We present the first large-scale quantitative MS-based profiling of cell envelopes and cytoplasmic proteins derived from a diverse panel of WHO gonococcal reference strains possessing all existing AMR profiles and intended for quality assurance in laboratory investigations. We describe nine novel gonorrhea vaccine candidates and six potential global proteomic AMR markers. A reference proteomics databank for gonococcal vaccine and AMR research endeavors is provided, which enables microbiological, clinical, or epidemiological projects and enhances the utility of the WHO reference strains.
Keywords: Absolute Quantification, Bacteria, Pathogens, Membranes, Drug Resistance, Antibiotics, Gonorrhea, Neisseria gonorrhoeae, TMT, Vaccine
Graphical Abstract
Highlights
Quantitative proteomes of the 2016 WHO Neisseria gonorrhoeae reference strains.
Novel gonorrhea vaccine candidates and potential global proteomic AMR markers.
First large-scale proteomic profiling of gonorrhea vaccine candidates and AMR.
A reference proteomics databank for gonococcal vaccine and AMR research endeavors.
Abstract
The sexually transmitted disease gonorrhea (causative agent: Neisseria gonorrhoeae) remains an urgent public health threat globally because of its reproductive health repercussions, high incidence, widespread antimicrobial resistance (AMR), and absence of a vaccine. To mine gonorrhea antigens and enhance our understanding of gonococcal AMR at the proteome level, we performed the first large-scale proteomic profiling of a diverse panel (n = 15) of gonococcal strains, including the 2016 World Health Organization (WHO) reference strains. These strains show all existing AMR profiles - established through phenotypic characterization and reference genome publication - and are intended for quality assurance in laboratory investigations. Herein, these isolates were subjected to subcellular fractionation and labeling with tandem mass tags coupled to mass spectrometry and multi-combinatorial bioinformatics. Our analyses detected 904 and 723 common proteins in cell envelope and cytoplasmic subproteomes, respectively. We identified nine novel gonorrhea vaccine candidates. Expression and conservation of new and previously selected antigens were investigated. In addition, established gonococcal AMR determinants were evaluated for the first time using quantitative proteomics. Six new proteins, WHO_F_00238, WHO_F_00635c, WHO_F_00745, WHO_F_01139, WHO_F_01144c, and WHO_F_01126, were differentially expressed in all strains, suggesting that they represent global proteomic AMR markers, indicate a predisposition toward developing or compensating gonococcal AMR, and/or act as new antimicrobial targets. Finally, phenotypic clustering based on the isolates' defined antibiograms and common differentially expressed proteins yielded seven matching clusters between established and proteome-derived AMR signatures. Together, our investigations provide a reference proteomics data bank for gonococcal vaccine and AMR research endeavors, which enables microbiological, clinical, or epidemiological projects and enhances the utility of the WHO reference strains.
Neisseria gonorrhoeae is an obligate human pathogen and the causative agent of the sexually transmitted disease gonorrhea. Gonorrhea is a global public health concern. In 2012 the World Health Organization (WHO)1 estimated over 78 million new urogenital cases per year in adults (15–49 years of age) worldwide (1, 2). The spread of gonorrhea is facilitated by the high prevalence of asymptomatic infections. Urogenital gonorrhea is asymptomatic in up to 10–15% of infected men and up to 50% of infected women. Pharyngeal and rectal infections, which have increased in prevalence in both sexes and are predominant among men who have sex with men, are primarily asymptomatic (3, 4). Untreated or inappropriately treated gonorrhea can result in serious consequences on reproductive and neonatal health. Women are disproportionately affected, as gonococcal infection can ascend from the cervix to the uterus, Fallopian tubes, ovaries, and surrounding tissue, causing pelvic inflammatory disease. Long-term sequelae include ectopic pregnancy, chronic pelvic pain, and infertility. Furthermore, gonorrhea is strongly associated with an increased risk of both the acquisition and transmission of HIV (5).
Antimicrobial therapy is the only mainstay in the effective management and control of gonorrhea. However, N. gonorrhoeae exhibits an extraordinary capacity to develop antimicrobial resistance (AMR) through mutations and acquisition of AMR genes. The evolution of AMR in N. gonorrhoeae has overcome every therapeutic option since the “miracle drug” penicillin was introduced for gonorrhea treatment. Currently, a dual antimicrobial therapy (mainly ceftriaxone and azithromycin) is recommended for treatment of uncomplicated infections (6). Of grave concern over the past decade is the proliferation of resistance or decreased susceptibility to ceftriaxone worldwide. Azithromycin resistance has also emerged in most settings (7). The first failure of one of the recommended dual antimicrobial therapies against pharyngeal gonorrhea was reported in 2016 (8), and the first N. gonorrhoeae isolates with resistance to ceftriaxone combined with high-level resistance to azithromycin were identified in the United Kingdom (9, 10) and Australia (11, 12) in early 2018. In consideration of dwindling treatment options, scarce therapeutic alternatives, disease prevalence and morbidity, and lack of a vaccine(s), N. gonorrhoeae has been categorized by the WHO as a high priority pathogen globally and by the Centers for Disease Control and Prevention (CDC) as an urgent level threat in the United States (13).
Developing an effective gonococcal vaccine is essential because this is the only sustainable solution to quell the spread of gonococcal AMR and gonorrhea in general. The battle against penicillin-nonsusceptible Streptococcus pneumoniae exemplifies a successful vaccination strategy. Introduction of a pneumococcal conjugate vaccine in 2010 reduced the number of infections over 45% (14). Unfortunately, despite its public health importance, gonorrhea vaccine development remains in its infancy. Since 1970, only three small-scale vaccine trials using whole cell (15), pilin (16), and porin proteins (17) have been launched. All were unsuccessful in developing immunity against reinfection with gonorrhea. However, recent breakthroughs, including the development of small animal models for evaluating gonorrhea vaccines (18, 19), increased knowledge about N. gonorrhoeae immune evasion mechanisms (20–26), and the development of an effective vaccine for the closely related N. meningitidis serogroup B, which provided a low level of cross-protection against gonococcal infection (27), have reinvigorated the interest in gonococcal vaccine development (28).
Proteomic technology offers a powerful toolbox to enable vaccine antigen mining (28–32) and AMR proteome analysis (33–35), and to provide insights into host-pathogen interactions (36–39). Proteomic approaches have an advantage over genomics in drug and vaccine discovery endeavors by delivering information pertaining to protein abundance, post-translational modification(s), structure-function relationships, and protein-protein interactions (40–42). In addition, subcellular fractionation steps preceding proteomic applications reduce sample complexity, increase the likelihood of discovering low-abundance proteins, and aid in defining protein localization, all of which provide further insights into the proteins' functions and interactomes (43, 44). For N. gonorrhoeae, proteomic approaches have begun to deliver proteinaceous vaccine candidates (29, 30, 39, 45) and to support elucidation of AMR patterns (46, 47). Current off-gel proteomics, such as isobaric tag labeling (isobaric tagging for absolute quantification, iTRAQ; and tandem mass tags, TMT) coupled with high-pressure liquid chromatography and mass spectrometry techniques (LC-MS/MS), demonstrate superb protein separation and identification and enable detection of proteins in the low femtomole to high attomole range with precision and reliability (29, 48, 49).
To address the need for discovery of additional gonorrhea vaccine and drug candidates and to enhance our understanding of AMR at the proteome level, herein we examined the 2016 WHO N. gonorrhoeae reference strains (50) and the FA6140 strain (51) using a global quantitative proteomic approach. The WHO panel consists of 14 N. gonorrhoeae reference strains strictly selected and validated internationally to represent the N. gonorrhoeae species. All known gonococcal phenotypic and genetic AMR determinants are included for use as quality control strains during phenotypic and genetic laboratory testing. Eight of the strains were initially included in the 2008 WHO reference strains [WHO F, G, K, L, M, N, O, and P; (52)] to which 6 novel strains (U, V, W, X, Y, and Z) were added to constitute the 2016 WHO reference strains (50). All WHO panel strains have been subjected to extensive phenotypic, genomic, and genetic analyses to establish diagnostic markers, molecular epidemiological characteristics, reference genomes, and AMR profiles (phenotypic and genetic) for all antimicrobials currently and previously used for gonorrhea treatment, in addition to novel antimicrobials considered for future interventions. This panel includes WHO X, the first extensively drug-resistant gonococcal strain identified with high-level resistance to ceftriaxone, as well as additional strains with different levels of resistance to ceftriaxone, azithromycin and any additional therapeutic antimicrobials. Complete genomes with detailed annotations are available for all panel strains, providing a fundamental resource for future molecular studies. Accordingly, the well-characterized 2016 WHO reference strains (50) are ideally suited to provide detailed descriptions of the global N. gonorrhoeae proteome, a greater understanding of gonococcal AMR at the proteome level, and a source for the identification of broadly conserved novel vaccine candidates. In addition to the WHO panel strains, we have included in our investigations N. gonorrhoeae FA6140, which is a penicillin-resistant, β-lactamase-negative isolate that was originally described after a local epidemic outbreak of 199 gonococcal cases in Durham, North Carolina, USA in 1983 (51). It serves as a model for gonococcal AMR studies and has facilitated the characterization of mutations in genes encoding the “multiple transferable resistance” repressor MtrR (53), ribosomal protein S10 (54), and penicillin-binding protein 2 (55) and their impact on AMR.
Our study is the first to investigate the global proteomic profiles of 15 N. gonorrhoeae reference strains using subcellular fractionation to separate cytoplasmic (C) and cell envelope (CE) associated proteomes, which were measured with tandem mass tags coupled to liquid chromatography and tandem mass spectrometry [TMT-LC-MS/MS; (56)], a highly reproducible and sensitive technique. These proteomic studies achieved our three major objectives. First, to enhance progress on gonorrhea vaccine development, novel vaccine candidates were identified, and the expression profiles of currently proposed antigens were established in diverse clinical isolates. Second, to broaden our understanding of AMR, proteomic signatures associated with AMR were defined by conducting a pairwise analysis of differentially expressed proteins to compare FA6140 and the 2016 WHO panel to WHO F, which possesses the largest genome and is susceptible to all relevant antimicrobials (50). Third, to facilitate the use of the 2016 WHO panel in various types of basic research and quality assurance, the complete reference proteomes of all tested strains were defined.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The 2016 WHO N. gonorrhoeae reference strains [n = 14; (50, 52)] and the N. gonorrhoeae FA6140 strain (51) were used in this study. The AMR profiles of all isolates were described previously (50). Gonococcal strains were cultured from frozen stocks (−80 °C) onto gonococcal base agar (GCB) medium (Difco, Sparks, MD) with Kellogg's supplements I and II, diluted 1:100 and 1:1,000, respectively (57). After incubation at 37 °C in a 5% CO2 enriched atmosphere for 18–20 h, nonpiliated and transparent colonies were subcultured onto GCB and incubated as described above. To initiate growth in liquid medium, nonpiliated colonies were collected from GCB and suspended to the Optical Density (OD600) of 0.1 in prewarmed GCB liquid (GCBL) medium supplemented as described above with the addition of 0.042% sodium bicarbonate. Suspensions were incubated at 37 °C with shaking at 220 rpm.
Subcellular Fractionation and TMT Labeling
All 15 N. gonorrhoeae strains were simultaneously cultured in GCBL as described above. Cells were collected by centrifugation (20 min, 6000 × g) when the OD600 of each culture reached 0.6–0.8, resuspended in PBS and lysed by passage through a French Press. The cell debris was removed by centrifugation and the crude CE fraction was separated from the C proteins using a sodium carbonate extraction procedure and subsequent ultracentrifugation steps. The fraction enriched with CE proteins was reconstituted in PBS supplemented with 0.1% SDS (29, 30). Experiments were conducted in two biological replicates. Sample quality and the overall subproteome profiles were examined by SDS-PAGE coupled with Colloidal Coomassie staining (58, 59). The total protein amount in each fraction was assessed using a Protein Assay Kit (Bio Rad, Hercules, CA). Each CE and C fraction containing 100 μg of protein in 25 μl volume of triethylammonium bicarbonate buffer was reduced with 11.3 mm tris(2-carboxyethyl)phosphine hydrochloride and the cysteines were alkylated using 20.1 mm iodoacetamide. Proteins were digested using trypsin (Promega, Madison, WI) at a 1:40 ratio. TMT reagents (ThermoFisher Scientific, Waltham, MA) were dissolved in acetonitrile (ACN) and used to label proteins in CE and C fractions as follows for the 10-plex experiment (ref 90111, Thermo Fisher Scientific): WHO F strain: TMT10-126, WHO K strain: TMT10-127C, WHO G strain: TMT10-127N, WHO M strain: TMT10-128C, WHO L strain: TMT10-128N, WHO O strain: TMT10-129C, WHO N strain: TMT10-129N, WHO U strain: TMT10-130C, WHO P strain: TMT10-130N, WHO V strain: TMT10-131; for the 6-plex experiment (ref 90402, Thermo Fisher Scientific): WHO F strain: TMT6-126, WHO W strain: TMT6-127, WHO X strain: TMT6-128, WHO Y strain: TMT6-129, WHO Z strain: TMT6-130, FA6140 strain: TMT6-131. Mixtures were incubated for 1 h at room temperature. The reaction was quenched by addition of 8 μl of 5% hydroxylamine. Samples were pooled, dried in a vacuum concentrator and stored at −80 °C before separation by high pressure liquid chromatography (HPLC) and MS analysis.
Sample Fractionation and MS Analysis
Samples were fractionated by strong cation exchange (SCX) with a Paradigm (Michrom Biosciences, San Diego, CA) HPLC with mobile phases of 5 mm potassium phosphate monobasic in 30% ACN/70% water (v/v) pH 2.7 (buffer A) and 5 mm potassium phosphate monobasic in 30% ACN/70% water (v/v) pH 2.7 with 500 mm potassium chloride (buffer B). The sample was brought up in buffer A (200 μl). The peptides were separated using a 2.1 mm × 100 mm Polysulfoethyl A column (PolyLC) over 60 min at a flow rate of 200 μl/min. The separation profile was as follows: hold 2% B for 5 min, 2% to 8% B in 0.1 min, 8% to 18% B in 14.9 min, 18% to 34% B in 12 min, 34% to 60% B in 18 min, 60% to 98% B in 0.1 min and hold for 10 min. Fractions were collected in 96-well microtiter plates at 1 min/fraction. Sixty fractions were pooled into 12 and dried using a speed vac. The samples were desalted using Oasis HLB 1cc cartridges. The cartridges were washed with 70% ACN/0.1% trifluoroacetic acid (TFA) and equilibrated with 0.1% TFA. Samples were loaded onto the cartridge in 0.1% TFA, washed with 0.1% TFA, and eluted in 1 ml 70% ACN/0.1% TFA. The samples were dried by vacuum centrifugation.
