent syncope with weakness and myalgia. Neuromuscul. Disord. 22, 162–165 (2012). [DOI] [PubMed] [Google Scholar]
- 142.Pederson BA et al. Abnormal cardiac development in the absence of heart glycogen. Mol. Cell. Biol. 24, 7179–7187 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Whelan WJ The initiation of glycogen synthesis. Bioessays 5, 136–140 (1986). [DOI] [PubMed] [Google Scholar]
- 144.Testoni G et al. Lack of glycogenin causes glycogen accumulation and muscle function impairment. Cell Metab. 26, 256–266. e4 (2017). [DOI] [PubMed] [Google Scholar]
- 145.Malfatti E et al. A new muscle glycogen storage disease associated with glycogenin-1 deficiency. Ann. Neurol. 76, 891–898 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hedberg-Oldfors C et al. Polyglucosan myopathy and functional characterization of a novel GYG1 mutation. Acta Neurol. Scand. 137, 308–315 (2018). [DOI] [PubMed] [Google Scholar]
- 147.Crooke ST & Bennett CF Progress in antisense oligonucleotide therapeutics. Annu. Rev. Pharmacol. Toxicol. 36, 107–129 (1996). [DOI] [PubMed] [Google Scholar]
- 148.Bennett CF & Swayze EE RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 50, 259–293 (2010). [DOI] [PubMed] [Google Scholar]
- 149.Schoch KM & Miller T M. Antisense oligonucleotides: translation from mouse models to human neurodegenerative diseases. Neuron 94, 1056–1070 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Smith RA et al. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 116, 2290–2296 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.DeVos SL et al. Antisense reduction of tau in adult mice protects against seizures. J. Neurosci. 33, 12887–12897 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kordasiewicz HB et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron 74, 1031–1044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lagier-Tourenne C et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl Acad. Sci. USA 110, E4530–E4539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Passini MA et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med 3, 72ra–18. (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Geary RS, Norris D, Yu R & Bennett CF Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 87, 46–51 (2015). [DOI] [PubMed] [Google Scholar]
- 156.Hua Y et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 24, 1634–1644 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hua Y, Vickers TA, Okunola HL, Bennett CF & Krainer AR Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 82, 834–848 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stein CA & Castanotto D FDA-approved oligonucleotide therapies in 2017. Mol. Ther. 25, 1069–1075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Davidson BL & McCray P B. Current prospects for RNA interference-based therapies. Nat. Rev. Genet. 12, 329–340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.MacRae IJ et al. Structural basis for doublestranded RNA processing by Dicer. Science 311, 195–198 (2006). [DOI] [PubMed] [Google Scholar]
- 161.McBride JL et al. Preclinical safety of RNAi- mediated HTT suppression in the rhesus macaque as < potential therapy for Huntington’s disease. Mol. Ther. 19, 2152–2162 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Harper SQ et al. RNA interference improves motor and neuropathological abnormalities in a Huntington’ disease mouse model. Proc. Natl Acad. Sci. USA 102, 5820–5825 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Machida Y et al. rAAV-mediated shRNA ameliorated neuropathology in Huntington disease model mouse. Biochem. Biophys. Res. Commun. 343, 190–197 (2006). [DOI] [PubMed] [Google Scholar]
- 164.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mali P, Esvelt KM & Church GM Cas9 as a versatile tool for engineering biology. Nat. Methods 10, 957–963 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Doudna JA & Charpentier E The new frontier of genome engineering with CRISPR-Cas9. Science 346 1258096 (2014). [DOI] [PubMed] [Google Scholar]
- 167.Chapman JR, Taylor MR & Boulton SJ Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 (2012). [DOI] [PubMed] [Google Scholar]
- 168.Rothkamm K, Krüger I, Thompson LH & Löbrich M Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol 23, 5706–5715 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Swiech L et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 33, 102–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Platt RJ et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shinmyo Y et al. CRISPR/Cas9-mediated gene knockout in the mouse brain using in utero electroporation. Sci. Rep 6, 20611 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Feng W et al. Chd7 is indispensable for mammalian brain development through activation of a neuronal differentiation programme. Nat. Commun. 8, 14758 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Solmesky LJ et al. A novel image-based high- throughput screening assay discovers therapeutic candidates for adult polyglucosan body disease. Biochem. J. 474, 3403–3420 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kakhlon O et al. Polyglucosan neurotoxicity caused by glycogen branching enzyme deficiency can be reversed by inhibition of glycogen synthase. J. Neurochem. 127, 101–113 (2013). [DOI] [PubMed] [Google Scholar]
- 176.Ashe KM et al. Inhibition of glycogen biosynthesis via mTORC1 suppression as an adjunct therapy for Pompe disease. Mol. Genet. Metab. 100, 309–315 (2010). [DOI] [PubMed] [Google Scholar]