Abstract
EBV and KSHV are etiologic agents of multiple types of lymphomas and carcinomas. The frequency of EBV+ or KSHV+ malignancies arising in immunocompromised individuals reflects the intricate evolutionary balance established between these viruses and their immunocompetent hosts. However, the specific mechanisms by which these pathogens drive tumorigenesis remain poorly understood. In recent years an enormous array of cellular and viral noncoding RNAs (ncRNAs) have been discovered, and host ncRNAs have been revealed as contributory factors to every single cancer hallmark cellular process. As new evidence emerges that gammaherpesvirus ncRNAs similarly alter host cell pathways, viral factors dysregulate host ncRNA expression, and novel viral ncRNAs have yet to be discovered, we examine the contribution of small, non-miRNA ncRNAs and long ncRNAs in gammaherpesvirus tumorigenesis.
1. Gammaherpesviruses, noncoding RNAs, and cancer.
It is estimated that viral infections contribute to 15–20% of all human cancers. Among these, the highly ubiquitous gammaherpesviruses Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV) play a major role, with a strong association to the development of a range of malignancies. These include Burkitt’s B cell lymphoma, diffuse large B cell lymphoma (DLBCL), Hodgkin’s Disease, nasopharyngeal carcinoma (NPC), and gastric carcinoma (GC), which are linked to EBV infection; Kaposi’s sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman’s disease (MCD), which are linked to KSHV infection; and post-transplant lymphoproliferative disease (PTLD), which can result from either EBV or KSHV infection [reviewed in 1]. Although the lifelong chronic infections associated with these agents are typically asymptomatic, gammaherpesvirus infections that occur coincident with dysregulated host immune responses or environmental cofactors can lead to lymphoproliferative and neoplastic disorders. Thus the variety and incidence of gammaherpesvirus-associated tumors reflects both (a) the inherent oncogenic capacity of these viruses and (b) the delicate counterpoise between these viruses and their hosts.
While small RNA or DNA viruses can evade some innate immune effectors to establish an infectious foothold, most of these pathogens are ultimately cleared by the immune system. In contrast, large dsDNA gammaherpesviruses must necessarily be equipped with sufficient genetic armaments to navigate their complex lifestyles. In addition to evading the initial wave of innate immunity, gammaherpesviruses must subvert adaptive B and T cell immune responses, replicate to a sufficient level for successful host dissemination, establish lifelong latent infection, and when called upon, reactivate replication to a level sufficient for inter-host dissemination. Accordingly, these viruses must be capable of hijacking host cell machinery that in some situations requires an elaborate cascade of viral gene expression to replicate viral DNA and produce new virus particles, and in other situations requires silencing of most viral genes to establish the exquisitely restricted gene expression program that is requisite for stable maintenance of long-term latency.
To maintain the flexibility to carry out this wide range of activities, gammaherpesviruses have acquired the ability to either commandeer host cell pathways or mimic molecular strategies used by the host. Indeed, for each discovery of a new host cell pathway, gammaherpesviruses have proven again and again the ability to modulate those pathways for their advantage. Likewise, for nearly every discovery of a molecular strategy used by host cells, gammaherpesviruses have been found to have either circumvented or adopted a similar strategy. Such is the case with our emerging understanding of noncoding RNAs (ncRNAs). In the last 15 years, advances in high throughput sequencing and bioinformatics have led to a new appreciation of the abundance and variety of ncRNA species expressed in mammalian cells. Among these are (i) the class of translationally repressive 21–23 nt microRNAs (miRNAs), which are well characterized elsewhere and will not be covered here 2, (ii) a variety of small RNAs ranging in size from 25 to 300 nts which include small nucleolar RNAs (snoRNAs) and PIWI-interacting RNAs (piRNAs) 3,4, and (iii) the very large and diverse class of long noncoding RNAs (lncRNAs) that are currently defined more by their length (200 nt or larger) than by any specific common structural or functional feature 5,6. Together there are thought to be more than 25,000 host ncRNA species 7.
Recently a plethora of new reports have revealed important roles for other ncRNAs in nearly every cellular process, and most importantly, additional genomic and mechanistic studies have begun to link dysregulation of ncRNAs to a wide range of cancers. Thus it stands to reason that gammaherpesviruses must exploit similar strategies during latent infection, likely both by encoding unique viral ncRNAs and by altering the expression or function of essential host ncRNAs. At present, the list of known EBV and KSHV ncRNAs is small. However, new genomics platforms and more in-depth analyses of herpesvirus genomes have begun to reveal the presence of numerous previously unknown noncoding transcripts. Such viral ncRNAs have become the subject of more intense scrutiny as our understanding of the potential functions of these molecules has begun to evolve, and as new technologies for examination of ncRNA function have emerged.
2. Host noncoding RNAs in cancer.
Mounting evidence has recently implicated ncRNAs as key players in central regulatory pathways such as proliferation, apoptosis and immune recognition that are frequently dysregulated in cancer 8–10. Although a direct mechanistic link between an individual ncRNA and a specific tumorigenic event has not yet been elucidated, it would seem only a matter of time before such a finding is uncovered: numerous studies have now demonstrated altered expression of small and long ncRNAs in specific malignancies, and links between altered ncRNA expression and tumor cell growth have been recently elucidated.
2a. Host small noncoding RNAs.
Although small ncRNAs were generally considered to be subtle molecular control elements, recent unbiased screens for cancer-associated genes have led to increased interest in these molecules as potential tumor-promoting molecules. For example, 60–300 nt snoRNAs form small nucleolar ribonucleoprotein complexes that guide post-transcriptional modification and processing of other RNAs 11–13. However, the C/D box snoRNA U50 may also hold tumor suppressor activity, as downregulation of U50 significantly correlates with some forms of prostate and breast cancers 14,15, and the U50 locus is an important translocation breakpoint in diffuse large B cell lymphoma 16. In contrast, increased expression of the H/ACA box SNORA42 is frequently associated with non-small cell lung cancer 17 and correlates with poor clinical prognosis 18. Likewise, although the 25–33 nt piRNAs form complexes with PIWI proteins ostensibly to silence transposable genetic elements 19, overexpression of piRNA-823 in multiple myeloma increases global DNA methyltransferase activity and promotes angiogenesis 20. Similarly, piRNA-651 is upregulated in some gastric, lung, breast, liver, and cervical cancers, and inhibition of piRNA-651 leads to cell cycle arrest and reduced proliferation 21.
2b. Host long noncoding RNAs.
