The three-dimensional crystal structure of the glutathione reductase from Streptococcus pneumoniae is reported at 2.56 Å resolution.
Keywords: glutathione reductase, X-ray crystallography, Streptococcus pneumoniae
Abstract
The glutathione reductase (GR) from Streptococcus pneumoniae is a flavoenzyme that catalyzes the reduction of oxidized glutathione (GSSG) to its reduced form (GSH) in the cytoplasm of this bacterium. The maintenance of an intracellular pool of GSH is critical for the detoxification of reactive oxygen and nitrogen species and for intracellular metal tolerance to ions such as zinc. Here, S. pneumoniae GR (SpGR) was overexpressed and purified and its crystal structure determined at 2.56 Å resolution. SpGR shows overall structural similarity to other characterized GRs, with a dimeric structure that includes an antiparallel β-sheet at the dimer interface. This observation, in conjunction with comparisons with the interface structures of other GR enzymes, allows the classification of these enzymes into three classes. Analyses of the kinetic properties of SpGR revealed a significantly higher value for K m(GSSG) (231.2 ± 24.7 µM) in comparison to other characterized GR enzymes.
1. Introduction
Glutathione (γ-glutamylcysteinyl-glycine) is a thiol-containing tripeptide that is present at high concentrations (1–10 mM) in eukaryotic and many prokaryotic species (Masip et al., 2006 ▸). Glutathione can exist in a thiol-reduced state (GSH) or in an oxidized state (GSSG), which consists of two GSH molecules linked together by a disulfide bond. GSH serves as a reductive cofactor for the antioxidant glutaredoxin enzymes (Ritz & Beckwith, 2001 ▸), with this activity converting glutathione from the reduced to the oxidized form. The ratio of GSH to GSSG in the bacterial cytoplasm is tightly regulated by the enzyme glutathione reductase (GR), which employs NADPH as the electron donor:
The Gram-positive bacterium Streptococcus pneumoniae (the pneumococcus) is the world’s foremost human bacterial pathogen. It is the leading cause of bacterial pneumonia, which accounts for 15% of all childhood disease mortality (Rudan et al., 2008 ▸, 2013 ▸). The acquisition of nutrients is essential to the ability of the pneumococcus to colonize and to mediate in vivo virulence. Analyses of genome sequences have revealed that S. pneumoniae lacks the genes required for de novo glutathione biosynthesis (Lanie et al., 2007 ▸). Instead, S. pneumoniae acquires GSH (∼10.9 ± 0.3 mM) from the extracellular environment via an ATP-binding cassette (ABC) transporter and the substrate-binding protein GshT (Potter et al., 2012 ▸; Begg et al., 2015 ▸). Isogenic deletion strains lacking either gshT or gor, which encodes GR, showed increased sensitivity to oxidative stress, albeit to differing extents (Potter et al., 2012 ▸). Further, the ΔgshT and Δgor strains were observed to be hypersensitive to intoxication by the divalent metal ions copper, zinc and cadmium (Potter et al., 2012 ▸; Begg et al., 2015 ▸).
Murine models of invasive pneumococcal infection have observed that infection triggers an innate immune response that includes increased levels of reactive oxygen and nitrogen species (ROS/RNS) and significant fluxes in the abundance of host metal ions such as zinc (Zn2+; McDevitt et al., 2011 ▸). This latter observation has been implicated in potentially exposing invading bacteria to zinc stress either directly or as a component of phagocytic cell killing (Botella et al., 2011 ▸; Martin et al., 2017 ▸; Eijkelkamp et al., 2014 ▸). Glutathione has been implicated in playing crucial roles in protection against these host-mediated stresses. ROS and RNS can be detoxified by GSH via direct interaction or by enzymatic processes, such as that catalyzed by glutathione peroxidase. Glutathione also forms metal complexes and thereby can contribute to the intracellular metal tolerance of ions such as zinc by forming a GSH–Zn2+ complex to moderate the abundance of the uncomplexed ion (Díaz-Cruz et al., 1998 ▸). These protective processes are dependent on cytoplasmic GSH, the concentration of which is dictated by S. pneumoniae GR.
The crystal structures of GR enzymes from Escherichia coli (Mittl & Schulz, 1994 ▸), Saccharomyces cerevisiae (Yu & Zhou, 2007 ▸), Plasmodium falciparum (Sarma et al., 2003 ▸; Böhme et al., 2000 ▸) and Homo sapiens (Schulz et al., 1978 ▸; Karplus & Schulz, 1989 ▸; Savvides & Karplus, 1996 ▸; Berkholz et al., 2008 ▸) have been described in conjunction with their kinetic properties (Mittl & Schulz, 1994 ▸; Savvides & Karplus, 1996 ▸; Yu & Zhou, 2007 ▸). All three enzymes are dimeric and bind one molecule of FAD per subunit. Here, we report the structural and kinetic characterization of the GR enzyme from S. pneumoniae (SpGR). Comparison of the SpGR structure with those from other sources reveals close similarities in the overall structures of these enzymes, with secondary-structural differences apparent at the dimer interface.
