Abstract
Exposure to environmentally relevant chemicals that activate the xenobiotic receptors aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR), and peroxisome proliferator-activated receptor alpha (PPARα) in rodent test systems often leads to increases in oxidative stress (OS) that contributes to liver cancer induction. We hypothesized that activation of the oxidant-induced transcription factor Nrf2 could be used as a surrogate endpoint for increases in OS. We examined the relationships between activation of xenobiotic receptors and Nrf2 using previously characterized gene expression biomarkers that accurately predict modulation. Using a correlation approach (Running Fisher Test), the biomarkers were compared with microarray profiles in a mouse liver gene expression compendium. Out of the 163 chemicals examined, 47% from 53 studies activated Nrf2. We found consistent coupling between CAR and Nrf2 activation. Out of the 41 chemicals from 32 studies that activated CAR, 90% also activated Nrf2. CAR was activated earlier and at lower doses than Nrf2, indicating CAR activation preceded Nrf2 activation. Nrf2 activation by 2 CAR activators was abolished in CAR-null mice. We hypothesized that Nrf2 is activated by reactive oxygen species from the increased activity of enzymes encoded by Cyp2b family members. However, Nrf2 was similarly activated in the livers of both TCPOBOP-treated wild-type and Cyp2b9/10/13-null mice. This study provides evidence that Nrf2 activation (1) often occurs after exposure to xenobiotic chemicals, (2) is tightly linked to activation of CAR, and (3) does not require induction of 3 Cyp2b genes secondary to CAR activation.
Keywords: Nrf2, Keap1, constitutive androstane receptor, peroxisome proliferator-activated receptor, transcript profiling, liver cancer, aryl hydrocarbon receptor, oxidative stress, Cyp2b
Oxidative stress (OS) plays roles in chemical-dependent tumor promotion (Goetz and Luch, 2008; Roberts et al., 2010). Induction of OS has been observed following exposure to chemicals that activate a number of xenobiotic receptors including the aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR), and peroxisome proliferator-activated receptor α (PPARα). Chemicals that activate 1 or more of these receptors cause liver cancer through mechanisms that do not involve direct damage to DNA (nongenotoxic) and generally act at the promotion stage by altering hepatocyte fate, most notably through increases in cell proliferation (Budinsky et al., 2014; Corton et al., 2018; Elcombe et al., 2014). AhR is a member of the basic region helix-loop-helix/period-aryl hydrocarbon nuclear translocator-simple-minded family of transcription factors that is activated by 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD; also generically referred to as dioxin), as well as dioxin-like chemicals including other polychlorinated dibenzo-p-dioxins, polychlorinated dibenzofurans, and polychlorinated biphenyls. CAR and PPARα are members of the nuclear receptor superfamily, and are activated by a large number of structurally heterogeneous pharmaceutical and industrial chemicals (Hernandez et al., 2009; LeBlanc et al., 2012; Yamada et al., 2006). Although PPARα functions to maintain lipid homeostasis in part through regulation of fatty acid oxidation (Corton et al., 2014), AhR and CAR play critical roles in regulating enzymes involved in xenobiotic metabolism including members of the cytochrome P450 (CYP) family (Budinsky et al., 2014; Elcombe et al., 2014).
Cellular oxidants induce the activation of nuclear factor erythroid 2-related factor 2 (NF-E2/related factor 2, Nrf2) and regulated genes, which are often considered to act as a first line of defense from cellular damage due to unchecked OS (Kensler et al., 2007). These genes encode a number of protective enzymes involved in xenobiotic detoxification, antioxidative response, and proteome maintenance (Kensler et al., 2007). Activation of Nrf2 is regulated by the actin-associated kelch-domain 1 protein (Keap1), an adaptor component of Cullin 3 (Cul3)-base ubiquitin E3 ligase. In the absence of OS, Keap1 sequesters Nrf2 in the cytoplasm and facilitates Nrf2 ubiquitination and degradation. The control of the Nrf2/Keap1 regulon depends on a multifaceted protein, known as p62 (also, SQSTM1, sequestosome1, A170) that regulates cellular redox homeostasis by binding to Keap1 and facilitating Keap1 degradation in an autophagy-dependent manner (Komatsu et al., 2010). Under conditions of Keap1 degradation including increased reactive oxygen species (ROS) or electrophiles, Nrf2 accumulates in the cytoplasm and translocates to the nucleus, where it heterodimerizes with members of the small Maf transcription factor family. Binding of Nrf2-Maf heterodimers to antioxidant response elements (AREs) in gene promoters leads to the induction of a battery of ARE-regulated genes (Itoh et al. 1997). Dysregulation of the Nrf2 pathway is implicated in various diseases, including cancers and inflammatory conditions (Cominacini et al., 2015; Esteras et al., 2016; Leinonen et al., 2015).
Chemical-induced OS occurs through a number of mechanisms. PPARα activators regulate the expression of enzymes that produce hydrogen peroxide as a byproduct of metabolism, including the peroxisomal, mitochondrial, and microsomal oxidases such as fatty acyl-CoA oxidase in hepatocytes (Becuwe and Dauça, 2005; Misra and Reddy, 2014). Exposure to PPARα activators also leads to activation of NADPH oxidase, which plays an important role in generating the superoxide radical in response to Kupffer cell activators (De Minicis et al., 2006; Ruysn et al., 2000). There is evidence that activators of AhR or CAR induce OS in part through induction of genes encoding CYP enzymes. In reactions catalyzed by CYP enzymes, 2 electrons are sequentially transferred from NADPH-dependent CYP oxidoreductase to each atom of bound oxygen on the CYP, resulting in the production of oxygenated substrate and water (Guengerich and Lieber, 1985; Poulos and Raag, 1992). Tight coupling normally exists between oxygen reduction and mono-oxygenation, but some reactive oxygen may be released as either superoxide or hydrogen peroxide in the course of electron transfer. The monooxygenase-dependent production of ROS in liver microsomes, supported by NADPH, is a well-known phenomenon (Gillette et al., 1957) that clearly contributes to the total cellular production of reactive oxygen in liver (Bondy and Naderi, 1994). Substrate-independent ROS production has been demonstrated for CYP2E (Dai et al., 1993; Ekstrom et al., 1986) and implied for CYP2B and CYP3A (Ahmed et al., 1995) in cell free extracts. Although CYP induction may lead to induction of OS, no studies have been carried out which systematically link exposure to chemicals that activate CYP family members and downstream effects on the ROS sensor, Nrf2.
The large amount of microarray data in publically available databases such as gene expression omnibus (GEO) is for the most part, an unexploited resource that can be reanalyzed for novel insights into chemical-induced effects. Our lab has developed computational procedures to construct, validate and use in testing, gene expression biomarkers that can accurately predict activation of transcription factors that control gene expression in the liver and other tissues (Oshida et al., 2015a,b,c, 2016a,b; Ryan et al., 2016). The biomarkers are comprised of lists of genes that require the transcription factor for regulation. The genes are selected by a weight of evidence approach by comparing statistically filtered gene lists from multiple microarray studies ideally when chemical comparisons are carried out in wild-type (WT) and transcription factor-null mice. We have created and used biomarkers for Ahr (Oshida et al., 2015a), CAR (Oshida et al., 2015b), and PPARα (Oshida et al., 2015c), as well as the STAT5b transcription factor that controls expression of sex-specific genes in the liver (Oshida et al., 2016a,b). In a previous study (Rooney et al., 2018), we characterized and validated a biomarker that accurately predicts Nrf2 modulation and used the biomarker to identify a strong linkage between Nrf2 activation and feminization of the liver transcriptome through suppression of STAT5b.
In this study, we used the Nrf2 biomarker to identify chemicals that modulate Nrf2 in a mouse liver microarray compendium. In an examination of phenobarbital (PB), the prototypical activator of CAR, we found a strong relationship between activation of CAR and activation of Nrf2. A global analysis of chemicals in our compendium showed that most chemicals that activate CAR also activate Nrf2 implying that these chemicals induce OS. Other chemicals that activate AhR or PPAR also activate Nrf2. We tested whether the activation of Nrf2 was due to OS produced as a byproduct of the activity of 1 or more of the Cyp2b family member genes activated by CAR.
MATERIALS AND METHODS
Strategy for identification of chemicals that affect Nrf2 and comparison to biomarker results for xenobiotic-activated transcription factors
The computational methods used in this study have been previously described in detail (Rooney et al., 2018). These include Nrf2 biomarker development and characterization, mouse liver gene expression database annotation, and methods used to identify factors that modulate Nrf2. Briefly, evaluation of effects of different factors on Nrf2 required a gene expression biomarker of Nrf2-dependent genes and an annotated database of gene expression profiles of statistically filtered genes (also called biosets). The Nrf2 biomarker is a list of genes with associated fold-change values that reflect average differences in expression after chemical or genetic activation and that require Nrf2 for these gene expression changes. A commercially available gene expression database provided by Illumina (BaseSpace Correlation Engine [BSCE]; https://www.illumina.com/products/by-type/informatics-products/basespace-correlation-engine.html; formally NextBio) was used as the starting point to create a compendium of mouse liver biosets. The BSCE database contains over approximately 21 600 highly curated, publically available, omic-scale studies across 15 species including approximately 134 000 lists of statistically filtered genes (as of October, 2017). Each list (bioset) is compared with all other biosets in the database using a fold-change rank-based statistical algorithm called the Running Fisher test, which assesses the overlap in regulated genes and whether those genes are regulated in a similar or opposite manner. In this study, biosets from mouse liver were evaluated. Available information about each bioset was extracted from BSCE and used to populate a compendium of information about the experiments. Each bioset was further annotated using information derived from the original GEO submission or the original publication. Each bioset was annotated for the type of factor (eg, chemical) and the name of the factor (eg, PB) examined. To assess Nrf2 activation or suppression, the Nrf2 biomarker was uploaded into the BSCE database and compared with all biosets in the database using the Running Fisher algorithm. Results of the tests were exported and used to populate the annotated compendium with −log(p-value)s of the comparisons. We have previously used this analysis strategy to accurately identify factors that activate or suppress individual transcription factors (AhR, CAR, and PPARα) using the same database (Oshida et al., 2015a,b,c). In this study, we used these procedures to examine the relationships between modulation of Nrf2 and that of xenobiotic-activated transcription factors.