LncRNAs are a very large and loosely defined collection of transcripts that are greaterthan 200 nt with no appreciable coding potential. hough our understanding of the mechanisms by which lncRNAs exert their influence over cellular processes is still evolving, several themes have emerged including functioning as molecular scaffolds for DNA, RNA and/or proteins as a means to alter chromatin, regulate mRNAs, regulate protein function, or alter the stoichiometry of miRNA/mRNA interactions. Moreover, lncRNAs have been shown to be involved in all cancer hallmark pathways 10, and altered expression of numerous host lncRNAs have been detected in a wide spectrum of tumor samples 22, suggesting the potential for the direct involvement of dysregulation of these molecules in tumorigenesis. Indeed, several examples to support this possibility have recently been revealed. For example, increased expression of the 2.16 kb lncRNA HOTAIR is a significant predictor of breast cancer metastases and death 23. Similarly, the 6.5 kb lncRNA MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) was first identified as an upregulated gene in lung cancer metastases 24, and marked increases in MALAT1 expression have been subsequently observed in lung, bladder, breast, and cervical cancers, suggesting that dysregulation of MALAT1 may have significant implications for the progression of numerous types of tumors.
Thus these and numerous other host ncRNAs are now being revealed as important players in the larger picture of tumor initiation and promotion. Even as our understanding of the biochemistry and function of these types of molecules in cellular processes increases, our understanding of fascinating classes of small RNAs continue to emerge, including vault RNAs 25, Y RNAs 26, and extracellular RNAs (exRNAs) 27. As only a very small percentage of these host molecules have thus far been examined, it seems likely that future research will elucidate many more host ncRNAs that contribute to tumorigenesis.
3. Gammaherpesvirus noncoding RNAs.
Through co-evolution with their mammalian hosts, gammaherpesviruses appear to have adapted the use of ncRNAs to their advantage, both through expression of their own unique ncRNA molecules and dysregulation of host ncRNAs. The EBV EBERs, first described in 1979, were among the first ncRNAs ever reported 28,29. Since that time, an array of different ncRNA molecules have been revealed to be encoded by gammherpesviruses, including a large number of miRNAs, an assortment of unique small noncoding RNAs and lncRNAs. Among these, miRNAs have thus far received the most attention and are described elsewhere 30–34. Here, we will focus on other gammaherpesvirus-encoded small and long ncRNAs. Thus far, only a small number of these molecules have been studied; however, the themes derived from those studies (outlined in Fig. 1), along with the new discovery of numerous novel gammaherpesvirus transcripts, illustrate the vast potential for the contribution of ncRNAs to the oncogenicity of these viruses.
Figure 1. Virus and host ncRNAs are central players in the tumorigenic attributes of gammaherpesviruses.
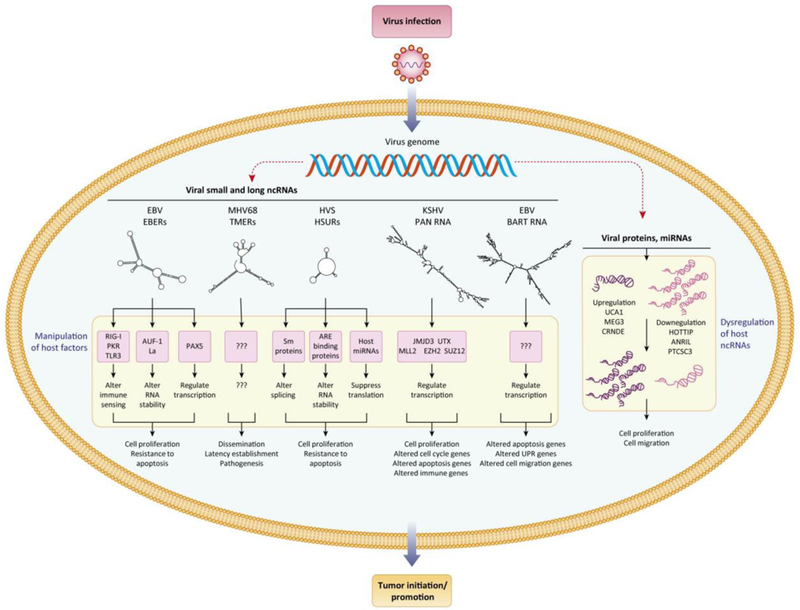
Gammaherpesviruses promote infection and pathogenesis through (a) utilization of viral ncRNAs that manipulate host cell factors and (b) dysregulation of the expression and stability of host ncRNAs. Viral ncRNAs coordinate interactions with host proteins, DNA and other ncRNAs to alter immune sensing, RNA stability, RNA splicing, transcription, and translation pathways. Such host cell modifications can drive hallmark cancer processes including promoting cell proliferation, blocking apoptosis, and altering immune response networks. Viral miRNAs and proteins can suppress or enhance the expression and stability of specific host nc NAs, including numerous lncRNAs that have already been linked to cancer.
3a. Gammaherpesvirus small noncoding RNAs: multifunctional RNAs expressed in all latently infected cells and tumors.
The Epstein-Barr virus encoded RNAs (EBERs).
The 167 nt EBER1 and 172 nt EBER2 are the two most abundant viral transcripts in latent EBV-infected cells 28,29. Unlike most ncRNAs, EBER1 and EBER2 are non-polyadenylated RNAs transcribed by RNA polymerase III 28,35,36. Despite the restricted viral gene expression programs operational during EBV latency, EBER transcripts accumulate to high levels in the nucleus of latently infected cells and tumor cells 37, suggesting that these molecules may be key players in chronic infection and tumorigenesis. As is typical for most ncRNAs, the ascribed cellular functions of the EBERs have been derived primarily from the discovery of their protein interactions, including with: the RNA binding proteins La, L22, and AU-rich element binding factor (AUF)-1 38; the antiviral immune sensors protein kinase R (PKR) 39, retinoic acid inducible gene- (RIG-I) 40, and toll-like receptor 3 (TLR3) 41; and the transcription factor PAX-5 42.
Not surprisingly, this array of molecular interactions may directly impact cellular functions that contribute to the tumorigenic potential of EBV. For example, numerous studies have demonstrated that EBV-negative BL B cell lines, NPC epithelial cell lines, and GC epithelial cell lines expressing EBERs are resistant to apoptosis, display increased proliferation, and demonstrate a tumorigenic phenotype 43–52. Moreover, transgenic mice constitutively expressing EBER1 in the lymphoid compartment develop lymphoid hyperplasia or B cell lymphoma at a high frequency 53.
