Abstract
Numerous databases containing information about DNA, RNA, and protein variations are available. Gene-specific variant databases (locus-specific variation databases, LSDBs) are typically curated and maintained for single genes or groups of genes for a certain disease(s). These databases are widely considered as the most reliable information source for a particular gene/protein/disease, but it should also be made clear they may have widely varying contents, infrastructure, and quality. Quality is very important to evaluate because these databases may affect health decision-making, research, and clinical practice. The Human Variome Project (HVP) established a Working Group for Variant Database Quality Assessment. The basic principle was to develop a simple system that nevertheless provides a good overview of the quality of a database. The HVP quality evaluation criteria that resulted are divided into four main components: data quality, technical quality, accessibility, and timeliness. This report elaborates on the developed quality criteria and how implementation of the quality scheme can be achieved. Examples are provided for the current status of the quality items in two different databases, BTKbase, an LSDB, and ClinVar, a central archive of submissions about variants and their clinical significance.
Keywords: genetic variation, database quality, quality scheme, components of quality, Human Variome Project, locus-specific variation databases, LSDB, gene variant databases
Introduction
Thousands of gene, RNA, and protein variation databases have been developed over the last two decades. They have widely varying content, resources, infrastructure, and quality. Assessment of their quality is ever more important as use increases and information is compared with other public databases. One fast increasing application area is interpretation of variations found by large-scale sequencing. For example, The U.S. Food and Drug Administration (FDA) is discussing how to leverage public databases to evaluate clinical performance of genetic tests http://www.fda.gov/downloads/MedicalDevices/NewsEvents/WorkshopsConferences/UCM427869.pdf).
Gene variant databases (locus-specific variation databases, LSDBs) are typically curated and maintained for single genes or groups of genes related to certain disease(s). These resources are widely used and many are considered as the most reliable information source for a particular gene/protein/disease. Several recommendations and guidelines have been published for the establishment, curation, contents, and other features of these databases [Cotton et al., 2008, 2009; den Dunnen et al., 2009; Kaput et al., 2009; Kohonen-Corish et al., 2010; Patrinos et al., 2011; Celli et al., 2012; Howard et al., 2012; Patrinos et al., 2012; Vihinen et al., 2012].
Computer science and information technology (IT) communities have discussed and developed general criteria for database platform/system quality [Kuhn et al., 1994; Wang and Strong, 1996; Rittberger and Rittberger, 1997; Hoxmeier, 1998; Blakeslee and Rumble Jr, 2003]; however, there are no widely accepted systematic criteria and evaluation systems. Each domain has developed its own. The most relevant scheme for variation databases is BioDBcore, which has developed a list of core attributes of biological databases [Gaudet et al., 2011]. However, BioDBcore was explicitly not developed as a quality scheme.
Despite the large number of biological and medical databases, there are a few papers focused on their quality, assessment, and control. There are some exceptions, for example, in controlling data deposition quality in the PRIDE proteomics repository [Csordas et al., 2012], and developing methods for assessing annotation quality at UniProtKB [Bell et al., 2012]. Otherwise, databases such as ClinVar [Landrum et al., 2014] typically include discussion of validation and standardization in more general papers.
Accreditation systems are in place for genetic laboratories in many countries, but not for the generated data. OECD has published guidelines for qualitative assurance in molecular genetic testing (http://www.oecd.org/sti/biotech/38839788.pdf) and the U. S. College of American Pathologists (CAP) has documented standards for evaluating next-generation sequencing (NGS) tests [Aziz et al., 2015] but evaluation of the database(s) used to evaluate alleles was not addressed in detail.
The criteria described in computer science and information technology quality papers are more suitable to assess database platforms, such as LOVD [Fokkema et al., 2011], MUTbase [Riikonen and Vihinen, 1999], UMD [Beroud et al., 2000], and others, rather than to evaluate the content itself. For some platforms, there might be little difference between the databases due to limited customization and flexibility, but using customizable platforms such as LOVD, which can host multiple databases with different curators in the same installation, the quality of the content may differ depending on the curator(s).
The Human Variome Project (HVP) has an established process for development and acceptance of guidelines and standards for variation data [Smith and Vihinen, 2015]. The HVP established a Working Group for Variant Database Quality Assessment in 2012. This group reviewed the literature and utilized the expertise of the Working Group to identify additional components pertinent to database quality. The basic principle was to develop a simple system that can nevertheless provide a good overview of the quality of a database. The HVP guideline went to consortium members for consultation to obtain the views of the community before finalizing the document.
The outcome of the group was the first HVP accepted guideline (http://www.humanvariomeproject.org/finish/19/255.html), which details the four main components of data quality, technical quality, accessibility, and timeliness (see also [Smedley et al., 2010]) and suggests approaches for assessment. This paper elaborates on those quality criteria and reviews implementation of the quality scheme in two representative databases.