2. Materials and methods
2.1. Macromolecule production
Recombinant SpGR was generated by PCR-amplification of the SPD_0685 gene from S. pneumoniae serotype 2 strain D39 using ligation-independent cloning and the oligonucleotides GR_LIC1F (5′-TGGGTGGTGGATTTCCTAGAGAATATGATATCATTGCTATCGG) and GR_LIC1R (5′-TTGGAAGTATAAATTTCCACGCATGGTTACAAATTCTTC) to insert the gene into an N-terminal dodecahistidine tag-containing vector, pCAMnLIC01, to generate pCAMnLIC01-GR.
Heterologous expression of recombinant SpGR was performed in E. coli BL21(DE3) cells grown in lysogeny broth at 37°C. The expression of recombinant protein was induced by 0.5 mM IPTG at an optical density (OD) of 0.6. The cells were incubated at 25°C for 20 h and were then harvested by high-speed centrifugation. The cell pellet was suspended in buffer A (20 mM Tris pH 7.5, 150 mM NaCl) and disrupted using a TS series benchtop cell disruptor (Constant Systems). The His-tagged SpGR protein was purified using a two-step protocol consisting of immobilized metal-affinity chromatography (IMAC) followed by size-exclusion chromatography. The cell lysate was applied onto Ni-Sepharose 6 Fast Flow resin (GE Healthcare Life Sciences) equilibrated with buffer A and eluted with buffer A containing 300 mM imidazole. The eluted SpGR protein was incubated with 2 mM FAD for 2 h at 4°C before application onto a HiLoad 16/600 Superdex 200 size-exclusion column (GE Healthcare Life Sciences) pre-equilibrated with buffer A. The purified SpGR protein was concentrated to ∼20 mg ml−1 as determined by a BCA assay (ThermoFisher Scientific) and was stored at −80°C until further use. Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
| Source organism | S. pneumoniae |
| DNA source | S. pneumoniae serotype 2 strain D39 |
| Forward primer | TGGGTGGTGGATTTCCTAGAGAATATGATATCATTGCTATCGG |
| Reverse primer | TTGGAAGTATAAATTTCCACGCATGGTTACAAATTCTTC |
| Expression vector | pCAMnLIC01 |
| Expression host | E. coli strain BL21(DE3) |
| Complete amino-acid sequence of the construct produced | MREYDIIAIGGGSGGIATMNRAGEHGAQAAVIEEKKLGGTCVNVGCVPKKIMWYGAQIAETFHQFGEDYGFKTTDLNFDFATLRRNRESYIDRARSSYDGSFKRNGVDLIEGHAEFVDSHTVSVNGELIRAKHIVIATGAHPSIPNIPGAELGGSSDDVFAWEELPESVAILGAGYIAVELAGVLHTFGVKTDLFVRRDRPLRGFDSYIVEGLVKEMERTNLPLHTHKVPVKLEKTTDGITIHFEDGTSHTASQVIWATGRRPNVKGLQLEKAGVTLNERGFIQVDEYQNTVVEGIYALGDVTGEKELTPVAIKAGRTLSERLFNGKTTAKMDYSTIPTVVFSHPAIGTVGLTEEQAIKEYGQDQIKVYKSSFASMYSACTRNRQESRFKLITAGSEEKVVGLHGIGYGVDEMIQGFAVAIKMGATKADFDATVAIHPTSSEEFVTMR |
2.2. Crystallization
Initial crystallization trials were carried out with commercially available screens (Molecular Dimensions and Qiagen) by the sitting-drop vapour-diffusion method using equal volumes (0.2 µl) of protein sample (10–20 mg ml−1 in 20 mM Tris, 0.15 M NaCl pH 7.5) and reservoir solution dispensed using a Crystal Gryphon robot (Art Robbins Instruments). Initial crystals grew after four weeks in 0.2 M potassium thiocyanate, 0.1 M bis-Tris propane pH 7.5, 20%(w/v) PEG 3350. Optimization of this condition yielded crystals of approximate dimensions 50 × 150 × 150 µm that were grown by hanging-drop vapour diffusion at 20°C by mixing equal volumes (1 µl) of protein solution (20 mg ml−1 in 20 mM Tris, 0.15 M NaCl pH 7.5) and reservoir solution (0.1 M bis-Tris propane pH 7.0, 0.2 M potassium thiocyanate, 14% PEG 3350, 0.1 mM NADP+) and equilibrating the drops against 0.5 ml reservoir solution. A single SpGR crystal was transferred to cryoprotectant solution (0.1 M bis-Tris propane pH 7.0, 0.2 M potassium thiocyanate, 14% PEG 3350, 0.1 mM NADP+, 21% glucose) and flash-cooled in liquid nitrogen. Crystallization information is summarized in Table 2 ▸.