While these studies clearly demonstrate the potential for the EBV EBERs to contribute to tumor initiation and/or promotion, defining the contribution of EBERs to tumorigenesis in the context of EBV infection remains a difficult and controversial problem, in part due to the necessary reliance of the field on in vitro studies. For example, while two groups have reported that loss of EBER from EBV strains did not affect primary B cell transformation in vitro 54,55, another study demonstrated a dose-dependent reduction of EBER-deficient EBV required to transform cells and significantly slower proliferation of resulting clones 56. These conflicting results illustrate the complications of studying individual viral genes in the context of intact viruses, which frequently have redundant mechanisms for achieving similar strategic outcomes. Moreover, none of these experiments address the function of the EBERs in in vivo tumorigenesis. Thus, although these latter findings demonstrate the potential of the EBERs to contribute to transformation, and underscore the molecular and cellular data indicating potential EBER functions that promote tumor cell fitness, much further work remains to determine the true function of the EBERs in tumorigenesis.
The murine gammaherpesvirus tRNA-miRNA encoded RNAs (TMERs).
Like its human virus counterparts, murine gammaherpesvirus 68 (MHV68) establishes lifelong latent infection in B cells and causes lymphoproliferative disease and B-cell lymphoma in immunocompromised hosts, making it an excellent model for the study of EBV and KSHV oncogenic factors. MHV68 encodes eight pol III-transcribed 200–250 nt ncRNAs 57–60 known as TMERs 61. Each MER harbors a tRNA-like element (vtRNA) followed by one or two downstream pre-miRNA hairpins. Through noncanonical processing, the TMERs yield up to 8 different vtRNAs and up to 28 mature miRNAs 58,62–65. Like the EBV EBERs, the MHV68 TMERs are highly expressed in latently infected cells and in virus-associated tumors 57,66, suggesting that these ncRNAs may play similar roles in chronic infection and oncogenesis. Consistent with this concept, targeted mutation of all eight TMERs reduces latent infection 61, particularly within the memory B cell population 61. Notably, during long term infection, a similar TMER mutant virus displays an increased load of cells carrying viral genome 67, suggesting that the TMERs may regulate the balance of lytic and latent infection. Strikingly, TMER mutation also completely ablates MHV68-induced lethal pneumonia 61, demonstrating the substantial potential for such gammaherpesvirus ncRNAs to contribute to in vivo pathogenesis.
Relatively little is known about the molecular mechanisms by which the TMERs function. However, it is now apparent that the T ERs are not simply primary miRNA transcripts, but instead may carry out biologically important functions through the use of alternately processed intermediates. To varying degrees, processing of each TMER yields intermediates that retain the vtRNA but lack one or both pre-miRNA hairpins 62,63,68. Some of these species are highly stable, supporting the concept that individual TMERs may encode multiple functional RNA elements. For example, an MHV68 mutant lacking TMER4 expression replicates normally in vivo, but is highly impaired for hematogenous dissemination 68,69, resulting in a substantial reduction in the establishment of peripheral latency. While an intermediate containing TMER4 vtRNA and one hairpin is sufficient to convey wild-type functionality, mutation of miRNA seed sequences within the remaining hairpin have no effect. Thus, as is the case for many ncRNAs, the secondary structure of the TMER4 hairpin is likely a key determinant of function. The TMER vtRNA sequences themselves may also hold some biological activity, as restoration of the TMER1 vtRNA alone in a combined TMER mutant herpesvirus is sufficient for partial recovery of lethal pneumonia 70. Thus the TMERs likely act as multifunctional ncRNA elements that directly participate at multiple stages of virus infection including dissemination and latency, and clearly contribute to the genesis of disease.
The Herpesvirus saimiri U RNAs (HSURs).
Herpesvirus saimiri (HVS) is a gammaherpesvirus that establishes asymptomatic lifelong latency in squirrel monkeys, its endemic hosts. In contrast, HVS infection leads to aggressive T cell leukemias and lymphomas in New World monkeys 71, and transforms marmoset T cells in vitro 72. HVS encodes seven small ncRNAs called Herpesvirus saimiri U RNAs (HSURs), which are pol II-derived, but nonpolyadenylated. HSURs range in size from 75 to 143 nt, and like the EBERs and TMERs are abundantly expressed in latently infected cells and tumor cells 73–77.
The HSURs appear to structurally and functionally mimic a subset of cellular small nuclear RNAs (snRNAs) that regulate splicing through their association with Sm proteins in RNPs 75. The 5’ ends of HSUR1 and 2 also interact extensively with AU-rich element (ARE) binding proteins 78,79, which normally regulate the stability of several subsets of cellular mRNAs. Interestingly, HSUR interaction with ARE binding proteins results in the upregulation of a small subset of genes that are hallmarks of T cell and NK cell activation 80.
The high degree of HSUR conservation among HVS strains implies an important role for these molecules during infection, but their specific functions during in vivo infection and tumorigenesis remain largely unknown. Although the HS Rs are dispensable for in vitro transformation of T cells 81,82, cells transformed with HVS lacking HSUR1 and 2 have a significantly slower growth phenotype 82, indicating that HSURs may also enhance the proliferative capacity of infected cells. Moreover, HSUR1 and 2 contain near perfect miRNA seed sequence matches for host miRNAs miR-16, miR-27 and miR-142–3p 83, and recent evidence indicates that HSUR2 directly recruits these miRNAs to host mRNA targets of HSUR2 binding, resulting in target mRNA repression 84. Consistent with a potential role for the HSURs in HVS tumorigenesis, HSUR2 mRNA targets include mRNAs in retinoblastoma, p53, and apoptosis pathways, and HSUR2-mediated repression of host mRNAs confers resistance to apoptosis.
3b. Gammaherpesvirus long noncoding RNAs: regulators of chromatin and transcription.
The KSHV polyadenylated nuclear (PAN) RNA.
To date, comparatively less is known about the existence and function of viral long ncRNAs as compared to viral small ncRNAs. KSHV expresses several potential noncoding transcripts antisense to known ORFs 95–101. Among these, the 10 kb antisense-to-latency transcript (ALT) is of great interest due it position antisense to the major KSHV latency locus. ALT was first revealed through genome-wide tiled microarray studies 96, and has since been validated and resolved through additional molecular approaches 102. However, as is the case for most other KSHV potential lncRNAs, the function of the ALT during infection and pathogenesis remains unknown.