Cyp2b9/10/13-null mice
All studies were performed according to NIH guidelines for the humane use of research animals and preapproved by the Clemson University Animal Care and Use Committee. WT and Cyp2b9/10/13-null mice are on the C57Bl6/J-background (Jackson Laboratory, Bar Harbor, Maine; 000664). All 5 Cyp2b subfamily members are found on chromosome 7 with 3 members primarily expressed in the liver (Cyp2b9, Cyp2b10, and Cyp2b13) found in tandem repeat (Damari et al., 2012; Kumar et al., 2017; Peng et al., 2012). The 3 Cyp2b subfamily members were targeted by injecting Cas9 mRNA, 20 nt guide sequences specific to the target sites, and with an 83 nt scaffold sequence common to all the sgRNAs into the cytoplasm of the mouse blastocyst (Horii et al., 2015) as described previously in Kumar et al. (2017). An 8- to 10-week-old WT or Cyp2b9/10/13-null male and female mice (n = 4–6) were injected with autoclaved corn oil or TCPOBOP (Sigma) dissolved in autoclaved corn oil at 3 mg/kg. Forty-eight hours later mice were weighed and euthanized by CO2 asphyxiation, livers excised, weighed, diced, and snap frozen for RNA extraction or microsome preparation, and stored at −80°C.
Western blots
Microsomes were prepared as described previously (van der Hoeven and Coon, 1974) by differential centrifugation following homogenization in ice with a Dounce homogenizer. Protein concentrations were determined with the Bio-Rad protein assay (Bio-Rad) according to the manufacturer’s instructions. Thirty μg of microsomal protein was separated by polyacrylamide gel electrophoresis in a 10% gel and transferred to 0.45 μm nitrocellulose (Bio-Rad) where the blot was blocked using 1% skim milk/0.1% Tween 20 dissolved in phosphate buffered saline. Previously characterized rabbit antimouse Cyp2b10 antibody (Mota et al., 2010, 2011) was used to confirm the loss of the Cyp2b9, 10, and 13 genes in mouse liver. Rabbit antimouse β-actin (Sigma Aldrich, St Louis, Missouri) was used to ensure equal loading of samples. The secondary antibodies used to detect the primary antibodies were an alkaline-phosphatase coupled goat antirabbit IgG for the Cyp2b10 antibody and alkaline phosphatase coupled goat antimouse IgG for the Actin antibodies (Bio-Rad). Bands were visualized using a chemilluminescent kit according to the manufacturer’s directions (Bio-Rad).
Dose response modeling of transcriptional data
Raw data (.cel files) from Geter et al. (2014) were downloaded from GEO (GSE54597), normalized by Robust Multi-array Average in R, and fold change values were calculated for each treated sample compared with the average of the same sex controls. Fold change values were fed into the ToxCast data analysis pipeline (tcpl) and, where possible, were fit with hill curves (https://www.epa.gov/sites/production/files/2015-08/documents/pipeline_overview.pdf; accessed May 30, 2017). AC50 (half maximal) concentrations were calculated from the resulting models. AC50 values for the 10 Cyps with the greatest fold induction after treatment with PB were compared with AC50 values for genes in the Nrf2 biomarker.
Evaluation of gene expression by reverse transcription-quantitative polymerase chain reaction
Expression of selected genes was quantified using reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis. Briefly, 1 µg of total RNA was reverse transcribed with the iScript Reverse Transcription Supermix kit per manufacturer instructions (Bio-Rad Laboratories, Hercules California). cDNA was amplified in custom printed 384-well PrimePCR assay plates (Bio-Rad) with Sso Advanced Universal SYBR Green Supermix (Bio-Rad). Reference genes (Ppia, Pum1) were selected from a reference gene panel (Reference Genes M384, Mouse. Bio-Rad) as the 2 genes with the best target stability. The list of the specific PrimePCR assays used is found in Supplementary Material 1. Gene expression was analyzed in Qbase+ (Biogazelle, Zwijnaarde, Belgium). Fold changes for each gene were analyzed for significance via a global ANOVA across both strains and treatments, and where applicable, individual differences assessed by Tukey’s HSD.
Additional computational analyses
Heat maps were generated using Eisen Treeview software (http://rana.lbl.gov/EisenSoftware.htm).
Statistical analyses
All gene lists used in this study were from the BSCE database which derives statistically filtered gene lists based on standard pairwise comparisons (p-value < .05) and are described in detail in Kuperschmidt et al. (2010). These gene lists were derived from treatment vs. control comparisons with at least 3 biological replicates per group. The Nrf2 biomarker or other gene lists (as in Figure 1 and Supplementary Material 2), were compared with a filtered gene list in a pairwise manner using the Running Fisher test in BSCE resulting in a p-value of the correlation between the 2 lists of genes. In the case of the expression of the Cyp2b10 gene, the values listed come from the list of statistically filtered genes (p < .05) described earlier. Regression analyses were carried out in Excel.
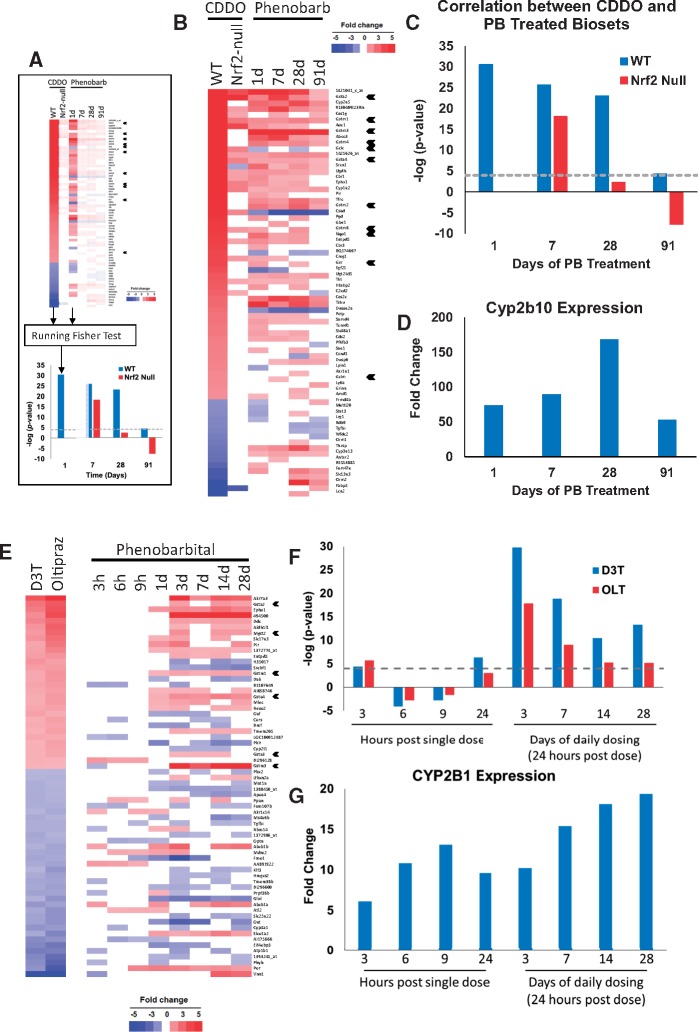
Figure 1.
Exposure to PB alters expression of Nrf2-regulated genes. A, Using the Running Fisher test to determine correlation between 2 gene lists (biosets). The p-value of the correlation test is converted to a −log(p-value) and to a negative number if there is negative correlation. B, Expression of genes after exposure to CDDO-Im or PB. Biosets were from a PB study (0.05% (w/v) in drinking water for 1, 7, 28, or 91 days; from GSE45465) and from WT or Nrf2-null mice treated with CDDO-Im (30 µM) for 6 h (Yates et al., 2009). Genes regulated by CDDO-Im in WT mice and by 1 or more of the PB biosets are shown. Nqo1, glutathione synthesis genes, and Gst family member genes are indicated with arrowheads. C, Significance of the correlation between biosets from the studies described in (B). D, Changes in the expression of the CAR marker gene Cyp2b10 derived from the PB microarray study. The gene expression changes shown were significantly different from the corresponding controls (p < .05). E, Expression of genes after exposure to D3T, oltipraz or PB. Rats were given 100 mg/kg PB by gavage and sacrificed at the indicated times (from the TG-GATES study) or treated with D3T (600 mg/kg) or oltipraz (500 mg/kg) for 24 days (Phan et al., 2009). The genes examined were selected as they exhibited consistent expression by D3T and oltipraz and were altered in expression in at least 3 or more of the 8 PB treatments. GST family member genes are indicated with arrowheads. F, Significance of the correlation between biosets derived from the PB time course study and the Nrf2 activators described in (E). G, Changes in the expression of the CAR marker gene CYP2B1 derived from the microarray study. The gene expression changes shown were significantly different from the corresponding controls (p < .05).
RESULTS
Exposure to PB Alters the Expression of Nrf2-Regulated Genes
CAR activators including PB increase OS in the livers of mice and rats (summarized in Elcombe et al., 2014). To gain insights into whether PB exposure leads to Nrf2 activation, a correlation analysis was carried out between lists of statistically significant genes (biosets) derived from the livers of mice exposed to PB or Nrf2 activators. The Running Fisher test (Kuperschmidt et al., 2010; Rooney et al., 2018) was used to derive a p-value of the correlation between a pair of gene lists (Figure 1A). Biosets from the livers of mice exposed to the potent Nrf2 activator CDDO-Im for 6 h at 30 μmol/kg body weight in WT and Nrf2-null backgrounds (Yates et al., 2009) were compared with those from mice treated with 0.05% (w/v) PB in the drinking water for up to 91 days (from GSE45465). The expression of the overlapping genes is shown in Figure 1B. Many of the genes with increased expression by PB and CDDO-Im are well-known targets of Nrf2 including glutathione synthesis genes, Gst family members, and Nqo1. Figure 1C shows the −log(p-value)s of the correlations between the 2 CDDO-Im biosets and the 1, 7, 28, and 91 days PB treatment groups. There was positive correlation between all PB groups and the bioset from CDDO-Im in WT mice (p-values = 1E-30–1E-4), while there was inconsistent correlation with the bioset from Nrf2-null mice treated with CDDO-Im. The CAR target gene Cyp2b10 exhibited sustained activation throughout the time of the exposures (Figure 1D). A similar analysis carried out using biosets from oltipraz-treated WT and Nrf2-null mice from our previous study (Rooney et al., 2018) showed a similar relationship between PB exposure and oltipraz in WT but not Nrf2-null mice (Supplementary Material 2). These results indicate that PB alters the expression of genes that are also regulated by Nrf2.
