Cohen et al. show that, despite extensive genomic cobinding, PRC1 is essential for epidermal integrity, whereas PRC2 is dispensable.
Keywords: Polycomb complex, PRC1, PRC2, skin, epidermis, cell adhesion, epigenetics
Abstract
Polycomb-repressive complex 1 (PRC1) and PRC2 are critical chromatin regulators of gene expression and tissue development. Here, we show that despite extensive genomic cobinding, PRC1 is essential for epidermal integrity, whereas PRC2 is dispensable. Loss of PRC1 resulted in blistering skin, reminiscent of human skin fragility syndromes. Conversely, PRC1 does not restrict epidermal stratification during skin morphogenesis, whereas PRC2 does. Molecular dissection demonstrated that PRC1 functions with PRC2 to silence/dampen expression of adhesion genes. In contrast, PRC1 promotes expression of critical epidermal adhesion genes independently of PRC2-mediated H3K27me3. Together, we demonstrate a functional link between epigenetic regulation and skin diseases.
Tissue development and function rely on the ability of different cell types to maintain their cell identity and give rise to differentiated progeny through cell type-specific gene expression programs. The Polycomb group (PcG) proteins are conserved chromatin modifiers that regulate gene expression to control tissue development and function (Schuettengruber et al. 2017). Polycomb-mediated transcriptional control is mediated by two principal multisubunit complexes: Polycomb-repressive complex 1 (PRC1) and PRC2 (Simon and Kingston 2013). Mammalian PRC2 contains EED, SUZ12, and EZH1/2 core subunits and establishes dimethylation and trimethylation on histone H3 Lys27 (H3K27me2/3) (Cao et al. 2002; Margueron and Reinberg 2011). There are several mammalian PRC1 complexes, all of which contain an E3 ubiquitin ligase, RING1A/B, that catalyzes monoubiquitination on histone H2A Lys119 (H2AK119ub) (de Napoles et al. 2004; Wang et al. 2004a). At targeted loci, PRC1 and PRC2 typically repress gene expression (Simon and Kingston 2013), although evidence also exists for PRC1 or PRC2 promoting gene expression (Gao et al. 2014; Xu et al. 2015; Cohen et al. 2018). PRC1 and PRC2 usually colocalize at genomic binding sites, and initial studies suggested that they function together in a hierarchical manner to establish gene silencing (Simon and Kingston 2013), with in vivo studies demonstrating similar requirements for PRC1 and PRC2 core components in early mouse embryonic development (Voncken et al. 2003; Pasini et al. 2004) and Drosophila pattern formation (Lewis 1978; Margueron and Reinberg 2011). However, recent work has demonstrated that PRC1 and PRC2 can be recruited to chromatin and modify chromatin independently of each other (for review, see Blackledge et al. 2015). Thus, it is unclear to what extent PRC1 and PRC2 functions overlap in somatic tissue control.
Here we study PRC1 and PRC2 in the developing skin epidermis, a tissue that provides essential barrier functions (Blanpain and Fuchs 2009). During development and homeostasis, epidermal stem cells (SCs) located in the basal layer move upward to the suprabasal layer, where they undergo terminal differentiation. The newly formed stratified epithelium relies on its architecture and structural integrity to maintain epidermal gene expression programs and protective functions. The main adhesive structures responsible for epidermal integrity are hemidesmosomes and desmosomes. The hemidesmosomes are responsible for anchoring basal layer cells to the basement membrane, and the desmosomes are responsible for epidermal cell–cell adhesion (Green and Jones 1996). Impairment of these structures is a cause of skin fragility syndrome, a group of skin disorders characterized by the tendency to develop skin blistering, erosions, or wounds upon minimal mechanical stress (Lopez-Pajares et al. 2013; Has and Bruckner-Tuderman 2014). Here we identify critical roles for PRC1 in the maintenance of epidermal integrity, whereas PRC2 function is dispensable. Through side-by-side analysis of PRC1-null and PRC2-null epidermis coupled with chromatin and transcriptional profiling data analyses, we determined how common and distinct functions of PRC1 and PRC2 preserve epidermal integrity.
