Abstract
N-Glycosylation is a fundamentally important protein modification with a major impact on glycoprotein characteristics such as serum half-life and receptor interaction. More than half of the proteins in human serum are glycosylated, and the relative abundances of protein glycoforms often reflect alterations in health and disease. Several analytical methods are currently capable of analyzing the total serum N-glycosylation in a high-throughput manner.
Here we evaluate and compare the performance of three high-throughput released N-glycome analysis methods. Included were hydrophilic-interaction ultra-high-performance liquid chromatography with fluorescence detection (HILIC-UHPLC-FLD) with 2-aminobenzamide labeling of the glycans, multiplexed capillary gel electrophoresis with laser-induced fluorescence detection (xCGE-LIF) with 8-aminopyrene-1,3,6-trisulfonic acid labeling, and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) with linkage-specific sialic acid esterification. All methods assessed the same panel of serum samples, which were obtained at multiple time points during the pregnancies and postpartum periods of healthy women and patients with rheumatoid arthritis (RA). We compared the analytical methods on their technical performance as well as on their ability to describe serum protein N-glycosylation changes throughout pregnancy, with RA, and with RA disease activity.
Overall, the methods proved to be similar in their detection and relative quantification of serum protein N-glycosylation. However, the non-MS methods showed superior repeatability over MALDI-TOF-MS and allowed the best structural separation of low-complexity N-glycans. MALDI-TOF-MS achieved the highest throughput and provided compositional information on higher-complexity N-glycans. Consequentially, MALDI-TOF-MS could establish the linkage-specific sialylation differences within pregnancy and RA, whereas HILIC-UHPLC-FLD and xCGE-LIF demonstrated differences in α1,3- and α1,6-branch galactosylation. While the combination of methods proved to be the most beneficial for the analysis of total serum protein N-glycosylation, informed method choices can be made for the glycosylation analysis of single proteins or samples of varying complexity.
Keywords: Glycomics, Glycosylation, High Throughput Screening, Plasma or Serum Analysis, Electrophoresis, Chromatography, Mass Spectrometry, Pregnancy, Rheumatoid Arthritis
Glycosylation is a critical and ubiquitous co- and posttranslational protein modification which affects a wide variety of biological functions (1, 2). Manipulation of protein N-glycosylation has shown to be effective for influencing protein half-life and receptor interaction in molecules as diverse as gamma-immunoglobulins (IgG) and alpha-immunoglobulins (IgA), as well as erythropoietin (3–5). Furthermore, glycans of pathogens and cancer cells, as well as their receptors, are promising targets for both small molecule drugs and biopharmaceuticals (6–8). Longitudinal studies have shown a remarkable stability of the total plasma protein N-glycome of individuals over a several-year period (9, 10), while population studies have revealed associations of protein N-glycosylation with aging, sex, inflammation, body-mass index, metabolism, and a variety of cancers and autoimmune disorders (11–13). Longitudinal glycosylation profiling of single proteins and complex biofluids provides an opportunity for early detection of systemic alterations and may serve to stratify patient populations (14–16).
The last decade saw major developments in analytical methodologies for achieving N-glycosylation analysis (17, 18). For example, the in-depth study of glycans and glycopeptides is facilitated by hydrophilic interaction liquid or reverse-phase chromatography coupled to mass spectrometry (19), and materials such as porous graphitic carbon display a high degree of isomeric separation of native and permethylated glycans as well (20, 21). However, of the methods available for glycan analysis only several have demonstrated the high-throughput (HTP) capability to profile the several thousands of samples making up many of the current-day clinical cohorts. The largest glycomics profiling studies thus far, all of them comprising more than 2,000 cases, were performed with hydrophilic-interaction (ultra-)high-performance liquid chromatography with fluorescence detection (HILIC-(U)HPLC-FLD) (22–24), multiplexed capillary gel electrophoresis with laser-induced fluorescence detection (xCGE-LIF) (25), and matrix-assisted laser desorption/ionization (time-of-flight) mass-spectrometry (MALDI-(TOF)-MS) (11) and have assessed the released N-glycans from total serum/plasma or single glycoproteins such as IgG and alpha-1-antitrypsin. However, while each of the analytical methods proved informative for the analysis of N-glycosylation of complex mixtures, none of them provided full structural characterization of glycan species without follow-up experiments such as exoglycosidase digestion and/or tandem-MS (26, 27).
For glycans of relatively low complexity, such as found on the fragment-crystallizable (Fc) portion of IgG, comparative analysis has revealed highly similar findings between the aforementioned analytical methods (28–30). However, the study of IgG-Fc glycosylation does not comprise structures of higher antennarity, i.e. tri- and tetraantennary species, nor the high levels of terminal N-acetylneuraminic acids found on most serum proteins other than IgG (27, 31). In addition, many biological sources show more heterogeneous glycosylation than human IgG, and information on the comparative performance of HTP glycomics methods on such complex samples is still missing.
Here, we have studied the performance of the latest generation of HTP N-glycome analysis methodologies, focusing on HILIC-UHPLC-FLD, xCGE-LIF, and MALDI-TOF-MS. With this study we aimed to assess their respective suitability for total serum protein N-glycome (TSNG) analysis, explore the overlap and orthogonality of the information between the methods, and determine their strengths and weaknesses for revealing different types of N-glycan properties. To answer these questions in a clinically relevant setting, all methods were challenged with analyzing the same subset of the pregnancy-induced amelioration of rheumatoid arthritis (PARA) cohort, a longitudinal study aimed at exploration of the temporary improvement of rheumatoid arthritis (RA) severity experienced by women during pregnancy (32).
EXPERIMENTAL PROCEDURES
Study Population
The research presented here was performed using serum samples from the PARA study, a prospective cohort to study the interaction of pregnancy and RA (32). In the current research, serum samples were included from 36 RA patient pregnancies, obtained prior to conception, at the third trimester of pregnancy, and after 26 weeks following partum. At every time point, disease activity was assessed using the 28-joint disease activity score (DAS) with three variables based on the C-reactive protein (CRP) level (mg/l) (DAS28(3)-CRP). In addition, serum samples from 32 apparently healthy pregnancies (without adverse obstetric histories) were included to serve as controls, for these including serum collected at the third trimester of pregnancy and later than 26 weeks postpartum. All pregnancies were completed, and all patients fulfilled the 1987 American College of Rheumatology (ACR) criteria for RA. The study was in compliance with the Helsinki Declaration and was approved by the Ethics Review Board at the Erasmus University Medical Center, Rotterdam, The Netherlands.
Of each of the 178 clinical samples, 200 μl serum were distributed in randomized order across two 96-well deep-well plates (polypropylene; NUNC, Rochester, NY). Per plate, an additional two positions were filled with phosphate-buffered saline solution to serve as blank, five positions with identical plasma as technical standard (Visucon-F frozen normal control plasma; Affinity Biologicals, Ancaster, ON), and three positions were left to facilitate the inclusion of standards local to each laboratory. These local standards were used for technical validation at the individual laboratories only and were not evaluated for method comparison. From these master plates, 40 μl sample were divided into five pairs of PCR plates (polypropylene; Greiner Bio-One, Frickenhausen, Germany), which were distributed among the participating laboratories.
HILIC-UHPLC-FLD analysis
Sample preparation and measurement by HILIC-UHPLC-FLD was performed by participants 1, 2, and 3, as previously reported (26, 33–35). The procedures are described in full detail in the supplementary information (respectively, supplemental Methods M1, M2, and M3). To summarize, N-glycans were released enzymatically from their protein backbones by overnight peptide-N-glycosidase F (PNGase F) treatment, labeled with 2-AB by reductive amination, enriched by HILIC solid-phase extraction, and subsequently analyzed by HILIC-UHPLC-FLD. For peak annotation, the sample retention times were calibrated on an external UHPLC run of dextran ladder. The hereby obtained Glucose Unit (GU) values per signal were used to connect to a database of previously established assignments (supplemental Table S1) (36).
xCGE-LIF Analysis
xCGE-LIF sample preparation and measurement were performed as previously described (supplemental Method M4) (10, 37, 38). Briefly, N-glycans were released from the serum proteins by PNGase F, fluorescently labeled by reductive amination with 8-aminopyrene-1,3,6-trisulfonic acid, enriched by HILIC solid-phase extraction, and analyzed by a multiplexed capillary gel electrophoresis system with laser-induced fluorescence detection. Each sample was internally calibrated by a co-migrating fluorescent standard, and the resulting migration times were annotated with glycan structures on basis of established database values (supplemental Table S2) (37). Automated migration time normalization, peak picking, integration, and database matching were performed by glyXtool (37).
MALDI-TOF-MS Analysis
MALDI-TOF-MS sample preparation and measurement were performed as described previously (supplemental Method M5) (39, 40). In short, after N-glycan release by PNGase F, an automated platform was employed to derivatize the sialic acids by ethyl esterification (of α2,6-linked sialic acids and lactonization of α2,3-linked sialic acids), to perform GHP HILIC solid-phase extraction, and to spot samples on a MALDI target (39). MALDI-TOF-MS analysis was performed in reflectron positive mode, with an accumulation of 10,000 shots per spot in a random walking pattern. Obtained signals were annotated to be [M+Na]+ glycan compositions on basis of the signal-to-noise ratio, the ppm error, and the isotopic ratio (supplemental Table S3).