To determine if Nrf2-regulated genes were also altered by PB in the rat liver, PB exposures were compared with those of 2 Nrf2 activators, dithiol-3-thione (D3T) or oltipraz. PB biosets were derived from the Japanese TG-GATES study (http://toxico.nibio.go.jp/) in which rats were treated with a single gavage dose of PB (100 mg/kg) and gene expression measured at 3-, 6-, 12-, or 24-h postexposure, or in a repeated daily gavage dose study for 3, 7, 14, or 28 days. The biosets of the Nrf2 activators were from the livers of rats fed diets containing D3T (600 mg/kg diet) or oltipraz (500 mg/kg diet) for 24 days (from GSE8880; Phan et al., 2009). The expression of genes commonly regulated by D3T and oltipraz is shown in Figure 1E. Changes in expression of the overlapping genes regulated by PB, including a number of known Nrf2 target genes, show a lack of obvious similarity to the D3T or oltipraz biosets before 3 days, as compared with the increased similarity at 3 days or longer exposures. Between 3 and 24 h after a single exposure the p-values of the correlations were > 1E-6 (Figure 1F), whereas daily exposures for 3 days or longer exhibited correlations that were generally more significant than single exposures (>1E-29). In contrast to the induction of the Nrf2-regulated genes, the CAR target gene Cyp2b1 was induced at the earliest time point, and exhibited sustained activation of > approximately 6-fold at 3 h and later (Figure 1G). Similar kinetics of expression of the Nrf2-regulated genes was seen when comparing these 2 Nrf2 activators to another CAR activator, phenytoin, as well as to 2 chemicals that activate Cyp2b1 and that are assumed to be CAR activators, hexachlorobenzene and diltiazem, all from the TG-GATES study (Supplementary Material 2). In each case, Cyp2b1 was induced before there was appreciable correlation between the biosets derived from the Nrf2 activators and the CAR activators. Overall, these results indicate that many Nrf2 target genes were induced by CAR activators in the rat liver, and their induction lags behind the induction of the CAR target gene, Cyp2b1.
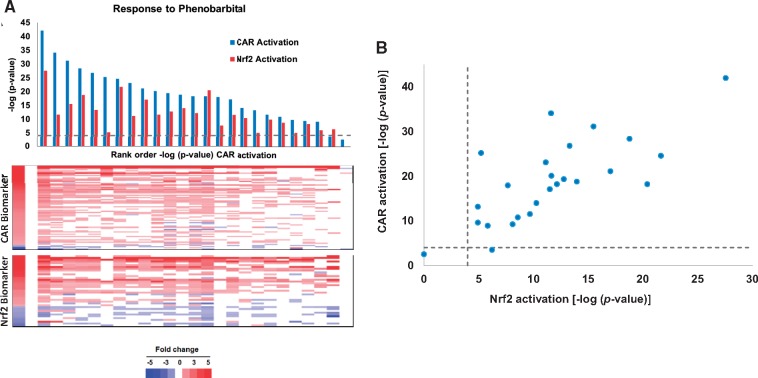
To further test whether PB exposure leads to Nrf2 activation, a previously characterized Nrf2 biomarker was used to assess Nrf2 activation after PB exposure. This Nrf2 biomarker was found to accurately predict Nrf2 activation (balanced accuracy = 96%) (Rooney et al., 2018). Nrf2 activation was evaluated in 25 biosets from 12 studies, in which transcriptional changes were examined in the livers of WT mice treated with PB. These studies were mostly those of mice treated with 0.05% (w/v) PB in water from 1 to 91 days of exposure (details of the exposure conditions are found in Supplementary Material 1). The biosets were also compared with a CAR biomarker, which was previously determined to have a balanced accuracy of 97% (Oshida et al., 2015b). The −log(p-value)s of the correlation of the PB biosets to the CAR biomarker were rank-ordered and compared with the −log(p-value)s of the correlation to the Nrf2 biomarker (Figure 2A). As expected, CAR was activated in the majority of PB treatments (p-value ≤ 1E-4 for 23 of the 25 biosets). Nrf2 was significantly activated (p-value ≤ 1E-4) for all but 1 of the PB treatments. The PB-induced expression changes in the CAR and Nrf2 biomarker genes (Figure 2a, bottom) shows the remarkable increase in similarity in expression direction and relative fold-change with the biomarker gene expression values as the −log(p-value)s increase. A plot of the −log(p-value)s for Nrf2 activation versus CAR activation shows that in general, the greater the significance of the correlation between the PB bioset and the CAR biomarker, the greater the significance of the correlation between that bioset and the Nrf2 biomarker (Figure 2B). The R2 of the comparison was 0.345 (p-value = 5.0E-45). These results indicate that PB exposure consistently leads to alteration of Nrf2-regulated genes and thus, Nrf2 activation.
Figure 2.
Coordinate activation of CAR and Nrf2 by PB across studies. A, (Top) Significance of the correlation between biomarkers for CAR or Nrf2 and 25 biosets from the livers of WT mice treated with PB from 12 studies (GSE16777; GSE34423; GSE40120; GSE40773; GSE43977; GSE44783; GSE45465; GSE51355; GSE55084; GSE57055; GSE60684; GSE6721). Biosets were rank ordered by the significance of the similarity of the biosets to the CAR biomarker. (Bottom) Expression of the genes in the biomarkers across the studies. B, Relationships between the −log(p-value)s of the correlations described in (A).
Nrf2 Activation by Chemical Activators of AhR, CAR, and PPARα
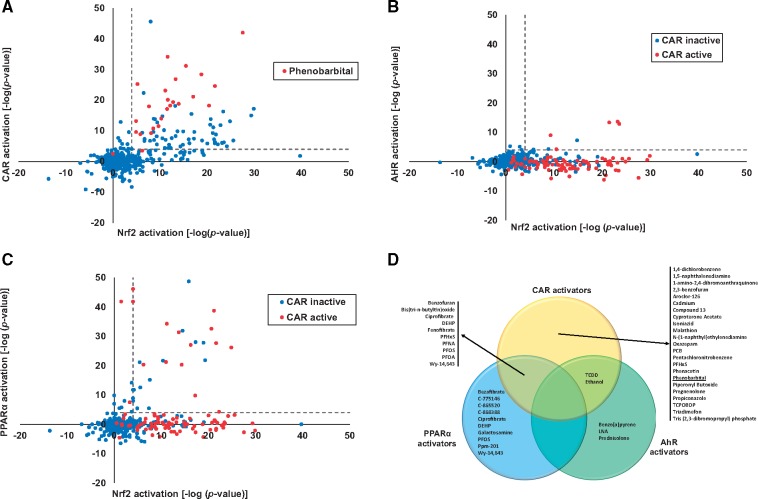
Given the consistent relationship between activation of CAR and Nrf2 by PB, we hypothesized that other CAR activators would also activate Nrf2. To test this hypothesis, a total of 577 biosets encompassing 179 chemicals from the livers of treated mice were evaluated for activation of CAR and Nrf2. Details of the studies are found in Supplementary Material 1. Figure 3A shows a positive trend for activation of both CAR and Nrf2 for a subset of chemicals in the compendium. Out of the 89 biosets in which there was significant CAR activation (p-value ≤ 1E-4), 90% (80 biosets) also led to Nrf2 activation (p-value ≤ 1E-4). After removing the 23 biosets from PB-treated mice, 86% of the biosets from the remaining chemicals led to Nrf2 activation. Only 4 chemicals activated CAR but not Nrf2 (arsenite, 100 ppb in water for 5 weeks; cobaltous chloride, 60 mg/kg for 48 h; penicillin, 1 mg/kg BW for the lifetime of the mice; and propylene glycol mono-t-butyl ether, 1200 ppm in air for 13 weeks). A number of these exposures may have been under conditions that would not lead to Nrf2 activation (doses too low or times of exposure too short).
Figure 3.
Nrf2 activation by chemical activators of xenobiotic receptors. A, Activation of Nrf2 by chemical activators of CAR in the mouse liver compendium. A total of 471 biosets from chemically treated WT mice representing 167 chemicals were examined for activation of CAR or Nrf2 based on biomarker correlation. The biosets from PB studies are indicated in red. B, Activation of AhR and Nrf2. Biosets described in (A) were examined for activation of AhR and Nrf2 using the respective biomarkers. The conditions in red (5 with significant AhR activation) are those with significant activation of CAR as assessed by the CAR biomarker. C, Activation of PPARα and Nrf2. Biosets described in (A) were examined for activation of PPARα and Nrf2 using the respective biomarkers. The exposure conditions in red (13 with significant PPARα activation) are those with significant activation of CAR as assessed by the CAR biomarker. D, Summary of chemicals that activate Nrf2 and 1 or more xenobiotic receptors. Chemicals which result in activation of Nrf2 and activate one or more of the indicated receptors were grouped in the Venn diagram. Details of each experiment are found in Supplementary Material 1.
Increases in OS have also been observed after exposure to compounds that activate AhR or PPARα. Nrf2 activation associated with increases in OS has not been measured across diverse exposure scenarios that activate 1 or more of these receptors. The relationships between activation of AhR or PPARα and Nrf2 were examined using previously described biomarkers for AhR and PPARα, which had balanced accuracies of 95% and 98%, respectively (Oshida et al., 2015a,c). Figures 3B and C show the relationships between activation of Nrf2 and the activation of either AhR or PPARα, respectively. R2 value for the test in the PPARα versus Nrf2 comparison was 0.082 (p-value = 2.46E-10), and the R2 value for the test in the AhR versus Nrf2 comparison was 0.0009 (p-value = .51), There were a number of biosets which exhibited activation of both AhR and Nrf2 or both PPARα and Nrf2. However, many of these biosets also exhibited CAR activation (Figs. 3B and 3C, red points). Out of the 6 biosets in which there was AhR activation but no activation of CAR or PPARα (p-value ≤ 1E-4), half of the biosets (3 biosets consisting of 3 chemicals) also led to Nrf2 activation (p-value ≤ 1E-4). After removing those biosets which also had CAR activation, 63% of the biosets (15 out of the 24 biosets consisting of 10 chemicals) exhibited both PPARα and Nrf2 activation (p-value ≤ 1E-4). There were a number of chemicals that activated PPARα but not Nrf2 (Supplementary Material 1). Overall, this global view of diverse chemical treatments in mouse liver showed very consistent relationships between activation of CAR and Nrf2 and to lesser extents, between activation of AhR and Nrf2 or PPARα and Nrf2.
