Abstract
Recent efforts aimed at integrating in vitro high-throughput screening (HTS) data into chemical toxicity assessments are necessitating increased understanding of concordance between chemical-induced responses observed in vitro versus in vivo. This investigation set out to (1) measure concordance between in vitro HTS data and transcriptomic responses observed in vivo, focusing on the liver, and (2) identify attributes that can influence concordance. Signal response profiles from 130 substances were compared between in vitro data produced through Tox21 and liver transcriptomic data through DrugMatrix, collected from rats exposed to a chemical for ≤5 days. A global in vitro-to-in vivo comparative analysis based on pathway-level responses resulted in an overall average percent agreement of 79%, ranging on a per-chemical basis between 41% and 100%. Whereas concordance amongst inactive chemicals was high (89%), concordance amongst chemicals showing in vitro activity was only 13%, suggesting that follow-up in vivo and/or orthogonal in vitro assays would improve interpretations of in vitro activity. Attributes identified to influence concordance included experimental design attributes (eg, cell type), target pathways, and physicochemical properties (eg, logP). The attribute that most consistently increased concordance was dose applicability, evaluated by filtering for experimental doses administered to rats that were within 10-fold of those related to likely bioactivity, derived using Tox21 data and high-throughput toxicokinetic modeling. Together, findings suggest that in vitro screening approaches to predict in vivo toxicity are viable particularly when certain attributes are considered, including whether activity versus inactivity is observed, experimental design, chemical properties, and dose applicability.
Keywords: DrugMatrix, high-throughput screening, in vitro-to-in vivo, Tox21, toxicokinetics, transcriptomics
A great deal of discussion within the toxicology community focuses on the need to adapt existing toxicity testing paradigms to decrease reliance upon traditional animal testing and increase reliance upon alternative methods, including in vitro high-throughput screening (HTS) and predictive toxicology based on computational approaches. This dialog is reflected in recent conference sessions and workshops (eg, Society of Toxicology [SOT] FutureTox meeting series [I/II/III] [SOT, 2017]) and reports by the National Academy of Sciences (NAS, 2007, 2017), with many international organizations dedicated to the advancement of alternative toxicological methods. The high number of chemicals for which little or no toxicological information is available necessitates testing strategies that move toward alternative methods that are economically efficient, maintain requisite underlying biology, and result in decreased animal testing. The development of in vitro HTS assays has the potential to address, in part, these posed challenges. With this development comes the necessary task of confirming/validating that toxicity responses observed in vitro can be used to predict in vivo toxicity.
In vitro HTS assays represent automated methods that allow for the rapid testing of select bioactivities at the molecular- or cellular-level across a large number of chemicals (EPA, 2018). The models being used in HTS assays range from a monolayer of cells to individual proteins, all of which are manipulated and carried out by robotics. These HTS methods are used to detect chemical-induced changes in bioactivity based on various experimental endpoints, including antibody binding, cytotoxicity, mitochondrial membrane potential, reporter gene transcription, etc. Results from these screening efforts can identify compounds that modulate specific biological pathways, which then enhances the understanding of biochemical interactions or the potential roles of a compound in biological processes (EPA, 2018). The utility of in vitro HTS assay toward predicting biochemical interactions/changes in biological processes in vivo remains under discussion largely due to the simpler nature of the biology captured by HTS assays (NAS, 2017). It is therefore important to understand which types of in vitro endpoints, derived under specific experimental conditions, are better suited to predict or inform select mechanisms of in vivo toxicity.
Previous studies have evaluated in vitro-to-in vivo (ie, in vitro-in vivo) response concordance with varied results. A cancer-related concordance study found an association between rodent hepatic liver lesions and human nuclear receptor-based assays (Shah et al., 2011); whereas two studies using in vitro bioactivity data found poor prediction of cancer in rodents (Cox et al., 2016) and for cancer hazard classification (Becker et al., 2017). Non-cancer biological endpoints have also been evaluated for response concordance. For instance, in vitro HTS response profiles have been shown to correlate with in vivo developmental toxicity endpoints (Sipes et al., 2011) and have shown strong bioactivity across chemicals with known male reproductive developmental phenotypes (Leung et al., 2016). Combining in vitro HTS data with chemical structure descriptors has shown utility in predicting liver lesions (Liu et al., 2015) and other organ-specific outcomes (Liu et al., 2017). Another study found that in vitro HTS data were no better at predicting in vivo toxicity than chemical descriptors alone (Thomas et al., 2012). Clearly, mixed results have been generated comparing in vitro HTS data to in vivo toxicity, demonstrating that further research is needed on this topic.
The majority of studies evaluating in vitro-in vivo response concordance, to date, have compared relatively simple biological assays to apical responses based on complex biological states or diseases. To place these comparisons in the context of adverse outcome pathways (AOPs) (Browne et al., 2017), analyses have largely compared in vitro molecular interactions (eg, molecular initiating events) against in vivo organ responses, whereas bypassing cellular response event(s) (ie, key events). This gap from in vitro molecular interactions to alterations in pathology may be so wide as to render such comparisons difficult, and at times, unfeasible with limited data. Instead, it may be more reasonable to carry out comparisons at molecular and cellular response levels. Indeed, a recent study found that transcriptomic responses evaluated at the pathway-level (representing a cellular response) showed similarities within in vitro mouse, rat, and human hepatocytes; in vivo mouse and rat liver; and in vivo zebrafish embryos (Driessen et al., 2015). Similarly, we recently compared Tox21 HTS assay data to in vivo transcriptomic responses at the pathway-level in the mouse intestine using hexavalent chromium as a case study (Rager et al., 2017). A comparison of these molecular/cellular responses showed both similar and different signaling profiles between datasets, suggesting that there may be domains of applicability in using HTS data to inform in vivo toxicity (Rager et al., 2017). This study therefore set out to expand upon these more mechanistic-based comparative strategies by evaluating in vitro HTS assay targets (representing molecular interactions) against their associated pathway alterations (representing cellular responses) using an expanded in vivo transcriptomic database.
Here, we present a comparison between chemical responses within the Tox21 HTS database and liver transcriptomic data available through the DrugMatrix database, as evaluated at the pathway-level. The liver was selected as the organ of interest, as it represents the most common target organ in animal toxicity studies (Ballet, 1997). Overall in vitro-in vivo concordance was first evaluated for 130 chemicals, and then attributes that potentially influence concordance were assessed. These attributes included specific design aspects of in vitro assays and in vivo experimentation, pathway targets, chemical-specific attributes (ie, physicochemical properties), and the applicability of chemical doses used for in vivo-in vitro comparisons. Findings provide increased understanding of the potential ranges of applicability for the use of in vitro assay data to predict or provide mechanistic context for in vivo toxicity responses.
MATERIALS AND METHODS
Organization of Tox21 in vitro HTS bioactivity data
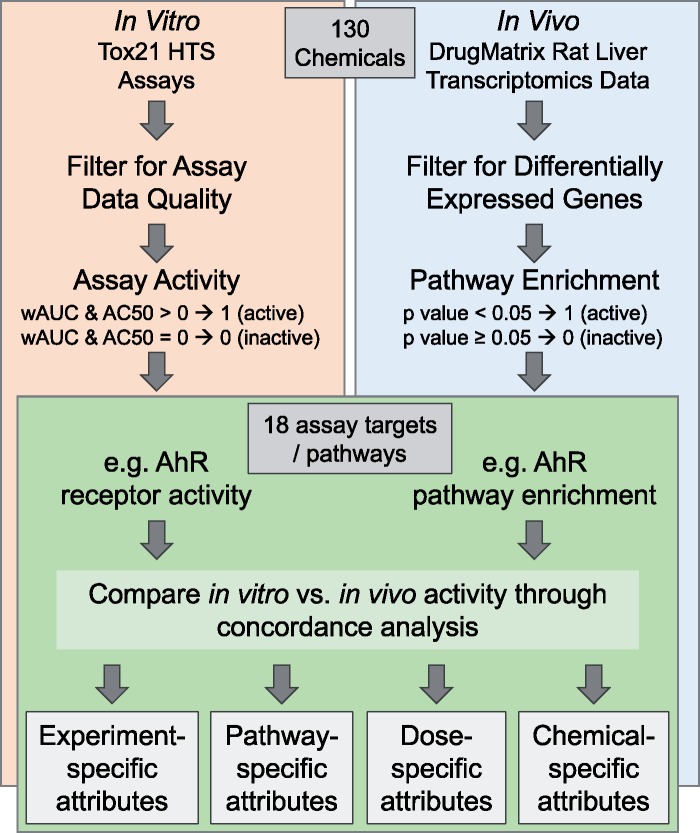
An overview of the steps used to organize data from Tox21 and DrugMatrix are provided in Figure 1. The Tox21 in vitro HTS database is organized by multiple agencies, including the National Toxicology Program (NTP), aimed at developing and making publicly available in vitro HTS assay data from large chemical toxicity screens (Hsieh et al., 2015; NTP, 2017b). Tox21 assay data currently consist of 43 in vitro assay endpoints tested across >10 000 chemicals, ranging from environmental and industrial chemicals to pharmaceutical agents (Hsieh et al., 2015). Tox21 data were obtained through the NTP Tox21 activity profiler (NTP, 2017b) for chemicals that were also within the DrugMatrix database. Data were downloaded based on chemical CASRN using default parameters, with the additional selections to download all available activity data, which allowed for the selection of parameters required to distinguish between inactive/active versus inconsistent/flagged bioactivity results. Data were specifically filtered to exclude failed purity testing and assay endpoint data that were flagged based on ‘autofluorescent,’ ‘cytotoxicity,’ ‘not_supported_by_ch2,’ ‘not_tested,’ and ‘weaknoisy_in_rep.’ These filters resulted in data that met suggested processing standards, as previously defined (Hsieh, 2016; Hsieh et al., 2015), that accounted for important interferences, including the cytotoxicity burst interference phenomenon (Judson et al., 2016).