Quality Assessment Criteria
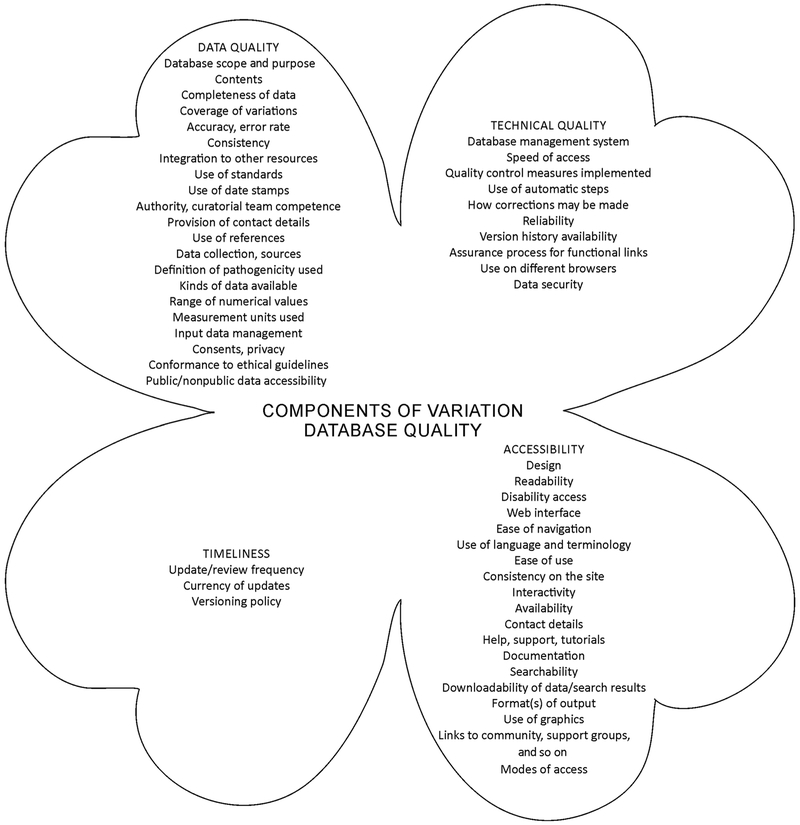
Criteria for quality assessment are divided into four major areas, each of which has several components (Fig. 1). These items are described briefly below.
Figure 1.
The four major components of variation database quality and their subcomponents.
Data Quality
The first major area is data quality. The data should be complete and accurate; however, there are several other quality issues on top of that. HVP is currently working toward guidelines for minimal requirements of variation databases. Once available, they should be applied to the quality scheme.
Database scope and purpose
End users need to know for what purpose the data were collected and what is the scope of the database. Variation data are typically used for several purposes including interpretation of variants found in patients, development of prediction methods, or generation of benchmark datasets. The applicability to a particular use depends on what data have been collected and how they have been processed.
Contents
Variation databases contain varying ranges of data types. Some resources include just details of the genetic variation, possibly with RNA and protein level changes, usually as predictions. Some databases contain patient data, even very detailed files. For ethical and privacy reasons, the contents of the latter databases are usually not publicly accessible. In the case of databases with clinical information, the extent of the nonpublic data needs to be disclosed so that users can contact curators in case they would like to have access to more detailed contents.
Completeness of data
It is essential that the dataset be as complete as possible and contain all the data items available for each case. Some estimate of the missing data is desirable.
Coverage of variations
One of the most important aspects users are interested in is whether the database contains all the known cases. Thus, data coverage is a major quality criterion.
Accuracy (error rate)
The curated data should be as accurate as possible with the smallest possible error rate. Unfortunately, even the original publications for variants sometimes contain errors.
Consistency
The language and spelling should be consistent throughout the database. This is important both for human readers as well as computer software to find all relevant cases. Variations should be annotated based on the same reference sequences throughout the database. Also, the type of curation, that is, how the data are collected and annotated, should remain the same throughout the database. If it changes over time, differences should be annotated.
Integration to other resources
Variation databases should not be used in isolation, instead combined and integrated with other information sources. From the point of view of quality the quantity, quality and type of integrated information are important.
Use of standards
Several standards have been developed for variation databases. These include systematic gene symbols and names, for human those available through the HUGO Gene Nomenclature Committee (HGNC) [Gray et al., 2015]. Reference sequences have to be specified including version numbers unless Locus Reference Genomic (LRG) [Dalgleish et al., 2010] entries are used. LRGs are recommended for human sequences as they are stable and allow unambiguous mapping of positions. For variant descriptions the Human Genome Variation Society (HGVS) nomenclature [den Dunnen and Antonarakis, 2001] should be used. Chromosomal aberrations should be described using the International System for Human Cytogenetic Nomenclature (ISCN2013) [Shaffer et al., 2013]. The standards used for describing the data within the database should be described. It is recommended to use ontologies and controlled vocabularies whenever possible: the Human Phenotype Ontology (HPO) [Robinson et al., 2008] for descriptions of phenotypes and diseases; and Variation Ontology (VariO) [Vihinen, 2014a] for description of effects, mechanisms and consequences of variants the Variation Ontology (VariO) [Vihinen, 2014a].
Use of date stamps
The dates when data were entered into the database as well as when last modified should be available.
Authority, curatorial team competence
The database curator team should have competence on the genes and diseases to be able to provide an authoritative database.
Provision of contact details
Contact information has to be available on the database homepage so users can contact database curators to obtain further details, for queries about included data, and so on.
Use of references
The cases and details in the database should cite publications and other public records when available. The correctness and completeness of these references are important components of database quality.
Data collection, sources
Curators need to provide details about how the data have been collected, what are inclusion/exclusion criteria, as well as what are the sources of the data.
Definition of pathogenicity used
If information on pathogenicity of the variations is provided, the definitions of the pathogenicity metrics or classification must also be available on the database Website.
Kinds of data available
The database description has to include description of the data types included, and whether variant data came from NGS pipelines, single-nucleotide variants from gene panels, or other types of experiments.
Range of numerical values
Minimum and maximum values for all numerical data should be provided.