Results and Discussion
Loss of PRC1, but not PRC2, in the developing skin epithelium leads to skin fragility
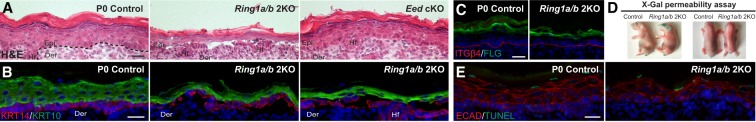
We previously generated PRC1 conditional knockout (cKO) mice in which the core PRC1 complex subunits RING1A/B were ablated in epidermal progenitors (Cohen et al. 2018). We crossed Ring1a-null Ring1b-floxed mice with Krt14-Cre mice that express cre recombinase in epidermal progenitors (Krt14-Cre; Ring1a−/− Ring1bflox/flox = Ring1a/b double knockout) starting at embryonic day 12 (E12). Histological analysis of postnatal day 0 (P0) pups revealed that the Ring1a/b double-knockout epidermis is fragile, being susceptible to damage upon minimal mechanical stress (Fig. 1A), similar to common human skin diseases showing skin fragility (Lopez-Pajares et al. 2013; Has and Bruckner-Tuderman 2014). These alterations were not observed in Eed cKO mice, in which the PRC2 core subunit EED was ablated in epidermal progenitors (Fig. 1A). Immunofluorescence (IF) analysis using the basal layer marker keratin 14 (KRT14) and the differentiating suprabasal layers marker KRT10 demonstrated that skin fragility in Ring1a/b double-knockout mice occurred between the basal layer and the underling basement membrane and between the basal and the first suprabasal layer (Fig. 1B).
Figure 1.
Ring1a/b double-knockout epithelium displays fragile skin epidermis. (A) Hematoxylin and eosin (H&E) staining. Bar, 25 µm. (B) IF staining for the basal layer marker KRT14 (red) and the differentiated suprabasal layer marker KRT10 (green). Bar, 25 µm. (C) IF staining for the late differentiation marker filaggrin (FLG; green). The basement membrane is labeled by ITGβ4 (red). Bar, 25 µm. (D) X-gal skin permeability assay in newborn Ring1a/b double-knockout and control mice. (E) TUNEL assay for apoptosis (green). Skin epithelium is labeled by E-cadherin (ECAD; red). Bar, 25 µm. (Epi) Epidermis; (Hf) hair follicle; (Der) dermis.
To determine whether the observed skin fragility was due to misregulation of the epidermal stratification program, we analyzed Ring1a/b double-knockout embryonic skin at E16.5, when only early differentiated layers are formed, confirming complete epidermal loss of RING1B and H2AK119ub (Supplemental Fig. S1A,B). Expression of the early differentiation marker KRT10 was detected in Ring1a/b double knockout and was restricted to differentiating suprabasal layers, as seen in controls (Supplemental Fig. S1C). There was no precocious induction of the late differentiation marker filaggrin (FLG) or accelerated acquisition of epidermal barrier function in Ring1a/b double-knockout epidermis at E16.5 (Supplemental Fig. S1D,E), as reported for the skin of PRC2-null E16.5 embryos (Ezhkova et al. 2009; Dauber et al. 2016). At later stages of morphogenesis, the epidermal stratification program progressed in Ring1a/b double-knockout skin, with FLG being detected in the upper suprabasal layers of Ring1a/b double-knockout mice, similarly to controls (Fig. 1C). Ring1a/b double-knockout mice formed a functional epidermal barrier (Fig. 1D). There was no change in cell proliferation (Cohen et al. 2018), and no aberrant cell death was detected in Ring1a/b double-knockout epidermis during epidermal morphogenesis (Fig. 1E; Supplemental Fig. S1F). Thus, the skin fragility phenotype in Ring1a/b double-knockout epidermis is not due to an arrested epidermal stratification program.