Desalted SCX fractions were analyzed by liquid chromatography electrospray ionization mass spectrometry (LC/ESI MS/MS) with a Thermo Scientific Easy-nLC II (Thermo Scientific) nano HPLC system coupled to a hybrid Orbitrap Elite ETD (Thermo Scientific) mass spectrometer. In-line de-salting was accomplished using a reversed-phase trap column (100 μm × 20 mm) packed with Magic C18AQ (5-μm 200Å resin; Michrom Bioresources) followed by peptide separations on a reversed-phase column (75 μm × 250 mm) packed with Magic C18AQ (5-μm 100Å resin; Michrom Bioresources) directly mounted on the electrospray ion source. A 90-min gradient from 7% to 35% ACN in 0.1% formic acid at a flow rate of 400 nL/min was used for chromatographic separations. The heated capillary temperature was set to 300 °C and a spray voltage of 2750 V was applied to the electrospray tip. The Orbitrap Elite instrument was operated in the data-dependent mode, switching automatically between MS survey scans in the Orbitrap [automatic gain control (AGC) target value 1,000,000; resolution 120,000; and injection time 250 msec] with MS/MS spectra acquisition in the Orbitrap (AGC target value of 50,000; 15,000 resolution; and injection time 250 msec). The 15 most intense ions from the Fourier-transform full scan were selected for fragmentation in the higher-energy C-trap dissociation (HCD) cell by higher-energy collisional dissociation with a normalized collision energy of 40%. Selected ions were dynamically excluded for 30 s with a list size of 500 and exclusion mass by mass width ± 10ppm. HPLC and MS/MS analyses were performed in the Proteomic Facility at the Fred Hutchinson Cancer Center, Seattle.
Proteomic Data Analysis
Data analysis was performed using Proteome Discoverer 1.4 (Thermo Scientific). The data were searched against WHO_F_CDS (supplemental Material S1; 2449 entries) with the common Repository of Adventitious Proteins (cRAP, http://www.thegpm.org/crap/) fasta file. Trypsin was set as the enzyme with maximum missed cleavages set to 2. The precursor ion tolerance was set to 10 ppm and the fragment ion tolerance was set to 0.8 Da. Variable modifications included TMT 6Plex (+229.163 Da) on any N-Terminus, oxidation on methionine (+15.995 Da), carbamidomethyl on cysteine (+57.021 Da), and TMT 6Plex on lysine (+229.163 Da).
Experimental Design and Statistical Rationale
All experiments described above were performed using the 15 bacterial strains in biological duplicates. Data were searched using Sequest HT. All search results were run through Percolator for scoring. Quantification was performed using the canned TMT 6plex or TMT 10plex methods through Proteome Discoverer with stringent criteria for protein identification including 1% False Discovery Rate (FDR), at least one unique peptide per protein, each identified peptide restricted to a single protein, and the score for every detected peptide of ≥1. Differential protein expression between CE and C fractions was determined by comparing the normalized total reporter ion intensities of groups using the WHO F protein expression profile as a reference.
Bioinformatic Analysis
To detect potential homologous proteins, amino acid sequences of each identified N. gonorrhoeae vaccine candidate were downloaded and compared against the GenBank proteome database (https://www.ncbi.nlm.nih.gov/genbank/) using our in-house designed program based on the Reciprocal Best Blast Hit approach (60) using BLASTP with the following parameters: percentage identity ≥50%, and E-value ≤1.0 e-5. The NCBI codes for the bacterial species included in this study are listed in supplemental Table S7.
Differential protein expression in four proteomics data sets (CE and C fractions in two biological replicates) was designated by fold changes ≥1.5 or ≤0.667 in reference to strain WHO F. Because of the variable nature of protein expression in N. gonorrhoeae, we took a conservative approach to designate protein expression and a protein was categorized as “upregulated” or “downregulated” solely when the fold change abundance was higher than 1.5 or lower than 0.667, respectively, to that of WHO F consistently in two biological experiments. A protein was designated as “ubiquitous” when its abundance was between 0.667–1.5-fold compared with WHO F in both experiments, or “variable” when its levels were not consistent between experiments.
A comprehensive assessment of predicted subcellular protein localization was accomplished by using the CELLO (61), PsortB 3.0.2 (62), SOSUI-GramN (63), SignalP 4.1 (64), LipoP 1.0 (65), and TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) prediction algorithms and a majority voting strategy. Furthermore, for proteins whose subcellular localization was not predicted using the aforementioned algorithms, we relied on the difference between their unique peptide counts in the CE and C fractions as follows:
where “i” is the sequential number assigned for samples and “x” is the total number of peptides detected in each fraction. Cytoplasmic proteins had more of their unique peptides detected in the C fraction (UPCD < 0), whereas membrane proteins had unique peptides enriched in the CE fraction (UPCD > 0). Proteins with UPCD = 0 were excluded from analysis using this UPCD formula. Proteins were categorized as follows: outer membrane, periplasmic, inner membrane, C proteins, and proteins with unknown localization.
The phenotypic and proteotypic clusters of all strains were constructed using as variables both their AMR (50) and proteomic profiles obtained in this study. These clusters were designed based on the Hamming distance between tested strains, which counts how many elements differ between two vectors, and is equivalent to Manhattan distance on binary data. Average linkage was used to determine distances between clusters.
Graphs were generated with GraphPad Prism version 7 for Mac (GraphPad Software). The proteotypes of strains that belong to the same phenotypic cluster were compared, highlighting proteins that are significantly up- or downregulated with respect to those proteins of WHO F.
RESULTS AND DISCUSSION
Study Rationale
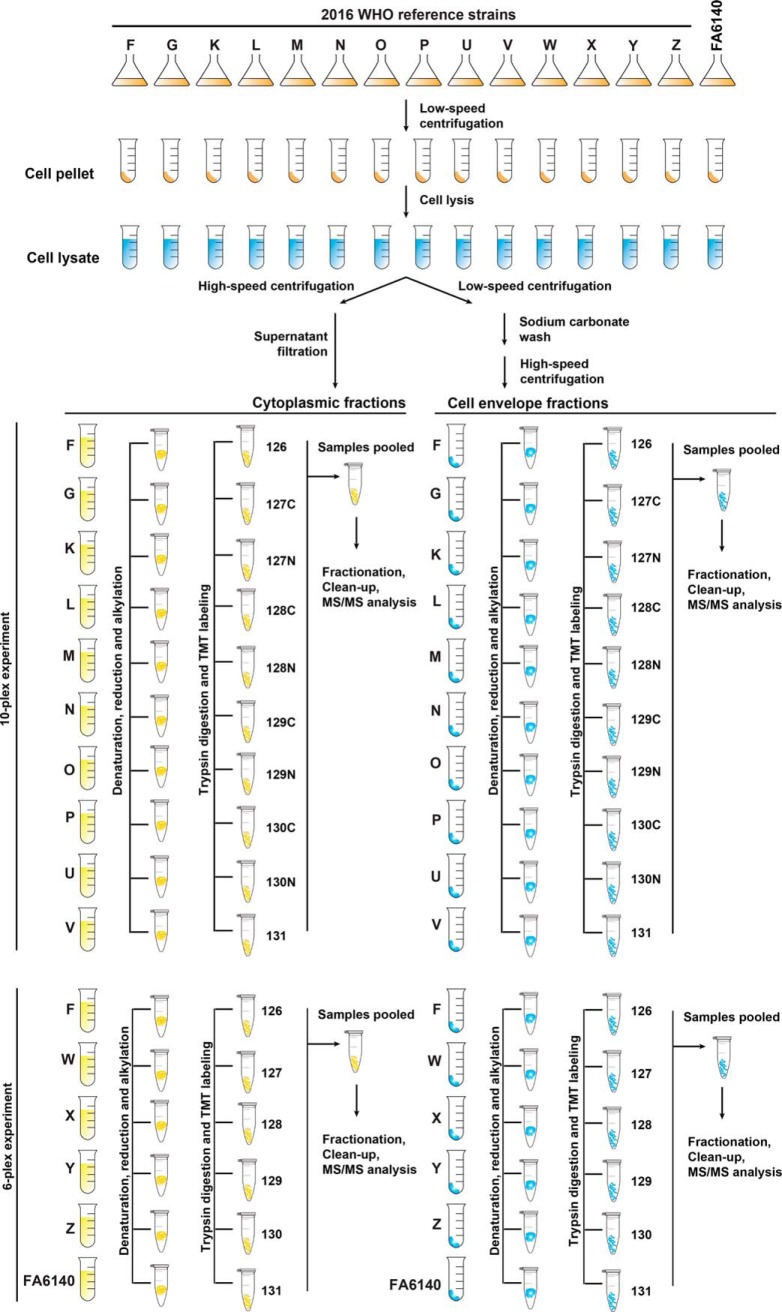
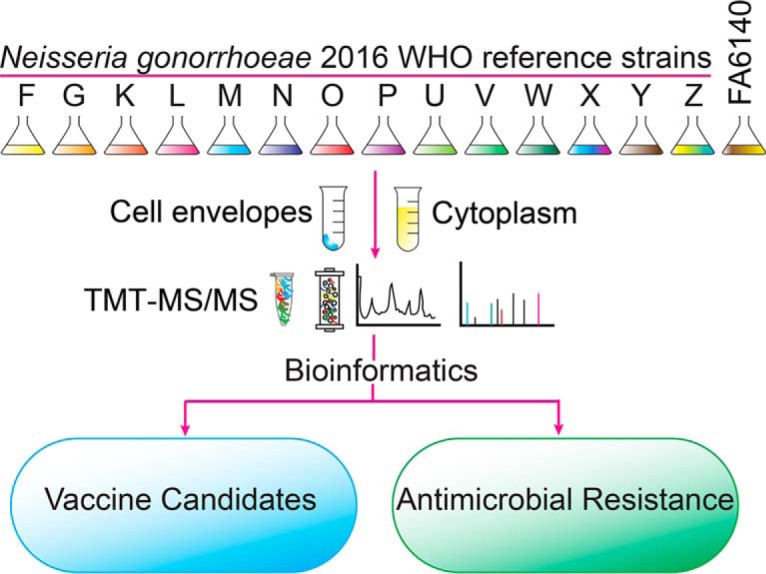
In our study design (Fig. 1), all 15 strains were cultured concurrently to mid-logarithmic growth, harvested, and subjected to subcellular fractionation to separate CE (outer membrane, periplasmic, inner membrane) and C proteins. We utilized TMT reagent technology for protein identification and quantitation as it provides a highly sensitive method for peptide labeling (56) and allows up to 10 biological samples to be analyzed in a single experiment (67). TMT-labeling, two-dimensional liquid chromatography fractionation, and subsequent MS/MS analyses were conducted on every 6-plex and 10-plex experiment pertaining to the CE and C fractions derived from each strain (Fig. 1). We selected WHO F as the reference strain for protein identification and quantitation because it has the largest genome (2,292,467 bp) and proteome (2,449 ORFs) among the 2016 WHO reference strains (50) and FA6140 (68), and it is susceptible to most antimicrobials currently or historically used for gonorrhea treatment.
Fig. 1.
Experimental paradigm of quantitative proteomic profiling of the N. gonorrhoeae 2016 WHO reference strains and the FA6140 strain. All gonococci were cultured concurrently in liquid medium until reaching mid-logarithmic growth. Bacterial cells were harvested, lysed, and subjected to subcellular fractionation to separate the crude cell envelope (CE) and cytoplasmic (C) proteomes. CE proteins were enriched using a sodium carbonate wash and ultracentrifugation. The obtained CE and C protein samples (100 μg) were denatured, reduced, alkylated, trypsinized, and the peptides from each strain were labeled using 10-plex and 6-plex Tandem mass tag (TMT) reagents, as indicated. Finally, samples were pooled, fractionated by strong cation exchange, and analyzed by liquid chromatography electrospray ionization mass spectrometry. Experiments were performed in biological duplicates.
Subcellular fractionation experiments coupled with proteomics repeatedly show cytoplasmic proteins associated with the membranes, which are commonly regarded as “contaminating” or “moonlighting” proteins (29, 30, 69, 70). Therefore, to focus solely on the enriched proteins in individual subproteomes, we first eliminated C and CE proteins that were detected in the CE and C protein fractions, respectively, from further analyses. Complete lists of all identified proteins are in supplemental Tables S1–S2. Subsequently, we performed two-armed proteomic data analyses: 1) for vaccine antigen mining, we focused on common proteins identified in all strains in the CE fraction with the overarching goal to discover omnipresent N. gonorrhoeae proteins; 2) to profile AMR signatures, we performed a pairwise comparison of individual strains to WHO F to enhance the discovery of strain-specific feature(s).
Overview of Cell Envelope and Cytoplasmic Proteomes
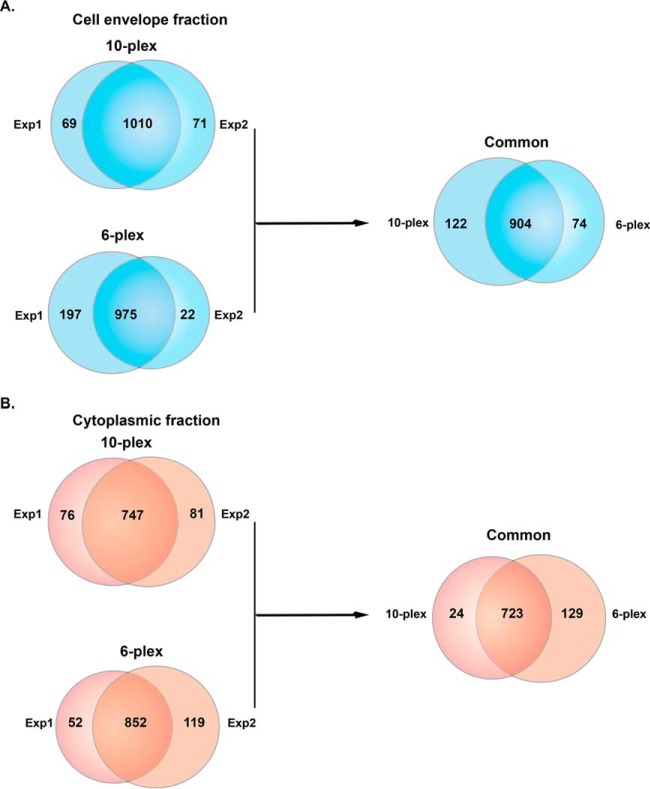
The 10-plex biological replicate experiments identified a total of 1150 proteins in the CE fraction of all ten strains, of which 1010 were common in both sets (Fig. 2A). In the two 6-plex experiments, 1194 proteins were identified; 975 were shared in all six isolates (Fig. 2A). Taken together, the 10-plex and the 6-plex experiments resulted in identification of 1100 proteins in the CE fractions, of which 904 were common among all examined N. gonorrhoeae strains (Fig. 2A). The proteome coverage per strain ranged from 41.22% (981 proteins) for WHO Y to 45.32% (1042 proteins) for WHO G (supplemental Table S3).
Fig. 2.