Data Preprocessing and analysis
Signal numbers were unified between the participants on basis of the structural annotation, principally based on the nomenclature of participant 1 (supplemental Table S4). For each of the methods, signal areas were normalized to the total sum of area per sample (total area normalization). Derived traits were calculated on basis of known enzymatic glycosylation pathways and glycoprotein populations (for derived trait calculations see supplemental Table S5, for a legend describing the derived traits see supplemental Table S6) (27, 31, 41–44).
Throughout data analysis, we made use of R 3.1.2 in RStudio 0.98.1091 (RStudio Team, Boston, MA) (45). The repeatability of each method was assessed by calculating the mean, S.D. and cv for each signal within the technical control samples (supplemental Table S7). Correlation between glycosylation features in the clinical data was established by calculating the Pearson correlation, and the results hereof were expressed in heat map format (supplemental Figs. S3-S5). Boxplots and scatterplots were similarly created in R using the total-area-normalized glycosylation values.
For the association analyses with pregnancy, RA and DAS28(3)-CRP, glycosylation (and derived trait) averages were centered to 0 and scaled to represent single S.D. variations. Pregnancy was defined as binary variable (both preconception and 26+ weeks postpartum = 0; third trimester of pregnancy = 1), as was RA (0 = control, 1 = RA). Mixed logistic regression was employed to test the association of glycosylation (independent) with pregnancy (dependent), correcting for interindividual effects by assigning a random intercept per individual (supplemental Table S8). Mixed logistic regression was additionally used to test the association between glycosylation (independent) and RA (dependent), correcting for pregnancy by modeling a random intercept per time point (supplemental Table S9). Linear regression was used to test the direct association between glycosylation (independent) and the linear variable DAS28-CRP (dependent), while mixed linear regression was used to differentiate the intraindividual effects (modeling a random intercept per individual) or interindividual effects (modeling a random intercept per time point) (supplemental Table S10).
The study-wide false discovery rate was controlled to be 5% by application of the Benjamini–Hochberg procedure (46), leading to an overall significance threshold of α = 1.7·10−2 (supplemental Table S11).
Nomenclature
In text, N-glycan structures have followed the Oxford nomenclature (26): A = number of antennary N-acetylglucosamines, G = number of galactoses, M = number of mannose residues (D1–3 indicating isomeric species), F = number of fucoses (an F at the beginning of the structure denoting core fucosylation), S = number of N-acetylneuraminic acids, and B = bisection. Numbers in brackets indicate the linkage of the following trait, e.g. S[3,6]2 declares two sialic acids, one being α2,3-linked and one being α2,6-linked.
N-Glycan compositions have followed H = hexose, N = N-acetylhexosamine, F = fucose (deoxyhexose), S = unspecified N-acetylneuraminic acid (sialic acid), L = (lactonized) α2,3-linked N-acetylneuraminic acid, and E = (ethyl esterified) α2,6-linked N-acetylneuraminic acid.
Figures have been annotated with glycan cartoons following the recommendations of the Consortium for Functional Glycomics (47) and designed using GlycoWorkbench 2.1 (build 146) (48).
RESULTS
To qualitatively compare glycomics analytical methodologies, we measured the released TSNGs of 36 RA patient pregnancies at three time points (before conception, at the third trimester of pregnancy, and 26 weeks postpartum) and 32 pregnancies of healthy controls at two time points (at the third trimester and 26+ weeks postpartum) (Table I). In addition, a repeat measurement of a standard plasma sample was included in the study to establish intra- and interplate variation. The methodologies used on the released glycan samples were as follows: HILIC-UHPLC-FLD after 2-aminobenzamide labeling, xCGE-LIF after 8-aminopyrene-1,3,6-trisulfonic acid labeling, and MALDI-TOF-MS after ethyl esterification of the sialic acids (Table II). The HILIC-UHPLC-FLD analysis was performed by three independent laboratories applying their standard protocols (supplemental Methods).
Table I. Characteristics of pregnancies included in the study.
ACPA, anti-citrullinated protein antibody; RU, rheumatoid factor.
| Pregnancies |
||
|---|---|---|
| Control (n = 32) | RA (n = 36) | |
| Age at delivery in years, mean (S.D.) | 32.1 (4.4) | 32.5 (4.0) |
| Duration of pregnancy in weeks, mean (S.D.) | 40.1 (1.4) | 39.2 (1.9) |
| Disease duration at first visit in years, mean (S.D.) | – | 7.3 (5.8) |
| ACPA positive patients, n (%) | – | 22 (61%) |
| RF positive patients, n (%) | – | 25 (69.4%) |
| Erosive disease, n (%) | – | 14 (38.9%) |
| Disease activity score (DAS28(3)-CRP) at preconception, mean (S.D.) | – | 3.8 (1.0) |
Table II. Participating laboratories and methodologies used in the study.
| Affiliation (participant) | Methodology | Labeling or modification | Clean-up | Automation | Annotation | References |
|---|---|---|---|---|---|---|
| NIBRT (1) | HILIC-UHPLC-FLD | 2-AB labeling | Solid-phase hydrazide bead capture; Hypersep Diol cartridge | Sample preparation | GU value with database | (26, 33) |
| Genos (2) | HILIC-UHPLC-FLD | 2-AB labeling | GHP filter plate | – | GU value with database | (26, 33) |
| Ludger (3) | HILIC-UHPLC-FLD | 2-AB labeling | LudgerClean T1 cartridge | Sample preparation | GU value with database; LC-MS composition analysis | (72) |
| MPI (4) | xCGE-LIF | APTS labeling | glyXera glyXbeads | Normalization; integration; database matching; quality control | Normalized migration time value with database | (10, 37, 38) |
| LUMC (5) | MALDI-TOF-MS | Ethyl esterification of sialic acids | GHP filter plate | Sample preparation; integration; quality control | mass error; isotope distribution | (39, 40) |
Signal Detection
HILIC-UHPLC-FLD allowed the integration of 46 signals, with peak identities inferred by matching standardized retention times via GU values to database entries (supplemental Table S1) (36). xCGE-LIF enabled the integration of 49 signals, and peak identity was inferred by matching migration times via database entries (supplemental Table S2) (37). For MALDI-TOF-MS, 61 signals were detected that passed the established quality criteria (supplemental Fig. S1; supplemental Table S3). The analyses broadly detected the same N-glycan species, with some variation per method in structural and compositional overlap (supplemental Table S4), and the assignments were in line with previously established serum and plasma N-glycosylation features (42–44).
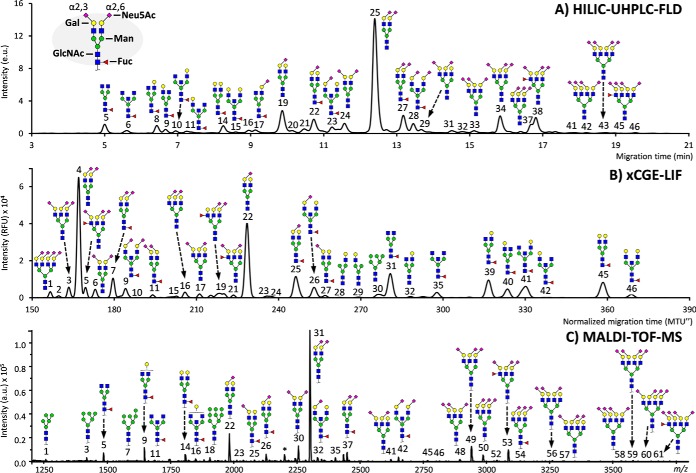
All analytical methods showed comparable results in their representation of the overall serum and plasma profile (Fig. 1). Clearly defined for all methods were the two largest signals, mainly belonging to A2G2S[6,6]2 (H5N4E2; for HILIC-UHPLC-FLD, xCGE-LIF and MALDI-TOF-MS peaks 25, 4, and 31, respectively) and A2G2S[6]1 (H5N4E1; peaks 19, 22, and 22) (supplemental Table S4). In addition, all methods demonstrated the high-definition detection of asialylated diantennary glycans, e.g. FA2 (H3N4F1; peaks 5, 31, 5) and FA2G2 (H5N4F1; peaks 14, 45, 14). However, the non-MS methods allowed distinction between the galactose-linkage sites of the monogalactosylated species, e.g. FA2[3]G1 (peaks 8, 39, –) and FA2[6]G1 (peaks 9, 40, –), whereas MALDI-TOF-MS could only detect the common composition belonging to these structures, namely H4N4F1 (peak 9). Similarly, while the presence of a bisecting N-acetylglycosamine led to unique retention/migration times, the corresponding mass spectrometric compositions also apply to triantennary structures with incomplete galactosylation. For example, the MALDI-TOF-MS composition H3N5F1 could align to FA2B (peaks 6, 35, 11) or FA3 (not detected), although these are not commonly found in high abundance in human serum (27, 31). On the other hand, MS proved capable of individually detecting different sizes of high-mannose and hybrid-type N-glycans, whereas these species overlapped with diantennary glycans in the non-MS methods (supplemental Table S4).