Table 2. Crystallization.
| Method | Hanging drop |
| Plate type | VDXm plates |
| Temperature (K) | 293 |
| Protein concentration (mg ml−1) | 16 |
| Buffer composition of protein solution | 20 mM Tris–HCl pH 7.5, 150 mM NaCl |
| Composition of reservoir solution | 0.2 M potassium thiocyanate, 0.1 M bis-Tris propane pH 7.0, 14%(w/v) PEG 3350, 0.1 mM NADP+ |
| Volume and ratio of drop | 2 µl, 1:1 |
| Volume of reservoir (µl) | 500 |
2.3. Data collection and processing
Diffraction data were recorded on an ADSC Quantum 315r detector at 100 K (in a nitrogen stream) on beamline MX2 at the Australian Synchrotron at a wavelength of 0.954 Å. Data were processed with XDS (Kabsch, 2010 ▸) and were merged and scaled with AIMLESS (Evans & Murshudov, 2013 ▸). Data-collection and processing statistics are summarized in Table 3 ▸
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | MX2 beamline, Australian Synchrotron (Aragão et al., 2018 ▸) |
| Wavelength (Å) | 0.954 |
| Temperature (K) | 100 |
| Detector | ADSC Quantum 315r |
| Crystal-to-detector distance (mm) | 400 |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 180 |
| Exposure time per image (s) | 0.5 |
| Space group | P21 |
| a, b, c (Å) | 96.75, 172.67, 135.43 |
| α, β, γ (°) | 90, 91.05, 90 |
| Mosaicity (°) | 0.21 |
| Resolution range (Å) | 50–2.56 (2.60–2.56) |
| Total No. of reflections | 531768 |
| No. of unique reflections | 141364 |
| Completeness (%) | 98.9 (84.1) |
| Multiplicity | 3.8 (3.4) |
| 〈I/σ(I)〉 | 8.7 (1.5) |
| R p.i.m. (%) | 6.2 (39.0) |
| CC1/2 | 0.994 (0.641) |
| Overall B factor from Wilson plot (Å2) | 39.3 |
2.4. Structure solution and refinement
The structure of SpGR was solved by molecular replacement with Phaser (McCoy et al., 2007 ▸). A search model was generated with CHAINSAW (Stein, 2008 ▸) from the structure of E. coli GR (EcGR; 61% sequence identity; PDB entry 1ger; Mittl & Schulz, 1994 ▸). Refinement was carried out using REFMAC5 (Murshudov et al., 2011 ▸) with iterative cycles of model building in Coot (Emsley & Cowtan, 2004 ▸). Tight noncrystallographic symmetry (NCS) restraints were applied during the early stages of refinement. These were relaxed and removed entirely as refinement progressed. Model geometry was analysed by MolProbity (Chen et al., 2010 ▸). Refinement statistics are summarized in Table 4 ▸.
Table 4. Structure solution and refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 50–2.56 (2.62–2.56) |
| Completeness (%) | 98.7 |
| σ Cutoff | None |
| No. of reflections, working set | 134126 (8684) |
| No. of reflections, test set | 7170 (434) |
| Final R cryst | 0.180 (0.326) |
| Final R free | 0.233 (0.338) |
| Cruickshank DPI | 0.229 |
| No. of non-H atoms | |
| Protein | 27516 |
| Ligand | 430 |
| Solvent | 1766 |
| Total | 29712 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Angles (°) | 1.079 |
| Average B factor (Å2) | 43.34 |
| Ramachandran plot | |
| Most favoured (%) | 96.8 |
| Allowed (%) | 99.7 |
| PDB code | 6du7 |
3. Results and discussion
3.1. Overall structure of SpGR
SpGR crystallized in space group P21 with eight molecules in the asymmetric unit (arranged as four homodimers; Fig. 2b). All residues (1–448) were modelled in chains A, B, C, D, F and G, while chains E and H included 447 and 446 residues, respectively. Refinement of the model to 2.56 Å resolution converged with residuals of R = 18.0% and R free = 23.3% with excellent statistics and model geometry. The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with accession code 6du7 (Table 4 ▸).
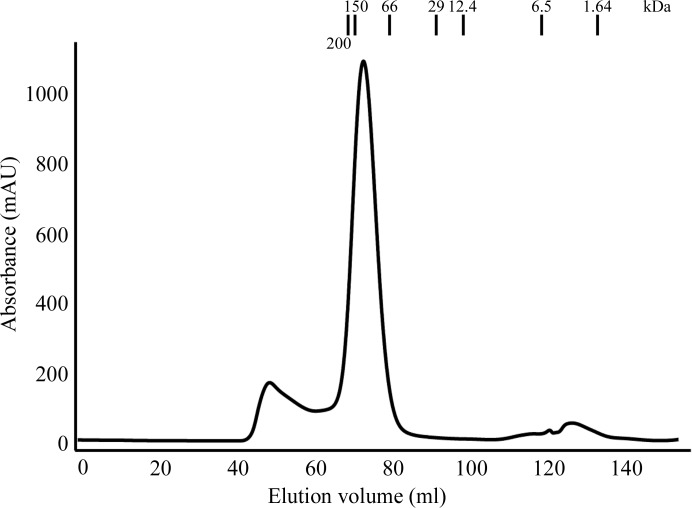
The observation of dimeric SpGR assemblies in the crystal is consistent with the analysis of the purified protein by size-exclusion chromatography (Fig. 1 ▸), which showed that SpGR eluted at an elution volume corresponding to a molecular weight of approximately 100 kDa. These data confirm that SpGR was also present as a dimer in solution (Fig. 1 ▸) and are consistent with the structural characterization of GR enzymes from E. coli (EcGR; Mittl & Schulz, 1994 ▸), H. sapiens (hGR; Schulz et al., 1978 ▸; Karplus & Schulz, 1989 ▸; Savvides & Karplus, 1996 ▸; Berkholz et al., 2008 ▸), P. falciparum (PfGR; Sarma et al., 2003 ▸; Böhme et al., 2000 ▸) and S. cerevisiae (yGR; Yu & Zhou, 2007 ▸). There are no significant differences between the backbone structures of the eight molecules in the asymmetric unit, as shown by r.m.s.d. values ranging from 0.2 to 0.4 Å for the superposition of 446 common Cα atoms between each combination of monomer pairs.
