Abstract
DHHC (Asp-His-His-Cys) palmitoyltransferases are eukaryotic integral membrane enzymes that catalyze protein palmitoylation, which is important in a range of physiological processes, including small guanosine triphosphatase (GTPase) signaling, cell adhesion, and neuronal receptor scaffolding. We present crystal structures of two DHHC palmitoyltransferases and a covalent intermediate mimic. The active site resides at the membrane-cytosol interface, which allows the enzyme to catalyze thioester-exchange chemistry by using fatty acyl–coenzyme A and explains why membrane-proximal cysteines are candidates for palmitoylation. The acyl chain binds in a cavity formed by the transmembrane domain. We propose a mechanism for acyl chain–length selectivity in DHHC enzymes on the basis of cavity mutants with preferences for shorter and longer acyl chains.
Introduction
Covalent attachment to lipids is a prevalent form of posttranslational modification (1) that influences membrane association of peripheral membrane proteins, protein targeting to membrane subdomains, protein-protein associations, and protein trafficking (2, 3). Of the main different forms of protein lipidation, attachment of a 14-carbon myristoyl group to the N terminus or a 15-carbon farnesyl or 20-carbon geranylgeranyl to the C terminus results in a relatively stable modification (4). By contrast, attachment of a fatty acyl group to an internal cysteine as a thioester (5, 6), known as protein S-acylation, is readily reversible through the action of cellular thioesterases (7, 8), making S-acylation a potentially dynamic form of lipidation (9).
Protein S-acylation is more commonly referred to as protein palmitoylation owing to the prevalence of the 16-carbon palmitate among the acyl chains that are attached to substrate proteins (10). However, for at least a subset of proteins, modification by fatty acyl chains longer or shorter than 16 carbons has been shown (10–12). The readily reversible nature of protein palmitoylation enables dynamic modulation of the hydrophobicity of substrate proteins. Protein palmitoylation plays critical roles in a wide range of physiological processes such as Ras signaling (13), localization of neuronal scaffolding proteins (14), intracellular trafficking (15), activity of ion channels (16), and host-pathogen interactions (17, 18). Since their discovery, an increasing number of proteins have been added to the repertoire of cellular proteins that are palmitoylated, with a recent estimate of close to 1000 proteins in humans (19). Although bioinformatic analyses of protein sequences proximal to the target cysteine have had some success in predicting palmitoylation sites, there are currently no reported consensus sequences for palmitoylation (20). Examination of experimentally identified palmitoylation sites and their sequence context, both in terms of physicochemical properties as well as predicted structure, is strongly indicative of the fact that one of the criteria for a cysteine to be palmitoylated is proximity to the membrane (20). Protein palmitoylation is connected to diseases, especially cancers and neuropsychiatric disorders (21), and it has been proposed that developing inhibitors of DHHC20, an enzyme that palmitoylates epidermal growth factor receptor (EGFR), can provide a therapeutic avenue for treating cancers that are resistant to EGFR-targeted therapy (22).
Although palmitoylation as a posttranslational modification was discovered in 1979 (5), the enzymes that catalyze protein palmitoylation were only discovered in 2002 (23, 24). These are low-abundance, polytopic eukaryotic integral membrane enzymes known as DHHC-palmitoyl transferases, so named because they contain a signature Asp-His-His-Cys (DHHC) motif within a cysteine-rich domain in an intracellular loop (fig. S1). Localization studies suggest that DHHC enzymes reside at a variety of cellular compartments, most prominently the Golgi complex (25). Beyond the shared cysteine-rich domain, there is considerable diversity in the DHHC family—some possess ankyrin repeats (24), a few have six transmembrane (TM) helices (26) instead of the canonical four, and at least one of them forms a functional heterodimer with an auxiliary subunit (23). Studies of yeast Erf2/Erf4 (homolog of mammalian DHHC9/GCP16) (27) and mammalian DHHC2 and DHHC3 (28) indicate that palmitate transfer to substrates occurs in two steps. First, autoacylation of a cysteine within the enzyme with palmitoyl–coenzyme A (CoA) forms a palmitoylated intermediate. This intermediate has been isolated in vitro, and, in the absence of a substrate, the autopalmitoylated enzyme undergoes slow hydrolysis. However, in presence of a protein substrate, the palmitate is transferred to a cysteine on the substrate in a transpalmitoylation reaction that regenerates the DHHC enzyme (28) (Fig. 1A). The specific roles of the conserved residues in the cysteine-rich domain that includes the DHHC motif are poorly understood. Genetic and biochemical analyses indicates that DHHC enzymes bind two Zn2+ ions (29) at two zinc finger–like domains, but the function of these Zn2+ ions in DHHC enzymes is unknown. Moreover, fatty acyl–CoA selectivity varies between DHHC enzymes (28, 30). Nevertheless, nothing is known about the site on the enzyme where the acyl-CoA binds and, thus, the determinants for fatty acid chain–length selectivity.
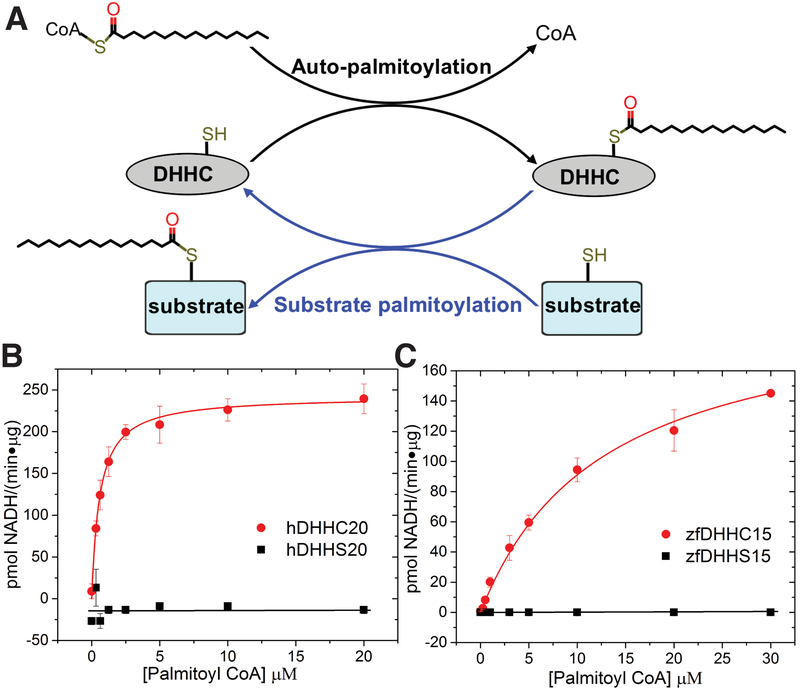
Figure 1. Functional characterization of DHHC enzymes.
(A) The proposed two-step catalytic mechanism of the DHHC enzymes in literature, where they first undergo self-acylation to form an acylated intermediate (shown in black) and subsequently transfer the acyl chain to a protein substrate in the second step (shown in blue). (B) Determination of kinetic parameters for the autopalmitoylation of hDHHC20 measured using the coupled-enzyme assay (fig. S3A). Shown is a Michaelis-Menten fit to palmitoyl-CoA titration with hDHHC20 (red circle) and hDHHS20 (black square). Km = 0.58 ± 0.04 μM; kcat = 16.5 ± 0.3 min−1. (C) Same as (B) for zfDHHC15. Km = 10 ± 1 μM; kcat = 14.6 ± 0.8 min−1. Data are mean ± SEM of three or more replicate measurements.
Functional characterization of DHHC15 and DHHC20 constructs
We carried out an extensive search for appropriate constructs of members of the DHHC family for crystallization using fluorescence size-exclusion chromatography (FSEC), screening for protein stability, yield, and monodispersity of size-exclusion chromatographic profile (31). Of the initial hits, human DHHC20 (hDHHC20) and zebrafish DHHC15 (zfDHHC15) were promising candidates (figs. S2 and S4). Because there was no biochemical characterization of either DHHC20 or DHHC15 in the literature, we tested whether they are bona fide palmitoyltransferase enzymes. We first used a coupled-enzyme assay that interrogates DHHC autoacylation by utilizing the released free CoA to generate an equivalent amount of reduced nicotinamide adenine dinucleotide (NADH) (32) (Fig.1A), which is detected by its fluorescence (fig. S3A). Both hDHHC20 and zfDHHC15 showed robust activity (Fig. 1, B and C, and figs. S3C and S4E). Mutants in which the active site cysteine was changed to a serine, designated as hDHHS20 and zfDHHS15, showed very little activity. To demonstrate the capability of the enzymes for catalyzing palmitoylation of protein substrates, we used the Legionella effector protein GobX (17) and human SNAP25b (30) as substrates for hDHHC20 and zfDHHC15, respectively. In an in vitro assay with purified proteins, hDHHC20 and zfDHHC15 displayed robust palmitoyl-transfer activity, whereas the catalytically inactive mutants, hDHHS20 and zfDHHS15, displayed the same level of activity as the “no enzyme” control (fig. S5). These experiments demonstrated that our preparations of hDHHC20 and zfDHHC15 have robust autoacylation activity in a fully reconstituted in vitro assay and are competent for substrate palmitoylation. Purified hDHHC20 and zfDHHC15 contain two Zn2+ ions per protein (figs. S2D and S4F), similar to what has been observed with DHHC3 (33) and Swf1, a yeast DHHC enzyme (29).
Crystallization and overall structural organization of DHHC enzymes
We obtained crystals of wild-type hDHHC20 from lipidic cubic phase (LCP) and crystals of the mutant, zfDHHS15, from hanging drop conditions. These crystals yielded diffraction data that enabled us to solve the structure using Zn–single-wavelength anomalous dispersion (SAD) data from the Zn2+ ions in the protein (hDHHC20) and with a combination of Zn-SAD and sulfur-SAD for zfDHHS15 (fig. S6, F to I). The hDHHC20 protein crystallizes in two different crystal forms in LCP: a P63 form with an antiparallel dimer and a P21 form with a loose parallel dimer in the asymmetric unit (fig. S6, A and B). Conversely, zfDHHS15 crystallizes in P42212 space group with an antiparallel dimer in the asymmetric unit (fig. S6C). We obtained x-ray diffraction data of zfDHHS15 crystals to 2.54-Å resolution. The hDHHC20 protein was reductively methylated for obtaining optimal diffraction data. The hexagonal form of hDHHC20 diffracted to 2.44-Å resolution. The DHHC structures display similar monomer structures, with a root mean square deviation (RMSD) of superposition (Cα) between the monomers of the two forms of hDHHC20 being 0.52 Å and between the monomer of zfDHHS15 and the monomer of hDHHC20 (P63 form) being 0.74 Å (fig. S6, D and E). We will focus our discussion on the hexagonal form of hDHHC20, with comments on zfDHHS15 where appropriate. The final models contain residues 5 to 299 of hDHHC20 and residues 8 to 298 of zfDHHS15 (residues 42 and 43 and 199 to 202 are absent from the zfDHHS15 model) (tables S1 and S2).
Protein palmitoylation almost exclusively occurs at the cytoplasmic face of organellar and plasma membranes, and thus the predicted topology of DHHC enzymes would place the DHHC loop in the cytoplasm. Based on this and the fact that the same hDHHC20 protein crystallized in lipid-rich phase in two different forms, we interpret the different dimeric interfaces as adventitious protein-protein interactions. Such dimer formation has been seen in other membrane-protein structures (34, 35). To rule out hDHHC20 or zfDHHC15 adopting an inverted topology, we carried out a fluorescence protease protection assay (36) to determine the physiological topology of both hDHHC20 and zfDHHC15. Both hDHHC20 and zfDHHC15 are putative Golgi resident proteins, and C-terminal green fluorescent protein (GFP)–tagged constructs localize to the Golgi. Upon permeabilization of the plasma membrane with digitonin and addition of trypsin, the fluorescent tag on the C terminus is not protected, whereas a control with a fluorescent tag in the lumen of the Golgi is (figs. S7 and S8). Given that both hDHHC20 and zfDHHC15 have four TM helices, with the DHHC loop located between TM2 and TM3, this experiment rules out the possibility of any physiologically relevant topology of hDHHC20 or zfDHHC15 that positions the DHHC loop in the lumen of the Golgi.