Venn diagrams illustrating the distribution of proteins identified in cell envelope and cytoplasmic fractions in two independent proteomic experiments. A, Cell envelope proteomes derived from the 2016 WHO reference strains and FA6140 were analyzed in 10-plex and 6-plex experiments performed in biological duplicates. A total of 1079 and 1081 proteins was identified in Experiments 1 and 2, respectively, and 1010 common proteins were found in both 10-plex experiments. The 6-plex TMT labeling revealed 975 common proteins as well as 197 and 22 unique proteins in Experiments 1 and 2, respectively. Further analyses were applied to 904 proteins mutually identified in both 10-plex and 6-plex experiments. B, The proteomic profiling of cytoplasmic fractions yielded 904 proteins shared among all 10 strains, of which 747 were common in both experiments. The 6-plex TMT identified 904 and 971 proteins in Experiments 1 and 2, respectively; of which 852 were common between replicates. In further analyses solely the 723 proteins shared between both experiments were included. Exp 1 - experiment 1; Exp 2 - experiment 2.
Proteomics of the C fraction in the 10-plex set conducted in biological replicates yielded 904 proteins that were shared among all 10 strains, of which 747 were common in both experiments (Fig. 2B). The two 6-plex experiments identified 1023 shared proteins, with 852 common among the two replicates (Fig. 2B). Cumulatively, C fraction profiling resulted in identification of 876 proteins with 723 common in all 15 N. gonorrhoeae strains (Fig. 2B). Proteome coverage ranged from 31.37% (746 proteins) in WHO U to 38.43% (852 proteins) in FA6140 (supplemental Table S3).
Subsequently, we allocated common proteins that were identified in all 15 N. gonorrhoeae strains to outer membrane, inner membrane, periplasm, cytoplasm, or unknown localization categories based on PSORTb 3.0.2 (62), SOSUIGramN (63), and CELLO (61) predictions and the majority-voting strategy. We used these software packages to take advantage of their different algorithms and statistical approaches for the prediction of protein subcellular localization. As expected from our subcellular fractionation approach (30, 49, 69), the CE fraction was enriched in membrane proteins compared with the C sample, with outer membrane (26 versus 8), periplasmic (51 versus 38), and inner membrane proteins (145 versus 6) that were also identified with considerably higher peptide counts (Fig. 3A–3C, and supplemental Tables S1–S2 and S4–S5). The C preparations yielded 592 cytoplasmic proteins that were identified with greater peptide counts compared with the cytoplasmic proteins associated with the CE fraction (Fig. 3D, supplemental Tables S5–S6). Furthermore, to increase the discovery of potential vaccine candidates, we searched the 149 proteins of unknown localization identified in the CE fraction (Fig. 3E) for the presence of signal peptides and transmembrane motifs using SignalP 4.1 (64), LipoP 1.0 (65), and TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The results of all software programs and the majority votes strategy revealed six additional CE proteins, four of which were found in the majority of examined strains (supplemental Table S6). In addition, literature searches for experimental evidence of protein surface exposure allowed assignment of BamE [NGO1780; (71)], SliC [NGO1063; (72)], MetQ [NGO2139; (30, 73)], Ng-MIP [NGO1225; (30, 74)], and BamG [NGO1985; (75)] to the cell surface. Subcellular location predictions for all proteins are listed in supplemental Table S8.
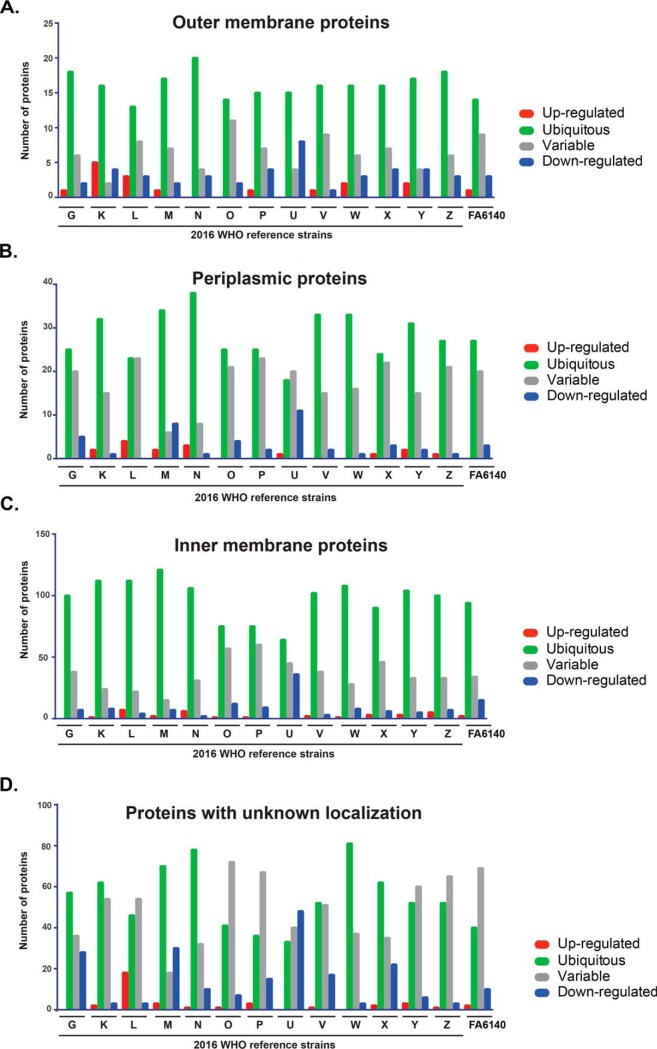
Fig. 3.
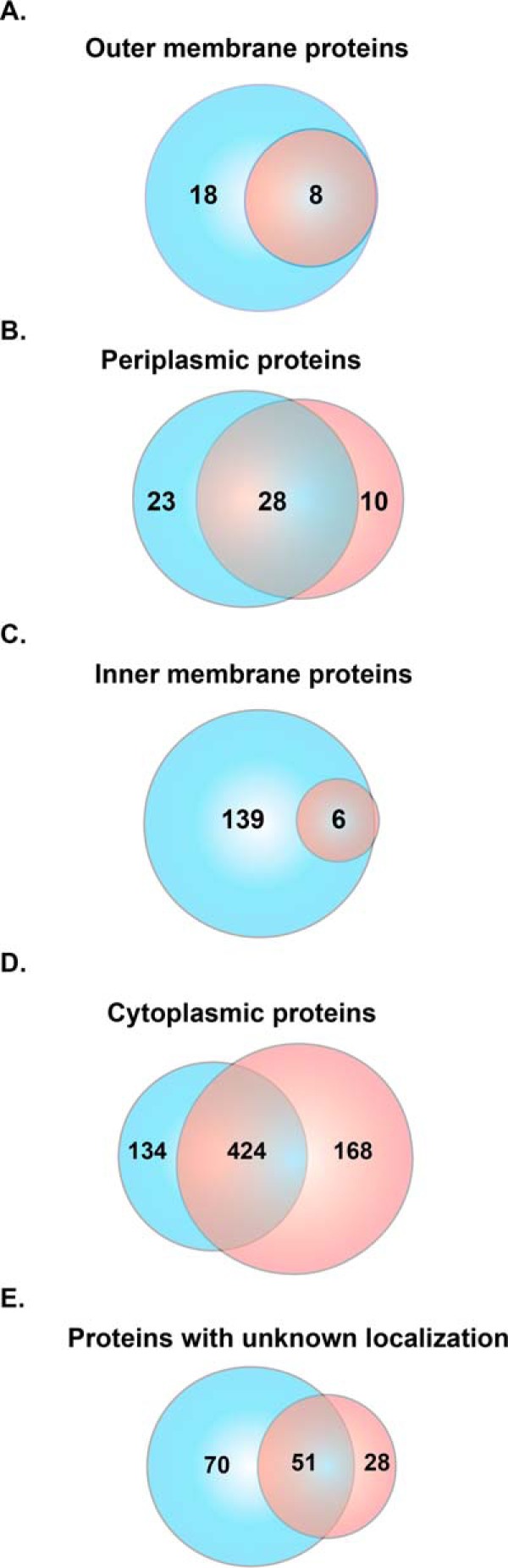
Subcellular localization of proteins identified in cell envelope and cytoplasmic subproteomes. Proteins identified in the cell envelope (blue circle) and cytoplasmic (red circle) fractions were subjected to comprehensive assessments of subcellular localization using different prediction algorithms and were allocated into the outer membrane (A), periplasm (B), inner membrane (C), cytoplasm (D), or unknown localization (E).
Expression Patterns of Common Identified Proteins Compared with WHO F
Proteins were categorized as ubiquitous, up- or downregulated, or variable based on their abundance in relation to the corresponding protein in WHO F in biological duplicate experiments. We investigated expression patterns of detected proteins in both subproteome fractions that were shared among all strains (Figs. 4–5, supplemental Tables S4–S5). Annotated cell envelope proteins were predominantly ubiquitous in the CE fraction. The proportion of ubiquitous CE outer membrane proteins (n = 26) ranged from 73% (n = 19 in WHO N) to 46.15% (n = 12 in WHO L; Fig. 4A). Ubiquitous periplasmic proteins ranged from 76.47% (n = 39 in WHO N) to 35.2% (n = 18 in WHO U) of the total number of proteins annotated to localize to the periplasm (n = 51; Fig. 4B). Finally, between 83.4% (n = 121 in WHO M) and 44.1% (n = 64 in WHO U) of inner membrane proteins (n = 145) were ubiquitously expressed (Fig. 4C). Upregulated outer membrane (0–19.23%), periplasmic (0–7.8%) and inner membrane (0–4.8%) proteins made up the smallest proportion of proteins in the CE fraction (supplemental Table S4), whereas downregulated outer membrane (3.85–30.77%), periplasmic (0–23.53%) and inner membrane (1.38–24.8%) proteins were moderately prevalent (supplemental Table S4). Further analysis of the CE fraction detected 121 common proteins with unknown localization (Fig. 3E, supplemental Table S4). Ubiquitous expression was the dominant pattern for these proteins in WHO G, K, M, N, V, W, and X and ranged from 42.97% (n = 52, WHO V) to 66.94% (n = 81, WHO W). Variable expression of proteins with unknown localization dominated in WHO L, O, P, Y, Z, and FA6140. WHO U had the highest number of downregulated proteins (39.67%, n = 48) with respect to those in WHO F (Fig. 4D).
Fig. 4.
Expression patterns of common proteins identified in the cell envelope fraction. Outer membrane (A), periplasmic (B), inner membrane (C), or proteins with unknown localization (D) are shown. Expression of each protein in each gonococcal strain was compared with the protein level in the reference WHO F isolate. Protein expression is categorized as ubiquitous (green bars); upregulated (red bar); downregulated (blue bar); and variable (gray bar).
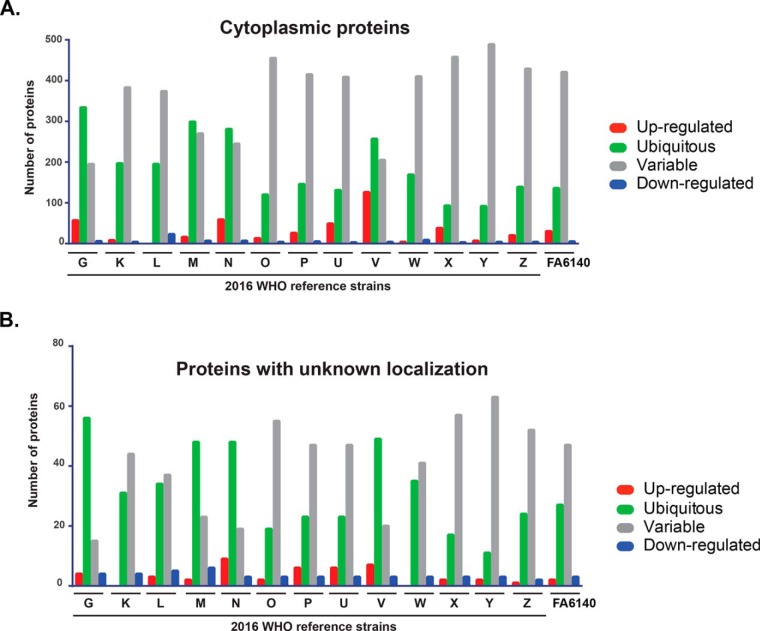
Fig. 5.
Expression patterns of common proteins identified in the cytoplasmic proteome. Cytoplasmic (A) and proteins with unknown localization (B) are shown. Protein levels in individual gonococcal strains were compared with the protein level in the reference WHO F isolate. Protein expression is categorized as ubiquitous (green bars); upregulated (red bar); downregulated (blue bar); and variable (gray bar).
In contrast to the CE expression pattern, we observed a striking increase in the number of variably expressed cytoplasmic proteins in the C fraction of analyzed strains compared with WHO F (Fig. 5A, supplemental Table S5). The percentage of variable proteins ranged from 32.9% to 82.6% for WHO G and WHO Y, respectively (supplemental Table S5). Ubiquitous proteins were the next most common category and oscillated from 15.5% in WHO Y to 56.4% in WHO G. The third group contained upregulated proteins (0–21.28%), and downregulated proteins ranged from 0.5–3.88% (supplemental Table S5). For proteins with no assigned localization, variable expression was the most prevalent pattern in WHO K, L, O, P, U, W, X, Y, Z, and FA6140 (Fig. 5B), ranging from 79.75% (n = 63, WHO Y) to 46.83% (n = 37, WHO L). Ubiquitous proteins were the dominating group in WHO G, M, N, and V (Fig. 5B). Finally, up- and downregulated proteins constituted up to 11.39% and 7.59%, respectively, of the total proteins with unknown localization in the C fraction.
Together, the first quantitative proteomic profiling of the 15 N. gonorrhoeae strains demonstrated distinct differences in their proteomes and showed that a pattern of ubiquitous protein expression was prevalent in the CE fraction, whereas variably expressed proteins were the dominant group in the C subproteome.
Antigen Mining Decision Tree
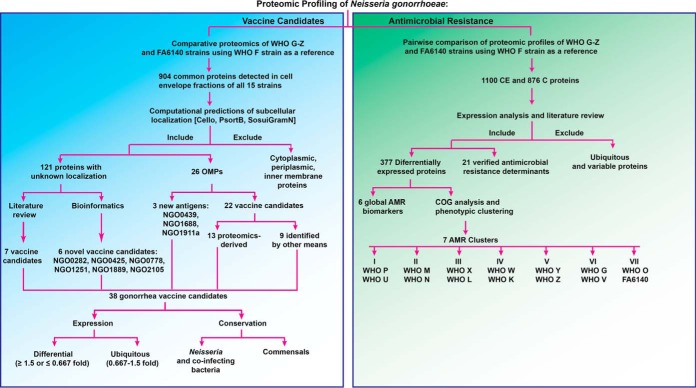
To identify novel gonorrhea antigens and to gain information about expression of previously identified vaccine candidates (26, 30, 49, 76), we designed an antigen mining decision tree (Fig. 6). We included in this process 25 outer membrane proteins (all outer membrane proteins identified except RmpM) and 121 proteins of unknown localization identified in CE proteomic profiling (Fig. 6 and supplemental Table S1). The latter group of proteins was subjected to signal sequence and transmembrane motif analyses to increase the coverage of potential vaccine candidates. Together, these investigations yielded nine novel antigens including NGO0282, NGO0425, NGO0439, NGO0778, NGO1251, NGO1688, NGO1889, NGO1911a, and NGO2105 in addition to previously discovered proteomics-derived antigens [(29, 30); Table I] and vaccine candidates identified by other means (Table II).
