Abstract
Osteoblast differentiation is controlled by transcription factor RUNX2 which temporally activates or represses several bone-related genes, including those encoding extracellular matrix proteins or factors that control cell-cell and cell-matrix interactions. Cell-cell communication in the many skeletal pericellular micro-niches is critical for bone development and involves paracrine secretion of growth factors and morphogens. This paracrine signaling is in part regulated by ‘A Disintegrin And Metalloproteinase’ (ADAM) proteins. These cell membrane-associated metalloproteinases support proteolytic release (‘shedding’) of protein ectodomains residing at the cell surface. We analyzed microarray and RNA-sequencing data for Adam genes and show that Adam17, Adam10 and Adam9 are stimulated during BMP2 mediated induction of osteogenic differentiation and are robustly expressed in human osteoblastic cells. ADAM17, which was initially identified as a tumor necrosis factor alpha (TNFα) converting enzyme also called (TACE), regulates TNFα-signaling pathway, which inhibits osteoblast differentiation. We demonstrate that Adam17 expression is suppressed by RUNX2 during osteoblast differentiation through the proximal Adam17 promoter region (−0.4 kb) containing two functional RUNX2 binding motifs. Adam17 downregulation during osteoblast differentiation is paralleled by increased RUNX2 expression, cytoplasmic-nuclear translocation and enhanced binding to the Adam17 proximal promoter. Forced expression of Adam17 reduces Runx2 and Alpl expression, indicating that Adam17 may negatively modulate osteoblast differentiation. These findings suggest a novel regulatory mechanism involving a reciprocal Runx2-Adam17 negative feedback loop to regulate progression through osteoblast differentiation. Our results suggest that RUNX2 may control paracrine signaling through regulation of ectodomain shedding at the cell surface of osteoblasts by directly suppressing Adam17 expression.
Keywords: Osteoblast differentiation, RUNX2, transcriptional regulation, ADAM genes, ADAM17
INTRODUCTION
Differentiation of osteoblastic lineage cells requires the complex genetic and biochemical interplay of gene regulatory signaling pathways, including BMPs, WNTs, FGFs, PTH, IGF, GPNMB/osteoactivin, CTGF/CCN2 and several key transcription factors (e.g., RUNX2, OSX/SP7, DLX5, ATF4, SATB2) (Stein et al., 2004; Long, 2011; Yang et al., 2011; Zhu et al., 2011; Artigas et al., 2014; Shimizu et al., 2014; Sondag et al., 2014; Hendesi et al., 2015; Hurley et al., 2016; Li et al., 2016). The essential role of RUNX2 during osteoblast maturation is reflected by severe bone phenotypes resulting from genetic mutations that abrogate the normal functions of RUNX2, its partner CBFB, or its downstream target Osterix/SP7 in mouse models and human disease (Stein et al., 2004; Liu et al., 2011; Long, 2011; Yoshida et al., 2012; Okura et al., 2014; Lim et al., 2015; Qin et al., 2015; Takarada et al., 2016). Consistent with the central role of RUNX2 in skeletal development and bone formation, the expression of the Runx2 gene is tightly controlled by chromatin-dependent mechanisms, multiple protein/DNA interactions and three-dimensional chromatin loops (Hovhannisyan et al., 2013; Barutcu et al., 2014; Kawane et al., 2014; Tai et al., 2014; Rojas et al., 2015; Aguilar et al., 2016). In addition, Runx2 expression is post-transcriptionally controlled by multiple miRNAs that regulate osteoblast differentiation (Kapinas et al., 2010; Zhang et al., 2011; Lian et al., 2012; Zhang et al., 2012; Smith et al., 2016) and the mitotic partitioning of Runx2 mRNA in proliferating osteoblasts (Varela et al., 2016). The broad biological functions of RUNX2 in osteoblasts are reflected by its intrinsic ability to activate a large cohort of target genes in a cell-stage specific manner (Teplyuk et al., 2009a; Teplyuk et al., 2009b; Hawse et al., 2011; Purcell et al., 2012; van der Deen et al., 2012; McGee-Lawrence et al., 2013a; McGee-Lawrence et al., 2013b; Yang et al., 2013; Meyer et al., 2014; Wu et al., 2014).
The ADAM (A Disintegrin and Metalloproteinase) proteins are a family of transmembrane metalloproteinases containing an extracellular catalytic domain implicated in ectodomain shedding of different cell surface proteins (i.e, growth factors, cytokines, receptors and cell adhesion molecules). The ADAM proteins are closely related to ADAMTS (A Disintegrin And Metalloproteinase with Thrombospontin motifs), a group of secreted metalloproteinases that mediate proteolytic processing or degradation of specific extracellular matrix (ECM) molecules (e.g. pro-collagen and aggrecan). ADAM proteins may affect the bone micro-niche through juxtacrine and paracrine effects by cleaving extracellular regions of cell surface associated proteins (ectodomain shedding). For example, these proteins have potent effects on multiple regulatory pathways including those involving fibroblast growth factor receptor 2 (FGFR2), insulin-like growth factor binding protein 5 (IGFBP5), interleukin-6 receptor (IL6R) and Notch receptors in osteoblasts (Inoue et al., 1998; Dallas et al., 1999; Mohan et al., 2002; Pan et al., 2004; Franchimont et al., 2005; Chan et al., 2012; Tan et al., 2016). Other roles that ADAMs perform in the bone micro-environment include effects on Notch modulators, receptor activator of NF-kappaB ligand (RANKL), as well as other mechanisms in osteoclasts, chondrocytes and bone-metastatic tumor cells (Hikita et al., 2006; Karadag et al., 2006; Hall et al., 2013; Zhou et al., 2014). To gain insight into the biological roles of ADAM proteins in osteoblastogenesis and bone formation, it is necessary to assess which Adam genes are expressed in bone cells.
Previous studies have shown that ADAM (ADAM8, 9, 10, 12, 15, 17 and 19) and ADAMTS (ADAMTS1, 4 and 5) family members are expressed in osteoblastic cells and bone tissue (Harris et. al., 1997; Kurisaki et. al., 1998; Miles et. al., 2000; Mohan et. al., 2002; Verrier et. al., 2004; Lind et. al., 2005; Nakamura et. al., 2005; Rehn et. al., 2007) and has been demonstrated that some of them shown significant changes in its expression levels during osteoblast differentiation (Inoue et. al., 1998; Govoni et. al., 2006). Moreover, Adamts4 and Adamts5 genes have been reported to be direct downstream targets of RUNX2 in chondrocytes (Thirunavukkarasu et. al., 2006; Thirunavukkarasu et. al., 2007; Lin et. al., 2009; Kadri et. al., 2010; Tetsunaga et. al., 2011). Collectively, these data suggest that Adam and Adamts genes are differentially expressed during osteoblast differentiation and that RUNX2 may transcriptionally control the expression of members of the ADAM and ADAMTS families. Particularly, conditional inactivation of Adam17 gene in osteochondroprogenitor cells results in several defects including increased osteoblast numbers, suggesting potential functions related to bone formation (Horiuchi et. al., 2009). ADAM17, also known as TNF-α converting enzyme (TACE), is a membrane-anchored metalloproteinase that cleaves diverse cell surface proteins (i.e, cytokines, cell adhesion proteins and cell growth factor receptors) including, interestingly, the type II membrane-bound precursor of TNFα, a known inhibitor of osteoblast differentiation (Gilbert et al., 2000). Because of pleiotropic defects observed in Adam17 conditional knockout mice, we dissected molecular mechanisms controlling Adam17 functions using cell culture models.
In this study, we show that osteoblastic cell types express multiple members of the ADAM family and the relatively abundantly expressed members ADAM9, ADAM10 and ADAM17 exhibit significant changes in gene expression levels during osteogenic differentiation. We show that RUNX2 directly attenuates Adam17 gene expression via selective recruitment of RUNX2 through a functional binding site contained in the proximal promoter of the Adam17 gene and that the C-terminal transactivation region of RUNX2 is essential for repression of the Adam17 promoter. Our study suggests that RUNX2 may control ectodomain shedding in the pericellular micro-environment of osteoblasts to control the cell-cell communication through the regulation of Adam17 gene expression.
MATERIALS AND METHODS
Cell Culture
Mouse MC3T3–E1 osteoblasts, mouse pre-myogenic mesenchymal C2C12 precursor cells, rat osteosarcoma ROS17/2.8 cells and human osteosarcoma cell lines (SAOS-2, MG63, U2OS, G292, HOS and 143B cells) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Immortalized mouse Runx2 null (Runx2−/−) calvarial osteoprogenitor cells were described previously (Bae et al., 2007). Cells were maintained in culture medium supplemented with 5–15% fetal bovine serum (FBS) (HyClone Laboratories Inc, Logan, UT, USA) plus 2mM L-glutamine and a penicillin-streptomycin cocktail at 37°C and 5% CO2 humidified atmosphere MC3T3–E1 and Runx2−/− cells were cultured in αMEM medium (Gibco, Grand Island, NY, USA) supplemented with 10% FBS. C2C12 cells were cultured in DMEM medium (Gibco, Grand Island, NY, USA) supplemented with 10% FBS. ROS17/2.8 cell were grown in F12 medium (Gibco, Grand Island, NY, USA) supplemented with 5% FBS. SAOS cells were maintained in McCoy’s medium (Sigma-Aldrich, St Louis, MO, USA) supplemented with 15% FBS. U2OS and G292 cells were cultured in McCoy’s medium with 10% FBS. MG63 and HOS cells were grown in DMEM medium with 10% FBS. 143B cells were maintained in DMEM medium, 1mM sodium pyruvate, 100 μg/ml of bromodeoxyuridine and 10% FBS. The growth medium was changed every 2 days. Primary osteoblastic outgrowth cultures for RNA-seq analysis were obtained from distal femur or proximal tibia bone specimens obtained as surgical waste samples with approval of the Mayo Clinic Institutional Review Board. Samples were manually minced and crushed with a scalpel and cells were allowed to grow for one passage until confluence in standard media using procedures described previously (Lewallen et al., 2016).