Figure 1.
Flowchart of steps used to evaluate in vitro-in vivo response concordance through the comparison of in vitro Tox21 bioactivity (left) against in vivo pathway-level changes in the rat liver (right). Abbreviation: AhR, aryl hydrocarbon receptor.
Assay bioactivity data included wAUC (weighted area under the curve) and AC50 (concentration at which the activity reaches 50% of its maximal values for an assay-chemical pair) values. These values were derived from methods outlined by Hsieh et al. (2015). Briefly, wAUCs were calculated as the ratio of the area under the curve over the range of concentrations tested multiplied by the point of departure (POD), representing the concentration at which the response was above assay-specific noise thresholds, with all concentration values converted using negative log10 to make higher POD values relate to higher wAUCs. For this study, assay bioactivity results were simplified to binary calls of inactive (0) or active (1), with inactive assay endpoints showing quality-filtered wAUC and AC50 values of 0 and active assays showing quality-filtered wAUC and AC50 values >0. Example assay endpoint activity plots were generated using the Tox21 Curve Browser (NTP, 2017c).
Organization of in vivo transcriptomic profiles from the DrugMatrix database
DrugMatrix is a publicly available toxicogenomic reference database that contains microarray data from tissues of rats administered pharmaceutical agents, environmental chemicals, or other substances. Initially developed by Incyte Genomics, Inc. and Iconix Pharmaceuticals, Inc., the DrugMatrix database was acquired by NTP in 2010 and data were made publicly available to the larger scientific community. This database has demonstrated utility toward predictive toxicity in preclinical drug safety and toxicity assessments. For example, chemical-induced transcriptomic signatures from DrugMatrix have been used for biomarker identification and mechanistic analyses to better understand mechanisms of action and toxicity (observed at the phenotypic-level) at earlier time points than historically used during drug development processes (Ganter et al., 2006). The current investigation focused on the DrugMatrix transcriptomic profiles in the liver (representing the tissue with the most data available) for a subset of chemicals selected based on their inclusion within the Tox21 database. Because the majority (∼80%) of Tox21 assays were conducted between 12- and 24-h durations (with all assays conducted at 1–40 h), in vivo data collected 24 h post-exposure were prioritized for analysis. In cases where 24-h exposure data were not available, 3- or 5-day exposure data were used. Chemicals were assessed at 1–3 doses in addition to the vehicle control at a single time-point.
The DrugMatrix database consisted of transcriptomic profiles from liver RNA samples hybridized to the Affymetrix GeneChip Rat Genome 230 2.0 array, comprising >31 000 probesets, representing >28 000 rat genes. Genes that were differentially expressed by chemical treatment conditions within the liver were identified using previously established data processing methods and statistical results generated and made publicly available by NTP (2017a). The current investigation specifically used results provided online for all statistical comparisons (NTP, 2017a). In brief, array data were passed through several quality control procedures, including background requirements, minimal noise requirements, and a lack of array correlation to tissue reference standards. Data were normalized using an algorithm detailed within the DrugMatrix Calculations White Paper (NIEHS/NTP, 2011), based on the assumption that unchanged probe signals represent the majority of the signal measurements and forms the center of a nonlinear curve fit to a reference template. This curve fits the mode of the signal distribution for gene sets against the gene set expected value. Resulting normalized probeset signal intensities were statistically assessed through the comparison of tissues from treatment groups (three animals per condition) and control groups (10–20 animals). Expression change significance was calculated using the t statistic with an Empirical Bayes method of estimating variance, as previously published (Baldi and Long, 2001).
For the purposes of the current investigation, probesets representing significantly differentially expressed genes (DEGs) met the following criteria: fold change ≥ ±1.5 (average exposed/average control) and p < .01 (exposed vs control). This represents a relatively relaxed filter, which prioritized the detection of gene changes after filtering for noise and baseline statistical requirements. Gene lists were further refined, as detailed in the pathway enrichment analysis section. Probeset annotation information was updated to reflect the most recent rat genome annotation release through Affymetrix (v36).
Pathway enrichment analysis of in vivo DEGs
DEGs in the rat liver were evaluated for enrichment of canonical pathways, as organized through the Ingenuity Systems Knowledgebase. To parallel the in vitro assay agonist versus antagonist activities, gene lists were analyzed separately for genes with exposure-induced increased expression and decreased expression. In a small portion of the instances (∼10%) when the number of genes with increased or decreased expression was >1000, pathway enrichment analyses focused on the 1000 unique DEGs with the largest magnitude of fold change. This strategy was employed to highlight the responses of highest magnitude whereas reducing the impact of overt toxicity resulting in a higher number of enriched pathways. An additional non-directional pathway analysis was also carried out using lists of genes filtered in the same manner as described above, except that genes showing up- and down-regulated expression levels were analyzed collectively.
Significantly enriched pathways were identified using the Fisher’s exact test in R package ‘piano’ (v3.4.1) (Varemo et al., 2013) with a p value requirement of <.05. This statistical filter was selected as it resulted in greater in vitro-in vivo concordance compared with using a multiple test corrected p value filter, and it parallels previous pathway-level investigations using transcriptomic data (Farmahin et al., 2017; Rager et al., 2017). All pathways that were significantly enriched were then considered ‘active’ and assigned a binary call of 1, and all pathways that were not significantly enriched were considered ‘inactive’ and assigned a binary call of 0.
Comparing in vitro bioactivity versus in vivo pathway alterations
To conduct an in vitro-in vivo response concordance analysis, the Tox21 assay endpoint responses were compared against pathway-level responses, assessed through in vivo transcriptomic profiles. The majority of Tox21 HTS assay endpoints focus on a single receptor which represent the apex or initiator of a larger canonical pathway. As such, activation/repression of these receptors is suggestive of activation/repression of corresponding canonical pathways. Tox21 assay endpoints that were not focused on a single receptor can also represent canonical pathways given the nature of the stress that the assay is measuring. Tox21 assay endpoints were thereby mapped to corresponding canonical pathways based on the receptor used in the assay (eg, aryl hydrocarbon receptor [AhR] assay endpoint assigned to the AhR signaling pathway) or the nature of the stress being investigated (eg, mitotoxicity assay endpoint assigned to the mitochondrial dysfunction pathway). To improve the comparison, the directionality of pathway changes was considered by matching the agonism (increase) or antagonism (decrease) assay endpoints to pathway enrichment based on genes with increased expression or pathway enrichment based on genes with decreased expression, respectively. For completeness, an additional comparison was carried out that did not take directionality into account. This non-directional comparison included all Tox21 assay endpoints regardless of whether they probed for agonist or antagonist activity, and pathways were identified as enriched analyzing all up- and down-regulated genes collectively. A total of 40 Tox21 assay endpoints were mapped to 18 canonical pathways (Supplementary Table 1). Three assay endpoints were not able to be mapped to available canonical pathways, specifically tox21-aromatase-antagonist-p1, tox21-hse-bla-agonist-p1, and tox21-ror-cho-antagonist-p1, and were thus excluded from the analysis.
The degree of concordance was assessed between in vitro assay endpoint activity and in vivo pathway activity. For each chemical (and corresponding dose used in the DrugMatrix experimentation), the status of concordance was determined based on the binary activity values, with concordant instances representing inactive matches (0 in vitro and 0 in vivo) or active matches (1 in vitro and 1 in vivo). Instances representing discordant in vitro and in vivo activities were identified as those with binary activity values that did not match (0 in vitro and 1 in vivo; 1 in vitro and 0 in vivo). Next, the number of instance comparisons were aggregated into a 2 × 2 contingency table indicating the total number in each of the four categories (ie, 0 and 0; 1 and 1; 0 and 1; 1 and 0).
Two measures were calculated to assess concordance between the in vitro and in vivo datasets: (1) percent agreement, and (2) the Cohen’s kappa statistic. Percent agreement was defined as the number of agreements (ie, instances when in vitro and in vivo activities were either both active or both inactive) divided by the total number (ie, all instances used to compare in vitro vs in vivo activities) multiplied by 100%, similar to previous concordance analyses (McHugh, 2012). The Cohen’s kappa statistic results in a potential range of values between −1 and 1, with −1 indicative of the theoretical case of complete disagreement, 0 indicative of the agreement being no better than selecting values at random, and 1 indicative of complete agreement (McHugh, 2012). Within values greater than 0, the quality of agreement has previously been defined as kappa <0.2 = poor, 0.21–0.4 = fair, 0.41–0.6 = moderate, 0.61–0.8 = good, and 0.81–1.0 = very good, as used for the evaluation of agreement between two different measuring or rating techniques (Kwiecien et al., 2011). Because the Cohen’s kappa statistic shows limitations when using skewed data (Feinstein and Cicchetti, 1990; McHugh, 2012; Uebersax, 1987), and the current comparative analysis showed an abundance of inactive measures, results largely focused on percent agreement as the concordance measure.
All data processing, organization, and statistical analyses were conducted using R (Team, 2017). The Cohen’s kappa statistic was determined with the cohen.kappa() function in R package ‘psych’ (Revelle, 2017). Changes in concordance that were associated with whether or not activity was observed in vitro were assessed using the same statistical approaches used in evaluating effects of experimental design attributes and pathway targets, as described in the following section. For visualization purposes, heat maps showing in vitro-in vivo activity versus inactivity were generated using the heatmap.2() function in R package ‘gplots’ (Warnes et al., 2016).
Assessment of experimental design attributes and pathway targets that influence in vitro-in vivo response concordance
The potential influence of experimental design on in vitro-in vivo response concordance was investigated. Tox21 design attributes that were evaluated included duration of exposure, tissue endpoint target type, origin of cells, and species of cells. Experimental design attributes within the DrugMatrix database included duration of exposure, route of administration, and vehicle of administration. Concordance changes associated with specific pathways were also investigated.