Fig. 6.
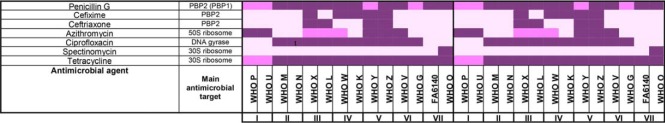
Decision tree designed for proteomic mining of Neisseria gonorrhoeae vaccine candidates and antimicrobial resistance (AMR) determinants. For vaccine antigen mining, we focused on 904 common proteins identified in all strains in the CE fraction with the overarching goal to discover omnipresent N. gonorrhoeae proteins. Computational predictions were employed to predict protein subcellular localization. Solely 26 OMPs and 121 proteins with unknown localization were included in further analyses. Bioinformatics and literature review yielded six novel vaccine candidates in the latter group of proteins. The OMPs constituted three new antigens and 22 previously proposed gonorrhea vaccine candidates including 13 proteomics-derived. Finally, the 38 identified gonorrhea vaccine candidates were subjected to expression and conservation analyses. To profile AMR signatures, we performed a pairwise comparison of individual strains to the antimicrobial susceptible WHO F strain to enhance the discovery of strain-specific feature(s). The maximum number of identified proteins reached 1100 and 876 for CE and C fractions, respectively. Expression profiling and literature review yielded 398 differentially expressed proteins including 21 verified AMR determinants. Ubiquitous and variably expressed proteins were excluded from the analysis. To link AMR phenotypes with proteomic signatures, COG analysis and phenotypic clustering of gonococcal strains based on their defined antibiograms and the 377 differentially expressed proteins were performed generating seven phenotypic clusters that matched between established and proteome-derived AMR signatures. OMPs - outer membrane proteins; CE - cell envelope; C - cytoplasm; COG - Cluster of Orthologous Genes.
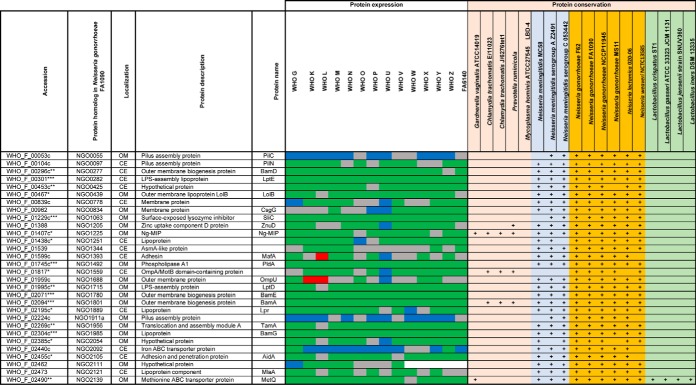
Table I. Expression and homologs of proteomics-derived Neisseria gonorrhoeae vaccine candidates#.
#Color legends for protein expression: Ubiquitous (green), up-regulated (red), down-regulated (blue), variable (grey). Ubiquitous expression among all 15 strains is marked with (***), 14 strains with (**), 12–13 strains with (*). Abbreviations: OM: Outer membrane, CE: cell envelope, +: protein homolog detected.
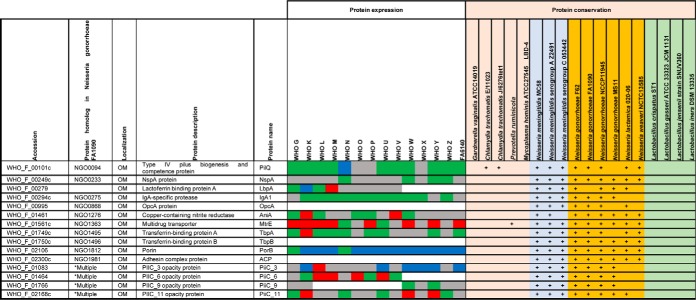
Table II. Expression and homologs of Neisseria gonorrhoeae vaccine candidates identified by other means#.
#Color legends for protein expression in cell envelope fraction: Ubiquitous (green), up-regulated (red), down-regulated (blue), and variable (grey). *Multiple protein homologs in Neisseria gonorrhoeae FA1090 are detected: NGO0066a, NGO0070, NGO0950a, NGO1040a, NGO1073a, NGO1277a, NGO1463a, NGO1513, NGO1553a, NGO1861a, NGO2060a. Abbreviations: OM: Outer membrane, +: protein homolog detected.
Further bioinformatics and literature searches were performed to gain insights into the new proteomics-derived vaccine candidates. The putative lipoprotein NGO0282 is a homolog of the outer membrane localized LptE, which is a component of the trans-envelope LptA-G machinery involved in the transport of lipopolysaccharide/lipooligosaccharide (LPS/LOS) molecules to the E. coli and N. meningitidis outer membrane, respectively. LptE's chaperone-like role in LptD biogenesis is conserved in both bacteria but LptE works in concert with LptD to translocate LPS to the cell surface only in E. coli (77, 78). LptD is essential for E. coli and N. gonorrhoeae viability but is dispensable for N. meningitidis (79–81). Therefore, the function of N. gonorrhoeae LptE in both LOS transport and LptD biogenesis needs to be elucidated. NGO0425 contains a tetratricopeptide repeat-like domain and a transmembrane helix. Together with its E. coli homolog, YfgM, NGO0425 belongs to the UPF0070 family. In E. coli, YfgM was proposed to act within the β-barrel trafficking chaperone network and its depletion in Δskp and ΔsurA knockout backgrounds contributed to further alterations in outer membrane integrity (82). The vaccine candidate protein NGO0439 is homologous to the E. coli outer membrane protein LolB, which is involved in lipoprotein trafficking to the outer membrane (83, 84). We consider N. gonorrhoeae LolB to be a vaccine candidate antigen because its surface localization should be experimentally verified. Differences in the localization of homologous proteins exist. For instance, fHbp is a surface-displayed lipoprotein in N. meningitidis but not in N. gonorrhoeae (85), whereas BamE is on the surface of gonococci but faces the periplasmic side of the outer membrane in E. coli (71). Protein NGO1688, annotated as OmpU, is a putative iron uptake outer membrane protein that is positively regulated by the oxygen-sensing transcription factor, FNR (86). NGO1911a is a predicted pilus assembly protein that is associated with the adhesin PilY (87). Finally, NGO0778, NGO1251 (a putative lipoprotein), and NGO1889 are hypothetical proteins. NGO1889 belongs to the LprI family (PFO7007) that comprises bacterial proteins of ∼120 amino acids in length that contain four conserved cysteine residues. LprI from Mycobacterium tuberculosis acts as a lysozyme inhibitor (88), providing the exciting possibility that N. gonorrhoeae LprI contributes to residual lysozyme resistance observed in gonococci deprived of surface-exposed lysozyme inhibitors SliC and ACP (72, 89). Lastly, NGO2105 contains peptidase S6 (residues 43–310) and autotransporter (residues 1215–1468) domains potentially involved in proteolytic activity and auto-translocation, respectively, suggesting that this is a newly identified autotransporter protein in N. gonorrhoeae. In support of this notion, the NGO2105 locus, also known as adhesion and penetration protein or “NEIS1959 (iga2)” in the PubMLST database, encodes IgA2 protease (AidA) and has homologs in other Neisseria (Table I) as well as Haemophilus influenzae (90).
Together, our investigations yielded nine novel gonorrhea vaccine candidates, including proteins with implications in pathogenesis such as IgA2 (AidA) and LprI, and provided valuable information regarding the expression patterns of previously selected vaccine candidates.
Expression and Homologs of Gonorrhea Vaccine Candidates
We first evaluated expression profiles of extensively studied gonorrhea vaccine candidates including MtrE (91–93), PorB (94, 95), PilQ (96), TbpA (97, 98), Opa (99, 100), and AniA (19, 101–103). MtrE was upregulated in 8 isolates (Table II). Compared with WHO F, PorB was present at similar levels only in WHO G and N and was downregulated in 12 strains. PilQ (96) was ubiquitously expressed in 10 strains, whereas expression of Opa proteins was widely variable, as expected (104, 105). The TbpA level was similar in 8 strains; however, we did not detect TbpB (106). Nor did we detect ACP (107, 108) or OpcA (109, 110) under the standard growth conditions used in our studies, which suggested that they might be specifically regulated. AniA was present at different levels in 7 strains, ubiquitous in five, and upregulated in two isolates. Immunoblotting experiments with anti-AniA antisera corroborated these findings (103). The cellular pool of NspA (111) varied in ten isolates, whereas lactoferrin binding protein LbpA (112) was variable in five strains and was ubiquitous in WHO L and G (Table II).
Strikingly, most of the proteome-derived vaccine candidates showed ubiquitous expression among numerous strains (Table I). SliC, PldA, BamE, BamA, and BamG were ubiquitous in all 15 isolates. Similar results for these proteins were obtained by iTRAQ-MS/MS applied to the proteomic profiling of cell envelopes and outer membrane vesicles (OMVs) isolated from four different strains of N. gonorrhoeae (29). Further, LolB, Ng-MIP, NGO1559, and NGO2054 were unvaryingly expressed in at least 12 isolates. Among the novel vaccine candidates identified in our study, LptE, LolB, IgA2, and NGO1251 were found ubiquitous in at least 13 strains. In addition, LprI and NGO0778 were similarly expressed in 12 and 9 isolates, respectively. In support of our proteomics data, immunoblotting analyses demonstrated similar cellular levels of BamA, MetQ, TamA, LptD, NGO2054 (30), BamE-D (71), SliC (72), and BamG (75) in whole cell lysates of the 2016 WHO strains as well as geographically and temporally diverse clinical isolates of N. gonorrhoeae from Baltimore (n = 5) and Seattle (n = 13). Our previous studies showed that PorB, PilQ, BamA-D, SliC, MafA, PldA, MetQ, IgA1 protease, and LptD are cargo proteins present at similar levels in naturally released gonococcal OMVs (29, 71, 72), which further highlights their potential as vaccine antigens considering the success of N. meningitidis OMV-based vaccines (26, 113).
Finally, we examined the presence of homologs of the gonorrhea vaccine candidates among non-gonococcal Neisseria species, other commensal bacteria (114, 115), and co-infecting microbes (116–119) that inhabit the same ecological niche as N. gonorrhoeae. Antigens conserved between these pathogens and preferably not in commensals have the potential to eradicate several sexually transmitted infections, if formulated into a protective vaccine(s). Our comparative analyses showed that all the proteomics-based antigens have homologous proteins in the majority of investigated N. meningitidis strains, and none are present in M. hominis (Table I). In addition, Ng-MIP-like proteins exist in C. trachomatis, G. vaginalis, and P. ruminicola; BamA and NGO1559 homologs were found in C. trachomatis and P. ruminicola. MetQ, a methionine transporter (73), was the only proteomics-derived vaccine candidate with homologs across all examined bacteria except for C. trachomatis, M. hominis, and P. ruminicola. Further, we detected protein homologs of PilQ in two C. trachomatis strains and MtrE and ZnuD in P. ruminicola; these three proteins were absent in commensal species.
Together, our investigations provide pioneering information into newly identified and existing gonorrhea vaccine candidates. We have established each candidate's expression pattern in diverse N. gonorrhoeae isolates and identified homologs among other pathogenic and/or commensal bacteria that share the same ecological niche. Stable expression in the WHO gonococcal panel coupled to presence in N. meningitidis and co-infecting agents—but rarely in urogenital commensals—further highlights the importance of including these antigens in gonorrhea vaccine(s).
Proteomics Profiling of N. gonorrhoeae Antimicrobial Resistance
Various genome-based AMR determinants have been deciphered in the gonococcus over the past decades (51, 120–125). However, many AMR determinants remain to be identified and characterized, e.g. the chromosomally encoded penicillin and cephalosporin resistance determinant “factor X” (126–128) and the AMR mechanisms that contribute to a large proportion of azithromycin resistance (129). The uncertainty behind these AMR determinants illustrates the need for alternative approaches to enhance our understanding of gonococcal AMR complexity. At the proteomic level, only two studies have attempted to address this challenge, both of which used 2D-SDS-PAGE exclusively (47, 130). Therefore, we focused on identifying proteomic AMR signatures that exist in the absence of antimicrobial pressure during standard in vitro growth conditions by performing a pairwise comparison of all identified proteins in each individual strain to the WHO F reference strain (Fig. 6, supplemental Tables S1–S2). As expected, we identified different numbers of proteins in the CE and C fractions in each comparison set because of differences in the number of open reading frames (ORFs) between the gonococcal strains (supplemental Table S3). A maximum of 1100 and 876 CE and C proteins were identified, respectively (Fig. 6, supplemental Tables S1–S2). Similarly to our previous analysis, we excluded typical cytoplasmic proteins from the CE subproteome and cell envelope proteins from the C fraction. We solely focused on 398 proteins with significantly different expression in the examined strains compared with the fully antimicrobial-susceptible strain WHO F with the rationale that these proteins may provide clues about the proteomic basis of AMR (Fig. 6). For instance, we identified MtrE as upregulated in many strains with increased resistance to numerous antimicrobials even in the absence of antimicrobial exposure, which represents an upregulation of the multidrug MtrCDE efflux pump and possibly additional efflux pumps for which MtrE acts as the outer membrane channel (29, 92, 131–133). Overall, we identified 162 and 95 proteins with known and unpredicted subcellular locations, respectively (Tables III–V). Peptide counting performed for the latter group of proteins yielded 55 and 36 proteins that are likely localized to the CE and C, respectively, and four proteins with ambiguous localization. Next, we separated 21 proteins that have been previously verified as N. gonorrhoeae AMR determinants (Fig. 6, Table III) from new potential proteomic AMR signatures (Tables IV–V).
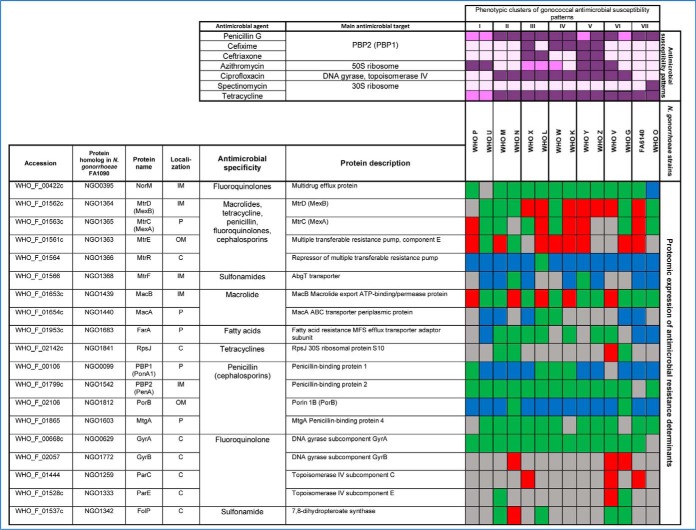
Table III. Proteomic signature of previously verified gonococcal antimicrobial resistance determinants#.
