Abstract
Lactic acid bacteria (LAB) is one of the main classes of acid-producing organisms in the food industry, and they play a vital part in many food and feed fermentations. We isolated and performed molecular identification of LAB from raw camel's milk and assessed their protective effects against pathogenicity induced by Staphylococcus aureus and Escherichia coli. Fourteen samples of camel's milk were obtained from several districts under aseptic conditions. Bacteria isolation was performed by plating the samples on selective media. Isolates were identified by amplification of the 16S ribosomal RNA by PCR and sequencing. A total of 32 isolates were randomly picked, eight of which were analysed in this study. On the basis of phenotypic and genotypic methods, isolated LAB was included Leuconostoc mesenteroides, Lactobacillus plantarum, Weissella paramesenteroides and Weissella confuse. Antagonistic activity of isolated LAB against two pathogenic bacteria showed that they had more inhibitory activity against S. aureus subsp. aureus PTCC 1431 than E. coli ATCC 25922. This study discovered that raw camel's milk obtained from three districts of Kerman province contain LAB bacteria that have antagonistic properties on S. aureus.
Keywords: Camel, lactic acid bacteria, milk, probiotic
Introduction
Lactic acid bacteria (LAB) are comprehensively dispersed in nature and occur normally as indigenous microflora in raw milk that assumes an essential part in several food and feed fermentations. LAB are a group of non–spore-forming Gram-positive bacteria that produce lactic acid as the main end product among the fermentation of carbohydrates and are utilized as starter culture [1]. Strains of the genera Lactobacillus [2], Enterococcus [3] and Bifidobacterium [4] are the most commonly studied and most widely used probiotic bacteria. Probiotics are defined as live microorganisms that, when administered in adequate amounts, confer a health benefit to the host [5]. Nowadays there is an expanding need for new strains of LAB that convey probiotic qualities and that have an impact on well-being as well as on human and animal health. Such LAB can be acquired from different natural ecologic niches which stay unexploited. Also, LAB are a focal point of intensive universal research for their fundamental part in most fermented food as well as for their capacity to create different antimicrobial compounds advancing probiotic properties, reduction of serum cholesterol, stabilization of gut microflora, alleviation of lactose intolerance and stimulation of the immune system [6].
Camel's milk, which has a high probiotic potential, is a source from which LAB can be isolated. In vitro activity of camel's milk against Gram-positive and -negative bacteria has been broadly reported [7]. On the one hand, Staphylococcus aureus, an important Gram-positive cocci, is responsible for several infections like endocarditis, septic arthritis, pneumonia and mastitis in animals; however, it may occur as a commensal [8]. Infections caused by S. aureus can spread through skin-to-skin contact with an infected individual by producing hyaluronidase that destroys tissues, through contact with pus from an infected wound and through contact with objects such as clothing, sheets, towels or athletic equipment used by an infected individual. On the other hand, Escherichia coli is a Gram-negative bacterium that is established in the intestinal tract [9]. Pathogenic strains of this bacilli are differentiated from normal flora by their possession of virulence factors like exotoxins [10].
We thus undertook to isolate and identify LAB from raw camel's milk from different locations in Kerman province, Iran. In addition, we assessed the natural protection of camel's milk against S. aureus and E. coli ATCC 25922.
Materials and Methods
Camel's milk samples
Fourteen samples of raw camel's milk were obtained randomly from apparently healthy camels owned by nomads from different districts of Kerman province, Iran. The milk samples were obtained directly from the udder. The udder was washed with distilled water before collection and dried with single-service towels. The initial three streams of milk were discarded. Samples were then gathered directly into sterile tubes, stored in an icebox and transferred to the lab within 24 hours.
Isolation and enumeration of LAB
To make an initial dilution (10−1), 10 mL camel's milk was homogenized with 90 mL of saline water (8.5 g/L). The suspension was utilized for making appropriate serial dilutions up to 10−8 by incorporating 1 mL into 9 mL of sterile saline water in sterile tubes. One milliliter of these dilutions was pour-plated in M17 agar, de Man Rogosa (MRS) agar [11] and kanamycin aesculin azide agar (Merck, Germany). After incubation at 37°C for 48 hours under anaerobic condition (using anaerocult A gas packs; Merck), individual different colonies were phenotypically selected. Colony enumeration was conducted after incubation and was recorded as CFU per liter of milk.
Identification of isolates
All isolates were microscopically investigated for cellular arrangement, Gram staining and cell morphology. Only Gram-positive and catalase-negative isolates were identified at the species level. Biochemical tests were used in the identification study as described elsewhere [12], [13].
