Abstract
Biological processes require close cooperation of multiple transcription factors that integrate different signals. Thyroid hormone receptors (TRs) induce Krüppel-like factor 9 (KLF9) to regulate neurogenesis. Here, we show that triiodothyronine (T3) also works through TR to induce KLF9 in HepG2 liver cells, mouse liver, and mouse and human primary hepatocytes and sought to understand TR/KLF9 network function in the hepatocyte lineage and stem cells. Knockdown experiments reveal that KLF9 regulates hundreds of HepG2 target genes and modulates T3 response. Together, T3 and KLF9 target genes influence pathways implicated in stem cell self-renewal and differentiation, including Notch signaling, and we verify that T3 and KLF9 cooperate to regulate key Notch pathway genes and work independently to regulate others. T3 also induces KLF9 in human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSC) and this effect persists during differentiation to definitive endoderm and hiPSC-derived hepatocytes. Microarray analysis reveals that T3 regulates hundreds of hESC and hiPSC target genes that cluster into many of the same pathways implicated in TR and KLF9 regulation in HepG2 cells. KLF9 knockdown confirms that TR and KLF9 cooperate to regulate Notch pathway genes in hESC and hiPSC, albeit in a partly cell-specific manner. Broader analysis of T3 responsive hESC/hiPSC genes suggests that TRs regulate multiple early steps in ESC differentiation. We propose that TRs cooperate with KLF9 to regulate hepatocyte proliferation and differentiation and early stages of organogenesis and that TRs exert widespread and important influences on ESC biology.
Keywords: Thyroid receptor, Krüppel-like factor 9, Induced pluripotent stem cell, Human embryonic stem cell, Notch
INTRODUCTION
Thyroid hormone (TH) receptors (TRs α and β) belong to the nuclear receptor superfamily. TRs occupy thyroid response elements and alter expression of genes in response to the active form of TH (T3). Primary events involved in TH-dependent changes in gene expression are well understood; T3 induces TR conformational changes that facilitate interactions with coregulators which enhance or repress TR-regulated gene transcription [1, 2]. Downstream consequences of TR-dependent induction of genes that encode other transcription factors (TFs) and interplay between TRs and these TFs are less clear.
The Krüppel-like factors (KLFs) are a family of conserved zinc finger TFs that act as transcriptional activators or repressors in a context-dependent manner and are implicated in transcriptional networking and regulation of balance between pluripotency, self-renewal, and differentiation in mouse embryonic stem cells (mESCs) [3, 4]. Expression of one member of this family, Krüppel-like factor 9 (KLF9), is often associated with differentiated states or early stages of differentiation processes [5–7]. Furthermore, TR actions on neurite extension and branching and neuronal differentiation of mammalian and amphibian cells are mediated by induction of KLF9 [5, 8, 9]. Thus, at least one crucial developmental effect of TR signaling involves KLF9. In mESC, simultaneous depletion of three KLFs, KLF2, KLF4, and KLF5 inhibits self-renewal and triggers cell differentiation [10]. KLF9 and KLF4 bind to the Notch1 gene and exert opposite effects on its transcription, thereby influencing the Notch signaling pathway [6]. Notch signaling works with Wnt, FGF, TGFb/BMP, and Hedgehog signaling pathways [11–15] and converges upon a core transcriptional network that involves Oct4, Nanog, and Sox2 to regulate stem cell maintenance, differentiation, and cellular homeostasis[16]. Thus, alterations in KLF9 levels could greatly influence cell differentiation processes through changes in Notch signaling and other pathways.
There are reasons to suspect that the TR/KLF9 axis is active in many cellular contexts, in addition to neural development. First TRs and KLF9 are detected in multiple tissue types and differentiating cells at different stages of development [4, 17–19]. Second, T3 induces KLF9 in non-neuronal cell types such as epithelial and erythroid cells [20, 21]. Third, TRs and KLF9 play similar roles in particular tissues; both TRa and KLF9 are active in intestinal stem cell regulation [22–24]. Thus, some T3 actions that are ascribed to TR could be results of KLF9 induction. Presently, little is known about the existence and possible roles of TR/KLF9 networking in non-neuronal contexts.
