This Outlook discusses Cohen et al.’s finding that despite extensive genomic cobinding, PRC1 is essential for epidermal integrity, whereas PRC2 is dispensable.
Keywords: Polycomb complex, PRC1, PRC2, skin, epidermis, cell adhesion, epigenetics
Abstract
All of the cells in our body share largely identical DNA, yet functionally distinct cells are generated to give rise to different tissues and organs. A fundamental question in biology is how different cell fates are specified and maintained. Epigenetic mechanisms hold a key answer to the question. Without changing the sequence of DNA but through modifying DNA, histones, or RNA, epigenetic mechanisms can decide which genes to express and which to suppress. Polycomb group (PcG) proteins are a group of evolutionarily conserved proteins that can regulate gene expression through histone modification. Although PcG proteins have been traditionally described as epigenetic repressors, emerging evidence suggests a more complex scenario in which PcG proteins can have a dynamic effect on gene expression. In this issue of Genes & Development, Cohen and colleagues (pp. 55–60) studied the function of Polycomb-repressive complex 1 (PRC1) in mouse skin development and identified PRC1's unique function independent of PRC2. Notably, the total loss of PRC1 but not canonical PRC1 in the skin leads to widespread down-regulation of genes involved in cell adhesion and cytoskeleton organization, resulting in skin fragility. This new study lays a foundation to examine the role of PRC1 in activating gene expression.
Polycomb group (PcG) proteins interact with each other to form chromatin-associated repressive complexes: Polycomb-repressive complex 1 (PRC1) and PRC2. The PRC2 complex is comprised of three core components, EZH1/2, EED, and SUZ12, which establish histone H3 dimethylation and trimethylation marks on Lys27 (H3K27me2/3) (Margueron and Reinberg 2011). Several mammalian PRC1 complexes have been identified, and each of them contains an E3 ubiquitin ligase (RING1A or RING1B), which catalyzes histone H2A ubiquitination on Lys119 (H2AK119ub) (Blackledge et al. 2015). Epidermal-specific knockout (cKO) of Ezh1/2, Eed, or Suz12, all essential components of PRC2, leads to premature epidermal cell differentiation, ectopic Merkel cell formation, and postnatal failure of hair follicle development (Dauber et al. 2016). At the molecular level, PRC2 functions to repress gene expression, consistent with the prevailing view obtained from other systems (Ezhkova et al. 2011; Blackledge et al. 2015). Here, Cohen et al. (2019) used a similar strategy to conditionally ablate the PRC1 core subunits Ring1a and Ring1b in the skin using epithelial-specific Krt14-Cre. Unlike the premature epidermal barrier formation phenotype observed in PRC2 subunit cKO animals, Ring1a/b double knockout showed relatively normal differentiation and a functional epidermal barrier, as determined by immunohistochemistry staining of differentiation markers and dye permeability assay, respectively. However, Ring1a/b double-knockout epidermis is fragile and susceptible to damage upon application of minimal mechanical stress, mimicking defects observed in skin fragility models (Has and Bruckner-Tuderman 2014). Transmission electron microscopy revealed that the defects in Ring1a/b double knockout are on the hemidesmosomes, which attach the basal epithelial cells to the basement membrane, as well as on the desmosomes, which mediate cell–cell interaction of epithelial cells. Compromised hemidesmosome and desmosome structures thus lead to the epidermal fragility observed in Ring1a/b double-knockout mice.
To dissect the molecular mechanism underlying the adhesion defects in Ring1a/b double-knockout epidermis, Cohen et al. (2019) used RNA sequencing (RNA-seq) to analyze the whole transcriptome of Ring1a/b double knockout and compared it with the transcriptome of PRC2 core component Eed cKO. Among the derepressed genes, nonskin epithelium genes were shared between Ring1a/b double knockout and Eed cKO. A group of genes related to actin cytoskeleton organization and actin filament-based processes was up-regulated in Ring1a/b double knockout but was not changed or was only slightly changed in Eed cKO. Some genes from this group are negative regulators of cell adhesion. However, it was more intriguing when down-regulated genes were examined. First, significantly more genes were down-regulated in Ring1a/b double knockout than Eed cKO (628 genes vs. 88 genes), indicating that PRC1 could promote gene expression as well as suppress gene expression, while PRC2 mainly suppresses gene expression. Second, those down-regulated genes in Ring1a/b double knockout were enriched for regulators of cell adhesion and actin cytoskeleton organization, many of which are key adhesion molecules in skin epithelium. These results could explain the observed skin fragility phenotype seen in Ring1a/b double knockout. Finally, when they genetically deleted Pcgf2/4, which are key components of canonical PRC1, no signs of skin fragility were observed, lending a support to the notion that noncanonical PRC1 is responsible for the observed defects of Ring1a/b deletion. Indeed, ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) and ChIP-PCR signals of RYBP, a component that is exclusive to noncanonical PRC1, colocalized with the signals of histone H3K27ac, a chromatin mark associated with gene activation. These results revealed that PRC1 exists as distinct complexes in the skin epithelium and that canonical PRC1 functions differently than noncanonical PRC1 (Fig. 1).