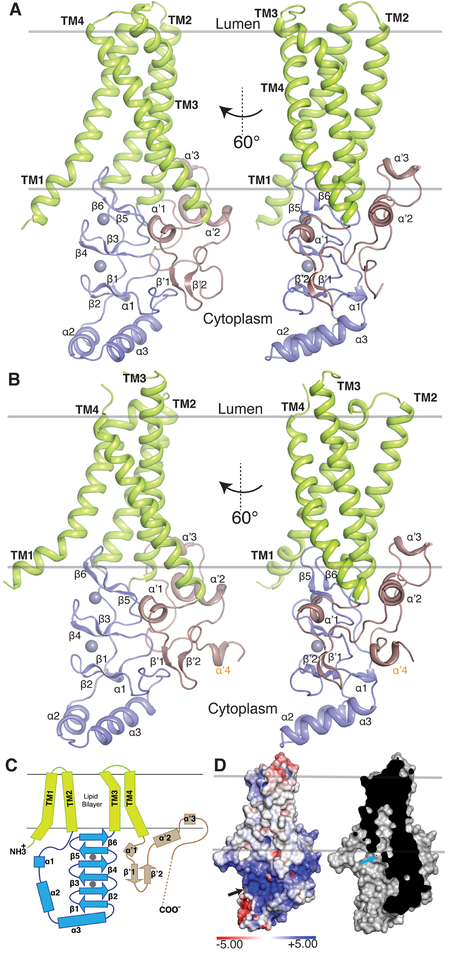
The four TM helices of hDHHC20 and zfDHHS15 adopt a tepee-like organization in the membrane, coming close together on the lumenal side and splaying apart on the cytoplasmic side, where substrate engagement and catalysis take place (Fig. 2). TM1 and TM4 are the most tilted with respect to the transmembrane normal. On the lumenal side, short loops connect TM1 and TM2, and TM3 and TM4. On the cytoplasmic side, the DHHC cysteine-rich domain connects TM2 and TM3. The N-terminal part of this domain consists of a helix-turn-helix that is connected to TM2 by a flexible linker. The C-terminal part of the domain has three stacked β hairpins arranged roughly in parallel with the bilayer and terminates in a short linker that is connected to TM3. The C-terminal domain of hDHHC20 is also at the cytoplasmic face and begins with a short helical segment (αʹ1) that wedges in between TM3 and the β hairpins in the DHHC domain. This domain is connected to a β hairpin that protrudes into the cytoplasm, but comes back to the membrane proximal region and ends in an amphipathic helix (αʹ2) that rests against TM3 and TM4. On the C-terminal side of the amphipathic helix, a hydrophobic loop with a short α-helical segment (αʹ3) inserts into the putative membrane bilayer region and makes extensive contacts with TM2 and TM3. Together αʹ2 and αʹ3 form a supporting belt on the cytoplasmic side of TM3 and TM4 (Fig. 2, A to C).
Figure 2. Overall structure of a DHHC enzyme.
(A) Cartoon representation of hDHHC20. The four TM helices are shown in green, the DHHC cysteine-rich domain in blue, and the C-terminal domain in brown. The two spheres represent the Zn2+ ions. (B) Same as (A) for zfDHHS15. An extra helix in the C-terminal domain is labeled in orange. (C) Diagram of the secondary structure elements of hDHHC20, showing helices as rectangles and β sheets as arrows. The two gray circles represent Zn2+ ions, and the dashed line indicates disordered C-terminal domain not observed in the crystal structure. Colors are the same as in (A). (D) Molecular surface representation (left) of hDHHC20 colored by electrostatic potential, showing a distinctive basic patch (black arrow) and a cross section (right) showing a cavity (cyan arrow) that exists above the active site. Electrostatic surface potential was generated using an Adaptive Poisson-Boltzmann Solver (APBS) server with default settings. Positively (blue) and negatively (red) charged surfaces are displayed at the contour levels of +5 and –5 kBT/e, respectively, where kB is the Boltzmann constant, T is temperature, and e is the charge on an electron.
Structure of the DHHC cysteine-rich domain and organization of the active site
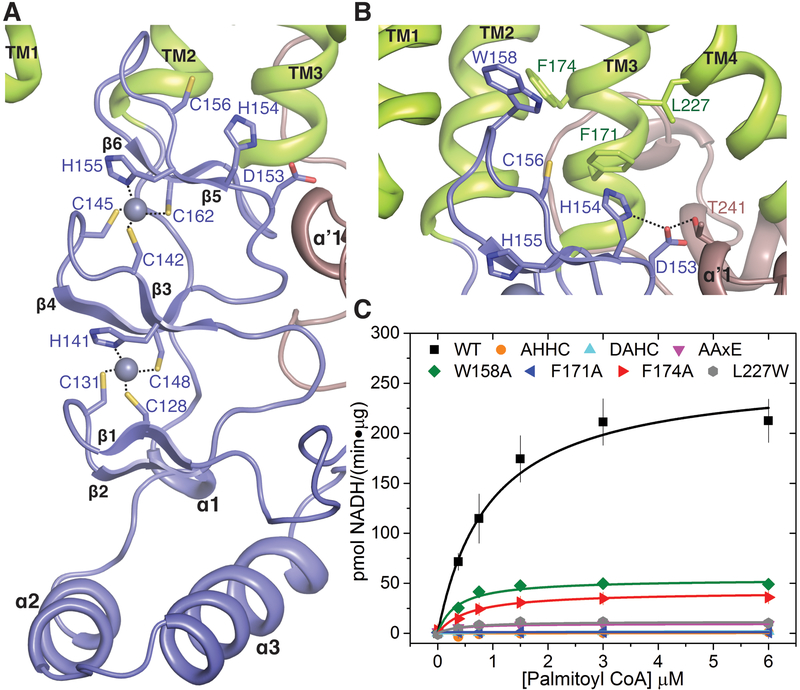
The cysteine-rich domain binds two Zn2+ ions, which is consistent with data from other DHHC members suggesting that this domain contains zinc finger motifs (29). The Zn2+ ions presumably play a structural role, as they are positioned between three parallel layers of β hairpins (Fig. 3A). Three cysteines and a histidine form the tetrahedral coordination environment of each Zn2+, resembling canonical zinc finger motifs (37). Neither of the Zn2+ ions coordinate the active site cysteine and thus do not directly participate in catalysis (38). However, mutations of the coordinating residues have been shown to drastically change the stability and/or catalytic activity of DHHC3 (33), supporting a role for the Zn2+ ions in positioning the catalytic cysteine optimally. Consequently, these residues are conserved among all members of the DHHC family (fig. S1). Additionally, a role for the zinc fingers in the binding of protein substrates can also not be ruled out (39).
Figure 3. Organization of the active site of DHHC20.
(A) Close-up of the Zn2+ binding region showing coordination of each Zn2+ ion by three cysteines and one histidine in a CCHC configuration. (B) Close-up of the DHHC enzyme active site showing the catalytic Cys156 pointing upward toward a hydrophobic groove. Also shown are the aspartic acid and the first histidine of the DHHC motif and the Thr241 of the TTXE motif. Hydrogen-bonding interactions are shown with dotted lines. Trp158, Phe171, and Phe174, which form the base of the acyl-binding cavity in Fig. 2D, are also shown in stick rendition. (C) Analysis of the enzymatic activity of selected mutants of the active site residues shown in (B). The coupled-enzyme assay was used, and Michaelis-Menten fits are shown. AHHC, DAHC, and F171A mutant curves all overlay with essentially no enzyme activity. Data are mean ± SEM of two independent measurements.
The active site is situated at the membrane-aqueous interface in agreement with the observation that proteins are palmitoylated at membrane-proximal cysteines (20). The catalytic DHHC motif (the two histidines are His154 and His155) is located on the β hairpins (β5-β6) that coordinate the Zn2+ ions (Fig. 3B). The aspartic acid and the His154 are on the top face of the β hairpin, protruding in the direction of the membrane, and presumably form part of a catalytic triad (40) where the aspartic acid forms a hydrogen-bonding interaction with the histidine (Fig. 3B). The His155 side chain coordinates a Zn2+ ion on the other face of the β hairpin and presumably positions the catalytic cysteine for nucleophilic attack. In the zfDHHS15 structure, the side chain of the active site serine residue rotates to form a hydrogen-bonding interaction with a main-chain amide nitrogen (fig. S9). However, the putative position of the catalytic cysteine in the wild-type enzyme will likely bring it within hydrogen-bonding distance of His154. The structure suggests that the aspartic acid and the first histidine of the DHHC motif serve crucial roles in activating the nucleophilic cysteine. Mutating these residues individually to alanine generates mutants with no discernible catalytic activity (Fig. 3C).
This organization of the active site is such that the catalytic cysteine of the DHHC motif protrudes in the direction of the membrane bilayer (Fig. 3B). Given that DHHC enzymes catalyze via a two-step reaction mechanism and that the acylated enzyme is an intermediate species with a long hydrophobic moiety attached to it, this suggests that the active site is preorganized to stabilize the acylated intermediate by inserting the acyl chain into the hydrophobic part of the bilayer. hDHHC20 has two positively charged patches on the cytoplasmic side (Fig. 2D, left); one of them arises from the binding of the two positively charged zinc ions. Another basic patch containing Arg126 and Lys135 showed electron density that we modeled as adenosine 5′-diphosphate with an additional phosphate at the 3′ end (5′-diphosphoadenosine 3′-phosphate) (fig. S10, A to C). This was based on consideration of components in the crystallization solution and other possible carryover contaminants such as adenosine 5′-triphosphate (ATP). Interestingly, this moiety is contained in the terminal part of CoA and thus could represent a byproduct of catalysis (fig. S10E). Because we did not add CoA during the protein purification, presumably this ligand was carried through the purification process. Notably, acyl-CoA has a long pantothenyl linker connecting thioester and the ribose ring. The extended form of this linker can easily span the length between the active site and the 5′-diphosphoadenosine 3′-phosphate binding site in the structure (fig. S10D). Based on these considerations, we suggest the 5′-diphosphoadenosine 3′-phosphate binding site to be the place where the terminal part of the coenzyme headgroup of palmitoyl-CoA binds.
Structure of the C-terminal domain
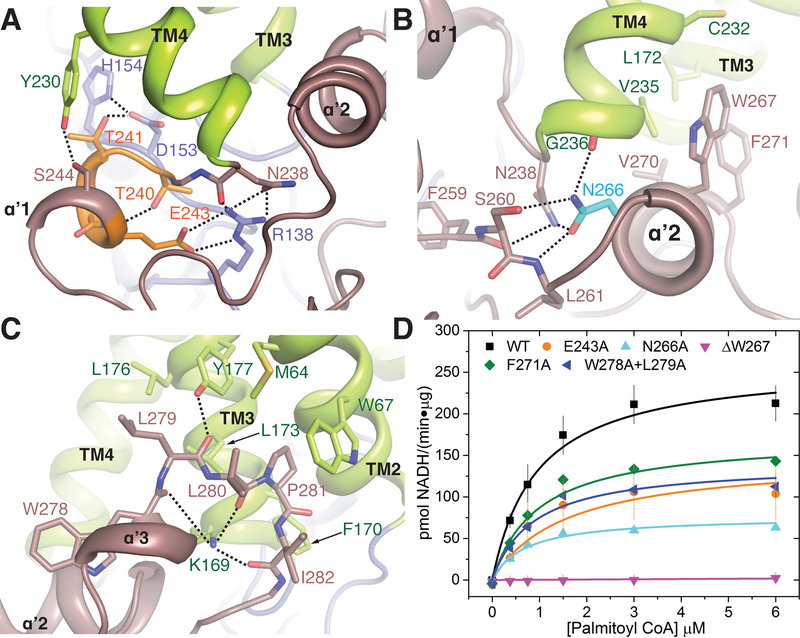
Although the C-terminal domain in DHHC enzymes is more variable compared to the cysteine-rich domain, it contains sequence motifs that are conserved to varying extents throughout the family. The most highly conserved among them is the TTXE (Thr-Thr-X-Glu) motif that resides in the short helical segment (αʹ1) immediately following TM4, where X is any amino acid residue. It makes intimate contact with the DHHC domain (Figs. 3B and 4A and fig. S1). The Thr240 of the conserved TTXE motif caps a main-chain amide nitrogen, and Thr241 forms a hydrogen bond with the aspartic acid side chain of the catalytic DHHC motif (Figs. 3B and 4A and fig. S9B). Consequently, an AAXE (Ala-Ala-X-Glu) mutant has drastically reduced enzymatic activity (Fig. 3C).