Osteoblast differentiation
For in vitro differentiation studies, MC3T3–E1 cells were plated in 100-mm or 60-mm plates or in six well plates and grown in regular medium up to confluence. Confluent cells were treated with 0.1 μM dexamethasone, 10 mM β-glycerophosphate, and 50 Ug/ml ascorbic acid in fresh regular media and cultured for 22 days. Media was changed every two days for the remainder of the experiment and cells were harvested at selected time points (at days 0, 4, 8, 12 16 and 22 of their osteogenic differentiation) for western blot, RT-PCR and immunohistochemistry. Cell cycle exit was determined by monitoring cyclin D1 (CCND1) expression. The capacity of cells to differentiate into the osteoblastic lineage was evaluated by monitoring Runx2 and alkaline phosphatase (Alpl) mRNA levels, as well as RUNX2 levels and ALPL activity.
DNA transfection and adenoviral infection
Subconfluent Runx2−/− cells or MC3T3–E1 cells were transiently transfected with pcDNA-Runx2, deletion mutant Runx2 1–361 (Runx2 ΔC), pcDNA-Adam17 or pcDNA-empty vector (control) using Lipofectamin 2000® reagent (Invitrogene, Carlsbad, CA, USA) according to the manufacturer’s recommendations. Adenoviral delivery of vector containing the cDNA of Runx2 coupled to Green Fluorescent Protein (GFP) (Runx2-IRES-gfp) under control of the CMV5 promoter was used as previously described (Pratap et al., 2003). Preparation and purification of virus were performed according to the manufacturer’s protocols (Promega, Madison, WI, USA). For control of infection, the same Adenovirus vector carrying gfp was used. The Adenovirus Runx2 (Adv-Runx2) contains both the gfp cassette and the Runx2 cDNA in forward orientation (+) and Adenovirus Vector (Adv-Vector) contains the gfp cassette in the forward orientation (+) and the Runx2 cDNA in reverse orientation (−) (Pratap et al., 2003). Runx2−/− cells were plated for infections in 60 mm plates at a density of 3 × 105 cells/plate and cultured in DMEM with 10% FBS. After 24 h, cells were infected at 60–70% of confluence with 30 × 1010 OPU/ml (optical particle unit) of each virus in 900 μl of DMEM supplemented with 1% FBS for 4 h. Upon addition of 600 μl of media containing 1% FBS, cells were incubated for an additional 10 h. After adenoviral infection, cells were grown for 24 to 72 hrs. Infection efficiencies were assessed by activity of GFP using a Nikon Diaphot inverted fluorescence microscope.
Western blot analysis
RUNX2, ADAM17, CCND1 and β-actin/ACTB were analyzed by western blot analysis as described previously (Galindo et al., 2005; Galindo et al., 2007). Briefly, equal amounts of total cellular protein collected in the presence of the proteasome inhibitor MG132 (Calbiochem, San Diego, CA, USA) and Complete cocktail of protease inhibitor (Roche Diagnostics, Mannhein, Germany) were resolved in 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Perkin Elmer, Boston, MA, USA). Blots were incubated with a 1:2,000 dilution of each primary antibody for 1 h. RUNX2-specific mouse monoclonal antibody 8G5 (MBL International, Woburn, MA, USA), ADAM17 rabbit polyclonal antibody (Anti-TACE 807–823) (Calbiochem, San Diego, CA, USA), CCND1 mouse monoclonal antibody DCS-6 (sc-20044) (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) and β-actin goat polyclonal antibody C-11 (sc-1615) (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) were acquired commercially. Membranes were incubated with the respective horseradish peroxidase-conjugated secondary antibody (sc-2005 anti-mouse or sc-2004 anti-rabbit or sc-2020 anti-goat) (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) for 1 h. Immuno-reactive protein bands were visualized on a Kodak BioMax Light film (Carestream Healt Inc, Rochester, NY, USA) using a chemiluminescence detection kit (Thermo Scientific, Rockford, IL, USA). Signal intensities were quantified by densitometry.
RNA-sequencing (RNA-seq) and semi-quantitative PCR
RNA-sequencing analysis of select samples from human osteosarcoma cells and osteoblastic bone-derived cells was performed using Illumina 2000 instrumentation and subsequently analyzed using a standardized bioinformatics pipeline (Bioinformatics Core at Mayo Clinic) as described in detail previously (Dudakovic et al., 2014). Expression values are denoted as reads per kilobase per million mapped reads (RPKM). Expression of select genes was visualized and validated by semi-quantitative PCR. Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s specifications. Total RNA (5 μg/lane) was separated in a 1% agarose-formaldehyde gel. Ethidium bromide staining of the gels was used to assess RNA quality of samples. Purified RNA (3 μg) was treated with RQ1RNase-Free DNase (Promega, Madison, WI, USA) and subjected to reverse transcription using random subjected hexamer primers (Promega, Madison, WI, USA) with M-MLV reverse transcriptase (Promega, Madison, WI, USA) according to the manufacturer’s recommendations. Gene expression was assessed by PCR using the following specific mouse (m) and human (h) gene primers (0.5 pmol/μl): m/r/hRunx2: F 5’-CCGCACGACAACCGCACCAT-3’, R 5’-CGCTCCGGCCCACAAATCTC-3’; m/hAdam9: F 5’- CAGACTGCTGTGAGAGAAG-3’, R 5’-CATTCCTGCAGTTCCACCA-3’; m/hAdam10: F 5’- CCTACGAATGAAGAGGGAC-3’, R 5’-ATCACAGCTTCTCGTGTTCC-3’; m/r/hAdam17: F 5’-GACATGAATGGCAAATGTGA-3’, R 5’-TGGACAAGAATGCTGAAAGGA-3’; mAlpl: F 5’-TCCATCCTGCGCTGGGCCAA-3’, R 5’-GGCCAGCAGTTCAGTGCGGT-3’; m/r/hGadph: F 5’-CCTTCATTGACCTCAACTA-3’, R 5’-GGCCATCCACAGTCTTCT-3’). PCR amplification of cDNAs was carried out using 1× PCR buffer (Promega, Madison, WI, USA) containing 0.2 mM dNTPs (Promega, Madison, WI, USA), 1.5 mM MgCL (Promega, Madison, WI, USA), 0.06 U/μl of Taq polymerase (Invitrogen, Carlsbad, CA, USA) by incubation for 5 min at 94°C and 20–30 amplification cycles of synthesis were applied to avoid product saturation (1 min at 94°C, 1 min at 52–62°C, and 1 min at 72°C), followed by a final extension step at 72°C for 6 min. Aliquots of the resulting products (5 μl) were visualized in 1% agarose gels by ethidium bromide staining.
Alkaline phosphatase (AP) activity
Differentiating MC3T3–E1 cells in 6-well plate or 60 mm plate were washed with PBS and then fixed with 4% paraformaldehyde for 30 sec. AP activity was detected by colorimetric reaction using the AP liquid substrate nitro blue tetrazolium and 5-bromo-4-chloro-3-idolyl phosphate (NBT/BCIP) (Roche Diagnostics, Mannhein, Germany). AP staining solution (NTB 0.4 mg/ml and BCIP 0.19 mg/ml in 100 mM Tris buffer, 50 mM MgSO4, pH 9.5) was added to each well and staining was carried out at 37 °C for 25 min. NBT/BCIP colorimetric reactions were stopped by aspirating the staining solution and rinsing the cells twice in PBS. AP positive cells were detected and photographed under a Zeiss Axiostar Plus light microscope.
Immunohistochemistry
MC3T3–E1 cells were washed twice with PBS and collected in a 1.5 ml tube using a scraper followed by centrifugation at 380 × g for 2 min. The cell pellets were fixed with formalin for 24 h, dehydrated and embedded in paraffin using standard procedures. Paraffin-embedded cells were sectioned (5 μm), adhered to glass slides, and rehydrated, and antigens were recovered by treatment with retrieval buffer (1mM Tris, 0.5 mM EGTA, pH 9.0). Sections were blocked with PBS supplemented with 1% bovine serum albumin. Then sections were incubated with 1:100 dilution of RUNX2 rabbit polyclonal antibody M-70 (sc-10758) (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) or ADAM17 rabbit polyclonal antibody, washed and then incubated with 1:200 dilution of the indicated biotylinated secondary antibody. Finally, antibodies bind to specific antigens were detected using a biotin-streptavidin detection system. Samples were observed under a Zeiss Axiostar Plus light microscope.
Luciferase reporter assays
For reporter assays, MC3T3–E1 cells were seeded at 8 × 104 cells/well in a six-well plate and transiently transfected 24–48 hours after plating at a cell density of 60–70% with 1 μg of the previously described construct Tace promoter/pGL2 luciferase reporter plasmid which contains the 2.304-kb mouse proximal Tace promoter fused to the firefly luciferase reporter (Charbonneau et. al., 2007). Alternatively, cells were transiently transfected with a series of Tace promoter 5′ deletants: pTace-1567, −903, −410, and −121, which were generated by digestion of the pTace-Luc vector with the appropriate restriction enzymes (Charbonneau et. al., 2007). Cells were co-transfected with 10 ng SV40/Ranilla luciferase plasmid (pRL-SV40) as an internal control. The promoterless pGL2 luciferase parent vector was used as a negative control. Lipofectamine 2000 was used as a transfection agent according to the manufacturer’s protocol and transfections were performed in absence of FBS and antibiotics. Medium was changed after 4 h to normal growth medium with FBS and antibiotics. Twenty-four hours after transfection, cells were harvested in 1× passive lysis buffer and luciferase activity was measured in cell lysates using the Dual Luciferase Reporter Assay System® kit (Promega, Madison, WI, USA) following the manufacturer’s instructions. Luminescent signal was quantified by a luminometer (Synergy® 2SL BioTek), and each measurement from the firefly luciferase construct was normalized using Renilla luciferase values.