#Color legends for protein expression: Ubiquitous (green), up-regulated (red), down-regulated (blue), variable (grey). Color legends for phenotype against antimicrobial agents are as follows: resistant (dark purple), intermediate susceptible/resistant (purple), susceptible (light purple). Abbreviations: OM: Outer membrane, IM: inner membrane, C: cytoplasmic, P: periplasmic.
Table IV. New potential proteomic-derived antimicrobial resistance signatures with defined subcellular localizations#.
#Color legends for protein expression: up-regulated (red), down-regulated (blue), and undetected (white). Abbreviations: OM: outer membrane, IM: inner membrane, C: cytoplasmic, COG: cluster of orthologous genes, PBP: penicillin binding protein. Color legends for phenotype against antimicrobial agents are as follows: resistant (dark purple), intermediate susceptible/resistant (purple), susceptible (light purple).
Table V. New potential proteomic-derived antimicrobial resistance signatures with undefined localization#.
#Color legends for protein expression: Up-regulated (red), down-regulated (blue). Color legends for phenotype against antimicrobial agents are as follows: resistant (dark purple), intermediate susceptible/resistant (purple), susceptible (light purple). Abbreviations: UPCD: unique peptide count difference; PBP: penicillin binding protein.
Proteomic Signature Of Previously Verified Gonococcal Antimicrobial Resistance Determinants
Our proteomic analysis detected subcomponents of all five efflux pumps described in N. gonorrhoeae, i.e. MtrCDE, MtrF, FarAB, MacAB, and NorM (Table III). The outer membrane-barrel protein MtrE serves as the channel for the tripartite MtrCDE pump and likely fulfills this same function in the MacAB and FarAB efflux pumps (131, 133). The MtrCDE complex is the most studied efflux pump system in N. gonorrhoeae. The multiple transferable resistance (mtr) locus contains the mtrCDE operon (134) that is negatively regulated by the repressor MtrR (135). Mutations that abrogate mtrR activity result in an overexpression of the MtrCDE efflux pump and decreased susceptibility to numerous antimicrobials, e.g. macrolides, penicillins, cephalosporins, and tetracycline (53, 133). AMR mutations in the mtrR promoter were previously identified in WHO G, K, M, O, P, V, W, X, Y, and Z and within the mtrR gene (G45D or a frameshift mutation resulting in a truncated peptide) in WHO K, L, M, P, and W (50), suggesting an overexpression of the MtrCDE efflux pump. Our proteomic profiling also verified that the levels of MtrE were significantly increased or variable in all isolates except for two strains lacking any type of mtrR AMR determinant (WHO U and N; Table III). Accordingly, MtrE proved to be an effective indicator of expression of the MtrCDE efflux pump. Our findings were further supported by the downregulation of MtrR in all examined strains except WHO L (Table III). WHO L is also the only examined strain that contains an mtr120 mutation, which generates a novel promoter for mtrCDE transcription that further enhances the expression of the MtrCDE efflux pump (50, 136). The second efflux system that showed differential expression was the MacAB efflux pump, which can decrease macrolide susceptibility (137). MacA expression varied across the isolates. Expression of the inner membrane component, MacB, was enhanced in the azithromycin resistant strains WHO P and V, but also in the azithromycin susceptible strains WHO N and K, as well as WHO L, which is intermediately susceptible to azithromycin. Interestingly, MacB was the most highly expressed in WHO V, the only strain with high-level azithromycin resistance [MIC>256 μg/ml; because of the 23S rRNA A2059G mutation in all four alleles (50)], which indicates that overexpression of the MacAB efflux pump may contribute to the high MICs of azithromycin and other macrolides in WHO V. The FarA component of the FarAB efflux pump system, which exports long-chain fatty acids and other hydrophobic agents (138), was not overexpressed in any of the examined strains and was instead ubiquitously expressed in seven WHO strains (M, N, K, L, X, Z, and V) and downregulated in WHO U, O, and FA6140. Our proteomic profiling also revealed that the NorM and MtrF efflux pumps, which can decrease the susceptibility to fluoroquinolones and sulfonamides, respectively (139, 140), were not overexpressed in any of the examined strains.
Among other established AMR determinants that were differentially expressed was the major porin of N. gonorrhoeae, PorB (141, 142), which was downregulated in all strains except for WHO G and N (Table III). This downregulation suggests reduced import of antimicrobials such as penicillins, cephalosporins and tetracyclines, which can contribute to a decreased antimicrobial susceptibility. Furthermore, WHO F, G, and N express PorB1a, which is associated with a lack of the AMR determinant penB and consequently high-level chromosomally mediated resistance to penicillins and cephalosporins (125). All other strains express PorB1b. All WHO strains with PorB1b (n = 11), except WHO U, contained the AMR determinant penB. Consequently, our proteomic data suggest that penB may also be associated with a decreased expression of PorB1b in addition to the previously documented decreased penetration through PorB1b, resulting in a decreased susceptibility to several antimicrobials. The expression of penicillin-binding protein 1 (PBP1) was significantly downregulated in nine out of the twelve WHO strains that possess the ponA1 resistance determinant, which encodes a L421P amino acid substitution in PBP1 that contributes to high-level chromosomally mediated penicillin resistance (50). Accordingly, our proteomic data indicate that the decreased PBP1 expression, in addition to the PBP1 L421P amino acid alteration might contribute to high-level chromosomally mediated penicillin resistance. In contrast, PBP2 (the main lethal target for penicillins and cephalosporins) was ubiquitously expressed in 13 of the 14 strains. Similarly, GyrA expression was ubiquitous in 13 of the 14 strains. No association between GyrA expression and the main fluoroquinolone resistance mutations [amino acids S91 and D95 (50)] was identified. GyrB and/or the second fluoroquinolone target, ParC, were overexpressed in the ciprofloxacin-resistant WHO strains G, N, V and X, which may suggest that these strains upregulate GyrB and/or ParC to compensate for the mutated main fluoroquinolone target GyrA (Table III).
New Potential Proteomic-derived Antimicrobial Resistance Signatures
In the CE fraction, two hypothetical proteins predicted to localize to the inner membrane, WHO_F_00238c and WHO_F_01126, were downregulated in all examined strains compared with the antimicrobial-susceptible WHO F strain (Table IV). WHO_F_00238c, which corresponds to NGO0222 in the FA1090 genome, is a small protein with a predicted molecular weight of 8.32 kDa that contains two predicted transmembrane domains but no signal peptide. WHO_F_01126 lacks a homologous protein in FA1090. This is also a small protein (5.39 kDa) with no peptides predicted to be recognized by signal peptidase I or II. In the C fraction, no protein was differentially expressed in all strains, but two cytoplasmic proteins, NGO0597 and NGO0701, were upregulated in all strains except WHO L. NGO0597 is a nucleoside diphosphate kinase (Ndk; 15.4 KDa) involved in DNA and RNA synthesis (143), regulation of gene transcription (144), and peptide chain elongation during translation (145), all processes that are targets for different antimicrobials. NdK is secreted from Pseudomonas aeruginosa (144), M. tuberculosis (146), and Leishmania (147) to modulate interaction with host cells, block phagosome maturation in macrophages (146, 148), and promote host cell apoptosis and necrosis (149). It remains to be investigated whether the gonococcal Ndk is secreted during infection and whether it may serve as an anti-virulence or antimicrobial target. Finally, we detected two proteins with undefined subcellular localization displaying global differential expression. WHO_F_01139 and WHO_F_01144c, which have no homologs in the FA1090 genome, were downregulated in all strains. Our use of UPCD predicted WHO_F_01139 to localize to the cell envelope. WHO_F_01139 is a putative lipoprotein (16.9 KDa) with a predicted signal peptide II domain. Based on UPCD, in addition to the lack of a predicted signal peptide and the absence of transmembrane domains, we predict the hypothetical protein WHO_F_01144c (7.4 kDa) is cytoplasmic. The impact of these six proteins on AMR is yet to be elucidated; however, our data suggest that they may represent general proteomic markers for gonococcal AMR, a predisposition toward developing or compensating for gonococcal AMR, and/or new antimicrobial targets.
Phenotypic Clustering Based on Antibiograms and Common Differentially Expressed Proteins
To link AMR phenotypes with proteomic signatures, we performed phenotypic clustering of gonococcal strains based on their defined antibiograms (50, 53–55, 150) and common differentially expressed proteins (Tables IV–V). We additionally investigated each protein's Cluster of Orthologous Genes (COG) annotations and inferred the functional relevance to the observed phenotypes. These analyses generated seven phenotypic clusters that matched between established and proteome-derived AMR signatures (I-VII; Tables IV–VI).
Table VI. Common differentially expressed proteins in phenotypically clustered Neisseria gonorrhoeae strains.
| Cluster I |
Cluster II |
Cluster III |
Cluster IV |
Cluster V |
Cluster VI |
Cluster VII |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Up-regulated | Down-regulated | Up-regulated | Down-regulated | Up-regulated | Down-regulated | Up-regulated | Down-regulated | Up-regulated | Down-regulated | Up-regulated | Down-regulated | Up-regulated | Down-regulated | |
| Amino acid metabolism | NGO0185 | NGO0269 NGO0679 | NGO1752 | NGO0779 | NGO0779 | NGO0185 | NGO0192 | |||||||
| NGO0269 | ||||||||||||||
| NGO0679 | ||||||||||||||
| Cell envelope biogenesis | NGO0085 | NGO2071 | ||||||||||||
| NGO0456 | ||||||||||||||
| LolA | ||||||||||||||
| Coenzyme metabolism | NGO2074 | NGO2056 | NGO2056 | MdaB | MdaB | NGO0059 | NGO2074 | NGO2056 | NGO2056 | |||||
| NGO0360 | NGO0360 | |||||||||||||
| DNA repair | NGO1707 | NGO0078 | WHO_F_02198c | NGO0078 | ||||||||||
| NGO0407 | NGO0407 | |||||||||||||
| UvrB | UvrB | |||||||||||||
| Energy production | WHO_F_02308c | NGO0143 | WHO_F_02308 | |||||||||||
| Hypothetical protein | NGO1239 | WHO_F_00875c, | NGO1942 | WHO_F_00875c | WHO_F_00875c | NGO1141 | NGO1239 | NGO1628 | NGO1299 | |||||
| NGO1958 | NGO1299 | NGO2097 | NGO1299 | NGO1966 | NGO1945 | |||||||||
| NGO1969 | NGO1945 | NGO7315 | NGO1969 | |||||||||||
| NGO1970 | NGO1967 | NGO1970 | ||||||||||||
| NGO2089 | NGO1969 | NGO2089 | ||||||||||||
| NGO1970 | ||||||||||||||
| NGO2089 | ||||||||||||||
| Inorganic ion metabolism | NGO0952 | NGO1059 | NGO0952 | NGO1411 WHO_F_00481c | NGO2137 | WHO_F_00481 | NGO0291 | |||||||
| NGO2137 | NGO1188 | |||||||||||||
| Intracellular trafficking and secretion | PilE | WHO_F_02394c | PilE | NGO0456 | NGO0452 | |||||||||
| NGO0454 | ||||||||||||||
| PilE | ||||||||||||||
| Lipid metabolism | NGO1243 | NGO1763 | NGO0624 | NGO1507 | NGO1507 | NGO1507 | NGO1243 | |||||||
| NGO1906 | NGO1055 | NGO1710 | ||||||||||||
| NGO1243 | ||||||||||||||
| Protein turnover | NGO0399 NGO1655 | NGO0926 | NGO0057 | NGO0057 | ||||||||||
| Translation and Ribosomal biogenesis | S15, S19, EngA, RNA helicase, Ribonuclease E L1, L2, L22 , NGO0657 | NGO0803 | NGO1870 | |||||||||||
| NGO1870 | ||||||||||||||
Cluster I strains, WHO P and U, exhibit resistance to azithromycin and the majority of upregulated proteins identified were involved in ribosomal biogenesis: 30S ribosomal proteins S15 and S19; 50S ribosomal proteins L1, L2, and L22; the small GTPase EngA; pseudouridine synthase; RNA helicase; and ribonuclease E. In contrast, proteins involved in cell envelope biogenesis - PilE, LolA, and PglB - were downregulated in both strains, which may be associated with the strains' decreased susceptibility to penicillin G.
Cluster II strains, WHO M and N, exhibit resistance to penicillin G, tetracycline, and ciprofloxacin. Upregulated proteins included DNA repair factors (DnaE, a putative type I-site specific deoxyribonuclease NGO0407, and exonuclease UvrB). Proteins involved in amino acid metabolism (NGO0269, NGO0679); and translation (NGO0803, NGO1870) were also upregulated, which suggested strains in Cluster II possess compensatory mechanisms for ciprofloxacin and tetracycline resistance, respectively. Hypothetical proteins represented most downregulated proteins and included WHO_F_00875c, NGO1299, NGO1945, NGO1967, NGO1969, NGO1970, and NGO2089.
Cluster III, IV, and V are comprised of WHO X and L, WHO W and K, and WHO Y and Z, respectively, and exhibit resistance to at least four different antimicrobials, with ciprofloxacin and tetracycline in common (Table IV-V). Three proteins in common between strains in Cluster III and IV were identified. Of these, homoserine dehydrogenase and holo-ACP synthase—involved in amino acid and lipid metabolism, respectively—were downregulated, whereas thioredoxin, which is involved in defense against oxidative stress and protein turnover, was upregulated (151, 152). The NADP quinone reductase (MdaB, modulator of drug activity B) was upregulated in Cluster IV and V strains. In E. coli, MdaB protects against polyketide compound toxicity (153), whereas overproduction of this protein defends P. aeruginosa from oxidative stress (154).
Strains in Cluster VI (WHO V and G) are resistant to penicillin G (WHO G intermediate susceptible), tetracycline and ciprofloxacin. Differentially regulated proteins in this cluster were strikingly like Cluster II, with 13 proteins in common (Table IV–VI). Seven proteins involved in DNA repair, amino acid metabolism and translation were upregulated, further strengthening a possible compensatory mechanism for the resistance to ciprofloxacin and tetracycline. Six proteins functioning in coenzyme metabolism (NGO2056) and with unknown functions (NGO1299, NGO1945, NGO1969, NGO1970, NGO2089) were downregulated (Table VI).
Finally, the cluster VII strains (WHO O and FA6140), displaying resistance to penicillin G and tetracycline, had eight common differentially expressed proteins (Table VI). Among these proteins, two metabolic coenzymes (NGO0360 and NGO2056) and a putative cytochrome b561 involved in energy production were downregulated. This cluster possessed a similar expression profile to strains in Cluster I that are intermediately susceptibility to penicillin G and tetracycline. Finally, NGO2071, a putative integral inner membrane protein; NGO0291, a potassium proton/antiporter; NGO452, annotated as a putative FimT protein; PilW (NGO0454); and PilE were also downregulated in the cluster VII strains (Table VI).