The distribution of receptor activation by those chemicals that activate Nrf2 is shown in Figure 3D. There were 46 chemicals that activated Nrf2 and 1 or more of the receptors while 37 chemicals activated Nrf2 only (Supplementary Material 1). There were some chemicals such as di-(2-ethylhexyl)phthalate, perfluorooctane sulfonate (PFOS), and WY, which were examined under multiple conditions and depending on the exposure conditions were found to activate PPARα only or PPARα and CAR. As described earlier, the greatest number of chemicals that activated Nrf2 was those that also activated CAR. The second largest group was those chemicals that activated PPARα and Nrf2.
Activation of CAR-Dependent Cyp2b Genes Occurs at Lower Doses of PB than Nrf2-Dependent Genes
The dose-dependent relationships of Nrf2 and CAR activation were examined using a study, in which PB was administered to male and female mice for 2 or 7 days (Geter et al., 2014; GSE54597). In all 4 sex-time comparisons, there was consistent CAR activation at the 14.1 mg/kg dose level and above, whereas Nrf2 activation only occurred consistently at 62.4 mg/kg and above (Figure 4A, top). The expression changes of genes in the biomarkers show the induction of many CAR biomarker genes at lower doses than those in the Nrf2 biomarker (Figure 4A, bottom). This was especially apparent for Cyp2b10, which was strongly expressed at 15 mg/kg and above. These results indicate that many of the CAR genes were induced at lower doses than those in the Nrf2 biomarker.
Figure 4.
Dose-dependent activation of Nrf2 by PB. A, (Top) Significance of the correlation between CAR or Nrf2 biomarkers and the biosets derived from the livers of male and female mice treated for 2 or 7 days with PB (Geter et al., 2014). (Bottom) Expression of the genes in the biomarkers across the treatments. Positions of Cyp genes are indicated. B, Transcriptional AC50 levels (mg/kg-day) for genes in the CAR or Nrf2 biomarkers. The 4 sex-time groups are indicated. Cyp2b10 AC50 levels are shown indicated as red circles.
To more precisely determine any differences in the dose-response characteristics of genes regulated by CAR and Nrf2, Hill models were fit to the transcriptional data using the ToxCast Data Analysis Pipeline, and AC50 values were determined from the resulting models. In both male and female mice, the average AC50 values for the 10 CAR-regulated Cyp genes with greatest induction after 2 days of PB treatment were lower than the average AC50 values for genes in the Nrf2 biomarker that showed dose response relationships (Figure 4B). After 7 days of PB treatment, there was no difference between the average AC50 values in either sex. These studies indicate that after 2 days of exposure to PB, CAR-regulated Cyp genes were activated at lower doses than those regulated by Nrf2.
Divergent Nuclear Receptor Dependence of Nrf2 Activation by Fibrates and Perfluorinated Chemicals
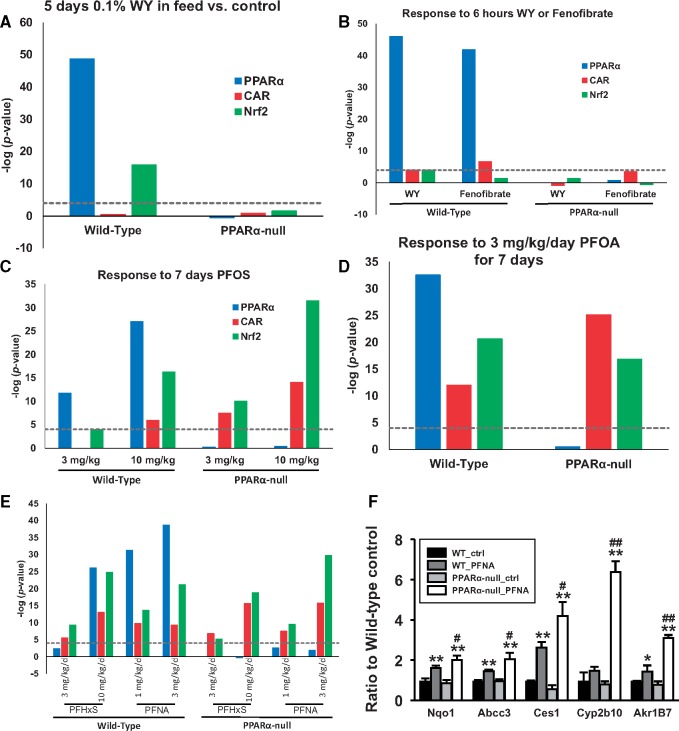
Chemicals associated with PPARα and Nrf2 activation fell into 2 groups: those that activated only PPARα such as fibrates and those that activated both PPARα and CAR such as the perfluorinated compounds. The dependence on PPARα for Nrf2 activation by these 2 groups of chemicals was examined by assessing activation of PPARα, CAR, and Nrf2 in chemically treated WT and PPARα-null mice. Figure 5A shows the activation profiles of (WY) (from GSE8295), in which mice were treated for 5 days with 0.1% w/w in the feed. Figure 5B shows the activation profiles of WY-14,163 WY or fenofibrate (from GSE8396), in which mice were treated for 6 h with 400 μl of 10 mg/ml in 0.5% carboxymethyl cellulose. In the WT mice, WY activated PPARα and Nrf2 but had inconsistent effects on CAR. Activation of all transcription factors was abolished in PPARα-null mice. Fenofibrate activated PPARα and CAR but not Nrf2 in WT mice; activation was abolished in the PPARα-null mice.
Figure 5.
Divergent nuclear receptor dependence of Nrf2 activation by fibrates and perfluorinated compounds. Activation of PPARα, CAR, and Nrf2 biomarkers were examined in the livers of WT and PPARα-null mice after treatment with fibrates and perfluorinated compounds. A, Mice were treated with WY for 5 days at 0.1% w/w in the feed (from GSE8295). B, Mice were treated with WY or fenofibrate for 6 h with 400 μl of 10 mg/ml in 0.5% carboxymethyl cellulose (from GSE8396). C, Mice were treated with PFOS at 3 and 10 mg/kg for 7 days (from GSE22871). D, Mice were treated with PFOA at 3 mg/kg for 7 days (from GSE9786). E, Mice were treated with PFHxS at 3 and 10 mg/kg or PFNA at 1 and 3 mg/kg for 7 days (from GSE55756). F, Expression of Nrf2 and CAR biomarker genes by qPCR in the livers of WT and PPARα-null mice treated with 10 mg/kg/day for 7 days with PFNA (Rosen et al., 2017; GSE55756). Values for Cyp2b10 and Akr1b7 came from Oshida et al. (2015b) and are shown for comparison. Significantly different between control and chemical treatment within strains: *p ≤ .05, **p ≤ .01. Significantly different between PFNA-treated in WT and PFNA-treated in nullizygous mice: #p ≤ .05, ##p ≤ .01.
We examined the effects of 4 perfluorinated compounds (perfluorooctanoic acid (PFOA), PFOS, perfluorohexanesulfonic acid (PFHxS), and perfluorononanoic acid (PFNA)) on receptor activation. The biosets came from three 7 day gavage studies: (1) PFOS at 3 and 10 mg/kg (GSE22871) (Figure 5C); (2) PFOA at 3 mg/kg (GSE9786) (Figure 5D); (3) PFHxS at 3 and 10 mg/kg or PFNA at 1 and 3 mg/kg (GSE55756) (Figure 5E). Activation of PPARα by all 4 chemicals was observed in WT but not PPARα-null mice, as expected (Figs. 5C–E). Except for PFOS at 3 mg/kg, CAR and Nrf2 activation were observed in both wild-type and PPARα-null mice.
Expression of Nrf2 and CAR marker genes was examined by RT-qPCR in WT and PPARα-null mice after exposure to PFNA (Figure 5F). The marker genes for Nrf2 included Nqo1, Ces1, and Abcc3, which are included in the Nrf2 biomarker (Rooney et al., 2018). Also shown is the expression of the CAR biomarker signature gene Cyp2b10 (Oshida et al., 2015b) and Akr1b7, a target for CAR activators (Liu et al., 2009). PFNA significantly increased the expression of all 3 Nrf2 marker genes in both WT and PPARα-null mice. PFNA increased the expression of Akr1b7 in both strains while Cyp2b10 was increased statistically in PPARα-null mice only. These results demonstrate that although WY increases Nrf2 activation in the absence of CAR activation and dependent on PPARα, perfluorinated chemicals increased Nrf2 activation independently of PPARα, possibly through CAR.
Activation of Nrf2 by PB and TCPOBOP Is CAR-Dependent
The dependence on CAR for Nrf2 activation was examined after exposure to CAR activators. In the first study, WT mice, CAR/pregnane X receptor (PXR)-null mice, and CAR/PXR-null mice which expressed the human CAR and PXR (humanized mice) were treated for up to 91 days with 0.05% PB (wt/vol) in drinking water (Luisier et al., 2014). CAR and PXR are often activated simultaneously after chemical exposure (Oladimeji et al., 2016). Reversibility of responses was examined in mice treated for 91 days followed by a 28 days recovery time (119 days time point). In WT mice activation of CAR and Nrf2 was observed at all treatment times except after the 28 days recovery period (119 days; Figure 6A). In CAR/PXR-null mice, CAR activation was abolished, as expected. Nrf2 activation was observed at the 2 earlier (1 and 7 days) time points but not the later time points. The pattern of consistent CAR and Nrf2 coactivation was restored in the humanized mice. The pattern of activation indicates that CAR and/or PXR were required for Nrf2 activation induced by PB. The activation of Nrf2 in the CAR/PXR-null mice at the earlier time points could point to increases in OS. The doses of PB in the livers of the CAR/PXR-null mice were up to approximately 3-fold higher than in the WT mice, yet there was no apparent liver damage under these conditions (Luisier et al., 2014).
Figure 6.
Activation of Nrf2 by PB and TCPOBOP is CAR-dependent. A, Assessment of CAR and Nrf2 activation in the livers of WT, CAR/PXR-null and humanized CAR/PXR (hCAR/hPXR) mice treated for up to 91 days with 0.05% PB (wt/vol) in drinking water (Luisier et al., 2014). Reversibility of responses was examined in mice treated for 91 days followed by a 28 days recovery time (119 days time point). B, Assessment of CAR and Nrf2 activation in the livers of WT, CAR-null and hCAR mice exposed to PB, TCPOBOP or CITCO for 3 days (data from GSE40120). C, Gene expression was examined in the livers from WT or CAR-null mice by qPCR after exposure to PB (0.085% w/w diet for 28 days; Ross et al., 2009). Expression analysis of Cyp2b10 and Akr1b7 has been previously published (Oshida et al., 2015b) and are shown for comparison. Significantly different between control and chemical treatment within strains: *p ≤ .05, **p ≤ .01. Significantly different between PB-treated in WT and PB-treated in nullizygous mice: #p ≤ .05, ##p ≤ .01.