Figure 4. C-terminal domain interactions.
(A) Interactions of the Thr240, Thr241, and Glu243 of the conserved TTXE motif with the active site and the rest of the protein. (B) Interactions of the amphipathic helix of the PaCCT domain with TM3 and TM4. Asn266 is one of the most conserved residues on the C-terminal domain of DHHC enzymes. (C) Penetration of the C-terminal short helical stretch αʹ3 together with the hydrophobic loop, shown in brown, into the bilayer, and their interactions with the cytoplasmic ends of TM2 and TM3. Relevant residues are shown in stick rendition. (D) Mutational analysis of selected residues on the C terminus involved in interactions shown in (A), (B), and (C). Data are mean ± SEM of two independent measurements.
Montoro et al. (41) have identified a motif at the C terminus of DHHC enzymes that they have named the palmitoyltransferase conserved C-terminus (PaCCT) motif. This motif is conserved in the majority of DHHC enzymes, and, in the 16-residue motif, the third and the eleventh residues are the most conserved. The third residue is usually a phenylalanine or tyrosine and, in hDHHC20, this residue, Phe259, forms a local packing core at the C terminus and is engaged in a number of hydrophobic and π-stacking interactions. The other conserved residue in this motif, Asn266, is at the beginning of the amphipathic helix (αʹ2) and simultaneously forms a capping interaction with TM4 and a hydrogen-bonding interaction with the backbone of Leu261 and the sidechain of Ser260. These interactions engage all of the hydrogen-bonding capabilities of the amide side chain and presumably hold this part of the structure together. Mutating Asn266 to alanine severely compromises catalytic activity (Fig. 4, B and D, and table S3).
One of the most prominent features of the C-terminal domain is the presence of an amphipathic helix (αʹ2) that makes contact with TM3 and TM4 and likely provides local stability. Sequence conservation indicates that the amphipathic helix is conserved across many members of the family, pointing to its importance for other DHHC enzymes as well. Deletion of a single phenylalanine residue in this region in DHHC21 has been genetically mapped to a depilated phenotype in mice that results in hair loss with thinner and shorter hairs (42). The homologous residue in hDHHC20 is Trp267. Deletion of Trp267 in hDHHC20 results in decreased yield of the recombinant protein and almost negligible catalytic activity (Fig. 4, B and D). The amphipathic helix is followed by a short helix (αʹ3) and a hydrophobic loop. The conformation of the loop is stabilized by highly conserved lysine and proline residues, and it inserts hydrophobic residues into the putative lipid bilayer and forms additional contacts with TM3 and TM2 (Fig. 4C). Mutation of the conserved Trp278 and Leu279 to a W278A/L279A double mutant reduces hDHHC20 activity by almost half (Fig 4D and table S3).
Acyl chain binding site
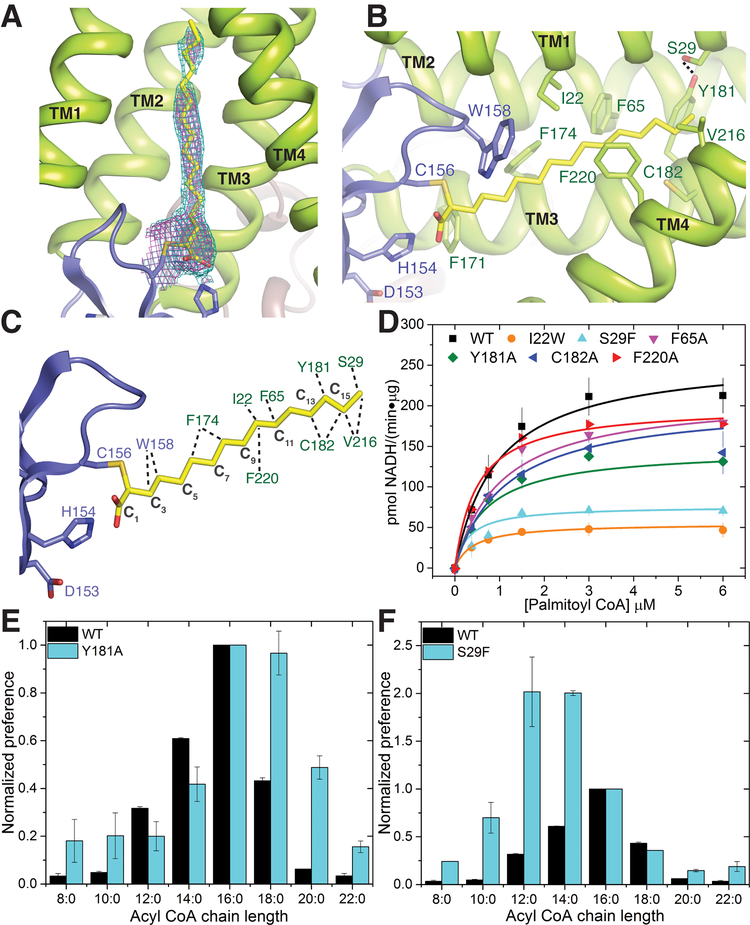
Starting at the active site residues and pointing inward into the bilayer, both hDHHC20 and zfDHHC15 have a hydrophobic cavity (Fig. 2D). In the hDHHC20 structure, we saw weak density in the cavity that was suggestive of a fatty acid, but was not strong enough to build a model. We thus utilized the widely used covalent inhibitor 2-bromopalmitate (2-BP) to purify an acylated intermediate mimic. This inhibitor covalently modifies DHHC enzymes at the active site cysteine through a nucleophilic displacement reaction (43, 44) (fig. S3B), resulting in alkylation of the cysteine through attachment at the α position of palmitic acid. Our initial biochemical and mass spectrometric data showed that, aside from the active site cysteine (Cys156), another cysteine (Cys263) was also consistently labeled with palmitate (fig. S11). Consequently, we used a C263A mutant (cysteine replaced with alanine at position 263) for preparation and crystallization of the covalently modified hDHHC20. We detected very low activity of the purified 2-BP–treated C263A mutant of hDHHC20, suggesting that the catalytic cysteine was mostly labeled in our preparation (fig. S3C). It crystallized under the same conditions and in the same hexagonal space group as hDHHC20 (table S1). The overall structures are very similar (Figs. 2A and 5A), and an omit map revealed clear density for the palmitate chain covalently attached to the active site cysteine, allowing us to build the complete acyl chain (Fig. 5A). In the zfDHHS15 structure, we saw continuous density in this cavity, which we modeled as a palmitic acid (fig. S12).
Figure 5. The acyl chain binding groove.
(A) 2Fo-Fc electron density map at contour level 1.0 σ (magenta) and 0.7 σ (cyan) of the palmitate chain covalently attached to Cys156 in the hDHHC20–2-BP structure (see text). (B) Close-up view of the acyl chain binding groove in the 2-BP structure showing the residues lining the groove. (C). Residues in the groove that interact with the palmitate chain identified by using the small-probe contact dot surfaces (67). (D) Mutational analysis of the residues in the acyl binding groove shown in (C). Data are mean ± SEM of two independent measurements. (E) Acyl-CoA chain–length selectivity in wild-type and Y181A mutant of hDHHC20, as determined by the autoacylation assay. The x axis shows the carbon-chain lengths of different acyl-CoA donors, and the y axis shows normalized activity (initial velocity) of wild-type or mutant versions of hDHHC20. Each data set is individually normalized to 1 for the activity with regard to palmitoyl-CoA. (F) Acyl chain–length selectivity of S29F hDHHC20. Selectivity data are mean ± SEM of two independent measurements.
The structure of the 2-BP–treated C263A hDHHC20 mutant (hereafter referred to as 2-BP structure) reveals crucial insights into the mechanism of acyl-chain binding and recognition in hDHHC20. The acyl chain inserts into the hydrophobic cavity seen both in the hDHHC20 and zfDHHC15 structures (Fig.2D and fig. S9) and is contacted by several residues lining the cavity (Fig. 5, B and C, and fig. S12, B and C). The acyl chains have very similar conformation in the cavity, and we will focus our description on the hDHHC20 2-BP structure. Notably, all four TM helices contribute to interactions with the acyl chain, albeit to different extents. The cavity is constricted near the acyl headgroup by Trp158 and Phe171. These are two of the most highly conserved residues across the DHHC family members (fig. S1). Mutation of Phe171 to alanine generates a catalytically inactive mutant, whereas mutation of Trp158 to alanine causes a considerable loss in enzymatic activity (Fig. 3C and table S3). Further into the membrane, two other residues contact the acyl chain, Phe174 and Leu227, conserved as hydrophobic residues in most DHHC members. Mutation of the leucine to a more bulky and rigid tryptophan severely compromises enzymatic activity (Fig. 3C and table S3).
Toward the top of the cavity where it tapers to a narrow end, residues from all four TM helices contact the acyl chain. Some of these residues that line the acyl binding cavity are highly conserved across all DHHC members, e.g., Trp28 in hDHHC20. Intriguingly, there are other residues that show only subgroup-specific conservation (fig. S1). As a case study, Ile22 is conserved among a subset of DHHC members and faces the acyl binding cavity in the structure, but does not make a close contact (Fig. 5B). The I22W mutant (isoleucine replaced with tryptophan at position 22) has drastically lower activity, presumably owing to the more bulky and rigid aromatic side chain of tryptophan, which makes a disfavorable contact with the acyl chain (Fig. 5D).
Acyl chain–length selectivity of DHHC enzymes and designing mutants with altered preferences
It has been shown that different DHHC members show different degrees of selectivity for acyl-CoA donors with different chain lengths (28, 30). Toward the tapering end of the acyl binding cavity in the 2-BP structure, Tyr181 forms a hydrogen-bonding interaction with Ser29, effectively closing off the cavity (Fig. 5B). Whereas wild-type hDHHC20 shows a preference for acyl-CoAs with a palmitoyl (16-carbon) chain, mutating Tyr181 to a less bulky alanine results in marked increase in preference for stearoyl (C18)–CoA (Fig. 5E). Mutating Ser29 to a bulkier phenylalanine increases the preference of hDHHC20 for short-chain acyl-CoA (Fig. 5F). The S29F (serine replaced with phenylalanine at position 29) mutation not only breaks the hydrogen-bonding interaction but also replaces the Tyr-Ser pair in hDHHC20 with a considerably bulkier Tyr-Phe pair in the cavity.
These results also lend insights into a recent report that investigated the acyl chain–length selectivities of two closely related DHHC enzymes, DHHC3 and DHHC7 (30). The homologous pair of Tyr181-Ser29 in hDHHC20 is Ile182-Phe53 in DHHC3 and Ser185-Leu56 in DHHC7. DHHC3 has a higher selectivity for palmitoyl (C16) over stearoyl (C18) compared to DHHC7. Presumably this is due to the placement of two large residues (isoleucine and phenylalanine) toward the end of the cavity, which impedes longer–chain length acyl-CoA. In comparison, the less sterically demanding Ser-Leu pair leads to a more relaxed acyl chain–length preference in DHHC7. Consistent with this hypothesis, the I182S (isoleucine replaced with serine at position 182) mutant of DHHC3 has higher preference for stearoyl (C18)–CoA, mirroring that observed in DHHC7 (30).
