Transformational advances in transmission electron microscopy at cryogenic temperature (cryo-EM) is enabling atomic-level insight into the three-dimensional structure and therefore the function of biological macromolecules in solution. Contemporary cryo-EM promises to resolve problems in structural biology that were intractable just a few years ago.
Keywords: cryo-EM, single-particle analysis, three-dimensional reconstruction, phase plates, direct detectors
Abstract
Structural biology is going through a revolution as a result of transformational advances in the field of cryo-electron microscopy (cryo-EM) driven by the development of direct electron detectors and ultrastable electron microscopes. High-resolution cryo-EM images of isolated biomolecules (single particles) suspended in a thin layer of vitrified buffer are subjected to powerful image-processing algorithms, enabling near-atomic resolution structures to be determined in unprecedented numbers. Prior to these advances, electron crystallography of two-dimensional crystals and helical assemblies of proteins had established the feasibility of atomic resolution structure determination using cryo-EM. Atomic resolution single-particle analysis, without the need for crystals, now promises to resolve problems in structural biology that were intractable just a few years ago.
1. Transmission electron microscopy: a pillar of modern structural biology
The fabrication of a transmission electron microscope by Knoll and Ruska in 1931 made it possible to visualize the morphology of an individual cell of a living organism for the first time. In the decades that followed, the discipline of transmission electron microscopy (TEM) made profound contributions to the development of cellular and molecular biology. For example, the first electron micrograph of a virus particle, a bacteriophage, was recorded in 1940 (Ruska, 1940 ▸; Peankuch & Kausche, 1940 ▸); that of an intact chemically fixed cell was produced by Porter et al. (1945 ▸), revealing the mitochondria, the Golgi apparatus and the endoplasmic reticulum; the first image of an animal virus particle, the RNA-containing retrovirus Rous sarcoma virus, was recorded by Claude et al. (1947 ▸); and an image of DNA-containing Herpes simplex virus 1 (HSV 1) was presented by Morgan et al. (1954 ▸). Subsequently, the development of negative staining, a technique in which the specimen is embedded in heavy-metal ions (see, for example, the early work by Hall, 1955 ▸), helped to resolve the morphology and architecture of a variety of biological specimens. This was followed a few years later by the pioneering studies of De Rosier & Klug (1968 ▸), in which they generated a three-dimensional reconstruction of the T4 bacteriophage tail using images of negatively stained specimens by exploiting the helical symmetry in the arrangement of subunits within the tail. This work ushered in the dawn of three-dimensional structural analysis of biological assemblies, which is now capable of revealing near-atomic resolution details.
In the past five years, a transformational development in TEM at cryogenic temperatures (cryo-EM) has revealed near-atomic resolution images of biological molecules and macromolecular complexes that were previously inaccessible. This advance has materialized through a combination of ultrastable electron microscopes, highly sensitive electron detectors and powerful image-processing algorithms. Traditionally, X-ray crystallography has been the method of choice for revealing atomic resolution structures of biomolecules. However, this method requires the generation of well ordered three-dimensional crystals, for which sizeable amounts of a pure, stable and homogeneous sample are necessary. Polymers, multiprotein complexes and membrane proteins have been difficult to study using X-ray crystallography because of difficulties in sample preparation and crystallization. Solution and solid-state NMR spectroscopy are also used for three-dimensional structure determination, but these methods are constrained by fundamental limitations imposed by protein size. On the other hand, single-particle cryo-EM, without the need for crystals, has become a powerful procedure for potentially imaging biological molecules at near-atomic resolution, as shown by the explosion in the number of such structures solved in the last few years (Fig. 1 ▸).
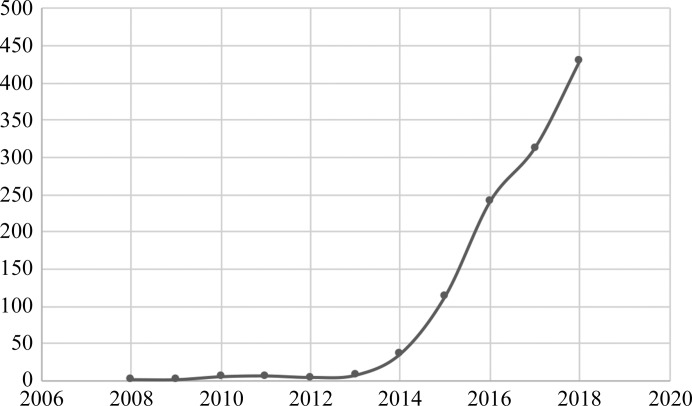
Figure 1.
The explosive growth in the number of structures solved by cryo-EM in the last ten years with a resolution of 4 Å or better as accessed from https://www.ebi.ac.uk/pdbe/emdb/statistics_num_res.html/.
2. The pathway to modern cryo-EM
The development of procedures for high-resolution imaging by TEM required that a number of roadblocks be overcome. These included the fact that electrons interact strongly with matter, so that the biological specimen needs to be imaged under high vacuum, requiring adequate preservation. The specimen also needs to be protected from radiation damage produced by the high-energy electrons, so that the process of imaging does not destroy the sample. It follows that images must be recorded with a low dose of electrons, exacerbating the signal-to-noise problem inherent to biological specimens composed primarily of light atoms. Other important issues include the need for a thin specimen so that the specimen can be treated as a phase object implicit in the three-dimensional reconstruction process and in which multiple scattering of electrons is minimized. Loss of high-resolution information owing to specimen movement during exposure is another critical factor that was firmly established soon after the new generation of electron detectors was commissioned and needed crucial consideration.
In early TEM studies that used chemically fixed, isolated organelles and tissues, specimen preparation preserved only ultrastructural details and could reveal only gross morphologies. Introduced around the 1950s, the embedding of biomolecules in heavy-metal salts (stains) such as uranyl acetate simultaneously preserves the specimen and enhances the contrast in the images, albeit reversed owing to the fact that the heavy-atom stain scatters electrons much more strongly than the protein. The stain mostly accumulates in solvent-accessible regions and thereby the image only yields information on the overall shape of the embedded biomolecule at modest resolution, typically ∼20 Å. Additionally, artifacts owing to non-uniform drying of the stain or collapse of the embedded specimen can severely compromise the information generated. Notwithstanding these issues, negative staining remains a popular technique to rapidly screen samples owing to the ease of specimen preparation (Ohi et al., 2004 ▸) and is frequently used as part of the workflow in further cryo-EM work, for instance to generate initial three-dimensional reference models. In many instances these can display structural details at the protein-domain level.