To statistically evaluate whether these experimental attributes potentially impacted response concordance, a chi-square test for equality of proportions with Yates’s continuity correction was used (R function ‘prop.test’). This test compared the proportion of concordant cases for each category of the attribute (or overall trend for continuous attributes) to the overall proportion of concordant cases. Because many chemical/dose instances were inactive both in vitro and in vivo, a separate analysis was also carried out using only chemical/dose instances that showed activity in vitro, in vivo, or both (ie, instances of at least one activity).
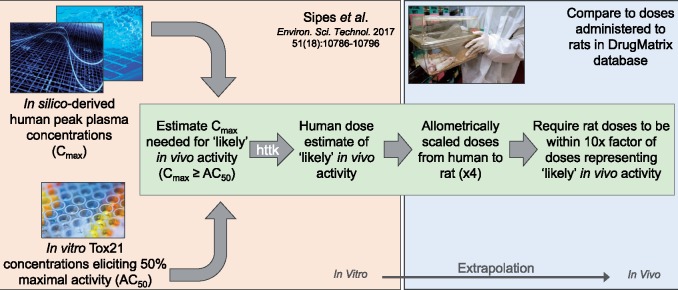
Assessment of in silico-derived dose applicability
In evaluating the potential effects of chemical dose administration on in vitro-in vivo response concordance, data were used from a recent study by Sipes et al. that implemented in silico approaches to estimate chemical concentrations in plasma that correspond to doses that elicit in vitro bioactivity (Sipes et al., 2017). In brief, peak human plasma concentrations (Cmax) that were estimated to be equivalent to or higher than concentrations in Tox21 data eliciting 50% maximal activity (AC50) were identified as chemical concentrations ‘likely’ to elicit in vivo activity. These Cmax values were then converted into corresponding estimates of daily doses in humans using high-throughput toxicokinetic modeling, made possible through the R package ‘httk’ (Pearce et al., 2017). Human daily doses corresponding to ‘likely’ in vivo activity were acquired from Sipes et al. (2017) (specifically, Sipes et al.Supplementary Table 5; all available likely doses), and then converted in this study into approximate daily doses in the rat through allometric scaling. Scaling was specifically achieved using a dosimetric adjustment factor of four, as recommended in the U.S. EPA’s Reference Dose and Reference Concentrations Guidelines (EPA, 2002). A filter criteria was then set to evaluate dose-specific attributes, which required that only data be used from experimental doses that were within a 10-fold factor of doses corresponding to ‘likely’ in vivo activity in at least one instance (ie, for at least one Tox21 assay AC50 value for that chemical) (Figure 2). Experimental conditions meeting these criteria were then considered to show in silico-derived dose applicability and were evaluated against results from experimental conditions that did not meet these criteria.
Figure 2.
Flowchart of steps used in filtering experimental conditions to evaluate in silico-derived dose applicability. Abbreviation: httk, high-throughput toxicokinetics.
In vivo dose estimate data were available through Sipes et al. (2017) for 3134 of the 5733 in vitro-in vivo response instance comparisons used in this study, due to limited toxicokinetic data availability and also the requirement for an in vitro activity concentration to calculate equivalent doses. Thus, data could not be pre-filtered prior to other attribute analyses without drastically reducing the number of in vitro-in vivo response comparisons. However, it should be noted that for the chemicals with data available in Sipes et al. (2017), the majority (47 of 75; 63%) had Tox21 AC50 values estimated to show applicability to concentrations used in DrugMatrix, suggesting that many of the other chemicals without toxicokinetic data may demonstrate in vitro-in vivo dose applicability as well. Paralleling the experimental attributes statistical analysis detailed above, a chi-square test for equality of proportions with Yates’s continuity correction was carried out to evaluate whether the in silico-derived dose applicability factor influenced in vitro-in vivo response concordance.
Assessment of chemical properties that influence in vitro-in vivo response concordance
The potential influence of chemical-specific attributes on in vitro-in vivo response concordance was considered through the evaluation of physicochemical properties, obtained from the U.S. EPA Chemistry Dashboard (EPA, 2017). Experimentally derived values were used when available, and predicted values were used in instances that lacked experimentally derived values. In cases with more than one experimentally derived value available, the average was used. The chemical-specific attributes consisted of a much larger database to statistically evaluate in comparison with the aforementioned experimental/dose-specific attributes, and there were potential interactions and dependencies between components across this large database. A different statistical approach was therefore implemented, namely through random forest modeling.
Random forest modeling represents an ensemble of bootstrapped decision trees and has been shown to be a powerful machine-learning approach to build models with large numbers of predictor variables with possible interactions, dependencies, and correlations (Breiman, 2001). The bootstrapped, ensemble nature of a random forest model allows quantitative estimation of the total importance of each predictor in the model. However, that same ensemble nature presents difficulty in interpreting the nature of the relationship between the predictors and the response variable, compared with a single decision tree model. To combine the robust results of random forest modeling with the interpretability of a single decision tree model, a two-stage process was used. At the first stage, random forest modeling was used to evaluate the importance of each chemical-specific attribute component in predicting response concordance. At the second stage, these most-important attributes were used to construct a decision tree model, to elucidate more details surrounding the attribute data ranges and response concordance. As this was a chemical-specific analysis, in vitro-in vivo concordance was expressed as percent agreement summarized on a per-chemical basis for each dose tested.
At the first stage, random forest modeling was carried out using the R package ‘randomForest’ (Liaw and Wiener, 2002). Because highly correlated predictors may distort random-forest importance measures (Toloşi and Lengauer, 2011), the R package ‘caret’ (Kuhn et al., 2017) was used to identify and exclude highly correlated attributes (using a threshold correlation coefficient of 0.75). As a significance check, five randomly generated noise predictors were added to the model: two from normal distributions with zero means and standard deviations of 1 and 5, and three from binomial distributions where a value of 1 occurred with a probability 0.5, 0.1, and 0.9, similar to previous random forest modeling approaches (Wambaugh et al., 2014). Any predictive importance of these noise variables occurs purely by chance; therefore, the importance of the noise variables can be used to gauge the significance of the importance of the chemical-specific attributes. A random forest model was then built to include 5000 trees with percent agreement as the response variable and chemical-specific attributes and noise variables as the predictors. Importance of the predictor variables was summarized based on the permutation-corrected percentage increase in mean squared error of the response when each predictor variable was randomly permuted (Altmann et al., 2010). The permutation-corrected variable importance was computed using the method implemented in the ‘pRF’ R package (Chakravarthy, 2016). Briefly, the null distribution of this quantity was estimated by repeatedly (200 times) randomly permuting the response variable, re-fitting the random forest, and determining the importance metric for each predictor variable. Then, a p value for each variable importance was derived by comparing the observed importance (with non-permuted response value) to the estimated null distribution. Smaller p values indicate that the observed variable importance is less likely to arise when there is no real relationship between predictor and response. Chemical-specific attributes with p < .01 were identified as the most-important predictors. At the second stage, a single decision tree was built using only the most-important attributes. Decision tree modeling was performed using R package ‘rpart’ (Therneau et al., 2017).
RESULTS
Tox21 and DrugMatrix Data Overview
Two large toxicology databases were probed for the investigation of in vitro-in vivo response concordance at the mechanistic-level: (1) the in vitro database, Tox21, and, (2) the in vivo database, DrugMatrix. After applying filters for the chemical purity status and data quality in Tox21 and focusing on chemicals with liver transcriptomic signatures in DrugMatrix obtained after 1–5 days of exposure, 130 unique chemicals with 167 unique chemical-dose pairs were available for the full in vitro to in vivo signal response concordance analysis (Supplementary Table 2). For these chemicals, all in vitro Tox21 experimental attributes and wAUC values are provided in Supplementary Table 3. All in vivo DrugMatrix experimental attributes are provided in Supplementary Table 4, along with the numbers of array probeset IDs and corresponding genes identified as significantly differentially expressed in the rat liver for each exposure condition. These differentially expressed genes were analyzed for enrichment of canonical pathways mapping to Tox21 target endpoints (as detailed in Supplementary Table 1), and pathways that were associated with differentially expressed genes were identified (Supplementary Table 5).
In Vitro-In Vivo Concordance Analysis
In vitro-in vivo signal response activities resulting from chemical exposure were compared through the evaluation of Tox21 activity and mapped pathway enrichment across differentially expressed genes identified through the DrugMatrix database. To detail, for every chemical in the Tox21 database that was also included in the DrugMatrix database, Tox21 activity was considered, with 1 indicating activity and 0 indicating inactivity. Mapped pathway enrichment results from the liver transcriptomic responses were compared against these in vitro activities in a directional-specific manner, with 1 indicating pathway enrichment in the direction corresponding to the Tox21 activity (eg, in vitro agonist endpoints corresponding to genes with increased expression) and 0 indicating that a pathway was not enriched in the direction corresponding to the Tox21 activity. An example of such a comparison is provided in Figure 3.
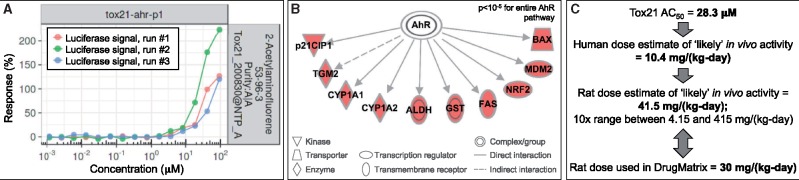
Figure 3.