Genotypic identification using 16S ribosomal RNA
The genomic DNA of the presumptive LAB strain was isolated utilizing the DNA extraction and purification kit according to the manufacturer's instructions (SinaClon, Karaj, Iran). The oligonucleotides used for PCR amplification of 16S ribosomal RNA (rRNA) gene were as follows: forward, 5′-AGAGTTTGATCMTGGCTCAG-3′, and reverse, 5′-TACGGYTACCTTGTTACGACTT-3′ [14], with an expected product size of 1500 bp. Sequencing of PCR product was made by Takapozist (Tehran, Iran; on behalf of Bioneer, Daejeon, Korea). All the sequencing results were translated by LaserGene programming (DNASTAR, Madison, WI, USA) including MegAlign, EditSeq and SeqMan software.
Proteolytic activity in reconstituted skim milk
All the bacterial strains were incubated overnight at 37°C in MRS broth (Merck, Germany). After that, bacterial strains were cultured on MRS agar plates containing 10% skim milk. Subsequently, the plates were incubated at 30°C for 5 days. Proteolytic activity of the strains was considered positive when a halo was observed around the growth site of the bacteria [15].
Determining antagonistic activity of isolated LAB using in vitro tests
The antagonistic activity of LAB isolated in the study against E. coli ATCC 25922 and S. aureus PTCC 1431 was determined using disc diffusion and agar spot methods [16]. Before directing the test, the possible LAB strains were cultured in MRS broth medium and incubated at 37°C for 24 hours. Then 4 μL of each bacterial isolate were spotted on the surface of MRS agar medium and incubated at 37°C for another 24 hours. Overnight culture of each strains was inoculated (1% v/v) in 15 mL of soft nutrient agar (containing 0.7% agar) and poured onto the inoculated MRS agar plates [17]. Additionally, sterile Whatman paper discs (6.0 mm) were set on the agar plate, and each disc was promptly inoculated with 15 μL of a broth culture of every inhibitor bacteria. After overnight incubation at 37°C, the antimicrobial activity of tested bacteria was determined by measuring the diameter of the inhibition (clear) zones surrounding the colonies. A clear zone of more than 1 mm around a spot was scored as positive [16]. Each test was performed twice. The inhibitory effect of MRS was tested as a negative control on each plate.
Results
A total of 32 isolates were randomly chosen from 14 collected samples. They were all Gram-positive bacteria; in addition, a considerable number of yeast and catalase-positive bacteria were observed. Only eight isolates were stable after subculture and purification; these were consequently studied. After phenotypical examination, the isolates were classified into three groups of Lactobacillus, Leuconostoc and Weissella genera. LAB were counted in camel's milk utilizing ordinary media by the classic method. Concerning presumptive Leuconostoc, the number varied from 4.7 × 102 to 4.9 × 107 CFU/mL with an average of 6.2 × 106 CFU/mL. The presumptive lactobacilli levels ranged from 1.5 × 102 to 5.1 × 107 CFU/mL with an average of 5.9 × 106 CFU/mL. The presumptive Weissella counts ranged from 4.9 × 102 to 5.8 × 107 CFU/mL with an average of 7.3 × 106 CFU/mL.
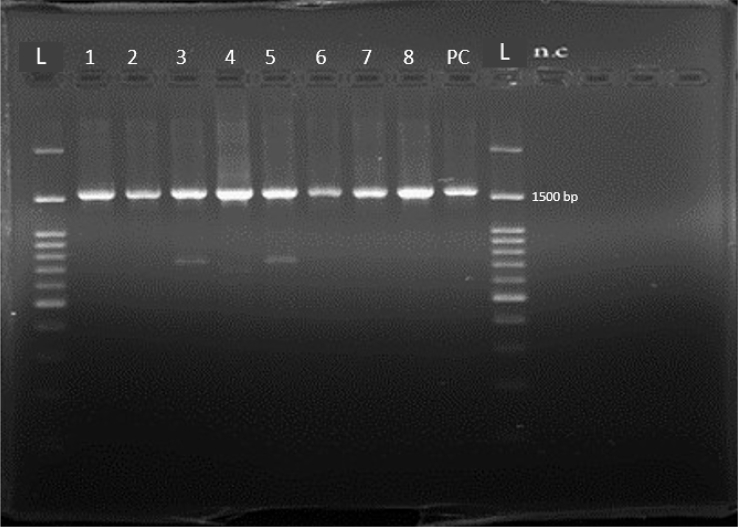
Representative isolates from every profile were examined for their 16S rRNA sequences (Fig. 1). Homology searches of the sequences uncovered (with 97–100% homology) that profile 1 belonged to Leuconostoc mesenteroides strain CAU1131 (accession number MH734173), profile 2 belonged to Leuconostoc mesenteroides strain JCM11043 (accession number MH734174), profile 3 belonged to Lactobacillus plantarum (accession number MH734175), profile 4 belonged to Weissella Paramesenteroides strain HYM110 (accession number MH734176), profile 5 belonged to Lactobacillus plantarum strain LAB12 (accession number MH734177), profile 6 belonged to Weissella paramesenteroides strain HYM110 (accession number MH734178), profile 7 belonged to Lactobacillus plantarum strain CAU2522 (accession number MH734179) and profile 8 belonged to Weissella confuse strain JS-7-1 (accession number MH734180). The results of proteolytic activity were reported negative in skim milk medium (Table 1).