Mechanism of palmitoylation
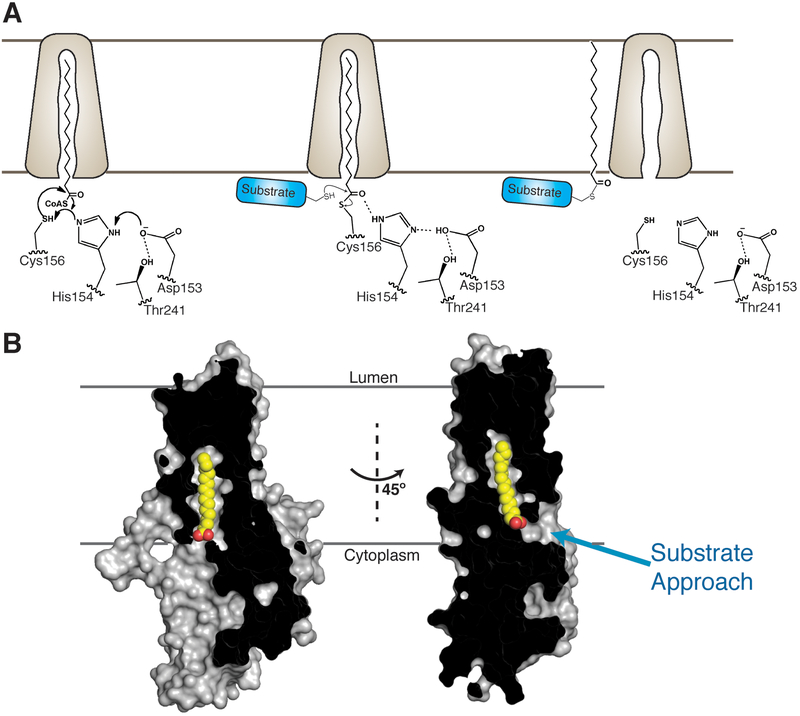
Although protein palmitoylation was discovered almost 40 years ago (5), there has been little understanding about the structural chemistry of enzyme-catalyzed protein palmitoylation. All of the DHHC enzymes that have been biochemically characterized to date utilize a two-step reaction mechanism (27, 28), with the first step being autoacylation of the active site cysteine. We show that the aspartic acid and His154 of the DHHC motif form a hydrogen-bonded pair that can accept a proton from the cysteine within the DHHC motif during catalysis, enabling nucleophilic attack on the carbonyl thioester of palmitoyl-CoA to generate the acyl-enzyme intermediate (Fig. 6A). The structures reported here do not give indication as to how the palmitoyl-CoA enters the binding cavity. However, palmitoyl-CoA partitions into the lipid bilayer and likely distorts the local structure of the membrane (45, 46). Given that interfaces of membrane proteins with surrounding lipids are also sites of distortion in the lipid bilayer and, thus, possible sites of accumulation of palmitoyl-CoA, we speculate that palmitoyl-CoA inserts into the cavity formed by DHHC enzymes from within the membrane.
Figure 6. Proposed reaction mechanism and substrate approach.
(A) Proposed reaction mechanism of DHHC enzymes that follow a two-step mechanism. (B) Molecular surface rendition of the 2-BP–treated enzyme with the acyl chain shown in yellow and red spheres. The putative direction of substrate approach is shown with a cyan arrow.
In the second step of the reaction, the acyl-enzyme intermediate transfers the palmitoyl group to the substrate cysteine. The structure of the 2-BP–treated DHHC20 suggests that the carbonyl oxygen of the acyl-enzyme thioester is positioned close to His154. We speculate that the protonated His154 activates the acyl-enzyme thioester for the subsequent palmitoyl transfer to the substrate by providing a proton to the carbonyl oxygen. The cysteine residue in each substrate that accepts the palmitoyl group lies in a different sequence context and, thus, different chemical and structural microenvironments. Consequently, activating the acyl group of the acyl-enzyme intermediate with an adjacent proton donor would be an effective strategy for catalysis of the second step. Residue His154, which accepts a proton in the first step, is optimally positioned for this role.The structure also shows that the organization of the active site is such that all but one side of the acyl-enzyme thioester is shielded by hydrophobic residues. This only leaves the front side of the acylated DHHC for approach of the cysteine residue on the substrate that reacts with the thioester in the second step (Fig.6B). However, engagement with the substrate may incur conformational changes and open up alternative directions of approach that are not obvious in the current set of structures.
Protein S-acylation has been shown to be heterogeneous. Analyses of S-acylated proteins from native sources have revealed that proteins can be modified not only with palmitic acid but also with other fatty acids, such as stearic acid and oleic acid (10, 11, 47). In the case of influenza hemagglutinin, specific cysteines have been shown to be modified by palmitoylation and stearoylation, pointing to the specific roles that distinct acylation states play in the function of cellular proteins (48). Recently, it has been shown that stearoylation of transferrin receptor 1 by DHHC6, specifically, modulates mitochondrial morphology (49). In vitro experiments have shown that for the two enzymes DHHC2 and DHHC3, DHHC2 can utilize a broad range of fatty acids with diverse structures such as palmitate, arachidonate, and palmitoleate, whereas DHHC3 has reduced activity for fatty acids longer than palmitic acid (28). It was also shown that the selectivity at the autoacylation state reflects the selectivity of fatty acyl labeling of the protein substrate (28). The structures of unacylated zfDHHS15 and hDHHC20 and of 2-BP hDHHC20 reveal that the fatty acyl chain fits into a cavity in the bilayer formed by the transmembrane helices of the enzyme. The residues lining this cavity form numerous contacts with the fatty acyl chain. However, although there are highly conserved residues, i.e., Trp158 and Phe171, lining this cavity that form important contacts with the acyl chain, there is considerable sequence diversity in the other residues that line this cavity between different subgroups of DHHC enzymes (fig. S1). These residues vary not only in their size, but also in their polarity, thus changing the chemical properties of the cavity in a complex manner. These can have a notable effect on the fatty acyl chain–length preference of different DHHC enzymes, and, as structures of more DHHC enzymes become available, presumably they will reveal further insights into their respective fatty acid chain–length selectivity.
DHHC20 overexpression was previously shown to cause cellular transformation (50). More recently, hDHHC20 has been proposed as a target for developing therapeutics against cancers that are resistant to EGFR-targeted therapy (22). The structures presented here are plausible starting points for developing a structure-guided program to hDHHC20 inhibitors that could lead to such a therapeutic intervention. It is worthwhile to note in this context that the entire field of protein palmitoylation suffers from the lack of small molecule probes and inhibitors for the DHHC family of enzymes, let alone the lack of specific modulators of individual DHHC members (51). Members of the DHHC family all use palmitoyl (acyl)–CoA and the same enzymatic chemistry to transfer an acyl group to substrate proteins. From this perspective, they are very similar to members of the family of protein kinases that use ATP to transfer a phosphate group to substrate proteins. Thus, using strategies like bump-hole pairs, where structural knowledge is used to design engineered versions of DHHC enzymes that can use orthogonal acyl-CoAs, could provide transformative insights into the biology of protein palmitoylation and help unravel the complex network of DHHC-substrate interactions, akin to the field of protein kinases and histone-modification enzymes (52–54). We demonstrate the feasibility of such an approach by designing mutants that can shift the acyl chain–length selectivity profile of the mutants toward the shorter or longer side of palmitoyl (C16)–CoA.
Materials and Methods
Molecular biology and cloning and yeast transformation
The codon optimized zebrafish DHHC15 (zfDHHC15) sequence was cloned into a modified version of pPICZ-C vector with an His10 tag followed by a GFP coding sequence and finally, by PreScission cleavage site at the N terminus of the zfDHHC15 encoding DNA sequence. Further, site directed mutagenesis was performed to mutate Cys153 to serine to get the zfDHHS15 expression construct. The optimized vector harboring zfDHHC15 or zfDHHS15 gene was transformed into Pichia pastoris HIS + Cells SDM1163 cells. Pichia were transformed using standard methods and the transformants were selected on YPDS plates containing 400–800 μg/mL zeocin.
The human DHHC20 (hDHHC20) expression construct was made similarly with N-terminal His10 tag, GFP and PreScission cleavage sequences without any modification/truncation of the hDHHC20 sequence and was transformed into Pichia following the same protocol.
A vector containing the sequence for human Snap25b was obtained from Addgene (Plasmid #53235) and the gene was cloned into pET28-Prx by digestion/ligation using NdeI/BamHI. The original TEV cleavage site was mutated to a PreScission cleavage site.
Protein expression and purification
Cell culture and lysis
Large scale culture of Pichia, induction of protein expression and cell lysis were carried out as described previously (55). Briefly, 100–200 mL BMGY (0.1 M Potassium phosphate, pH 6.0, 3.4 g/L yeast nitrogen base, 1% glycerol, 0.4 μg/mL biotin) with 500 μg/mL Zeocin cultures of Pichia were grown overnight at 30°C with vigorous shaking. 1/10th volume of these starter cultures, which usually reached cell densities of OD600 ~20, were then used to inoculate 1.5–2 liter cultures in the same media but without the Zeocin. In 24–36 hours, the cells were pelleted by centrifugation at 1500 g, 4°C. for 10 min and thoroughly washed with BMMY (0.1 M Potassium phosphate, pH 6.0, 3.4 g/L yeast nitrogen base, 1% methanol, 0.4 μg/mL biotin). Cells were finally re-suspended in 1.5–2 liters of BMMY media and protein expression induced at 22–24°C for 24–36 hours. Cells were harvested by centrifugation at 6000 g, 4°C. for 20 min. The cell pellets were scooped by a spatula and frozen by immersing into liquid N2 followed by storage at –80°C. Cells were lysed by cryo-milling using Retsch MM400 millers with liquid N2 for cooling.
Purification of zfDHHC15 and zfDHHS15
Lysed and frozen yeast powder was dissolved in the lysis buffer (1 g cells per 3 mL of lysis buffer) containing 50 mM Tris-HCl pH-7.5, 150 mM NaCl, 1 mM TCEP, 0.1 mg/mL deoxyribonuclease I, 0.1 mg/mL pepstatin, 1 μg/mL leupeptin, 1 μg/mL aprotinin, 1 mM benzamidine and 0.1 mg/mL soy trypsin inhibitor. The pH of the cell lysate was adjusted to 7.8 followed by addition of the 0.14 g of DDM (n-dodecyl-β-D-maltopyranoside) per g of cells. The proteins were extracted from the cells by stirring at 4°C. for 3 hours. The pH of the cell lysate was adjusted to 7.5 and centrifuged at 38,000 g, 4°C. for 30 min. The supernatant was allowed to bind with 2.5 mL of Talon (Clontech) resin pre-equilibrated with the equilibration buffer (50 mM Tris-Cl pH 7.5, 150 mM NaCl, 1 mM TCEP and 1 mM DDM) for 3 hours at 4°C. The protein bound Talon resin was collected in a Bio-Rad Econo column and washed with 10 bed volumes of wash buffer 1 (20 mM Tris-Cl pH 7.5, 150 mM NaCl, 1 mM TCEP and 2 mM of Thio-DM (n-Decyl- β-D-Thiomaltopyranoside) followed by additional wash with 10 bed volumes of wash buffer 2 (wash buffer 1+ 20 mM imidazole). An additional wash was performed with 10 bed volumes of wash buffer 1 in order to completely wash away any traces of imidazole. Talon resin was resuspended in 10 mL of wash buffer 1. His10 tagged GFP was removed from the N terminus of zfDHHS15 by incubation of the resin slurry with PreScission protease overnight at 4°C. Flow-through containing cleaved protein was concentrated with 50 kDa molecular weight cut-off (MWCO) 15 mL concentrators (Millipore). The concentrated protein sample was further purified by size exclusion chromatography on Superdex 200 column in buffer containing 20 mM Tris-Cl pH 7.5, 150 mM NaCl, 1 mM TCEP and 2 mM Thio-DM. The peak fractions from the size exclusion chromatogram were collected and concentrated with 50 kDa MWCO concentrator up to 20 mg/mL and used for crystallization.