In the second study, PB, the mouse-selective CAR activator TCPOBOP, and the human-selective CAR activator CITCO were administered to WT, CAR-null, and CAR-null mice carrying the human CAR (hCAR mice) for 3 days (from GSE40120). PB and TCPOBOP in WT mice resulted in activation of CAR and Nrf2 while CITCO had no effects (Figure 6B). Effects of all compounds were abolished in CAR-null mice. In hCAR mice, CITCO and PB activated CAR, and Nrf2 was not significantly activated.
Expression of Nrf2 and CAR marker genes was examined RT-qPCR in WT and CAR-null mice after exposure to PB (0.085% w/w diet for 28 days) (Figure 6C). The Nrf2- and CAR-regulated genes exhibited increased expression after PB exposure that was abolished in CAR-null mice. Overall, the data demonstrates that Nrf2 activation by PB and TCPOBOP is CAR-dependent.
Is Activation of Nrf2 by TCPOBOP Dependent on Cyp2b Expression?
We hypothesized that Nrf2 activation is dependent on Cyp enzyme-induced OS. A comprehensive analysis of the expression of Cyp genes altered by PB in the mouse liver has not been carried out. Figure 7A (left) shows the pattern of expression of all 41 Cyp genes that were altered by PB in the 25 biosets described in Figure 2A. The 5 genes which exhibited the highest average fold-change increases included Cyp2b10 (127-fold), Cyp2c65 (54-fold), Cyp2c55 (33-fold), Cyp2b9 (15-fold), and Cyp2b13 (9-fold) (Figure 7A, right). The average expression of the top 5 Cyps was plotted versus −log(p-value) of the Nrf2 biomarker correlation to the PB biosets. The results show that in general as the average expression of the 5 Cyps increases so does Nrf2 activation (R2 = 0.551; p-value = 2.17E-05) (Figure 7A, inset). When the analysis was extended to include all chemicals, the plot shows that an average expression fold-change ≥ 25 resulted in 62 out of 64 (97%) biosets with Nrf2 activation (Figure 7B). Dropping the cutoff to fold-change ≥ 10 resulted in 104 out of 121 (86%) biosets with Nrf2 activation. Most of these chemical exposures led to CAR activation (Figure 7B, red dots). Thus, expression of these Cyps in many of the chemical exposures appears to be a predictor of Nrf2 activation, as would be expected if there was a mechanistic link between the level of Cyp induction and Nrf2 activation.
Figure 7.
Relationship between expression of Cyp genes and activation of Nrf2 by CAR activators. A, Heat map showing the expression of Cyp family members after exposure to PB from the 25 biosets in the same order as Figure 2. The graph shows the average expression of each Cyp across the 25 biosets. (Inset) Relationship between average expression of the top 5 ranking Cyp genes and Nrf2 activation for the 25 biosets from PB-treated mice. B, Relationship between average expression of the top 5 ranking Cyp genes and Nrf2 activation in the livers of mice treated with 135 chemicals from 318 biosets.
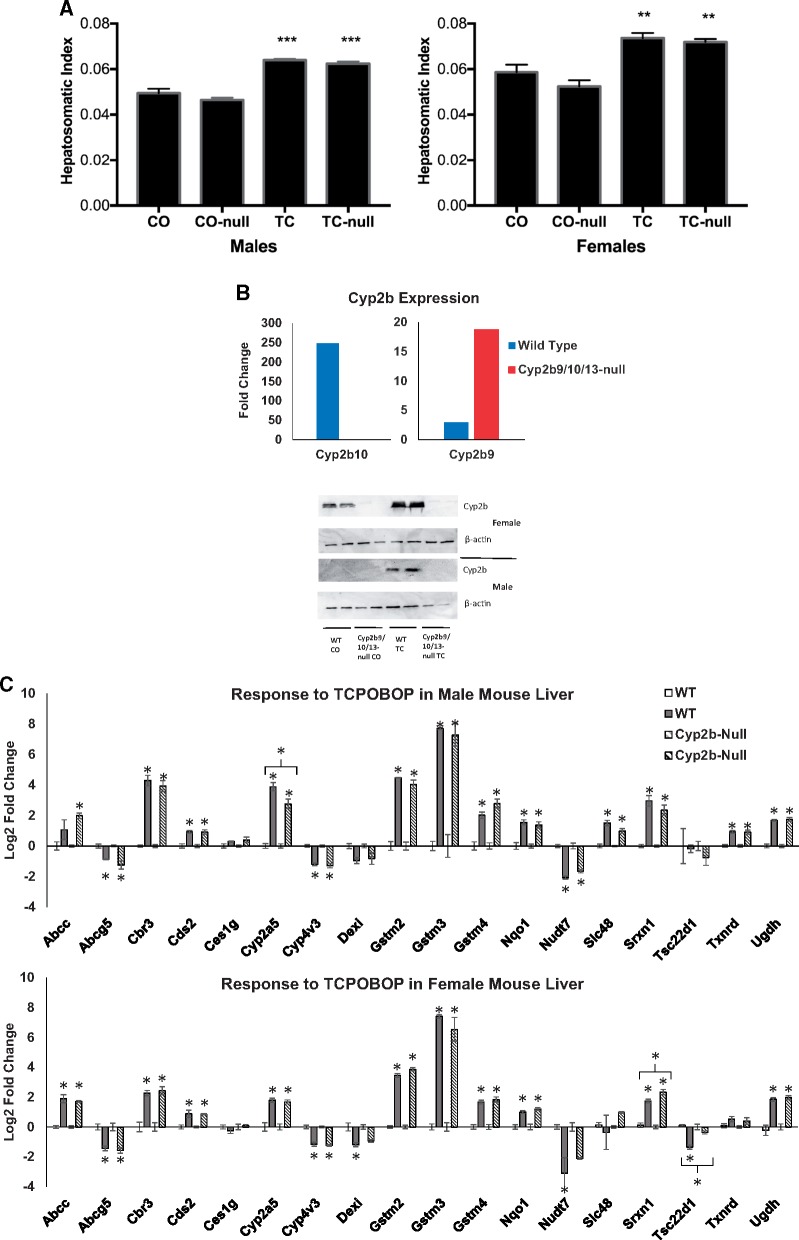
The role of Cyp2b family members on Nrf2 activation was tested directly by examining Nrf2-responsive genes in the livers of a novel nullizygous mouse, in which a 287 kilobase pair (kb) Cyp2b cluster on mouse chromosome 7 was deleted using Crispr-Cas9 technology (Kumar et al., 2017). This strain of mice lacks intact Cyp2b9, Cyp2b13, and Cyp2b10 genes. The lack of the 3 genes had little effect on the expression and activity of other Cyps (Kumar et al., 2017). Males and females of the Cyp2b9/10/13-null strain and their WT counterparts were treated with 1 injection (i.p.) of TCPOBOP (3 mg/kg, in corn oil) or corn oil alone and sacrificed 48 h later. The expected increases in liver to body weight ratios were not different between the strains (Figure 8A). Increases in the expression of the Cyp2b10 gene were abolished in the null strain as expected (Figure 8B, top). Unexpectedly, there were higher levels of activation of Cyp2b9 in the null mice compared with the WT controls. Deletion of the 287 kb section of Cyp2b10-Cyp2b13-Cyp2b9 does not include the promoter region of Cyp2b10 or the final 1000 bp of the 3’-end of the Cyp2b9 gene, the last gene in the region (Kumar et al., 2017). Therefore, it is possible that some increase in Cyp2b9 could be measured due to activation of the Cyp2b10 promoter. Using a polyclonal antibody that recognizes Cyp2b9, 10, 13, and 19 subfamily members (Damiri et al., 2012), Western blots showed that the induction observed in WT mice was abolished in the null mice (Figure 8B, bottom). qPCR assessment of gene expression changes for a subset of Nrf2 biomarker genes confirmed activation of the Nrf2 pathway by TCPOBOP in both genotypes and both sexes (Figure 8C), but overall there were only 3 genes, which exhibited differences between strains. Cyp2a5 exhibited decreased expression in the male null mice. In female null mice, Srxn1 exhibited increased expression and Tsc22d1 exhibited a loss of down-regulation. Overall, the results indicate that activation of Nrf2 by TCPOBOP does not require these 3 Cyp2b family members.
Figure 8.
Activation of Nrf2 by TCPOBOP in WT and Cyp2b9/10/13-null mice. Male and female mice were treated with 1 dose of TCPOBOP (3 mg/kg) for 48 h. A, Liver to body weight changes (hepatosomatic index). **p < .01. ***p < .0001. Significantly different between controls and TCPOBOP-treated mice. B, (Top) Cyp2b gene expression changes after exposure to TCPOBOP. Fold-change values are relative to control treated mice. The gene expression changes shown were significantly different from corresponding controls (p < .05). (Bottom) Cyp2b protein expression after exposure to TCPOBOP as determined by Western blot. C, qPCR analysis of Nrf2 biomarker genes. Significantly different from corresponding control: *p < .05, **p < .01.