In this study, we show that TR can induce KLF9 in multiple cell types of hepatocyte origin and stem cells and sought to understand roles of the TR/KLF9 signaling network in these contexts using in vitro cell models. We demonstrate that TR activation leads to KLF9 induction in HepG2 cells, nontrans-formed liver cells, human induced pluripotent stem cells (hiPSC), and in human embryonic stem cells (hESCs) and this effect persists during hiPSC and hESC differentiation to definitive endoderm and mature hepatocytes. Dissection of TR/KLF9 effects reveals important roles in key signaling pathways, including the Notch pathway, in HepG2 and ESCs. T3 effects upon KLF9 in ESCs occur in the context of widespread TR-dependent effects on genes that are implicated in early stages of ESC differentiation. Our data therefore suggest that the TR/KLF9 axis plays important roles throughout several stages of the hepatocyte lineage and in the choice between stem cell renewal and differentiation.
MATERIALS AND METHODS
Reagents
Triiodothyronine (T3) was from Sigma-Aldrich (Sigma, St. Louis, MO, http://www.sigmaaldrich.com).
Cell Culture
Human HepG2 cells expressing TRα (HepG2-TRα) or TRβ (HepG2-TRβ) were maintained as described [25]. hESC line KCL034 and hiPSC lines iKCL004 and iKCL011 were maintained in either TeSR2 (Stem Cell Technologies, Vancouver, BC; http://www.stemcell.com/) or Nutristem medium (Stemgent, Cambridge, MA, https://www.stemgent.com) on Matrigel-coated six-well plates in the absence of feeder cells. Matrigel-coated plates were prepared by incubating 1 ml of 0.33 mg/ml growth factor reduced Matrigel (BD Biosciences, San Jose, CA, 354230, http://www.bdbiosciences.com) in Dulbecco’s modified Eagle’s medium (DMEM)/F-12 (Life Technologies, Carlsbad, CA, http://www.lifetechnologies.com) per well for 1 hour at 37°C.
Animals
Experiments were approved by Methodist Hospital IACUC following NIH guidelines for ethical use of animals in biomedical research. C57B/6J mice were purchased from Jackson Laboratory (Bar Harbor, ME, http://www.jax.org) at 9 weeks of age. Animals were maintained on a 12:12 hours light dark cycle, with food and water available ad libitum and were divided into two groups (n = 4): control and T3. Animals were treated for 3 days by oral gavage ± 1 mg/kg T3. Three days after, animals were killed and liver tissue collected for RNA purification.
Isolation of Primary Mouse Hepatocytes
Primary mouse hepatocytes were isolated from male C57B/6J mice using Life Technologies Protocol (Life Technologies, Carlsbad, CA, http://www.lifetechnologies.com), plated in collagen-coated plates (Invitrogen, Carlsbad, CA, http://www.invitrogen.com/) and incubated at 37° C for 2–3 hours using Williams’ Medium E, + 5 ml penicillin-streptomycin (×100), and 5% fetal bovine serum (FBS). Medium was then changed to HepatoZYME-SFM (Invitrogen, Carlsbad, CA, http://www.invitrogen.com/). Cells were treated with 1 nM, 10 nM, or 100 nM T3 for 16 hours.
Primary Human Hepatocytes
Primary human hepatocytes were a gift from Prof. Dhawan, Hepatocyte Biology and Transplantation Group, King’s College London. Hepatocytes were isolated from donor organs rejected for transplantation and consented for research. The research was undertaken with full institutional ethical approval and conducted according to the principles of the Declaration of Helsinki. The procedure is based on papers by Berry and Friend, modified by Seglen, on hepatocyte isolation from rat livers [26, 27]. Briefly, liver is perfused with 500 ml oxygenated HBSS-EGTA (Lonza, Walkersville, MD, www.lonza.com) prior to perfusion with oxygenated EMEM (Lonza, Walkersville, MD, www.lonza.com) supplemented with collagenase-P for 15 minutes. Tissue is filtered and centrifuged at 50g for 5 minutes twice. The pellet contains hepatocytes separated from nonparenchymal cells and dead cells and are used fresh or cryopreserved in University of Wisconsin solution supplemented with 10% DMSO and 4% glucose, using a controlled-rate freezer.