Fig. 1.
The respective profiles of the same total plasma protein N-glycan standard as recorded by HILIC-UHPLC-FLD, xCGE-LIF and MALDI-TOF-MS. (A) Chromatogram as obtained by HILIC-UHPLC-FLD after 2-aminobenzamide labeling. (B) Electropherogram as obtained by xCGE-LIF after APTS labeling. (C) Mass spectrum as obtained by MALDI-TOF-MS after ethyl esterification, with species assigned as [M+Na]+. Signals of all recordings have been annotated to the best of knowledge, making use of the detections across the methods as well as established literature on biochemical pathways and plasma/serum N-glycosylation. The display of linkage has been restricted to the N-acetylneuraminic acids (sialic acids), which was principally acquired by MALDI-TOF-MS. Branching differences (galactose arm, bisection, fucose position) were only distinguishable by HILIC-UHPLC-FLD and xCGE-LIF. For full assignments of the signals, see supplemental Tables S1–S3, as well as supplemental Fig. S1.
The two main species belonging to the triantennary structures were also detected by all analyses, i.e. A3G3S[3,6,6]3 (H6N5L1E2; peaks 34, 3, 49) and A3F1GS[3,6,6]3 (H6N5F1L1E2; peaks 38, 5, 53). However, while the specific sialic acid linkage variants of the triantennary glycan compositions could be discriminated by MALDI-TOF-MS, e.g. H6N5L3, H6N5L2E1, H6N5L1E2, and H6N5E3, the accompanying structures could not clearly be assigned to individual signals for HILIC-UHPLC-FLD and xCGE-LIF without additional steps of linkage-specific enzymatic removal of the sialic acids and sample remeasurement. This situation was similar for the tetraantennary compositions.
Based on the structural information and separation achieved by the various methods, we constructed a series of derived traits to describe single glyco-enzymatic steps and presumed protein-specific glycosylation patterns (supplemental Tables S5 and S6) (31, 39).
Method Repeatability
All methods showed robust detection of the plasma standard analyzed in 10 replicates, with UHPLC generally displaying the least variation and MALDI-TOF-MS the most (supplemental Fig. S2 and supplemental Table S7). Specifically, the main peak (A2G2S[6,6]2 or H5N4E2) showed for HILIC-UHPLC-FLD a cv of 1.9% (participant 2, which displayed the lowest variation among the UHPLC methods), for xCGE-LIF of 5.1%, and for MALDI-TOF-MS of 5.8%. Further examples include the cvs of FA2 (H3N4F1), respectively, being 1.7%, 13.8%, and 17.7% and of A3G3S[3,6,6]3 being 0.9%, 10.3%, and 13.2%. Overall, the average cvs of the 10 most abundant peaks for each method were 1.6%, 6.9%, and 11.5%. For MALDI-TOF-MS most derived traits showed a higher repeatability than the individual peaks making up the traits, exemplified by the 50 most abundant traits having a mean cv of 2.6%, while for the non-MS methods the cvs of derived traits remained similar to their constituent peaks.
Intermethod Signal Correlation
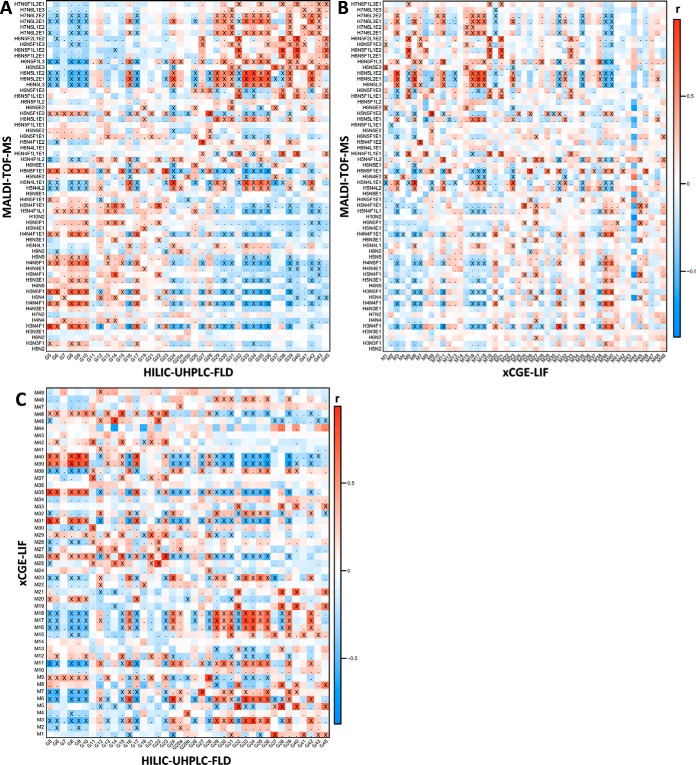
Using the clinical cohort data, we could explore which signals displayed similar behavior across the orthogonal methods. To achieve this, Pearson correlation coefficients were calculated between the methods for all single signals (Fig. 2), as well as for the derived traits (supplemental Figs. S3–S6). Note that the correlations obtained in this manner are the result of similarity of N-glycan behavior across pregnancy, RA, and disease activity thereof, as well as of other biological and technical sources of variation that were unaccounted for in this study. As such, positive correlation within and between methods occurred when signals contained the same glycan structure (single isolated hit) or when different glycans underwent the same enzymatic modification (multiple hits sharing a single property such as antennary fucosylation). Negative correlation could similarly arise due the relationship between enzymatic substrates and products (e.g. the process of galactosylation induces negative correlation between galactosylated and nongalactosylated species) and protein-abundance changes (e.g. an increase in all diantennary species such as present on immunoglobulins will lead to a relative decrease in all tri- and tetraantennary glycans).
Fig. 2.
Heat map visualizing the Pearson correlation between signals from MALDI-TOF-MS, HILIC-UHPLC-FLD (participant 2) and xCGE-LIF. (A) HILIC-UHPLC-FLD (horizontal) with MALDI-TOF-MS (vertical). (B) xCGE-LIF (horizontal) with MALDI-TOF-MS (vertical). (C) HILIC-UHPLC-FLD (horizontal) with xCGE-LIF (vertical). Signal correlations were calculated on clinical data, and represent similarity of behavior with biological phenotypes such as pregnancy and RA as well as technical variation. Crosses (X) indicate correlations significant below a p value of 1·10−5, whereas dots (.) indicate correlation below a p value of 0.05. For similar heat maps between non-MS methods and derived traits see supplemental Figs. S4–S6. H = hexose, N = N-acetylhexosamine, F = deoxyhexose (fucose), L = (lactonized) α2,3-linked N-acetylneuraminic acid, and E = (ethyl esterified) α2,6-linked N-acetylneuraminic acid.
Between the UHPLC methods a good correlation was observed for peaks containing the same glycans, examples being peak 5 (FA2; mean r = 0.90 S.D. ± 0.07), peak 27 (FA2G2S[6,6]2; r = 0.93 ± 0.01) and peak 34 (A3G3S[3,6,6]3; r = 0.90 ± 0.02) (supplemental Figs. S4A–4C). Signals with lower correlation across the methods were generally of low intensity. Strong correlation of the aforementioned signals was also visible with the corresponding xCGE-LIF annotation, i.e. UHPLC peak 5 with xCGE-LIF peak 31 (FA2; r = 0.96), 27 with 7 (FA2G2S[6,6]2; r = 0.87), and 34 with 3 (A3G3S[3,6,6]3; r = 0.88) (supplemental Fig. S4D). Next to single signals, the derived traits showed to be highly comparable between methods as well (supplemental Fig. S5).
Complementarity of Methods for Signal Assignment
When comparing the signals of the non-MS methods with MALDI-TOF-MS, an advantage of analyzing the sample set with two vastly orthogonal methods becomes apparent (Fig. 2). For instance, in several cases ambiguously annotated non-MS signals could be attributed to specific glycan structures by making use of the correlation with MS. One example hereof is chromatographic peak 28, which, while theoretically encompassing FA2BG2S[6,6]2, FA2BG2S[3,6]2 and FA2BG2S[3,3]2, strongly correlated with MALDI-TOF-MS composition H5N5F1E2 (FA2BG2S[6,6]2; r = 0.92) and to a much lesser degree with H5N5F1L1E1 (FA2BG2S[3,6]2; r = 0.27) and H5N5F1L2 (FA2BG2S[3,3]2; not detected) (Fig. 2A, supplemental Fig. S3). Similarly, by correlation xCGE-LIF peak 1 is likely to contain H6N5E3 (A3G3S[6,6,6]3; r = 0.68), even though this was not principally annotated for the electrophoretic signal (Fig. 2B). On the other hand, structural characteristics could be attributed to MALDI-TOF-MS compositions on basis of the assignments from the non-MS methods. For instance, the fucosylated triantennary compositions with at least one α2,3-linked sialic acid correlated strongly with antennary-fucosylated but not with core-fucosylated structures, e.g. H6N5F1L1E2 with UHPLC peak 38 and xCGE-LIF peak 5 (A3F1G3S[3,6,6]3; respectively r = 0.86 and r = 0.82), and not with UHPLC peak 36 (FA3G3S[3,6,6]3; r = 0.10). While core-fucosylated structures are still likely present in the MALDI-TOF-MS composition, it does appear that the main differences observed within the cohort originated instead from antennary fucosylation.