Purification of hDHHC20 and hDHHS20
20–24 g of milled cell powder was suspended in ~65 mL of Tris-buffered saline and stirred using a magnetic stirrer for 20–30 min to make a homogeneous slurry. The final buffer composition was 40 mM Tris.HCl, 270mM NaCl, 5 mM βME. Protease inhibitors (Benzamidine HCl, PMSF, AEBSF, Aprotinin, Pepstatin, Leupeptin) and DNase were added and the pH of the cell slurry adjusted to ~7.5. Solid DDM was added to give a final concentration of 2% (40mM) and the protein extracted from the membranes by vigorous stirring at 4°C for 3 hours. Cell debris was pelleted by centrifugation at 38,000 g for 30 min at 4°C. The pH of the supernatant was adjusted to 7.5 and then incubated with 2 mL of Talon resin for 2–3 hours at 4°C.
Protein-bound resin was washed with 30 mL of 50 mM Hepes, pH 7.5, 250 mM NaCl, 5 mM βME, 2 mM TCEP, 1 mM DDM buffer containing 5 mM imidazole. The resin was further washed with same volume of buffer containing 25 mM imidazole prior to elution with ~5–7 mL of buffer with 200 mM imidazole. All affinity chromatography was done by gravity flow at ambient temperature with buffers chilled on ice. The bright green eluate was then concentrated in a 100 kDa molecular weight cut-off centrifugal concentrator to ~0.3 mL. 1/10 volume PreScission protease (~1 mg/mL) was then added and rotated overnight at 4°C. The PreScission protease cleaved protein was injected into a Superdex 200 Increase size exclusion column at 4°C. to separate hDHHC20 from the GFP tag and the PreScission protease. The size exclusion buffer was 50 mM Hepes pH 7.5, 150 mM NaCl, 2 mM TCEP, 0.5 mM DDM. The fractions containing the protein were pooled and concentrated using a 50 kDa MWCO 0.5 mL Amicon centrifugal concentrator. Prior to any assays, protein concentration was determined using the 660 nm protein assay kit (ThermoFisher). In cases where the GFP tag was not removed, the overnight PreScission cleavage step was omitted and the concentrated protein applied to the Superdex 200 increase column (GE Healthcare).
Reductive methylation of hDHHC20 for crystallization
hDHHC20 for crystallization was prepared as above to the step of overnight PreScission digest. The next morning, instead of applying the GFP-cleaved protein to the gel filtration column, it was diluted to ~12 mL in 50 mM Hepes, pH 7.5, 250 mM NaCl, 2 mM TCEP, 0.5 mM DDM buffer lacking any imidazole. The GFP tag was then removed by binding it to ~1 mL Talon resin and the flowthrough containing the hDHHC20 protein was concentrated in 50 kDa MWCO 4 mL concentrator to ~0.5 mL. This protein was again diluted to ~12 mL and re-concentrated to ~1 mL to remove any imidazole. The protein was then reductively methylated using formaldehyde and dimethylamineborane (DMAB) complex following standard protocols (56). The next morning, the methylation reaction was quenched with 100 μl of 1 M Tris HCl, pH 8.0 and 5 μl of 1 M dithiothreitol (DTT) for 1 hour at 4°C. The protein was concentrated to ~0.25 mL and applied to Superdex 200 Increase size-exclusion column as previously mentioned. Peak fractions were collected and concentrated as before. All wash buffers for the crystallization sample of DHHC20 contained 0.1 mg/mL POPC:POPG:POPA (3:1:1) lipids and 1 mM DDM. This was lowered in the size exclusion chromatography buffer to 0.05 mg/mL lipids and 0.5 mM DDM.
Irreversibly inhibited hDHHC20 preparation and purification
To prepare an irreversibly inhibited protein with a covalently attached lipid chain, 2-bromopalmitate (2-BP) dissolved in methanol was added to the Pichia cells at a final concentration of 300 μM during induction. However, we decided to add the DHHC inhibitor only after the cells had been induced for at least 12 hours as 2-BP is toxic to cells and we wanted to ensure that it did not affect protein expression initially. Five hours prior to harvest, we again added 2-BP to a final concentration of ~100 μM. We first attempted this with the wild-type hDHHC20 and checked the covalent modification with mass spectrometry. Unexpectedly we found that apart from the active site cysteine, 2-bromopalmitate also modifies Cys263. Therefore, to reduce any possible heterogeneity during crystallization, we used a C263A mutant for the 2-BP modified DHHC20. hDHHC20 C263A protein was prepared as for wild-type hDHHC20. Enzymatic activity of the 2-BP treated sample was checked and found to be essentially nonexistent as expected.
Purification of GobX protein
GobX protein expression vectors were a gift from M. Machner (NIH). BL21(DE3) Gold cells were transformed and plated on LB-agar with Ampicillin. Next morning the entire plate was used to start two 1 liter cultures in LB media. Bacteria were grown to an OD600 of ~0.6 at 37°C. with vigorous shaking. Temperature was reduced to 30°C. and protein expression was induced with a final IPTG concentration of 1mM. Cells were harvested ~5 hours later. Protease inhibitors (PMST, AEBSF) were added to the cells prior to flash freezing in liquid N2. Next morning the cells were thawed in 40 mM Tris-HCl, pH 7.2, 270 mM NaCl, 10 mM βME, 20% glycerol, DNAse, and more protease inhibitors. Powder lysozyme was added to facilitate cell lysis. Once the cells had thawed to slurry, the cell suspension was diluted such that it now contained 10% glycerol. DDM was then added to 1% final concentration.
Cells were broken by sonication on ice. During the sonication process, 1/100 volume PMSF was added after every 1 min. The cell debris were removed by centrifugation at 38,000 g for 30 min at 4°C. The supernatant was incubated with 2 mL of HisPur Ni-NTA resin for 1–2 hours at 4°C rotator. Protein bound resin was packed on a Bio-Rad Econo column and washed with ~30 mL Tris-buffered saline containing 10% glycerol and 10 mM imidazole. The resin was further washed with same buffer containing 50 mM imidazole before finally eluting it with ~8 mL 300 mM imidazole buffer. The eluate was concentrated in 10 kDa MWCO concentrator to ~0.5 mL and diluted 100-fold to 50 mL in 50 mM TrisHCl, pH 8.0, 10% glycerol, 1 mM DDM buffer. The diluted protein was applied to a 1 mL HiTrap MonoQ column using peristaltic pump. GobX protein was eluted off the MonoQ with a two-gradient program between 50 mM TrisHCl, pH 8.0, 10% glycerol, 10 mM NaCl and same buffer with 1 M NaCl on an AKTA explorer at 4°C. DDM is omitted from the MonoQ column elution buffers. Two major peaks are eluted – the first is a cleaved version of the GobX and the second is the full-length GobX. The second peak fractions were pooled, concentrated in a 10 kDa MWCO Amicon spin concentrator to ~5–7 mg/mL and flash frozen in liquid N2 until later use in palmitoylation assays.
Purification of Snap25b
E. coli BL21(DE3)-Gold was transformed with Snap25b in pET28-Prx vector and plated onto LB-Kanamycin plates. A single colony was used to inoculate 5mL of LB (with 50 μg/mL of Kanamycin) and cells were grown at 37°C for 20 hours. 2 mL of this starter culture was used to inoculate 1 L of LB (with 50 μg/mL of Kanamycin) and cells were grown until OD600 was around 1.0. Cells were cooled to 30°C. and induced with 0.5 mM of IPTG. After 5 hours, cells were harvested by centrifugation at 8000 g for 10 min at 4°C. Cell pellets were flash frozen in liquid nitrogen and stored at –80°C.
For a typical purification, 10 g of frozen pellet were dissolved by stirring at 4°C (30–40 min) in 100mL of lysis buffer containing: 50 mM Tris-HCl pH 8.0, 250 mM NaCl, 5% glycerol, 5 mM βME, protease inhibitor cocktail (0.1 mg/mL deoxyribonuclease I, 0.1 mg/mL pepstatin, 1 μg/mL leupeptin, 1 μg/mL aprotinin, 1 mM benzamidine and 0.1 mg/mL soy trypsin inhibitor and 1 mM PMSF) and 10 mg lysozyme. Resuspended cells were disrupted by sonication, stirred for 20 min at 4°C and subsequently centrifuged at 38,000 g for 30 min at 4°C. Pellets were discarded and supernatant was batch-bound to 5 mL of Ni-NTA resin (previously equilibrated with binding buffer containing 50 mM Tris-HCl pH 8.0, 250 mM NaCl, 5% glycerol, 5 mM β-mercaptoethanol) by rotation at 4°C. for 1 hour. Protein-bound resin was washed successively with 10 bed volumes of binding buffer, 10 bed volumes of binding buffer with 10 mM imidazole and finally, 10 bed volumes of binding buffer with 30 mM imidazole. Protein was finally eluted with binding buffer containing 250 mM imidazole. Eluted protein was incubated at 4°C with rotation for 20 hours in the presence of 1 mg/mL of PreScission protease to remove the His6 tag. Completeness of cleavage was assessed by gel shift using 12% acrylamide SDS-PAGE after Coomassie staining. Completed cleavage reaction was dialyzed against 4 liters of binding buffer for 15 hours at 4°C to remove imidazole. The dialyzed protein solution was re-applied to 5 mL of Ni-NTA resin equilibrated with binding buffer and incubated with rotation at 4°C for 1 hour. Resin was loaded onto a column and flow-through was collected. The concentration of purified protein was determined using the 660nm Assay Kit (Thermo Fisher).
Protein crystallization, optimization, and data collection
hDHHC20 crystallization and optimization
hDHHC20 protein at ~15–20 mg/mL was incorporated into monoolein using established protocols (57) and crystallization trials were set up on Laminex 96-well plastic sandwich plates (Molecular Dimensions) using a Mosquito crystallization robot (TTP Labtech). An initial hit was obtained with 0.1 M Hepes, pH 7.0 and 30% PEG 300 at 20°C. Optimization of crystals was carried out using varying salts, buffers and pH. Of the salts, only sodium malonate and potassium di-hydrogen phosphate at 50 mM gave crystals that diffracted consistently. Further improvements in size of crystals were achieved through screening diol additives with 1–5% of 2,5-Hexanediol being the best. Better diffraction was observed when fresh DTT was included as an additive at concentrations of 25–50 mM. Addition of lipids during the protein preparation improved the crystallizability of DHHC20. While we had initially observed rectangular crystals (indexed as P21), inclusion of lipids during purification led to us to obtain hexagonal crystals (indexed as P63) as well. Further improvements in diffraction were also achieved by methylating the protein. The final optimized crystallization screen was a pH and PEG 300 grid with MES (pH 5.5–6.5), MOPS (pH 6.5–7.5), and HEPES (pH 6.5–7.5) buffers and PEG 300 from ~25–35%. We also expanded the salt from potassium di-hydrogen phosphate to include the sodium salt as well, both at 50 mM final concentration.
As diffraction from crystals obtained in the sandwich plates hardly extended to better than 4 Å and the harvesting itself was challenging, we decided to switch to a microbatch format. In this format, 100 nl of the LCP bolus was set on 96-well Microbatch plates (Hampton Research) followed by 3.24 μl crystallization solution and 0.36 μl of a 25% 2,5-Hexanediol and 0.5 DTT (freshly prepared) mixture. Plates were incubated at 20°C. The crystals generally took longer to appear than sandwich plates (a week versus a few days) and also grew larger and thicker. Crystals were directly fished out of the wells using Mitegen 100 micron loops and frozen in liquid N2.