Endoderm Differentiation
hESC (KCL034) and hiPSC (iKCL004 and iKCl011) were differentiated as a monolayer into definitive endoderm [28]. Undifferentiated cells, at 80% confluence, were induced to differentiate by culturing in RPMI-based serum-free medium + 10% serum-free defined medium (SFD), Wnt3a (40 ng/ml), and Activin A (100 ng/ml) for 1 day. For the next 2 days, media were switched to RPMI supplemented with BMP4 (0.5 ng/ml), basic fibroblast growth factor (bFGF) (10 ng/ml), Activin A (100 ng/ml), and vascular endothelialgrowth factor (VEGF) (10 ng/ml). The last 2 days, cells were maintained in SFD + BMP4 (0.5 ng/ml), bFGF (10 ng/ml), Activin A (100 ng/ml), and VEGF (10 ng/ml). SFD serum-free medium consists of 75% Iscove’s modified Dulbecco’s medium (IMDM) (Invitrogen, Carlsbad, CA, www.invitrogen.com/), 25% Ham’s F-12 (Mediatech, Inc., Cell-gro, Manassas, VA 20109, http://www.cellgro.com), 0.53 N2-Supplememt (Gibco/Life Technologies, Carlsbad, CA, http://www.lifetechnologies.com/ipac/en/home/brands/gibco.html), 0.53 B27 without retinoic acid, 0.1% bovine serum albumin (BSA) (Sigma, St. Louis, MO, http://www.sigmaaldrich.com), 50 μg/ml ascorbic acid, and 4.5 × 10−4 M monothioglycerol.
Precommercial iCell highly purified iPSC-derived human hepatocytes (iHep) were purchased from Cellular Dynamics International (CDI; Madison, WI, http://www.cellulardynamics.com). Cells were maintained according to iCell Hepatocytes User’s Guide (CDI; Madison, WI, http://www.cellulardynamics.com).
Quantitative Real-Time PCR
Total RNA was prepared using the RNeasy mini-kits (Qiagen, Venlo, Limburg, http://www.qiagen.com). For HepG2, Aurum Total RNA kit (Bio-Rad, Hercules, CA, http://www.bio-rad.com/) was used. Reverse transcription reactions were performed using 1 μg total RNA with an iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA, http://www.bio-rad.com). Quantitative real-time PCR (qPCR) was performed with the Roche LightCycler 480 RT PCR Instrument using SYBR Green Mastermix (Roche, Mannheim, Germany, www.roche.com). Primer sequences are available per request. Data were collected and analyzed using comparative threshold cycle method with β-actin and 18S rRNA as reference. Experiments were performed at least three times, and mean ± SD was calculated and statistical analysis was performed using Prism curve-fitting program (GraphPad Prism, version 6.01).
RNA Interference
HepG2-TRβ cells were plated in 10% FBS-DMEM/F-12 media and grown to 50% confluence. Cells were transfected with TRβ or KLF9 ON-TARGET plus SMART pool siRNA (Dharmacon, Waltham, MA, http://www.thermoscientificbio.com/Dharmacon/) at 50 nM final concentration. Positive and negative non-targeting control siRNAs were also from Dharmacon. After 3 days, cells were treated with 100 nM T3 for 8 or 24 hours and RNA or protein prepared.
hESC (KCL034) and hiPSC (iKCL004 and iKCL011) were plated at 125,000/well of six-well dish and transfected with 5 μM nontargeting control or siKLF9 with DharmaFECT1 according to manufacturer’s protocol 2 days later. Cells were exposed to 100 nM T3 for 18 hours 2 (for RNA) or 3 (for protein isolation) days post-transfection.