Fig. 1.
PCR amplification of 16S rRNA sequences used in this study by universal primers. NC, negative control; PC, positive control; rRNA, ribosomal RNA.
Table 1.
Antagonistic and proteolytic activity of isolated lactic acid bacteria using in vitro tests
| Isolate no. | Isolate | Staphylococcus aureus subsp. aureus PTCC 1431 | Escherichia coli O157 H7 | Proteolytic activity |
|---|---|---|---|---|
| 1 | Leuconostoc mesenteroides strain CAU1131 | + | + | − |
| 2 | Leuconostoc mesenteroides strain JCM11043 | + | − | − |
| 3 | Lactobacillus plantarum | + | + | − |
| 4 | Weissella paramesenteroides strain HYM110 | + | − | − |
| 5 | Lactobacillus plantarum strain LAB12 | + | − | − |
| 6 | Weissella paramesenteroides strain HYM110 | + | − | − |
| 7 | Lactobacillus plantarum strain CAU2522 | + | + | − |
| 8 | Weissella confuse strain JS-7-1 | − | − | − |
Table 1 shows the inhibitory activity of isolated LAB against two pathogenic bacteria in the present investigation. Overall, the examined isolates had antagonistic effect against pathogenic bacteria because the inhibition was measured positive if the diameter of the clear zone around the colonies was 0.5 mm or larger. Isolates 1 to 7 had maximal inhibitory activity against S. aureus subsp. aureus PTCC 1431; isolates 1, 3 and 7 had inhibitory activity against E. coli ATCC 25922 (Table 1).
Discussion
Intestinal pathogenic bacteria are one of the most main causes of food poisoning and diarrhoea, particularly in developing countries. Because of drug resistance in these bacteria, the therapeutic effects of these infections are decreased and need to be replaced with new strategies. In this study, we were able to detect and isolate different LAB such as those of the Leuconostoc, Lactobacillus and Weissella genera.
Several studies have suggested the potential of probiotics to improve the host's health in many ways, such as by boosting the immune system through the formation of lymphocytes in many organs, by ameliorating the effects of dyslipidaemia and type 2 diabetes and by presenting antimicrobial effect in vivo, among other functional properties [18], [19]. Studies have shown that probiotic dairy products containing LAB may be a good way to enhance the blood lipid profile and could be used to improve antioxidant defences [19], [20], [21].
Because of the bactericidal impact of protease-sensitive bacteriocins, LAB have appeared to have inhibitory activities, mostly towards Gram-positive pathogens and closely related bacteria [22]. S. aureus causes food poisoning by releasing enterotoxins into sustenance and toxic shock syndrome by releasing pyrogenic exotoxins into the circulatory system. These conditions have started a search for naturally produced biopreservatives. One area that has created much interest is the utilization of antimicrobial metabolites from LAB used in food fermentation. Some of the metabolites of these bacteria, including proteinaceous substances bacteriocins, diacetyls, lactic acid and hydrogen peroxide, have an antimicrobial effect against many pathogenic bacteria and food spoilage [23], [24]. LAB were also able to control the growth of Gram-negative pathogens, including foodborne pathogens [25]. This report showed that most of the isolates had an inhibitory effect on S. aureus subsp. aureus PTCC 1431, while only few had an inhibitory effect on E. coli ATCC 25922. In addition, the proteolytic activity was negative. Proteolytic activity is one speculative marker of the health-promoting advantages that could be claimed in the consumption of fermented products.