The initial hexagonal crystals were obtained in 100 mM HEPES, pH 7.3, 50 mM NaH2PO4, 25 mM DTT, 1.25% 2,5-hexanediol after approximately one week. Data sets were collected from hexagonal crystals at 9.665 keV at 10% transmission with 0.25° rotation and 0.2 s exposure. A total of 720 frames spanning 180° were collected. The best diffracting hexagonal crystals were obtained in 50 mM MES, pH 5.8, 50 mM KH2PO4, 27.7% PEG 300, 25 mM DTT, 1.25% 2,5-Hexanediol and grew to optimal size in 3.5 months. A native data set of 360 frames was collected at Se-edge of 12.66 keV with 10% transmission, 0.25° rotation and 0.5 s exposure. These data sets were collected at the NE-CAT 24ID-C beamline at the Advanced Photon Source (APS) at Argonne National Laboratory in Chicago, IL.
The best P21 crystal was obtained in 50 mM MES, pH 6.5, 50 mM NaH2PO4, 30.3% PEG 300, 50 mM DTT, 2.5% 2,5-hexanediol and grew to optimal size in 3 months. Complete data set was collected at 9.665 keV with 10% transmission, 0.3° rotation, and 0.2 s exposure. A total of 1500 frames were collected. The data set for the 2-BP modified hDHHC20 crystals was obtained from a single crystal obtained after 1.5 months in 100 mM HEPES, pH 7.03, 50 mM KH2PO4, 31% PEG 300, 50 mM DTT, and 2.5% 2,5-hexanediol. Data set of 900 frames was obtained with 20% transmission, 0.2° rotation, and 0.2 s exposure at a 12.66 keV beam. These data sets were collected at the GM/CA-CAT 23ID-D and 23ID-B beamlines respectively at the APS at Argonne National Laboratory in Chicago, IL.
hDHHC20 structure determination
Diffraction data from three different crystals of the P63 form were merged together to improve redundancy for the phasing. The data sets were processed using XDS and AIMLESS in the CCP4 suite (58). Initial phases were obtained using AutoSol in the PHENIX Suite (59). Four predicted zinc heavy atom sites were identified and provided a phasing power to generate a map clearly showing molecular boundaries and secondary structure elements. An initial model was built using AutoBuild and manually modified using COOT (60). The resulting structure was used as a search model for molecular replacement against the native data set. This structure was also used as a starting molecular replacement model for solution of the P21 form and the 2-BP treated hDHHC20. Restraints of the 2-BP and CoA head group were generated using the eLBOW module within the PHENIX suite and edited manually. Iterative model building and refinement was carried out using COOT and PHENIX. The coordinates and structure factors of the human hDHHC20 in P63 and P21 space groups and 2-BP modified structure have been deposited to the Protein Data Bank (PDB) with the accession code of 6BMN, 6BMM and 6BML, respectively.
DHHS15 crystallization and data collection
ZfDHHS15 protein crystallization was performed using the hanging drop vapor diffusion method. Initial crystal hits were obtained with 0.1 M KCl, 20% PEG 400 and 100 mM HEPES, pH 6.5 at 4°C. These crystals were further optimized to obtain better diffraction quality crystals. The best diffracting crystals of zfDHHS15 grew from 0.1 M KCl, 20% PEG 400, 100 mM HEPES pH 6.5 and 30% ethylene glycol at 4°C. The optimum size of the better diffracting crystals was obtained in 7 days. Crystals were exposed to the cryo-solution (30% ethylene glycol with all other remaining same as reservoir) by exchanging reservoir solution with cryo-solution. Ethylene glycol percentage was gradually increased from 20% to 30% in increments of 5%. Each step was incubated at 4°C for 6–7 hrs. Crystals were harvested using Hampton nylon loops and immediately flash-frozen in liquid nitrogen. Three different types of data were collected in order to solve the structure. Two different single-wavelength anomalous dispersion data sets at 3.31-Å resolution from single crystals were collected at the Zn absorption edge wavelength, 1.2820 Å, on beamline 24ID-C (NE-CAT) at the Advanced Photon Source (APS), Argonne National Laboratory. These data sets were merged together, integrated with XDS and scaled with XSCALE. Additionally, S-SAD data were collected at 1.71 Å from 10 different single crystals at 23ID-B (GM-CA) at APS. These data sets were merged together and integrated with XDS and scaled with XSCALE. A third native data set of 2.54-Å resolution was acquired at x-ray wavelength 0.997 Å using single crystals on beamline 22ID (SER-CAT) at the APS. The native data set was processed with HKL 2000.
DHHS15 structure determination
The structure was solved using the Zn-SAD data in P42212 space group using the data collected at 24ID-C beamline, integrated using XDS. Data from two crystals were used. They were scaled using XSCALE. Four Zn atoms were located using hkl2map and subsequently refined using experimental phasing methods in Phaser. An electron density map was generated after NCS averaging and density modification (using Parrot, part of the CCP4 suite of crystallographic software) and an overall model was built into it. At this stage, a highly redundant S-SAD data was collected at 23ID-GM-CA (wavelength 1.71 Å, data redundancy ~100) using 10 crystals. Using the model phases, sulfur atoms were located using anomalous difference Fourier methods and the sulfur sites were refined using Phaser. A total of 48 sulfur sites were found (24 per monomer, 9 Met and 15 Cys). This density modified map was used for iterative model building and refinement in PHENIX. A partial model of the zfDHHS15 was obtained with Rwork of 32.2 and Rfree of 33.7%. This partial model was used for molecular replacement with native data set of 2.45 Å resolution with Phaser module in PHENIX. Non-crystallographic symmetry was used during refinement of the model. The native data set was further refined using PHENIX as well as Refmac in ccp4 followed by manual examination and rebuilding of the refined coordinates in coot using both 2mFo – DFc and mFo – DFc maps, as well as omit maps generated through PHENIX. The restraint parameters for palmitate, dodecylmaltoside and POPC were generated through ELBOW program of PHENIX was used during refinement. The final data collection and refinement statistics are shown in tables S2. The final model had the following Ramachandran statistics, 97.8% in favored region, 2.17% in allowed region, 0.2% of rotamer outliers and 0% of Ramachandran outliers.
Biochemical assays
zfDHHC15
Coupled-enzyme assay
zfDHHC15 autoacylation activity was measured using a coupled-enzyme assay (31) in 384-well low-volume plates (Thermo Fisher). Plates were read in a Tecan M1000Pro fluorimeter with 340 nm excitation and 465 nm emission. The final reaction volume (20 μL) contained 20 nM zfDHHC15/zfDHHS15, 0.25 mM oxidized Nicotinamide adenine dinucleotide (NAD+), 0.2 mM thiamine pyrophosphate (TPP), 2 mM 2-oxoglutarate, 1 mM EDTA, 1 mM DTT, 2-oxoglutarate dehydrogenase, 0.6 mM DDM detergent in 50 mM sodium phosphate buffer at pH 6.8. The reaction was initiated by the addition of palmitoyl-CoA and monitored for 30 min at 30°C. The linear part of the progress curve was used to determine initial rates.
zfDHHC15 concentration was determined using the 660 nm assay (Thermo Fisher). Km and Vmax were estimated using non-linear least square fitting to the Michaelis-Menten model (Graphpad Prism 6). Kcat was determined by dividing the estimated Vmax by the total enzyme concentration. Each Km and kcat value is the average and standard error of three independent measurements.
Snap25 palmitate transfer assay
For in vitro Snap25b acylation assay, affinity purified GFP-zfDHHC15 (50 nM) and GFP-zfDHHS15 (50 nM) were incubated with affinity purified Snap25b (500 nM) in reaction buffer (50 mM HEPES, pH 7.0, 200 mM NaCl, 0.2 μM DTT, 1 mM EDTA, 0.3 mM DDM) for 10 min at room temperature (24°C). Reaction was started by addition of {N-[(7-nitro-2–1,3-benzoxadiazol-4-yl)-methyl]amino}palmitoyl-CoA (NBD PalCoA) to a final concentration of 1 μM. NBD PalCoA is a fluorescent derivative of palmitoyl-CoA. Samples were taken at different time points and reaction quenched with non-reducing SDS sample buffer. Samples were separated in 12% SDS-PAGE gels and fluorescence detected using a Chemidoc with Blue Epi illumination and a 530/25 filter.
hDHHC20
Coupled-enzyme assay
hDHHC20 kinetic assays were done using an established fluorescence based coupled-enzyme assay (32) in 96-well format at 30°C. Plates were read in a Tecan M1000Pro plate reader with 340 nm excitation and 465 nm emission at 30 s intervals. All assays were done in 25 mM MES, pH 6.5, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 0.3 mM DDM. Final NAD+, TPP, and 2-oxoglutarate concentrations were 0.25 mM, 0.2 mM, and 2 mM respectively. Although the original assay used αKDH purchased from Sigma, we were dissatisfied with the activity and availability of the enzyme. Therefore, we prepared the αKDH in-house from beef heart using published protocol (61) αKDH quantity required for the assay was determined empirically by titration until the slope of the reaction progression curve did not change further. Palmitoyl-CoA and other acyl-CoA were usually kept below 20 μM as at higher concentrations we observed reduced enzyme activity. DHHC20 enzyme concentration in the assay to determine kinetic parameters was usually 2 nM to 5 nM. At higher DHHC20 concentrations it was difficult to get enough time points for low palmitoyl-CoA concentrations to determine the initial velocity. Km and kcat were determined using Michaelis-Menten equation using non-linear least squares fit in OriginLab. Curve-fitting was weighted to the error and was iteratively done to minimize reduced chi-square. Adjusted r-square values for most fits are >0.9 except for the inactive mutants where the data are not described by the Michaelis-Menten kinetics.
For assays where initial slopes at low palmitoyl-CoA concentrations were not necessary or a stronger signal was more important, DHHC20 enzyme concentrations were increased to 10–100 nM. In such cases the acyl-CoA concentrations were usually fixed at 5 or 10 μM.
Protein substrate palmitoylation assay
GobX protein palmitoylation assays were carried out in 96-well plates at ambient temperature (~24°C). Reaction condition was 50 mM HEPES, pH 7.0, 200 mM NaCl, 0.2 μM DTT, 1 mM EDTA, 0.3 mM DDM. eGPF-DHHC20 and GobX concentrations were 0.2 and ~30 μM respectively (we used the eGFP-tagged DHHC20 because the DHHC20 and GobX protein overlap in the gel). After mixing the enzyme and substrate, trans-palmitoylation reaction was initiated by the addition of the fluorescent NBD–palmitoyl-CoA to a final concentration of 1 μM. 10 μl of reaction mixture was pipetted out at different time points and the reaction quenched by adding it to 2x non-reducing SDS-sample buffer. Quenched reactions were run on a SDS-PAGE and the NBD fluorescence imaged on a gel imager (Bio-Rad). Protein signal was obtained from Coomassie staining or the use of a stain-free gel (Bio-Rad).