Chromatin immunoprecipitation (ChIP) analysis
ChIP assays studies were performed as described previously (van der Deen et al., 2008). Pre-cleared chromatin fragments (200 to 300 bp) obtained from MC3T3–E1 cells were immunoprecipitated overnight with agitation using anti-RUNX2 M70 polyclonal antibodies. The PCR primers used to evaluate the proximal (−400/−167), middle (−930/−731) and distal (−1651/−1422) regions of the mouse Adam17 promoter by Quantitative real-time PCR were: (−400/−167): F 5’-GGACAGAGGCGAGAGAGAGA-3’, R 5’-GCTGAGAGCGGCTTAACTC-3’; (−930/−731): F 5’-GCAAGACATTCCACAACGAA-3’, R 5’-AGTGAACAGGAGCGACCATC-3’; (−1651/−1422): F 5’-AGTGGCACTCAGCCTTCCTA-3’, R 5’-GTACTCAACCCCTTGGGTCA-3’. Q-PCR was performed using the Brilliant II SYBR Green Q-PCR Master Mix (Agilent Technologies, Santa Clara, CA, USA) in an MX3000P spectrofluorometric thermal cycler (Stratagene-Agilent, Santa Clara, CA, USA) according to the manufacturer’s recommendations.
Statistical analysis
Data were represented as mean ± SEM with a minimum of three independent samples and analyzed using Student’s t-test. P value of less than 0.05 was considered statistically significant.
RESULTS
Identification of BMP-2-responsive and abundantly expressed ADAM genes in the osteogenic cell lineage.
To identify Adam genes that would be involved in the osteoblast differentiation process, we determined gene expression patterns of these genes at early stages of osteogenic lineage commitment in C2C12 mesenchymal progenitor cells stimulated with the Bone Morphogenetic Protein 2 (BMP-2). This cell culture model system focuses on the first stages of osteogenic lineage-commitment (1 to 24 hr) in response to BMP2/BMPR/SMAD signaling and the initial activation of osteoblast specific gene expression programs as evidenced by microarray gene expression profiling (Balint et al., 2003). Examination of these published data revealed that three Adam genes are upregulated (Adam17, Adam10 and Adam9) between 4–12 h after BMP-2 treatment (Fig. 1A) and three genes are downregulated (Adam8, Adam15 and Adam19) (Fig. 1B). Expression analysis of ADAM genes by RNA-seq reveals that the same group of BMP-2 responsive genes are the most abundantly expressed ADAM genes in primary human osteoblastic bone derived cells (hOBs) and collectively cover the ~97% of all ADAM transcripts in this cells (Fig. 1C). Further expression analysis of the three BMP2- upregulated ADAMs in other cell types revealed that expression of ADAM17, ADAM10 and ADAM9 is rather constant in human primary osteoblastic bone-derived cells (i.e., hOB1, hOB2 and hOB3), but its expression appears to be more variable in osteosarcoma cell types (Fig. 1D–F). Hence, cells in the osteogenic lineage express a limited number of ADAMs genes, and their expression may be selectively modulated in osteosarcoma cell types.
Figure 1. Adam17, Adam10 and Adam9 gene expression is up regulated during early stage of commitment and differentiation to the osteoblast phenotype and stably expressed in human immature osteoblastic cells.
To identify genes that immediately respond to osteogenic stimuli, we retrieved microarray data were retrieved for experiments with mouse C2C12 mesenchymal cells that were treated with 300 ng/ml of BMP-2 and analysed at distinct time points (0, 4, 8, 12, 16, 20, and 24 h). Data on ADAM genes were filtered for genes that show more than a 2 fold change in gene expresseion. This analysis revealed that the Adam17, Adam10 and Adam9 genes are upregulated for more than 2-fold (A), while three others are downregulated (Adam8, Adam15, Adam19) (B). (C) The bar graph shows average expression values (in RPKM; STD as error bar for n=3 human donors) that were rank ordered for relative expression based on RNA-seq analysis of human osteoblastic bone-derived cells from three different donors. The pie chart presented in the inset shows that the six most highly expressed ADAM genes (including the BMP-responsive ADAMs ADAM17, ADAM10 and ADAM9, which are presented in color) account for almost all (~97%) ADAM-related transcripts. (D) Results of RNA-seq analysis for each individual donor and select osteosarcoma cell lines (SaOS-2, MG63 and U2OS) as indicated. (E) Visual presentation and validation of gene expression data using semi-quantitative RT-PCR and ethidium bromide staining. ADAM17, ADAM10 and ADAM9 gene expression was assessed as indicated in human SAOS-2, MG63, U2OS, HOS, G292 and 143B immature osteoblast cells. The data shown are representative of three experiments with similar outcomes. The graphs show quantification of the RT-PCR data relative to Gapdh mRNA (D). All data are presented as mean ± SEM from three independent experiments.
RUNX2 modulates Adam17 gene expression
Because conditional in vivo inactivation of Adam17 gene exhibited several bone-related defects including increased osteoblast numbers and because ADAM17 protein is an important regulator of TNFα-signaling pathway that inhibits osteoblast differentiation, we focused our subsequent analyses on the molecular mechanism that control the expression of Adam17 during osteoblast differentiation.
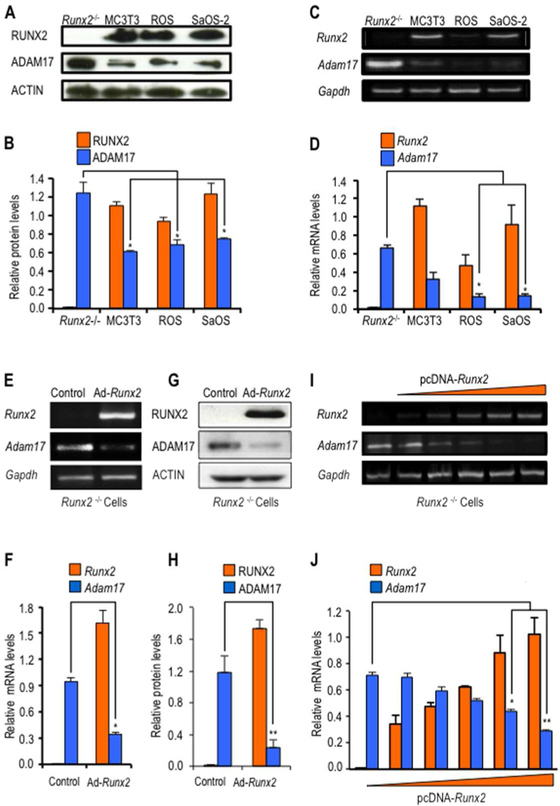
We first investigated whether there is a functional coupling between RUNX2 and Adam17 expression in Runx2-null calvarial cells compared to several osteoblastic cell lines. In agreement with our bioinformatics analyses, both Runx2 and Adam17 are expressed at detectable mRNA and protein levels in osteoblastic cell types (MC3T3–E1, ROS17/2.8 and SAOS-2 cells). Interestingly, depletion of RUNX2 expression in Runx2 null cells results in significant increases of Adam17 expression (Fig. 2A–D). This finding suggests that Adam17 expression is negatively regulated by RUNX2. This possibility is supported by a robust decrease in Adam17 expression induced by overexpression of Runx2 in the same Runx2-null cells (Fig. 2 E–H). Importantly, dosing experiments using different amounts of expression vector reveal that Adam17 expression is inversely proportional to the amount of exogenous Runx2 expressed in Runx2 null cells (Fig. 2 I and J). Hence, our data indicate that RUNX2 suppresses Adam17 mRNA and protein expression levels in osteogenic lineage cells.
Figure 2. Adam17 gene expression is negatively regulated by RUNX2.
Adam17 and Runx2 expression was assessed in mouse Runx2-null osteoprogenitor cells (Runx2−/−) and mouse MC3T3–E1 pre-osteoblasts cells, as well as rat ROS17/2.8 and human SAOS-2 osteosarcoma cells. Protein and mRNA levels were evaluated by western blot analysis (A, and down graph B) and RT-PCR (C, and down graph D), respectively. Runx2-null cells were infected with an adenovirus vector expressing RUNX2 or GFP (control) as indicated (E and G). Alternativelly, cells were transiently transfected with different concentrations (0.5, 1, 2.5, 5, and 10 μg of DNA) of pcDNA-Runx2 or pcDNA-empty vector (control) (I). Runx2-null cells expressing Adam17 and Runx2 mRNA (E, I, and down graph F and J) and protein levels (G, and down graph H) were evaluated by RT-PCR and western blot analysis, respectively. The data shown are representative of three experiments with similar outcomes. Adam17 and Runx2 mRNA and protein values were normalized to Gapdh and Actin, respectively. All data are presented as mean ± SEM from three independent experiments. *P<0.05 and **P<0.01.
Adam17 gene is directly regulated by RUNX2 at the promoter level in committed osteoblasts
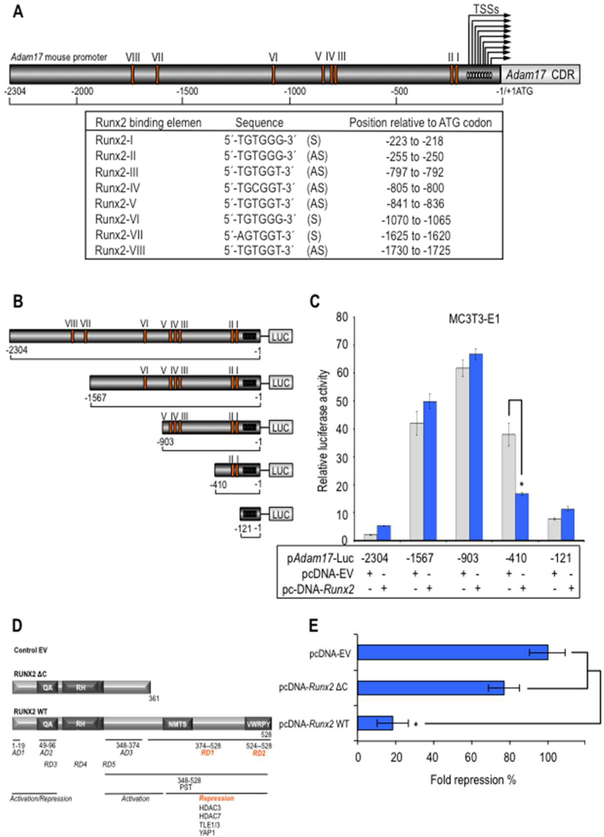
Considering the inverse correlation between the expression levels of Runx2 and Adam17, we hypothesized that Adam17 can be a direct transcriptional target of RUNX2 during osteoblast differentiation. In silico analysis of the mouse, rat and human Adam17 promoter sequences identified several potential RUNX2 binding sites. The mouse promoter exhibits nine transcription start site (TSS) (Mizui et al., 1999) whereas the analysis of rat and human promoters showed a single TSS (data non shown). We evaluate the presence of RUNX2-binding motifs in the Adam17 promoter using the consensus RUNX2 motif 5’-(T/A/C)G(T/A/C)GG(T/G) that was previously validated in a genomic-wide occupancy study (van der Deen et al., 2012). The mouse Adam17 gene promoter contains at least eight RUNX2 consensus motifs (Fig. 3A) whereas the analysis of rat and human promoters showed eight and three RUNX2 motifs, respectively (data not shown). The motifs 5’-TGTGGT and 5-AGTGGT, that are perfect matches respect to consensus sequence, represent 4/8 of the total putative RUNX2 motifs observed in the mouse Adam17 promoter. Interestingly, a variable number of a one-mismatch 5’-TGTGGG RUNX2 motifs are localized close to the TSSs in the mouse Adam17 promoter (sites Runx2 I, II and VI). Moreover, one site (‘Runx2 I’) is highly conserved in the rat and human Adam17 promoters (data not shown). This one-mismatch motif is the second most frequently observed RUNX2 motif in genomic promoters, and a large fraction of these gene promoters (~35%) is not co-occupied by RNAPII (van der Deen et al., 2012), consistent with the model that this sequence motif may support Runx2-mediated gene repression.