Figure 1.
Purification of SpGR. The elution profile of purified SpGR relative to the indicated protein strandards analysed by size-exclusion chromatography. The molecular weight of the SpGR protein is estimated to be 100 kDa, which corresponds to a homodimer.
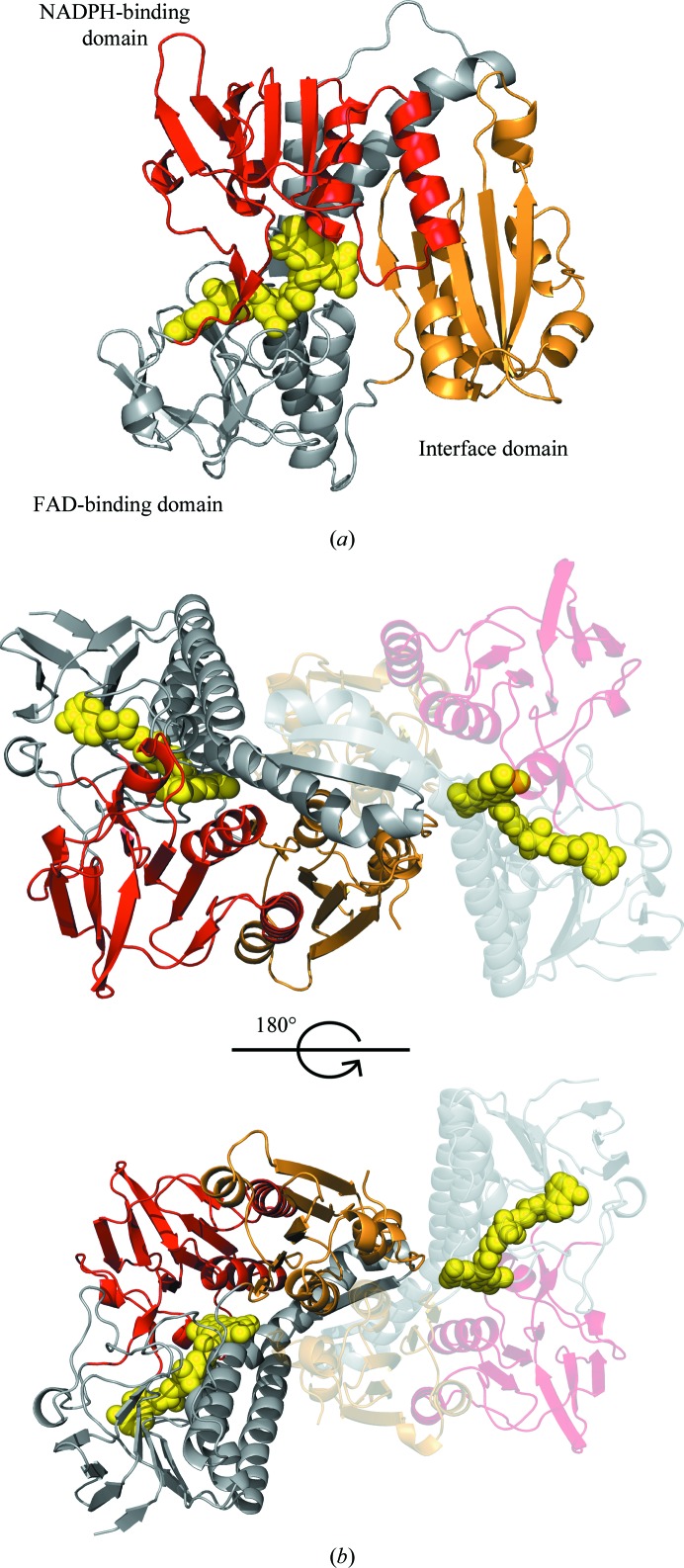
The SpGR monomer consists of three distinct domains: an NADPH-binding domain (residues 139–263), an FAD-binding domain (residues 1–138 and 264–334) and a dimerization domain (residues 335–448) (Fig. 2 ▸ a). The NADPH-binding and FAD-binding domains both show Rossmann folds, and the dimerization domain comprises five α-helices and five β-strands. GR enzymes contain a redox-active disulfide bond that is reduced by NADPH via FAD (Pai & Schulz, 1983 ▸), which is observed between residues Cys41 and Cys46 in the SpGR structure, and is positioned adjacent to the isoalloxazine ring of the FAD cofactor. The FAD cofactor is bound in an extended conformation within the FAD-binding domain.
Figure 2.
Structure of the glutathione reductase from S. pneumoniae. (a) The structure of the SpGR monomer is shown in cartoon representation with the FAD-binding domain in grey, the NADPH-binding domain in red and the interface domain in gold. The FAD cofactor is represented as yellow spheres. (b) Structure of the SpGR dimer: both monomers are coloured according to their domain structures as in (a).