DHHC20 mutant analyses
The Venus-hDHHC20 construct was inserted in the multiple cloning site of pEG-Bacmam vector (62) under the control of the CMV promoter. All mutagenesis was carried out using the Quikchange strategy. Venus-hDHHC20 wild-type and mutants were expressed in HEK293T cells grown in DMEM with 2 mM glutamine, 2% FBS, and 100 units/mL penicillin/streptomycin. 10 μg of DNA was complexed with 30 μg of polyethyleneimine (PEI) and used to transfect 100 mm plates of ~80% confluent HEK293T cells (63). Cells were harvested after 48 hours and washed with phosphate-buffered saline. The cell pellets were then stored at –20°C. for later use. In parallel 1 μg of the same DNA was also used to transfect HEK293T cells in 12-well plates. FSEC was used to assess the expression level and the gel filtration profile of the DHHC20 mutants. Following satisfactory assessment of FSEC profile, frozen HEK293T pellets were thawed on ice and re-suspended in 500 μl of 40 mM TrisHCl, pH 7.2, 270 mM NaCl, 5 mM 2-ME, 5 mM MgCl2 containing Dnase and protease inhibitors. 100 μl of a 0.3 M stock DDM was added to the cell suspension and rotated for 1–2 hours at 4°C. Cell debris was removed by centrifugation at 21,000 g for 10 min at 4°C. The supernatant was then applied to ~30–40 μl of TALON resin in a microfuge tube and incubated on a rotator for ~1 hour at 4°C. Resin was first washed with 800 μl of same buffer containing 5 mM imidazole and 1 mM DDM but no MgCl2. Following a second wash with 500 μl of buffer containing 25 mM imidazole, protein was eluted by ~40 μl of 20 mM TrisHCl, pH 7.2, 135 mM NaCl, 2 mM TCEP, 5 mM 2-ME buffer containing 300 mM imidazole.
Protein concentration was estimated by running a stain-free SDS-PAGE (Bio-Rad) gel with previously purified Venus-hDHHC20 standards (8, 4, 1, 0.25 μM) from which a linear calibration curve could be constructed. Venus-hDHHC20 concentrations for most mutants were usually 2.5–6 μM. This translates to protein yields of ~7–16 μg per 100 mm cell culture dish. Of the mutants, AAxE and the ΔW267 mutants were the worst expressing. Therefore, in the case of these mutants, cells were transfected in 150 mm dishes (with 30 μg DNA) instead of 100 mm dishes. For the AAxE mutant, two such 150 mm dishes (equivalent to six 100 mm dishes) gave protein yields of ~14 μg. For the ΔW267 mutant, three 150 mm dishes (equivalent to nine 100 mm dishes) gave protein yields of ~3 μg.
Coupled-enzyme assays to measure Michaelis-Menten constants were carried out as described above. For the acyl-CoA selectivity experiments, enzyme and acyl-CoA concentrations were kept fixed at 10 nM and 10 μM respectively.
Mass spectrometry of irreversibly inhibited hDHHC20
For mass spectrometry measurements, purified wild-type hDHHC20–2-BP sample was run on SDS-PAGE under reducing conditions. Gel was stained by coomassie brilliant blue, destained, and thoroughly washed in deionized water. Gel band (~5 μg protein) was excised, treated with the reducing agent TCEP and the free cysteines blocked with N-ethylmaleimide. Protein was digested with chymotrypsin and applied to a C4 reverse phase column run at a flow-rate of 300 nL/min on a Ultimate 3000 HPLC (Thermo-Dionex) connected to a Orbitrap Elite mass spectrometer (Thermo Scientific).
Fluorescence protease protection assay
hDHHC20
DHHC20 cells tagged with N- or C-terminal Venus YFP were expressed in Cos7 cells using PEI transfection in 4-well or 8-well chambered microscope containers (Lab-Tek). Along with the Venus tagged DHHC20, we also co-expressed Cerulean CFP as a cytoplasmic marker and mCherry-TGN38 as a Golgi-marker with the mCherry RFP residing in the Golgi lumen. 24 hours post transfection, cells were washed with KHM (110 mM Potassium acetate, 20 mM HEPES, pH 7.2, and 3 mM MgCl2) buffer and then maintained in the same. Approximately a minute after initiation of time series imaging, cell plasma membrane was selectively permeabilized by the addition of digitonin dissolved in KHM to a final concentration of 22–25 μM. The permeabilization was followed by the disappearance of the cytoplasmic marker. Following disappearance of the cytoplasmic marker, trypsin prepared in KHM buffer was added to a final concentration of 1–2 mM and the cells further imaged for another minute or two until no change of any signal occurred. Control experiments were also done in which trypsin was added in the absence of digitonin. In these experiments, no significant loss in fluorescence was observed thus ruling out any pH or non-specific effects of trypsin on the cells.
Imaging was carried out on a Zeiss LSM780 microscope equipped with Argon laser (458, 514 nm) and HeNe-laser (594 nm) with a Plan-Apochromat 63x/1.40 Oil objective and a 458/514/594 multiband filter set. Cerulean CFP, Venus YFP, and mCherry RFP were excited with the 458, 514, and 594 nm lasers respectively. Emission was collected from 462–506 nm, 516–587 nm, and 605–685 nm for Cerulean, Venus, and mCherry respectively. Images were analyzed using FIJI (64).
zfDHHC15
Cos7 cells were seeded onto 8-well chamber plates (Lab-Tek) in 400 μl complete media and allowed them to grow and adhere to the surface of the plate for 16–18 hours. These cells were transfected with 100 ng of each vector GFP-zebrafish DHHC15, mCherry-TGN38, and BFP empty vector in serum free media with Lipofectamine 3000 transfection agent (Invitrogen). These transfected cells allowed to grow for 12–16 hours. The cells were permeabilized with 20 μM of digitonin in KHM buffer (110 mM potassium acetate, 2 mM Magnesium chloride and 50 mM HEPES, pH 7.5) for 30 s followed by trypsinization of the cells with 4 mM of trypsin in KHM buffer for 50 s during time dependent imaging experiment. Imaging data of adhered cells, from pre-permeabilized state to trypsinised state were acquired with confocal laser microscopy system equipped with a Plan Apochromat 63X/1.4NA oil immersion objective lens (LSM 780 Exciter, Carl Zeiss) at 37°C. Time-lapsed imaging was performed with the same instrument. Further, images were processed and fluorescence intensity was calculated using LSM and ImageJ softwares.
Chemicals and Reagents
PEG 300 and 2,5-Hexanediol were obtained from Sigma-Aldrich. PEG 400 was from USB. The buffers and salts were either from Sigma-Aldrich or Fisher Scientific. 2-bromopalmitate was from Sigma-Aldrich. All detergents were from Anatrace. All lipids and acyl coenzyme A was from Avanti Polar Lipids Inc. DTT and IPTG were from GoldBio. Digitonin (high purity) was from Calbiochem. Trypsin was obtained from Worthington Biochemicals. Methanol free 16% formaldehyde was obtained from Pierce-ThermoFisher and the DMAB complex from Sigma-Aldrich.
Sequence alignment
Multiple sequence alignment was carried out using MUSCLE (65). Figure for sequence alignment with secondary structure was generated using ESPRIPT (66).
Supplementary Material
Acknowledgments
The authors thank M. Machner (NIH) for the GobX protein expression plasmid, E. Gouaux (Oregon Health & Science University) for the pEG-Bacmam vector, Y. Li (NIH) for mass spectrometry, C. Williamson (NIH) for help with confocal microscopy, and J. Swan (NIH) for help with the figure accompanying the print version of the manuscript. The authors thank J. Bonifacino (NIH), S. Buchanan (NIH), W. Prinz (NIH), G. Storz (NIH), K. Swartz (NIH), and W. Yang (NIH) for comments on the manuscript. This work was supported by the Intramural Research Program of the NIH, the National Institutes of Child Health and Human Development (1ZIAHD008928 to M.S.R., P.K., C.-J.L., R.V., and A.B.), and the National Institute of Neurological Disorders and Stroke (to R.V. and A.B.). Crystals for this project were screened and data sets collected at various stages at the following beamlines: NE-CAT, GM/CA, and SER-CAT (22-ID) at the Advanced Photon Source, Argonne National Laboratory. NE-CAT is funded by the National Institute of General Medical Sciences from the NIH (P41 GM103403). The Pilatus 6M detector on 24-ID-C beamline (NE-CAT) is funded by a NIH-ORIP HEI grant (S10RR029205). GM/CA has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). The Eiger 16M detector (GM/CA) was funded by an NIH, Office of Research Infrastructure Programs, High-End Instrumentation Grant (1S10OD012289–01A1). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE), Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02–06CH11357. The structures have been deposited to the Protein Data Bank (PDB) with the accession codes 6BML (hDHHC20–2-BP treated), 6BMM (hDHHC20, P21 form), 6BMN (hDHHC20, P63 form), and 6BMS (zfDHHS15). All data needed to evaluate the conclusions in the paper are present in the main text or supplementary materials. A.B. conceived the project. A.B., M.S.R., and P.K. designed experiments. M.R. performed expression, purification, crystallization, data collection, and biochemical and imaging experiments of DHHC20. P.K. performed expression, purification, crystallization, data collection, crystallography, and imaging experiments of zfDHHC15. C.-J.L. performed crystallography of hDHHC20, and R.V. performed biochemical assays of zfDHHC15. K.R.R. assisted with crystallography. A.B. contributed to crystallization, data collection, and crystallography. M.S.R., P.K., C.-J.L., R.V., K.R.R., and A.B. analyzed data. A.B. wrote the manuscript with help from M.S.R. C.-J.L., and P.K. helped in figure preparation. The authors declare no competing financial interests.