Figure 3. Adam17 promoter has a proximal region containing RUNX2 binding sites supporting transcriptional repression.
Adam17 gene promoter from mouse was analyzed for the presence of the genome-wide consensus Runx motif (5’-[A/C]CC[A/T/G]C[A/T/G]-3’) previously established for us (van der Deen et al., 2012) (A). S and AS indicated sense- and antisense-stranded DNA. Arrows indicated transcription start sites (TSS) (−161 to −55, relative to ATG codon). MC3T3–E1 cells were transfected with either a luciferase reporter gene under transcriptional control of the promoter region of mouse Adam17 (pAdam17 2.3-Kb) or with a series of deletion mutants spanning the mouse Adam17 gene promoter that overexpress will-type Runx2 or empty vector control pcDNA 3.1 (EV) for Runx2 expression construct (B and C). Effect of will type Runx2 (Runx2 WT) or deletion mutant Runx2 1–361 (Runx2 ΔC) expression or empty vector control pcDNA 3.1 (EV) on 0.4-Kb Adam17 proximal promoter was also determined by a luciferase activity assay (D and E). The promoter activity was determined by a luciferase activity assay after 24 h of transient transfection and normalized by cotransfection with Renilla luciferase. All data are presented as mean ± SEM from three independent experiments. *P<0.05.
To determine the functional contribution of RUNX2 to regulation of Adam17 promoter, we performed transient transfection assays with Adam17 mouse promoter fragments spanning 2304 bp of 5’ sequence (containing eight RUNX2 motifs) in a luciferase reporter (pAdam17-Luc, originally described as pTace-Luc) (Charbonneau et al., 2007), in pre-osteoblast MC3T3–E1 cells. Specifically, we identify promoter regions implicated in RUNX2 transcriptional regulation using a series of 5’ deletion constructs of the pAdam17-Luc vector (Fig. 3B). Deletion of nt −2304 to −903 results in an increase of Adam17 promoter activity, suggesting the presence of repressive elements in the 5’ region of the distal-promoter (Fig. 3C). However, deletion of nt −903 to −121 decreases promoter activity, thus also suggesting the presence of activating elements at the proximal-promoter. Interestingly, co-expression of Runx2 does not significantly activate or repress Adam17 promoter at the putative RUNX2 binding sites VIII-III localized between nt −2304 to −410, but RUNX2 significantly attenuates promoter activity of the −410 bp deletion construct (nt −410 to −121) containing the RUNX2 I and II binding elements (Fig. 3C). These results indicate that there are at least two functional RUNX2 binding sites located in the −410/−121 region at the proximal Adam17 promoter that can orchestrate the RUNX2-mediated repression of this regulatory region.
Because the C-terminal domain of RUNX2 is required for its gene repressive functions, we next analyze the contribution of this region to repress activity of the −410 bp Adam17 gene promoter in MC3T3–E1 pre-osteoblastic cells (Fig. 3D). Interestingly, only wild type Runx2 but not the mutant version of this transcription factor (Runx2-Δ361) which lacks the C-terminal region results in significant reductions of the −410 Adam17 promoter in osteoblastic cells, indicating that Adam17 gene repression by RUNX2 requires the C-terminal region (Fig. 3E).
Adam17 gene expression is reduced during osteoblastic differentiation
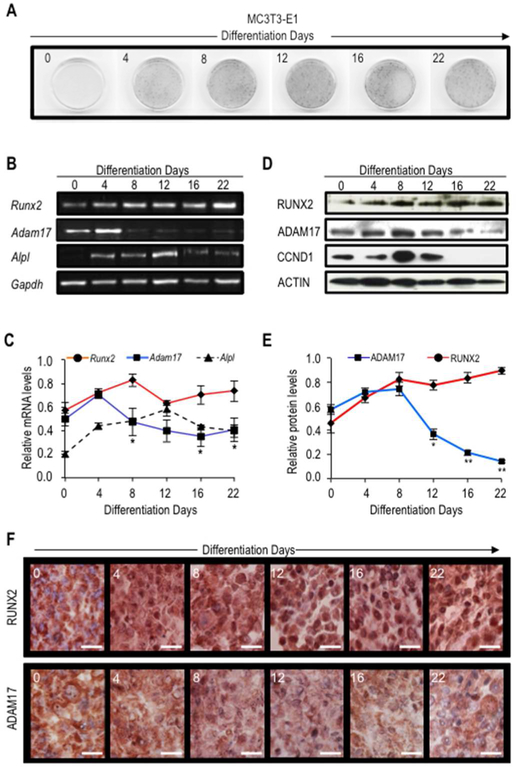
To address whether Adam17 downregulation is biological coupled to the physiologic upregulation of RUNX2 observed during MC3T3–E1 osteoblast differentiation, we analyzed endogenous expression of Adam17 in relation to RUNX2 expression levels. Osteogenic differentiation was monitored by ALPL activity (a cognate RUNX2-target gene) (Fig. 4A). We found a significant reduction of Adam17 mRNA levels that precedes the ADAM17 decrease in both protein levels and immune staining after day 4–8, in parallel with the early elevation in Runx2 mRNA and RUNX2 protein levels as well as RUNX2 protein nuclear immune staining (4–8 days) (Fig. 4B–F). These patterns are associated with the subsequent cessation in cell proliferation and subsequent initiation of osteoblast differentiation, as evidenced by the absence of CCND1 expression after day 12 and elevation of Alpl expression between days 4–12 (Fig. 4D).
Figure 4. Adam17 expression is down-regulated during osteogenic differentiation.
MC3T3–E1 cells were induced to differentiate with ascorbic acid and β-glycerophosphate for 28 days. Cells were fixed in paraformaldehy for histochemical detection of alkaline phosphatase at the indicated days of differentiation (day 0, 4, 8, 12, 16, 22 and 28) (A). Adam17 and Runx2 mRNA and protein levels as well as ADAM17 and RUNX2 cellular localization were determined by RT-PCR (B, and down graph C), western blot (D, and down graph E) and immunostaining (F), respectively. Expression of osteoblast phenotypic genes alkaline phosphatase (Alpl) as well as cell cycle marker cyclin D (Ccnd1) were additionally examined by RT-PCR and western blot, respectively. The data shown are representative of three experiments with similar outcomes. Runx2 and Adam17 mRNA (C, right graph) and protein (E, left graph) values were normalized to Gapdh and Actin, respectively. All data are presented as mean ± SEM from three independent experiments. *P<0.05 and **P<0.01.
Proximal promoter of the Adam17 selectively associated with RUNX2 during osteoblastic differentiation
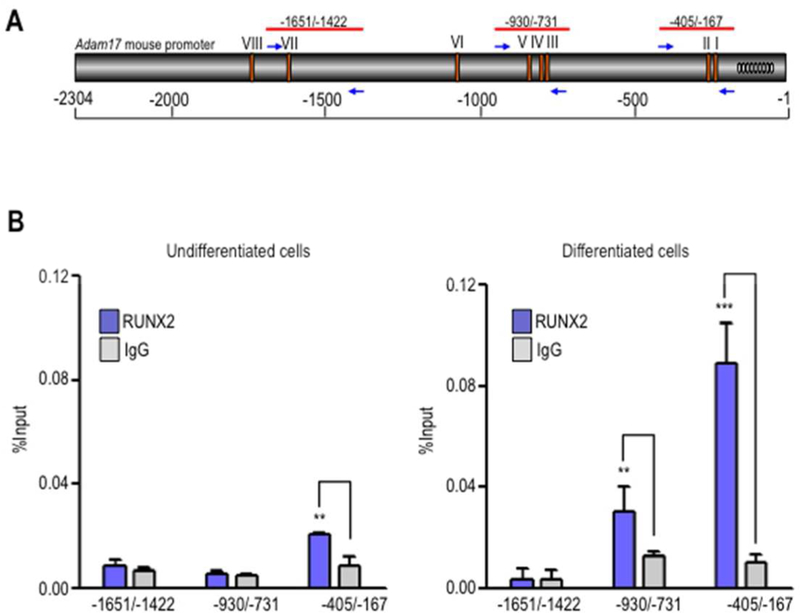
To determine whether RUNX2 bind to the endogenous Adam17 gene proximal promoter during osteogenic differentiation, we performed chromatin immunoprecipitation (ChIP) analysis under in vitro culture conditions that reproduce osteoblast differentiation. We tested RUNX2 binding to sites I and II at the proximal promoter (−405/−167), as well as to others in two selected regions upstream of this region (−930/−731 and −1651/−1422) that contain sites III-V and site VII, respectively (Fig. 5A). The data provide further in vivo evidence showing that RUNX2 selectively binds to the proximal promoter of the Adam17 gene, containing the RUNX2 I and II binding elements in pre-osteoblast MC3T3–E1 cells (Fig. 5B). ChIP analyses also revealed that RUNX2 differentially binds to the 5’-proximal promoter of Adam17 and that occupancy of sites I-II and sites III-IV increases significantly in differentiating osteoblasts, while the distal upstream region was not enriched in the precipitated DNA samples (Fig. 5C). Taken together, our findings demonstrate that Adam17 gene repression during osteoblastic differentiation is associated with RUNX2 binding at the Adam17 gene promoter.