DISCUSSION
Although it is generally accepted that chemical-induced OS plays roles in carcinogenesis (Goetz and Luch, 2008; Roberts et al., 2010), the role of OS in hepatocarcinogenesis induced by chemicals that activate AhR, CAR, and PPARα is less clear (Budinsky et al., 2014; Corton et al., 2018; Elcombe et al., 2014). The global microarray analysis strategy employed in the present study identified consistent relationships between activation of these receptors and assumed increases in OS indirectly measured by activation of genes regulated by Nrf2. In particular, we found strong evidence for linkages between CAR and Nrf2 activation. PB, the prototypical activator of CAR, was shown to regulate genes in the mouse liver that were also regulated in the same direction by activators of Nrf2 including CDDO-Im and oltipraz, in WT but not Nrf2-null mice (Figs. 1B and 1C;Supplementary Material 2). In the rat liver, PB also caused modulation of genes that were altered by the Nrf2 inducers, D3T and oltipraz (Figs. 1E and 1F), demonstrating that the induction of Nrf2-regulated genes by PB is common to 2 species susceptible to CAR-induced liver tumors. Using the gene expression biomarker, we found that Nrf2 was regulated in the livers of mice treated with PB. Out of the 25 biosets from PB-treated mice, in which CAR was activated, all but one (96%) had significant activation of Nrf2 (Figure 2). The activation of Nrf2 was also found after exposure to 34 structurally diverse chemicals that activated CAR. For these chemicals, represented by 66 biosets, in which there was activation of CAR, 86% also exhibited significant activation of Nrf2 (Figure 3A). The chemicals that activated CAR and Nrf2 included many that are well known to cause liver cancer including perfluorinated chemicals and conazole fungicides (Figure 3D). CAR was shown to be required for Nrf2 activation. Nrf2 was activated in all WT and CAR/PXR humanized mice treated with PB but not consistently activated in PB-treated CAR/PXR-null mice (Figure 6A). PB and TCPOBOP were found to induce Nrf2 in WT but not CAR-null mice (Figure 6B). The strong linkage between CAR and Nrf2 activation across multiple chemicals is supported by previous studies, in which CAR and Nrf2 target genes were coordinately induced (Aleksunes and Klaassen, 2012; Knight et al., 2008). Induction of the Nrf2 target genes Nqo1 and Cyp2a5 by PB were found to be dependent on Nrf2 (Ashino et al., 2014). Our global microarray analysis approach that incorporates the principles of weight of evidence revealed a consistent relationship between activation of CAR and Nrf2.
Our findings allow us to put OS and Nrf2 activation into the context of the CAR liver cancer mode of action (MOA). The proposed model is shown in Figure 9 and includes key events required for sustained CAR activation to increase liver tumors (Elcombe et al., 2014). The key events after CAR activation include altered gene expression specific to CAR, cell proliferation, and clonal expansion of initiated hepatocytes (Figure 9, top line). We hypothesized that chemicals that activate CAR induce the enzyme activity of 1 or more Cyp family members resulting in increases in the levels of oxidants detected by the Keap1-Nrf2 regulon that can damage DNA. Furthermore, we provide evidence below that the OS is mechanistically involved in hepatocarcinogenesis.
Figure 9.
Model for Nrf2 induction by CAR activators and relationship to the MOA for liver cancer. See text for explanation.
The model predicts that chemical-induced increases in ROS and subsequent activation of Nrf2 require prior expression of Cyp enzymes that would be time- and dose-dependent. Consistent with this, experiments carried out in the rat liver showed that there was a lag in time after initial exposure of PB before Nrf2-regulated genes were appreciably induced. Although the Cyp2b1 gene was induced by PB as early as 3 h, Nrf2-regulated genes were not consistently induced until after 1 day (Figs. 1F and 1G). A number of other known or putative chemical activators of CAR were examined in the TG-GATES dataset including phenytoin, hexachlorobenzene, and diltiazem. For each chemical, activation of Nrf2-dependent genes was not apparent until sometime between 1 and 3 days, well after induction of Cyp2b1 (Supplementary Figure 1). Furthermore, dose-response modeling demonstrated that CAR-regulated Cyp genes, were induced at doses lower than those in the Nrf2 biomarker after 2 days of exposure (Figs. 4A and 4B). Taken together, these observations indicate that CAR was activated at earlier times and lower doses than Nrf2, presumably, because induction of 1 or more Cyp genes and oxidant producing encoded proteins was required to achieve a threshold of OS necessary to activate Nrf2.
The dependence of Nrf2 activation on Cyp expression was tested genetically in a unique model, in which a cluster of 3 Cyp2b genes on mouse chromosome 7 were deleted by Crispr-Cas9 technology (Kumar et al., 2017). This strain lacks functional Cyp2b9, Cyp2b13, and Cyp2b10 genes that were among the top 5 genes with greatest fold-change increases by PB across 24 biosets (Figure 7A). Increased expression of these top 5 genes was associated with increased Nrf2 activation across diverse chemical exposures (Figure 7B). Both WT mice and Cyp2b9/10/13-null mice exposed to TCPOBOP exhibited similar induction of Nrf2-dependent genes based on qPCR of 18 genes in the Nrf2 biomarker (Figure 8). Thus, induction of Nrf2 by TCPOBOP did not require these 3 Cyp2b genes. These results lead us to speculate that the increases in OS are due to other Cyp protein family members, the cumulative effects of many Cyp protein family members, or other induced proteins. It is possible that the large increases in Cyp2b gene expression do not result in parallel increases in Cyp protein expression and activity, as there are many examples of lack of parallel increases in expression and activity (Molina-Ortiz et al., 2014; Nishiyama et al., 2016; Ohtsuki et al., 2012). Furthermore, increases in Cyp enzymatic activity do not always result in increases in OS (Ekstrom et al., 1986). In summary, this is the first study which tested the dependence of specific Cyp protein family members in activating Nrf2 using a transgenic mouse model.
Despite the lack of genetic dependence of OS induction and Nrf2 activation on the proteins encoded by the 3 Cyp genes, there remains strong biochemical evidence for the involvement of Cyp enzymes in mediating the increases in OS. PB exposure led to oxidation of genomic DNA as assessed by measurement of 8-hydroxy-2'-deoxyguanosine, a biomarker of DNA oxidation in vivo, which was inhibited by the inhibitor ketoconazole (Imaoka et al., 2004). Two CAR inducers PB and Aroclor 1254 significantly increased hydrogen peroxide generation, the OS marker malondialdehyde, and NADPH oxidation in vitro; the chemicals also significantly enhanced formation of malondialdehyde in vivo, in both liver and plasma (Dostalek et al., 2007). PB-mediated increases in liver and plasma F2-isoprostanes, a marker for OS could be ablated by the CYP inhibitor 1-aminobenztriazole, implicating the PB-induced CYP(s) in the F2-isoprostane elevation (Dostalek et al., 2007). The CAR activator propiconazole increased ROS levels in mouse liver cells that were inhibited by diphenylene iodonium chloride, a CYP reductase inhibitor (Nesnow et al., 2011). Over-expression of rat CYP2B1 in mouse liver in the absence of chemical exposure led to enhanced spontaneous tumor formation (Lehman-McKeeman et al., 1999), although whether overexpression of CYP2B1 caused increases in ROS and DNA damage in this animal model is not known. Overall, these studies point to a role of Cyps but not necessarily Cyp2b family members in the induction of ROS, DNA damage, and hepatocarcinogenesis after exposure to CAR activators. However, the proteins responsible for the increases in OS remain unknown.
Although considered in the Elcombe et al. (2014) MOA analysis, increases in OS was not classified as a key event involved in PB-induced liver tumors. However, the consistent linkage between activation of CAR and Nrf2 as well as OS-induced DNA damage discussed above points to the importance of OS in CAR-mediated hepatocarcinogenesis. If OS was mechanistically important in the CAR MOA, then chemopreventative agents that decrease OS or markers of OS should also decrease the number of foci or tumors induced by CAR activators. We tested this hypothesis by examining the effects of diverse chemopreventative agents some of which are known antioxidants and Nrf2 activators on liver carcinogenesis. In each of these studies, initiation was carried out using diethylnitrosamine followed by promotion with PB. Out of 17 studies evaluated, 14 showed consistent decreases in markers of OS and the size and/or number of liver tumors or preneoplastic lesions compared with PB treatment alone (Table 1). These studies provide strong evidence that exposure to agents with antioxidant activity suppress a key event in PB-mediated liver cancer, namely increases in OS. Future reanalysis of the CAR MOA should reconsider OS as a key or modulating event.
Table 1.
Suppression of Phenobarbital-Induced Oxidative Stress and Hepatocarcinogenesis by Chemopreventative Agents
| PMID | Sex | Compound or Mixture | PMID of Papers Showing That The Compound or Mixture Activates Nrf2 | Compound Administered Before or After Initiator? | Phenobarbital Administered | Effect on Marker of Oxidative Damage | Effects On Tumor Area and Number | Correlation Between Oxidative Stress and Liver Tumor Formation? |
|---|---|---|---|---|---|---|---|---|
| 9045889 | Male | S-Methyl methanethiosulfonate | No information | After | 0.05% w/v in drinking water | Decrease in lipid peroxidation in rabbit erythrocyte membrane ghosts and rat liver; no significance indicated | Decrease in incidence, multiplicity and average area | Yes |
| 9121950 | Female | Vitamin E | 20153624 | After | 0.5g/100g feed in diet | Decrease in TBARS | No change in area and number | Equivocal |
| 11341044 | Female | High fish oil | 24157545; 24478369 | After | 0.05% in diet | Increase in TBARS | Decrease in area and number | Opposite |
| 15112340 | Male | 1α, 25-dihydroxyvitamin D3 | 27484042 | After | 0.05% in diet | Decrease in lipid peroxidation; no significance indicated | Decrease in incidence and number | Yes |
| 15924350 | Male | Apigenin | 27656262 | After | 0.05% w/v in drinking water | Decrease in lipid peroxidation and protein carbonylation | Decrease in number of tumor bearing rats and % of tumor nodules; significance not indicated | Yes |
| 16188247 | Male | Methanolic extract of Solanum trilobatum | No information | After | 0.05% in diet | Decrease in TBARS | Decrease in number, volume, and size | Yes |
| 16830150 | Male | Vanadium | Inhibition of Nrf2 nuclear accumulation (20599494, 19367690); vanadium(III)-L-cysteine induced Nrf2 (26573721) | Before | 0.05% in diet | Decrease in 8-OHdG | Decrease in nodule multiplicity | Yes |
| 17300697 | Male | Extract of Solanum trilobatum | No information | After | 0.05% in diet | Decrease in TBARS | Decrease in mean nodular volume | Yes |
| 17658503 | Male | Star anise | No information | After | 0.05% w/v in drinking water | Decrease in lipid peroxidation | Decrease in area and number | Yes |
| 21445586 | Male | Enzymatically modified isoquercitrin | No information | After | 500 ppm in diet | No difference in TBARS and 8-OHdG | Decrease in number and area | Equivocal |
| 23665940 | Male | Orphenadrine | No information | After | 60 ppm in drinking water | Increase in TBARS; significance not indicated | Increase in number and area | Yes |
| 24025784 | Male | Piperonyl butoxide | 19690152 | After | 60 ppm in drinking water | Increase in TBARS; significance not indicated | Increase in number; significance not indicated | Yes |
| 24060683 | Male | Brucine | No information | Before | 0.05% w/v in drinking water | Decrease in TBARS | Decrease in incidence, total number and average number/liver, volume | Yes |
| 24418717 | Male | Indole-3-carbinol | 15826493; 21396975 | After | 60 ppm in drinking water | Increase in TBARS; significance not indicated | Increase in number and area; significance not indicated | Yes |
| 24511000 | Male | Lipid-soluble tea polyphenols | Green tea polyphenols activated Nrf2 (11768769) | Before | 0.05% w/v in drinking water | Decrease in 8-OHdG | Decrease in number and size | Yes |
| 24660901 | Male | Lycopene | 15657364 | After | 0.05% w/v in drinking water | Decrease in MDA | Decrease in nodule incidence, mean of nodule, mean nodular volume | Yes |
| 24761908 | Male | Extract of Cleistocalyx nervosum | No information | Before | 500 ppm in drinking water | Decrease in TBARS | Decrease in total number of foci | Yes |
Each of the studies (PubMed ID in first column) examined the effects in rats of a chemopreventative agent on oxidative stress and hepatocarcinogenesis caused by diethylnitrosamine initiation followed by chronic treatment with PB. The list of PubMed IDs in the fourth column is not meant to be exhaustive. The final assessment of correlation is based on whether indicators of oxidative stress and measures of liver cancer were both increasing, both decreasing, changing in opposite directions (opposite), or were equivocal because one of the indicators of oxidative stress or liver cancer was not altered.