Our proteomic findings elucidate many differentially regulated proteins as potential general proteomic markers for gonococcal AMR, a predisposition toward developing or compensating for gonococcal AMR, and/or new antimicrobial targets, e.g. NGO0222, WHO_F_01126, NGO0597, NGO0701, WHO_F_01139, WHO_F_01144c. Deeper analysis of gonococcal proteotypes that relied on AMR-based phenotypic clustering identified additional proteomic markers potentially associated with (or compensating for) AMR in clusters I, II, VI, and VII. Further studies should examine the proteomic profiles of wild type and AMR gonococcal strains during exposure to varying levels of different antimicrobials. In line with this, the expression of eight outer membrane proteins was enhanced in ampicillin resistant E. coli strains upon exposure to the minimal inhibitory concentration of ampicillin (33). Additionally, the functional role(s) of the differentially regulated hypothetical proteins potentially involved in gonococcal AMR need to be elucidated, which would help decode the intricate AMR network and promote the design of ways to curb the spread of AMR among N. gonorrhoeae strains.
CONCLUSIONS
The present study provides the first global quantitative proteomic characterization of the 2016 WHO N. gonorrhoeae reference strains (50) and FA6140 (51) to identify new vaccine candidates, gain information about expression of previously identified antigens, and enhance our understanding of AMR in N. gonorrhoeae. To our knowledge, this is also the largest quantitative proteomics study performed on bacterial subproteomes to date. Importantly, nine novel vaccine candidates have been identified, significantly broadening the gonorrhea antigen repertoire. Further, expression of 21 previously verified AMR determinants at the proteome level was investigated and six new proteomic signatures were identified that may be associated with AMR or may indicate a strain's likelihood of developing or compensating for the physiological consequences of gonococcal AMR. The proteomic signatures we identified may also represent new antimicrobial targets. Expression patterns of antimicrobial targets and AMR determinants provide proteomic signatures that can complement, verify, and enhance our phenotypic- and genetic-derived understanding of gonococcal AMR complexity. Considering that commensal Neisseria species are among the most highly abundant bacteria of the human oropharyngeal microbiota, in addition to the substantial genetic, molecular and phenotypic diversity existing within the Neisseria genus, further analyses of the conservation of gonococcal vaccine candidates and AMR determinants in additional commensal Neisseria species are crucial.
Cumulatively, our studies provide a wealth of information regarding gonococcal proteomic profiles and will contribute to ongoing efforts in vaccine/drug development as well as elucidation of AMR mechanisms in N. gonorrhoeae.
DATA AVAILABILITY
The raw mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (66) partner repository with the data set identifier PXD008412.
Supplementary Material
Acknowledgments
We thank Benjamin I. Baarda for critical reading of this manuscript.
Footnotes
* This work was supported by the National Institute of Allergy and Infectious Diseases R01-AI117235 to A.E.S., and the World Health Organization, Geneva, Switzerland (2015) and the Foundation for Medical Research at Örebro University Hospital, Sweden (2016) to M.U.
 This article contains supplemental Tables.
This article contains supplemental Tables.
1 The abbreviations used are:
- WHO
- World Health Organization
- ACN
- acetonitrile
- AGC
- automatic gain control
- AMR
- antimicrobial resistance
- C
- cytoplasmic
- CDC
- centers for disease control and prevention
- CE
- cell envelope
- COG
- cluster of orthologous genes
- cRAP
- common repository of adventitious proteins
- FDR
- false discovery rate
- GCB
- gonococcal base agar
- GCBL
- gonococcal base liquid medium
- KEGG
- Kyoto encyclopedia of genes and genomes
- LPS
- lipopolysaccharide
- OMV
- outer membrane vesicle
- ORF
- open reading frame.
REFERENCES
- 1. Newman L, Rowley J, Vander Hoorn S, Wijesooriya NS, Unemo M, Low N, et al. (2015) Global Estimates of the Prevalence and Incidence of Four Curable Sexually Transmitted Infections in 2012 Based on Systematic Review and Global Reporting. PloS One. 10, e0143304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. (CDC) CfDCP. Gonorrhea statistics 2017 [Available from: https://www.cdc.gov/std/stats16/gonorrhea.htm.
- 3. Kent C. K., Chaw J. K., Wong W., Liska S., Gibson S., Hubbard G., et al. (2005) Prevalence of rectal, urethral, and pharyngeal chlamydia and gonorrhea detected in 2 clinical settings among men who have sex with men: San Francisco, California, 2003. Clin. Infect. Dis. 41, 67–74 [DOI] [PubMed] [Google Scholar]
- 4. Newman L. M., Dowell D., Bernstein K., Donnelly J., Martins S., Stenger M., et al. (2012) A tale of two gonorrhea epidemics: results from the STD surveillance network. Public Health Rep. 127, 282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cohen M. S., Hoffman I. F., Royce R. A., Kazembe P., Dyer J. R., Daly C. C., et al. (1997) Reduction of concentration of HIV-1 in semen after treatment of urethritis: implications for prevention of sexual transmission of HIV-1. AIDSCAP Malawi Research Group. Lancet 349, 1868–1873 [DOI] [PubMed] [Google Scholar]
- 6. Unemo M., Bradshaw C. S., Hocking J. S., de Vries H. J. C., Francis S. C., Mabey D., et al. (2017) Sexually transmitted infections: challenges ahead. Lancet Infect. Dis. 17, e235–e279 [DOI] [PubMed] [Google Scholar]
- 7. Wi T, Lahra MM, Ndowa F, Bala M, Dillon JR, Ramon-Pardo P, et al. (2017) Antimicrobial resistance in Neisseria gonorrhoeae: Global surveillance and a call for international collaborative action. PLoS Med. 14, e1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fifer H., Natarajan U., Jones L., Alexander S., Hughes G., Golparian D., et al. (2016) Failure of Dual Antimicrobial Therapy in Treatment of Gonorrhea. N. Engl. J. Med. 374, 2504–2506 [DOI] [PubMed] [Google Scholar]
- 9. England PH. (2018) UK case of Neisseria gonorrhoeae with high-level resistance to azithromycin and resistance to ceftriaxone acquired abroad. 29 March 2018. Contract No.: 11 [Google Scholar]
- 10. Eyre DW, Sanderson ND, Lord E, Regisford-Reimmer N, Chau K, Barker L, et al. Gonorrhoea treatment failure caused by a Neisseria gonorrhoeae strain with combined ceftriaxone and high-level azithromycin resistance, England, February 2018. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murphy B. Multi-drug resistant gonorrhoea Australia: Australian Government Department of Health; 2018. [updated 17 April 2018] [Google Scholar]
- 12. Whiley D. M., Jennison A., Pearson J., and Lahra M. M. (2018) Genetic characterisation of Neisseria gonorrhoeae resistant to both ceftriaxone and azithromycin. Lancet Infect. Dis. 18, 717–718 [DOI] [PubMed] [Google Scholar]
- 13. Alirol E, Wi TE, Bala M, Bazzo ML, Chen XS, Deal C, et al. (2017) Multidrug-resistant gonorrhea: A research and development roadmap to discover new medicines. PLoS Med. 14, e1002366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hampton L. M., Farley M. M., Schaffner W., Thomas A., Reingold A., Harrison L. H., et al. (2012) Prevention of antibiotic-nonsusceptible Streptococcus pneumoniae with conjugate vaccines. J. Infect. Dis. 205, 401–411 [DOI] [PubMed] [Google Scholar]
- 15. Greenberg L., Diena B. B., Ashton F. A., Wallace R., Kenny C. P., Znamirowski R., et al. (1974) Gonococcal vaccine studies in Inuvik. Can. J. Public Health 65, 29–33 [PubMed] [Google Scholar]
- 16. Boslego J. W., Tramont E. C., Chung R. C., McChesney D. G., Ciak J., Sadoff J. C., et al. (1991) Efficacy trial of a parenteral gonococcal pilus vaccine in men. Vaccine 9, 154–162 [DOI] [PubMed] [Google Scholar]
- 17. Tramont EC. (1989) Gonococcal vaccines. Clin. Microbiol. Rev. 2, S74–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taylor-Robinson D., Furr P. M., and Hetherington C. M. (1990) Neisseria gonorrhoeae colonises the genital tract of oestradiol-treated germ-free female mice. Microb. Pathog. 9, 369–373 [DOI] [PubMed] [Google Scholar]
- 19. Rice P. A., Shafer W. M., Ram S., and Jerse A. E. (2017) Neisseria gonorrhoeae: Drug Resistance, mouse models, and vaccine development. Ann. Rev. Microbiol. 71, 665–686 [DOI] [PubMed] [Google Scholar]
- 20. Normark S., Albiger B., and Jonsson A. B. (2002) Gonococci cause immunosuppression by engaging a coinhibitory receptor on T lymphocytes. Nat. Immunol. 3, 210–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lewis L. A., Choudhury B., Balthazar J. T., Martin L. E., Ram S., Rice P. A., et al. (2009) Phosphoethanolamine substitution of lipid A and resistance of Neisseria gonorrhoeae to cationic antimicrobial peptides and complement-mediated killing by normal human serum. Infection Immunity. 77, 1112–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Packiam M., Wu H., Veit S. J., Mavrogiorgos N., Jerse A. E., and Ingalls R. R. (2012) Protective role of Toll-like receptor 4 in experimental gonococcal infection of female mice. Mucosal Immunol. 5, 19–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Packiam M., Yedery R. D., Begum A. A., Carlson R. W., Ganguly J., Sempowski G. D., et al. (2014) Phosphoethanolamine decoration of Neisseria gonorrhoeae lipid A plays a dual immunostimulatory and protective role during experimental genital tract infection. Infection Immunity. 82, 2170–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sintsova A., Wong H., MacDonald K. S., Kaul R., Virji M., and Gray-Owen S. D. (2015) Selection for a CEACAM receptor-specific binding phenotype during Neisseria gonorrhoeae infection of the human genital tract. Infection Immunity. 83, 1372–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y., Liu W., and Russell M. W. (2014) Suppression of host adaptive immune responses by Neisseria gonorrhoeae: role of interleukin 10 and type 1 regulatory T cells. Mucosal Immunol. 7, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu Y., Hammer L. A., Liu W., Hobbs M. M., Zielke R. A., Sikora A. E., et al. (2017) Experimental vaccine induces Th1-driven immune responses and resistance to Neisseria gonorrhoeae infection in a murine model. Mucosal Immunol. 10, 1594–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Petousis-Harris H., Paynter J., Morgan J., Saxton P., McArdle B., Goodyear-Smith F., et al. (2017) Effectiveness of a group B outer membrane vesicle meningococcal vaccine against gonorrhoea in New Zealand: a retrospective case-control study. Lancet 390, 1603–1610 [DOI] [PubMed] [Google Scholar]
- 28. Unemo M., and Sikora A. E. (2017) Infection: Proof of principle for effectiveness of a gonorrhoea vaccine. Nat. Rev. Urol. 14, 643–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zielke R. A., Wierzbicki I. H., Weber J. V., Gafken P. R., and Sikora A. E. (2014) Quantitative proteomics of the Neisseria gonorrhoeae cell envelope and membrane vesicles for the discovery of potential therapeutic targets. Mol. Cell. Proteomics 13, 1299–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zielke R. A., Wierzbicki I. H., Baarda B. I., Gafken P. R., Soge O. O., Holmes K. K., et al. (2016) Proteomics-driven Antigen Discovery for Development of Vaccines Against Gonorrhea. Mol. Cell. Proteomics 15, 2338–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baarda B. I., and Sikora A. E. (2015) Proteomics of Neisseria gonorrhoeae: the treasure hunt for countermeasures against an old disease. Front. Microbiol. 6, 1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Otto A., Becher D., and Schmidt F. (2014) Quantitative proteomics in the field of microbiology. Proteomics 14, 547–565 [DOI] [PubMed] [Google Scholar]
- 33. Xu C., Lin X., Ren H., Zhang Y., Wang S., and Peng X. (2006) Analysis of outer membrane proteome of Escherichia coli related to resistance to ampicillin and tetracycline. Proteomics 6, 462–473 [DOI] [PubMed] [Google Scholar]
- 34. Al-Haroni M., Skaug N., Bakken V., and Cash P. (2008) Proteomic analysis of ampicillin-resistant oral Fusobacterium nucleatum. Oral Microbiol. Immunol. 23, 36–42 [DOI] [PubMed] [Google Scholar]
- 35. Giddey A. D., de Kock E., Nakedi K. C., Garnett S., Nel A. J., Soares N. C., et al. (2017) A temporal proteome dynamics study reveals the molecular basis of induced phenotypic resistance in Mycobacterium smegmatis at sub-lethal rifampicin concentrations. Sci. Reports 7, 43858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Len A. C., Harty D. W., and Jacques N. A. (2004) Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology 150, 1339–1351 [DOI] [PubMed] [Google Scholar]
- 37. Liebeke M., Dorries K., Zuhlke D., Bernhardt J., Fuchs S., Pane-Farre J., et al. (2011) A metabolomics and proteomics study of the adaptation of Staphylococcus aureus to glucose starvation. Mol. bioSystems. 7, 1241–1253 [DOI] [PubMed] [Google Scholar]
- 38. Alreshidi M. M., Dunstan R. H., Macdonald M. M., Smith N. D., Gottfries J., and Roberts T. K. (2015) Metabolomic and proteomic responses of Staphylococcus aureus to prolonged cold stress. J. Proteomics 121, 44–55 [DOI] [PubMed] [Google Scholar]
- 39. Baarda B. I., Emerson S., Proteau P. J., and Sikora A. E. (2017) Deciphering the function of new gonococcal vaccine antigens using phenotypic microarrays. J Bacteriol. 199, e00037-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Christodoulides M. (2014) Neisseria proteomics for antigen discovery and vaccine development. 11, 573–591 [DOI] [PubMed] [Google Scholar]
- 41. Otto A., Bernhardt J., Hecker M., and Becher D. (2012) Global relative and absolute quantitation in microbial proteomics. Curr. Opin. Microbiol. 15, 364–372 [DOI] [PubMed] [Google Scholar]
- 42. Adamczyk-Poplawska M., Markowicz S., and Jagusztyn-Krynicka E. K. (2011) Proteomics for development of vaccine. J. Proteomics 74, 2596–2616 [DOI] [PubMed] [Google Scholar]
- 43. Zielke RA, Sikora AE (2014) Isolation of cell envelopes and naturally released membrane vesicles of Neisseria gonorrhoeae. Curr. Protoc. Microbiol. 34, 4A.3.1–17 [DOI] [PubMed] [Google Scholar]
- 44. Olaya-Abril A., Jimenez-Munguia I., Gomez-Gascon L., and Rodriguez-Ortega M. J. (2014) Surfomics: shaving live organisms for a fast proteomic identification of surface proteins. J. Proteomics 97, 164–176 [DOI] [PubMed] [Google Scholar]