The SpGR coordinates were used to perform a secondary-structure search of the PDB with PDBeFold (http://www.ebi.ac.uk/msd-srv/ssm; Krissinel & Henrick, 2004 ▸) and revealed close structural similarities between the SpGR structure and those of EcGR, hGR, yGR and PfGR, in addition to those of the GR enzymes from Vibrio parahaemolyticus (VpGR; PDB entry 5u1o), Yersinia pestis (YpGR; PDB entry 5vdn) and S. mutans (SmGR; PDB entry 5v36), all of which are unpublished (Table 5 ▸). In addition, the SpGR structure is closely similar to that of the glutathione amide reductase from Chromatium gracile (Table 5 ▸; Van Petegem et al., 2007 ▸).
Table 5. Structural comparisons between SpGR and homologues.
| Organism | PDB code | R.m.s.d. (Å) | N align | Sequence identity (%) |
|---|---|---|---|---|
| Vibrio parahaemolyticus | 5u1o | 0.74 | 444 | 59 |
| Streptococcus mutans | 5v36 | 0.95 | 441 | 60 |
| Escherichia coli | 1ger | 0.94 | 445 | 61 |
| Homo sapiens | 1gra | 1.23 | 444 | 51 |
| Yersinia pestis | 5vdn | 0.95 | 444 | 59 |
| Plasmodium falciparum | 1onf | 1.11 | 425 | 44 |
| Saccharomyces cerevisiae | 2hqm | 1.06 | 436 | 47 |
| Chromatium gracile † | 2r9z | 1.21 | 437 | 49 |
Glutathione amide reductase (Van Petegem et al., 2007 ▸).
3.2. Structure of the subunit interface
The subunit interface of SpGR shows a total buried surface area of 3502 Å2 per monomer, comprising ∼16% of the surface area of each protomer. Two main structural regions contribute to dimer formation: residues 46–93 from the FAD-binding domain (buried surface area of 1276 Å2) and residues 408–448 at the C-terminus of the dimerization domain (buried surface area of 1481 Å2).
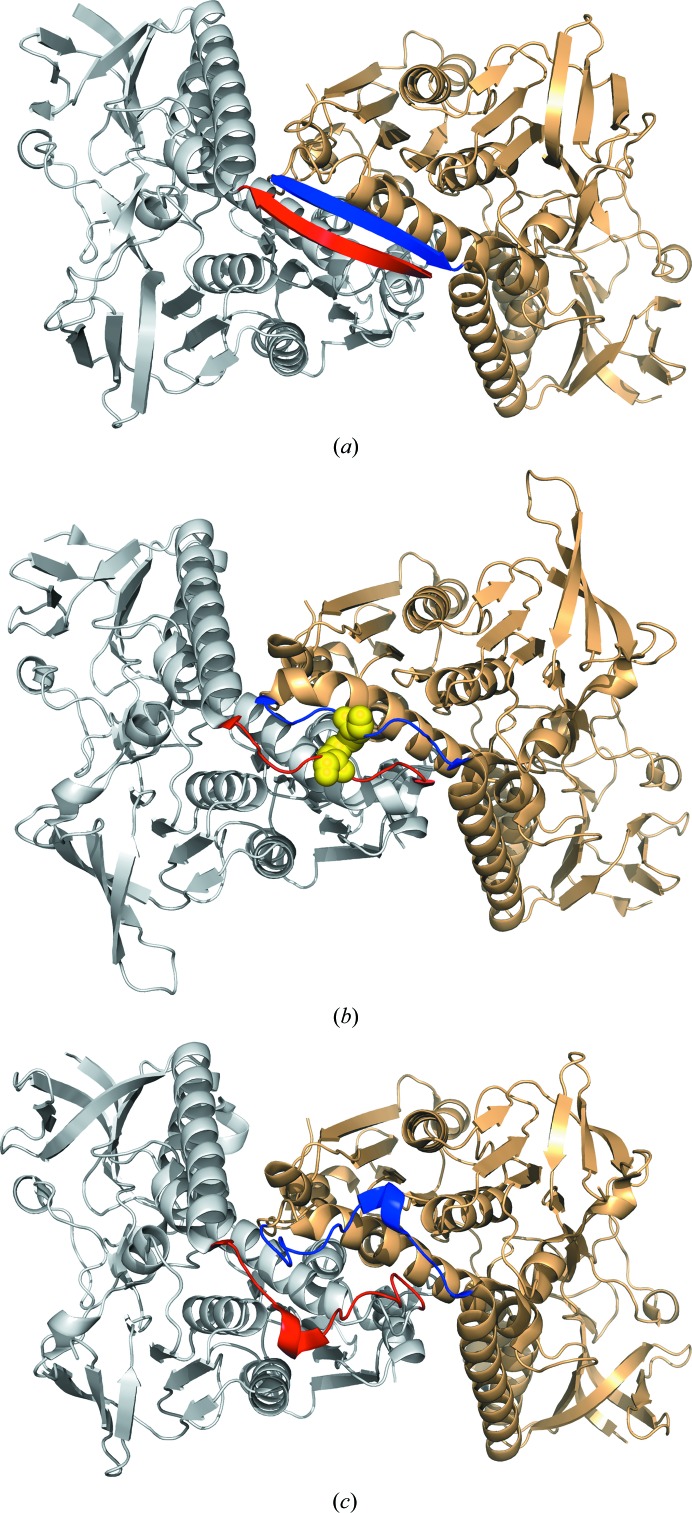
Residues 70–79 from each monomer align at the dimer interface in an antiparallel β-sheet structure (Fig. 3 ▸). This closely mirrors the structure of the dimerization interface of EcGR, but is distinct from those observed for yGR and hGR. In hGR an intersubunit disulfide bridge between residues Cys90 and Cys90′ is present in this position, while the yGR structure features a short α-helix connected by two loose loops owing to an insertion of six residues in this region of the structure (Fig. 3 ▸). The determination of the SpGR structure described here divides GR enzymes into three classes based on the structures of the dimer interfaces (Fig. 3 ▸). These are disulfide-linked (hGR), looped (yGR) and β-sheet (EcGR, SpGR, PfGR and other bacterial homologues). The bacterial enzymes therefore represent the major class (as defined by the structures of their dimer interfaces), all of which share the same structural features in this region (Fig. 3 ▸). Despite these structural differences, the composition of the GR dimer interface does not appear to determine the enzyme activity. Mutagenesis of the EcGR protein to introduce an intersubunit disulfide bond, similar to that observed in the hGR enzyme, did not have an impact on its catalytic activity or thermal stability (Scrutton et al., 1988 ▸).