Measurement units used
All parameters must be provided in correct units and the units described in the database. This applies especially to clinical details.
Input data management
The data need to be consistent with submission mechanisms so that all the details provided to the system are available in the database. Input data also need to be validated to ensure they are appropriate for entry into the database.
Consents, privacy
The curators must have appropriate consents for the cases coming from their own laboratories and require submitters from other laboratories to take care of necessary and appropriate consents. It is important to protect the privacy of the subjects.
Conformance to ethical guidelines
All ethical issues mandated by the home institute and country of the database hosting provider have to be professionally met. There are published guidelines for LSDB curators [Povey et al., 2010] that are currently under revision at HVP to make them better suited for LSDB practice.
Public/nonpublic data accessibility
Most of the existing variation databases are publicly available and others are accessible only by consortia members or paid customers. The availability of the data has to be described clearly on the database home page. The recommendation is to provide as much freely available data as possible for the full benefit of the scientific and clinical communities.
Technical Quality
Technical quality refers to the quality of the database implementation including details of the management system, system automation, server reliability, and other aspects.
Database management system
A dedicated database management system should be used for storing and sharing variation information. These systems have several benefits, the most important being systematic data description as well as ease of search of data. The most commonly used variant database management system is LOVD [Fokkema et al., 2011], the others include MUTbase [Riikonen and Vihinen, 1999], and UMD [Beroud et al., 2000]. Use of generic database management systems or static Web pages is not recommended.
Speed of access
Any requested data should be retrieved from the database quickly. Users of databases are not willing to wait. The speed is dependent on a number of factors including the database implementation and management system, system load, and internet connection.
Quality control measures implemented
What measures have been implemented to guarantee the quality in the database? LSDBs contain numerous details and all of them should be correct (see previous section). It is very easy to introduce errors and thus systems for increasing accuracy are of importance. Details of such measures are needed for quality assessment.
Use of automatic steps
Some parts of data submission and generation can be automated, depending on the database management system. One of the most common automated features is generation, checking, and correction of systematic HGVS variant names with Mutalyzer [Wildeman et al., 2008].
How corrections may be made
The process for making corrections needs to be documented.
Reliability
An important aspect of the technical reliability is the uptime of the service—how often servers are down due to problems or maintenance and if known beforehand are they announced at the Web site?
Version history availability
Details of database version history are required, especially if the old versions are stored. Any significant alterations to the database should be described.
Assurance process for functional links
Variation databases can contain numerous links to other resources. These links need to be frequently checked as resources may move or alter their links. This is a process that can often be automated and this is to be recommended.
Use on different browsers
The database Web site should be implemented so that it can be browsed with any of the widely used tools, thus browser-specific features should be avoided.
Data security
The database needs to be secured in different ways. These include operating system, data backups, firewall, and other security measures to access the actual database. The security measures should not affect normal use of the database.
Accessibility
Accessibility of the database covers a number of components including user-friendliness, consistency, and availability. A key use of the term relates to accessibility for users with disabilities (http://www.w3.org/standards/webdesign/accessibility).
Design
The design of the database and its Web pages should allow easy and intuitive navigation within the resource without need for accessing help instructions (which need to be in place).
Readability
This relates both to the readability of the contents as well as that of the Web service. To avoid semantic ambiguities, the language used has to be systematic. When designing the contents and mode of presentation, database curators should bear in mind the most obvious use cases and adjust accordingly.
Disability access
Web sites should take account of and conform to guidelines to make them accessible to those with disabilities, such as those developed by the World Wide Web Consortium (W3C) (http://www.w3.org/TR/2008/REC-WCAG20-20081211/).
Web interface
Web services should be user-friendly and readable, meaning easy to access and use, fast response time, logical interface, and so on. The user interface should be relatively simple and be consistent at different levels. Colors, text types, and so on can be used for hierarchy and clarity. Color combinations should be selected with care to prevent items from becoming invisible to the color blind.
Ease of navigation
The Web pages must be divided into logical sections to allow navigation. Links must be clearly visible.
Use of language and terminology
In addition to contents also, the language has to be consistent and correct. Ontologies and other standards should be used for systematics.
Ease of use
Database users are interested in finding information relevant for them; thus, it should be made easy to find that information. Databases may contain plenty of useful features and data items that are less frequently requested but highly important for some users. These data need to be logically linked. Optimal solutions may not necessarily provide, for example, all search items at a time, instead group them logically. By considering the most common user scenarios, the system can be designed for different kinds of queries, users, and so on.
Consistency on the site
The database interface has to be consistent so as to be logically browsable and searchable.
Interactivity
The user interface has to be interactive and thus effective.
Availability
Variation databases should be made freely available. It is not recommended to require registration as this slows the use of the data and may prevent or hamper programmatic access.
How to contact
In case of problems with the resource contact details must be provided.
Help, support, tutorials
Material to help and support users should be provided on the Web page.
Documentation
Several details related to the database should be provided including purpose, scope, motivation, copyright statement, user licenses, disclaimer, database policy, data items, annotation guidelines, and others. HVP has released a guideline for Disclaimer Statements for Gene/Disease Specific Databases (http://www.humanvariomeproject.org/finish/19/273.html).
Searchability
Apart from the smallest databases that contain just a handful of cases, search engines, or possibilities for searching the contents of the database need to be implemented. These should provide as much flexibility to the user as possible.
Downloadability of data/search results
In the interest of open access to data, the database should provide the possibility of downloading database contents and results of searches.
Format(s) of output
For further analyses, it is necessary to provide details for the output formats. Where widely accepted standards exist, for example, for NGS data, these should be used.