Perturbed cell–cell adhesion in the absence of PRC1 in the developing skin epidermis
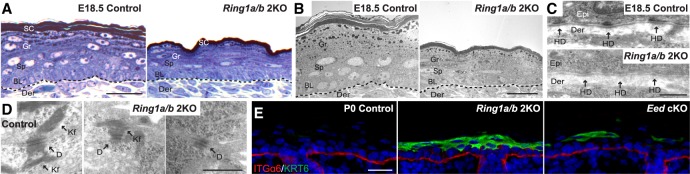
Ultrastructural analysis of skins collected from E18.5 Ring1a/b double-knockout embryos (thereby excluding tissue damage that might occur at birth) revealed the presence of keratohyalin granules and lamellar bodies in the granular layer epidermis as well as the formation of cornified layers of the stratum corneum (Fig. 2A,B; Supplemental Fig. S2A), confirming that the epidermal stratification program was generally not impaired in Ring1a/b double-knockout epidermis. Transmission electron microscopy (TEM) analysis revealed that hemidesmosome structures at the basement membrane in Ring1a/b double-knockout mice were poorly developed, with diffused appearance and weak staining of the intracellular components composing the inner and outer plaques and the subbasal dense plates (Fig. 2C). While desmosomes localized normally to cell membranes, desmosome structure was morphologically abnormal in Ring1a/b double-knockout epidermis (Fig. 2D). Desmosomes between the basal and suprabasal layers in Ring1a/b double-knockout epidermis were smaller and less obvious than controls, with reduced and disorganized tonofilaments (Fig. 2D). Eed cKO mice demonstrated apparently normal hemidesmosomes, whereas desmosomes were slightly smaller but well developed and comparable with controls (Supplemental Fig. S2B,C). Thus, the epidermal fragility observed in Ring1a/b double-knockout mice may result from impaired hemidesmosome and desmosome functions, leading to poor cell–cell adhesion and tissue anchorage.
Figure 2.
Perturbed cell adhesion in Ring1a/b double-knockout skin epidermis. (A) Toluidine blue staining of semithin skin sections. Bar, 25 µm. (B–D) TEM ultrastructural analyses of control and Ring1a/b double-knockout skins. (B) Representative region of skin epidermis. Bar, 10 µm. (C) High magnification of epidermal basement membrane region. Bar, 0.5 µm. (D) High magnification of cell–cell adhesion region between basal and spinous layers of skin epidermis. Bar, 0.5 µm. (E) IF staining for KRT6 (green). The basement membrane is labeled by ITGα6 (red). Bar, 25 µm. (Der) Dermis; (BL) basal layer; (Sp) spinous layer; (Gr) granular layer; (SC) stratum corneum; (HD) hemidesmosome; (D) desmosome; (Kf) keratin intermediate filaments.
IF analysis of KRT6, normally restricted to hair follicles (HFs) but induced in the interfollicular epidermis upon wounding or in hyperproliferative disorders (Kirfel et al. 2003), revealed the induction of KRT6 in Ring1a/b double-knockout mice throughout the skin epidermis (Fig. 2E). Patchy induction of KRT6 was also evident in Eed cKO epidermis (Fig. 2E). ChIP-seq (chromatin immunoprecipitation combined with high-throughput sequencing) data showed that Krt6a and Krt6b genes are not targets of PRC1-mediated H2AK119ub in epidermal progenitors (Supplemental Fig. S2D). Taken together, the induction of KRT6 in Ring1a/b double-knockout epidermis supports the impairment of cell adhesion components identified in our TEM analysis. The patchy induction of KRT6 in Eed cKO epidermis suggests that some common pathways involved in cell adhesion may be misregulated in both PRC1-null and PRC2-null epidermis.