In the 1970s remarkable breakthroughs in specimen preparation were made using unstained two-dimensional, one-molecule-thick protein crystals. These studies established that protein structure determination at high resolution was possible using TEM. In the mid-1970s Taylor and Glaeser examined catalase crystals flash-cooled in liquid nitrogen and recorded low-dose diffraction patterns that extended to ∼3 Å resolution using specimen holders operated at −120°C (Taylor & Glaeser, 1974 ▸, 1976 ▸). These experiments directly demonstrated structural preservation, and allowed data acquisition to hitherto unprecedented resolutions. Around the same time, Unwin and Henderson, working at room temperature, used glucose embedding to preserve naturally occurring two-dimensional crystals of the integral membrane protein bacteriorhodopsin from the purple membrane of the archaebacterium Halobacterium salinarum (Unwin & Henderson, 1975 ▸; Henderson & Unwin, 1975 ▸). Formally, the specimen preparation is analogous to that used to prepare heavy-metal-stained samples. The crystals, thus preserved, are susceptible to radiation damage, but by using low-dose recording techniques electron diffraction patterns to ∼3.5 Å resolution could again be recorded. This result attested to the success of this preparation method at room temperature in maintaining a high degree of order similar to that observed in the frozen hydrated preparations of Taylor and Glaeser. Combining amplitudes measured from the diffraction patterns and phases from the computed Fourier transform of the recorded images, Unwin and Henderson produced an ∼7 Å resolution density map of bacteriorhodopsin in projection, looking down the membrane plane (Unwin & Henderson, 1975 ▸). The projection map suggested the presence of transmembrane helices running perpendicular to the lipid bilayer. Later, a detailed three-dimensional density map of the protein was generated, the first of its kind using TEM, by including electron crystallographic data from tilted specimens (Henderson & Unwin, 1975 ▸). For this purpose, tilting the thin (one-subunit-thick) two-dimensional crystal generated the multiple views of the protein necessary for three-dimensional reconstruction, the mathematical treatment having been laid down earlier by De Rosier & Klug (1968 ▸) in the reconstruction procedure they applied to images of the T4 bacteriophage tail helix. The map produced by Unwin and Henderson, at ∼9 Å resolution, was sufficient to delineate the characteristic seven transmembrane helices of bacteriorhodopsin, an architecture that is common to all members of the GPCR family, which includes the closely related mammalian rhodopsin. Subsequently, electron crystallographic data from two-dimensional crystals of bacteriorhodpsin and a number of other proteins at cryogenic temperatures preserved in sugars reached resolutions that were sufficient to allow de novo building of three-dimensional atomic models of the polypeptide chain (Henderson et al., 1990 ▸; Kühlbrandt et al., 1994 ▸; Nogales et al., 1998 ▸; Murata et al., 2000 ▸). The growth of this technique was hindered by the fact that the production of highly ordered two-dimensional crystals is not straightforward and that there are other technical issues that compromise high-resolution data collection. These are mainly the physical difficulty of tilting the specimen to high angles and the difficulty of keeping the crystals atomically flat, both of which are factors that lead to anisotropic resolution loss. Notwithstanding, until a few years ago electron crystallography of two-dimensional protein crystals examined at cryogenic temperatures remained one of the few methods for very high-resolution protein structure determination using TEM.
In the early 1980s, the pioneering work of Dubochet and coworkers ushered in the age of modern transmission electron microscopy (cryo-EM) by developing a method to encase biological specimens in a thin layer of vitrified, amorphous buffer. In 1980, Brueggeller and Mayer first described a procedure to achieve the vitrification of water or aqueous solution droplets by using a cryogen (Brüggeller & Mayer, 1980 ▸). However, it was Dubochet and coworkers who established the simpler procedure for vitrification appropriate for biological specimens that was rapidly adopted as the method of choice for examining unstained biomolecules under near-native conditions without any cryopreservation (Dubochet & McDowall, 1981 ▸). The core of this method involves spreading a thin film of water on a support that is very quickly immersed in liquid ethane (Dubochet et al., 1988 ▸). The small size of the biomolecule in aqueous suspension and the speed of cooling act as critical factors facilitating the vitrification process. In the commonly followed setup, a thin film of buffer containing the sample is created on an EM grid covered with a holey (fenestrated) carbon film by blotting the grid with a filter paper. The grid is then rapidly plunge-frozen in liquid ethane by a semi-automatic guillotine device, trapping the sample in vitrified buffer.
The susceptibility of biological samples to radiation damage necessitates the recording of only low-dose images that have a low signal-to-noise ratio (SNR). A true in-focus, cryo-EM image of a biological specimen has almost negligible contrast. Traditionally, to achieve recordable signal, the contrast is enhanced by deliberate defocusing, producing the so-called phase contrast. This feature is associated with the phase-contrast transfer function of the microscope describing sinusoidal modulation of the phase shifts of the scattered waves arising from the applied defocus and spherical aberration. This leads to variation in the intensity upon interference of the scattered wave with the unscattered wave on the image plane and hence to contrast in the image. Over several decades, by following Dubochet’s vitrification procedure, a steady improvement in the attained resolutions of three-dimensional reconstructions, approaching values of nanometres or better, was achieved, especially for those of symmetric objects (see, for example, Tao & Zhang, 2000 ▸). The demonstration that unstained, frozen hydrated biological samples preserved in vitreous ice can indeed be imaged at near-atomic resolution was accomplished first for protein–lipid two-dimensional crystals of the integral membrane protein human aquaporin 1 water channel (AQP1; 3.7 Å resolution; Ren et al., 2001 ▸) and for helical crystals of the nicotinic acetylcholine receptor (nAChR) channel (4.0 Å resolution; Miyazawa et al., 2003 ▸). These analyses generated density maps which could be fitted by de novo tracing of the polypeptide chains. Topologically, the helical crystals can be thought of as a two-dimensional crystal wrapped onto a cylindrical surface, and the diffraction can be analyzed by a Fourier–Bessel decomposition. From the perspective of data acquisition, helical specimens present an easier case than two-dimensional crystals, as tilting is not required to generate many different views of the repeating unit. Specimens with high-order point-group symmetry, such as icosahedral viruses, also facilitate structural analysis, and near-atomic resolution density maps of a number of spherical viruses were generated using vitrified specimens (Grigorieff & Harrison, 2011 ▸).