Use of graphics
Graphics can be used for many purposes on variation database Web sites. The graphics should be purposeful and special attention paid to their clarity and intuitiveness.
Links to community, support groups, and so on
The database site should link to research organizations and societies in the field, especially those for disease groups and patient organizations.
Modes of access
In addition to browser use are there other ways to access the Web services, for example, API (application programmatic access)? These facilitate easy access to data sets and are encouraged.
Timeliness
Timeliness refers to the quality of containing up-to-date information in the variation database.
Update/review frequency
Information about how often the database is updated or reviewed. In case of rare diseases, new cases may be identified only occasionally.
Currency of updates
This item relates to how often new data are included to the database (to be distinguished from when new public releases are made). Does the database contain information about the latest variations published?
Versioning policy
Information about versioning, and distribution of old versions, if supported, should be provided.
Assessing Database Quality
Here, we have aimed at compiling and listing items and components of variation databases. These need to be implemented into a quality scheme, and this is currently being discussed at HVP. The quality scheme could enable ranking of the databases, for example, in the style of stars used for ranking quality of hotels, perhaps under the major categories listed, or could be color coded. In addition to overall ranking, it would be useful to provide a breakdown to more detailed quality components. This information would allow (prospective) database users to obtain an idea about the quality of databases as well as provide guidelines and encouragement for database curators to improve their services. The accrediting body, such as HVP, could provide an electronic seal (perhaps at different grades) for display on the database home page.
Many of the quality components (Fig. 1) need to be more specifically defined to enable numerical ranking. This remains the task of the quality assessment implementation. Plenty of technical information required for the quality audit of a database might be provided by the database administrators and some of it could be collected automatically. A standard file format for externally reporting quality information could be developed, supporting self-assessment, and saving time for the quality evaluators. Although popular platforms (e.g., LOVD) could employ scripts to automatically collect a number of facts, there will remain a number of quality items that need to be evaluated manually. A dedicated team is needed for that purpose.
The quality items described above need to be translated into quantitative quality measures by a group of individuals knowledgeable about variations, databases, and Web services. Certain database quality items could be collected from curators. Tools need to be developed, for example, for automatic data collection from databases, and then the actual quality assessment step can be performed. The final assessment has to be performed by an impartial body that does not have conflict of interest. The results of this process will provide the details of quality of a database. Users could then use the results of the evaluation to choose appropriate database(s), and the curators could obtain valuable feedback about how to improve their services. The assessment results should be visible on the database home page. The process would need to be repeated at frequent intervals to be able to reflect the current quality status of variation databases.
Case Study: BTKbase
BTKbase (http://structure.bmc.lu.se/idbase/BTKbase/) is an LSDB for Bruton agammablobulinemia tyrosine kinase (BTK) variations causing X-linked agammaglobulinemia (XLA) originally released 1995 [Vihinen et al., 1995] and then maintained ever since the last reports being [Väliaho et al., 2006; Schaafsma and Vihinen, 2015]. There are currently 1,357 public entries. Previously, BTK-base used the MUTbase database management system [Riikonen and Vihinen, 1999], which was originally developed for primary immunodeficiency variation databases (IDbases) [Piirilä et al., 2006] at http://structure.bmc.lu.se/idbase/, recently the variant descriptions were moved to the LOVD system. Here, the quality issues as implemented currently in the BTKbase are discussed.
Data Quality
Database scope, purpose, and contents are briefly described on the Web page. Note that BTKbase is more than just the database in LOVD. The database homepage at http://structure.bmc.lu.se/idbase/BTKbase/ contains more information than is possible to include in a standard LOVD database. Most of the entries in BTKbase originate from literature, during the curation process all the relevant data items included to the database are compiled from articles and other sources. The database especially serves the community of primary immunodeficiency researchers and along with the other IDbases was developed under the auspices of the European Society of ImmunoDeficiencies (ESID) as a community resource. After the initial interest, however, the number of direct submissions is rather low. The database is updated periodically, the latest version is up-to-date.
The database has had a number of quality checks and controls. The accuracy was further improved while moving into the LOVD database management software. Several consistency checks and corrections were made during this process. Because during the 20 years the database has existed there have been numerous curators and the software and submission routines have changed, inconsistencies were noticed and corrected. Where possible, variant descriptions were checked with the Mutalyzer tool, but currently checks of gross deletions are limited by the maximum size of custom reference sequences (four million base pairs). The LOVD software also contains consistency and other quality checks. The variations are systematically described using the HGVS format and the Variation Ontology variation type annotations [Vihinen, 2014a]. The language used for, for example, symptoms, country of origin, and so on is English, and thus standardized. The LOVD software has predefined values for certain fields of which the user has to choose from.
There are extensive links to other resources. For instance, literature references can be accessed through links to PubMed, gene accessions through links to HGNC and Entrez Gene, and diseases are linked to OMIM. Links to other BTK resources include those to HGMD, GeneCards and GeneTests, and the BTKbase pages. The use of HGNC standards for gene names and symbols, and HGVS format for variant descriptions are mandatory. Reference sequences are taken from LRG and NCBI/RefSeq.
Date stamps are generated for the created_date and edited_date fields. These fields are available for the gene, transcript, variant, individual, and screening data.
The curators are experts in the disease. Prof. C. I. Edvard Smith is one of authors of the paper indicating BTK to be in charge of XLA when containing variations [Vetrie et al., 1993]. He has studied the gene and disease ever since. Prof. Vihinen originally established the database along with the other IDbases. Contact details are available for being in touch with the curators.