In contrast, the KSHV polyadenylated nuclear (PAN) RNA is the most well-studied gammaherpesvirus lncRNA to date [reviewed in 103]. PAN RNA is a pol II-transcribed, 1.08 kb capped transcript that was initially identified in KSHV-infected Kaposi’s sarcoma lesions and primary effusion lymphoma (PEL) cell lines 100,104–106. PAN RNA is abundantly expressed throughout lytic replication, during which time it accumulates to a remarkably high level 100. Accumulation appears to result largely from a 79 nt ENE element that acts as an intramolecular clamp for the PAN RNA poly(A) tail and prevents nuclear exonuclease activity 107,108. Notably, this finding led to the discovery of other lncRNAs (eg, host MALAT1) which carry stabilizing ENE-like elements 109–111 and the identification of a nuclear RNA decay pathway mediated by the poly(A) binding protein PABPN1 112. Additional factors also appear to block PAN RNA decay, including virus-mediated nuclear localization of the cytoplasmic poly(A) binding protein PABPC1 113,114, direct binding to the viral protein ORF57 115,116, and interaction with the nuclear mRNA export protein ALYREF 115,117–119.
In the context of KSHV infection, PAN RNA has been implicated in several functions including as a regulator of transcription and chromatin remodeling. Knockdown of PAN RNA in KSHV-infected cells, or infection of cells using a KSHV mutant carrying a partial PAN RNA deletion, results in significantly reduced viral gene expression and subsequent virus production 113,120,121. PAN RNA associates with numerous viral gene promoters, as well as a subset of cellular gene promoters 121–123. Like HOTAIR and other cellular lncRNAs, PAN RNA may regulate transcription at these sites through recruitment of histone modifying enzymes 121,123. Importantly, although these interactions clearly influence viral fitness, it is apparent that that the presence of this incredibly abundant ncRNA also impacts cellular function. Indeed, PAN RNA associates with the promoters of multiple host genes, including those encoding proteins involved in cell cycle, cell death, and immune function 121–123. Moreover, PAN RNA expression is sufficient to induce proliferation of several different cell types123, strongly suggesting that this KSHV lncRNA could contribute to tumorigenesis.
The EBV BamH1 rightward transcript (BART) RNAs.
The EBV BART RNAs were initially identified as highly abundant transcripts expressed in nasopharyngeal carcinoma samples and cell lines 85–87. Subsequent work has reported BART RNA expression in all EBV-associated diseases, including highly abundant expression in gastric carcinoma tissues 88. Although the BART RNAs harbor several open reading frames (ORFs), BART-derived proteins have not been detected from endogenous translation 89,90. Instead, the BART RNAs appear to serve dual roles as pri-miRNA transcripts that produce up to 44 miRNAs 91,92, and as lncRNAs, at least some of which localize primarily to the nucleus 89,93,94. It is likely that many lncRNA-associated functions of the BART RNAs have yet to be revealed. However, recent evidence indicates that at least one spliced BART isoform significantly alters epithelial cell transcription independent of miRNA formation, resulting of the reduction of expression of multiple genes, including genes whose products promote the induction of apoptosis, the unfolded protein response, and cell migration 94. Notably, the set of host cell genes perturbed by this nuclear BART RNA overlapped with a large subset of genes that was significantly altered following EBV infection, indicating that the BART lncRNAs may play a central role in the host cell transcriptional changes observed in EBV-associated epithelial tumors.
3c. Discovery of new gammaherpesvirus RNAs.
Our growing understanding of the functions of these known ncRNAs provides important insight into their potential contributions to gammaherpesvirus infection and disease. However, it can be argued that these examples represent only a small fraction of all gammaherpesvirus ncRNAs. As has been the case for mammalian genomes, the application of tiled microarray and next generation sequencing technologies to herpesviruses genomes has revealed unexpectedly widespread transcription 95,124–127. Many of these regions do not contain canonical ORFs and thus may harbor previously unknown ncRNA genes. However, the high density of gammaherpesvirus genomes, with multiple overlapping transcripts and the potential for complex alternative splicing, has greatly hampered efforts to fully define the viral transcriptomes. Thus while short sequencing reads of such regions has revealed new genomic transcript features, global delineation of specific transcripts within these regions has thus far been impossible.
Fortunately, recent advances in sequencing technology and computational pipelines have facilitated the demarcation of individual overlapping transcripts within these regions. In particular, single molecule real time (SMRT) long-read sequencing has provided a critically important step forward by allowing reproducible reads lengths over 10 kb. While individual SMRT reads may reflect bona fide full - length transcript isoforms, sequencing limitations including 5’ truncations nevertheless necessitate validation of each output structure. Although this can be accomplished for individual isoforms using laborious classical molecular techniques, the use of alternative genomics platforms coupled with the computational pipeline TRIMD has enabled global bioinformatic validation of SMRT sequencing transcript structures 127.
Application of this approach to EBV-infected B cells has already yielded a plethora of new information about the EBV transcriptome 127, including more precise determination of the 5’ and 3’ ends of nearly two-thirds of previously annotated EBV transcripts, and the identification of 296 novel polyadenylated EBV transcripts. Of these, 65 are predicted to be noncoding. Likewise, the application of this approach to MHV68 has led to the first global annotation of MHV68 transcripts, including the discovery of at least 29 putative ncRNAs (REF in revision). These findings reveal the breadth of ncRNAs encoded by gammaherpesviruses and give insight into the depth to which these viruses may utilize such ncRNAs to modulate the host.
4. Dysregulation of host ncRNAs by gammaherpesviruses.
As exemplified by the ncRNAs described above, gammaherpesviruses implement a remarkable array of tactics to alter host cell functions for their benefit. To date, most functional outcomes have been measured in terms of modulation of host protein functions. However, with our recent gains in knowledge about the enormous breadth of host ncRNA molecules and functions, we should have every expectation that these gammaherpesvirus also target host ncRNAs for their advantage. Recent work with KSHV supports this concept, and further indicates that such alterations of host ncRNAs may also contribute to tumorigenic phenotypes (Fig. 1).
LncRNA expression profiling of endothelial cells revealed that the expression of hundreds of host lncRNAs is dysregulated during KSHV infection, including 325 with increased and 533 decreased expression 128. Despite the limited available information about the function of most lncRNAs, at least 54 of these dysregulated transcripts have been previously shown to be aberrantly expressed in human cancers. These include the well-studied lncRNA HOTTIP, which is upregulated in numerous malignancies 129, ANRIL, an oncogenic lnc NA that promotes proliferation in numerous cancers 130, UCA1, an oncogenic lncRNA that is upregulated in several tumors 131, and MEG3, a tumor suppressor lncRNA that is lost in a wide range of malignances132.