3. Single-particle analysis reaching near-atomic resolution
In general, most biomolecules are asymmetric and many do not readily crystallize. In order to achieve three-dimensional reconstruction for such systems, the ‘single-particle analysis’ (SPA) procedure was developed following the pioneering work of Frank and coworkers (Frank, 1975 ▸). In this procedure, three-dimensional reconstructions are generated by combining information from two-dimensional images of isolated, identical copies of a biomolecule in different orientation, yielding independent projected views of the imaged object. Typically, many thousands of images are combined to generate a three-dimensional reconstruction. Frank and coworkers investigated the huge asymmetric ribosomal complex of Escherichia coli (50S and 30S subunits) at progressively higher resolution, reaching a three-dimensional reconstruction at ∼6.7 Å resolution (Villa et al., 2009 ▸). En route to their seminal single-particle work, Frank and coworkers created the SPIDER program package (Frank et al., 1981 ▸) that preceded many of the currently popular software packages, for example EMAN (Tang et al., 2007 ▸), IMAGIC (van Heel et al., 1996 ▸), Bsoft (Heymann & Belnap, 2007 ▸), RELION (Scheres, 2012 ▸, 2016 ▸) and Frealign (Grigorieff, 2007 ▸), that are now being used in the field of SPA. Historically, prior to SPIDER, a few other digital image-processing software packages were available, which included, for example, SEMPER (Saxton et al., 1979 ▸), MDPP (Smith, 1978 ▸) etc. The endeavors of Frank and coworkers were foundational, as they established a versatile image-processing technique that has become synonymous with cryo-EM. In the past few years, courtesy of the new generation of direct detectors, powerful ultrastable microscopes and sophisticated advances in image processing, biomolecular structures derived using SPA are reaching near-atomic resolution in ever-increasing numbers. This has led to what is now being popularly termed the ‘resolution revolution’ (Kühlbrandt, 2014 ▸).
The first step in SPA is the manual or automated selection of images of isolated particles in a variety of orientations suspended in a thin layer of amorphous ice optimized for the given particle size and buffer conditions. Typically, phase contrast, introduced by deliberately defocusing the objective lens, is used to enhance the contrast of the images. This process in turn leads to modulation of the spatial frequencies by the so-called contrast transfer function (CTF; Erickson & Klug, 1971 ▸). After translational and rotational alignment of the selected images, correcting for the effect of the contrast transfer function (for example phase flipping of spatial frequencies), groups of images that correspond to similar views are averaged to generate higher signal-to-noise ratio (SNR) two-dimensional class averages. These are iteratively refined to yield final classes that initiate the three-dimensional reconstruction process. Many of the current reconstruction software packages carry out this process by invoking the fundamental central projection theorem. This states that a Fourier transform of each two-dimensional projection is a central slice of the three-dimensional Fourier transform of the object under study. Next, and most importantly, the relative orientations of a sufficiently large set of particle images that describe a wide range of views are determined by one of several methods. The two-dimensional Fourier transforms of each pair of projection images share a common line of intersection in the three-dimensional Fourier transform, which can be used to assign the relative orientation by real-space (van Heel, 1987 ▸) or reciprocal-space (Crowther, 1971 ▸) methods. Next, the associated central section slices are ‘positioned’ according to the determined relative orientation and combined to build up the three-dimensional Fourier transform that leads to a three-dimensional reconstruction. De novo reference models are experimentally generated, using methods such as random conical tilt (RCT) reconstruction (Radermacher et al., 1987 ▸), wherein information from pairs of images in groups of randomly oriented particles recorded at a known high tilt and then at no tilt are combined. Given that in such a protocol the geometry of image recording defines the orientation parameters, a reliable model, albeit generally at a low resolution, can be generated. This can serve as an initial reference model and traditionally has produced validated final structures. Indeed, random conical tilt is the method of choice in cases where the particles display only preferred orientation. In addition, computer-generated initial reference models approximating the correct structure of the object under examination are also used. In general, one needs to be very careful in choosing the reference model because the issue of bias introduced into the analysis is paramount (Henderson, 2013 ▸).
This initial reference map can be refined by the so-called ‘projection-matching’ procedure, for which the procedure derived by Frank and coworkers (Penczek et al., 1994 ▸) is commonly used. In such a process, two-dimensional reprojections of the three-dimensional density map generated by uniformly sampling the orientation space are compared with individual raw particle images by, for example, cross-correlation. An iterative process can then refine the orientation assignments for the various particle images, leading to an improvement in the three-dimensional model and the resolution achieved. In this context, the algorithm of the maximum-likelihood (Sigworth, 1998 ▸) orientation assignment has been a very notable advance, wherein instead of a single orientation assignment, a probabilistic distribution of orientations for each image dependent on the level of noise is applied. Apart from usage in the XMIPP (Sorzano et al., 2004 ▸) and FREALIGN (Grigorieff, 2007 ▸) reconstruction software, this protocol underpins the reconstruction algorithm used in RELION, which currently appears to be the most widely used software and has been shown to lead to robust solutions.
Any reconstruction process by SPA proceeds, by definition, under the assumption that the imaged sample is a homogeneous population of chemically and structurally identical objects. However, many ‘purified’ biological samples are actually heterogeneous, for example owing to the simultaneous presence of multiple conformations, the variable nature or stoichiometry of a bound ligand, or variability in the size and/or composition in the case of a protein complex. In such a case, the achieved final resolution will be compromised unless the images are separated into homogeneous sets. If the projection images can be visually separated into distinct groups, then the RCT approach, in combination with the corresponding tilt images, can be applied to generate multiple three-dimensional initial models. These can then be used to generate refined three-dimensional models for the groups of coexisting populations. The automated maximum-likelihood algorithm, such as that used in RELION, lends itself naturally to tackling this three-dimensional classification problem and has proven to be extremely useful in generating multiple structures that speak to conformational and structural dynamics of the molecular assembly (see, for example, Chen et al., 2016 ▸). The attained resolution of a macromolecular complex can often be compromised owing, for example, to localized flexibility, symmetry mismatch, variable stoichiometry in subunit binding etc. In such cases, localized reconstruction has proven to be effective (Ilca et al., 2015 ▸; Amunts et al., 2015 ▸), wherein the localized areas are masked out and extracted, and treated as single particles.
4. Microscope and detector technology enabling rapid progress in three-dimensional resolution of cryo-EM structures
The progress in high-resolution SPA accelerated significantly in the early 2010s with the advent of 300 kV high-throughput ultrastable FEI TEMs such as Polara and additionally with highly coherent parallel illumination, as applied in the design of the FEI Titan Krios. The use of charge-coupled device (CCD) cameras greatly facilitated image processing owing to direct digital image recording, which is especially necessary for large-volume, automatic data collection; however, their performance is poorer than electron-sensitive film at high resolution. Not surprisingly, using these new-generation microscopes, the majority of sub-4 Å resolution structures were initially determined using film (see, for example, Grigorieff & Harrison, 2011 ▸). These included the 3.3 Å resolution structure of icosahedral Aquareovirus by Zhou and coworkers (Zhang et al., 2010 ▸), which is the highest resolution three-dimensional structure determined using analog detector technology, allowing facile de novo fitting of atomic models.