Example in vitro-in vivo response comparison for the compound, 2-acetylaminofluorene, and its relation to altered aryl hydrocarbon receptor (AhR) signaling. A, In vitro Tox21 response profiles for the tox21-ahr-agonist-p1 assay showed activity in the agonist direction resulting from 2-acetylaminofluorene treatment. B, In vivo transcriptomic responses in the rat liver from the DrugMatrix database showed increased expression levels for genes encoding proteins (shown in red) regulated by AhR signaling. C, Comparisons between the Tox21 AC50 value for the tox21-ahr-agonist-p1 assay, related in vivo dose estimates, and the dose used in the DrugMatrix database showed that the in vitro activity concentration was estimated to fall within the 10× range of in vivo dose applicability. Together, this example in vitro-in vivo comparative response instance was noted as an in vitro active (activity call of 1) and in vivo active (activity call of 1) showing in silico-derived dose applicability. (For interpretation of the reference to color in this figure legend, the reader is referred to the web version of this article.)
Considering the 167 chemical-dose pairs evaluated across 40 Tox21 assay endpoints mapping to 18 in vivo canonical pathways, there were 5733 total instances of comparison between the in vitro and in vivo experiments (Supplementary Table 6A). Because several chemicals in the DrugMatrix database were evaluated across two or three doses at a particular time-point, coupled with varying Tox21 assay endpoint data availability, the resulting number of comparisons did not simply equate to the product between the number of chemical and assay endpoints.
The global comparison of the 5733 intersecting instances between in vitro Tox21 and in vivo DrugMatrix databases yielded 96 instances of activity in both, 649 instances of in vitro activity and in vivo inactivity, 554 occurrences of in vitro inactivity and in vivo activity, and 4434 instances of inactivity in both (Figure 4, Supplementary Table 6A). Thus, most response signals (ie, 77% of the comparisons) were indicative of inactivity resulting from chemical exposure. The percent agreement across this global comparison was high, at an average of 79%. Statistical evaluation of this contingency table resulted in a Cohen’s kappa concordance statistic of 0.02. As described in the Materials and Methods section, this statistic represents an overall poor agreement among the in vitro and in vivo datasets. However, this statistic should be interpreted with caution, as it shows limitations in cases of skewed data (Feinstein and Cicchetti, 1990; McHugh, 2012; Uebersax, 1987). Concordance comparisons for each chemical are shown in Supplementary Table 6A. On a per-chemical basis (separated according to experimental dose), the percent agreement values ranged between 41 and 100%. The Cohen’s kappa statistic ranged on a per-chemical basis between −0.25 and 0.76, representing agreement ranging from ‘poor’ to ‘good’ (Kwiecien et al., 2011).
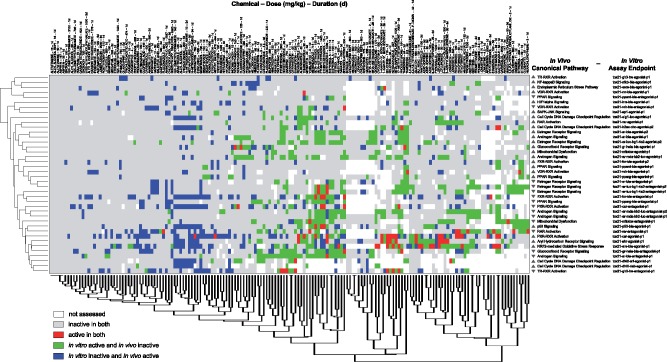
Figure 4.
Global in vitro-in vivo activity comparisons for all chemicals and doses under investigation. Comparisons were based on in vitro Tox21 activity and in vivo liver transcriptomic changes from DrugMatrix showing enrichment for 18 mapped canonical pathways (right). Data are organized based on complete linkage clustering using Euclidean distance measures. Note the red cell for 2-acetylaminofluorene, for which data is shown in Figure 3. (For interpretation of the reference to color in this figure legend, the reader is referred to the web version of this article.)
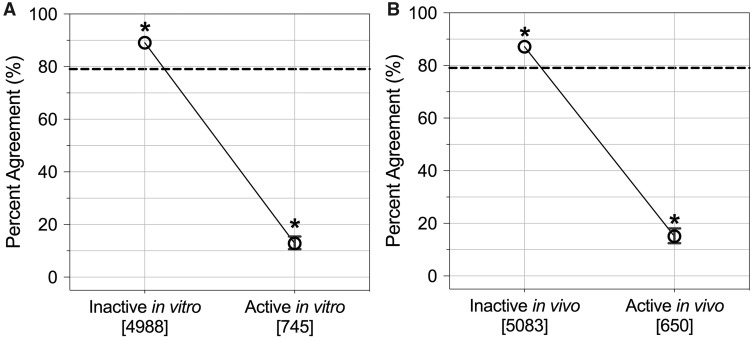
These comparisons also showed that, when inactivity was observed through in vitro HTS, 89% of the comparative instances showed inactivity in vivo. Conversely, when activity was observed through in vitro HTS, 13% of the comparative instances showed activity in vivo, representing a significant decrease in concordance for instances of in vitro activity (Figure 5A). When considering the in vivo data, concordance was also high in instances of in vivo activity (87%) and low in instances of in vivo inactivity (18%) (Figure 5B). These findings, together, support the notion that in vitro-in vivo response concordance is, on average, higher when inactivity is observed.
Figure 5.
In vitro-in vivo response concordance separated according to whether or not activity was observed in vitro. Average percent agreement values are plotted with error bars showing upper and lower 95th percentiles; note that these are not visible when error bars are small enough to overlap with the average percent agreement symbol. Counts of in vitro-in vivo comparative instances are provided in brackets. *p < .05 (significance between each category and overall average percent agreement). The horizontal dashed line indicates overall average percent agreement.
Because the global response comparisons were highly influenced by instances of inactivity in both in vitro and in vivo systems, it was important to carry out additional concordance analyses focusing on comparisons that included at least one instance of activity (ie, activity observed in vitro and/or in vivo). Percent agreement values showed an overall average agreement of 7.4%. This agreement value was much lower than that produced from the global comparison, as the global comparison was largely driven by the instances of matching inactivity. Percent agreement also ranged on a per-chemical basis, between 0% and 66.7% for instances of at least one activity (Supplementary Table 6B). A Cohen’s kappa statistic for these comparisons could not be calculated, as one of the contingency table cells was effectively removed.
An additional in vitro-in vivo concordance analysis was carried out to further substantiate overall findings by implementing a non-directional comparative approach. Here, agonist and antagonist in vitro assays were compared against pathway enrichment results based on genes showing differential expression in any direction. This non-directional in vitro-in vivo comparison resulted in a Cohen’s kappa concordance statistic of −0.0085 and percent agreement of 73%, which was slightly less than the concordance values obtained in the previously described analysis that considered response directionality (Supplementary Figure 1). Because the previously detailed directional-based comparative approach yielded better concordance, it was used throughout the rest of the study to evaluate attributes influencing in vitro-in vivo response concordance.
Experimental Design Components That Increase Response Concordance
To identify specific experimental design attributes associated with changes in response concordance, chi-square tests with Yates’s continuity correction were carried out, comparing the percent agreement in each category to the overall average percent agreement. Analyses were carried out on all in vitro-in vivo comparative instances (ie, global comparison), as well as instance comparisons showing at least one activity, as the global comparison contained largely inactive instances (n = 4288 out of 5733 comparisons) and thus heavily skewed the results toward different attributes, favoring those that were associated with inactivity. For the Tox21 assay design attribute of exposure duration, concordance was found to significantly increase as duration decreased; however, this trend was not apparent when evaluating comparisons with instances of at least one activity (Table 1). When evaluating tissue origin of cells, cells from the cervix and embryonic kidney showed significantly higher concordance than average, which was largely indicative of inactivity associated with exposure. In the comparisons with at least one activity, cells from the liver showed significantly higher concordance than the overall average. The different endpoint target types and species of cells did not exhibit concordance significantly different from the overall average. The only DrugMatrix design attribute to show significantly higher than average concordance was water as the vehicle of administration, apparent only in the global analysis. No duration of exposure or route of administration exhibited differences in concordance (Table 1).
Table 1.
Specific Attributes That Increase In Vitro-In Vivo Response Concordance Between Tox21 HTS Assays and Rat Liver Pathway-Level Responses As Identified Through Transcriptomic Analysis of DrugMatrix Data
| Global Comparisonsa | Comparisons With Instances of At Least One Activity | |
|---|---|---|
| In vitro Tox21 assay-specific attributes | ||
| Duration of exposure | Shorter durations (eg, 1–6 h) | — |
| Endpoint target type | — | — |
| Species of cells | — | — |
| Tissue origin of cells | Cervix, embryonic kidney | Liver |
| In vivo DrugMatrix experiment-specific attributes | ||
| Duration of exposure | — | — |
| Route of administration | — | — |
| Vehicle of administration | Water | — |
| In vitro and in vivo attribute | ||
| Pathway targets | Endoplasmic reticulum stress pathway, HIF1α signaling, NF-κ signaling, PPAR signaling, SAPK-JNK signaling, VDR-RXR activation | AhR signaling, NRF2-mediated oxidative stress, PXR-RXR activation |
| In silico-derived dose range of applicability | Within 10× range of dose applicability | Within 10× range of dose applicability |
| Chemical-specific attributesb | logP, logKoa, and water solubility; | logP, logKoa, and Henry’s law constant; |
| Specific data ranges include: logP<4.85 (logP<2.615 for greatest increased concordance), logKoa≥4.275, water solubility≥2 × 10−3 mol/l | Specific data ranges include: logP<5, logKoa≥8.855 | |
Note that global comparison findings were largely driven by instances of inactivity observed both in vitro and in vivo.
bThese represent the most significant chemical-specific attributes (p < .01). More specific data ranges are provided in Figure 8.