Interestingly, altered expression of many of these lncRNAs was dependent on KSHV miRNA targeting 128, and retrospective analyses of Argonaute crosslinking immuno precipitation (CLIP) data sets from KSHV- or EBV-infected cells revealed thousands of host lncRNAs within these viral and host miRNA targetomes respectively, suggesting that miRNA/lncRNA interaction are a novel paradigm of gene expression133. Some lncRNAs including ANRIL were also significantly dysregulated by KSHV latency proteins, demonstrating that at least in some cases this virus utilizes multiple strategies to alter host lncRNA expression.
The true benefits of modulation of host lncRNAs for gammaherpesviruses remain to be understood. However, even from these initial studies it is clear that altered expression could impact tumorigenicity: knockdown of KSHV-upregulated UCA1 reduced both proliferation and cell migration induced by KSHV. Thus these findings demonstrate that dysregulated host lncRNA expression could have significant functional consequences for the infected cell, and strongly suggest that this is yet another common strategy by which gammaherpesviruses hijack host cell processes.
5. Concluding Remarks.
In 2005, the discovery of tens of thousands of host ncRNAs 7 forever altered our perception of the mammalian transcriptome. Since that time, it has become apparent that at least a subset of these ncRNAs play important roles in nearly every important cellular process, including all of those that are cancer hallmarks 10. Viruses themselves have long demonstrated the ability to utilize their highly efficient toolkits to surreptitiously disseminate through hosts, enter permissive cells, and commandeer crucial host pathways for their advantage. We now know that viruses carry out these lines of attack in part through the use of their own unique ncRNAs and by altering the expression of host ncRNAs. Yet, the development of new genomics platforms and computational pipelines is revealing that we have perhaps only begun to understand the depth to which these viruses utilize ncRNAs to their advantage (see Outstanding Questions). We are now at the forefront of the study of ncRNA biology. As this exploding field continues to elucidate the molecular mechanism by which ncRNAs act and the critical role that host ncRNAs play in tumorigenesis, the coming years should bring an exciting new understanding of the very likely role that these molecules play in the genesis of gammaherpesvirus-associated tumors.
Outstanding questions.
Do gammaherpesvirus ncRNAs have a specific and essential function during chronic in vivo infection?
Is gammaherpesvirus manipulation of host ncRN s a viral strategy to modulate the host cell, or an indirect effect of virus alteration of host cell transcription and epigenetics?
Do gammaherpesvirus ncRNAs directly mimic host cell ncRNA functions through the use of similar secondary structure?
Can discovery of gammaherpesvirus ncRNA functions be manipulated for new therapeutic interventions or to prevent infection?
Do gammaherpesviruses directly regulate host ncRNA processing machinery for their advantage?
Highlights:
The gammaherpesviruses EBV and KSHV are ubiquitous human pathogens that are associated with a variety of malignancies including several types of lymphomas and carcinomas.
Host cell noncoding RNAs (ncRNAs) play central roles in all cancer hallmark cellular processes.
Dysregulation of host cell ncRNAs is associated with the development of multiple types of malignancies.
Gammaherpesviruses encode numerous types of noncoding RNAs, including both short and long ncRNAs.
Gammaherpesvirus ncRNAs modulate multiple host cell functions, resulting in the induction of cell proliferation and cell migration, blocking apoptosis, and immune evasion.
Gammaherpesvirus ncRNAs are central players in latency and pathogenesis.
Gammaherpesvirus factors dysregulate host cell ncRNAs.
Numerous gammaherpesvirus ncRNAs have recently been discovered.
Acknowledgements.
S.A.T, R.R. and E.K.F, were supported by NIH P01CA214091. S.A.T. was also supported by NIH R01AI108407.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Jha HC, Banerjee S & Robertson ES The Role of Gammaherpesviruses in Cancer Pathogenesis. Pathogens 5, 18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwasaki YW, Siomi MC & Siomi H PIWI-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem 84, 405–433 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Matera AG, Terns RM & Terns MP Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol 8, 209–220 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Ulitsky I & Bartel DP lincRNAs: Genomics, Evolution, and Mechanisms. Cell 154, 26–46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derrien T et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res 22, 1775–1789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carninci P et al. The transcriptional landscape of the mammalian genome. Science 309, 1559–1563 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D & Weinberg RA The Hallmarks of Cancer. Cell 100, 57–70 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D & Weinberg RA Hallmarks of Cancer: The Next Generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Schmitt AM & Chang HY Long Noncoding RNAs in Cancer Pathways. Cancer Cell 29, 452–463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M & Kiss T Site-Specific Ribose Methylation of Preribosomal RNA: A Novel Function for Small Nucleolar RNAs. Cell 85, 1077–1088 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Tollervey D & Kiss T Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol 9, 337–342 (1997) [DOI] [PubMed] [Google Scholar]
- 13.Weinstein LB & Steitz JA Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol 11, 378–384 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Dong X-Y et al. SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum. Mol. Genet 17, 1031–1042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong X-Y et al. Implication of snoRNA U50 in human breast cancer. J. Genet. Genomics 36, 447–454 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka R et al. Intronic U50 small-nucleolar-RNA (snoRNA) host gene of no protein-coding potential is mapped at the chromosome breakpoint t(3;6)(q27;q15) of human B-cell lymphoma. Genes ells 5, 277–287 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Liao J et al. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol. Cancer 9, 198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mei Y-P. et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 31, 2794–2804 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moyano M & Stefani G piRNA involvement in genome stability and human cancer. J. Hematol. Oncol.J Hematol Oncol 8, 38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan H et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia 29, 196–206 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Cheng J et al. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin. Chim. Acta 412, 1621–1625 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Prensner JR & Chinnaiyan AM The Emergence of lncRNAs in Cancer Biology. Cancer Discov 1, 391–407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta RA et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji P et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Stadler PF et al. Evolution of Vault RNAs. Mol. Biol. Evol 26, 1975–1991 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Kowalski MP & Krude T Functional roles of non-coding Y RNAs. Int. J. Biochem. Cell Biol 66, 20–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patton JG et al. Biogenesis, delivery, and function of extracellular RNA. J. Extracell. Vesicles 4, 27494 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lerner MR, Andrews NC, Miller G & Steitz JA Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. U. S. A 78, 805–809 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rymo L Identification of transcribed regions of Epstein-Barr virus DNA in Burkitt lymphoma-derived cells. J. Virol 32, 8–18 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boss IW & Renne R Viral miRNAs: tools for immune evasion. Curr. Opin. Microbiol 13, 540–545 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cullen BR Herpesvirus microRNAs: Phenotypes and functions. Curr. Opin. Virol 1, 211–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feldman ER & Tibbetts SA Emerging Roles of Herpesvirus microRNAs During In Vivo Infection and Pathogenesis. Curr. Pathobiol. Rep 3, 209–217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottwein E Kaposi’s Sarcoma-Associated Herpesvirus microRNAs. Front. Microbiol 3, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grundhoff A & Sullivan CS Virus-encoded microRNAs. Virology 411, 325–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arrand JR & Rymo L Characterization of the major Epstein-Barr virus-specific RNA in Burkitt lymphoma-derived cells. J. Virol 41, 376–389 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosa MD, Gottlieb E, Lerner MR & Steitz JA Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol. Cell. Biol 1, 785–796 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howe JG & Steitz JA Localization of Epstein-Barr virus-encoded small RNAs by in situ hybridization. Proc. Natl. Acad. Sci 83, 9006–9010 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee N, Pimienta G & Steitz JA AUF1/hnRNP D is a novel protein partner of the EBER1 noncoding RNA of Epstein-Barr virus. RNA 18, 2073–2082 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharp TV et al. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res 21,4483–4490 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samanta M, Iwakiri D, Kanda T, Imaizumi T & Takada K EB virus‐encoded RNAs are recognized by RIG‐I and activate signaling to induce type I IFN. EMBO J 25, 4207–4214 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwakiri D et al. Epstein-Barr virus (EBV)–encoded small RNA is released from EBV-infected cells and activates signaling from toll-like receptor 3. J. Exp. Med 206, 2091–2099 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee N, Moss WN, Yario TA & Steitz JA EBV Noncoding RNA Binds Nascent RNA to Drive Host PAX5 to Viral DNA. Cell 160, 607–618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houmani JL, Davis CI & Ruf IK Growth-Promoting Properties of Epstein-Barr Virus EBER-1 RNA Correlate with Ribosomal Protein L22 Binding. J. Virol 83, 9844–9853 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwakiri D, Eizuru Y, Tokunaga M & Takada K Autocrine Growth of Epstein-Barr Virus-Positive Gastric Carcinoma Cells Mediated by an Epstein-Barr Virus-Encoded Small RNA. Cancer Res 63, 7062–7067 (2003). [PubMed] [Google Scholar]
- 45.Iwakiri D, Sheen T-S, Chen J-Y, Huang DP & Takada K Epstein–Barr virus-encoded small RNA induces insulin-like growth factor 1 and supports growth of nasopharyngeal carcinoma-derived cell lines. Oncogene 24, 1767 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Komano J, Maruo S, Kurozumi K, Oda T & Takada K Oncogenic Role of Epstein-Barr Virus-Encoded RNAs in Burkitt’s Lymphoma Cell Line Akata. J. Virol 73, 9827–9831 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruf IK, Rhyne PW, Yang C, Cleveland JL & Sample JT Epstein-Barr Virus Small RNAs Potentiate Tumorigenicity of Burkitt Lymphoma Cells Independently of an Effect on Apoptosis. J.Virol 74, 10223–10228 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu N, Tanabe-Tochikura A, Kuroiwa Y & Takada K Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt’s lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J. Virol 68, 6069–6073 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsang CM et al. Epstein-Barr virus infection in immortalized nasopharyngeal epithelial cells: Regulation of infection and phenotypic characterization. Int. J. Cancer 127, 1570–1583 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Wong HL et al. Stable expression of EBERs in immortalized nasopharyngeal epithelial cells confers resistance to apoptotic stress. Mol. Carcinog 44, 92–101 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto N, Takizawa T, Iwanaga Y, Shimizu N. & Yamamoto N Malignant transformation of B lymphoma cell line BJAB by Epstein – Barr virus-encoded small RNAs. FEBS Lett 484, 153–158 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Yoshizaki T et al. Oncogenic role of Epstein-Barr virus-encoded small RNAs (EBERs) in nasopharyngeal carcinoma. Auris. Nasus. Larynx 34, 73–78 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Repellin CE, Tsimbouri PM, Philbey AW & Wilson JB Lymphoid Hyperplasia and Lymphoma in Transgenic Mice Expressing the Small Non-Coding RNA, EBER1 of Epstein-Barr Virus. PLOS ONE 5, e9092 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gregorovic G et al. Cellular Gene Expression That Correlates with EBER Expression in Epstein-Barr Virus-Infected Lymphoblastoid Cell Lines. J. Virol 85, 3535–3545 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swaminathan S, Tomkinson B & Kieff E Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc. Natl. Acad. Sci. U. S. A 88, 1546–1550 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yajima M, Kanda T & Takada K Critical Role of Epstein-Barr Virus (EBV)-Encoded RNA in Efficient EBV-Induced B-Lymphocyte Growth Transformation. J. Virol 79, 4298–4307 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bowden RJ, Simas JP, Davis AJ & Efstathiou S Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J. Gen. Virol 78 ( Pt 7), 1675–1687 (1997). [DOI] [PubMed] [Google Scholar]
- 58.Pfeffer S et al. Identification of microRNAs of the herpesvirus family. Nat Meth 2, 269–276 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Simas JP, Bowden RJ, Paige V & Efstathiou S Four tRNA-like sequences and a serpin homologue encoded by murine gammaherpesvirus 68 are dispensable for lytic replication in vitro and latency in vivo. J. Gen. Virol 79 ( Pt 1), 149–153 (1998). [DOI] [PubMed] [Google Scholar]