Figure 1.
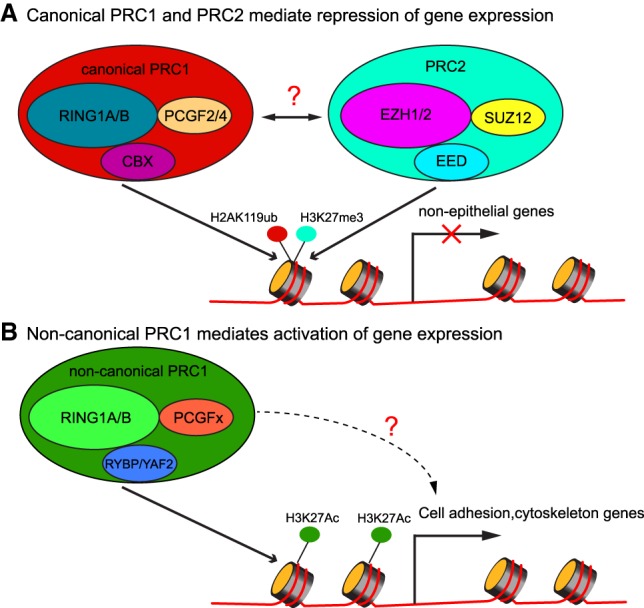
Schematic illustration of PRC1- and PRC2-mediated gene expression regulation. (A) Canonical PRC1 and PRC2 deposit histone H2AK119ub and histone H3K27me3 marks, respectively, and are correlated with repression of gene expression, mostly guarding against the expression of nonepithelial genes. (B) Noncanonical PRC1 colocalizes with histone H3K27ac, a mark for gene expression activation, and promotes the expression of cell adhesion and cytoskeleton genes.
The traditional view is that PRC1 and PRC2 cooperate to bind and compact chromatin and repress gene expression (Fig. 1A; Blackledge et al. 2015). Consistent with this view, Cohen et al. (2019) did find that the up-regulated cell adhesion and cytoskeleton organization genes were cobound by PRC1 and PRC2, indicated by RING1B and H3K27me3 ChIP-seq. However, in the down-regulated cell adhesion genes, their promoter often showed RING1B signals but not H3K27me3 or H2AK119ub signature, suggesting that those genes are direct targets of noncanonical PRC1 but not canonical PRC1 or PRC2. Although the underlying mechanism remains to be determined, these data suggest that noncanonical PRC1 may promote the expression of those genes (Fig. 1B). This possibility is particularly intriguing given the functional demarcation between the total PRC1 and canonical PRC1 based on the genetic evidence and because PRC1 has been widely recognized for its repressive function. Furthermore, because noncanonical PRC1 does not have CBX proteins as components (Blackledge et al. 2015) that recognize H3K27me3 marks, its recruitment to chromatin is most likely independent of PRC2. Future investigation will be required to elucidate the molecular basis of noncanonical PRC1-mediated gene activation and how it is recruited to chromatin. From the perspective of skin biology, it is fascinating that the loss of PRC1 directly interferes with the expression of cell adhesion and cytoskeleton genes, leading to the defects of skin fragility. These data formally link PRC1-mediated epigenetic regulation to cell adhesion and cytoskeleton organization, a fundamental property of epithelial cells. Altogether, this exciting study now lays a foundation to probe the mechanism of noncanonical PRC1 in promoting gene expression in mammalian somatic tissues.
Acknowledgments
Work in the Yi laboratory was supported by grants from National Institute of Health AR059697, AR059697-07S1, AR066703, and AR071435.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.322651.118.
References
- Blackledge NP, Rose NR, Klose RJ. 2015. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol 16: 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen I, Zhao D, Menon G, Nakayama M, Koseki H, Zheng D, Ezhkova E. 2019. PRC1 preserves epidermal tissue integrity independently of PRC2. Genes Dev (this issue) 10.1101/gad.319939.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber KL, Perdigoto CN, Valdes VJ, Santoriello FJ, Cohen I, Ezhkova E. 2016. Dissecting the roles of Polycomb repressive complex 2 subunits in the control of skin development. J Invest Dermatol 136: 1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien W-H, Stokes N, Pasolli HA, Silva JM, Fuchs E. 2011. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 25: 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Has C, Bruckner-Tuderman L. 2014. The genetics of skin fragility. Annu Rev Genomics Hum Genet 15: 245–268. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]