At this point, it was recognized that a significant impediment to achieving high resolution was the fact that the recorded signals in cryo-EM images were at best about 10% of their true values (Henderson & Glaeser, 1985 ▸). This limited the sizes of the biomolecules that could be studied. Amongst the many contributing factors, the sensitivity (detection quantum efficiency; DQE) of a digital recording device is a key factor, and the discovery of devices with high DQE remained a key focus. It was the development and commercial availability of direct electron detectors (DEDs) that engendered a transformational advance in cryo-EM imaging. In a CCD, the electron-detection process via a scintillator is indirect, leading to a marked diminution of the high-resolution signal. Direct electron detectors, developed on the basis of seminal contributions by a number of groups, for example Deptuch et al. (2007 ▸) and Faruqi & Henderson (2007 ▸), involve reduced radiation-sensitive monolithic active pixel sensors using complementary metal-oxide semiconductor technology (CMOS/MAP), which allows direct quantitation of impinging electrons with high DQE and very little noise.
Using a DED recorder, Grigorieff and coworkers (Brilot et al., 2012 ▸) made a fundamental observation that revealed the primary reason for the loss of contrast in traditional low-dose, single-exposure imaging as follows. DED cameras can be operated with a rolling shutter owing to a very rapid readout mechanism, so that a single 1 s exposure can be subdivided into multiple frames, typically 20–40 frames, each at a user-decided dose (for example ∼1–2 e Å−2) in a so-called movie-mode acquisition. Analysis of these rapidly acquired individual images revealed beam-induced movement of the particles between frames, leading to blurring in the traditional composite or summed image. This blurring can be minimized by computational alignment of the individual frames, leading to a summed image that rescues high-resolution information and has an enhanced SNR. Not surprisingly, this led to a pronounced improvement in resolution. The first report of the determination of a three-dimensional structure by cryo-EM was by Milazzo et al. (2011 ▸), who used a DE12 direct detector (Direct Electron, San Diego, USA). Since then, direct detectors have replaced film for essentially all high-resolution image analysis and there has been an avalanche of near-atomic resolution structures of biomolecules determined using SPA. These reconstruction exercises have also been facilitated (i) by the development of powerful image-processing software, as discussed above, (ii) by automating the process of image recording, for example Leginon (Carragher et al., 2000 ▸) and SerialEM (Mastronarde, 2005 ▸), enabling high-throughput data collection, and (iii) by ancillary improvements in the microscope stage and grid handling, allowing frozen grids to be examined contamination-free for several days. These factors have led to three-dimensional structure determinations of progressively smaller molecules using fewer images, essentially fulfilling the predictions made by Glaeser and Henderson in the last millennium (Henderson, 1995 ▸; Glaeser, 1999 ▸). In this context, the seminal paper by Scheres and coworkers entitled Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles (Bai et al., 2013 ▸), represented the proverbial coming of age of cryo-EM in the atomic resolution domain, which until then was solely occupied by X-ray crystallography. A continuous stream of publications has followed, some reporting structures at an unprecedented resolution of near 2 Å, for example Bartesaghi et al. (2015 ▸) and Banerjee et al. (2016 ▸). These results have led cryo-EM to also be hailed as a bona fide technique for application to drug discovery (Banerjee et al., 2016 ▸; Merk et al., 2016 ▸)
As in the case of X-ray crystallography, owing to the ease of biochemical handling, soluble proteins and macromolecular complexes have been the main focus of recent structural study by cryo-EM. However, as we have noted, it was a membrane protein, bacteriorhodpsin, that ushered in the high-resolution investigation of the three-dimensional structure of biomolecules. Thus, it was just a question of time before near-atomic resolution single-particle analysis of noncrystalline membrane-protein preparations was achieved, as for example in the pioneering studies on detergent-solubilized TRPV (Liao et al., 2013 ▸), a magnesium transporter (Matthies et al., 2016 ▸) and a class B GPCR (Liang et al., 2017 ▸). Such achievements are particularly noteworthy since ∼70% of clinically important drug targets are membrane proteins that are usually refractory to the overexpression of folded proteins and/or are difficult to crystallize. Therefore, cryo-EM holds a distinct advantage in combination with VPP technology (see below) in the field of membrane proteins.
5. New advances in cryo-EM
5.1. Phase-plate technology
In traditionally recorded phase-contrast images, details of high-resolution information are attenuated together with non-uniform enhancement of low-resolution features owing to the effect of the contrast-transfer function. For many years, defocus-independent modification of the phase shift between unscattered and scattered waves has been explored as a means to enhance contrast and overcome the limitations imposed by defocus phase contrast. For this purpose, the most popular systems studied have been phase plates, which are designed to introduce a fixed phase shift between the central unscattered and scattered beams. Typically, these are thin films placed at the back focal plane of the objective lens. The two most actively studied phase-plate arrangements have been the Zernike phase plate (ZPP; Danev et al., 2009 ▸) and the Volta phase plate (VPP; Danev et al., 2014 ▸). The ZPP consists of a thin (∼25 µm) amorphous carbon film with a thickness attuned to the accelerating voltage to generate a phase shift of π/2 and containing a small (∼1 µm) hole at the center to allow a phase-unmodified central beam to pass through. The effectiveness of the ZPP is compromised because of the low usage time per plate owing to contamination, the difficulty associated with precise centering of the hole for the unscattered beam and fringe artifacts at the edge of the hole. The VPP is considerably simpler, just being a thin (∼10 µm) carbon film typically kept at an elevated temperature of ∼200°C. The carbon film, after being aged (pre-irradiated) for a few days under illumination, creates a phase advance for the unscattered beam that reaches saturation. The genesis of the phase shift is postulated to be owing to a negative Volta potential generated between the surface and the interior of the carbon film upon electron illumination. Compared with the ZPP, the prominent advantage of the VPP is that for a continuous carbon film there is no need for the exact centering of the hole, which makes automatic data collection possible, an image-recording operation that has become routine/mandatory for large-volume high-resolution image analysis. In particular, such in-focus imaging leads to an approximately twofold enhancement of SNR owing to the fact that the low spatial frequencies are transferred with equal weight and the high-frequency signals are not attenuated. However, there remains the disadvantage of recording images accurately in focus or analytically correcting for the contrast-transfer function, e.g. by using software developed by Mindell & Grigorieff (2003 ▸), of very low residual underfocused images. To mitigate this, Volta phase-plate recording at small (∼500 nm) underfocus and corrected by CTF estimation (Rohou & Grigorieff, 2015 ▸) has recently been applied successfully (Khoshouei et al., 2017 ▸). This seminal work on the 64 kDa hemoglobin molecule, extending an earlier study on an ∼250 kDa human PrX-3 dodecamer (Khoshouei et al., 2016 ▸), has made feasible the determination of near-atomic resolution structures of low-contrast, small-molecular-weight biomolecules by cryo-EM that were hitherto beyond reach by traditional methods.