Figure 3.
The structures of the dimer interfaces for GR enzymes (highlighted in red and blue). (a) SpGR (this work), (b) hGR (PDB entry 3dk4), (c) yGR (PDB entry 2hqm).
The catalytic mechanism and the location of the substrate-binding sites in hGR have been extensively investigated through X-ray crystallographic analyses of crystals soaked with NADPH and glutathione (Karplus & Schulz, 1989 ▸). A comparison of the SpGR structure with that of hGR shows that the dimer interface contributes to the structure of the GSSG binding pocket, which is composed of residues from both monomers. The binding pocket for NADPH is separated from the GSSG binding site by the Cys41–Cys46 disulfide linkage. Despite the fact that the SpGR crystallization conditions contained NADP+, we did not observe evidence for bound NADP+ in the electron density. The presence of the disulfide linkage in the SpGR structure described here confirms that this structure was solved in the oxidized (resting) state. Extensive analyses of the activities of GR enzymes from other sources have demonstrated the importance of this linkage in enzyme catalysis, where it mediates hydride transfer from NADPH to GSSG (Karplus & Schulz, 1989 ▸).
3.3. Kinetic analyses of the SpGR enzyme
Michaelis–Menten analyses of the glutathione reductase activity of SpGR for the substrates NADPH and GSSG revealed the kinetic parameters K m(NADPH) = 23.2 ± 3.3 µM, K m(GSSG) = 231.2 ± 24.7 µM and V max = 319.3 ± 15.9 µmol min−1 mg −1 (Table 6 ▸). The values of V max and K m(NADPH) for SpGR are consistent with those for the other characterized enzymes in this family; however, the determined value of K m(GSSG) for SpGR is significantly higher than those reported for the majority of kinetically characterized GRs (Table 6 ▸). The only exception is the GR enzyme from spinach, which has a comparable K m(GSSG) to that of SpGR.
Table 6. Kinetic parameters for GR enzymes from various organisms.
| Organism | V max (µmol min−1 mg−1) | K m(NADPH) (µM) | K m(GSSG) (µM) |
|---|---|---|---|
| Streptococcus pneumoniae † | 319 ± 16 | 23.2 ± 3.3 | 231 ± 25 |
| Homo sapiens ‡ | 147§ | 8.5 | 65 |
| Escherichia coli ¶ | 360.6§ | 16 | 66 |
| Saccharomyces cerevisiae †† | 166.7§ | 15 | 74.6 |
| Plasmodium falciparum ‡‡ | 204§ | 4.8 ± 0.4 | 69 ± 2 |
| Calf liver§§ | 190§ | 21.1 ± 1 | 101 ± 7 |
| Rat liver¶¶ | 207§ | 8.2 ± 0.8 | 26.3 ± 5.7 |
| Phaeodactylum tricornutum § | 190§ | 14 | 60 |
| Pea | 26§ | 3 | 62 |
| Spinach† | 246§ | 2.78 ± 0.34 | 196 ± 40 |
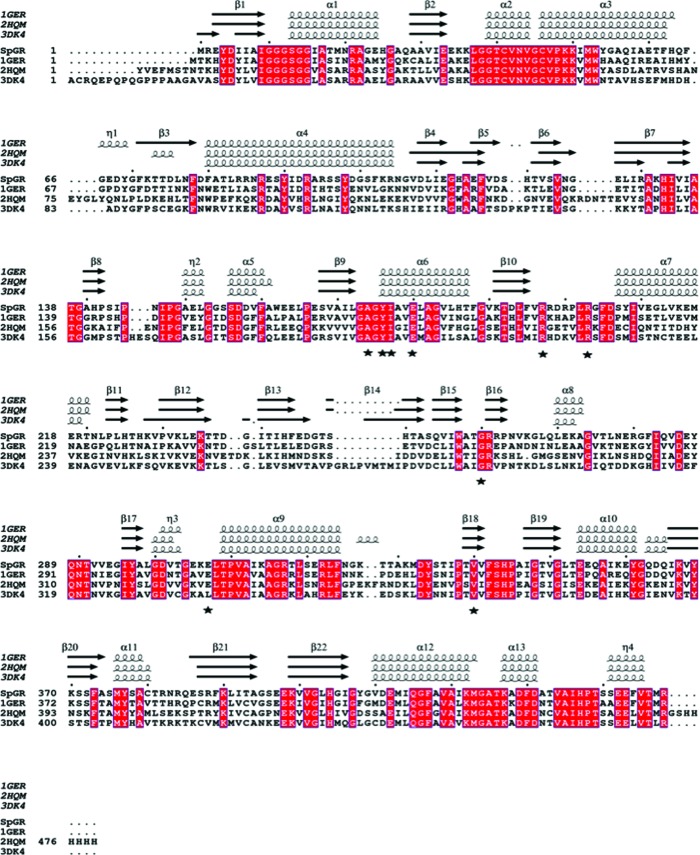