- 45. Zielke R. A., Wierzbicki I. H., Baarda B. I., Gafken P. R., Soge O. O., Holmes K. K., Jerse A. E., Unemo M., and Sikora A. E. (2016) Proteomics-driven Antigen Discovery for Development of Vaccines Against Gonorrhea. Mol Cell Proteomics 15, 2338–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yoo J. S., Seong W. K., Kim T. S., Park Y. K., Oh H. B., and Yoo C. K. (2007) Comparative proteome analysis of the outer membrane proteins of in vitro-induced multi-drug resistant Neisseria gonorrhoeae. Microbiol. Immunol. 51, 1171–1177 [DOI] [PubMed] [Google Scholar]
- 47. Nabu S., Lawung R., Isarankura-Na-Ayudhya P., Isarankura-Na-Ayudhya C., Roytrakul S., and Prachayasittikul V. (2014) Reference map and comparative proteomic analysis of Neisseria gonorrhoeae displaying high resistance against spectinomycin. J. Med. Microbiol. 63, 371–385 [DOI] [PubMed] [Google Scholar]
- 48. Smith S. M. (2017) Strategies for the purification of membrane proteins. Meth. Mol. Biol. 1485, 389–400 [DOI] [PubMed] [Google Scholar]
- 49. Zielke R. A., Gafken P. R., and Sikora A. E. (2014) Quantitative proteomic analysis of the cell envelopes and native membrane vesicles derived from gram-negative bacteria. Curr. Protoc. Microbiol. 34, 1F.3.1–16 [DOI] [PubMed] [Google Scholar]
- 50. Unemo M., Golparian D., Sanchez-Buso L., Grad Y., Jacobsson S., Ohnishi M., Lahra M. M., Limnios A., Sikora A. E., Wi T., and Harris S. R. (2016) The novel 2016 WHO Neisseria gonorrhoeae reference strains for global quality assurance of laboratory investigations: phenotypic, genetic and reference genome characterization. J. Antimicrob. Chemother. 71, 3096–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Faruki H., Kohmescher R. N., McKinney W. P., and Sparling P. F. (1985) A community-based outbreak of infection with penicillin-resistant Neisseria gonorrhoeae not producing penicillinase (chromosomally mediated resistance). N. Engl. J. Med. 313, 607–611 [DOI] [PubMed] [Google Scholar]
- 52. Unemo M., Fasth O., Fredlund H., Limnios A., and Tapsall J. (2009) Phenotypic and genetic characterization of the 2008 WHO Neisseria gonorrhoeae reference strain panel intended for global quality assurance and quality control of gonococcal antimicrobial resistance surveillance for public health purposes. J. Antimicrobial Chemotherapy 63, 1142–1151 [DOI] [PubMed] [Google Scholar]
- 53. Veal W. L., Nicholas R. A., and Shafer W. M. (2002) Overexpression of the MtrC-MtrD-MtrE efflux pump because of an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J. Bacteriol. 184, 5619–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hu M., Nandi S., Davies C., and Nicholas R. A. (2005) High-level chromosomally mediated tetracycline resistance in Neisseria gonorrhoeae results from a point mutation in the rpsJ gene encoding ribosomal protein S10 in combination with the mtrR and penB resistance determinants. Antimicrobial Agents Chemotherapy 49, 4327–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Powell A. J., Tomberg J., Deacon A. M., Nicholas R. A., and Davies C. (2009) Crystal structures of penicillin-binding protein 2 from penicillin-susceptible and -resistant strains of Neisseria gonorrhoeae reveal an unexpectedly subtle mechanism for antibiotic resistance. J. Biol. Chem. 284, 1202–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thompson A., Schafer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., et al. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 57. Spence JM, Wright L, Clark VL (2008) Laboratory maintenance of Neisseria gonorrhoeae. Curr. Protoc. Microbiol. Chapter 4, Unit 4A 1 [DOI] [PubMed] [Google Scholar]
- 58. Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G. M., Carnemolla B., et al. (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327–1333 [DOI] [PubMed] [Google Scholar]
- 59. Shevchenko A., Tomas H., Havlis J., Olsen J. V., and Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protocols. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 60. Moreno-Hagelsieb G., and Latimer K. (2008) Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 24, 319–324 [DOI] [PubMed] [Google Scholar]
- 61. Yu C. S., Chen Y. C., Lu C. H., and Hwang J. K. (2006) Prediction of protein subcellular localization. Proteins 64, 643–651 [DOI] [PubMed] [Google Scholar]
- 62. Yu N. Y., Wagner J. R., Laird M. R., Melli G., Rey S., Lo R., et al. (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Imai K., Asakawa N., Tsuji T., Akazawa F., Ino A., Sonoyama M., et al. (2008) SOSUI-GramN: high performance prediction for sub-cellular localization of proteins in gram-negative bacteria. Bioinformation 2, 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 65. Juncker A. S., Willenbrock H., Von Heijne G., Brunak S., Nielsen H., and Krogh A. (2003) Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12, 1652–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vizcaino J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Senko M. W., Remes P. M., Canterbury J. D., Mathur R., Song Q., Eliuk S. M., Mullen C., Earley L., Hardman M., Blethrow J.D., Bui H., Specht A., Lange O., Denisov E., Makarov A., Horning S., and Zabrouskov V. (2013) Novel parallelized quadrupole/linear ion trap/Orbitrap tribrid mass spectrometer improving proteome coverage and peptide identification rates. Anal. Chem. 85, 11710–11714 [DOI] [PubMed] [Google Scholar]
- 68. Abrams AJ, Trees DL, Nicholas RA (2015) Complete genome sequences of three Neisseria gonorrhoeae laboratory reference strains, determined using PacBio Single-Molecule Real-Time technology. Genome Announc. 3, e01052–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Papanastasiou M., Orfanoudaki G., Koukaki M., Kountourakis N., Sardis M. F., Aivaliotis M., Karamanou S., and Economou A. (2013) The Escherichia coli peripheral inner membrane proteome. Mol. Cell. Proteomics 12, 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jan A. T. (2017) Outer Membrane Vesicles (OMVs) of Gram-negative Bacteria: A Perspective Update. Front Microbiol. 8, 1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zielke R. A., Wierzbicki I. H., Baarda B. I., Gafken P. R., Soge O. O., Holmes K. K., Jerse A. E., Unemo M., and Sikora A. E. (2016) Proteomics-driven antigen discovery for development of vaccines against gonorrhea. Mol. Cell. Proteomics 15, 2338–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sikora A. E., Wierzbicki I. H., Zielke R. A., Ryner R. F., Korotkov K. V., Buchanan S. K., and Noinaj N. (2018) Structural and functional insights into the role of BamD and BamE within the beta-barrel assembly machinery in Neisseria gonorrhoeae. J. Biol.Chem. 293, 1106–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zielke R. A., Le Van A., Baarda B. I., Herrera M. F., Acosta C. J., Jerse A. E., and Sikora S. E. (2018) SliC is a surface-displayed lipoprotein that is required for the anti-lysozyme strategy during Neisseria gonorrhoeae infection. PLoS Pathogens 14, e1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Semchenko E. A., Day C. J., and Seib K. L. (2017) MetQ of Neisseria gonorrhoeae is a surface-expressed antigen that elicits bactericidal and functional blocking antibodies. Infection Immunity 85, e00898–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Leuzzi R., Serino L., Scarselli M., Savino S., Fontana M. R., Monaci E., Taddei A., Fischer G., Rappuoli R., and Pizza M. (2005) Ng-MIP, a surface-exposed lipoprotein of Neisseria gonorrhoeae, has a peptidyl-prolyl cis/trans isomerase (PPIase) activity and is involved in persistence in macrophages. Mol. Microbiol. 58, 669–681 [DOI] [PubMed] [Google Scholar]
- 76. Wierzbicki IH, A ZR, Jerse AE, Begum AA, Unemo M, Sikora AE, editors. NGO1985 is a surface-exposed outer membrane lipoprotein involved in Neisseria gonorrhoeae cell envelope homeostasis and a novel gonorrhea vaccine candidate. International Pathogenic Neisseria Conference; 2016; Manchester, UK [Google Scholar]
- 77. Liu Y., Hammer L. A., Liu W., Hobbs M. M., Zielke R. A., Sikora A. E., Jerse A. E., Egilmez N .K., and Russell M. W. (2017) Experimental vaccine induces Th1-driven immune responses and resistance to Neisseria gonorrhoeae infection in a murine model. Mucosal Immunol. 5, 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lomholt H., Lind I., and Kilian M. (1995) Neisseria gonorrhoeae IgA1 proteases share epitopes recognized by neutralizing antibodies. Vaccine 13, 1213–1219 [DOI] [PubMed] [Google Scholar]
- 79. Malojcic G., Andres D., Grabowicz M., George A. H., Ruiz N., Silhavy T. J., and Kahne D. (2014) LptE binds to and alters the physical state of LPS to catalyze its assembly at the cell surface. Proc. Natl. Acad. Sci. U.S.A. 111, 9467–9472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bos M. P., and Tommassen J. (2011) The LptD chaperone LptE is not directly involved in lipopolysaccharide transport in Neisseria meningitidis. J. Biol. Chem. 286, 28688–28696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bos M. P., Tefsen B., Geurtsen J., and Tommassen J. (2004) Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl. Acad. Sci. U.S.A. 101, 9417–9422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zielke R. A., Simmons R. S., Park B. R., Nonogaki M., Emerson S., and Sikora A. E. (2014) The type II secretion pathway in Vibrio cholerae is characterized by growth-phase dependent expression of exoprotein genes and is positively regulated by sigmaE. Infection Immunity 82, 2788–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Braun M., and Silhavy T. J. (2002) Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol. 45, 1289–1302 [DOI] [PubMed] [Google Scholar]
- 84. Gotzke H., Palombo I., Muheim C., Perrody E., Genevaux P., Kudva R., Müller M., and Daley D. O. (2014) YfgM is an ancillary subunit of the SecYEG translocon in Escherichia coli. J. Biol. Chem. 289, 19089–19097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Matsuyama S., Yokota N., and Tokuda H. (1997) A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 16, 6947–6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Grabowicz M., and Silhavy T. J. (2017) Redefining the essential trafficking pathway for outer membrane lipoproteins. Proc. Natl. Acad. Sci. U.S.A. 114, 4769–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jongerius I., Lavender H., Tan L., Ruivo N., Exley R. M., Caesar J. J., Lea S. M., Johnson S., and Tang C. M. (2013) Distinct binding and immunogenic properties of the gonococcal homologue of meningococcal factor h binding protein. PLoS Pathog. 9, e1003528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Whitehead R. N., Overton T. W., Snyder L. A., McGowan S. J., Smith H., Cole J. A., and Saunders N. J. (2007) The small FNR regulon of Neisseria gonorrhoeae: comparison with the larger Escherichia coli FNR regulon and interaction with the NarQ-NarP regulon. BMC Genomics 8, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Alm R. A., Hallinan J. P., Watson A. A., and Mattick J. S. (1996) Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol. Microbiol. 22, 161–173 [DOI] [PubMed] [Google Scholar]
- 90. Sethi D., Mahajan S., Singh C., Lama A., Hade M. D., Gupta P., and Dikshit K. L (2016) Lipoprotein LprI of Mycobacterium tuberculosis Acts as a Lysozyme Inhibitor. J.Biol. Chem. 291, 2938–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ragland S. A., Humbert M. V., Christodoulides M., and Criss A. K. (2018) Neisseria gonorrhoeae employs two protein inhibitors to evade killing by human lysozyme. PLoS pathogens. 14, e1007080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. van Ulsen P., and Tommassen J. (2006) Protein secretion and secreted proteins in pathogenic Neisseriaceae. FEMS Microbiol Rev. 30, 292–319 [DOI] [PubMed] [Google Scholar]
- 93. Jerse A. E., Sharma N. D., Simms A. N., Crow E. T., Snyder L. A., and Shafer W. M. (2003) A gonococcal efflux pump system enhances bacterial survival in a female mouse model of genital tract infection. Infection Immunity 71, 5576–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Delahay R. M., Robertson B. D., Balthazar J. T., Shafer W. M., and Ison C. A. (1997) Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic agents. Microbiology 143, 2127–2133 [DOI] [PubMed] [Google Scholar]
- 95. Warner D. M., Folster J. P., Shafer W. M., and Jerse A. E. (2007) Regulation of the MtrC-MtrD-MtrE efflux-pump system modulates the in vivo fitness of Neisseria gonorrhoeae. J Infect Dis. 196, 1804–1812 [DOI] [PubMed] [Google Scholar]
- 96. Edwards J. L., Brown E. J., Uk-Nham S., Cannon J. G., Blake M. S., and Apicella M. A. (2002) A co-operative interaction between Neisseria gonorrhoeae and complement receptor 3 mediates infection of primary cervical epithelial cells. Cell. Microbiol. 4, 571–584 [DOI] [PubMed] [Google Scholar]
- 97. Ling A. E., Bash M. C., Lynn F., Lister N. A., Zhu P., Garland S. M., Fairley C. K., and Tabrizi S. N. (2007) Evaluation of PorB variable region typing of Neisseria gonorrhoeae using PCR-ELISA in samples collected from men who have sex with men. J. Clin. Lab. Anal. 21, 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Haghi F., Peerayeh S. N., Siadat S. D., and Zeighami H. (2012) Recombinant outer membrane secretin PilQ(406–770) as a vaccine candidate for serogroup B Neisseria meningitidis. Vaccine 30, 1710–1714 [DOI] [PubMed] [Google Scholar]
- 99. Price G. A., Masri H. P., Hollander A. M., Russell M. W., and Cornelissen C. N. (2007) Gonococcal transferrin binding protein chimeras induce bactericidal and growth inhibitory antibodies in mice. Vaccine 25, 7247–7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Noto J. M., and Cornelissen C. N. (2008) Identification of TbpA residues required for transferrin-iron utilization by Neisseria gonorrhoeae. Infection Immunity 76, 1960–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Callaghan M. J., Lewis S., Sadarangani M., Bailey S. E., Chan H., Ferguson D. J., Derrick J. P., Feavers I., Maiden M. C., and Pollard A. J. (2011) Potential of recombinant opa proteins as vaccine candidates against hyperinvasive meningococci. Infection Immunity 79, 2810–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Cole J. G., and Jerse A. E. (2009) Functional characterization of antibodies against Neisseria gonorrhoeae opacity protein loops. PloS One 4, e8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Boulanger M. J., and Murphy M. E. (2002) Crystal structure of the soluble domain of the major anaerobically induced outer membrane protein (AniA) from pathogenic Neisseria: a new class of copper-containing nitrite reductases. J. Mol. Biol. 315, 1111–1127 [DOI] [PubMed] [Google Scholar]