Pathway Targets That Increase Response Concordance
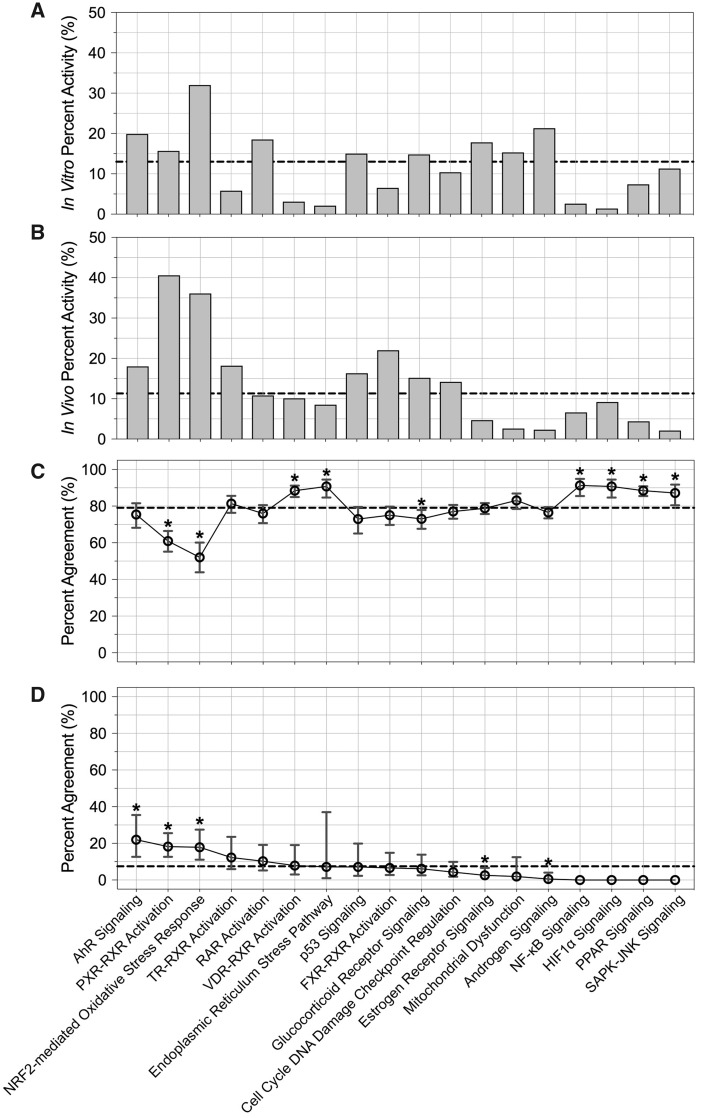
In assessing pathway targets associated with changes in response concordance, different pathways were identified as associated with increased concordance when evaluating all in vitro-in vivo response comparisons versus comparisons filtered to include at least one instance of activity, again demonstrating the large influence of inactivity on concordance trends. To detail, six pathways showed significantly increased concordance when evaluating all in vitro-in vivo instance comparisons: endoplasmic reticulum stress pathway, hypoxia-inducible factor 1 alpha (HIF1α) signaling, nuclear factor-kappa B (NF-κ) signaling, peroxisome proliferator-activated receptor (PPAR) signaling, stress-activated protein kinases-jun amino-terminal kinases (SAPK-JNK) signaling, and vitamin D receptor-retinoic acid receptor (VDR-RXR) activation. These pathways showed largely inactive responses to exposure, both in vitro and in vivo, further highlighting that the global comparative analysis is largely driven by inactivity (Table 1, Figs. 6A–C, Supplementary Table 7). When evaluating in vitro-in vivo comparisons with at least one instance of activity, other pathways were identified with significantly increased concordance, namely aryl hydrocarbon receptor (AhR) signaling, nuclear factor (erythroid-derived 2)-like 2 (NRF2)-mediated oxidative stress, and pregnane X receptor-RXR (PXR-RXR) (also known as CAR and/or nuclear receptor subfamily 1 group I member 3 [NR1I3]) activation (Table 1, Figure 6D, Supplementary Table 7). It is notable that these active pathways also clustered together when evaluating global concordance results using Euclidean distance measures (Figure 4).
Figure 6.
Relationships between pathway targets and (A) in vitro activity, (B) in vivo activity, (C) global response concordance, and (D) response concordance for comparisons including at least one instance of activity. Pathways are organized from those associated with the highest (left) to lowest (right) in vitro-in vivo percent agreement for comparisons showing at least one instance of activity. Dashed lines represent overall averages. *p < .05 (significance of difference between pathway percent agreement and overall average percent agreement). Horizontal dashed lines indicate overall average percent agreement.
In Silico-Derived Dose Applicability and Its Influence on Concordance
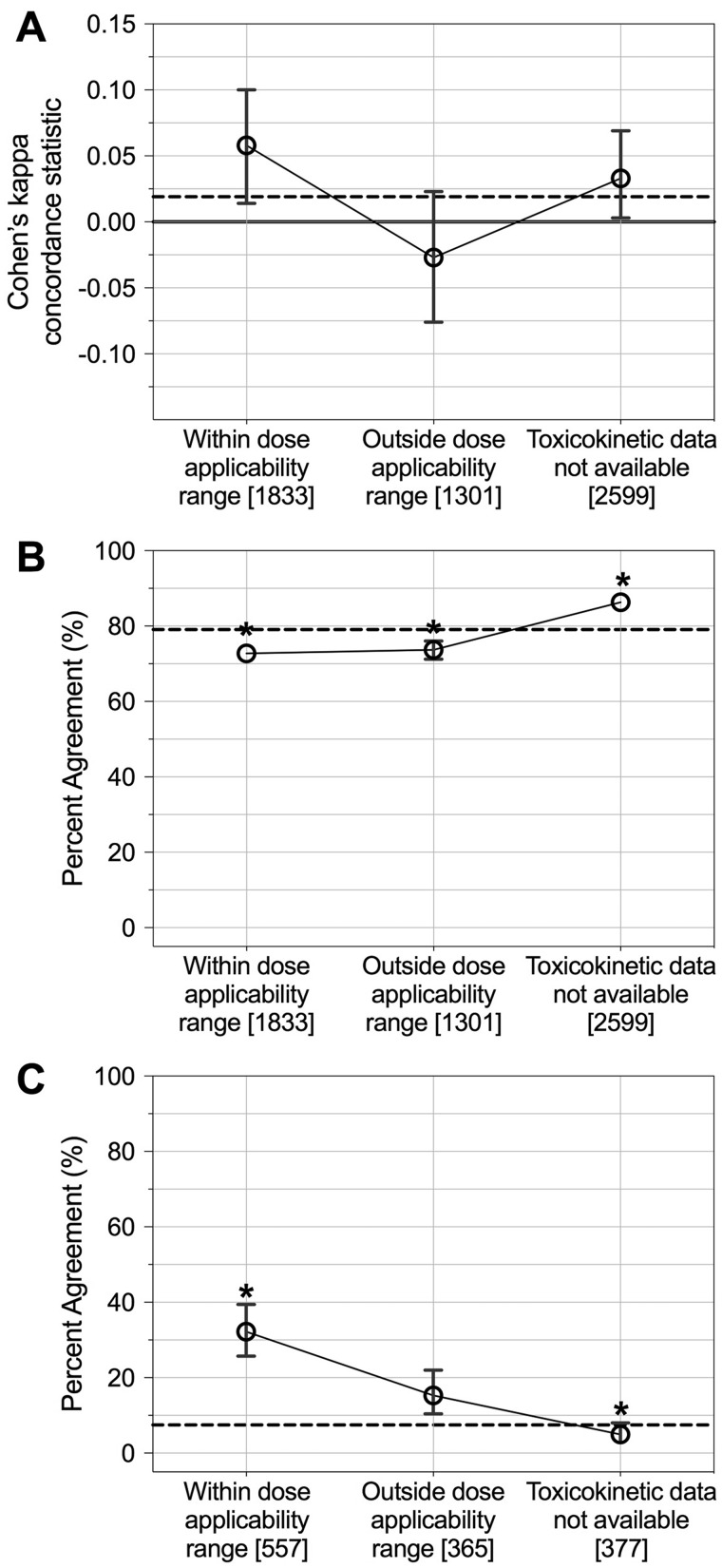
With the recent publication by Sipes et al. (2017), in vivo doses are available which are predicted to elicit responses observed in vitro, as probed for through the ToxCast/Tox21 database, and were used here to refine the current dataset to only select in vivo doses estimated to likely yield biological activity. Upon filtering the data, 47 unique chemicals had toxicokinetic data available and were evaluated in vivo at doses that fell within the 10× of the doses representing ‘likely’ in vivo activity, as defined in the Materials and Methods section (Supplementary Table 8, Figure 2). Filtering data for this in silico-derived dose applicability factor increased the overall, global in vitro-in vivo response concordance, with the Cohen’s kappa statistic increasing from 0.02 to 0.06 (Figure 7A). Evaluating potential changes in percent concordance, similar concordance values were measured globally (all within 10% of the overall 79% concordance) (Figure 7B); and filtering data for dose applicability resulted in significantly increased concordance for comparisons requiring at least one instance of activity (p < .001, 32.2% vs the overall 7.4%) (Figure 7C). Thus, dose applicability was identified to increase concordance, both globally and when instances of in vitro and in vivo inactivity were removed.
Figure 7.
In vitro-in vivo response concordance separated according to in silico-derived dose applicability. Overall, global concordance is shown using Cohen’s kappa concordance statistics in (A) and percent agreement in (B). Concordance for data filtered for comparisons showing at least one instance of activity is shown in (C). Average values are plotted with error bars showing upper and lower 95th percentiles; note that these are not visible when error bars are small enough to overlap with the average percent agreement symbol. Counts of in vitro-in vivo comparative instances are provided in brackets. p < .05 (significance between each category and overall average percent agreement). The horizontal dashed line indicates overall concordance.