5.2. Specimen preparation
A bottleneck to the rapid acquisition of usable cryo-EM images remains the production of a good-quality film of vitrified buffer with a thickness appropriate for the specimen under examination. Many variables are involved, such as the performance of the blotting process in the plunge-freezing device (for example Vitrobot from FEI or CP3 from Gatan), protein aggregation and denaturation at the air–water interface, and non-uniform spreading of the sample buffer, which is dependent on the nature and treatment of the support film. These variables can make reproducibility elusive and constitute a significant limitation (Glaeser et al., 2016 ▸). One path forward may be to eliminate the traditional blotting step. One of the new-generation cryo-grid preparation devices called ‘Spotiton’ uses a self-blotting nanowire mesh on specially treated grids that acts to wick small volumes (picolitres) of sample away from the holes containing the buffer-suspended specimen, leaving a thin film that is subsequently vitrified (Dandey et al., 2018 ▸). Polydimethylsiloxane (PDMS)-based microfluidic devices for spraying consistent-sized very thin films of sample have been used (Feng et al., 2017 ▸) that also possess the option for setting up time-resolved experiments directly on the grids (see, for example, Berriman & Unwin, 1994 ▸; Subramaniam et al., 1993 ▸). Finally, Arnold et al. (2017 ▸) have devised a technique to spread sample volumes as low as 3–20 nl by microcapillary to achieve a thin film on a holey grid. While the traditional method of filter-paper blotting remains popular, these efforts reflect a real need to streamline this critical step in the cryo-EM workflow.
5.3. Aberration-corrected microscopy
The most common lens aberration in TEM that influences high-resolution information in the images of low-contrast biological specimens is the spherical aberration (Cs) in the objective lens. In particular, the frequent sign reversals of the contrast transfer function associated with a defocused phase-contrast image need to be very accurately determined in order to ensure fidelity of image interpretation, and this can limit the achievable resolution. To enable high-resolution analysis, the use of a Cs-corrector TEM was proposed some time ago (Evans et al., 2008 ▸). In recent work (Fan et al., 2017 ▸), a Cs-corrector was used in combination with a Volta phase plate to establish the usability of overfocus images in near-atomic resolution structure determination. It is anticipated that a Cs-corrector will be a regular feature in future-generation microscopes equipped with modular optical systems.
6. Concluding remarks
In 2015 the journal Nature Methods chose single-particle cryo-EM as the Method of the Year (Eisenstein, 2016 ▸), and fittingly in 2017 the Nobel Prize in Chemistry was shared by Drs Richard Henderson, Joachim Frank and Jacques Dubochet for their seminal contributions to the growth of the cryo-EM discipline. Given the remarkable progress in cryo-EM, which has seen numerous difficult systems such as dynamic macromolecular complexes, membrane proteins and fibrous assemblies imaged at high resolution, perhaps a choice of Method of the Decade is now more appropriate. In this context, the acquisition of a sub-2.5 Å resolution structure by single-particle analysis for the first time by Subramaniam and coworkers (Bartesaghi et al., 2015 ▸) and more recently of glutamate dehydrogenase at 1.8 Å resolution (Merk et al., 2016 ▸) have raised the possibility that cryo-EM could be relevant to drug discovery, a field so far monopolized by X-ray crystallography (Peplow, 2017 ▸). In favorable cases, such an exercise becomes feasible even at lower resolutions, such as the visualization of proteasome inhibitors against the malarial proteasome (Li et al., 2016 ▸). In this context, given that ∼70% of drug targets are membrane proteins, success in resolving the structure of the TRPV channel at near-atomic resolution by SPA (Liao et al., 2013 ▸; Gao et al., 2016 ▸) is an important milestone in the growth of cryo-EM. Indeed, a near-atomic resolution structure of a member of the major drug-target GPCR family, a class B calcitonin receptor–G protein complex with a bound ligand, was recently solved by SPA and phase-plate technology (Liang et al., 2018 ▸). Phase-plate technology appears to be poised to have a profound effect on the cryo-EM field with the growth of the number of laboratories using this technology. The recent work to reveal a 3.2 Å resolution structure of 64 kDa hemoglobin using a VPP (Khoshouei et al., 2017 ▸) is extremely promising, and suggests that the postulated ∼38 kDa size limit (Henderson, 1995 ▸) may be well within reach.
Direct electron detectors are now obligatory for high-resolution cryo-EM, and the new generation of detectors possess a much improved DQE (e.g. K3 over K2). This trend is likely to continue, which will significantly reduce the volume of data and improve the quality necessary for very high-resolution analysis. The current top-of-the-line microscopes, such as the 300 kV Titan Krios, with essential maintenance contracts come with a heavy price tag that can be beyond the budget of most small cryo-EM laboratories and/or institutions establishing such a discipline. In this context, fabricating and marketing lower-end, e.g. 100 kV FEG, microscopes with high-end optics and robotic sample-loading capabilities may prove to be a good solution. Of interest are the ThermoFisher/FEI intermediate 200 kV microscopes Glacios and Talos Arctica. The latter has been shown to yield sub-3 Å resolution structures (Herzik et al., 2017 ▸).
Large macromolecular complexes such as the well studied ribosome that ushered in modern SPA are being discovered at a rapid pace (Alberts, 1998 ▸) and will remain attractive topics in SPA cryo-EM. The commonly observed heterogeneity in these systems will attract, for example, the development of more robust classification strategies (Sorzano et al., 2019 ▸) with the goal of improved resolution of the identified configurations. Clearly, substantial further investigations supported by novel algorithms are needed, as pointed out, for example, by Frank (2018 ▸). Collaborative study by his group implied that the functional pathway of complex molecular entities involves a very large number of states in the configuration landscape that currently are only approximated by accessing, for example, a handful of states. Such an exercise of possible high-resolution identification of a multitude of states imaged in the functional pathway will clearly further enhance the use of cryo-EM. At the other end of the size spectrum, successful efforts to solve near-atomic resolution structures by SPA of ‘small’ molecular-weight systems (Fig. 2 ▸) such as the 120 kDa γ-secretin (Bai et al., 2015 ▸), the 93 kDa isocitrate dehydrogenase (Merk et al., 2016 ▸) and the aforementioned 64 kDa hemoglobin leads to the expectation that typical ‘garden-variety’ proteins will be within reach sooner or later. Given that near-atomic resolution cryo-EM investigations in this ‘small-size’ domain are still in their infancy, the recent successes of cryo-EM notwithstanding, comments by some that cryo-EM may soon replace X-ray crystallography are arguably premature (Vénien-Bryan et al., 2017 ▸). In all such endeavors, it is essential that whenever possible the pathways followed in the cryo-EM analysis are informed by the insights gained from other complementary structural techniques, principally X-ray crystallography.