Literature references are used extensively, with links to PubMed. Data originate mainly from literature, but there are also some direct submissions. Submission of new variants and patients is done through a guided submission process defined by the LOVD system. The data in BTKbase are from XLA patients. XLA patients display a spectrum of phenotypes ranging from mild to severe. Details both for signs and symptoms as well as laboratory values are included in BTKbase for a number of patients.
The contents of the database have been published [Vihinen et al., 1997].
The range of numerical values is not provided. Units used in specific columns are given in the column headings, and are also indicated in the submission forms. Input of new data is done by using submission forms. Since most data come from already published cases, no new consents are necessary. Submitters of nonpublished data have to take care of getting patient’s consents. According to local authorities, the database does not need an ethical permission as all the cases are anonymous (not known even for curators) and mainly coming from published reports. Almost all data are publicly available, except a few confidential cases waiting to be published.
Technical Quality
For the previous versions of BTKbase MUTbase software was used, now the LOVD database management software is used (presently version 3.0 build 14). It is the most widely used system for LSDBs. Since the database is part of an LOVD installation hosted by LUMC, Leiden, the Netherlands, speed of access is dependent on that server and its network connection. Technical reliability depends on the servers in Leiden. Backups, firewalls, and strict access rights are taken care of by the database administrators at LUMC.
Data are manually curated with automated steps. New data are submitted through a submission form. Automated steps include the use of the submission forms, which include check of variant descriptions with Mutalyzer, and the use of VariOtator [Schaafsma and Vihinen, 2016] for annotating variants with Variation Ontology terms.
Corrections can be made by the curators, who do so whenever they are needed. Database version number is available. Older versions of the database are not available.
All links are to established services of which the URL tend not to change often. The developers of the LOVD software check for the correctness of the most common links. The LOVD software has been tested on all major browsers.
Accessibility
The design of the database is mainly dictated by the LOVD developers and the LOVD3 shared administrators, but there is room for user-defined columns. Parts in http://structure.bmc.lu.se/idbases were developed in cooperation with the primary immunodeficiency community. The data are presented in a systematic way, including use and links to ImmunoDeficiency Resource (IDR) [Samarghitean et al., 2007] for more general information about BTK and XLA, as well as PIDs more in general. BTKbase use the same systematics in regards to language and terms as used in IDR.
No special measures were taken for disability access. People can zoom in and out using the browser’s features. The LOVD graphic user interface has different views for the logical subsections for genes, transcripts, variants, individuals, diseases, screenings, submission, and documentation. There are search boxes for every field. Response time is in general good, downtime of the server has been minimal. The BTKbase Website (http://structure.bmc.lu.se/idbase/BTKbase) has figures and links to the LOVD database and other resources.
Both the BTKbase and LOVD BTK pages are divided into logical subsections that are easy to navigate by using the provided menus. On the IDR fact file pages, systematic and consistent terminology has been used. In the database, there are columns with VariO terms for annotation of the variant descriptions on DNA, RNA, and protein level.
LOVD provides search boxes for fields in the database. The IDR pages have a clear menu and links to other fact files and resources. These pages also provide tables and figures in which the data are grouped into useful and/or logical divisions.
Consistency on the site is part of the LOVD and IDR design. All views in LOVD are query based. BTKbase is freely accessible. Contact information is available. LOVD has extensive documentation as how to install and use the LOVD software. Support can be obtained through the LOVD developers and/or the database curators. Tutorials have not been developed for BTKbase but are available for LOVD.
Details related to the database can be found in the database documentation. Disclaimers, database policies, and so on can be found on the database homepages. Guidelines concerning annotation using the VariO have been published [Vihinen, 2014b, 2015). The built-in search possibilities of the LOVD software (all fields are searchable) offer the users much flexibility. Data in BTKbase are freely accessible to users, apart from some confidential cases to be published in the future. LOVD offers a standard output for downloads. LOVD and the BTKbase home pages offer some graphical displays. BTKbase offers links to research organizations, patient groups, clinical information, molecular information, and so on. Apart from the regular access through a browser, there is an API available that offers limited access to the LOVD database.
Timeliness
BTKbase is updated infrequently. For checking the status, there is the “Date last updated” field on the homepage. Since BTKbase is run basically without funding, it is not possible to update it constantly. Users can request updates.
Old versions are not available.
Case Study: ClinVar
ClinVar is an archive for interpretations of the clinical significance of variants across the genome [Landrum et al., 2014]. ClinVar staff do not curate the values of clinical significance in the database; all statements of clinical significance are provided by submitters, which include clinical testing laboratories, research laboratories, resources such as OMIM® and GeneReviews™, LSDBs, as well as expert panels and practice guidelines. Thus, ClinVar is not an LSDB but an archive aggregating data from multiple sources including LSDBs. ClinVar currently holds interpretations for more than 138,000 variants; for variants that affect a single gene, more than 4,600 genes are represented.
Data Quality
Data in ClinVar are public and freely available to all users for any purpose; submitters are required to indicate the purpose for which the data were collected (e.g., clinical testing, research) to aid the user in evaluating the appropriateness of data for their specific use. Other data that are required in a ClinVar submission include the variant, the disease or phenotype, the clinical significance, and limited information about the evidence, including the allele origin and the health status of individuals with the variant. Many more fields are optional in a submission, such as the age, ethnicity, and zygosity of individuals with the variant, and evidence about family history. However, since the data are fully public, submitters should not provide enough information to make individuals identifiable, and submitters should have appropriate consent to provide the data to ClinVar. Information describing the experimental method is optional in a ClinVar record and is infrequently provided by submitters.