Abbreviations: TBARS, thiobarbituric acid reactive substances; MDA, malondialdehyde.
Although we focused mostly on linkages between CAR and Nrf2, it is notable that we found Nrf2 was activated by a number of chemical activators of AhR and PPARα. Mechanistic linkages between activation of AhR or PPARα and OS have been previously described. Although not as consistent as the linkages between CAR and Nrf2, 63% and 50% of biosets, in which PPARα or AhR were activated also exhibited activation of Nrf2. Activation of Nrf2 by TCDD was found to be AhR-dependent (data not shown), although almost all gene expression changes by TCDD were abolished in the AhR-null mice (Tijet et al., 2006). The activation of Nrf2-regulated genes by TCDD was found to depend not only on AhR but on Nrf2 and was hypothesized to depend on increases in ROS generated from increased Cyp activity (Yeager et al., 2009). WY activated Nrf2 after 6 h of exposure in WT but not PPARα-null mice (Figs. 5A and 5B). WY may be typical of chemicals that activate Nrf2 through PPARα but not other receptors. This is in contrast to the perfluorinated compounds which activate PPARα, CAR, and Nrf2 (Figs. 5C–E). All of the perfluorinated compounds activated Nrf2 in a PPARα-independent manner. PPARα activators are thought to increase ROS through 2 main mechanisms including increases in hydrogen peroxide as a byproduct of increased fatty acid catabolism and by activation of NADPH oxidase (Corton et al., 2018). A number of studies have shown that when antioxidants are coadministered with PPARα activating chemicals, there were decreases in the size and incidence of liver tumors compared with the chemicals alone (Corton et al., 2014) similar to the studies with PB and chemopreventative agents discussed earlier. Overall, our findings indicate that Nrf2 is activated under conditions which also activate AhR or PPARα, although less consistently than when CAR is activated.
In summary, we have utilized an annotated microarray compendium applying a weight of evidence analysis to characterize the consistent activation of Nrf2 that occurs after exposure to almost all CAR activators. The chemicals which likely activate Nrf2 through AhR or PPARα were identified as well. We tested the hypothesis that proteins encoded by 3 Cyp2b family members were involved in mediating the increases in OS that leads to Nrf2 activation by a CAR activator. Contrary to our hypothesis, these Cyp2b family members do not appear to play a significant role in Nrf2 induction. It is possible that the increases in OS are due to other Cyp genes or other yet to be identified proteins induced by CAR. Strong evidence in the literature shows that inhibition of Cyp enzymatic activity led to decreases in markers of OS after CAR activator exposure, and that chemopreventative agents suppress both OS and liver cancer caused by PB. The question remains as to whether OS detected as Nrf2 activation in microarray profiles is an associative event, or as indicated in the examination of chemotherapeutic agents, should be considered a key event in the CAR MOA.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
This study was carried out as part of the EPA CSS cancer and steatosis AOP projects. Research support in part provided by National Institutes of Health grant (R15ES017321 to W.S.B.).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr Brian Chorley and Dr Steve Simmons for review of the article, Dr Ivan Rusyn for livers from studies carried out in his lab, Dr Xinxin Ding for valuable discussions, and Mr Naresh Vasani for assistance in carrying out the RT-PCR experiments.
REFERENCES
- Ahmed S. S., Napoli K. L., Strobel H. W. (1995). Oxygen radical formation during cytochrome P450-catalyzed cyclosporine metabolism in rat and human liver microsomes at varying hydrogen ion concentrations. Mol. Cell Biochem. 151, 131–140. [DOI] [PubMed] [Google Scholar]
- Aleksunes L. M., Klaassen C. D. (2012). Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARα-, and Nrf2-null mice. Drug Metab. Dispos. 40, 1366–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashino T., Ohkubo-Morita H., Yamamoto M., Yoshida T., Numazawa S. (2014). Possible involvement of nuclear factor erythroid 2-related factor 2 in the gene expression of Cyp2b10 and Cyp2a5. Redox. Biol. 2, 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becuwe P., Dauça M. (2005). Comparison of cytotoxicity induced by hypolipidemic drugs via reactive oxygen species in human and rodent liver cells. Int. J. Mol. Med. 16, 483–492. [PubMed] [Google Scholar]
- Bondy S. C., Naderi S. (1994). Contribution of hepatic cytochrome P450 systems to the generation of reactive oxygen species. Biochem. Pharmacol. 48, 155–159. [DOI] [PubMed] [Google Scholar]
- Budinsky R. A., Schrenk D., Simon T., Van den Berg M., Reichard J. F., Silkworth J. B., Aylward L. L., Brix A., Gasiewicz T., Kaminski N., et al. (2014). Mode of action and dose-response framework analysis for receptor-mediated toxicity: The aryl hydrocarbon receptor as a case study. Crit. Rev. Toxicol. 44, 83–119. [DOI] [PubMed] [Google Scholar]
- Cominacini L., Mozzini C., Garbin U., Pasini A., Stranieri C., Solani E., Vallerio P., Tinelli I. A., Fratta Pasini A. (2015). Endoplasmic reticulum stress and Nrf2 signaling in cardiovascular diseases. Free Radic. Biol. Med. 88, 233–242. [DOI] [PubMed] [Google Scholar]
- Corton J. C., Cunningham M. L., Hummer B. T., Lau C., Meek B., Peters J. M., Popp J. A., Rhomberg L., Seed J., Klaunig J. E. (2014). Mode of action framework analysis for receptor-mediated toxicity: The peroxisome proliferator-activated receptor alpha (PPARα) as a case study. Crit. Rev. Toxicol. 44, 1–49. [DOI] [PubMed] [Google Scholar]
- Corton J. C., Peters J. M., Klaunig J. E. (2018), The PPARα-dependent rodent liver tumor response is not relevant to humans: Addressing misconceptions. Arch. Toxicol. 92, 83–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y., Rashba-Step J., Cederbaum A. I. (1993). Stable expression of human cytochrome P4502E1 in HepG2 cells: Characterization of catalytic activities and production of reactive oxygen intermediates. Biochemistry 32, 6928–6937. [DOI] [PubMed] [Google Scholar]
- Damiri B., Holle E., Yu X., Baldwin W. S. (2012). Lentiviral-mediated RNAi knockdown yields a novel mouse model for studying Cyp2b function. Toxicol. Sci. 125, 368–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Minicis S., Bataller R., Brenner D. A. (2006). NADPH oxidase in the liver: Defensive, offensive, or fibrogenic? Gastroenterology 131, 272–275. [DOI] [PubMed] [Google Scholar]
- Dostalek M., Brooks J. D., Hardy K. D., Milne G. L., Moore M. M., Sharma S., Morrow J. D., Guengerich F. P. (2007). In vivo oxidative damage in rats is associated with barbiturate response but not other cytochrome P450 inducers. Mol. Pharmacol. 72, 1419–1424. [DOI] [PubMed] [Google Scholar]
- Ekstrom G., Cronholm T., Ingelman-Sundberg M. (1986). Hydroxyl-radical production and ethanol oxidation by liver microsomes isolated from ethanol-treated rats. Biochem. J. 233, 755–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcombe C. R., Peffer R. C., Wolf D. C., Bailey J., Bars R., Bell D., Cattley R. C., Ferguson S. S., Geter D., Goetz A., et al. (2014). Mode of action and human relevance analysis for nuclear receptor-mediated liver toxicity: A case study with phenobarbital as a model constitutive androstane receptor (CAR) activator. Crit. Rev. Toxicol. 44, 64–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteras N., Dinkova-Kostova A. T., Abramov A. Y. (2016). Nrf2 activation in the treatment of neurodegenerative diseases: A focus on its role in mitochondrial bioenergetics and function. Biol. Chem. 397, 383–400. [DOI] [PubMed] [Google Scholar]
- Geter D. R., Bhat V. S., Gollapudi B. B., Sura R., Hester S. D. (2014). Dose-response modeling of early molecular and cellular key events in the CAR-mediated hepatocarcinogenesis pathway. Toxicol. Sci. 138, 425–445. [DOI] [PubMed] [Google Scholar]
- Gillette J. R., Brodie B. B., La Du B. N. (1957). The oxidation of drugs by liver microsomes: On the role of TPNH and oxygen. J. Pharmacol. Exp. Ther. 119, 532–540. [PubMed] [Google Scholar]
- Goetz M. E., Luch A. (2008). Reactive species: A cell damaging rout assisting to chemical carcinogens. Cancer Lett. 266, 73–83. [DOI] [PubMed] [Google Scholar]
- Guengerich F. P., Liebler D. C. (1985). Enzymatic activation of chemicals to toxic metabolites. CRC Crit. Rev. Toxicol. 14, 259–307. [DOI] [PubMed] [Google Scholar]
- Hernandez J. P., Mota L. C., Baldwin W. S. (2009). Activation of CAR and PXR by dietary, environmental and occupational chemicals alters drug metabolism, intermediary metabolism, and cell proliferation. Curr. Pharmacog. Personal Med. 7, 81–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T., Arai Y., Yamazaki M., Morita S., Kimura M., Itoh M., Abe Y., Hatada I. (2015). Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci. Rep. 4, 4513.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaoka S., Osada M., Minamiyama Y., Yukimura T., Toyokuni S., Takemura S., Hiroi T., Funae Y. (2004). Role of phenobarbital-inducible cytochrome P450s as a source of active oxygen species in DNA-oxidation. Cancer Lett. 203, 117–125. [DOI] [PubMed] [Google Scholar]
- Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. (1997). An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236, 313–322. [DOI] [PubMed] [Google Scholar]
- Kensler T. W., Wakabayashi N., Biswal S. (2007). Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 47, 89–116. [DOI] [PubMed] [Google Scholar]