Western Blotting
Total proteins were separated with 4%–12% gradient Bis-Tris gels (Invitrogen, Grand Island, NY, http://www.invitrogen.com/), transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad, Hercules, CA, http://www.bio-rad.com/), and incubated with anti-KLF9 antibody (Santa Cruz Biotechnology, Dallas, TX, www.scbt.com) followed by anti-mouse IgG Ab conjugated with HRP (Santa Cruz Biotechnology, Santa Cruz Biotechnology, Dallas, TX, www.scbt.com). A Luminata Classico Western HRP Substrate (EMD Millipore, Billerica, MA, www.millipore. com) was used for protein detection.
Microarray Analysis
Human HT-12_v4 whole genome expression arrays were from Illumina (Illumina, San Diego, CA, http://www.illumina.com/). cRNA synthesis and labeling were performed using Illumina TotalPrep-96 RNA Amplification Kit (Ambion/Life Technologies, Carlsbad, CA, http://www.lifetechnologies.com/us/en/home/brands/ambion.html). Labeling in vitro transcription reaction was performed at 37° C for 14 hours. Biotinylated cRNA samples were hybridized to arrays at 58° C for 18 hours according to manufacturer’s protocol. Arrays were scanned using BeadArray Reader.
Unmodified microarray data obtained from GenomeStudio were background-subtracted and quantile-normalized using the lumi package [29] and analyzed with the limma package[30] within R [31]. Effect of KLF9 knockdown was determined through comparison between non-T3 treated control and KLF9 knockdown. T3-response was determined by comparing cells treated with T3 for 8 or 24 hours against their respective untreated controls. All analyses were corrected for multiple hypothesis testing [32], and effects determined as significant when more than or equal to twofold with an adjusted p-value ≤.05. To facilitate comparisons among datasets, all data were uploaded into a SQLite3 database (http://www.sqlite.org/).
Pathway Enrichment Analysis (GeneCodis)
We used the GeneCodis analysis (http://genecodis.cnb.csic.es/) to identify enriched pathways and functional themes. Gene-Codis integrates different information resources (GO, Panther pathways, SwissProt, etc.), searches, and arranges gene set annotation by statistical significance [33–35]. Genes of interest, defined as at least twofold differentially expressed according to microarray, were uploaded as standard human gene symbols and genes in the interaction network with false discovery rate (FDR) <0.05 were taken into consideration.
GeneMANIA
We used GeneMANIA (http://www.genemania.org) to find genes related to input genes, using a very large set of functional interaction data [36–38]. Inputs were differentially expressed genes underlying specific functional themes and pathways as identified by GeneCodis. We focus analysis on high confidence physical interactions (from various protein interaction databases included in GeneMANIA) and pathway interactions (from Reactome pathway database).
Ingenuity Pathway Analysis
Data were also analyzed using ingenuity pathway analysis (Ingenuity Systems; IPA, Redwood City, CA, http://www.ingenuity.com). Genes of interest, defined as at least twofold differentially expressed, were uploaded. Each gene identifier was mapped to its corresponding gene object in the ingenuity pathways knowledge base (IPKB). The IPKB, containing a large network of curated molecular interactions and pathways, was searched to find subnetworks enriched in genes of interest.
Immunostaining
hESC/iPSC-derived definitive endoderm cells grown as a monolayer were fixed in 3.8% paraformaldehyde for 20 minutes, permeabilized in 0.5% Triton X-100/phosphate buffered saline (PBS) for 5 minutes, washed in PBS, and incubated with goat anti-GATA4 polyclonal Ab (R&D Systems cat. no. 2606; Minneapolis, MN; www.rndsystems.com) overnight at +4°C. Samples were washed, incubated with rhodamine X-conjugated donkey anti-goat IgG antibodies, and washed again 3× in PBS. In the second wash 10 μg/ml Hoechst 33342 (Life Technologies, Carlsbad, CA, http://www.lifetechnologies.com) was added to visualize nuclei. Samples were mounted in Vectashield (Vector Laboratories, Burlingame, CA; www.vector-labs.com) and analyzed with an epifluorescence microscope (Nikon, model E50i) equipped with Retiga 400R cooled monochrome camera (QImaging, Surrey, BC; www.qimaging.com). Images were processed using AdobePhotoshop CS5 software.