Next to single glycans, derived traits showed good overlap between MS and non-MS methods, but a larger set of these could be constructed for MS due to the unambiguous compositional assignment of signals (supplemental Fig. S6).
Association with Pregnancy and RA
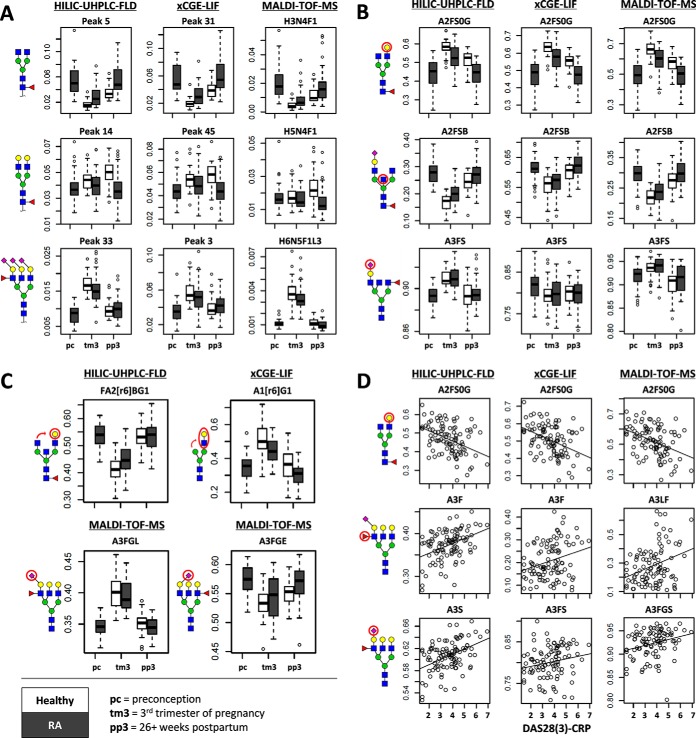
Within the PARA study, we compared for the different methods the glycosylation changes observed throughout pregnancy (assessed at preconception, the third trimester of pregnancy, and 26+ weeks postpartum) between healthy controls and RA patients, as well as the association with RA disease activity as expressed by the DAS28(3)-CRP value. The comparability between methods was assessed by the effect directions of the significant findings from mixed model regression analyses and are represented as box- and scatterplots (Fig. 3). For all analyses, glycosylation parameters were centered to zero and scaled to represent single S.D. changes. Effects were deemed significant under a study-wide false discovery rate of 5% (leading to a threshold α = 1.7·10−2).
Fig. 3.
Comparability of HILIC-UHPLC-FLD (participant 2) (left), xCGE-LIF (middle), and MALDI-TOF-MS (right) for the detection of clinical characteristics of pregnancy, RA and RA disease activity. (A) Comparability of single glycan signals (% area) with pregnancy (preconception = pc, third trimester of pregnancy = tm3, 26+ weeks postpartum = pp3), and RA (healthy = white, RA = gray). (B) Comparability of derived glycosylation traits with pregnancy and RA. (C) Derived trait differences detected uniquely by a single method with pregnancy and RA. (D) Association of derived glycosylation traits with RA disease activity (DAS28(3)-CRP). For a legend of the derived traits see supplemental Table S6.
Pregnancy showed to have a major effect on the TSNG, and congruent effects were observed by all methods (supplemental Table S8), both for the single glycans (Fig. 3A) and derived traits (Fig. 3B). Notable examples included the decrease of FA2 with pregnancy, as seen for HILIC-UHPLC-FLD (participant 2 throughout) peak 5 (β = −2.32 S.E. ± 0.38), xCGE-LIF peak 31 (β = −2.37 ± 0.39) and MALDI-TOF-MS composition H3N4F1 (β = −2.34 ± 0.43), and the consequentially increased overall galactosylation of (nonsialylated) diantennary fucosylated species representative of IgG-Fc (A2FS0G; respectively, β = 1.62 ± 0.27, β = 1.92 ± 0.31, and β = 1.67 ± 0.28) (31). Likewise, all methods indicated a profound decrease in bisection of sialylated diantennary species, likely representative of non-IgG-Fc immunoglobulin glycosylation (A2FSB, β = –3.10 ± 0.48, β = –1.55 ± 0.26, and β = –1.81 ± 0.29) (31).
Interestingly, one of the major N-glycan species to change with pregnancy, namely the fully α2,3-sialylated A3F1G3S[3,3,3]3, was only uniquely separable as the MALDI-TOF-MS composition H6N5F1L3 (β = 1.61 ± 0.26) but likely also drove the changes in the ambiguously assigned UHPLC signal 33 (β = 2.92 ± 0.43) and xCGE-LIF signal 3 (β = 1.31 ± 0.24). However, due to overlap in retention and migration times, the derived trait for α2,3-linked sialylation on triantennary species (A3GL) could only be established for MALDI-TOF-MS (β = 2.57 ± 0.40) (Fig. 3C). HILIC-UHPLC-FLD, on the other hand, uniquely showed with pregnancy a strong difference in the galactose position of monogalactosylated and bisected diantennary N-glycan species (e.g. relative 6-arm galactosylation, FA2[r6]BG1, β = –9.41 ± 2.42), whereas xCGE-LIF could uniquely detect a change in GlcNAc position of monoantennary species (e.g. relative 6-arm GlcNAc position, A1[r6]G1, β = 1.96 ± 0.31).
Differences between RA patients and healthy controls proved less pronounced than the differences during pregnancy but were similarly detected across methods. Mixed logistic regression was used for the comparison of glycosylation parameters with RA (healthy = 0, RA = 1), correcting for pregnancy by modeling a random intercept per time point (supplemental Table S9). RA patients showed to have a lower galactosylation of glycans commonly found on IgG-Fc (A2FS0G, for HILIC-UHPLC-FLD β = –1.89 ± 0.36, for xCGE-LIF β = –1.85 ± 0.36, and for MALDI-TOF-MS β = –1.78 ± 0.36), and higher bisection of sialylated fucosylated diantennary species (A2FSB, β = 1.25 ± 0.31, β = 0.53 ± 0.21, and β = 0.88 ± 0.26) (Fig. 3B). Also detected by HILIC-UHPLC-FLD and MALDI-TOF-MS was a higher bisection with RA of afucosylated nonsialylated diantennary species (A2F0S0B, respectively β = 1.11 ± 0.28 and β = 0.69 ± 0.20). Absent from MALDI-TOF-MS but present for the non-MS methods were differences in branching for lower complexity N-glycans, namely for HILIC-UHPLC-FLD the relative 6-branch galactosylation of monogalactosylated and sialylated diantennary species with bisection (FA2[r6]BG1S1, β = –0.90 ± 0.30) and for xCGE-LIF the antennary-branch of monoantennary species (A1[r6]G1, β = –1.05 ± 0.28) (Fig. 3C).
Lastly, linear regression was used to model the association of glycosylation (independent) with DAS28(3)-CRP (dependent). In addition, mixed linear regression was used with either a random intercept per individual to assess the association throughout pregnancy or with a random intercept per time point to assess the association between individuals. These different models provided highly congruent findings (supplemental Table S10). In all, for each method a strong negative association was seen between DAS28(3)-CRP and IgG-Fc-type galactosylation (A2FS0G, HILIC-UHPLC-FLD β = –0.51 ± 0.11, xCGE-LIF β = –0.51 ± 0.11, MALDI-TOF-MS β = –0.50 ± 0.11) (Fig. 3D). Increased with disease activity proved to be the fucosylation of triantennary structures (A3F, β = 0.50 ± 0.11, β = 0.33 ± 0.12, and β = 0.38 ± 0.12), as well as the sialylation thereof (A3S β = 0.52 ± 0.11, A3FS β = 0.23 ± 0.12 (trend), and A3FGS β = 0.41 ± 0.12). MALDI-TOF-MS analysis specifically showed that the increases in fucosylation and sialylation were similar for the tri- and tetraantennary species (e.g. A4FE β = 0.42 ± 0.12) and revealed that there were no sialic acid linkage-biases in these increases (e.g. A3EF β = 0.39 ± 0.12 versus A3LF β = 0.40 ± 0.12). HILIC-UHPLC-FLD, on the other hand, was again able to detect significant branching differences with changing disease activity (FA2[r6]G1S1 β = –0.34 ± 0.12), while xCGE-LIF showed higher resolution for the monoantennary species (e.g. A1F β = 0.48 ± 0.14).