PRC1 regulates the expression of cell adhesion and cytoskeleton organization genes
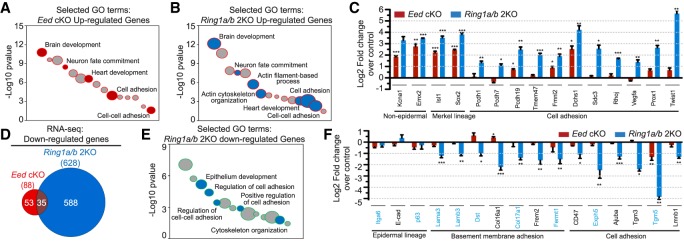
To gain insights into the molecular mechanisms underlying the observed adhesion defects in Ring1a/b double-knockout epidermis, we used RNA sequencing (RNA-seq) data from P0 Eed cKO, Ring1a/b double-knockout, and control samples enriched for epidermal SCs (Supplemental Fig. S3A,B; Cohen et al. 2018) and subjected the differentially expressed genes (Supplemental Table S1) to gene ontology (GO) analysis (Huang et al. 2009). Because the phenotypes of Ring1a/b double-knockout and Eed cKO mice were so different (adhesion defects in Ring1a/b double-knockout and precocious epidermal differentiation during skin morphogenesis in Eed cKO), we decided to use Eed cKO data for comparison purposes. Among genes that were up-regulated in Eed cKO and Ring1a/b double-knockout compared with controls, we observed common enrichment for genes of nonskin epithelium lineages as well as genes related to cell adhesion (Fig. 3A,B; Supplemental Table S2). Generally, the number of adhesion-related genes that were up-regulated in Ring1a/b double knockout was higher than those up-regulated in Eed cKO (Supplemental Table S2). In addition, genes up-regulated in Ring1a/b double-knockout epidermis were also enriched for GO terms related to actin cytoskeleton organization and actin filament-based processes (Fig. 3B; Supplemental Table S2). RT-qPCR analysis confirmed the RNA-seq findings (Fig. 3C) and showed that for genes related to cell adhesion that were up-regulated in Ring1a/b double-knockout epidermis, their expression was either largely unaffected or only weakly up-regulated in Eed cKO epidermis (Fig. 3C). Some of these up-regulated genes, such as Prox1 and Twist1, are known to negatively regulate cell adhesion and promote cell invasiveness (Yang et al. 2004; Dadras et al. 2008), but there was no epithelial-to-mesenchymal transition in Ring1a/b double-knockout epidermis (Supplemental Fig. S3C,D).
Figure 3.
PRC1 preserves transcriptional identity in skin epidermis. (A) GO analysis of genes up-regulated in Eed cKO versus control epidermis. Selected GO terms are in red. (B) GO analysis of genes up-regulated in Ring1a/b double-knockout versus control epidermis. Selected GO terms are in blue. (C,F) RT-qPCR analysis of FACS-isolated epidermal cells in Eed cKO and Ring1a/b double-knockout compared with control epidermal cells. Genes mutated in human diseases with blistering skin are labeled in blue. Data are mean ± SEM. n = 3. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001, two-sided t-test. (D) Venn diagram showing the overlap of significantly down-regulated genes between Eed cKO and Ring1a/b double-knockout epidermal cells. (E) GO analysis of genes down-regulated in Ring1a/b double-knockout versus control epidermis. Selected GO terms are in blue.
Next, we analyzed genes that were significantly down-regulated in Eed cKO or Ring1a/b double-knockout compared with controls (Supplemental Table S1). While 628 genes were significantly down-regulated in Ring1a/b double-knockout, only 88 genes were significantly down-regulated in Eed cKO compared with control epidermis, with small overlap between the two groups (Fig. 3D). Genes down-regulated in Ring1a/b double-knockout epidermis were also enriched for GO terms related to cell adhesion, cell–cell adhesion, and actin cytoskeleton organization (Fig. 3E; Supplemental Table S2). RT-qPCR showed that for genes related to cell adhesion that were down-regulated in Ring1a/b double-knockout epidermis, their expression was largely unaffected in Eed cKO epidermis (Fig. 3F). IF staining further showed that down-regulated genes analyzed also had reduced protein levels in Ring1a/b double-knockout epidermis (Supplemental Fig. S3E–H). Genes down-regulated in Ring1a/b double-knockout epidermis included Lama3, Dst, Col17a1, Fermt1, Exph5, and Tgm5, known to be mutated in common skin diseases characterized by skin blistering and/or poor cell adhesion and tissue anchorage (Has and Bruckner-Tuderman 2014). Moreover, many of Ring1a/b double-knockout down-regulated genes related to adhesion form functional groups together with those genes mutated in blistering skin diseases (Supplemental Fig. S4A). Thus, the up-regulation of cell adhesion/cytoskeleton organization genes that are normally silenced or poorly expressed, together with the down-regulation of critical epidermal cell adhesion genes, perturbs epidermal integrity and leads to skin fragility in Ring1a/b double-knockout epidermis.