- Knight T. R., Choudhuri S., Klaassen C. D. (2008). Induction of hepatic glutathione S-transferases in male mice by prototypes of various classes of microsomal enzyme inducers. Toxicol. Sci. 106, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y. S., Ueno I., Sakamoto A., Tong K. I., et al. (2010). The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12, 213–223. [DOI] [PubMed] [Google Scholar]
- Kumar R., Mota L. M., Litoff E. J., Rooney J. P., Boswell W. T., Courter E., Henderson C. M., Hernandez J. P., Corton J. C., Moore D. D., et al. (2017). Compensatory changes in CYP expression in three different toxicology mouse models: CAR-null, Cyp3a-null, and Cyp2b9/10/13-null mice. PLoS One 12, e0174355.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupershmidt I., Su Q. J., Grewal A., Sundaresh S., Halperin I., Flynn J., Shekar M., Wang H., Park J., Cui W., et al. (2010). Ontology-based meta-analysis of global collections of high-throughput public data. PLoS One 5, e13066.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc G. A., Norris D. O., Kloas W., Kullman S. W., Baldwin W. S., Greally J. M. Detailed Review Paper on the State of the Science on Novel In Vitro and In Vivo Screening and Testing Methods and Endpoints for Evaluating Endocrine Disruptors Series on Testing & Assessment: No. 178. Paris, Organisation for Economic Co-operation and Development: 213. February 2012. [Google Scholar]
- Lehman-McKeeman L. D., Caudill D., Vassallo J. D., Fix A. S. (1999). Increased spontaneous liver tumor susceptibility in cytochrome P450 2B1 (CYP2B1) transgenic mice. Toxicol. Sci. 48. [Google Scholar]
- Leinonen H. M., Kansanen E., Pölönen P., Heinäniemi M., Levonen A. L. (2015). Dysregulation of the Keap1-Nrf2 pathway in cancer. Biochem. Soc. Trans. 43, 645–649. [DOI] [PubMed] [Google Scholar]
- Liu M. J., Takahashi Y., Wada T., He J., Gao J., Tian Y., Li S., Xie W. (2009). The aldo-keto reductase Akr1b7 gene is a common transcriptional target of xenobiotic receptors pregnane X receptor and constitutive androstane receptor. Mol. Pharmacol. 76, 604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisier R., Lempiäinen H., Scherbichler N., Braeuning A., Geissler M., Dubost V., Müller A., Scheer N., Chibout S.-D., Hara H., et al. (2014). Phenobarbital induces cell cycle transcriptional responses in mouse liver humanized for constitutive androstane and pregnane X receptors. Toxicol. Sci. 139, 501–511. [DOI] [PubMed] [Google Scholar]
- Misra P., Reddy J. K. (2014). Peroxisome proliferator-activated receptor-α activation and excess energy burning in hepatocarcinogenesis. Biochimie 98, 63–74. [DOI] [PubMed] [Google Scholar]
- Molina-Ortiz D., Camacho-Carranza R., Gonzalez-Zamora J. F., Shalkow-Kalincovstein J., Cardenas C., Nosti-Palacios R., Vences M. A. (2014). Differential expression of cytochrome P450 enzymes in normal and tumor tissues from childhood rhabdomyosarcoma. PLoS One 9, e93261.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota L. C., Barfield C., Hernandez J. P., Baldwin W. S. (2011). Nonylphenol-mediated Cyp induction is PXR-dependent: The use of humanized mice and human hepatocytes suggests that hPXR is less sensitive than mouse PXR to nonylphenol treatment. Toxicol. Appl. Pharmacol. 252, 259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota L. C., Hernandez J. P., Baldwin W. S. (2010). Constitutive androstane receptor-null mice are sensitive to the toxic effects of parathion: Association with reduced cytochrome p450-mediated parathion metabolism. Drug Metab. Dispos. 38, 1582–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesnow S., Grindstaff R. D., Lambert G., Padgett W. T., Bruno M., Ge Y., Chen P. J., Wood C. E., Murphy L. (2011). Propiconazole increases reactive oxygen species levels in mouse hepatic cells in culture and in mouse liver by a cytochrome P450 enzyme mediated process. Chem. Biol. Interact. 194, 79–89. [DOI] [PubMed] [Google Scholar]
- Nishiyama Y., Nakayama S. M., Watanabe K. P., Kawai Y. K., Ohno M., Ikenaka Y., Ishizuka M. (2016). Strain differences in cytochrome P450 mRNA and protein expression, and enzymatic activity among Sprague Dawley, Wistar, Brown Norway and Dark Agouti rats. J. Vet. Med. Sci. 78, 675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S., Schaefer O., Kawakami H., Inoue T., Liehner S., Saito A., Ishiguro N., Kishimoto W., Ludwig-Schwellinger E., Ebner T., et al. (2012). Simultaneous absolute protein quantification of transporters, cytochromes P450, and UDP-glucuronosyltransferases as a novel approach for the characterization of individual human liver: Comparison with mRNA levels and activities. Drug Metab. Dispos. 40, 83–92. [DOI] [PubMed] [Google Scholar]
- Oladimeji P., Cui H., Zhang C., Chen T. (2016). Regulation of PXR and CAR by protein-protein interaction and signaling crosstalk. Expert Opin. Drug Metab. Toxicol. 12, 997–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshida K., Vasani N., Jones C., Moore T., Hester S., Nesnow S., Auerbach S., Geter D. R., Aleksunes L. M., Thomas R. S., et al. (2015b). Identification of chemical modulators of the constitutive activated receptor (CAR) in a gene expression compendium. Nucl. Recept. Signal 13, e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshida K., Vasani N., Thomas R. S., Applegate D., Gonzalez F. J., Aleksunes L. M., Klaassen C. D., Corton J. C. (2015a). Screening a mouse liver gene expression compendium identifies modulators of the aryl hydrocarbon receptor (AhR). Toxicology 336, 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshida K., Vasani N., Thomas R. S., Applegate D., Rosen M., Abbott B., Lau C., Guo G., Aleksunes L. M., Klaassen C., et al. (2015c). Identification of modulators of the nuclear receptor peroxisome proliferator-activated receptor alpha (PPARalpha) in a mouse liver gene expression compendium. PLoS One 10, e0112655.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshida K., Vasani N., Waxman D. J., Corton J. C. (2016a). Disruption of STAT5b-regulated sexual dimorphism of the liver transcriptome by diverse factors is a common event. PLoS One 11, e0148308.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshida K., Waxman D. J., Corton J. C. (2016b). Chemical and hormonal effects on STAT5b-dependent sexual dimorphism of the liver transcriptome. PLoS One 11, e0150284.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Yoo B., Gunewardena S. S., Lu H., Klaassen C. D., Zhong X.-B. (2012). RNA sequencing reveals dynamic changes of mRNA abundance of cytochromes p450 and their alternative transcripts during mouse liver development. Drug Metab. Dispos. 40, 1198–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan V., Olusegun George E., Tran Q. T., Goodwin S., Bodreddigari S., Sutter T. R. (2009). Analyzing microarray data with transitive directed acyclic graphs. J. Bioinform. Comput. Biol. 07, 135–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos T. L., Raag R. (1992). Cytochrome P450cam: Crystallography, oxygen activation, and electron transfer. FASEB J 6, 674–679. [DOI] [PubMed] [Google Scholar]
- Roberts R. A., Smith R. A., Safe S., Szabo C., Tjalkens R. B., Robertson F. M. (2010). Toxicological and pathophysiological roles of reactive oxygen and nitrogen species. Toxicology 276, 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney J. P., Oshida K., Vasani N., Vallanat B., Ryan N., Chorley C., Wu C., Aleksunes L. M., Klaassen C., Kensler T., et al. (2018). Activation of Nrf2 in the liver is associated with stress resistance mediated by suppression of the growth hormone-regulated STAT5b transcription factor. PLoS One. 13, e0200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen M. B., Das K. P., Rooney J., Abbott B., Lau C., Corton J. C. (2017). PPARα-independent transcriptional targets of perfluoroalkyl acids revealed by transcript profiling. Toxicology 387, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusyn I., Yamashina S., Segal B. H., Schoonhoven R., Holland S. M., Cattley R. C., Swenberg J. A., Thurman R. G. (2000). Oxidants from nicotinamide adenine dinucleotide phosphate oxidase are involved in triggering cell proliferation in the liver due to peroxisome proliferators. Cancer Res. 60, 4798–4803. [PubMed] [Google Scholar]
- Ryan N., Chorley B., Tice R. R., Judson R., Corton J. C. (2016). Moving toward integrating gene expression profiling into high-throughput testing: A gene expression biomarker accurately predicts estrogen receptor α modulation in a microarray compendium. Toxicol. Sci. 151, 88–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijet N., Boutros P. C., Moffat I. D., Okey A. B., Tuomisto J., Pohjanvirta R. (2006). Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol. Pharmacol. 69, 140–153. [DOI] [PubMed] [Google Scholar]
- van der Hoeven T. A., Coon M. J. (1974). Preparation and properties of partially purified cytochrome P450 and NADPH-cytochrome P450 reductase from rabbit liver microsomes. J. Biol. Chem. 249, 6302–6310. [PubMed] [Google Scholar]
- Yates M. S., Tran Q. T., Dolan P. M., Osburn W. O., Shin S., McCulloch C. C., Silkworth J. B., Taguchi K., Yamamoto M., Williams C. R., et al. (2009). Genetic versus chemoprotective activation of Nrf2 signaling: Overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis 30, 1024–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H., Ishii Y., Yamamoto M., Oguri K. (2006). Induction of the hepatic cytochrome P450 2B subfamily by xenobiotics: Research history, evolutionary aspect, relation to tumorigenesis, and mechanism. Curr. Drug Metab. 7, 397–409. [DOI] [PubMed] [Google Scholar]
- Yeager R. L., Reisman S. A., Aleksunes L. M., Klaassen C. D. (2009). Introducing the “TCDD-inducible AhR-Nrf2 gene battery”. Toxicol. Sci. 111, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.