DISCUSSION
Previous method comparisons involved the analysis of released N-glycans and glycopeptides of single proteins, examples being IgG, prostate-specific antigen, and transferrin (28–30, 49, 50), or the analysis of complex samples in a limited number (51). In the current study we have primarily focused on methodologies with demonstrated high-throughput capability. We have opted for the analysis of the released N-glycans from the total pool of serum glycoproteins, a sample type of interest for biomarker screening, and which may display N-glycosylation of considerable complexity (27, 31, 41).
We included in our comparison analytical methodologies that have displayed the throughput capacity for several thousands of serum N-glycome samples, as currently the case for HILIC-UHPLC-FLD, xCGE-LIF, and MALDI-(TOF-)MS (11, 22, 23, 25). Technical considerations for these studies include analyte stability and preparation and measurement throughput, as well as software solutions for the integration and analysis of the consequentially large datasets, all of which have been demonstrably addressed for the indicated techniques. To appreciate the technical possibilities and constraints of the methods, their mechanisms of separation need to be considered. 1) HILIC-UHPLC-FLD separates glycan structures on the basis of their hydrophilic interaction with a stationary phase, generally meaning that larger structures, as well as those with larger surface areas or charged N-acetylneuraminic acids, have increased retention times (52). 2) In xCGE-LIF glycans are separated along an electric field inside a polymer-filled capillary, according to their mass/charge (m/z) ratio and their size/shape (hydrodynamic diameter). Analytes with lower m/z (higher charges and/or lower masses) migrate faster through the capillary than those with higher m/z (10, 53). 3) MALDI-TOF-MS separates ionized glycans (here [M+Na]+) by m/z ratio, with the esterification procedure employed to stabilize sialic acids to prevent unfavorable negative charges on the sialylated species and to introduce an MS-detectable mass difference between α2,3- (lactonized) and α2,6-linked (ethyl esterified) sialic acids (40).
In this comparison study, we judged the relative performance of the HTP TSNG methods on the basis of technical replicate measurements, as well as by a set of samples from a longitudinal study on the improvement of disease activity within RA patients during pregnancy (32).
Throughput and Repeatability
Sample preparation throughput proved similar between the methods, each of them requiring (overnight) enzymatic N-glycan release, 1–2-h chemical derivatization at either the glycan-reducing end or at the sialic acid, and HILIC solid-phase extraction prior to analysis. Automated sample preparation was reported for both the HILIC-UHPLC-FLD and MALDI-TOF-MS workflows (33, 39), but the congruencies in protocols suggest that xCGE-LIF could make use of similar strategies. Aside from sample preparation, MALDI-TOF-MS showed to have the highest analytical throughput with an approximate throughput of 10 s per sample, whereas xCGE-LIF and UHPLC runs required 40 min to an hour. A major advantage for xCGE-LIF is the multiplexing capability (with up to 96 capillaries in parallel) allowing the simultaneous analysis of up to 96 samples, which reduces the effective analysis time per sample to less than 30 s.
The most repeatable method proved to be HILIC-UHPLC-FLD (albeit with variation between participants), whereas MALDI-TOF-MS showed the most technical variation. Interestingly, although the lower repeatability of the MALDI-TOF-MS method did indeed lead to larger S.D.s on the biological effects as well as a consequential decrease in statistical significance (an increase in p value) by typically one or more orders of magnitude, in practice very few findings were rejected in this study due to lack of statistical power. As such, the lower repeatability of MALDI-TOF-MS does not appear to hamper the glycomics analyses of larger cohorts, but the repeatability of the non-MS methods would definitely be of benefit for the quantification of small effect sizes.
Of interest, while MALDI-TOF-MS repeatability was higher for the derived traits, which often are groupings of chemically similar N-glycan species, no such improvement was observed for either HILIC-UHPLC-FLD or xCGE-LIF. This would suggest the presence of an MS-specific component of measurement error, possibly relative ionization efficiency or response linearity, both of which could be controlled for with the use of internal standards (54, 55).
Analyte Separation and Annotation
Whereas MALDI-TOF-MS does not principally separate all N-glycans but only those with differing chemical compositions, both HILIC-UHPLC-FLD and xCGE-LIF produce unique standardized retention times, respectively migration times, for glycans with different structures. However, while the MS analysis provided the resolution to separate the majority of possible compositions, for the non-MS methods a large portion of glycan structures were not separated from other analytes. Accordingly, only HILIC-UHPLC-FLD and xCGE-LIF were capable of revealing additional features in the low-complexity (IgG-Fc) regions of their chromatograms or electropherograms for the analyses of the TSNG, e.g. N-acetylglucosamine linkage (antennary or bisecting), galactose position (α1,3 or α1,6 branch), and fucose position (core or antennary). On the other hand, MALDI-TOF-MS proved the most informative for larger glycan structures (e.g. tri- and tetraantennary species), for instance on the exact number of LacNAc units (antennarity), the number of fucoses, and the number and linkage of N-acetylneuraminic acids. To generalize, in profiling mode HILIC-UHPLC-FLD and xCGE-LIF appeared optimal for high-density structural identification of a low-complexity sample, e.g. immunoglobulin glycosylation, be it from blood or most of the commonly used recombinant production systems, whereas MALDI-TOF-MS appeared preferentially suitable for the analysis of samples with high-complexity and/or larger N-glycan species.
Each of the described methods may obtain additional structural information on the glycans by including more experimental dimensions. One can think of on-line hyphenation of separation techniques, examples including LC-MS(/MS), CE-MS(/MS), and ion-mobility MS (56–58), or off-line approaches such as exoglycosidase digestion (26). While increasing the information content, these added dimensions also drastically increase analysis times and data complexity and have thus far only been reported for relatively small sample sets. However, with the advancements in development of rapid glycan preparations protocols, laboratory automation and evolution of big data analysis methods, the idea of HTP laboratories employing these innovative approaches in glycan analysis of larger sample sets on different glycoprotein levels might not be too far way. Today, more commonly achieved for HTP applications is the thorough structural annotation of one or a few samples, with the expectation that these are representative for the set, as is for instance achieved with exoglycosidase digestion or MS/MS experiments (26, 27). Interestingly, our study suggests that parallel glycomics analysis by orthogonal HTP methods as well allows the determination of many of the sample-relevant structural characteristics, without necessarily including a step of throughput-limiting serial hyphenation. A prominent example of this is the structure FA2BG2S2 correlating with composition H5N5F1E2, for which HILIC-UHPLC-FLD and xCGE-LIF determined the core-fucosylation and bisection while MALDI-TOF-MS determined the α2,6-linkage of the N-acetylneuraminic acids (supplemental Figs. S3B and S3C).
Clinical Observations
Associations found between N-glycosylation and pregnancy, RA, and RA disease activity (DAS28(3)-CRP) were very much in line with previous TSNG studies (59–62), and agreed with reports on single glycoproteins dominant in human serum (63–66). The main findings with pregnancy included an increased antennarity, nonsialylated FA2 galactosylation, and α2,3-linked sialylation as well as a decreased bisection of sialylated FA2. Individuals with RA proved to have on average a lower FA2 galactosylation and higher bisection compared with their healthy counterparts, and disease activity of RA associated positively with A3/A4 for both fucosylation and sialylation (without particular preference in linkage) and negatively with the galactosylation of FA2.
As with all analyses of the released TSNG, the here-reported differences may originate from changes in protein glycosylation, or from differences in the relative abundances of glycoproteins in serum. Nonetheless, the changing glycosylation phenotypes seem to reflect immune modulation of either IgG-Fc (predominantly nonsialylated FA2) (31, 64), IgG-Fab, and other plasma-cell-derived immunoglobulins (highly sialylated FA2) (31, 64) or acute-phase glycoproteins such as alpha-1-antitrypsin and alpha-1-acid glycoprotein (tri- and tetraantennary species) (31, 65, 67, 68). In addition, the previously reported MALDI-TOF-MS association of A3FGS with DAS28(3)-CRP was reproduced (62), although its protein of origin is as of yet unclear.
Interestingly, HILIC-UHPLC-FLD and xCGE-LIF allowed new findings on the glycosylation changes occurring with pregnancy and RA disease activity, in the form of branching differences on both monogalactosylated and monoantennary species. With pregnancy, for example, an increased galactosylation was detected of the α1,6-branch (as opposed to the α1,3-branch) of nonbisected monogalactosylated FA2 and a decreased α1,6-branched galactosylation of the bisected variant. The same phenotypes were inversely associated with RA disease activity, principally matching the DAS28(3)-CRP decrease observed with pregnancy (32). These observations could be the result of altered glycosyltransferase expression or regulation, but one attractive alternative explanation might be a shift of relative IgG-subclass abundances during pregnancy (69). As IgG2 and IgG3 display higher α1,3-branch galactosylation than α1,6-branch galactosylation, which is contrary to IgG1 and IgG4 (70), the reported relative increase of IgG2 and IgG3 with pregnancy would lead to our observed linkage change in the TSNG (69, 71).