Chemical-Specific Attributes That Influence Response Concordance
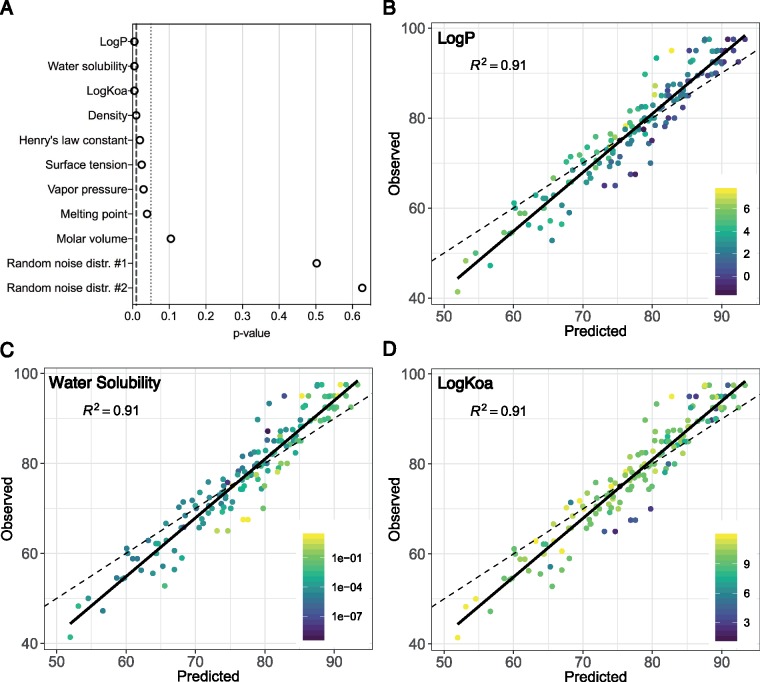
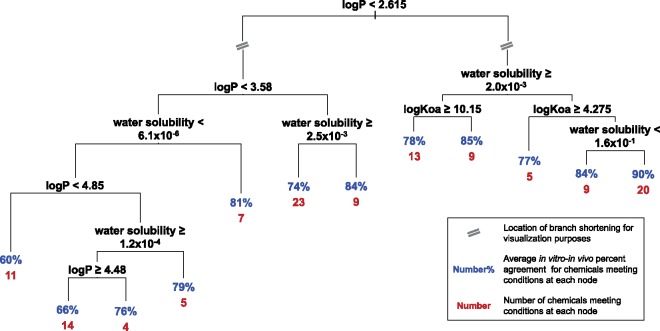
Because the in vitro-in vivo response concordances greatly varied across chemicals, chemical-specific attributes were also evaluated for potential impacts on concordance. Data were parsed into individual chemical-dose pairs, and random forest modeling was used to investigate the importance of the effects of physicochemical properties (Supplementary material, Table 9) on percent agreement between in vitro-in vivo responses. For the global in vitro-in vivo comparative analysis, the chemical-specific attributes with most significant importance (p < .01) were logP (octanol-water partition coefficient), logKoa (octanol-air partition coefficient), and water solubility (Figure 8A). To better visualize the results, the model-predicted versus observed concordance were plotted and colored according to the values of these three physicochemical properties (Figs. 8B–D). These results show, for instance, that chemicals with lower logP and higher water solubility generally show higher levels of response concordance. Decision tree modeling of these attributes identified specific ranges of the data related to increased response concordance (Table 1, Figure 9). As an example, chemicals with logP <4.785 showed higher in vitro-in vivo concordance, on average, in comparison with chemicals with logP ≥4.785. The highest concordance was identified for chemicals having logP < 2.615, water solubility ≥0.0027 and <0.156 mol/l, and logKoa≥4.276 (average percent agreement of 90.37%).
Figure 8.
Contribution of physicochemical properties toward global concordance. A, Variable importance plot (based on random forest modeling): the top nine variables are listed, from most (top) to least (bottom) importance significance. B–D, Relationships between the model-predicted versus observed concordance, colored according to the three most important predictors, logP, water solubility, and logKoa.
Figure 9.
Decision tree modeling of the most significant concordance predictors showing discrete ranges of predictor data associated with increased global concordance. At each node (split), the listed condition is true on the right branch, and false on the left branch. At each terminal node, the average percent agreement is shown in blue, and the number of chemicals at the node is shown in red. As an example, chemicals with logP <2.615 showed higher concordance, on average, in comparison with chemicals with logP >2.615. Note that one chemical did not have data for at least one of these physicochemical parameters and was thus excluded from this figure. Units are as follows: logP (unitless), logKoa (unitless), water solubility (mol/l). (For interpretation of the reference to color in this figure legend, the reader is referred to the web version of this article.)
For the comparative analysis with instances of at least one activity, similar chemical properties were identified to significantly contribute to the predictive model, with logP, logKoa, and Henry’s Law constant identified as the most significant (p < .01) contributing variables. Decision tree modeling results produced similar findings, with chemicals having logP < 5 and logKoa≥8.855 having increased in vitro-in vivo response concordance, on average. An additional analysis was also carried out to test whether the number of tested pathways (for each chemical) could contribute toward the predictive model of global concordance. Interestingly, the number of tested pathways did show significant variable importance (p < .05, with increasing number of tested pathways associated with increased concordance), though it was not amongst the top ranked variables (data not shown).
DISCUSSION
The growing prevalence of in vitro HTS data has supported the need for increased research surrounding the proper utilization of these data, particularly in predicting in vivo toxicology testing outcomes. Previous investigations have largely compared in vitro assay data to apical endpoints observed in vivo. This study set out to simplify these comparisons by evaluating the initiation of a molecular interaction (eg, receptor activation) and subsequent cellular responses (ie, pathway alterations) using in vitro Tox21 assay data and in vivo DrugMatrix transcriptomic profile data, respectively. In addition, attributes that could serve as considerations for informing ranges of applicability for in vitro assays were also investigated.
The global comparison between HTS bioactivity and liver pathway-level responses showed an overall percent agreement of 79%, representing a relatively high measure of agreement in biological activity. Notably, the majority (77%) of the in vitro-in vivo comparisons represented inactivity in both systems. A more statistically driven concordance measure that takes into account the difference between the expected and observed probability of agreement, the Cohen’s kappa statistic, was 0.02, which could be categorized as ‘poor’ based on previous definitions (Kwiecien et al., 2011). There are notable limitations in using the Cohen’s kappa statistic for skewed data, as these data can cause an imbalance of marginal totals which are used to calculate expected probabilities of agreement (Feinstein and Cicchetti, 1990; McHugh, 2012; Uebersax, 1987). Despite these potential limitations, the Cohen’s kappa statistic remains a widely used concordance statistic, and it was therefore included alongside percent agreement measures, paralleling published recommendations (McHugh, 2012). Consistent between these two concordance measures, in vitro-in vivo concordance was found to widely vary on a per-chemical basis.
In vitro-in vivo response concordance was also found to dramatically differ for substances according to whether activity versus inactivity was observed. To detail, when inactivity was observed in vitro, inactivity was also observed in vivo in 89% of the comparative instances. It is still important to identify these instances of in vivo inactivity, as these chemicals could be prioritized for use in drug/product development over those eliciting activity. However, concordance significantly decreased to only 13% for substances that showed activity through in vitro HTS. Furthermore, when any instance of activity was observed (in vitro and/or in vivo), concordance was only 7.4%. This finding suggests that follow-up in vivo and/or orthogonal in vitro assays should be conducted to increase confidence in interpreting instances of in vitro HTS activity; although future studies should expand these findings using different target tissues and experimental systems.
This investigation evaluated the influence of various experimental design parameters on in vitro-in vivo response concordance, different attributes were identified when assessing all comparative instances versus comparisons with inactivity removed. For instance, cervical and embryonic kidney cells showed increased concordance across all comparisons, largely resulting from inactive responses. When focusing on in vitro-in vivo comparisons containing activity, liver cells showed significantly increased concordance. Given that these comparisons used in vivo data from liver tissue, the increased concordance with in vitro data derived from liver cells makes biological sense, particularly when focusing on active responses. These changes in concordance were likely impacted by variable expression and functionality of genes/protein receptors that can differ according to cell type. It is therefore important to consider tissue origin of cells when using in vitro data to inform in vivo toxicity.
Due to the large range in biological targets covered by the HTS assay and transcriptomics databases, it was important to consider potential changes in concordance associated with different pathway targets. When assessing all in vitro-in vivo comparative instances, pathways that were largely inactive showed increased concordance (eg, HIF1α, NF-κ, and PPAR signaling). When data were filtered for comparisons including instances of activity, pathways that have recognized function or activity in the liver showed increased concordance, including AhR, PXR, and oxidative stress signaling. AhR and PXR pathways are commonly activated by environmental chemicals and pharmaceuticals, and are recognized to induce transcription/translation of phase I metabolizing enzymes in the liver (Denison and Nagy, 2003; Luo et al., 2004). In addition, oxidative stress signaling can occur in response to various liver activities, including xenobiotic metabolism, which can produce reactive oxygen species (Chen et al., 2013). Given that these pathways have higher in vitro-in vivo concordance, coupled with their known functionality in the liver, findings support the incorporation of biological target context when using in vitro data to inform mechanisms of in vivo toxicity.
A critical attribute shown to influence in vitro-in vivo response concordance was chemical dose, where the selection of doses estimated to likely cause in vivo activity based on in vitro AC50 values was found to increase concordance, regardless of whether or not data were filtered for activity. This finding highlights the importance of considering how concentrations used in vitro relate to in vivo toxicity and vice versa, which in part, can be addressed by the in silico toxicokinetic approaches used here. The incorporation of in silico toxicokinetic tools in chemical assessments is not necessarily new; yet there have been recent advances in pharmacokinetic parameter predictions that better capitulate experimentally derived data. These advances, coupled with improvements in human dosimetry models, result in in vitro-in vivo extrapolation estimates that are continually being improved and expanded upon (Kratochwil et al., 2017; Rotroff et al., 2010; Sipes et al., 2017; Wetmore et al., 2013). Incorporating such in silico modeling strategies to better gauge in vitro dose applicability clearly improves HTS data interpretation.