From the perspective of probiotic potential, L. mesenteroides, L. plantarum, W. paramesenteroides and W. confuse isolated from camel's milk were chosen as LAB. Yateem et al. [17] in a survey on camel's milk showed that the presence of L. lactis and L. pentosus act as probiotic LAB in raw camel's milk. An investigation was carried out for isolation and molecular identification of LAB from camel's milk and assessment of their probiotic properties in the Golestan province of Iran; the most frequently isolated LAB was enterococci, while Leuconostoc, Weissella, pediococci and lactobacilli were found less frequently [26]. In addition, these results demonstrated that antimicrobial activity of the examined isolates was remarkable against E. coli and Bacillus cereus and Pediococcus pentosaceus showed the most antibacterial activity. In a 2015 study, the protective effects of camel's milk towards pathogenicity induced by S. aureus and E. coli in Wistar rats were also examined [27]. Results showed that camel's milk had synergistic activity with ciprofloxacin against these two bacteria to decrease the dose of antibiotics and reduce bacterial resistance.
Brasca et al. (‘Metabolic characteristics of lactic acid bacteria from camel milk,’ paper presented at the 5th International Dairy Federation (IDF) Symposium on Cheese Ripening, 9–13 March 2008, Bern, Switzerland) purified 92 isolates of LAB utilizing frozen camel's milk, which classified as 44.56% and 55.43% of rods and cocci isolates, respectively, while Ashmaig et al. [28] isolated 24 LAB from 12 samples of gariss (fermented camel's milk) in the Sudan. The isolates were classified into 66.6% rods and 33.3% cocci. Likewise, Khay et al. [29] isolated a total of 450 cultures from 25 samples of dromedary milk collected from the Laâyoune region of Morocco. Of these, 30 were determined to be LAB. The presence of yeast in the examined samples is perhaps a result of udder skin contamination, as specified by earlier studies [17], [25].
In conclusion, the results obtained from this investigation recommend camel's milk as a source for the isolation of potentially probiotic LAB strains, which may have antagonistic properties against enteric pathogenic bacteria. More extensive investigations are expected to affirm these proposed medical advantages and to support the use of camel's milk.
Conflict of interest
None declared.
References
- 1.Singh G., Sharma R.R. Dominating species of lactobacilli and leuconostocs present among the lactic acid bacteria of milk of different cattles. Asian J Exp Sci. 2009;23:173–179. [Google Scholar]
- 2.Dahroud B.D., Mokarram R.R., Khiabani M.S., Hamishehkar H., Bialvaei A.Z., Yousefi M. Low intensity ultrasound increases the fermentation efficiency of Lactobacillus casei subsp. casei ATTC 39392. Int J Biol Macromol. 2016;86:462–467. doi: 10.1016/j.ijbiomac.2016.01.103. [DOI] [PubMed] [Google Scholar]
- 3.Ogier J.C., Serror P. Safety assessment of dairy microorganisms: the Enterococcus genus. Int J Food Microbiol. 2008;126:291–301. doi: 10.1016/j.ijfoodmicro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 4.Yateem A., Balba M., Al-Surrayai T., Al-Mutairi B., Al-Daher R. Isolation of lactic acid bacteria with probiotic potential from camel milk. Int J Dairy Sci. 2008;3:194–199. [Google Scholar]
- 5.Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 6.Khedid K., Faid M., Mokhtari A., Soulaymani A., Zinedine A. Characterization of lactic acid bacteria isolated from the one humped camel milk produced in Morocco. Microbiol Res. 2009;164:81–91. doi: 10.1016/j.micres.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Benkerroum N., Mekkaoui M., Bennani N., Hidane K. Antimicrobial activity of camel’s milk against pathogenic strains of Escherichia coli and Listeria monocytogenes. Int J Dairy Technol. 2004;57:39–43. [Google Scholar]
- 8.Cimolai N. MRSA and the environment: implications for comprehensive control measures. Eur J Clin Microbiol Infect Dis. 2008;27:481–493. doi: 10.1007/s10096-008-0471-0. [DOI] [PubMed] [Google Scholar]
- 9.Bialvaei A.Z., Kafil H.S., Asgharzadeh M., Aghazadeh M., Yousefi M. CTX-M extended-spectrum β-lactamase–producing Klebsiella spp., Salmonella spp., Shigella spp. and Escherichia coli isolates in Iranian hospitals. Braz J Microbiol. 2016;47:706–711. doi: 10.1016/j.bjm.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welinder-Olsson C., Kaijser B. Enterohemorrhagic Escherichia coli (EHEC) Scand J Infect Dis. 2005;37:405–416. doi: 10.1080/00365540510038523. [DOI] [PubMed] [Google Scholar]