Summary
In summary, we compared the performance of HILIC-UHPLC-FLD, xCGE-LIF, and MALDI-TOF-MS for the analysis of the released serum protein N-glycome. Next to providing the technical and biological translation between methods, we discussed their advantages and disadvantages, including their respective throughput and repeatability (Table III). In addition, we explored the differences in information content for various glycosylation types within the TSNG, and speculated upon the suitability of the methods to characterize different sample types. The combined analysis with orthogonal HTP methods proved to be highly informative for the study of the TSNG, and has led, next to confirming previous findings, to the discovery of new glycosylation traits associated with RA and disease activity thereof.
Table III. Overview of study results.
| Methodology | Information | Advantages | Disadvantages | Sample suitability |
|---|---|---|---|---|
| HILIC-UHPLC-FLD | Structural | Best repeatability; well-established database; good separation of A2 | A3/A4 signal overlap; no novel annotationa | Plasma cell glycoproteins or mixtures (immunoglobulins; A2 glycans) |
| xCGE-LIF | Structural | Throughput due to multiplexing; good separation of A2 | A3/A4 signal overlap; no novel annotationa | Plasma cell glycoproteins or mixtures (immunoglobulins; A2 glycans) |
| MALDI-TOF-MS | Compositional; sialic acid linkage | Best throughput; compositional annotation; infrequent signal overlap; good separation of A3/A4; derived traits | Lowest repeatability; no structural informationa | Hepatic cell glycoproteins (acute phase proteins; A3/A4 glycans); unknown samples |
aAdditional information may be obtained by subsequent experiments such as exoglycosidase sequencing and tandem MS.
Data Availability
The raw data used in the manuscript can be found online at https://data.mendeley.com/datasets/f73bxyjb6f/draft?a=cef35bea-088e-44c4-968e-4b598035fb74.
Supplementary Material
Acknowledgments
We would like to thank Gerda Vreeker, Marco Bladergroen, and Yuri van der Burgt for their assistance with measuring the MALDI-TOF-MS data.
Footnotes
*This work was supported by the European Union Seventh Framework Programme project HighGlycan (278535) and the Dutch Arthritis Foundation (NR-10-1-411). Competing financial interest: Richard Gardner, Archana Shubhakar, Daniel I. Spencer, and Daryl L. Fernandes are employed at Ludger, Ltd., Gordan Lauc is the owner and Irena Trbojević-Akmačić and Maja Pučić-Baković are employees of Genos Glycoscience Research Laboratory, and René Hennig and Erdmann Rapp are employed at glyXera GmbH, all of which perform commercial glycosylation analysis. Karli R. Reiding and Manfred Wuhrer are inventors of IP licensed to Ludger, Ltd. and glyXera GmbH.
 This article contains supplemental material Tables S1-S11 and Figs. S1-S6.
This article contains supplemental material Tables S1-S11 and Figs. S1-S6.
REFERENCES
- 1. Varki A. (2017) Biological roles of glycans. Glycobiology 27, 3–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hart G. W., and Copeland R. J. (2010) Glycomics hits the big time. Cell 143, 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thomann M., Schlothauer T., Dashivets T., Malik S., Avenal C., Bulau P., Rüger P., and Reusch D. (2015) In vitro glycoengineering of IgG1 and its effect on Fc receptor binding and ADCC activity. PLoS ONE 10, e0134949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rouwendal G. J., van der Lee M. M., Meyer S., Reiding K. R., Schouten J., de Roo G., Egging D. F., Leusen J. H., Boross P., Wuhrer M., Verheijden G. F., Dokter W. H., Timmers M., and Ubink R. (2016) A comparison of anti-HER2 IgA and IgG1 in vivo efficacy is facilitated by high N-glycan sialylation of the IgA. mAbs 8, 74–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oh M. J., Hua S., Kim B. J., Jeong H. N., Jeong S. H., Grimm R., Yoo J. S., and An H. J. (2013) Analytical platform for glycomic characterization of recombinant erythropoietin biotherapeutics and biosimilars by MS. Bioanalysis 5, 545–559 [DOI] [PubMed] [Google Scholar]
- 6. Pancera M., Shahzad-Ul-Hussan S., Doria-Rose N. A., McLellan J. S., Bailer R. T., Dai K., Loesgen S., Louder M. K., Staupe R. P., Yang Y., Zhang B., Parks R., Eudailey J., Lloyd K. E., Blinn J., Alam S. M., Haynes B. F., Amin M. N., Wang L. X., Burton D. R., Koff W. C., Nabel G. J., Mascola J. R., Bewley C. A., and Kwong P. D. (2013) Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat. Struct. Mol. Biol. 20, 804–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. von Itzstein M., Wu W. Y., Kok G. B., Pegg M. S., Dyason J. C., Jin B., Van Phan T., Smythe M. L., White H. F., Oliver S. W., and et al. (1993) Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 363, 418–423 [DOI] [PubMed] [Google Scholar]
- 8. Dalziel M., Crispin M., Scanlan C. N., Zitzmann N., and Dwek R. A. (2014) Emerging principles for the therapeutic exploitation of glycosylation. Science 343, 1235681. [DOI] [PubMed] [Google Scholar]
- 9. Gornik O., Wagner J., Pucić M., Knezević A., Redzic I., and Lauc G. (2009) Stability of N-glycan profiles in human plasma. Glycobiology 19, 1547–1553 [DOI] [PubMed] [Google Scholar]
- 10. Hennig R., Cajic S., Borowiak M., Hoffmann M., Kottler R., Reichl U., and Rapp E. (2016) Towards personalized diagnostics via longitudinal study of the human plasma N-glycome. Biochim. Biophys. Acta 1860, 1728–1738 [DOI] [PubMed] [Google Scholar]
- 11. Reiding K. R., Ruhaak L. R., Uh H. W., El Bouhaddani S., van den Akker E. B., Plomp R., McDonnell L. A., Houwing-Duistermaat J. J., Slagboom P. E., Beekman M., and Wuhrer M. (2017) Human plasma N-glycosylation as analyzed by matrix-assisted laser desorption/ionization-Fourier transform ion cyclotron resonance-MS associates with markers of inflammation and metabolic health. Mol. Cell. Proteomics 16, 228–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Albrecht S., Unwin L., Muniyappa M., and Rudd P. M. (2014) Glycosylation as a marker for inflammatory arthritis. Cancer Biomark. 14, 17–28 [DOI] [PubMed] [Google Scholar]
- 13. Vučković F., Theodoratou E., Thaci K., Timofeeva M., Vojta A., Štambuk J., Pučić-Baković M., Rudd P. M., Ðerek L., Servis D., Wennerström A., Farrington S. M., Perola M., Aulchenko Y., Dunlop M. G., Campbell H., and Lauc G. (2016) IgG Glycome in Colorectal Cancer. Clin. Cancer Res. 22, 3078–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kemna M. J., Plomp R., van Paassen P., Koeleman C. A. M., Jansen B. C., Damoiseaux J. G. M. C., Cohen Tervaert J. W., and Wuhrer M. (2017) Galactosylation and sialylation levels of IgG predict relapse in patients with PR3-ANCA associated vasculitis. EBioMedicine 17, 108–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Novokmet M., Lukić E., Vučković F., Ðurić Ž., Keser T., Rajšl K., Remondini D., Castellani G., Gašparović H., Gornik O., and Lauc G. (2014) Changes in IgG and total plasma protein glycomes in acute systemic inflammation. Sci. Rep. 4, 4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Verhelst X., Vanderschaeghe D., Castéra L., Raes T., Geerts A., Francoz C., Colman R., Durand F., Callewaert N., and Van Vlierberghe H. (2017) A Glycomics-based test predicts the development of hepatocellular carcinoma in cirrhosis. Clin. Cancer Res. 23, 2750–2758 [DOI] [PubMed] [Google Scholar]
- 17. Shubhakar A., Reiding K. R., Gardner R. A., Spencer D. I., Fernandes D. L., and Wuhrer M. (2015) High-throughput analysis and automation for glycomics studies. Chromatographia 78, 321–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gaunitz S., Nagy G., Pohl N. L., and Novotny M. V. (2017) Recent advances in the analysis of complex glycoproteins. Anal. Chem. 89, 389–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walker S. H., Carlisle B. C., and Muddiman D. C. (2012) Systematic comparison of reverse phase and hydrophilic interaction liquid chromatography platforms for the analysis of N-linked glycans. Anal. Chem. 84, 8198–8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ashwood C., Lin C. H., Thaysen-Andersen M., and Packer N. H. (2018) Discrimination of isomers of released N- and O-glycans using diagnostic product ions in negative ion PGC-LC-ESI-MS/MS. J. Am. Soc. Mass Spectrom. 29, 1194–1209 [DOI] [PubMed] [Google Scholar]