PRC1 regulates adhesion genes through PRC2-dependent and PRC2–independent mechanisms
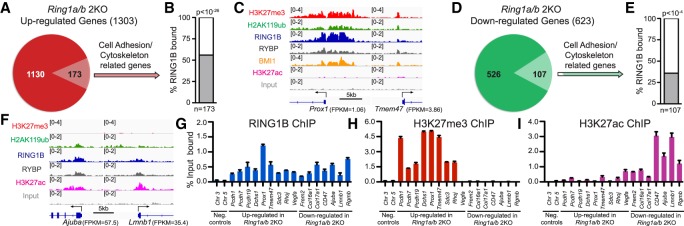
To understand how PRC1 controls the expression of genes related to cell adhesion and cytoskeleton organization, we merged RNA-seq with ChIP-seq data to reveal that >50% of the genes related to cell adhesion and cytoskeleton organization that were up-regulated in Ring1a/b double-knockout epidermis are direct targets of RING1B in control epidermis (Fig. 4A–C; Supplemental Table S3). The majority of these genes were also targets of PRC2-mediated H3K27me3 in control epidermis despite minimal changes in expression of these genes in Eed cKO epidermis (Fig. 4C; Supplemental Fig. S5A). These genes could be further divided into two groups. Group 1 genes were not expressed in control epidermis with mean expression of fragments per kilobase of transcript per million (FPKM) < 2, and group 2 genes were expressed at low to moderate levels in control epidermis (Supplemental Fig. S5B). In line with the differential expression levels between the two gene categories, group 2 genes had significantly lower levels of H3K27me3 around the transcription start site (TSS) compared with group 1 genes (Fig. 4C; Supplemental Fig. S5C). These data indicate that PRC1 and PRC2 cobind to repressed cell adhesion and cytoskeleton organization genes.
Figure 4.
PRC1 directly regulates cell adhesion and cytoskeleton organization genes. (A) Pie chart showing the portion of cell adhesion and cytoskeleton organization genes among Ring1a/b double-knockout up-regulated genes. (B) Percentage of up-regulated genes related to cell adhesion and cytoskeleton organization occupied by RING1B (C,F) Integrative Genomics Viewer (IGV) browser views of ChIP-seq data for the indicated genes. (D) Pie chart showing the portion of cell adhesion and cytoskeleton organization genes among Ring1a/b double-knockout down-regulated genes. (E) The percentage of down-regulated genes related to cell adhesion and cytoskeleton organization occupied by RING1B. (G–I) ChIP-qPCR showing the binding of RING1B (G), H3K27me3 (H), and H3K27ac (I) in control epidermis. Data are mean ± SEM. n = 2.
PRC1 complexes can be divided into canonical PRC1 (cPRC1) and noncanonical PRC1 (ncPRC1) complexes. cPRC1 complexes contain MEL18 (PCGF2) or BMI1 (PCGF4) and a CBX protein that recognizes H3K37me3, enabling cPRC1 recruitment to chromatin (Min et al. 2003; Wang et al. 2004b), whereas ncPRC1 complexes contain PCGF subunit (PCGF1–6) together with RYBP or YAF2 instead of the CBX subunit, and their recruitment to chromatin is not dependent on H3K27me3 (Gao et al. 2012; Blackledge et al. 2014; Cooper et al. 2014). The large overlap between PRC1 and PRC2 at repressed genes prompted us to test whether cPRC1 complexes are responsible for the observed phenotype. Analysis of ChIP-seq data of the key cPRC1 subunit BMI1 demonstrated that >50% of RING1B-bound genes related to cell adhesion and cytoskeleton organization that were up-regulated in Ring1a/b double-knockout were direct targets of BMI1 (Fig. 4C; Supplemental Fig. S5D). We therefore ablated the essential cPRC1 subunits MEL18 (PCGF2) and BMI1 (PCGF4) in the skin epithelium (Pcgf2/4 cKO). However, cPRC1-null epidermis was intact, without signs of fragile skin or induction of KRT6 (Supplemental Fig. S5E,F). In line with this, genes related to cell adhesion that were up-regulated in Ring1a/b double-knockout epidermis were largely unaffected or only weakly up-regulated in Pcgf2/4 cKO epidermis (Supplemental Fig. S5G). These indicate that ncPRC1 complexes are important in epidermal tissue control and to preserve epidermal integrity.