- 60.Virgin HW et al. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol 71, 5894–5904 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feldman ER et al. Virus-Encoded MicroRNAs Facilitate Gammaherpesvirus Latency and Pathogenesis In Vivo. mBio 5, e00981–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bogerd HP et al. A Mammalian Herpesvirus Uses Noncanonical Expression and Processing Mechanisms to Generate Viral MicroRNAs. Mol. Cell 37, 135–142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diebel KW, Smith AL & van Dyk LF Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters. RNA 16, 170–185 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reese TA et al. Identification of Novel MicroRNA-Like Molecules Generated from Herpesvirus and Host tRNA Transcripts. J Virol 84, 10344–10353 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu JY et al. Identification and Analysis of Expression of Novel MicroRNAs of Murine Gammaherpesvirus 68. J Virol 84, 10266–10275 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tarakanova VL et al. Murine Gammaherpesvirus 68 Infection Is Associated with Lymphoproliferative Disease and Lymphoma in BALB {beta}2 Microglobulin-Deficient Mice. J Virol 79, 14668–14679 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steer B et al. The small noncoding RNAs (sncRNAs) of murine gammaherpesvirus 68 (MHV-68) are involved in regulating the latent-to-lytic switch in vivo. Sci. Rep 6, 32128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feldman ER et al. A Gammaherpesvirus Noncoding RNA Is Essential for Hematogenous Kincaid Dissemination and Establishment of Peripheral Latency. mSphere 1, e00105–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69., Kincaid RP & Sullivan CS Lessons Learned from In Vivo Studies of a Viral Noncoding RNA. MSphere 1, e00026–16(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diebel KW et al. Gammaherpesvirus small noncoding RNAs are bifunctional elements that regulate infection and contribute to virulence in vivo. mBio 6, e01670–01614 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fleckenstein B & Desrosiers RC Herpesvirus saimiri and herpesvirus ateles. in The Herpesviruses 1, 253–332 (Plenum Press, 1982). [Google Scholar]
- 72.Schirm S, Müller I, Desrosiers RC & Fleckenstein B Herpesvirus saimiri DNA in a lymphoid cell line established by in vitro transformation. J. Virol 49, 938–946 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Albrecht J. & Fleckenstein B Nucleotide sequence of HSUR 6 and HSUR 7, two small RNAs of herpesvirus saimiri. Nucleic Acids Res 20, 1810 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee SI & Steitz J. Herpesvirus saimiri U RNAs are expressed and assembled into ribonucleoprotein particles in the absence of other viral genes. J. Virol 64, 3905–3915 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee SI, Murthy SCS, Trimble JJ, Desrosiers RC & Steitz JA Four novel U RNAs are encoded by a herpesvirus. Cell 54, 599–607 (1988). [DOI] [PubMed] [Google Scholar]
- 76.Murthy S, Kamine J & Desrosiers RC Viral-encoded small RNAs in herpes virus saimiri induced tumors. EMBO J 5, 1625–1632 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wassarman DA, Lee SI & Steitz JA Nucleotide sequence of HSUR 5 RNA from herpesvirus saimiri. Nucleic Acids Res 17, 1258 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cook HL, Mischo HE & Steitz JA The Herpesvirus saimiri Small Nuclear RNAs Recruit AU-Rich Element-Binding Proteins but Do Not Alter Host AU-Rich Element-Containing mRNA Levels in Virally Transformed T Cells. Mol. Cell. Biol 24, 4522–4533 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Myer VE, Lee SI & Steitz JA Viral small nuclear ribonucleoproteins bind a protein implicated in messenger RNA destabilization. Proc. Natl. Acad. Sci 89, 1296–1300 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cook HL et al. Small Nuclear RNAs Encoded by Herpesvirus saimiri Upregulate the Expression of Genes Linked to T Cell Activation in Virally Transformed T Cells. Curr. Biol 15, 974–979 (2005). [DOI] [PubMed] [Google Scholar]
- 81.Ensser A, Pfinder A, Müller-Fleckenstein I.&Fleckenstein B TheURNAGenesofHerpesvirus Saimiri (Strain C488) Are Dispensable for Transformation of Human T Cells In Vitro. J. Virol 73, 10551–10555 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Murthy SC, Trimble JJ & Desrosiers RC Deletion mutants of herpesvirus saimiri define an open reading frame necessary for transformation. J. Virol 63, 3307–3314 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cazalla D, Yario T & Steitz JA Down-Regulation of a Host MicroRNA by a Herpesvirus saimiri Noncoding RNA. Science 328, 1563–1566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gorbea C, Mosbruger T & Cazalla D A viral Sm-class RNA base-pairs with mRNAs and recruits microRNAs to inhibit apoptosis. Nature 550, 275–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gilligan K et al. Novel transcription from the Epstein-Barr virus terminal EcoRI fragment, DIJhet, in a nasopharyngeal carcinoma. J. Virol 64, 4948–4956 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hitt MM et al. EBV gene expression in an NPC-related tumour. EMBO J 8, 2639–2651 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Raab-Traub N, Hood R, Yang CS, Henry B & Pagano J. Epstein-Barr virus transcription in nasopharyngeal carcinoma. J. Virol 48, 580–590 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sugiura M et al. Transcriptional analysis of Epstein-Barr virus gene expression in EBV-positive gastric carcinoma: unique viral latency in the tumour cells. Br. J. Cancer 74, 625–631 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Al-Mozaini M et al. Epstein–Barrvirus BART gene expression. J. Gen. Virol 90, 307–316 (2009). [DOI] [PubMed] [Google Scholar]
- 90.Thornburg NJ, Kusano S & Raab-Traub N Identification of Epstein-Barr Virus RK-BARF0-Interacting Proteins and Characterization of Expression Pattern. J. Virol 78, 12848–12856 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cai X et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog 2, e23 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pfeffer S et al. Identification of virus-encoded microRNAs. Science 304, 734–736 (2004). [DOI] [PubMed] [Google Scholar]
- 93.Edwards RH, Marquitz AR & Raab-Traub N Epstein-Barr Virus BART MicroRNAs Are Produced from a Large Intron prior to Splicing. J. Virol 82, 9094–9106 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marquitz AR, Mathur A, Edwards RH & Raab-Traub N Host Gene Expression Is Regulated by Two Types of Noncoding RNAs Transcribed from the Epstein-Barr Virus BamHI A Rightward Transcript Region. J. Virol 89, 11256–11268 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Arias C et al. KSHV 2.0: Comprehensive Annotation of the Kaposi’s Sarcoma-Associated Herpesvirus Genome Using Next-Generation Sequencing Reveals Novel Genomic and Functional Features. PLoS Pathog 10, e1003847 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chandriani S, Xu Y & Ganem D The Lytic Transcriptome of Kaposi’s Sarcoma-Associated Herpesvirus Reveals Extensive Transcription of Noncoding Regions, Including Regions Antisense to Important Genes. J. Virol 84, 7934–7942 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dresang LR et al. Coupled transcriptome and proteome analysis of human lymphotropic tumor viruses: insights on the detection and discovery of viral genes. BMC Genomics 12, 625 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Majerciak V et al. A Viral Genome Landscape of RNA Polyadenylation from KSHV Latent to Lytic Infection. PLOS Pathog 9, e1003749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.aveliev AK, Zhu FX & Yuan Y Transcription Mapping and Expression Patterns of Genes in the Major Immediate-Early Region of Kaposi’s Sarcoma-Associated Herpesvirus. Virology 299, 301–314 (2002). [DOI] [PubMed] [Google Scholar]