References
- 1.Khoury GA, Baliban RC, Floudas CA, Proteome-wide posttranslational modification statistics: Frequency analysis and curation of the swiss-prot database. Sci. Rep. 1, 90 (2011).doi: 10.1038/srep00090; pmid: 22034591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniotti JL, Pedro MP, Valdez Taubas J, The role of S-acylation in protein trafficking. Traffic 18, 699–710 (2017).doi: 10.1111/tra.12510; pmid: 28837239 [DOI] [PubMed] [Google Scholar]
- 3.Linder ME, Deschenes RJ, Palmitoylation: Policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 (2007).doi: 10.1038/nrm2084; pmid: 1718336s2 [DOI] [PubMed] [Google Scholar]
- 4.Hentschel A, Zahedi RP, Ahrends R, Protein lipid modifications—More than just a greasy ballast. Proteomics 16, 759–782 (2016). doi: 10.1002/pmic.201500353; pmid: 26683279 [DOI] [PubMed] [Google Scholar]
- 5.Schmidt MF, Schlesinger MJ, Fatty acid binding to vesicular stomatitis virus glycoprotein: A new type of post-translational modification of the viral glycoprotein. Cell 17,813–819 (1979). doi: ; pmid: 226266 [DOI] [PubMed] [Google Scholar]
- 6.Schlesinger MJ, Magee AI, Schmidt MF, Fatty acid acylation of proteins in cultured cells. J. Biol. Chem. 255, 10021–10024 (1980). pmid: 7430112 [PubMed] [Google Scholar]
- 7.Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS, Identification of PSD-95 palmitoylating enzymes. Neuron 44, 987–996 (2004). doi: 10.1016/j.neuron.2004.12.005; pmid: [DOI] [PubMed] [Google Scholar]
- 8.Hernandez JL et al. , APT2 inhibition restores scribble localization and S-palmitoylation in snail-transformed cells. Cell Chem. Biol. 24, 87–97 (2017). doi: 10.1016/j.chembiol.2016.12.007; pmid: 28065656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF, Global profiling of dynamic protein palmitoylation. Nat. Methods 9, 84–89 (2011). doi: 10.1038/nmeth.1769; pmid: 22056678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muszbek L, Haramura G, Cluette-Brown JE, Van Cott EM, Laposata M, The pool of fatty acids covalently bound to platelet proteins by thioester linkages can be altered by exogenously supplied fatty acids. Lipids 34 (Suppl), S331–S337 (1999). doi: 10.1007/BF02562334; pmid: 10419194 [DOI] [PubMed] [Google Scholar]
- 11.Liang X et al. , Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J. Biol. Chem. 276, 30987–30994 (2001). doi: 10.1074/jbc.M104018200; pmid: 11423543 [DOI] [PubMed] [Google Scholar]
- 12.Berthiaume L, Resh MD, Biochemical characterization of a palmitoyl acyltransferase activity that palmitoylates myristoylated proteins. J. Biol. Chem. 270, 22399–22405 (1995). doi: 10.1074/jbc.270.38.22399; pmid: 7673226 [DOI] [PubMed] [Google Scholar]
- 13.Rocks O et al. , An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307, 1746–1752 (2005). doi: 10.1126/science.1105654; pmid: 15705808 [DOI] [PubMed] [Google Scholar]
- 14.El-Husseini AE et al. , Dual palmitoylation of PSD-95 mediates its vesiculotubular sorting, postsynaptic targeting, and ion channel clustering. J. Cell Biol. 148, 159–172 (2000). doi: 10.1083/jcb.148.1.159; pmid: 10629226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang K et al. , Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron 44, 977–986 (2004). doi: 10.1016/j.neuron.2004.11.027; pmid: 15603740 [DOI] [PubMed] [Google Scholar]
- 16.Hayashi T, Thomas GM, Huganir RL, Dual palmitoylation of NR2 subunits regulates NMDA receptor trafficking. Neuron 64, 213–226 (2009). doi: 10.1016/j.neuron.2009.08.017; pmid: 19874789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin YH et al. , Host cell-catalyzed S-palmitoylation mediates Golgi targeting of the Legionella ubiquitin ligase GobX. J. Biol. Chem. 290, 25766–25781 (2015). doi: 10.1074/jbc.M115.637397; pmid: 26316537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicks SW, Charron G, Hang HC, Galán JE, Subcellular targeting of Salmonella virulence proteins by host-mediated S-palmitoylation. Cell Host Microbe 10, 9–20 (2011). doi: 10.1016/j.chom.2011.06.003; pmid: 21767808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanc M et al. , SwissPalm: Protein Palmitoylation database. F1000Res. 4, 261 (2015). pmid: 26339475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy KD et al. , Physicochemical sequence characteristics that influence S-palmitoylation propensity. J. Biomol. Struct. Dyn. 35, 2337–2350 (2017). pmid: 27498722 [DOI] [PubMed] [Google Scholar]
- 21.Sanders SS et al., Curation of the mammalian palmitoylome indicates a pivotal role for palmitoylation in diseases and disorders of the nervous system and cancers. PLOS Comput. Biol 11, e1004405 (2015). doi: 10.1371/journal.pcbi.1004405; pmid: 26275289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Runkle KB et al. , Inhibition of DHHC20-mediated EGFR palmitoylation creates a dependence on EGFR signaling. Mol. Cell 62, 385–396 (2016). doi: 10.1016/j.molcel.2016.04.003; pmid: 27153536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobo S, Greentree WK, Linder ME, Deschenes RJ, Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277, 41268–41273 (2002). doi: 10.1074/jbc.M206573200; pmid: 12193598 [DOI] [PubMed] [Google Scholar]
- 24.Roth AF, Feng Y, Chen L, Davis NG, The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol 159, 23–28 (2002). doi: 10.1083/jcb.200206120; pmid: 12370247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohno Y, Kihara A, Sano T, Igarashi Y, Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta 1761, 474–483 (2006). doi: 10.1016/j.bbalip.2006.03.010; pmid: 16647879 [DOI] [PubMed] [Google Scholar]
- 26.Politis EG, Roth AF, Davis NG, Transmembrane topology of the protein palmitoyl transferase Akr1. J. Biol. Chem 280, 10156–10163 (2005). doi: 10.1074/jbc.M411946200; pmid: 15632165 [DOI] [PubMed] [Google Scholar]
- 27.Mitchell DA, Mitchell G, Ling Y, Budde C, Deschenes RJ, Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J. Biol. Chem. 285, 38104–38114 (2010). doi: 10.1074/jbc.M110.169102; pmid: 20851885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jennings BC, Linder ME, DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J. Biol. Chem 287, 7236–7245 (2012). doi: 10.1074/jbc.M111.337246; pmid: 22247542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.González Montoro A, Quiroga R, Valdez Taubas J, Zinc coordination by the DHHC cysteine-rich domain of the palmitoyltransferase Swf1. Biochem. J 454, 427–435 (2013). doi: 10.1042/BJ20121693; pmid: 23790227 [DOI] [PubMed] [Google Scholar]
- 30.Greaves J et al. , Molecular basis of fatty acid selectivity in the zDHHC family of S-acyltransferases revealed by click chemistry. Proc. Natl. Acad. Sci. U.S.A 114, E1365–E1374 (2017). doi: 10.1073/pnas.1612254114; pmid: 28167757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawate T, Gouaux E, Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006). doi: 10.1016/j.str.2006.01.013; pmid: 16615909 [DOI] [PubMed] [Google Scholar]
- 32.Hamel LD, Deschenes RJ, Mitchell DA, A fluorescencebased assay to monitor autopalmitoylation of zDHHC proteins applicable to high-throughput screening. Anal. Biochem 460,1–8 (2014). doi: 10.1016/j.ab.2014.05.013; pmid: 24878334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottlieb CD, Zhang S, Linder ME, The cysteine-rich domain of the DHHC3 palmitoyltransferase is palmitoylated and contains tightly bound zinc. J. Biol. Chem 290, 29259–29269 (2015). doi: 10.1074/jbc.M115.691147; pmid: 26487721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manglik A, Kobilka B, The role of protein dynamics in GPCR function: Insights from the b2AR and rhodopsin. Curr. Opin. Cell Biol 27, 136–143 (2014). doi: 10.1016/j.ceb.2014.01.008; pmid: 24534489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oswald C et al. , Intracellular allosteric antagonism of the CCR9 receptor. Nature 540, 462–465 (2016). doi: 10.1038/nature20606; pmid: 27926729 [DOI] [PubMed] [Google Scholar]
- 36.Lorenz H, Hailey DW, Lippincott-Schwartz J, Fluorescence protease protection of GFP chimeras to reveal protein topology and subcellular localization. Nat. Methods 3, 205–210 (2006).doi: 10.1038/nmeth857; pmid: 16489338 [DOI] [PubMed] [Google Scholar]
- 37.Krishna SS, Majumdar I, Grishin NV, Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 31, 532–550 (2003). doi: 10.1093/nar/gkg161;pmid:12527760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers LC, Jackow F, Verdine GL, Metal dependence of transcriptional switching in Escherichia coli Ada. J. Biol. Chem. 270, 6664–6670 (1995). doi: 10.1074/jbc.270.12.6664; pmid: 7896807 [DOI] [PubMed] [Google Scholar]
- 39.Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP, Sticky fingers: Zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 32, 63–70 (2007). doi: 10.1016/j.tibs.2006.12.007; pmid: 17210253 [DOI] [PubMed] [Google Scholar]
- 40.Dodson G, Wlodawer A, Catalytic triads and their relatives. Trends Biochem. Sci. 23, 347–352 (1998). doi: ; pmid: 9787641 [DOI] [PubMed] [Google Scholar]
- 41.González Montoro A, Quiroga R, Maccioni HJ, Valdez Taubas J, A novel motif at the C-terminus of palmitoyltransferases is essential for Swf1 and Pfa3 function in vivo. Biochem. J. 419, 301–308 (2009). doi: 10.1042/BJ20080921; pmid: 19138168 [DOI] [PubMed] [Google Scholar]
- 42.Mill P et al. , Palmitoylation regulates epidermal homeostasis and hair follicle differentiation. PLOS Genet. 5, e1000748 (2009). doi: 10.1371/journal.pgen.1000748; pmid: 19956733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coleman RA, Rao P, Fogelsong RJ, Bardes ES, 2-Bromopalmitoyl-CoA and 2-bromopalmitate: Promiscuous inhibitors of membrane-bound enzymes. Biochim. Biophys. Acta 1125, 203–209 (1992). doi: ; pmid: 1571364 [DOI] [PubMed] [Google Scholar]
- 44.Davda D et al. , Profiling targets of the irreversible palmitoylation inhibitor 2-bromopalmitate. ACS Chem. Biol. 8, 1912–1917 (2013). doi: 10.1021/cb400380s; pmid: 23844586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bánhegyi G et al. , Fatty acyl-CoA esters and the permeability of rat liver microsomal vesicles. Biochem. J. 320, 343–344 (1996). doi: 10.1042/bj3200343; pmid: 8947507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Requero MA, González M, Goñi FM, Alonso A, Fidelio G, Differential penetration of fatty acyl-coenzyme A and fatty acylcarnitines into phospholipid monolayers. FEBS Lett. 357, 75–78 (1995). doi: ; pmid: 8001684 [DOI] [PubMed] [Google Scholar]
- 47.O’Brien PJ, St Jules RS, Reddy TS, Bazan NG, Zatz M, Acylation of disc membrane rhodopsin may be nonenzymatic. J. Biol. Chem. 262, 5210–5215 (1987). pmid: 3558391 [PubMed] [Google Scholar]
- 48.Kordyukova LV, Serebryakova MV, Baratova LA, Veit M, S acylation of the hemagglutinin of influenza viruses: Mass spectrometry reveals site-specific attachment of stearic acid to a transmembrane cysteine. J. Virol. 82, 9288–9292 (2008). doi: ; pmid: 18596092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Senyilmaz D et al. , Regulation of mitochondrial morphology and function by stearoylation of TFR1. Nature 525, 124–128 (2015). doi: 10.1038/nature14601; pmid: 26214738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Draper JM, Smith CD, DHHC20: A human palmitoyl acyltransferase that causes cellular transformation. Mol. Membr. Biol. 27, 123–136 (2010). doi: 10.3109/09687681003616854; pmid: 20334580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chamberlain LH, Shipston MJ, The physiology of protein S-acylation. Physiol. Rev. 95, 341–376 (2015). doi: 10.1152/physrev.00032.2014; pmid: 25834228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shah K, Liu Y, Deirmengian C, Shokat KM, Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc. Natl. Acad. Sci. U.S.A 94, 3565–3570 (1997). doi: 10.1073/pnas.94.8.3565; pmid: 9108016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bishop AC et al. , A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407, 395–401 (2000). doi: 10.1038/35030148; pmid: 11014197 [DOI] [PubMed] [Google Scholar]
- 54.Baud MGJ et al. , A bump-and-hole approach to engineer controlled selectivity of BET bromodomain chemical probes. Science 346, 638–641 (2014). doi: 10.1126/science.1249830; pmid: 25323695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Long SB, Tao X, Campbell EB, MacKinnon R, Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 (2007). doi: 10.1038/nature06265; pmid: 18004376 [DOI] [PubMed] [Google Scholar]
- 56.Rayment I, Reductive alkylation of lysine residues to alter crystallization properties of proteins. Methods Enzymol. 276, 171–179 (1997). doi: [DOI] [PubMed] [Google Scholar]
- 57.Caffrey M, Cherezov V, Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009). doi: 10.1038/nprot.2009.31; pmid: 19390528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winn MD et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr 67, 235–242 (2011). doi: 10.1107/S0907444910045749; pmid: 21460441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams PD et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol. Crystallogr 66, 213–221 (2010). doi: 10.1107/S0907444909052925; pmid: 20124702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). doi: 10.1107/S0907444910007493; pmid: 20383002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stanley CJ, Perham RN, Purification of 2-oxo acid dehydrogenase multienzyme complexes from ox heart by a new method. Biochem. J. 191, 147–154 (1980). doi: 10.1042/bj1910147; pmid: 7470091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goehring A et al. , Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014). doi: 10.1038/nprot.2014.173; pmid: 25299155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Longo PA, Kavran JM, Kim MS, Leahy DJ, Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol. 529, 227–240 (2013). doi: ; pmid: 24011049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schindelin J et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). doi: 10.1038/nmeth.2019; pmid: 22743772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC, MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004). doi: ; pmid: 15318951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robert X, Gouet P, Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42 (W1), W320–W324 (2014). doi: 10.1093/nar/gku316; pmid: 24753421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Word JM et al. , Visualizing and quantifying molecular goodness-of-fit: Small-probe contact dots with explicit hydrogen atoms. J. Mol. Biol 285, 1711–1733 (1999). doi: 10.1006/jmbi.1998.2400; pmid: 9917407 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.