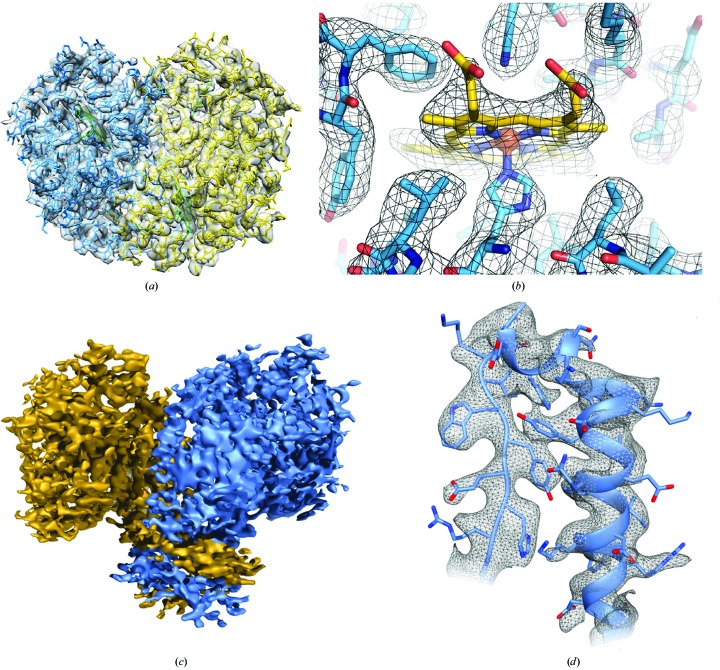
Figure 2.
Two example structures of proteins with a molecular mass of less than 100 kDa determined by SPA cryo-EM. The upper row shows (a) the density map of the 64 kDa hemoglobin at 3.2 Å resolution solved using a Volta phase plate (Khoshouei et al., 2017 ▸) and (b) a section of density near the heme group, while the bottom row shows (c) the map for the 93 kDa enzyme isocitrate dehydrogenase 1 and (d) the quality of the map, demonstrating side-chain density in an α-helical region (Merk et al., 2016 ▸). Parts (a) and (b) are reproduced from Khoshouei et al. (2017 ▸) under a Creative Commons Attribution 4.0 International License, and parts (c) and (d) are reproduced from Merk et al. (2016 ▸), with permission from Elsevier.
Acknowledgments
The author thanks Richard Kingston for careful reading of the manuscript and for helpful comments, and the anonymous referees for instructive comments.
References
- Alberts, B. (1998). Cell, 92, 291–294. [DOI] [PubMed]
- Amunts, A., Brown, A., Toots, J., Scheres, S. H. W. & Ramakrishnan, V. (2015). Science, 348, 95–98. [DOI] [PMC free article] [PubMed]
- Arnold, S. A., Albiez, S., Bieri, A., Syntychaki, A., Adaixo, R., McLeod, R. A., Goldie, K. N., Stahlberg, H. & Braun, T. (2017). J. Struct. Biol. 197, 220–226. [DOI] [PubMed]
- Bai, X.-C., Fernandez, I. S., McMullan, G. & Scheres, S. H. W. (2013). Elife, 2, e00461. [DOI] [PMC free article] [PubMed]
- Bai, X.-C., Yan, C., Yang, G., Lu, P., Ma, D., Sun, L., Zhou, R., Scheres, S. H. W. & Shi, Y. (2015). Nature (London), 525, 212–217. [DOI] [PMC free article] [PubMed]
- Banerjee, S., Bartesaghi, A., Merk, A., Rao, P., Bulfer, S. L., Yan, Y., Green, N., Mroczkowski, B., Neitz, R. J., Wipf, P., Falconieri, V., Deshaies, R. J., Milne, J. L., Huryn, D., Arkin, M. & Subramaniam, S. (2016). Science, 351, 871–875. [DOI] [PMC free article] [PubMed]
- Bartesaghi, A. Merk, A., Banerjee, S., Matthies, D., Wu, X., Milne, J. L. S. & Subramaniam, S. (2015). Science, 348, 1147–1151. [DOI] [PMC free article] [PubMed]
- Berriman, J. & Unwin, N. (1994). Ultramicroscopy, 56, 241–252. [DOI] [PubMed]
- Brilot, A. F., Chen, J. Z., Cheng, A., Pan, J., Harrison, S. C., Potter, C. S., Carragher, B., Henderson, R. & Grigorieff, R. (2012). J. Struct. Biol. 177, 630–637. [DOI] [PMC free article] [PubMed]
- Brüggeller, P. & Mayer, E. (1980). Nature (London), 288, 569–571.
- Carragher, B., Kisseberth, N., Kriegman, D., Milligan, R. A., Potter, C. S., Pulokas, J. & Reilein, A. (2000). J. Struct. Biol. 132, 33–45. [DOI] [PubMed]
- Chen, Y., Clarke, O. B., Kim, J., Stowe, S., Kim, Y.-K., Assur, Z., Cavalier, M., Godoy-Ruiz, R., von Alpen, D. C., Manzini, C., Blaner, W. S., Frank, J., Quadro, L., Weber, D. J., Shapiro, L., Hendrickson, W. A. & Mancia, F. (2016). Science, 353, aad8266. [DOI] [PMC free article] [PubMed]
- Claude, A., Porter, K. R. & Pickels, E. G. (1947). Cancer Res. 7, 421–430.