Because ncPRC1 promotes the expression of active lineage genes in the skin epithelium (Cohen et al. 2018), we also analyzed genes that are down-regulated in Ring1a/b double-knockout epidermal progenitors. We identified a significant overlap with RING1B binding, but not with H3K27me3, and Ring1a/b double-knockout down-regulated genes involved in cell adhesion and cytoskeleton organization (Fig. 4D–F; Supplemental Fig. S5H; Supplemental Table S3). Additionally, RING1B-bound genes that were down-regulated in Ring1a/b double-knockout had significantly lower H3K27me3 levels around their TSSs compared with RING1B-bound genes that were up-regulated in Ring1a/b double-knockout (Fig. 4C,F; Supplemental Fig. S5C).
To validate our ChIP-seq analysis, we performed ChIP-qPCR on genes that were either up-regulated or down-regulated in Ring1a/b double-knockout compared with control epidermis. For both classes of genes, our analysis confirmed the binding of RING1B as well as the ncPRC1 subunit RYBP (Fig. 4G; Supplemental Figs. S5I, S6A). H3K27me3 and H2AK119ub were present near the promoter regions of up-regulated genes at relatively high levels compared with down-regulated genes (Fig. 4C,F,H; Supplemental Figs. S5J, S6B,C), whereas H3K27ac, associated with transcriptional activation, was present near the promoter regions of down-regulated genes at higher levels compared with up-regulated genes (Fig. 4C,F,I). Thus, PRC1 directly regulates, through different mechanisms, both silent and active genes responsible for cell adhesion and cytoskeleton organization.
Here we revealed differential requirements for PRC1 and PRC2 in skin epidermis. PRC1 is essential for epidermal integrity, whereas PRC2 is dispensable. Conversely, our previous studies demonstrated that PRC2 temporally restricts the epidermal stratification program and is required to prevent precocious differentiation (Ezhkova et al. 2009; Dauber et al. 2016), whereas our current studies show that PRC1 is not required for these processes during morphogenesis. Differential requirements for PRC1 and PRC2 in somatic SCs have been reported in the developing HF and the adult intestine. In developing HFs, PRC2-null skin exhibits normal HF morphogenesis at P0 but developmental arrest and degenerate in adults (Ezhkova et al. 2011; Dauber et al. 2016). The decreased HF cell proliferation and induced apoptosis in PRC2-null HFs coincide with up-regulation of Cdkn2a/Cdkn2b genes, and silencing of Cdkn2a/Cdkn2b in PRC2-null HF cells can revert these defects in vitro (Ezhkova et al. 2011). In PRC1-null epithelium, however, HF morphogenesis is impaired at much earlier developmental stages, HF SCs are not properly specified in Ring1a/b double-knockout mice, and these defects are independent of Cdkn2a (Cohen et al. 2018). Similarly, in the adult mouse intestine, loss of PRC2 induces cell cycle arrest and differentiation skewing toward secretory cells in a Cdkn2a-dependent manner (Chiacchiera et al. 2016b), whereas PRC1 is required to preserve intestinal SC self-renewal independently of Cdkn2a (Chiacchiera et al. 2016a). Taken together, these findings demonstrate that loss of PRC1 or PRC2 in a tissue-specific context can lead to different biological outcomes.
The mechanisms underlying the distinct PRC1 and PRC2 phenotypes are presently unclear, given that PRC1 and PRC2 cobind to a large set of common target genes. Analysis of the fragile epidermis phenotype showed that PRC1 functions together with PRC2-mediated H3K27me3 to repress genes involved in cell adhesion and cytoskeleton organization. In most cases, however, PRC1 has a more prominent role in repression than PRC2, as loss of PRC2 was not sufficient for robust gene derepression. This is supported by our observation that loss of cPRC1, the main form of PRC1 complexes that interacts with PRC2-mediated H3K27me3, was also not sufficient for robust gene derepression. Furthermore, at other genomic targets, ncPRC1 can function independently of H3K27me3 to promote the expression of genes involved in cell adhesion and cytoskeleton organization. Understanding PRC1 and PRC2 functions in somatic SC systems is essential in light of the roles of individual PcG proteins in a large variety of human diseases, including cancers (for review, see Sauvageau and Sauvageau 2010).