Chemical-specific attributes were also identified to influence in vitro-in vivo concordance and included the physicochemical properties, logP and logKoa. These properties are interrelated and are commonly used as chemical solubility metrics that inform chemical concentration/suitability in study designs. As an example, logP has been used to evaluate the suitability of chemicals for testing in ToxCast/Tox21, where chemicals with logP <−1 or >7 were identified to show potential issues related to DMSO solubility and/or inability to transport through lipid bilayers (Richard et al., 2016). In this study, chemicals with logP <4.785 generally showed higher in vitro-in vivo concordance. Chemicals with logP < 2.615 showed the highest average concordance, and only three chemicals having logP <−1; thus, these values are generally within range of those suggested as suitable for HTS. LogP is also informative within in vivo study designs, as it can influence chemical protein binding and distribution. For example, logP is used as a criterion for the biopharmaceutics classification system, with drugs showing logP > 1.72 considered highly permeable and likely to be absorbed within the gastrointestinal tract (Dahan et al., 2009). Filtering for chemicals with lower logP would therefore eliminate highly permeable compounds; although additional studies are needed to confirm the applicability of this filter in different models/target tissues.
Incorporating the attributes identified in this study (summarized in Table 1) can facilitate researchers when interpreting in vitro HTS data to inform in vivo mechanisms of toxicity. For example, 2-acetylaminofluorene was found to elicit in vitro bioactivity in the Tox21 assay probing for AhR agonism. By gauging how the attributes associated with this specific chemical are related to the attributes that were found to increase in vitro-in vivo response concordance, an increased confidence can be ascribed to the in vitro data. Specifically, the Tox21 AhR agonism assay was conducted in liver cells and queried for AhR activation, representing a cell type and pathway associated with increased concordance in instances of in vitro activity. In addition, the concentration eliciting in vitro activity was estimated to be within a 10× range of the dose evaluated in the rat, meeting the dose applicability filter. Furthermore, the chemical attributes of 2-acetylaminofluorene, including logP, logKoa, density, and melting point, were within ranges associated with increased concordance. In this case, 2-acetylaminofluorene was found to up-regulate the AhR pathway in vivo, in the rat liver, based on the analysis of transcriptomic signatures from the DrugMatrix database. Further supporting these findings, 2-acetylaminofluorene is recognized to bind to AhR and cause cancer in the liver of rats (Cikryt et al., 1990; NIEHS/NTP, 2011). This case study shows how researchers can therefore use the attributes identified in the current evaluation to bolster understanding of mechanistic in vivo toxicity when using in vitro HTS bioactivity.
Whereas this study provides insight toward understanding in vitro-in vivo response concordance, it is not without limitations. Importantly, the in vivo transcriptomic data were only from the liver, representing a major site of chemical-induced toxicity. This inherently biased the concordance to assays whose receptors are most active in the liver, although the expression levels of all evaluated receptors were confirmed to be above background in these samples (data not shown). Future studies evaluating different organs, species, endpoints, and data comparison approaches will further inform this research area. An additional challenge within this investigation, as well as the larger HTS community, is the incorporation of potential chemical metabolism (Wilk-Zasadna et al., 2015). This study considered chemical metabolism, in particular during the in silico derivations of each chemicals’ Cmax related to parent compound clearance rates and associated doses required to elicit likely in vivo activity (Ring et al., 2017; Sipes et al., 2017). Still, in silico predictions of chemical metabolism have limitations and uncertainties (Kirchmair et al., 2015; Zhang et al., 2011), making future analyses that focus on the effects of chemical metabolism during in vitro-in vivo comparisons of high interest.
In conclusion, results from this study provide increased understanding of the potential ranges of applicability for using in vitro assay data to predict or inform mechanisms of in vivo toxicity, with a focus on pathway alterations in the rat liver. Future studies could additionally evaluate the molecular/cellular responses in relation to apical endpoints and further develop computational models to predict in vivo activity based on in vitro observations. This in vitro-in vivo concordance analysis, in itself, provides an important baseline understanding of the attributes that influence whether or not a mechanistic response observed in vitro may also be observed in vivo, and vice versa. Results support the need to consider whether in vitro activity versus inactivity is observed, as findings were heavily influenced by the large abundance of inactivity. Results also consistently showed that considerations of dose applicability using high-throughput toxicokinetic modeling and dose equivalent estimates increased in vitro-in vivo concordance. These findings highlight that in vitro-in vivo concordance varies according to chemical, and there are experimental, pathway, dose, and chemical-specific attributes that should be considered when using in vitro HTS data to predict in vivo chemical toxicity.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
This work was supported by the American Chemistry Council Long Range Research Initiative. WD Klaren was supported as a T32 training fellow and an externship opportunity funded by the Regulatory Science in Environmental Health training grant awarded to Texas A&M University (T32 ES026568). This work was supported in part by the National Institutes of Health (NIH), National Institute of Environmental Health Sciences (NIEHS).
Supplementary Material
REFERENCES
- Altmann A., Toloşi L., Sander O., Lengauer T. (2010). Permutation importance: A corrected feature importance measure. Bioinformatics 26, 1340–1347. [DOI] [PubMed] [Google Scholar]
- Baldi P., Long A. D. (2001). A Bayesian framework for the analysis of microarray expression data: Regularized t-test and statistical inferences of gene changes. Bioinformatics 17, 509–519. [DOI] [PubMed] [Google Scholar]
- Ballet F. (1997). Hepatotoxicity in drug development: Detection, significance and solutions. J. Hepatol. 26(Suppl 2), 26–36. [DOI] [PubMed] [Google Scholar]
- Becker R. A., Dreier D. A., Manibusan M. K., Tony Cox L. A., Simon T. W., Bus J. S. (2017). How well can carcinogenicity be predicted by high throughput “characteristics of carcinogens” mechanistic data? Regul. Toxicol. Pharmacol. 90, 185–196. [DOI] [PubMed] [Google Scholar]
- Breiman L. (2001). Random forests. Mach. Learn. 45, 5–32. [Google Scholar]
- Browne P., Noyes P. D., Casey W. M., Dix D. J. (2017). Application of adverse outcome pathways to U.S. EPA's endocrine disruptor screening program. Environ. Health Perspect. 125, 096001.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy A. (2016). pRF: Permutation Significance for Random Forests. R Package Version 1.2. Available at: https://CRAN.R-project.org/package=pRF. Accessed March 1, 2018.
- Chen Y., Dong H., Thompson D. C., Shertzer H. G., Nebert D. W., Vasiliou V. (2013). Glutathione defense mechanism in liver injury: Insights from animal models. Food Chem. Toxicol. 60, 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cikryt P., Kaiser T., Gottlicher M. (1990). Binding of aromatic amines to the rat hepatic Ah receptor in vitro and in vivo and to the 8S and 4S estrogen receptor of rat uterus and rat liver. Environ. Health Perspect. 88, 213–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A. T., Popken D. A., Kaplan A. M., Plunkett L. M., Becker R. A. (2016). How well can in vitro data predict in vivo effects of chemicals? Rodent carcinogenicity as a case study. Regul. Toxicol. Pharmacol. 77, 54–64. [DOI] [PubMed] [Google Scholar]
- Dahan A., Miller J. M., Amidon G. L. (2009). Prediction of solubility and permeability class membership: Provisional BCS classification of the world's top oral drugs. AAPS J. 11, 740–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M. S., Nagy S. R. (2003). Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annal. Rev. Pharmacol. Toxicol. 43, 309–343. [DOI] [PubMed] [Google Scholar]
- Driessen M., Vitins A. P., Pennings J. L., Kienhuis A. S., Water B., van der Ven L. T. (2015). A transcriptomics-based hepatotoxicity comparison between the zebrafish embryo and established human and rodent in vitro and in vivo models using cyclosporine A, amiodarone and acetaminophen. Toxicol. Lett. 232, 403–412. [DOI] [PubMed] [Google Scholar]
- EPA U. S. (2002). A Review of the Reference Dose and Reference Concentration Processes. U.S. EPA Risk Assessment Forum. EPA/630/P-02/002F, Washington, DC. [Google Scholar]
- EPA U. S. (2017). Chemistry Dashboard Available at: https://comptox.epa.gov/dashboard/. Accessed August 24, 2017.
- EPA U. (2018). Use of High Throughput Assays and Computational Tools in the Endocrine Disruptor Screening Program Available at: https://www.epa.gov/endocrine-disruption/use-high-throughput-assays-and-computational-tools-endocrine-disruptor. Accessed July 1, 2018.