Figure 5. RUNX2 binds to the Adam17 proximal promoter region in differentiating osteoblastic cells.
Diagram illustrates the location of the primers used in the ChIP experiments. Arrows indicate the direction of each primer, and the negative values indicate their position relative to ATG (A). MC3T3–E1 cells were induced to differentiate with ascorbic acid and β-glycerophosphate and cultured up to day 12. Undifferentiated (B) and differentiated (C) cells were cross-linked with 1% formaldehyde, and the sonicated chromatin fragments were immunoprecipitated using specific polyclonal antibodies against RUNX2 protein. The enrichment of Adam17 promoter sequences in the precipitated chromatin fragments was quantified by qPCR using the primers described in panel A. All data are presented as mean ± SEM from three independent experiments. **P<0.01 and ***P<0.001.
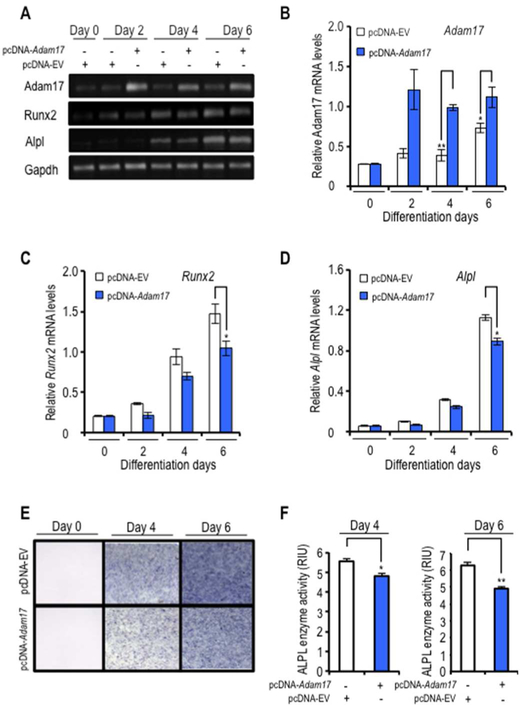
Adam17 overexpression attenuates osteoblast differentiation
To understand the biological relevance of Adam17 gene repression in osteoblast differentiation, we overexpressed exogenous Adam17 above normal physiological levels in differentiating MC3T3–E1 cells using a CMV-driven expression vector. Adam17 was efficiently expressed above the normal endogenous levels in osteoblasts from day 2–6 of differentiation (Fig. 6A and B). Moreover, Adam17 overexpression attenuates Runx2 expression and reduces both the expression and activity of the RUNX2-target gene Alpl that is normally increased during early stages of osteoblast differentiation (Fig. 6C–F). Thus, our data suggest that Adam17 expression regulates osteoblastic differentiation. Our collective findings are consistent with a novel regulatory model in which reciprocal feedback regulation between Runx2-Adam17 controls progression of osteoblast differentiation.
Figure 6. Adam17 expression affects osteoblastic differentiation.
MC3T3–E1 cells were transfected with a construct expressing Adam17 or control empty vector (EV). Cells were induced to differentiate at 24 h after transfection with ascorbic acid and β-glycerophosphate for 6 days. Expression of Adam17, Runx2 and alkaline phosphatase (Alpl) were examined by RT-PCR at day 0, 2, 4, and 6 of differentiation (A). Adam17 (B), Runx2 (C) and Alpl mRNA (D) values normalized to Gapdh. Additionally, cells were fixed in paraformaldehy for histochemical detection of alkaline phosphatase at the indicated days of differentiation (day 0, 4, and 6) (E). Histochemical staining of ALP activity was quantified using Image J processing software and values were expressed in relative intensity units (RIU) (F). All data are presented as mean ± SEM from three independent experiments. *P<0.05 and **P<0.01.
DISCUSSION
In this study, we provide evidence that expression of Adam17, which physiologically controls the presentation of proteins on the cell surface of osteoblasts, is a direct transcriptional target of RUNX2 during osteoblast differentiation. The role of ADAM17 in skeletal development has been previously analyzed in knockout mice in which Adam17 was conditionally disrupted in chondrocytes and osteochondroprogenitor cells (Horiuchi et al., 2009; Hall et al., 2013; Saito et al., 2013). These studies showed that mutant mice exhibit growth retardation, reduction on femur length with impaired growth of chondroblasts, elongated hypertrophic zone and accumulation of differentiated chondrocytes that produces a calcified matrix suggesting that ADAM17 regulates terminal differentiation of chondrocytes during endochondral ossification (Hall et al., 2013; Saito et al., 2013). Interestingly, a histomorphometric analysis revealed that osteoblast-related parameters, including numbers of osteoblast, were increased in conditional Adam17-deficient mouse suggesting that ADAM17 may also be involved in osteoblast differentiation (Horiuchi et. al., 2009).
We examined the role of RUNX2 in controlling Adam17 expression in osteoblasts using different mouse and human cellular models included the MC3T3–E1 pre-osteoblast cell line, human osteoblasts and the immortalized Runx2-null osteoprogenitor cells. Our data are consistent with microarray gene expression profiling with Runx2 null cells expressing wild type and mutant RUNX2 proteins, which revealed changes in the expression of several ADAM and ADAMTS members (Teplyuk et al., 2008). Moreover, examination of ChIP-microarray data sets for RUNX2 target genes in human osteoblastic cells (van der Deen et al., 2012) revealed additional genomic promoter interactions for RUNX2 on other promoters of ADAM and ADAMTS genes (data not shown). Thus, several members of both gene families could be potential downstream target genes of RUNX2, and our data clearly show that Adam17 is a prominent member controlled by RUNX2.
RUNX2 is a bifunctional transcription factor that may interact with a broad spectrum of co-activators and co-repressor, thus supporting activation or repression of RUNX2 target genes. Several repression domains (RD) have been characterized in different regions of RUNX2 protein, which define binding of specific co-repressors. Specifically, the RUNX2 C-terminal region exhibits at least two different repression domains capable of interacting with several proteins involved in gene repression like histone deacetylases (HDACs) (Bradley et al., 2015; Westendorf 2006; Ziros et al., 2008). The RUNX2 C-terminal region may support transcriptional repression of Adam17 gene trough interactions with HDACs or other chromatin related co-repressors. Interestingly, previous reports showed that RUNX2 can positively regulate expression of Adamts4 and Adamts5 in chondrocytes (Thirunavukkarasu et al., 2006; Thirunavukkarasu et al., 2007; Lin et al., 2009; Tetsunaga et al., 2011), perhaps suggesting that Runx2 may have a bifunctional role during osteogenesis.
ADAM17 was initially characterized as an enzyme anchored to the plasma membrane which is involved in proteolytic processing and the release of TNF-α to the extracellular milieu. Interestingly, TNF-α (TNF) inhibits extracellular matrix maturation and mineralization in osteoblasts by decreasing production of type I collagen, a key component of a mature skeletal matrix, and by evoking an attenuated response to vitamin D associated with reduced ALPL activity and osteocalcin/BGLAP release (Smith et al., 1987; Gowen et al., 1988). In addition, TNF-α also induces a reduction of the IGF-1 expression, a growth factor that is known to promote osteoblast differentiation, (Gilbert et al, 2000). Other studies have shown that TNF-α downregulated expression of BMPR and attenuated BMP-2-mediate Alpl and Oc/Bglap expression in human bone cells (Singhatanadgit et al., 2006). Moreover, TNF-α inhibits osteoblast differentiation through the inhibition of mineralized nodule formation and inhibition of Runx2 and Osx/Sp7 expression, both master genes of the osteoblast differentiation, and skeletal-specific genes (e.g., Alpl, Bsp/Ibsp, Oc/Bglap) (Gilbert et al., 2002; Gilbert et al., 2005; Lu et al., 2005; Gilbert et al., 2013). Hence, our results could be envisioned as part of a RUNX2-ADAM17-TNF axis. In this model, the osteogenic activation of RUNX2 stimulates osteoblast differentiation by preventing the ADAM17-mediated release of TNF which otherwise suppresses osteoblast maturation. Negative regulation of RUNX2 by ADAM17 may ensure that this axis only works when RUNX2 levels have reached a critical threshold.
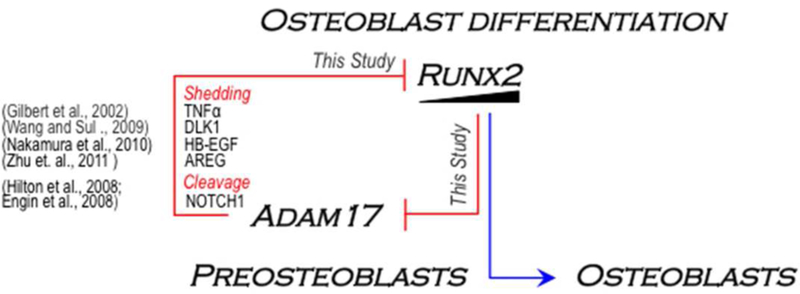
On the other hand, ADAM17 protein also releases a soluble form of the membrane-anchored DLK1 protein (named fetal antigen 1, FA1) in mesenchymal precursors (Abdallah et al., 2004; Taipaleenmäki et al., 2012; Abdallah et al., 2015) and pre-osteoblastic cells (Wang and Sul, 2009). Preosteoblastic cells treated with soluble DLK1 or overexpressing Dlk1 exhibit elevated Sox9 expression that prevents osteoblast differentiation and Runx2 expression (Wang and Sul, 2009). Hence, it is reasonable to propose that the results observed following Adam17 overexpression in osteoblastic cells may also be due, at least in part to enhanced DLK1 release that impairs osteoblast differentiation. Other well know substrates of ADAM17 are known to suppresses osteoblast differentiation and Runx2/RUNX2 expression/transcriptional activity, including HB-EGF and Amphiregulin (AREG), which are released by proteolytic cleavage, or the NOTCH1 receptor, which is regulated by ADAM17-dependent cleavage of it extracellular domain (Hilton et al., 2008; Engin et al., 2008; Nakamura et al., 2010; Zanotti and Canalis, 2010; Zhu et. al., 2011; Zunke and Rose-John, 2017). Taken together, we propose a molecular mechanism in which RUNX2 governs Adam17 gene expression during osteoblast differentiation (Fig. 7). This mechanism implies that RUNX2 may be controlling the cell-cell communication between osteoblastic cells in the bone micro-niche, by modulating the shedding and release of extracellular ligands and/or RIPping (regulated intramembrane proteolysis) of cognate receptors to support normal progression from precursor cells to mature osteoblast during osteogenesis.