- 100.Sun R, Lin SF, Gradoville L & Miller G Polyadenylylated nuclear RNA encoded by Kaposisarcoma-associated herpesvirus. Proc. Natl. Acad. Sci 93, 11883–11888 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Taylor JL, Bennett HN, Snyder BA, Moore PS&Chang Y Transcriptionalanalysisoflatent and inducible Kaposi’s sarcoma-associated herpesvirus transcripts in the K4 to K7 region. J. Virol 79, 15099–15106 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schifano JM, Corcoran K, Kelkar H & Dittmer DP Expression of the Antisense-to-Latency Transcript Long Noncoding RNA in Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol 91, e01698– 16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Conrad NK New insights into the expression and functions of the Kaposi’s sarcoma-associated herpesvirus long noncoding PAN RNA. Virus Res 212, 53–63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Staskus KA et al. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol 71, 715–719 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhong W & Ganem D Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8). J. Virol 71, 1207–1212 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhong W, Wang H, Herndier B & Ganem D Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. U. S. A 93, 6641–6646 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Conrad NK& Steitz JAA Kaposi’s sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J 24, 1831–1841 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA & Steitz JA Poly(A) Tail Recognition by a Viral RNA Element Through Assembly of a Triple Helix. Science 330, 1244–1247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brown JA, Valenstein ML, Yario TA, Tycowski KT & Steitz JA Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc. Natl. Acad. Sci 109, 19202–19207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tycowski KT, Shu M-D, Borah S, Shi M & Steitz JA Conservation of a Triple-Helix-Forming RNA Stability Element in Noncoding and Genomic RNAs of Diverse Viruses. Cell Rep 2, 26–32 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wilusz JE. et al. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 26, 2392–2407 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bresson SM & Conrad NK The Human Nuclear Poly(A)-Binding Protein Promotes RNA Hyperadenylation and Decay. PLOS Genet 9, e1003893 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Borah S, Darricarrère N, Darnell A, Myoung J & Steitz JA A Viral Nuclear Noncoding RNA Binds Re-localized Poly(A) Binding Protein and Is Required for Late KSHV Gene Expression. PLOS Pathog 7, e1002300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee YJ & Glaunsinger BA Aberrant Herpesvirus-Induced Polyadenylation Correlates With Cellular Messenger RNA Destruction. PLOS Biol 7, e1000107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Massimelli MJ et al. Stability of a Long Noncoding Viral RNA Depends on a 9-Nt Core Element at the RNA 5’ End to Interact with Viral ORF57 and Cellular PABPC1. Int. J. Biol. Sci 7, 1145–1160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sahin BB, Patel D & Conrad NK Kaposi’s Sarcoma-Associated Herpesvirus ORF57 Protein Binds and Protects a Nuclear Noncoding RNA from Cellular RNA Decay Pathways. PLOS Pathog 6, e1000799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li D-J, Verma D & Swaminathan S Binding of Cellular Export Factor REF/Aly by Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) ORF57 Protein Is Not Required for Efficient KSHV Lytic Replication. J. Virol 86, 9866–9874 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Massimelli MJ, Majerciak V, Kruhlak M & Zheng Z-M Interplay between olyadenylate-Binding Protein 1 and Kaposi’s Sarcoma-Associated Herpesvirus ORF57 in Accumulation of Polyadenylated Nuclear RNA, a Viral Long Noncoding RNA. J. Virol 87, 243–256 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stubbs SH, Hunter OV, Hoover A & Conrad NK Viral Factors Reveal a Role for REF/Aly in Nuclear RNA Stability. Mol. Cell. Biol 32, 1260–1270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ideue T, Hino K, Kitao S, Yokoi T & Hirose T Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA 15, 1578–1587 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rossetto CC & Pari G KSHV PAN RNA Associates with Demethylases UTX and JMJD3 to Activate Lytic Replication through a Physical Interaction with the Virus Genome. PLOS Pathog 8, e1002680 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rossetto CC & Pari GS Kaposi’s Sarcoma- ssociated Herpesvirus Noncoding Polyadenylated Nuclear RNA Interacts with Virus- and Host Cell-Encoded Proteins and Suppresses Expression of Genes Involved in Immune Modulation. J. Virol 85, 13290–13297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rossetto CC, Tarrant-Elorza M, Verma S, Purushothaman P & Pari GS Regulation of Viral and.Cellular Gene Expression by Kaposi’s Sarcoma-Associated Herpesvirus Polyadenylated Nuclear RNA. J. Virol 87, 5540–5553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cheng BYH et al. Tiled Microarray Identification of Novel Viral Transcript Structures and Distinct Transcriptional Profiles during Two Modes of Productive Murine Gammaherpesvirus 68 Infection. J Virol 86, 4340–4357 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gatherer D. et al. High-resolution human cytomegalovirus transcriptome. Proc. Natl. Acad. Sci. U. S A 108, 19755–19760 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Johnson LS, Willert EK & Virgin HW Redefining the Genetics of Murine Gammaherpesvirus 68 via Transcriptome-Based Annotation. Cell Host Microbe 7, 516–526 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.O’Grady T et al. Global Bidirectional Transcription of the Epstein-Barr Virus Genome during Reactivation. J. Virol 88, 1604–1616 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sethuraman S, Gay LA, Jain V, Haecker I & Renne R microRNA dependent and independent deregulation of long non-coding RNAs by an oncogenic herpesvirus. PLOS Pathog 13, e1006508 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lian Y et al. HOTTIP: a critical oncogenic long non-coding RNA in human cancers. Mol. Biosyst 12, 3247–3253 (2016). [DOI] [PubMed] [Google Scholar]
- 130.Congrains A, Kamide K, Ohishi M & Rakugi H ANRIL: Molecular Mechanisms and Implications in Human Health. Int. J. Mol. Sci 14, 1278–1292 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xue M, Chen W & Li X Urothelial cancer associated 1: a long noncoding RNA with a crucial role in cancer. J. Cancer Res. Clin. Oncol 142, 1407–1419 (2016). [DOI] [PubMed] [Google Scholar]
- 132.Zhou Y, Zhang X & Klibanski A MEG3 noncoding RNA: a tumor suppressor. J. Mol. Endocrinol 48, R45–R53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sethuraman S, Thomas M, Gay LA & Renne R Computational analysis of ribonomics datasets identifies long non-coding RNA targets of γ-herpesviral miRNAs. Nucleic Acids Res (2018). doi: 10.1093/nar/gky459 [DOI] [PMC free article] [PubMed] [Google Scholar]