- Farmahin R., Williams A., Kuo B., Chepelev N. L., Thomas R. S., Barton-Maclaren T. S., Curran I. H., Nong A., Wade M. G., Yauk C. L. (2017). Recommended approaches in the application of toxicogenomics to derive points of departure for chemical risk assessment. Arch. Toxicol. 91, 2045–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein A. R., Cicchetti D. V. (1990). High agreement but low kappa: I. The problems of two paradoxes. J. Clin. Epidemiol. 43, 543–549. [DOI] [PubMed] [Google Scholar]
- Ganter B., Snyder R. D., Halbert D. N., Lee M. D. (2006). Toxicogenomics in drug discovery and development: Mechanistic analysis of compound/class-dependent effects using the DrugMatrix database. Pharmacogenomics 7, 1025–1044. [DOI] [PubMed] [Google Scholar]
- Hsieh J. H. (2016). Accounting artifacts in high-throughput toxicity assays. Methods Mol. Biol. 1473, 143–152. [DOI] [PubMed] [Google Scholar]
- Hsieh J. H., Sedykh A., Huang R., Xia M., Tice R. R. (2015). A data analysis pipeline accounting for artifacts in Tox21 quantitative high-throughput screening assays. J. Biomol. Screen. 20, 887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R., Houck K., Martin M., Richard A. M., Knudsen T. B., Shah I., Little S., Wambaugh J., Setzer R. W., Kothiya P., et al. (2016). Analysis of the effects of cell stress and cytotoxicity on in vitro assay activity across a diverse chemical and assay space. Toxicol. Sci. 153, 409.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmair J., Goller A. H., Lang D., Kunze J., Testa B., Wilson I. D., Glen R. C., Schneider G. (2015). Predicting drug metabolism: Experiment and/or computation. Nat. Rev. Drug Discov. 14, 387–404. [DOI] [PubMed] [Google Scholar]
- Kratochwil N. A., Meille C., Fowler S., Klammers F., Ekiciler A., Molitor B., Simon S., Walter I., McGinnis C., Walther J., et al. (2017). Metabolic profiling of human long-term liver models and hepatic clearance predictions from in vitro data using nonlinear mixed-effects modeling. AAPS J. 19, 534–550. [DOI] [PubMed] [Google Scholar]
- Kuhn M., Wing J., Weston S., Williams A., Keefer C., Engelhardt A., Cooper T., Mayer Z., Kenkel B., Team R. C. (2017). caret: Classification and Regression Training. R Package Version 6.0-76. Available at: https://CRAN.R-project.org/package=caret. Last accessed July 1, 2018.
- Kwiecien R., Kopp-Schneider A., Blettner M. (2011). Concordance analysis: Part 16 of a series on evaluation of scientific publications. Dtsch. Arztebl. Int. 108, 515–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung M. C., Phuong J., Baker N. C., Sipes N. S., Klinefelter G. R., Martin M. T., McLaurin K. W., Setzer R. W., Darney S. P., Judson R. S., et al. (2016). Systems toxicology of male reproductive development: Profiling 774 chemicals for molecular targets and adverse outcomes. Environ. Health Perspect. 124, 1050–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A., Wiener M. (2002). Classification and regression by randomForest. R News 2, 18–22. [Google Scholar]
- Liu J., Mansouri K., Judson R. S., Martin M. T., Hong H., Chen M., Xu X., Thomas R. S., Shah I. (2015). Predicting hepatotoxicity using ToxCast in vitro bioactivity and chemical structure. Chem. Res. Toxicol. 28, 738–751. [DOI] [PubMed] [Google Scholar]
- Liu J., Patlewicz G., Williams A., Thomas R. S., Shah I. (2017). Predicting organ toxicity using in vitro bioactivity data and chemical structure. Chem. Res. Toxicol. 30, 2046–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Guenthner T., Gan L., Humphreys W. G. (2004). CYP3A4 induction by xenobiotics: Biochemistry, experimental methods and impact on drug discovery. Curr. Drug Metab. 5, 483–505. [DOI] [PubMed] [Google Scholar]
- McHugh M. L. (2012). Interrater reliability: The kappa statistic. Biochem. Med. (Zagreb) 22, 276–282. [PMC free article] [PubMed] [Google Scholar]
- NAS. (2007). Toxicity Testing in the 21st Century: A Vision and a Strategy. Committee on Toxicity Testing and Assessment of Environmental Agents, National Research Council, Washington, DC. [Google Scholar]
- NAS. (2017). Using 21st Century Science to Improve Risk-Related Evaluations. Committee on Incorporating 21st Century Science into Risk-Based Evaluations; Board on Environmental Studies and Toxicology; Division on Earth and Life Studies; National Academies of Sciences, Engineering, and Medicine, Washington, DC. [PubMed]
- NIEHS/NTP. (2011). DrugMatrix Calculations White Paper Available at: https://ntp.niehs.nih.gov/drugmatrix/projects/DrugMatrix/support/White_Paper/DrugMatrix_Calculations.pdf. Accessed April 4, 2017.
- NTP. (2011). 2-Acetylaminofluorene, report on carcinogens. Fourteenth edition. Rep. Carcinog. 12, 24–25.21829238 [Google Scholar]
- NTP. (2017a). DrugMatrix Available at: https://ntp.niehs.nih.gov/drugmatrix. Accessed April 5, 2017.
- NTP. (2017b). Tox21 Activity Profiler. National Toxicology Program. Available at: https://ntp.niehs.nih.gov/sandbox/tox21-activity-browser/. Accessed August 23, 2017.
- NTP. (2017c). Tox21 Concentration Response Browser Available at: https://sandbox.ntp.niehs.nih.gov/tox21-curve-visualization/. Accessed October 2, 2017.
- Pearce R. G., Setzer R. W., Strope C. L., Sipes N. S., Wambaugh J. F. (2017). httk: R package for high-throughput toxicokinetics. J. Stat. Soft. 79, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager J. E., Ring C. L., Fry R. C., Suh M., Proctor D. M., Haws L. C., Harris M. A., Thompson C. M. (2017). High-throughput screening data interpretation in the context of in vivo transcriptomic responses to oral Cr(VI) exposure. Toxicol. Sci. 1581, 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revelle W. (2017). psych: Procedures for Personality and Psychological Research. Version 1.7.5. Northwestern University, Evanston, IL: Available at: https://cran.r-project.org/web/packages/psych/index.html [Google Scholar]
- Richard A. M., Judson R. S., Houck K. A., Grulke C. M., Volarath P., Thillainadarajah I., Yang C., Rathman J., Martin M. T., Wambaugh J. F., et al. (2016). ToxCast chemical landscape: Paving the road to 21st century toxicology. Chem. Res. Toxicol. 29, 1225–1251. [DOI] [PubMed] [Google Scholar]
- Ring C. L., Pearce R. G., Setzer R. W., Wetmore B. A., Wambaugh J. F. (2017). Identifying populations sensitive to environmental chemicals by simulating toxicokinetic variability. Environ. Int. 106, 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotroff D. M., Wetmore B. A., Dix D. J., Ferguson S. S., Clewell H. J., Houck K. A., Lecluyse E. L., Andersen M. E., Judson R. S., Smith C. M., et al. (2010). Incorporating human dosimetry and exposure into high-throughput in vitro toxicity screening. Toxicol. Sci. 117, 348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah I., Houck K., Judson R. S., Kavlock R. J., Martin M. T., Reif D. M., Wambaugh J., Dix D. J. (2011). Using nuclear receptor activity to stratify hepatocarcinogens. PLoS One 6, e14584.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipes N. S., Martin M. T., Reif D. M., Kleinstreuer N. C., Judson R. S., Singh A. V., Chandler K. J., Dix D. J., Kavlock R. J., Knudsen T. B. (2011). Predictive models of prenatal developmental toxicity from ToxCast high-throughput screening data. Toxicol. Sci. 124, 109–127. [DOI] [PubMed] [Google Scholar]
- Sipes N. S., Wambaugh J. F., Pearce R., Auerbach S. S., Wetmore B. A., Hsieh J. H., Shapiro A. J., Svoboda D., DeVito M. J., Ferguson S. S. (2017). An intuitive approach for predicting potential human health risk with the Tox21 10k library. Environ. Sci. Technol. 51, 10786–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOT. (2017). Previous CCT Meetings and Webinars. Available at: http://www.toxicology.org/events/shm/cct/previous.asp. Accessed October 9, 2017. [Google Scholar]
- Team R. C. (2017). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: Available at: https://www.R-project.org [Google Scholar]
- Therneau T., Atkinson B., Ripley B. (2017). rpart: Recursive Partitioning and Regression Trees. R Package Version 4.1-11. Available at: https://CRAN.R-project.org/package=rpart
- Thomas R. S., Black M. B., Li L., Healy E., Chu T. M., Bao W., Andersen M. E., Wolfinger R. D. (2012). A comprehensive statistical analysis of predicting in vivo hazard using high-throughput in vitro screening. Toxicol. Sci. 128, 398–417. [DOI] [PubMed] [Google Scholar]
- Toloşi L., Lengauer T. (2011). Classification with correlated features: Unreliability of feature ranking and solutions. Bioinformatics 27, 1986–1994. [DOI] [PubMed] [Google Scholar]
- Uebersax J. S. (1987). Diversity of decision-making models and the measurement of interrater agreement. Psychol. Bull. 101, 140–146. [Google Scholar]
- Varemo L., Nielsen J., Nookaew I. (2013). Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 41, 4378–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wambaugh J. F., Wang A., Dionisio K. L., Frame A., Egeghy P., Judson R., Setzer R. W. (2014). High throughput heuristics for prioritizing human exposure to environmental chemicals. Environ. Sci. Technol. 48, 12760–12767. [DOI] [PubMed] [Google Scholar]
- Warnes G. R., Bolker B., Bonebakker L., Gentleman R., Liaw W. H., Lumley T., Maechler M., Magnusson A., Moeller S., Schwartz M. (2016). gplots: Various R Programming Tools for Plotting Data Available at: https://cran.r-project.org/web/packages/gplots/index.html
- Wetmore B. A., Wambaugh J. F., Ferguson S. S., Li L., Clewell H. J., Judson R. S., Freeman K., Bao W., Sochaski M. A., Chu T.-M., et al. (2013). Relative impact of incorporating pharmacokinetics on predicting in vivo hazard and mode of action from high-throughput in vitro toxicity assays. Toxicol. Sci. 132, 327–346. [DOI] [PubMed] [Google Scholar]
- Wilk-Zasadna I., Bernasconi C., Pelkonen O., Coecke S. (2015). Biotransformation in vitro: An essential consideration in the quantitative in vitro-to-in vivo extrapolation (QIVIVE) of toxicity data. Toxicology 332, 8–19. [DOI] [PubMed] [Google Scholar]
- Zhang T., Chen Q., Li L., Liu L. A., Wei D. Q. (2011). In silico prediction of cytochrome P450-mediated drug metabolism. Comb. Chem. High Throughput Screen. 14, 388–395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.