Figure 7. A cross-talk model of the RUNX2 and ADAM17 signaling axes in osteoblast differentiation control.
The current study provides evidence for a regulatory mechanism involving a reciprocal RUNX2-ADAM17 negative feedback loop to regulate progression of osteoblast differentiation. We have shown that RUNX2 represses Adam17 expression through its binding to the Adam17 proximal promoter and that Adam17 overexpression antagonizes Runx2 expression during osteoblast differentiation. Several substrates of ADAM17 (e.g. TNF-α, DLK1, EB-EGF, AREG and NOTCH1) have been shown to control osteoblast differentiation by suppressing either the expression or the activity of RUNX2. As a whole, our study and those from others suggest that RUNX2 may regulate progression of osteoblast differentiation through a negative modulation of both receptor ripping (NOTCH1) and shedding of various extracellular signals (e.g. TNF-α, DLK1, EB-EGF and AREG) derived from repression of Adam17 gene expression.
ACKNOWLEDGMENTS
We thank the members of our research groups, including Mariana Osorio, Francisco Villanueva, Andres Stevenson, Nelson Varela (University of Chile) for stimulating discussions. This study was supported by National Fund for Scientific and Technological Development (FONDECYT Chile), grant numbers 1095234 and 1130931 (to M.A.G.), 1130706 (to M.M.), 1171213 (to F.S-O.), 1160214 (to J.P.R.), 1110778 and 1150352 (to R.D.M.); Fund for Research Centers in Prioritary Areas (FONDAP Chile), grant number 15090007 (to M.M.); The Scientific and Technological Development Support Fund (FONDEF Chile), grant number ID16110148 (to F.S-O.); and Millennium Science Initiative from Ministry for the Economy, Development and Tourism, Chile, grant number P09/016-F (to M.A.G. and F.S-O.). Additional support was provided by PhD fellowship of the National Commission for Scientific and Technological Research (CONICYT Chile) (to S.J. and H.F.A.). This work was also supported by National Institutes of Health Grants R01 AR049069 (to A.J.v.W.) and P01 CA082834 (to G.S.S.).
Contract grant sponsor: Iniciativa Científica Milenio (to MAG and FS-O); Contract grant number: P09/016-F.
Contract grant sponsor: FONDECYT (to MAG); Contract grant numbers: 1095234, 1130931.
Contract grant sponsor: FONDECYT (to FS-O); Contract grant number: 1171213.
Contract grant sponsor: FONDECYT (to MM); Contract grant number: 1130706.
Contract grant sponsor: FONDECYT (to RDM); Contract grant number: 1110778, 1150352.
Contract grant sponsor: FONDECYT (to JPR); Contract grant number: 1160214.
Contract grant sponsor: FONDEF (to FS-O); Contract grant number: ID16110148.
Contract grant sponsor: FONDAP (to MM); Contract grant number: 15090007.
Contract grant sponsor: National Institutes of Health (to AJvW); Contract grant number: R01AR049069.
Contract grant sponsor: National Institutes of Health (to GSS); Contract grant number: P01 CA082834.
Footnotes
Conflict of interest: None
LITERATURE CITED
- Abdallah BM, Jensen CH, Gutierrez G, Leslie RG, Jensen TG, Kassem M. 2004. Regulation of human skeletal stem cells differentiation by Dlk1/Pref-1. J Bone Miner Res 19:841–852. [DOI] [PubMed] [Google Scholar]
- Abdallah BM, Jafari A, Zaher W, Qiu W, Kassem M. 2015. Skeletal (stromal) stem cells: an update on intracellular signaling pathways controlling osteoblast differentiation. Bone 70:28–36. [DOI] [PubMed] [Google Scholar]
- Aguilar R, Bustos FJ, Saez M, Rojas A, Allende ML, van Wijnen AJ, van Zundert B, Montecino M. 2016. Polycomb PRC2 complex mediates epigenetic silencing of a critical osteogenic master regulator in the hippocampus. Biochim Biophys Acta 1859:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artigas N, Ureña C, Rodríguez-Carballo E, Rosa JL, Ventura F. 2014. Mitogen-activated protein kinase (MAPK)-regulated interactions between Osterix and Runx2 are critical for the transcriptional osteogenic program. J Biol Chem 289:27105–27117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae JS, Gutierrez S, Narla R, Pratap J, Devados R, van Wijnen AJ, Stein JL, Stein GS, Lian JB, Javed A. 2007. Reconstitution of Runx2/Cbfa1-null cells identifies a requirement for BMP2 signaling through a Runx2 functional domain during osteoblast differentiation. J Cell Biochem 100:434–449. [DOI] [PubMed] [Google Scholar]
- Balint E, Lapointe D, Drissi H, van der Meijden C, Young DW, van Wijnen AJ, Stein JL, Stein GS, Lian JB. 2003. Phenotype discovery by gene expression profiling: mapping of biological processes linked to BMP-2-mediated osteoblast differentiation. J Cell Biochem 89:401–426. [DOI] [PubMed] [Google Scholar]
- Barutcu AR, Tai PW, Wu H, Gordon JA, Whitfield TW, Dobson JR, Imbalzano AN, Lian JB, van Wijnen AJ, Stein JL, Stein GS. 2014. The bone-specific Runx2-P1 promoter displays conserved three-dimensional chromatin structure with the syntenic Supt3h promoter. Nucleic Acids Res 42:10360–10372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley EW, Carpio LR, van Wijnen AJ, McGee-Lawrence ME, Westendorf JJ. 2015. Histone Deacetylases in Bone Development and Skeletal Disorders. Physiol Rev 95:1359–13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KM, Wong HL, Jin G, Liu B, Cao R, Cao Y, Lehti K, Tryggvason K, Zhou Z. 2012. MT1-MMP inactivates ADAM9 to regulate FGFR2 signaling and calvarial osteogenesis. Dev Cell 22:1176–1190. [DOI] [PubMed] [Google Scholar]
- Charbonneau M, Harper K, Grondin F, Pelmus M, McDonald PP, Dubois CM. 2007. Hypoxia-inducible factor mediates hypoxic and tumor necrosis factor alpha-induced increases in tumor necrosis factor-alpha converting enzyme/ADAM17 expression by synovial cells. J Biol Chem 282:33714–33724. [DOI] [PubMed] [Google Scholar]
- Dallas DJ, Genever PG, Patton AJ, Millichip MI, McKie N, Skerry TM. 1999. Localization of ADAM10 and Notch receptors in bone. Bone 25:9–15. [DOI] [PubMed] [Google Scholar]
- Dudakovic A, Camilleri E, Riester SM, Lewallen EA, Kvasha S, Chen X, Radel DJ, Anderson JM, Nair AA, Evans JM, Krych AJ, Smith J, Deyle DR, Stein JL, Stein GS, Im HJ, Cool SM, Westendorf JJ, Kakar S, Dietz AB, van Wijnen AJ. 2014. High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J Cell Biochem. 115:1816–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, Chen Y, Wang L, Zheng H, Sutton RE, Boyce BF, Lee B. 2008. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med 14:299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchimont N, Lambert C, Huynen P, Ribbens C, Relic B, Chariot A, Bours V, Piette J, Merville MP, Malaise M. 2005. Interleukin-6 receptor shedding is enhanced by interleukin-1beta and tumor necrosis factor alpha and is partially mediated by tumor necrosis factor alpha-converting enzyme in osteoblast-like cells. Arthritis Rheum 52:84–93. [DOI] [PubMed] [Google Scholar]
- Galindo M, Pratap J, Young DW, Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS, van Wijnen AJ. 2005. The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. J Biol Chem 280:20274–20285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo M, Kahler RA, Teplyuk NM, Stein JL, Lian JB, Stein GS, Westendorf JJ, van Wijnen AJ. 2007. Cell cycle related modulations in Runx2 protein levels are independent of lymphocyte enhancer-binding factor 1 (Lef1) in proliferating osteoblasts. J Mol Histol 38:501–506. [DOI] [PubMed] [Google Scholar]
- Gilbert L, He X, Farmer P, Boden S, Kozlowski M, Rubin J, Nanes MS. 2000. Inhibition of osteoblast differentiation by tumor necrosis factor-alpha. Endocrinology 141:3956–3964. [DOI] [PubMed] [Google Scholar]
- Gilbert L, He X, Farmer P, Rubin J, Drissi H, van Wijnen AJ, Lian JB, Stein GS, Nanes MS. 2002. Expression of the osteoblast differentiation factor RUNX2 (Cbfa1/AML3/Pebp2alphaA) is inhibited by tumor necrosis factor-alpha. J Biol Chem 277:2695–26701. [DOI] [PubMed] [Google Scholar]
- Gilbert LC, Rubin J, Nanes MS. 2005. The p55 TNF receptor mediates TNF inhibition of osteoblast differentiation independently of apoptosis. Am J Physiol Endocrinol Metab 288:1011–1018. [DOI] [PubMed] [Google Scholar]
- Gilbert LC, Chen H, Lu X, Nanes MS. 2013. Chronic low dose tumor necrosis factor-α (TNF) suppresses early bone accrual in young mice by inhibiting osteoblasts without affecting osteoclasts. Bone 56:174–183. [DOI] [PubMed] [Google Scholar]
- Govoni KE, Amaar YG, Kramer A, Winter E, Baylink DJ, Mohan S. 2006. Regulation of insulin-like growth factor binding protein-5, four and a half lim-2, and a disintegrin and metalloprotease-9 expression in osteoblasts. Growth Horm IGF Res 16:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen M, MacDonald BR, Russell RG. 1988. Actions of recombinant human gamma-interferon and tumor necrosis factor alpha on the proliferation and osteoblastic characteristics of human trabecular bone cells in vitro. Arthritis Rheum 31:1500–1507. [DOI] [PubMed] [Google Scholar]
- Hall KC, Hill D, Otero M, Plumb DA, Froemel D, Dragomir CL, Maretzky T, Boskey A, Crawford HC, Selleri L, Goldring MB, and Blobel CP. 2013. ADAM17 Controls Endochondral Ossification by Regulating Terminal Differentiation of Chondrocytes. Mol Cell Biol 33:3077–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris HA, Murrills RJ, Komm BS. 1997. Expression of meltrin-alpha mRNA is not restricted to fusagenic cells. J Cell Biochem 67:136–142. [DOI] [PubMed] [Google Scholar]