- 21. Zhou S., Huang Y., Dong X., Peng W., Veillon L., Kitagawa D. A. S., Aquino A. J. A., and Mechref Y. (2017) Isomeric separation of permethylated glycans by porous graphitic carbon (PGC)-LC-MS/MS at high temperatures. Anal. Chem. 89, 6590–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lauc G., Essafi A., Huffman J. E., Hayward C., Knežević A., Kattla J. J., Polašek O., Gornik O., Vitart V., Abrahams J. L., Pučić M., Novokmet M., Redžić I., Campbell S., Wild S. H., Borovečki F., Wang W., Kolčić I., Zgaga L., Gyllensten U., Wilson J. F., Wright A. F., Hastie N. D., Campbell H., Rudd P. M., and Rudan I. (2010) Genomics meets glycomics-the first GWAS study of human N-glycome identifies HNF1alpha as a master regulator of plasma protein fucosylation. PLoS Genet. 6, e1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruhaak L. R., Uh H. W., Beekman M., Hokke C. H., Westendorp R. G., Houwing-Duistermaat J., Wuhrer M., Deelder A. M., and Slagboom P. E. (2011) Plasma protein N-glycan profiles are associated with calendar age, familial longevity and health. J. Proteome Res. 10, 1667–1674 [DOI] [PubMed] [Google Scholar]
- 24. Lauc G., Huffman J. E., Pučić M., Zgaga L., Adamczyk B., Mužinić A., Novokmet M., Polašek O., Gornik O., Krištić J., Keser T., Vitart V., Scheijen B., Uh H. W., Molokhia M., Patrick A. L., McKeigue P., Kolčić I., Lukić I. K., Swann O., van Leeuwen F. N., Ruhaak L. R., Houwing-Duistermaat J. J., Slagboom P. E., Beekman M., de Craen A. J., Deelder A. M., Zeng Q., Wang W., Hastie N. D., Gyllensten U., Wilson J. F., Wuhrer M., Wright A. F., Rudd P. M., Hayward C., Aulchenko Y., Campbell H., and Rudan I. (2013) Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLoS Genet. 9, e1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruhaak L. R., Koeleman C. A., Uh H. W., Stam J. C., van Heemst D., Maier A. B., Houwing-Duistermaat J. J., Hensbergen P. J., Slagboom P. E., Deelder A. M., and Wuhrer M. (2013) Targeted biomarker discovery by high throughput glycosylation profiling of human plasma alpha1-antitrypsin and immunoglobulin A. PLoS ONE 8, e73082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saldova R., Asadi Shehni A., Haakensen V. D., Steinfeld I., Hilliard M., Kifer I., Helland A., Yakhini Z., BØrresen-Dale A. L., and Rudd P. M. (2014) Association of N-glycosylation with breast carcinoma and systemic features using high-resolution quantitative UPLC. J. Proteome Res. 13, 2314–2327 [DOI] [PubMed] [Google Scholar]
- 27. Stumpo K. A., and Reinhold V. N. (2010) The N-glycome of human plasma. J. Proteome Res. 9, 4823–4830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huffman J. E., Pučić-Baković M., Klarić L., Hennig R., Selman M. H., Vučković F., Novokmet M., Krištić J., Borowiak M., Muth T., Polašek O., Razdorov G., Gornik O., Plomp R., Theodoratou E., Wright A. F., Rudan I., Hayward C., Campbell H., Deelder A. M., Reichl U., Aulchenko Y. S., Rapp E., Wuhrer M., and Lauc G. (2014) Comparative performance of four methods for high-throughput glycosylation analysis of immunoglobulin G in genetic and epidemiological research. Mol. Cell. Proteomics 13, 1598–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reusch D., Haberger M., Maier B., Maier M., Kloseck R., Zimmermann B., Hook M., Szabo Z., Tep S., Wegstein J., Alt N., Bulau P., and Wuhrer M. (2015) Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles—Part 1: Separation-based methods. mAbs 7, 167–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reusch D., Haberger M., Falck D., Peter B., Maier B., Gassner J., Hook M., Wagner K., Bonnington L., Bulau P., and Wuhrer M. (2015) Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles—Part 2: Mass spectrometric methods. mAbs 7, 732–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clerc F., Reiding K. R., Jansen B. C., Kammeijer G. S., Bondt A., and Wuhrer M. (2016) Human plasma protein N-glycosylation. Glycoconj. J. 33, 309–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Man Y. A., Dolhain R. J., van de Geijn F. E., Willemsen S. P., and Hazes J. M. (2008) Disease activity of rheumatoid arthritis during pregnancy: results from a nationwide prospective study. Arthritis Rheum. 59, 1241–1248 [DOI] [PubMed] [Google Scholar]
- 33. Stockmann H., O'Flaherty R., Adamczyk B., Saldova R., and Rudd P. M. (2015) Automated, high-throughput serum glycoprofiling platform. Integr. Biol. (Camb.) 7, 1026–1032 [DOI] [PubMed] [Google Scholar]
- 34. Akmačić I. T., Ugrina I., Štambuk J., Gudelj I., Vučković F., Lauc G., and Pučić-Baković M. (2015) High-throughput glycomics: Optimization of sample preparation. Biochemistry 80, 934–942 [DOI] [PubMed] [Google Scholar]
- 35. Adamczyk B., Stöckmann H., O'Flaherty R., Karlsson N. G., and Rudd P. M. (2017) High-throughput analysis of the plasma N-glycome by UHPLC. Methods Mol. Biol. 1503, 97–108 [DOI] [PubMed] [Google Scholar]
- 36. Campbell M. P., Royle L., Radcliffe C. M., Dwek R. A., and Rudd P. M. (2008) GlycoBase and autoGU: Tools for HPLC-based glycan analysis. Bioinformatics 24, 1214–1216 [DOI] [PubMed] [Google Scholar]
- 37. Hennig R., Reichl U., and Rapp E. (2011) A software tool for automated high-throughput processing of CGE-LIF based glycoanalysis data, generated by a multiplexing capillary DNA sequencer. Glycoconj. J. 28, 331 [Google Scholar]
- 38. Hennig R., Rapp E., Kottler R., Cajic S., Borowiak M., and Reichl U. (2015) N-glycosylation fingerprinting of viral glycoproteins by xCGE-LIF. Methods Mol. Biol. 1331, 123–143 [DOI] [PubMed] [Google Scholar]
- 39. Bladergroen M. R., Reiding K. R., Hipgrave Ederveen A. L., Vreeker G. C., Clerc F., Holst S., Bondt A., Wuhrer M., and van der Burgt Y. E. (2015) Automation of high-throughput mass spectrometry-based plasma N-glycome analysis with linkage-specific sialic acid esterification. J. Proteome Res. 14, 4080–4086 [DOI] [PubMed] [Google Scholar]
- 40. Reiding K. R., Blank D., Kuijper D. M., Deelder A. M., and Wuhrer M. (2014) High-throughput profiling of protein N-glycosylation by MALDI-TOF-MS employing linkage-specific sialic acid esterification. Anal. Chem. 86, 5784–5793 [DOI] [PubMed] [Google Scholar]
- 41. Klein A. (2008) Human total serum N-glycome. Adv. Clin. Chem. 46, 51–85 [PubMed] [Google Scholar]
- 42. Nairn A. V., York W. S., Harris K., Hall E. M., Pierce J. M., and Moremen K. W. (2008) Regulation of glycan structures in animal tissues: transcript profiling of glycan-related genes. J. Biol. Chem. 283, 17298–17313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Freeze H. H. (2006) Genetic defects in the human glycome. Nat. Rev. Genet. 7, 537–551 [DOI] [PubMed] [Google Scholar]
- 44. Kang P., Mechref Y., and Novotny M. V. (2008) High-throughput solid-phase permethylation of glycans prior to mass spectrometry. Rapid Commun. Mass Spectrom. 22, 721–734 [DOI] [PubMed] [Google Scholar]
- 45. RCore Team. (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 46. Benjamini Y., and Hochberg Y. (1995) Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B Met. 57, 289–300 [Google Scholar]
- 47. Varki A., Cummings R. D., Aebi M., Packer N. H., Seeberger P. H., Esko J. D., Stanley P., Hart G., Darvill A., Kinoshita T., Prestegard J. J., Schnaar R. L., Freeze H. H., Marth J. D., Bertozzi C. R., Etzler M. E., Frank M., Vliegenthart J. F., Lütteke T., Perez S., Bolton E., Rudd P., Paulson J., Kanehisa M., Toukach P., Aoki-Kinoshita K. F., Dell A., Narimatsu H., York W., Taniguchi N., and Kornfeld S. (2015) Symbol nomenclature for graphical representations of glycans. Glycobiology 25, 1323–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ceroni A., Maass K., Geyer H., Geyer R., Dell A., and Haslam S. M. (2008) GlycoWorkbench: A tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 7, 1650–1659 [DOI] [PubMed] [Google Scholar]