Submitters also have the option of providing documentation for the criteria that they use to classify variants. This documentation, referred to in ClinVar as “assertion criteria,” is optional; however, it does increase the review status, represented graphically as a number of stars, for the submission.
Because ClinVar is dependent upon submissions, it is not possible for it to be a comprehensive database, either with respect to the number of variants represented or the evidence provided for each interpretation. That said, data in ClinVar can be compared with all variants registered in NCBI’s databases, especially through tools such as variation viewer (https://www.ncbi.nlm.nih.gov/variation/view/). Thus, access to a comprehensive list of reported variants is close at hand.
Accuracy of content depends on what was submitted. ClinVar does, however, validate data such as HGVS expressions for correct format and asserted sequence. The use of standards such as HGVS nomenclature for variants, HGNC for gene symbols, database identifiers like MIM numbers for diseases and HPO for observed phenotypes, and VariO terms for functional consequence allows specificity and consistency in data provided by diverse groups of submitters. ClinVar does provide reports back to submitters if content fails a validation step, and is adding automated checks when categories of gaps or errors are identified, for example, unexpected gene–disease relationships.
Links to related databases are provided; for example, most records in ClinVar link to the corresponding dbSNP or dbVar record. Links to other resources, including LSDBs, OMIM, and PubMed, are also provided.
ClinVar employs several date stamps, including the date a variant was first entered in the database, dates for each submission about that variant, and when the submitter last reinterpreted a variant.
Each group that submits data to ClinVar receives public attribution; submitting groups may also include a public contact person who is willing to be contacted by ClinVar users for more information about an interpretation.
Alternatively, citations may be provided as support for the interpretation of a variant identified elsewhere, such as in a clinical testing environment.
Technical Quality
ClinVar is maintained in a relational database management system (MS SQL Server). The database is backed up nightly, and archived to tape, as part of NCBI’s set of databases. Maintenance has built-in redundancy, with rollover to mirrored servers if there is an outage at the primary location. Web access to ClinVar is not based on the database that is used to process new records, but on redundant copies, with load balancing. These methods have been implemented to provide reliable, continuous access.
Response to any query should be within milliseconds.
ClinVar has been a developing suite of quality control measures. For example, NCBI developed internal tools to calculate HGVS expressions based on variant descriptions, and alignment of reference sequences to current assemblies or RefSeqGene/LRG sequences (https://www.ncbi.nlm.nih.gov/variation/hgvs/). This code base is used to validate variants submitted according to what looks like an HGVS name and to aggregate submissions based on different reference sequences (transcripts or genome assemblies). This code base is currently being reviewed and benchmarked against Mutalyzer. Gene symbols, sequence accessions, database identifiers, and terms used to interpret the effect of the variant are validated against local copies of data from public resources such as HGNC, INSDC, Ref-Seq, OMIM, HPO, Orphanet, VariO, and Sequence Ontology. Novel gene–disease relationships calculated from the genomic location of the variant are starting to be returned to submitters for review. Assertions of pathogenicity inconsistent with allele frequency are also under scrutiny.
ClinVar uses many automatic steps, including processing submission spreadsheets to XML format, loading subsets of data to the SQL database, aggregation of data, and generation of XML for reporting. In addition, automated processes are used to add value to submitted data, such as calculating HGVS expressions and genomic locations, identifying the corresponding dbSNP identifier, integrating information about allele frequency from 1000 Genomes and GO-ESP, and calculating molecular consequences for submitted variants based on NCBI’s gene annotation. Inconsistent interpretations from independent submitters are also identified and reported.
Corrections to data added by NCBI staff may be made at any time. Corrections to data provided by submitters may only be made by the submitting group. If any question is raised to ClinVar about content, ClinVar staff may contact a submitter to request an update.
Reliability
Given the redundancy built into ClinVar’s Web presence, the data are expected to be available 24 hr a day, 7 days a week. Any scheduled interruption that may reduce that redundancy is posted in advance.
The full ClinVar dataset is released as a monthly XML extract with a date stamp as the “version” of the database. Each ClinVar record is also accessioned and versioned. Individual submissions are assigned an accession with an SCV prefix; these SCV records are the basis of aggregate records provided by NCBI and assigned an accession with an RCV prefix. The history of individual records is accessible in the ClinVar XML file; support for Web access to previous versions of records is planned. Development is underway to accession and version variant-centric aggregate records as well.
NCBI’s quality assessment team periodically checks that representative links to standard resources are not broken. Any static page (one with supporting documentation for example) is tested for valid links before the page can go public.
Databases provided via NCBI must be functional on the set of browsers documented here: https://www.ncbi.nlm.nih.gov/guide/ browsers/. Each modification to the software displaying ClinVar’s data is tested on multiple platforms before the update can be released to public servers.
ClinVar’s database is backed up nightly. Original submission files are archived and backed up in file systems and in independent data archives. No external user can connect to the database directly; submissions are buffered though submission files or an independent submission system (https://submit.ncbi.nlm.nih.gov/) with appropriate authorization and authentication controls.
Accessibility
ClinVar provides ample documentation describing the purpose of the database, data standards that are used, how to access the data, disclaimers, statistics for the database, how to submit data, and how to search the database and use data that is presented on variant pages.