- Hawse JR, Cicek M, Grygo SB, Bruinsma ES, Rajamannan NM, van Wijnen AJ, Lian JB, Stein GS, Oursler MJ, Subramaniam M, Spelsberg TC. 2011. TIEG1/KLF10 modulates Runx2 expression and activity in osteoblasts. PLoS One 6:e19429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendesi H, Barbe MF, Safadi FF, Monroy MA, Popoff SN. 2015. Integrin mediated adhesion of osteoblasts to connective tissue growth factor (CTGF/CCN2) induces cytoskeleton reorganization and cell differentiation. PLoS One 10:e0115325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikita A, Yana I, Wakeyama H, Nakamura M, Kadono Y, Oshima Y, Nakamura K, Seiki M, Tanaka S. 2006. Negative regulation of osteoclastogenesis by ectodomain shedding of receptor activator of NF-kappaB ligand. J Biol Chem 281:36846–36855. [DOI] [PubMed] [Google Scholar]
- Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, Long F. 2008. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med 14:306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K, Kimura T, Miyamoto T, Miyamoto K, Akiyama H, Takaishi H, Morioka H, Nakamura T, Okada Y, Blobel CP, Toyama Y. 2009. Conditional inactivation of TACE by a Sox9 promoter leads to osteoporosis and increased granulopoiesis via dysregulation of IL-17 and G-CSF. J Immunol 182:2093–20101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovhannisyan H, Zhang Y, Hassan MQ, Wu H, Glackin C, Lian JB, Stein JL, Montecino M, Stein GS, van Wijnen AJ. 2013. Genomic occupancy of HLH, AP1 and Runx2 motifs within a nuclease sensitive site of the Runx2 gene. J Cell Physiol 228:313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley MM, Adams DJ, Wang L, Jiang X, Burt PM, Du E, Xiao L. 2016. Accelerated fracture healing in transgenic mice overexpressing an anabolic isoform of fibroblast growth factor 2. J Cell Biochem 117:599–611. [DOI] [PubMed] [Google Scholar]
- Inoue D, Reid M, Lum L, Krätzschmar J, Weskamp G, Myung YM, Baron R, Blobel CP. 1998. Cloning and initial characterization of mouse meltrin beta and analysis of the expression of four metalloprotease-disintegrins in bone cells. J Biol Chem 273:4180–4187. [DOI] [PubMed] [Google Scholar]
- Kadri A, Funck-Brentano T, Lin H, Ea HK, Hannouche D, Marty C, Lioté F, Geoffroy V, Cohen-Solal ME. 2010. Inhibition of bone resorption blunts osteoarthritis in mice with high bone remodelling. Ann Rheum Dis 69:1533–1538. [DOI] [PubMed] [Google Scholar]
- Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. 2010. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem 285:25221–25231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadag A, Zhou M, Croucher PI. 2006. ADAM-9 (MDC-9/meltrin-gamma), a member of the a disintegrin and metalloproteinase family, regulates myeloma-cell-induced interleukin-6 production in osteoblasts by direct interaction with the alpha(v)beta5 integrin. Blood 107:3271–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawane T, Komori H, Liu W, Moriishi T, Miyazaki T, Mori M, Matsuo Y, Takada Y, Izumi S, Jiang Q, Nishimura R, Kawai Y, Komori T. 2014. Dlx5 and mef2 regulate a novel runx2 enhancer for osteoblast-specific expression. J Bone Miner Res 29:1960–1969. [DOI] [PubMed] [Google Scholar]
- Kurisaki T, Masuda A, Osumi N, Nabeshima Y, Fujisawa-Sehara A. 1998. Spatially- and temporally-restricted expression of meltrin alpha (ADAM12) and beta (ADAM19) in mouse embryo. Mech Dev 73:211–215. [DOI] [PubMed] [Google Scholar]
- Lewallen EA, Jones DL, Dudakovic A, Thaler R, Paradise CR, Kremers HM, Abdel MP, Kakar S, Dietz AB, Cohen RC, Lewallen DG, van Wijnen AJ. 2016. Osteogenic potential of human adipose-tissue-derived mesenchymal stromal cells cultured on 3D-printed porous structured titanium. Gene. 581:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ge C, Franceschi RT. 2016. MAP Kinase-Dependent RUNX2 Phosphorylation Is Necessary for Epigenetic Modification of Chromatin During Osteoblast Differentiation. J Cell Physiol. Accepted Author Manuscript Aug 11. doi: 10.1002/jcp.25517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. 2012. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol 8:212–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KE, Park NR, Che X, Han MS, Jeong JH, Kim SY, Park CY, Akiyama H, Kim JE, Ryoo HM, Stein JL, Lian JB, Stein GS, Choi JY. 2015. Core binding factor β of osteoblasts maintains cortical bone mass via stabilization of Runx2 in mice. J Bone Miner Res 30:715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Seeto BL, Bartoszko JM, Khoury MA, Whetstone H, Ho L, Hsu C, Ali SA, Alman BA. 2009. Modulating hedgehog signaling can attenuate the severity of osteoarthritis. Nat Med 15:1421–1425. [DOI] [PubMed] [Google Scholar]
- Lind T, McKie N, Wendel M, Racey SN, Birch MA. 2005. The hyalectan degrading ADAMTS-1 enzyme is expressed by osteoblasts and up-regulated at regions of new bone formation. Bone 36:408–417. [DOI] [PubMed] [Google Scholar]
- Liu JC, Lengner CJ, Gaur T, Lou Y, Hussain S, Jones MD, Borodic B, Colby JL, Steinman HA, van Wijnen AJ, Stein JL, Jones SN, Stein GS, Lian JB. 2011. Runx2 protein expression utilizes the Runx2 P1 promoter to establish osteoprogenitor cell number for normal bone formation. J Biol Chem 286:30057–30070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F Building strong bones: molecular regulation of the osteoblast lineage. 2011. Nat Rev Mol Cell Biol. 13:27–38. [DOI] [PubMed] [Google Scholar]
- Lu X, Gilbert L, He X, Rubin J, Nanes MS. 2006. Transcriptional regulation of the osterix (Osx, Sp7) promoter by tumor necrosis factor identifies disparateeffects of mitogen-activated protein kinase and NF kappa B pathways. J Biol Chem 281:6297–62306 [DOI] [PubMed] [Google Scholar]
- McGee-Lawrence ME, Li X, Bledsoe KL, Wu H, Hawse JR, Subramaniam M, Razidlo DF, Stensgard BA, Stein GS, van Wijnen AJ, Lian JB, Hsu W, Westendorf JJ. 2013a. Runx2 protein represses Axin2 expression in osteoblasts and is required for craniosynostosis in Axin2-deficient mice. J Biol Chem 288:5291–5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee-Lawrence ME, Bradley EW, Dudakovic A, Carlson SW, Ryan ZC, Kumar R, Dadsetan M, Yaszemski MJ, Chen Q, An KN, Westendorf JJ. 2013b. Histone deacetylase 3 is required for maintenance of bone mass during aging. Bone 52:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, Pike JW. 2014. The RUNX2 cistrome in osteoblasts: characterization, down-regulation following differentiation, and relationship to gene expression. J Biol Chem 289:16016–16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles RR, Sluka JP, Halladay DL, Santerre RF, Hale LV, Bloem L, Thirunavukkarasu K, Galvin RJ, Hock JM, Onyia JE. 2000. ADAMTS-1: A cellular disintegrin and metalloprotease with thrombospondin motifs is a target for parathyroid hormone in bone. Endocrinology 141:4533–4542. [DOI] [PubMed] [Google Scholar]
- Mizui Y, Yamazaki K, Sagane K, Tanaka I. 1999. cDNA cloning of mouse tumor necrosis factor-alpha converting enzyme (TACE) and partial analysis of its promoter. Gene 233:67–74. [DOI] [PubMed] [Google Scholar]
- Mohan S, Thompson GR, Amaar YG, Hathaway G, Tschesche H, Baylink DJ. 2002. ADAM-9 is an insulin-like growth factor binding protein-5 protease produced and secreted by human osteoblasts. Biochemistry 41:15394–15403. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Sone S, Takahashi I, Mizoguchi I, Echigo S, Sasano Y. 2005. Expression of versican and ADAMTS1, 4, and 5 during bone development in the rat mandible and hind limb. J Histochem Cytochem 53:1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Toita H, Yoshimoto A, Nishimura D, Takagi T, Ogawa T, Takeya T, Ishida-Kitagawa N. 2010. Potential involvement of Twist2 and Erk in the regulation of osteoblastogenesis by HB-EGFEGFR signaling. Cell Struct Funct 35:53–61. [DOI] [PubMed] [Google Scholar]
- Okura H, Sato S, Kishikawa S, Kaneto S, Nakashima T, Yoshida N, Takayanagi H, Kiyono H. 2014. Runx2-I isoform contributes to fetal bone formation even in the absence of specific N-terminal amino acids. PLoS One 9:e108294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Farrugia AN, To LB, Findlay DM, Green J, Lynch K, Zannettino AC. 2004. The nitrogen-containing bisphosphonate, zoledronic acid, influences RANKL expression in human osteoblast-like cells by activating TNF-alpha converting enzyme (TACE). J Bone Miner Res 19:147–154. [DOI] [PubMed] [Google Scholar]
- Pratap J, Galindo M, Zaidi SK, Vradii D, Bhat BM, Robinson JA, Choi JY, Komori T, Stein JL, Lian JB, Stein GS, van Wijnen AJ. 2003. Cell growth regulatory role of Runx2 during proliferative expansion of preosteoblasts. Cancer Res 63:5357–5362. [PubMed] [Google Scholar]