Although we were unable to determine the structure of the SpGR enzyme in the presence of the substrates NADPH and GSSG, we predicted the binding locations of these molecules by superposition with the structure of hGR, which has been solved in complex with NADP+ and GSSG (PDB entry 3dk4; Berkholz et al., 2008 ▸). We observed that the structures of the respective NADPH and GSSG binding sites are almost identical in the EcGR, hGR, yGR and SpGR enzymes. The hGR complex structure defined nine residues that directly interact with the substrate NADPH (Ala195, Tyr197, Ile198, Glu201, Arg218, Arg224, Gly290, Leu337 and Val370; Karplus & Schulz, 1989 ▸), eight of which are conserved in the sequences of SpGR, EcGR and yGR (Fig. 4 ▸). The single substitution is that of Leu337 in hGR for Glu in SpGR, EcGR and yGR. Based on the kinetic data [K m(NADPH) = 23.2 ± 3.3 µM], this substitution does not appear to affect the affinity of binding of NADPH to SpGR. In the structure of hGR in complex with NADP+ and GSSG, the interaction of this residue with the substrate NADPH is side-chain-independent, being mediated by a hydrogen bond between its amide nitrogen and NADPH. The close structural similarities between the GSSG binding sites in the SpGR, hGR, EcGR and yGR enzymes indicate no obvious features that would explain the differences in the kinetic parameters [specifically K m(GSSG)] for these enzymes.
Figure 4.
Sequence alignment of SpGR against the EcGR, hGR and yGR homologues. The alignment was performed using T-Coffee and ESPript (http://tcoffee.crg.cat/apps/tcoffee/do:expresso; Robert & Gouet, 2014 ▸). The secondary-structural elements were identified from PDB entries 1ger, 2hqm and 3dk4 using ESPript and are displayed at the top of the alignment. The α-helices and β-sheets are denoted α and β, respectively. Conserved residues are indicated by white lettering on a red background. Resides that form the NADPH-binding site are indicated with black stars.
4. Conclusion
The crystal structure of SpGR determined at 2.56 Å resolution shows high structural conservation with GR enzymes from human, E. coli (and other bacterial organisms) and S. cerevisiae. Despite the significant structural conservation, kinetic studies reveal a weaker affinity of the enzyme for its substrate, GSSG. Further investigation of the links between the structural and kinetic properties of SpGR must await its structure solution in the presence of bound substrates.
Supplementary Material
PDB reference: glutathione reductase from Streptococcus pneumoniae, 6du7
Acknowledgments
Aspects of this research were undertaken on the Macromolecular Crystallography beamline MX2 at the Australian Synchrotron, Victoria, Australia and we thank the beamline staff for their enthusiastic and professional support.
Funding Statement
This work was funded by National Health and Medical Research Council grant GNT1080784 to Megan Maher, , and David Aragão.