The design for ClinVar’s variant pages is still under active development. ClinVar staff are working with NCBI’s usability group to assess the strengths and weaknesses of the current design and develop an improved Web display. Improvements to the search functions of ClinVar are also in development, to make searching more intuitive to users.
As a Website of the United States federal government, ClinVar adheres to standards required for effective access by the disabled. All ClinVar pages include a link in the page footer to “Write to the Help Desk” for users to send questions and report problems.
Data in ClinVar are available for download in several formats, including a comprehensive XML report, VCF file, and summary reports for genes and transcripts, all of which are updated monthly. A summary of search results in the Web interface may also be downloaded. Programmatic access to the database is also available through NCBI’s E-Utilities; the esearch, esummary, and efetch functions are provided for ClinVar (https://www.ncbi.nlm.nih.gov/ clinvar/docs/maintenance_use).
Timeliness
New data are submitted to ClinVar daily, including both published and unpublished data. However, whether the most recently published variants are available in ClinVar depends on whether they are provided by a submitter; ClinVar is not staffed to curate the literature for all genes. The ClinVar Website is updated weekly and a comprehensive release (noted above) is provided monthly. Files that are released monthly are archived on the ftp site and dated.
References
- Aziz N, Zhao Q, Bry L, Driscoll DK, Funke B, Gibson JS, Grody WW, Hegde MR, Hoeltge GA, Leonard DG, Merker JD, Nagarajan R, et al. 2015. College of American Pathologists’ laboratory standards for next-generation sequencing clinical tests. Arch Pathol Lab Med 139:481–493. [DOI] [PubMed] [Google Scholar]
- Bell MJ, Gillespie CS, Swan D, Lord P. 2012. An approach to describing and analysing bulk biological annotation quality: a case study using UniProtKB. Bioinformatics 28:i562–i568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroud C, Collod-Beroud G, Boileau C, Soussi T, Junien C. 2000. UMD (Universal mutation database): a generic software to build and analyze locus-specific databases. Hum Mutat 15:86–94. [DOI] [PubMed] [Google Scholar]
- Blakeslee DM, Rumble J Jr. 2003. The essentials of a database quality process. Data Sci J 12:35–46. [Google Scholar]
- Celli J, Dalgleish R, Vihinen M, Taschner PE, den Dunnen JT. 2012. Curating gene variant databases (LSDBs): toward a universal standard. Hum Mutat 33:291–297. [DOI] [PubMed] [Google Scholar]
- Cotton RG, Al Aqeel AI, Al-Mulla F, Carrera P, Claustres M, Ekong R, Hyland VJ, Macrae FA, Marafie MJ, Paalman MH, Patrinos GP, Qi M, et al. 2009. Capturing all disease-causing mutations for clinical and research use: toward an effortless system for the Human Variome Project. Gene Med 11:843–849. [DOI] [PubMed] [Google Scholar]
- Cotton RG, Auerbach AD, Beckmann JS, Blumenfeld OO, Brookes AJ, Brown AF, Carrera P, Cox DW, Gottlieb B, Greenblatt MS, Hilbert P, Lehvaslaiho H, et al. 2008. Recommendations for locus-specific databases and their curation. Hum Mutat 29:2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas A, Ovelleiro D, Wang R, Foster JM, Rios D, Vizcaino JA, Hermjakob H. 2012. PRIDE: quality control in a proteomics data repository. Database (Oxford) 2012:bas004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgleish R, Flicek P, Cunningham F, Astashyn A, Tully RE, Proctor G, Chen Y, McLaren WM, Larsson P, Vaughan BW, Beroud C, Dobson G, et al. 2010. Locus Reference Genomic sequences: an improved basis for describing human DNA variants. Genome Med 2:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. 2001. Nomenclature for the description of human sequence variations. Hum Genet 109:121–124. [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Sijmons RH, Andersen PS, Vihinen M, Beckmann JS, Rossetti S, Talbot CC Jr., Hardison RC, Povey S, Cotton RG. 2009. Sharing data between LSDBs and central repositories. Hum Mutat 30:493–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. 2011. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat 32:557–563. [DOI] [PubMed] [Google Scholar]
- Gaudet P, Bairoch A, Field D, Sansone SA, Taylor C, Attwood TK, Bateman A, Blake JA, Bult CJ, Cherry JM, Chisholm RL, Cochrane G, et al. 2011. Towards BioDBcore: a community-defined information specification for biological databases. Nucleic Acids Res 39:D7–D10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. 2015. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res 43:D1079–D1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard HJ, Beaudet A, Gil-da-Silva Lopes V, Lyne M, Suthers G, Van den Akker P, Wertheim-Tysarowska K, Willems P, Macrae F. 2012. Disease-specific databases: why we need them and some recommendations from the Human Variome Project Meeting, May 28, 2011. Am J Med Genet A 158A:2763–2766. [DOI] [PubMed] [Google Scholar]
- Hoxmeier JA. 1998. Typology of database quality factors. Software Qual J 7:179–193. [Google Scholar]
- Kaput J, Cotton RG, Hardman L, Watson M, Al Aqeel AI, Al-Aama JY, Al-Mulla F, Alonso S, Aretz S, Auerbach AD, Bapat B, Bernstein IT, et al. 2009. Planning the human variome project: the Spain report. Hum Mutat 30:496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohonen-Corish MR, Al-Aama JY, Auerbach AD, Axton M, Barash CI, Bernstein I, Beroud C, Burn J, Cunningham F, Cutting GR, den Dunnen JT, Greenblatt MS, et al. 2010. How to catch all those mutations—the report of the third Human Variome Project Meeting, UNESCO Paris, May 2010. Hum Mutat 31:1374–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn P, Deplanque R, Fluck E. 1994. Criteria of quality assessment for scientific databases. J Chem Inf Comput Sci 34:517–519. [Google Scholar]
- Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. 2014. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42:D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrinos GP, Al Aama J, Al Aqeel A, Al-Mulla F, Borg J, Devereux A, Felice AE, Macrae F, Marafie MJ, Petersen MB, Qi M, Ramesar RS, et al. 2011. Recommendations for genetic variation data capture in developing countries to ensure a comprehensive worldwide data collection. Hum Mutat 32:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrinos GP, Smith TD, Howard H, Al-Mulla F, Chouchane L, Hadjisavvas A, Hamed SA, Li XT, Marafie M, Ramesar RS, Ramos FJ, de Ravel T, et al. 2012. Human Variome Project country nodes: documenting genetic information within a country. Hum Mutat 33:1513–1519. [DOI] [PubMed] [Google Scholar]
- Piirilä H, Väliaho J, Vihinen M. 2006. Immunodeficiency mutation databases (IDbases). Hum Mutat 27:1200–1208. [DOI] [PubMed] [Google Scholar]
- Povey S, Al Aqeel AI, Cambon-Thomsen A, Dalgleish R, den Dunnen JT, Firth HV, Greenblatt MS, Barash CI, Parker M, Patrinos GP, Savige J, Sobrido MJ, et al. 2010. Practical guidelines addressing ethical issues pertaining to the curation of human locus-specific variation databases (LSDBs). Hum Mutat 31:1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riikonen P, Vihinen M. 1999. MUTbase: maintenance and analysis of distributed mutation databases. Bioinformatics 15:852–859. [DOI] [PubMed] [Google Scholar]
- Rittberger M, Rittberger W. 1997. Measuring quality in the production of databases. J Inf Sci 23:25–37. [Google Scholar]
- Robinson PN, Kohler S, Bauer S, Seelow D, Horn D, Mundlos S. 2008. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet 83:610–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarghitean C, Väliaho J, Vihinen M. 2007. IDR knowledge base for primary immunodeficiencies. Immunome Res 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaafsma GC, Vihinen M. 2015. Genetic variation in Bruton tyrosine kinase. In: Plebani A, Lougaris V, editors. Agammaglobulinemia. Switzerland: Springer; p 75–85. [Google Scholar]
- Schaafsma GC, Vihinen M. 2016. VariOtator, a software tool for variation annotation with the Variation Ontology. Hum Mutat. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Shaffer LG, McGowan-Jordan J, Schmid M. 2013. ISCN 2013: an International System for Human Cytogenetic Nomenclature. Basel, Switzerland: Karger. [Google Scholar]
- Smedley D, Schofield P, Chen CK, Aidinis V, Ainali C, Bard J, Balling R, Birney E, Blake A, Bongcam-Rudloff E, Brookes AJ, Cesareni G, et al. 2010. Finding and sharing: new approaches to registries of databases and services for the biomedical sciences. Database (Oxford) 2010:baq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TD, Vihinen M. 2015. Standard development at the Human Variome Project. Database (Oxford) pii:bav024 00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Väliaho J, Smith CIE, Vihinen M. 2006. BTKbase: the mutation database for X-linked agammaglobulinemia. Hum Mut 27:1209–1217. [DOI] [PubMed] [Google Scholar]
- Vetrie D, Vořechovský Sideras P, Holland J, Davies A, Flinter F, Hammarström L, Kinnon C, Levinsky R, Bobrow M, Smith CIE, Bentley DR. 1993. The gene involved in X-linked agammaglobulinaemia is a member of the src family of protein-tyrosine kinases. Nature 361:226–233. [DOI] [PubMed] [Google Scholar]
- Vihinen M 2014a. Variation Ontology for annotation of variation effects and mechanisms. Genome Res 24:356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihinen M 2014b. Variation Ontology: annotator guide. J Biomed Semantics 5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihinen M 2015. Types and effects of protein variations. Hum Genet 134:405–421. [DOI] [PubMed] [Google Scholar]
- Vihinen M, Belohradsky BH, Haire RN, Holinski-Feder E, Kwan SP, Lappalainen I, Lehväslaiho H, Lester T, Meindl A, Ochs HD, Oilila J, Vorechovsky I, Weiss M, Smith CI. 1997. BTKbase, mutation database for X-linked agammaglobulinemia (XLA). Nucleic Acids Res 25:166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihinen M, Cooper MD, de Saint Basile G, Fischer A, Good RA, Hendriks RW, Kinnon C, Kwan SP, Litman GW, Notarangelo LD. 1995. BTKbase: a database of XLA-causing mutations. International Study Group. Immunol Today 16:460–465. [DOI] [PubMed] [Google Scholar]
- Vihinen M, den Dunnen JT, Dalgleish R, Cotton RG. 2012. Guidelines for establishing locus specific databases. Hum Mutat 33:298–305. [DOI] [PubMed] [Google Scholar]
- Wang RY, Strong DM. 1996. Beyond accuracy: what data quality means to data consumers. J Manag Inf Syst 12:5–34. [Google Scholar]
- Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE. 2008. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat 29:6–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Most of the existing variation databases are publicly available and others are accessible only by consortia members or paid customers. The availability of the data has to be described clearly on the database home page. The recommendation is to provide as much freely available data as possible for the full benefit of the scientific and clinical communities.