Loss of PRC1 function in the skin results in compromised epidermal integrity, similar to skin fragility diseases in humans, which are characterized by trauma-induced skin blisters, erosions, and wounds (Has and Bruckner-Tuderman 2014). Typically, these diseases result from mutations in genes encoding for structural components of hemidesmosomes responsible for basement membrane zone anchoring or in components of desmosomes responsible for epidermal cell–cell adhesions (Lopez-Pajares et al. 2013). Our analyses of PRC1-null epidermis revealed that both hemidesmosome and desmosome structures were impaired. PRC1-null epidermal cells still executed an epidermal differentiation program that resulted in formation of functional protective barrier, indicating that loss of PRC1 affected cell function rather than cell differentiation. We observed massive alterations in the expression of genes related to cell adhesions, many of which were directly controlled by PRC1. Our study provides a link between PRC1, a key epigenetic transcriptional regulator, and skin fragility syndromes, demonstrating how changes in the epigenetic machinery can lead to disease pathogenesis.
Materials and methods
Mice
All mice were housed at the Center of Comparative Medicine and Surgery, Icahn School of Medicine at Mount Sinai, and cared for according to the Institutional Animal Care and Use Committee guidelines (protocol no. LA11-0020). Krt14-Cre mice (stock no. 004782) were obtained from the Jackson Laboratories. Ring1a−/− and Ring1bflox/flox were described previously (del Mar Lorente et al. 2000; Cales et al. 2008). Pcgf2flox/flox and Pcgf4flox/flox mice were generated in H. Koseki's laboratory and are discussed in another study (M.N. and H.K., unpubl.). Eedflox/flox mice were provided by Weipeng Mu and Terry Magnuson (Mu et al. 2014). Genotyping primers are listed in Supplemental Table S4.
Additional experimental details are in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank Ya-Chieh Hsu, Jose Silva, Sergei Ezhkov, and Venu Pothula for help and critical suggestions. We are grateful for the assistance and reagents provided by Julie Segre and Elaine Fuchs. We thank Miguel Vidal for the Ring1a−/− and Ring1bflox/flox mice, and Weipeng Mu and Terry Magnuson for the Eedflox/flox mice. We thank the Flow Cytometry Core Facility as well as Ronald Gordon from the Electron Microscopy Facility at the Icahn School of Medicine at Mount Sinai. Research reported here was supported by the National Institutes of Health (NIH)/National Institute of Arthritis and Musculoskeletal and Skin Diseases under award number R01 AR069078 (to E.E.), NIH/National Heart, Lung, and Blood Institute grant R01HL133120 (to D. Zheng), and the Tisch Cancer Institute P30 Cancer Support Grant (to E.E.).
Author contributions: I.C. and E.E. conceived and designed the experiments. I.C. performed the experiments. G.M. performed the TEM data analysis. D. Zhao and D. Zheng performed the bioinformatic analyses. H.K. provided the Ring1a−/− and Ring1bflox/flox mice. M.N. and H.K. provided the Pcgf2flox/flox and Pcgf4flox/flox mice. I.C., D. Zhao, D. Zheng, and E.E. analyzed the data. I.C. and E.E. wrote the manuscript with input from all of the other authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.319939.118.