- Crowther, R. A. (1971). Philos. Trans. R. Soc. B Biol. Sci. 261, 221–230. [DOI] [PubMed]
- Dandey, V. P., Wei, H., Zhang, Z., Tan, Y. Z., Acharya, P., Eng, E. T., Rice, W. J., Kahn, P. A., Potter, C. S. & Carragher, B. (2018). J. Struct. Biol. 202, 161–169. [DOI] [PMC free article] [PubMed]
- Danev, R., Buijsse, B., Khoshouei, M., Plitzko, J. M. & Baumeister, W. (2014). Proc. Natl Acad. Sci. USA, 111, 15635–15640. [DOI] [PMC free article] [PubMed]
- Danev, R., Glaeser, R. M. & Nagayama, K. (2009). Ultramicroscopy, 109, 312–325. [DOI] [PMC free article] [PubMed]
- Deptuch, G., Besson, A., Rehak, P., Szelezniak, M., Wall, J., Winter, M. & Zhu, Y. (2007). Ultramicroscopy, 107, 674–684. [DOI] [PubMed]
- De Rosier, D. J. & Klug, A. (1968). Nature (London), 217, 130–134. [DOI] [PubMed]
- Dubochet, J., Adrian, M., Chang, J.-J., Homo, J.-C., Lepault, J., McDowall, A. W. & Schultz, P. (1988). Q. Rev. Biophys. 21, 129–228. [DOI] [PubMed]
- Dubochet, J. & McDowall, A. W. (1981). J. Microsc. 124, 3–4.
- Eisenstein, M. (2016). Nature Methods, 13, 19–22. [DOI] [PubMed]
- Erickson, H. P. & Klug, A. (1971). Philos. Trans. R. Soc. B Biol. Sci. 261, 105–118.
- Evans, J. E., Hetherington, C., Kirkland, A., Chang, L.-Y., Stahlberg, H. & Browning, N. (2008). Ultramicroscopy, 108, 1636–1644. [DOI] [PMC free article] [PubMed]
- Fan, X., Zhao, L., Liu, C., Zhang, J. -C., Fan, K., Yan, X., Peng, H.-L., Lei, J. & Wang, H.-W. (2017). Structure, 25, 1623–1630. [DOI] [PubMed]
- Faruqi, A. R. & Henderson, R. (2007). Curr. Opin. Struct. Biol. 17, 549–555. [DOI] [PubMed]
- Feng, X., Fu, Z., Kaledhonkar, S., Jia, Y., Shah, B., Jin, A., Liu, Z., Sun, M., Chen, B., Grassucci, R. A., Ren, Y., Jiang, H., Frank, J. & Lin, Q. (2017). Structure, 25, 663–670. [DOI] [PMC free article] [PubMed]
- Frank, J. (1975). Ultramicroscopy, 1, 159–162. [DOI] [PubMed]
- Frank, J. (2018). Biochemistry, 57, 888. [DOI] [PMC free article] [PubMed]
- Frank, J., Shimkin, B. & Dowse, H. (1981). Ultramicroscopy, 6, 343–357.
- Gao, Y., Cao, E., Julius, D. & Cheng, Y. (2016). Nature (London), 534, 347–351. [DOI] [PMC free article] [PubMed]
- Glaeser, R. M. (1999). J. Struct. Biol. 128, 3–14. [DOI] [PubMed]
- Glaeser, R. M., Han, B.-G., Csencsits, R., Killilea, A., Pulk, A. & Cate, J. H. D. (2016). Biophys. J. 110, 749–755. [DOI] [PMC free article] [PubMed]
- Grigorieff, N. (2007). J. Struct. Biol. 157, 117–125. [DOI] [PubMed]
- Grigorieff, N. & Harrison, S. C. (2011). Curr. Opin. Struct. Biol. 21, 265–273. [DOI] [PMC free article] [PubMed]
- Hall, C. E. (1955). J. Cell Biol. 1, 1–12.
- Heel, M. van (1987). Ultramicroscopy, 21, 111–123. [DOI] [PubMed]
- Heel, M. van, Harauz, G., Orlova, E. V., Schmidt, R. & Schatz, M. (1996). J. Struct. Biol. 116, 17–24. [DOI] [PubMed]
- Henderson, R. (1995). Q. Rev. Biophys. 28, 171–193. [DOI] [PubMed]
- Henderson, R. (2013). Proc. Natl Acad. Sci. USA, 110, 18037–18041. [DOI] [PMC free article] [PubMed]
- Henderson, R., Baldwin, J. M., Ceska, T. A., Zemlin, F., Beckmann, E. & Downing, K. H. (1990). J. Mol. Biol. 213, 899–929. [DOI] [PubMed]
- Henderson, R. & Glaeser, R. M. (1985). Ultramicroscopy, 16, 139–150.
- Henderson, R. & Unwin, P. N. T. (1975). Nature (London), 257, 28–32. [DOI] [PubMed]
- Herzik, M. A. Jr, Wu, M. & Lander, G. C. (2017). Nature Methods, 14, 1075–1078. [DOI] [PMC free article] [PubMed]
- Heymann, J. B. & Belnap, D. M. (2007). J. Struct. Biol. 157, 3–18. [DOI] [PubMed]
- Ilca, S. L., Kotecha, A., Sun, X., Poranen, M. M., Stuart, D. I. & Huiskonen, J. T. (2015). Nature Commun. 6, 8843. [DOI] [PMC free article] [PubMed]
- Khoshouei, M., Radjainia, M., Baumeister, W. & Danev, R. (2017). Nature Commun. 8, 16099. [DOI] [PMC free article] [PubMed]
- Khoshouei, M., Radjainia, M., Phillips, A. J., Gerrard, J. A., Mitra, A. K., Plitzko, J. M., Baumeister, W. & Danev, R. (2016). Nature Commun. 7, 10534. [DOI] [PMC free article] [PubMed]
- Kühlbrandt, W. (2014). Science, 343, 1443–1444. [DOI] [PubMed]
- Kühlbrandt, W., Wang, D. N. & Fujiyoshi, Y. (1994). Nature (London), 367, 614–621. [DOI] [PubMed]
- Li, H., O’Donoghue, J., van der Linden, W. A., Xie, S. C., Yoo, E., Foe, I. T., Tilley, L., Craik, C. S., da Fonseca, P. C. A. & Bogyo, M. (2016). Nature (London), 530, 233–236. [DOI] [PMC free article] [PubMed]
- Liang, Y.-L., Khoshouei, M., Glukhova, A., Furness, S. G. B., Zhao, P., Clydesdale, L., Koole, C., Truong, T. T., Thal, D. M., Lei, S., Radjainia, M., Danev, R., Baumeister, W., Wang, M.-W., Miller, L. J., Christopoulos, A., Sexton, P. M. & Wootten, D. (2018). Nature (London), 555, 121–125. [DOI] [PubMed]