- 11.De Man J.C., Rogosa M., Sharpe M.E. A medium for the cultivation of lactobacilli. J Appl Microbiol. 1960;23:130–135. [Google Scholar]
- 12.Harrigan W.F., McCance M.E. Academic Press; London: 1976. A simple key for the identification of Gram-positive bacteria. [Google Scholar]
- 13.Manero A., Blanch A.R. Identification of Enterococcus spp. with a biochemical key. Appl Environ Microbiol. 1999;65:4425–4430. doi: 10.1128/aem.65.10.4425-4430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagacé L., Pitre M., Jacques M., Roy D. Identification of the bacterial community of maple sap by using amplified ribosomal DNA (rDNA) restriction analysis and rDNA sequencing. Appl Environ Microbiol. 2004;70:2052–2060. doi: 10.1128/AEM.70.4.2052-2060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shihata A., Shah N.P. Proteolytic profiles of yogurt and probiotic bacteria. Int Dairy J. 2000;10:401–408. [Google Scholar]
- 16.Jacobsen C.N., Nielsen V.R., Hayford A., Møller P.L., Michaelsen K., Paerregaard A. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol. 1999;65:4949–4956. doi: 10.1128/aem.65.11.4949-4956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yavuzdurmaz H. Izmir Institute of Technology; 2007. Isolation, characterization, determination of probiotic properties of lactic acid bacteria from human milk.http://library.iyte.edu.tr/tezler/master/gidamuh/T000712.pdf Unbublished MS thesis. Available at: [Google Scholar]
- 18.Balthazar C.F., Silva H.L., Esmerino E.A., Rocha R.S., Moraes J., Carmo M.A. The addition of inulin and Lactobacillus casei 01 in sheep milk ice cream. Food Chem. 2018;246:464–472. doi: 10.1016/j.foodchem.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Lollo P.C., Morato P.N., Moura C.S., Almada C.N., Felicio T.L., Esmerino E.A. Hypertension parameters are attenuated by the continuous consumption of probiotic Minas cheese. Food Res Int. 2015;76:611–617. doi: 10.1016/j.foodres.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 20.Moura C., Lollo P., Morato P., Esmerino E., Margalho L., Santos-Junior V. Assessment of antioxidant activity, lipid profile, general biochemical and immune system responses of Wistar rats fed with dairy dessert containing Lactobacillus acidophilus La-5. Food Res Int. 2016;90:275–280. doi: 10.1016/j.foodres.2016.10.042. [DOI] [PubMed] [Google Scholar]
- 21.Sperry M.F., Silva H.L., Balthazar C.F., Esmerino E.A., Verruck S., Prudencio E.S. Probiotic Minas Frescal cheese added with L. casei 01: physicochemical and bioactivity characterization and effects on hematological/biochemical parameters of hypertensive overweighted women—a randomized double-blind pilot trial. J Funct Foods. 2018;45:435–443. [Google Scholar]
- 22.Jack R.W., Tagg J.R., Ray B. Bacteriocins of Gram-positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barefoot S.F., Klaenhammer T.R. Detection and activity of lactacin B, a bacteriocin produced by Lactobacillus acidophilus. Appl Environ Microbiol. 1983;45:1808–1815. doi: 10.1128/aem.45.6.1808-1815.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daeschel M.A. Antimicrobial substances from lactic acid bacteria for use food preservatives. Food Technol. 1989;43:164–167. [Google Scholar]
- 25.Hamed E., Elattar A. Identification and some probiotic potential of lactic acid bacteria isolated from Egyptian camels milk. Life Sci J. 2013;10:1952–1961. [Google Scholar]
- 26.Davati N., Yazdi F.T., Zibaee S., Shahidi F., Edalatian M.R. Study of lactic acid bacteria community from raw milk of Iranian one humped camel and evaluation of their probiotic properties. Jundishapur J Microbiol. 2015;8 doi: 10.5812/jjm.8(5)2015.16750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yassin M.H., Soliman M.M., Mostafa S.A.E., Ali H.A.M. Antimicrobial effects of camel milk against some bacterial pathogens. J Food Nutr Res. 2015;3:162–168. [Google Scholar]
- 28.Ashmaig A., Hasan A., El Gaali E. Identification of lactic acid bacteria isolated from traditional Sudanese fermented camels milk (Gariss) Afr J Microbiol Res. 2009;3:451–457. [Google Scholar]
- 29.Khay E.O., Idaomar M., Castro L., Bernárdez P., Senhaji N., Abrini J. Antimicrobial activities of the bacteriocin-like substances produced by lactic acid bacteria isolated from Moroccan dromedary milk. Afr J Biotechnol. 2011;10:10447–10455. [Google Scholar]