References
- Aragão, D., Aishima, J., Cherukuvada, H., Clarken, R., Clift, M., Cowieson, N. P., Ericsson, D. J., Gee, C. L., Macedo, S., Mudie, N., Panjikar, S., Price, J. R., Riboldi-Tunnicliffe, A., Rostan, R., Williamson, R. & Caradoc-Davies, T. T. (2018). J. Synchrotron Rad. 25, 885–891. [DOI] [PMC free article] [PubMed]
- Arias, D. G., Marquez, V. E., Beccaria, A. J., Guerrero, S. A. & Iglesias, A. A. (2010). Protist, 161, 91–101. [DOI] [PubMed]
- Begg, S. L., Eijkelkamp, B. A., Luo, Z., Couñago, R. M., Morey, J. R., Maher, M. J., Ong, C. L., McEwan, A. G., Kobe, B., O’Mara, M. L., Paton, J. C. & McDevitt, C. A. (2015). Nature Commun. 6, 6418. [DOI] [PMC free article] [PubMed]
- Berkholz, D. S., Faber, H. R., Savvides, S. N. & Karplus, P. A. (2008). J. Mol. Biol. 382, 371–384. [DOI] [PMC free article] [PubMed]
- Böhme, C. C., Arscott, L. D., Becker, K., Schirmer, R. H. & Williams, C. H. Jr (2000). J. Biol. Chem. 275, 37317–37323. [DOI] [PubMed]
- Botella, H., Peyron, P., Levillain, F., Poincloux, R., Poquet, Y., Brandli, I., Wang, C., Tailleux, L., Tilleul, S., Charrière, G. M., Waddell, S. J., Foti, M., Lugo-Villarino, G., Gao, Q., Maridonneau-Parini, I., Butcher, P. D., Castagnoli, P. R., Gicquel, B., de Chastellier, C. & Neyrolles, O. (2011). Cell Host Microbe, 10, 248–259. [DOI] [PMC free article] [PubMed]
- Carlberg, I., Altmejd, B. & Mannervik, B. (1981). Biochim. Biophys. Acta, 677, 146–152. [DOI] [PubMed]
- Carlberg, I. & Mannervik, B. (1981). Anal. Biochem. 116, 531–536. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Connell, J. P. & Mullet, J. E. (1986). Plant Physiol. 82, 351–356. [DOI] [PMC free article] [PubMed]
- Díaz-Cruz, M. S., Mendieta, J., Monjonell, A., Tauler, R. & Esteban, M. (1998). J. Inorg. Biochem. 70, 91–98.
- Eijkelkamp, B. A., Morey, J. R., Ween, M. P., Ong, C. L., McEwan, A. G., Paton, J. C. & McDevitt, C. A. (2014). PLoS One, 9, e89427. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Halliwell, B. & Foyer, C. H. (1978). Planta, 139, 9–17. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Karplus, P. A. & Schulz, G. E. (1989). J. Mol. Biol. 210, 163–180. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Lanie, J. A., Ng, W.-L., Kazmierczak, K. M., Andrzejewski, T. M., Davidsen, T. M., Wayne, K. J., Tettelin, H., Glass, J. I. & Winkler, M. E. (2007). J. Bacteriol. 189, 38–51. [DOI] [PMC free article] [PubMed]
- Martin, J. E., Edmonds, K. A., Bruce, K. E., Campanello, G. C., Eijkelkamp, B. A., Brazel, E. B., McDevitt, C. A., Winkler, M. E. & Giedroc, D. P. (2017). Mol. Microbiol. 104, 636–651. [DOI] [PMC free article] [PubMed]
- Masip, L., Veeravalli, K. & Georgiou, G. (2006). Antioxid. Redox Signal. 8, 753–762. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McDevitt, C. A., Ogunniyi, A. D., Valkov, E., Lawrence, M. C., Kobe, B., McEwan, A. G. & Paton, J. C. (2011). PLoS Pathog. 7, e1002357. [DOI] [PMC free article] [PubMed]
- Mittl, P. R. & Schulz, G. E. (1994). Protein Sci. 3, 799–809. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Pai, E. F. & Schulz, G. E. (1983). J. Biol. Chem. 258, 1752–1757. [PubMed]
- Potter, A. J., Trappetti, C. & Paton, J. C. (2012). J. Bacteriol. 194, 6248–6254. [DOI] [PMC free article] [PubMed]
- Ritz, D. & Beckwith, J. (2001). Annu. Rev. Microbiol. 55, 21–48. [DOI] [PubMed]
- Robert, X. & Gouet, P. (2014). Nucleic Acids Res. 42, W320–W324. [DOI] [PMC free article] [PubMed]
- Rudan, I., Boschi-Pinto, C., Biloglav, Z., Mulholland, K. & Campbell, H. (2008). Bull. World Health Organ. 86, 408–416. [DOI] [PMC free article] [PubMed]
- Rudan, I., O’Brien, K. L., Nair, H., Liu, L., Theodoratou, E., Qazi, S., Lukšić, I., Fischer Walker, C. L., Black, R. E. & Campbell, H. (2013). J. Glob. Health, 3, 010401. [DOI] [PMC free article] [PubMed]
- Sarma, G. N., Savvides, S. N., Becker, K., Schirmer, M., Schirmer, R. H. & Karplus, P. A. (2003). J. Mol. Biol. 328, 893–907. [DOI] [PubMed]
- Savvides, S. N. & Karplus, P. A. (1996). J. Biol. Chem. 271, 8101–8107. [DOI] [PubMed]
- Schulz, G. E., Schirmer, R. H., Sachsenheimer, W. & Pai, E. F. (1978). Nature (London), 273, 120–124. [DOI] [PubMed]
- Scrutton, N. S., Berry, A. & Perham, R. N. (1988). FEBS Lett. 241, 46–50. [DOI] [PubMed]
- Stein, N. (2008). J. Appl. Cryst. 41, 641–643.
- Storey, B. T., Alvarez, J. G. & Thompson, K. A. (1998). Mol. Reprod. Dev. 49, 400–407. [DOI] [PubMed]
- Van Petegem, F., De Vos, D., Savvides, S., Vergauwen, B. & Van Beeumen, J. (2007). J. Mol. Biol. 374, 883–889. [DOI] [PubMed]
- Yu, J. & Zhou, C.-Z. (2007). Proteins, 68, 972–979. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: glutathione reductase from Streptococcus pneumoniae, 6du7