- 49. Leymarie N., Griffin P. J., Jonscher K., Kolarich D., Orlando R., McComb M., Zaia J., Aguilan J., Alley W. R., Altmann F., Ball L. E., Basumallick L., Bazemore-Walker C. R., Behnken H., Blank M. A., Brown K. J., Bunz S. C., Cairo C. W., Cipollo J. F., Daneshfar R., Desaire H., Drake R. R., Go E. P., Goldman R., Gruber C., Halim A., Hathout Y., Hensbergen P. J., Horn D. M., Hurum D., Jabs W., Larson G., Ly M., Mann B. F., Marx K., Mechref Y., Meyer B., Möginger U., Neusüß C., Nilsson J., Novotny M. V., Nyalwidhe J. O., Packer N. H., Pompach P., Reiz B., Resemann A., Rohrer J. S., Ruthenbeck A., Sanda M., Schulz J. M., Schweiger-Hufnagel U., Sihlbom C., Song E., Staples G. O., Suckau D., Tang H., Thaysen-Andersen M., Viner R. I., An Y., Valmu L., Wada Y., Watson M., Windwarder M., Whittal R., Wuhrer M., Zhu Y., and Zou C. (2013) Interlaboratory study on differential analysis of protein glycosylation by mass spectrometry: the ABRF glycoprotein research multi-institutional study 2012. Mol. Cell. Proteomics 12, 2935–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wada Y., Azadi P., Costello C. E., Dell A., Dwek R. A., Geyer H., Geyer R., Kakehi K., Karlsson N. G., Kato K., Kawasaki N., Khoo K. H., Kim S., Kondo A., Lattova E., Mechref Y., Miyoshi E., Nakamura K., Narimatsu H., Novotny M. V., Packer N. H., Perreault H., Peter-Katalinic J., Pohlentz G., Reinhold V. N., Rudd P. M., Suzuki A., and Taniguchi N. (2007) Comparison of the methods for profiling glycoprotein glycans—HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology 17, 411–422 [DOI] [PubMed] [Google Scholar]
- 51. Ito H., Kaji H., Togayachi A., Azadi P., Ishihara M., Geyer R., Galuska C., Geyer H., Kakehi K., Kinoshita M., Karlsson N. G., Jin C., Kato K., Yagi H., Kondo S., Kawasaki N., Hashii N., Kolarich D., Stavenhagen K., Packer N. H., Thaysen-Andersen M., Nakano M., Taniguchi N., Kurimoto A., Wada Y., Tajiri M., Yang P., Cao W., Li H., Rudd P. M., and Narimatsu H. (2016) Comparison of analytical methods for profiling N- and O-linked glycans from cultured cell lines: HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycoconj. J. 33, 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hemström P., and Irgum K. (2006) Hydrophilic interaction chromatography. J. Sep. Sci. 29, 1784–1821 [DOI] [PubMed] [Google Scholar]
- 53. Callewaert N., Geysens S., Molemans F., and Contreras R. (2001) Ultrasensitive profiling and sequencing of N-linked oligosaccharides using standard DNA-sequencing equipment. Glycobiology 11, 275–281 [DOI] [PubMed] [Google Scholar]
- 54. Gong B., Hoyt E., Lynaugh H., Burnina I., Moore R., Thompson A., and Li H. (2013) N-glycosylamine-mediated isotope labeling for mass spectrometry-based quantitative analysis of N-linked glycans. Anal. Bioanal. Chem. 405, 5825–5831 [DOI] [PubMed] [Google Scholar]
- 55. Alvarez-Manilla G., Warren N. L., Abney T., Atwood J. 3rd, Azadi P., York W. S., Pierce M., and Orlando R. (2007) Tools for glycomics: Relative quantitation of glycans by isotopic permethylation using 13CH3I. Glycobiology 17, 677–687 [DOI] [PubMed] [Google Scholar]
- 56. Zhou S., Dong X., Veillon L., Huang Y., and Mechref Y. (2017) LC-MS/MS analysis of permethylated N-glycans facilitating isomeric characterization. Anal. Bioanal. Chem. 409, 453–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhong X., Chen Z., Snovida S., Liu Y., Rogers J. C., and Li L. (2015) Capillary electrophoresis-electrospray ionization-mass spectrometry for quantitative analysis of glycans labeled with multiplex carbonyl-reactive tandem mass tags. Anal. Chem. 87, 6527–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Guttman M., and Lee K. K. (2016) Site-specific mapping of sialic acid linkage isomers by ion mobility spectrometry. Anal. Chem. 88, 5212–5217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jansen B. C., Bondt A., Reiding K. R., Lonardi E., de Jong C. J., Falck D., Kammeijer G. S., Dolhain R. J., Rombouts Y., and Wuhrer M. (2016) Pregnancy-associated serum N-glycome changes studied by high-throughput MALDI-TOF-MS. Sci. Rep. 6, 23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ruhaak L. R., Uh H. W., Deelder A. M., Dolhain R. E., and Wuhrer M. (2014) Total plasma N-glycome changes during pregnancy. J. Proteome Res. 13, 1657–1668 [DOI] [PubMed] [Google Scholar]
- 61. Parekh R. B., Dwek R. A., Sutton B. J., Fernandes D. L., Leung A., Stanworth D., Rademacher T. W., Mizuochi T., Taniguchi T., Matsuta K., and et al. (1985) Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature 316, 452–457 [DOI] [PubMed] [Google Scholar]
- 62. Reiding K. R., Vreeker G. C. M., Bondt A., Bladergroen M. R., Hazes J. M. W., van der Burgt Y. E. M., Wuhrer M., and Dolhain R. (2017) Serum protein N-glycosylation changes with rheumatoid arthritis disease activity during and after pregnancy. Front. Med. 4, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bondt A., Selman M. H., Deelder A. M., Hazes J. M., Willemsen S. P., Wuhrer M., and Dolhain R. J. (2013) Association between galactosylation of immunoglobulin G and improvement of rheumatoid arthritis during pregnancy is independent of sialylation. J. Proteome Res. 12, 4522–4531 [DOI] [PubMed] [Google Scholar]
- 64. Bondt A., Rombouts Y., Selman M. H., Hensbergen P. J., Reiding K. R., Hazes J. M., Dolhain R. J., and Wuhrer M. (2014) Immunoglobulin G (IgG) Fab glycosylation analysis using a new mass spectrometric high-throughput profiling method reveals pregnancy-associated changes. Mol. Cell. Proteomics 13, 3029–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brinkman-van der Linden E. C., de Haan P. F., Havenaar E. C., and van Dijk W. (1998) Inflammation-induced expression of sialyl LewisX is not restricted to alpha1-acid glycoprotein but also occurs to a lesser extent on alpha1-antichymotrypsin and haptoglobin. Glycoconj. J. 15, 177–182 [DOI] [PubMed] [Google Scholar]
- 66. Havenaar E. C., Axford J. S., Brinkman-van der Linden E. C., Alavi A., Van Ommen E. C., van het Hof B., Spector T., Mackiewicz A., and Van Dijk W. (1998) Severe rheumatoid arthritis prohibits the pregnancy-induced decrease in alpha3-fucosylation of alpha1-acid glycoprotein. Glycoconj. J. 15, 723–729 [DOI] [PubMed] [Google Scholar]
- 67. Kolarich D., Weber A., Turecek P. L., Schwarz H. P., and Altmann F. (2006) Comprehensive glyco-proteomic analysis of human alpha1-antitrypsin and its charge isoforms. Proteomics 6, 3369–3380 [DOI] [PubMed] [Google Scholar]
- 68. Lee J. Y., Lee H. K., Park G. W., Hwang H., Jeong H. K., Yun K. N., Ji E. S., Kim K. H., Kim J. S., Kim J. W., Yun S. H., Choi C. W., Kim S. I., Lim J. S., Jeong S. K., Paik Y. K., Lee S. Y., Park J., Kim S. Y., Choi Y. J., Kim Y. I., Seo J., Cho J. Y., Oh M. J., Seo N., An H. J., Kim J. Y., and Yoo J. S. (2016) Characterization of site-specific N-glycopeptide isoforms of alpha-1-acid glycoprotein from an interlaboratory study using LC-MS/MS. J. Proteome Res. 15, 4146–4164 [DOI] [PubMed] [Google Scholar]
- 69. Wilson R., Maclean M. A., Jenkins C., Kinnane D., Mooney J., and Walker J. J. (2001) Abnormal immunoglobulin subclass patterns in women with a history of recurrent miscarriage. Fertil. Steril. 76, 915–917 [DOI] [PubMed] [Google Scholar]
- 70. Jefferis R., Lund J., Mizutani H., Nakagawa H., Kawazoe Y., Arata Y., and Takahashi N. (1990) A comparative study of the N-linked oligosaccharide structures of human IgG subclass proteins. Biochem. J. 268, 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Larsson A., Palm M., Hansson L. O., Basu S., and Axelsson O. (2008) Reference values for alpha1-acid glycoprotein, alpha1-antitrypsin, albumin, haptoglobin, C-reactive protein, IgA, IgG and IgM during pregnancy. Acta Obstet. Gynecol. Scand. 87, 1084–1088 [DOI] [PubMed] [Google Scholar]
- 72. Ventham N. T., Gardner R. A., Kennedy N. A., Shubhakar A., Kalla R., Nimmo E. R., Fernandes D. L., Satsangi J., and Spencer D. I. (2015) Changes to serum sample tube and processing methodology does not cause intra-individual [corrected] variation in automated whole serum N-glycan profiling in health and disease. PLoS ONE 10, e0123028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data used in the manuscript can be found online at https://data.mendeley.com/datasets/f73bxyjb6f/draft?a=cef35bea-088e-44c4-968e-4b598035fb74.