- Purcell DJ, Khalid O, Ou CY, Little GH, Frenkel B, Baniwal SK, Stallcup MR. 2012. Recruitment of coregulator G9a by Runx2 for selective enhancement or suppression of transcription. J Cell Biochem 113:2406–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Jiang Q, Matsuo Y, Kawane T, Komori H, Moriishi T, Taniuchi I, Ito K, Kawai Y, Rokutanda S, Izumi S, Komori T. 2015. Cbfb regulates bone development by stabilizing Runx family proteins. J Bone Miner Res 30:706–714. [DOI] [PubMed] [Google Scholar]
- Rehn AP, Birch MA, Karlström E, Wendel M, Lind T. 2007. ADAMTS-1 increases the three-dimensional growth of osteoblasts through type I collagen processing. Bone 41:231–238. [DOI] [PubMed] [Google Scholar]
- Rojas A, Aguilar R, Henriquez B, Lian JB, Stein JL, Stein GS, van Wijnen AJ, van Zundert B, Allende ML, Montecino M. 2015. Epigenetic Control of the Bone-master Runx2 Gene during Osteoblastlineage Commitment by the Histone Demethylase JARID1B/KDM5B. J Biol Chem 290:28329–28342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Horiuchi K, Kimura T, Mizuno S, Yoda M, Morioka H, Akiyama H, Threadgill D, Okada Y, Toyama Y, Sato K. 2013. Conditional Inactivation of TNFα-Converting Enzyme in Chondrocytes Results in an Elongated Growth Plate and Shorter Long Bones. PLoS One 8:e54853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu E, Nakatani T, He Z, Partridge NC. 2014. Parathyroid hormone regulates histone deacetylase (HDAC) 4 through protein kinase A-mediated phosphorylation and dephosphorylation in osteoblastic cells. J Biol Chem 289:21340–21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhatanadgit W, Salih V, Olsen I. 2006. Bone morphogenetic protein receptors and bone morphogenetic protein signaling are controlled by tumor necrosis factor-alpha in human bone cells. Int J Biochem Cell Biol 38:1794–1807. [DOI] [PubMed] [Google Scholar]
- Smith DD, Gowen M, Mundy GR. 1987. Effects of interferon-gamma and other cytokines on collagen synthesis in fetal rat bone cultures. Endocrinology 120:2494–2499. [DOI] [PubMed] [Google Scholar]
- Smith SS, Dole NS, Franceschetti T, Hrdlicka HC, Delany AM. 2016. MicroRNA-433 Dampens Glucocorticoid Receptor Signaling, Impacting Circadian Rhythm and Osteoblastic Gene Expression. J Biol Chem 291:21717–21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondag GR, Salihoglu S, Lababidi SL, Crowder DC, Moussa FM, Abdelmagid SM, Safadi FF. 2014. Osteoactivin induces transdifferentiation of C2C12 myoblasts into osteoblasts. J Cell Physiol 229:955–966. [DOI] [PubMed] [Google Scholar]
- Stein GS, Lian JB, van Wijnen AJ, Stein JL, Montecino M, Javed A, Zaidi SK, Young DW, Choi JY, Pockwinse SM. 2004. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene 23:4315–4329. [DOI] [PubMed] [Google Scholar]
- Tai PW, Wu H, Gordon JA, Whitfield TW, Barutcu AR, van Wijnen AJ, Lian JB, Stein GS, Stein JL. 2014. Epigenetic landscape during osteoblastogenesis defines a differentiation-dependent Runx2 promoter region. Gene 550:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipaleenmäki H, Harkness L, Chen L, Larsen KH, Säämänen AM, Kassem M, Abdallah BM. 2012. The crosstalk between transforming growth factor-β1 and delta like-1 mediates early chondrogenesis during embryonic endochondral ossification. Stem Cells 30:304–313. [DOI] [PubMed] [Google Scholar]
- Takarada T, Nakazato R, Tsuchikane A, Fujikawa K, Iezaki T, Yoneda Y, Hinoi E. 2016. Genetic analysis of Runx2 function during intramembranous ossification. Development 143:211–218. [DOI] [PubMed] [Google Scholar]
- Tan Y, Fu R, Liu J, Wu Y, Wang B, Jiang N, Nie P, Cao H, Yang Z, Fang B. 2016. ADAM10 is essential for cranial neural crest-derived maxillofacial bone development. Biochem Biophys Res Commun 475:308–314. [DOI] [PubMed] [Google Scholar]
- Teplyuk NM, Galindo M, Teplyuk VI, Pratap J, Young DW, Lapointe D, Javed A, Stein JL, Lian JB, Stein GS, van Wijnen AJ. 2008. Runx2 Regulates G Protein-coupled Signaling Pathways to Control Growth of Osteoblast Progenitors. J Biol Chem 283: 27585–27597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplyuk NM, Haupt LM, Ling L, Dombrowski C, Mun FK, Nathan SS, Lian JB, Stein JL, Stein GS, Cool SM, van Wijnen AJ. 2009a. The osteogenic transcription factor Runx2 regulates components of the fibroblast growth factor/proteoglycan signaling axis in osteoblasts. J Cell Biochem 107:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplyuk NM, Zhang Y, Lou Y, Hawse JR, Hassan MQ, Teplyuk VI, Pratap J, Galindo M, Stein JL, Stein GS, Lian JB, van Wijnen AJ. 2009b. The osteogenic transcription factor runx2 controls genes involved in sterol/steroid metabolism, including CYP11A1 in osteoblasts. Mol Endocrinol 23:849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsunaga T, Nishida K, Furumatsu T, Naruse K, Hirohata S, Yoshida A, Saito T, Ozaki T. 2011. Regulation of mechanical stress-induced MMP-13 and ADAMTS-5 expression by RUNX-2 transcriptional factor in SW1353 chondrocyte-like cells. Osteoarthritis Cartilage 19:222–232. [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu K, Pei Y, Moore TL, Wang H, Yu XP, Geiser AG, Chandrasekhar S. 2006. Regulation of the human ADAMTS-4 promoter by transcription factors and cytokines. Biochem Biophys Res Commun 345:197–204. [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu K, Pei Y, Wei T. 2007. Characterization of the human ADAMTS-5 (aggrecanase-2) gene promoter. Mol Biol Rep 34:225–231. [DOI] [PubMed] [Google Scholar]
- van der Deen M, Hassan MQ, Pratap J, Teplyuk NM, Young DW, Javed A, Zaidi SK, Lian JB, Montecino M, Stein JL, Stein GS, van Wijnen AJ. 2008. Chromatin immunoprecipitation assays: application of ChIP-on-chip for defining dynamic transcriptional mechanisms in bone cells. Methods Mol Biol 455:165–176. [DOI] [PubMed] [Google Scholar]
- van der Deen M, Akech J, Lapointe D, Gupta S, Young DW, Montecino MA, Galindo M, Lian JB, Stein JL, Stein GS, van Wijnen AJ. 2012. Genomic promoter occupancy of runt-related transcription factor RUNX2 in Osteosarcoma cells identifies genes involved in cell adhesion and motility. J Biol Chem 287:4503–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela N, Aranguiz A, Lizama C, Sepulveda H, Antonelli M, Thaler R, Moreno RD, Montecino M, Stein GS, van Wijnen AJ, Galindo M. 2016. Mitotic Inheritance of mRNA Facilitates Translational Activation of the Osteogenic-Lineage Commitment Factor Runx2 in Progeny of Osteoblastic Cells. J Cell Physiol 231:1001–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier S, Hogan A, McKie N, Horton M. 2004. ADAM gene expression and regulation during human osteoclast formation. Bone 35:34–46. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sul HS. 2009. Pref-1 regulates mesenchymal cell commitment and differentiation through Sox 9. Cell Metab. 9:287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf JJ. 2006. Transcriptional co-repressors of Runx2. J Cell Biochem 98:54–64. [DOI] [PubMed] [Google Scholar]
- Wu H, Whitfield TW, Gordon JA, Dobson JR, Tai PW, van Wijnen AJ, Stein JL, Stein GS, Lian JB. 2014. Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biol 15:R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Okamura H, Nakashima Y, Haneji T. 2013. Histone demethylase Jmjd3 regulates osteoblast differentiation via transcription factors Runx2 and osterix. J Biol Chem 288:33530–33541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Xu H, Yu S, Cao H, Fan J, Ge C, Fransceschi RT, Dong HH, Xiao G. 2011. Foxo1 mediates insulin-like growth factor 1 (IGF1)/insulin regulation of osteocalcin expression by antagonizing Runx2 in osteoblasts. J Biol Chem 286:19149–19158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida CA, Komori H, Maruyama Z, Miyazaki T, Kawasaki K, Furuichi T, Fukuyama R, Mori M, Yamana K, Nakamura K, Liu W, Toyosawa S, Moriishi T, Kawaguchi H, Takada K, Komori T. 2012. SP7 inhibits osteoblast differentiation at a late stage in mice. PLoS One 7:e32364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S and Canalis E. 2010. Notch and the skeleton. Mol Cell Biol 30:886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, Stein GS. 2011. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A 108:9863–9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xie RL, Gordon J, LeBlanc K, Stein JL, Lian JB, van Wijnen AJ, Stein GS. 2012. Control of mesenchymal lineage progression by microRNAs targeting skeletal gene regulators Trps1 and Runx2. J Biol Chem 287:21926–21935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Fujiwara T, Ye S, Li X, Zhao H. 2014. Downregulation of Notch modulators, tetraspanin 5 and 10, inhibits osteoclastogenesis in vitro. Calcif Tissue Int 95:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Shimizu E, Zhang X, Partridge NC, Qin L. 2011. EGFR signaling suppresses osteoblast differentiation and inhibits expression of master osteoblastic transcription factors Runx2 and Osterix. J Cell Biochem 112:1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziros PG, Basdra EK, Papavassiliou AG. 2008. Runx2: of bone and stretch. Int J Biochem Cell Biol 40:1659–63. [DOI] [PubMed] [Google Scholar]
- Zunke F, Rose-John S. 2017. The shedding protease ADAM17: Physiology and pathophysiology. Biochim Biophys Acta Molecular Cell Research 1864:2059–2070 [DOI] [PubMed] [Google Scholar]