References
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, et al. 2014. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157: 1445–1459. 10.1016/j.cell.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Rose NR, Klose RJ. 2015. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol 16: 643–649. 10.1038/nrm4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. 2009. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol 10: 207–217. 10.1038/nrm2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cales C, Roman-Trufero M, Pavon L, Serrano I, Melgar T, Endoh M, Perez C, Koseki H, Vidal M. 2008. Inactivation of the polycomb group protein Ring1B unveils an antiproliferative role in hematopoietic cell expansion and cooperation with tumorigenesis associated with Ink4a deletion. Mol Cell Biol 28: 1018–1028. 10.1128/MCB.01136-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043. 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- Chiacchiera F, Rossi A, Jammula S, Piunti A, Scelfo A, Ordóñez-Morán P, Huelsken J, Koseki H, Pasini D. 2016a. Polycomb complex PRC1 preserves intestinal stem cell identity by sustaining Wnt/β-catenin transcriptional activity. Cell Stem Cell 18: 91–103. 10.1016/j.stem.2015.09.019 [DOI] [PubMed] [Google Scholar]
- Chiacchiera F, Rossi A, Jammula S, Zanotti M, Pasini D. 2016b. PRC2 preserves intestinal progenitors and restricts secretory lineage commitment. EMBO J 35: 2301–2314. 10.15252/embj.201694550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen I, Zhao D, Bar C, Valdes VJ, Dauber-Decker KL, Nguyen MB, Nakayama M, Rendl M, Bickmore WA, Koseki H, et al. 2018. PRC1 fine-tunes gene repression and activation to safeguard skin development and stem cell specification. Cell Stem Cell 22: 726–739.e7. 10.1016/j.stem.2018.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S, Koseki H, Klose R, et al. 2014. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep 7: 1456–1470. 10.1016/j.celrep.2014.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadras SS, Skrzypek A, Nguyen L, Shin JW, Schulz MM, Arbiser J, Mihm MC, Detmar M. 2008. Prox-1 promotes invasion of kaposiform hemangioendotheliomas. J Invest Dermatol 128: 2798–2806. 10.1038/jid.2008.176 [DOI] [PubMed] [Google Scholar]
- Dauber KL, Perdigoto CN, Valdes VJ, Santoriello FJ, Cohen I, Ezhkova E. 2016. Dissecting the roles of Polycomb repressive complex 2 subunits in the control of skin development. J Invest Dermatol 136: 1647–1655. 10.1016/j.jid.2016.02.809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Mar Lorente M, Marcos-Gutierrez C, Perez C, Schoorlemmer J, Ramirez A, Magin T, Vidal M. 2000. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 127: 5093–5100. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676. 10.1016/j.devcel.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. 2009. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136: 1122–1135. 10.1016/j.cell.2008.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. 2011. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 25: 485–498. 10.1101/gad.2019811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45: 344–356. 10.1016/j.molcel.2012.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Lee P, Stafford JM, von Schimmelmann M, Schaefer A, Reinberg D. 2014. An AUTS2–Polycomb complex activates gene expression in the CNS. Nature 516: 349–354. 10.1038/nature13921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KJ, Jones JC. 1996. Desmosomes and hemidesmosomes: structure and function of molecular components. FASEB J 10: 871–881. 10.1096/fasebj.10.8.8666164 [DOI] [PubMed] [Google Scholar]
- Has C, Bruckner-Tuderman L. 2014. The genetics of skin fragility. Annu Rev Genomics Hum Genet 15: 245–268. 10.1146/annurev-genom-090413-025540 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Kirfel J, Magin TM, Reichelt J. 2003. Keratins: a structural scaffold with emerging functions. Cell Mol Life Sci 60: 56–71. 10.1007/s000180300004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. 10.1038/276565a0 [DOI] [PubMed] [Google Scholar]
- Lopez-Pajares V, Yan K, Zarnegar BJ, Jameson KL, Khavari PA. 2013. Genetic pathways in disorders of epidermal differentiation. Trends Genet 29: 31–40. 10.1016/j.tig.2012.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM. 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17: 1823–1828. 10.1101/gad.269603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu W, Starmer J, Fedoriw AM, Yee D, Magnuson T. 2014. Repression of the soma-specific transcriptome by Polycomb-repressive complex 2 promotes male germ cell development. Genes Dev 28: 2056–2069. 10.1101/gad.246124.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23: 4061–4071. 10.1038/sj.emboj.7600402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. 2010. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 7: 299–313. 10.1016/j.stem.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. 2017. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell 171: 34–57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. 2013. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 49: 808–824. 10.1016/j.molcel.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M. 2003. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci 100: 2468–2473. 10.1073/pnas.0434312100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. 2004a. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878. 10.1038/nature02985 [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. 2004b. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell 14: 637–646. 10.1016/j.molcel.2004.05.009 [DOI] [PubMed] [Google Scholar]
- Xu J, Shao Z, Li D, Xie H, Kim W, Huang J, Taylor JE, Pinello L, Glass K, Jaffe JD, et al. 2015. Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol Cell 57: 304–316. 10.1016/j.molcel.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117: 927–939. 10.1016/j.cell.2004.06.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.