- Liang, Y.-L., Khoshouei, M., Radjainia, M., Zhang, Y., Glukhova, A., Tarrasch, J., Thal, D. M., Furness, S. G. B., Christopoulos, G., Coudrat, T., Danev, R., Baumeister, W., Miller, L. J., Christopoulos, A., Kobilka, B. K., Wootten, D., Skiniotis, G. & Sexton, P. M. (2017). Nature (London), 546, 118–123. [DOI] [PMC free article] [PubMed]
- Liao, M., Cao, E., Julius, D. & Cheng, Y. (2013). Nature (London), 504, 107–112. [DOI] [PMC free article] [PubMed]
- Mastronarde, D. N. (2005). J. Struct. Biol. 152, 36–51. [DOI] [PubMed]
- Matthies, D., Dalmas, O., Borgnia, M. J., Dominik, P. K., Merk, A., Rao, P., Reddy, B. G., Islam, S., Bartesaghi, A., Perozo, E. & Subramaniam, S. (2016). Cell, 164, 747–756. [DOI] [PMC free article] [PubMed]
- Merk, A., Bartesaghi, A., Banerjee, S., Falconieri, V., Rao, P., Davis, M. I., Pragani, R., Boxer, M. B., Earl, L. A., Milne, J. L. S. & Subramaniam, S. (2016). Cell, 165, 1698–1707. [DOI] [PMC free article] [PubMed]
- Milazzo, A. C., Cheng, A., Moeller, A., Lyumkis, D., Jacovetty, E., Polukas, J., Ellisman, M. H., Xuong, N.-H., Carragher, B. & Potter, C. S. (2011). J. Struct. Biol. 176, 404–408. [DOI] [PMC free article] [PubMed]
- Mindell, J. A. & Grigorieff, N. (2003). J. Struct. Biol. 142, 334–347. [DOI] [PubMed]
- Miyazawa, A., Fujiyoshi, Y. & Unwin, N. (2003). Nature (London), 423, 949–955. [DOI] [PubMed]
- Morgan, C., Ellison, S. A., Rose, H. M. & Moore, D. H. (1954). J. Exp. Med. 100, 195–202. [DOI] [PMC free article] [PubMed]
- Murata, K., Mitsuoka, K., Hirai, T., Walz, T., Agre, P., Heymann, J. B., Engel, A. & Fujiyoshi, Y. (2000). Nature (London), 407, 599–605. [DOI] [PubMed]
- Nogales, E., Wolf, S. G. & Downing, K. H. (1998). Nature (London), 391, 199–203. [DOI] [PubMed]
- Ohi, M., Li, Y., Cheng, Y. & Walz, T. (2004). Biol. Proced. Online, 6, 23–34. [DOI] [PMC free article] [PubMed]
- Peankuch, E. & Kausche, G. A. (1940). Naturwissenschaften, 28, 46.
- Penczek, P. A., Grassucci, R. A. & Frank, J. (1994). Ultramicroscopy, 53, 251–270. [DOI] [PubMed]
- Peplow, M. (2017). Nature Rev. Drug. Discov. 16, 815–817. [DOI] [PubMed]
- Porter, K. R., Claude, A. & Fullam, E. F. (1945). J. Exp. Med. 81, 233–246. [DOI] [PMC free article] [PubMed]
- Radermacher, M., Wagenknecht, T., Verschoor, A. & Frank, J. (1987). J. Microsc. 146, 113–136. [DOI] [PubMed]
- Ren, G., Reddy, V. S., Cheng, A., Melnyk, P. & Mitra, A. K. (2001). Proc. Natl Acad. Sci. USA, 98, 1398–1403. [DOI] [PMC free article] [PubMed]
- Rohou, A. & Grigorieff, N. (2015). J. Struct. Biol. 192, 216–221. [DOI] [PMC free article] [PubMed]
- Ruska, H. (1940). Naturwissenschaften, 28, 45–46.
- Saxton, W. O., Pitt, T. J. & Horner, M. (1979). Ultramicroscopy, 4, 343–353.
- Scheres, S. H. W. (2012). J. Struct. Biol. 180, 519–530. [DOI] [PMC free article] [PubMed]
- Scheres, S. H. W. (2016). Methods Enzymol. 579, 125–157. [DOI] [PubMed]
- Sigworth, F. J. (1998). J. Struct. Biol. 180, 519–530.
- Smith, P. R. (1978). Ultramicroscopy, 3, 153–160. [DOI] [PubMed]
- Sorzano, C. O. S., Marabini, R., Velázquez-Muriel, J., Bilbao-Castro, J. R., Scheres, S. H. W., Carazo, J. M. & Pascual-Montano, A. (2004). J. Struct. Biol. 148, 194–204. [DOI] [PubMed]
- Sorzano, C. O. S., Jiménez, A., Mota, J., Vilas, J. L., Maluenda, D., Martínez, M., Ramírez-Aportela, E., Majtner, T., Segura, J., Sánchez-García, R., Rancel, Y., del Caño, L., Conesa, P., Melero, R., Jonic, A., Vargas, J., Cazals, F., Freyburg, Z., Krieger, J., Bahar, I., Marabini, R. & Carazo, J. M. (2019). Acta Cryst. F75, 19–32. [DOI] [PMC free article] [PubMed]
- Subramaniam, S., Gerstein, M., Oesterhelt, D. & Henderson, R. (1993). EMBO J. 12, 1–8. [DOI] [PMC free article] [PubMed]
- Tang, G., Peng, L., Baldwin, P. R., Mann, D. S., Jiang, W., Rees, I. & Ludtke, S. J. (2007). J. Struct. Biol. 157, 38–46. [DOI] [PubMed]
- Tao, Y. & Zhang, W. (2000). Curr. Opin. Struct. Biol. 10, 616–622. [DOI] [PubMed]
- Taylor, K. A. & Glaeser, R. M. (1974). Science, 186, 1036–1037. [DOI] [PubMed]
- Taylor, K. A. & Glaeser, R. M. (1976). J. Ultrastruct. Res. 55, 448–456. [DOI] [PubMed]
- Unwin, P. N. T. & Henderson, R. (1975). J. Mol. Biol. 94, 425–440. [DOI] [PubMed]
- Vénien-Bryan, C., Li, Z., Vuillard, L. & Boutin, J. A. (2017). Acta Cryst. F73, 174–183. [DOI] [PMC free article] [PubMed]
- Villa, E., Sengupta, J., Trabuco, L. G., LeBarron, J., Baxter, W. T., Shaikh, T. R., Grassucci, R. A., Nissen, P., Ehrenberg, M., Schulten, K. & Frank, J. (2009). Proc. Natl Acad. Sci. USA, 106, 1063–1068. [DOI] [PMC free article] [PubMed]
- Zhang, X., Jin, L., Fang, Q., Hui, W. H. & Zhou, Z. H. (2010). Cell, 141, 472–482. [DOI] [PMC free article] [PubMed]