- 104. Shewell L. K., Ku S. C., Schulz B. L., Jen F. E., Mubaiwa T. D., Ketterer M. R., Apicella M. A., and Jennings M. P. (2013) Recombinant truncated AniA of pathogenic Neisseria elicits a non-native immune response and functional blocking antibodies. Biochem. Biophys. Res. Commun. 431, 215–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sikora A. E., Mills R. H., Weber J. V., Hamza A., Passow B. W., Romaine A., Williamson Z. A., Reed R. W., Zielke R. A., and Korotkov K. V. (2017) Peptide Inhibitors Targeting the Neisseria gonorrhoeae Pivotal Anaerobic Respiration Factor AniA. Antimicrobial Agents Chemotherapy 61, e00186–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Jerse A. E., Bash M. C., and Russell M. W. (2014) Vaccines against gonorrhea: current status and future challenges. Vaccine 32, 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bhat K. S., Gibbs C. P., Barrera O., Morrison S. G., Jahnig F., Stern A., Kupsch E. M., Meyer T. F., and Swanson J. (1991) The opacity proteins of Neisseria gonorrhoeae strain MS11 are encoded by a family of 11 complete genes. Mol. Microbiol. 5, 1889–1901 [DOI] [PubMed] [Google Scholar]
- 108. Noinaj N., Buchanan S. K., and Cornelissen C. N. (2012) The transferrin-iron import system from pathogenic Neisseria species. Mol. Microbiol. 86, 246–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Humbert M. V., Awanye A. M., Lian L. Y., Derrick J. P., and Christodoulides M. (2017) Structure of the Neisseria Adhesin Complex Protein (ACP) and its role as a novel lysozyme inhibitor. PLoS Pathog. 13, e1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hung M. C., Heckels J. E., and Christodoulides M. (2013) The adhesin complex protein (ACP) of Neisseria meningitidis is a new adhesin with vaccine potential. mBio. 4, mBio.00041-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Keiser P. B., Biggs-Cicatelli S., Moran E. E., Schmiel D. H., Pinto V. B., Burden R. E., Miller L. B., Moon J. E., Bowden R. A., Cummings J. F., and Zollinger W. D. (2011) A phase 1 study of a meningococcal native outer membrane vesicle vaccine made from a group B strain with deleted lpxL1 and synX, over-expressed factor H binding protein, two PorAs and stabilized OpcA expression. Vaccine 29, 1413–1420 [DOI] [PubMed] [Google Scholar]
- 112. Moore J., Bailey S. E., Benmechernene Z., Tzitzilonis C., Griffiths N. J., Virji M., and Derrick J. P. (2005) Recognition of saccharides by the OpcA, OpaD, and OpaB outer membrane proteins from Neisseria meningitidis. J. Biol. Chem. 280, 31489–31497 [DOI] [PubMed] [Google Scholar]
- 113. Li G., Jiao H., Jiang G., Wang J., Zhu L., Xie R., Yan H., Chen H., and Ji M. (2011) Neisseria gonorrhoeae NspA induces specific bactericidal and opsonic antibodies in mice. Clin. Vaccine Immunol. 18, 1817–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Pettersson A., Kortekaas J., Weynants V. E., Voet P., Poolman J. T., Bos M. P., and Tommassen J. (2006) Vaccine potential of the Neisseria meningitidis lactoferrin-binding proteins LbpA and LbpB. Vaccine 24, 3545–3557 [DOI] [PubMed] [Google Scholar]
- 115. Holst J., Oster P., Arnold R., Tatley M. V., Naess L. M., Aaberge I. S., Galloway Y., McNicholas A., O'Hallahan J., Rosenqvist E., and Black S. (2013) Vaccines against meningococcal serogroup B disease containing outer membrane vesicles (OMV): lessons from past programs and implications for the future. Hum. Vaccin. Immunother. 9, 1241–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Fredricks D. N., Fiedler T. L., and Marrazzo J. M. (2005) Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med. 353, 1899–1911 [DOI] [PubMed] [Google Scholar]
- 117. Nelson D. E., Dong Q., Van der Pol B., Toh E., Fan B., Katz B. P., Mi D., Rong R., Weinstock G. M., Sodergren E., and Fortenberry J. D. (2012) Bacterial communities of the coronal sulcus and distal urethra of adolescent males. PloS One. 7, e36298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lim R. B., Wong M. L., Cook A. R., Brun C., Chan R. K., Sen P., and Chio M. (2015) Determinants of Chlamydia, Gonorrhea, and Coinfection in Heterosexual Adolescents Attending the National Public Sexually Transmitted Infection Clinic in Singapore. Sexually Transmitted Diseases 42, 450–456 [DOI] [PubMed] [Google Scholar]
- 119. Oriel J. D., Reeve P., Thomas B. J., and Nicol C. S. (1975) Infection with Chlamydia group A in men with urethritis due to Neisseria gonorrhoeae. J. Infect. Dis. 131, 376–382 [DOI] [PubMed] [Google Scholar]
- 120. Moller B. R., Mardh P. A., Ahrons S., and Nussler E. (1981) Infection with Chlamydia trachomatis, Mycoplasma hominis and Neisseria gonorrhoeae in patients with acute pelvic inflammatory disease. Sexually Transmitted Diseases 8, 198–202 [PubMed] [Google Scholar]
- 121. Vonck R. A., Darville T., O'Connell C. M., and Jerse A. E. (2011) Chlamydial infection increases gonococcal colonization in a novel murine coinfection model. Infection Immunity 79, 1566–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Sparling P. F. (1972) Antibiotic resistance in Neisseria gonorrhoeae. Med. Clin. N. Am. 56, 1133–1144 [DOI] [PubMed] [Google Scholar]
- 123. Maness M. J., and Sparling P. F. (1973) Multiple antibiotic resistance due to a single mutation in Neisseria gonorrhoeae. J. Infect. Dis. 128, 321–330 [DOI] [PubMed] [Google Scholar]
- 124. Sarubbi F. A. Jr, Blackman E., and Sparling P. F. (1974) Genetic mapping of linked antibiotic resistance loci in Neisseria gonorrhoeae. J. Bacteriol. 120, 1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Sparling P. F., Sarubbi F. A. Jr, and Blackman E. (1975) Inheritance of low-level resistance to penicillin, tetracycline, and chloramphenicol in Neisseria gonorrhoeae. J. Bacteriol. 124, 740–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Biswas G., Comer S., and Sparling P. F. (1976) Chromosomal location of antibiotic resistance genes in Neisseria gonorrhoeae. J. Bacteriol. 125, 1207–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Unemo M., and Shafer W. M. (2014) Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin. Microbiol. Rev. 27, 587–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ohnishi M., Golparian D., Shimuta K., Saika T., Hoshina S., Iwasaku K., Nakayama S., Kitawaki J., and Unemo M. (2011) Is Neisseria gonorrhoeae initiating a future era of untreatable gonorrhea?: detailed characterization of the first strain with high-level resistance to ceftriaxone. Antimicrobial Agents Chemotherapy. 55, 3538–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lindberg R., Fredlund H., Nicholas R., and Unemo M. (2007) Neisseria gonorrhoeae isolates with reduced susceptibility to cefixime and ceftriaxone: association with genetic polymorphisms in penA, mtrR, porB1b, and ponA. Antimicrobial Agents Chemotherapy. 51, 2117–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zhao S., Duncan M., Tomberg J., Davies C., Unemo M., and Nicholas R. A. (2009) Genetics of chromosomally mediated intermediate resistance to ceftriaxone and cefixime in Neisseria gonorrhoeae. Antimicrobial Agents Chemotherapy. 53, 3744–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Grad Y. H., Harris S. R., Kirkcaldy R. D., Green A. G., Marks D. S., Bentley S. D., Trees D., and Lipsitch M. (2016) Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000–2013. J. Infect. Dis. 214, 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Nabu S., Lawung R., Isarankura-Na-Ayudhya P., Roytrakul S., Dolprasit S., Sengyee S., Isarankura-Na-Ayudhya C., and Prachayasittikul V. (2017) Comparative proteomics analysis of Neisseria gonorrhoeae strains in response to extended-spectrum cephalosporins. EXCLI J. 16, 1207–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Golparian D., Shafer W. M., Ohnishi M., and Unemo M. (2014) Importance of multidrug efflux pumps in the antimicrobial resistance property of clinical multidrug-resistant isolates of Neisseria gonorrhoeae. Antimicrobial Agents Chemotherapy 58, 3556–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Veal W. L., Yellen A., Balthazar J. T., Pan W., Spratt B. G., and Shafer W. M. (1998) Loss-of-function mutations in the mtr efflux system of Neisseria gonorrhoeae. Microbiology 144, 621–627 [DOI] [PubMed] [Google Scholar]
- 135. William M. Shafer EWY, Corinne Rouquette-Loughlin, Daniel Golparian, Ann E. Jerse, Magnus Unemo. Efflux Pumps in Neisseria gonorrhoeae: Contributions to Antimicrobial Resistance and Virulence. In: Xian-Zhi Li CAE, Helen I. Zgurskaya, editor. Efflux-Mediated Antimicrobial Resistance in Bacteria: Mechanisms, Regulation and Clinical Implications Springer; 2016. p. 439–469 [Google Scholar]
- 136. Hagman K. E., Pan W., Spratt B. G., Balthazar J. T., Judd R. C., and Shafer W. M. (1995) Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141, 611–622 [DOI] [PubMed] [Google Scholar]
- 137. Zarantonelli L., Borthagaray G., Lee E. H., and Shafer W. M. (1999) Decreased azithromycin susceptibility of Neisseria gonorrhoeae due to mtrR mutations. Antimicrobial Agents Chemotherapy 43, 2468–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Ohneck EA, Zalucki YM, Johnson PJ, Dhulipala V, Golparian D, Unemo M, Jerse A. E., and Shafer W. M. (2011) A novel mechanism of high-level, broad-spectrum antibiotic resistance caused by a single base pair change in Neisseria gonorrhoeae. mBio. 2, mBio.00187-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Rouquette-Loughlin C. E., Balthazar J. T., and Shafer W. M. (2005) Characterization of the MacA-MacB efflux system in Neisseria gonorrhoeae. J. Antimicrobial Chemotherapy 56, 856–860 [DOI] [PubMed] [Google Scholar]
- 140. Lee E. H., and Shafer W. M. (1999) The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol. Microbiol. 33, 839–845 [DOI] [PubMed] [Google Scholar]
- 141. Rouquette-Loughlin C., Dunham S. A., Kuhn M., Balthazar J. T., and Shafer W. M. (2003) The NorM efflux pump of Neisseria gonorrhoeae and Neisseria meningitidis recognizes antimicrobial cationic compounds. J Bacteriol. 185, 1101–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Su C. C., Bolla J. R., Kumar N., Radhakrishnan A., Long F., Delmar J. A., Chou T. H., Rajashankar K. R., Shafer W. M., and Yu E. (2015) Structure and function of Neisseria gonorrhoeae MtrF illuminates a class of antimetabolite efflux pumps. Cell Rep. 11, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Deo P., Chow S. H., Hay I. D., Kleifeld O., Costin A., Elgass K. D., Jiang J. H., Ramm G., Gabriel K., Dougan G., Lithgow T., Heinz E., and Naderer T. (2018) Outer membrane vesicles from Neisseria gonorrhoeae target PorB to mitochondria and induce apoptosis. PLoS Pathog. 14, e1006945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Zhu W., Tomberg J., Knilans K. J., Anderson J. E., McKinnon K. P., Sempowski G. D., Nicholas R. A., and Duncan J. A. (2018) Properly folded and functional PorB from Neisseria gonorrhoeae inhibits dendritic cell stimulation of CD4(+) T cell proliferation. J. Biol. Chem. 293, 11218–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Levit M. N., Abramczyk B. M., Stock J. B., and Postel E. H. (2002) Interactions between Escherichia coli nucleoside-diphosphate kinase and DNA. J. Biol. Chem. 277, 5163–5167 [DOI] [PubMed] [Google Scholar]
- 146. Yu H., Xiong J., Zhang R., Hu X., Qiu J., Zhang D., Xu X., Xin R., He X., Xie W., Sheng H., Chen Q., Zhang L., Rao X., and Zhang K. (2016) Ndk, a novel host-responsive regulator, negatively regulates bacterial virulence through quorum sensing in Pseudomonas aeruginosa. Sci. Reports. 6, 28684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Mukhopadhyay S., Shankar S., Walden W., and Chakrabarty A. M. (1997) Complex formation of the elongation factor Tu from Pseudomonas aeruginosa with nucleoside diphosphate kinase modulates ribosomal GTP synthesis and peptide chain elongation. J. Biol. Chem. 272, 17815–17820 [DOI] [PubMed] [Google Scholar]
- 148. Sun J., Wang X., Lau A., Liao T. Y., Bucci C., and Hmama Z. (2010) Mycobacterial nucleoside diphosphate kinase blocks phagosome maturation in murine RAW 264.7 macrophages. PloS One 5, e8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Kolli B. K., Kostal J., Zaborina O., Chakrabarty A. M., and Chang K. P. (2008) Leishmania-released nucleoside diphosphate kinase prevents ATP-mediated cytolysis of macrophages. Mol. Biochem. Parasitol. 158, 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Sun J., Singh V., Lau A., Stokes R. W., Obregon-Henao A., Orme I. M., Wong D., Av-Gay Y., and Hmama Z. (2013) Mycobacterium tuberculosis nucleoside diphosphate kinase inactivates small GTPases leading to evasion of innate immunity. PLoS Pathogens 9, e1003499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Chopra P., Singh A., Koul A., Ramachandran S., Drlica K., Tyagi A. K., and Singh Y. (2003) Cytotoxic activity of nucleoside diphosphate kinase secreted from Mycobacterium tuberculosis. Eur. J. Biochem. 270, 625–634 [DOI] [PubMed] [Google Scholar]
- 152. Tomberg J., Unemo M., Davies C., and Nicholas R. A. (2010) Molecular and structural analysis of mosaic variants of penicillin-binding protein 2 conferring decreased susceptibility to expanded-spectrum cephalosporins in Neisseria gonorrhoeae: role of epistatic mutations. Biochemistry 49, 8062–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Archibald F. S., and Duong M. N. (1986) Superoxide dismutase and oxygen toxicity defenses in the genus Neisseria. Infection Immunity 51, 631–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Belenky P., Ye J. D., Porter C. B., Cohen N. R., Lobritz M. A., Ferrante T., Jain S., Korry B. J., Schwarz E. G., Walker G. C., and Collins J. J. (2015) Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep. 13, 968–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Adams M. A., and Jia Z. (2006) Modulator of drug activity B from Escherichia coli: crystal structure of a prokaryotic homologue of DT-diaphorase. J. Mol. Biol. 359, 455–465 [DOI] [PubMed] [Google Scholar]
- 156. Green L. K., La Flamme A. C., and Ackerley D. F. (2014) Pseudomonas aeruginosa MdaB and WrbA are water-soluble two-electron quinone oxidoreductases with the potential to defend against oxidative stress. J. Microbiol. 52, 771–777 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (66) partner repository with the data set identifier PXD008412.