RESULTS
T3 Activates KLF9 in Hepatocytes
Our previous analysis of T3 response in parental HepG2 and HepG2 cells that stably express either TRα or TRβ [25] suggested that KLF9 is a T3 inducible-target in this cell background. We used qRT-PCR to verify T3-activation of KLF9 in these cells (Fig. 1A). T3 response was observed at 24 hours in parental HepG2, which express very low levels of TRβ [39] (Fig.1A) but more rapid T3 induction was observed in both HepG2-TR cell lines, occurring within one hour of T3 treatment and maintained up to 24 hours (Fig. 1A). T3-induction of KLF9 persisted after pretreatment with protein synthesis inhibitor cycloheximide (CHX) in HepG2-TRβ cells (Supporting Information Fig. S1A). Thus, T3 induction of KLF9 is a direct effect that does not require new protein synthesis. The specific role for TRβ was confirmed by siRNA (Supporting Information Fig. S1B, S1C).
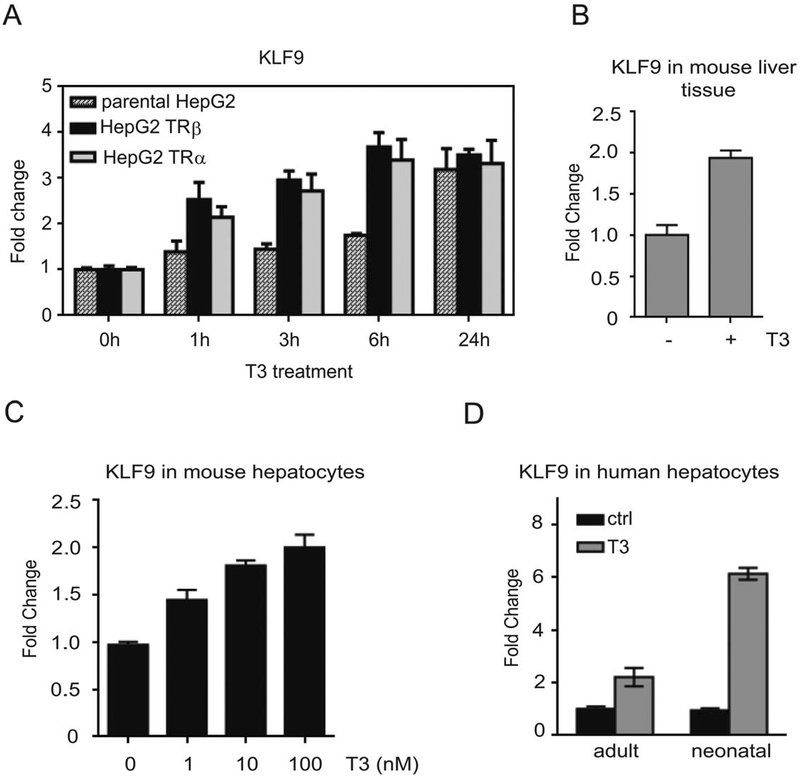
Figure 1.
T3 induces KLF9 in HepG2 cells and in hepatocytes. (A): Parental HepG2, HepG2-TRβ, and HepG2-TRα cells were treated with 100 nM T3 for the indicated times and KLF9 mRNA levels were determined by quantitative real-time PCR (qPCR). (B–D): Expression of KLF9 after T3 treatment was assessed by qPCR in mice liver tissue (B), in isolated mice hepatocytes treated with increasing concentrations of T3 (C), and human hepatocytes isolated from neonatal and adult livers (D).
Since HepG2 cells are a liver carcinoma cell line, we verified the effect of T3 on KLF9 expression in mouse liver. We treated 9 weeks old C57/Bl6 male mice ± T3 by daily oral gavage. After 3 days, we isolated livers and showed that T3 induced KLF9 about twofold (Fig. 1B). We confirmed that KLF9 was not expressed in cholangiocytes, as judged by absence of colocalization with the cholangiocyte marker CK19 (Supporting Information Fig. S2). We also confirmed that T3 induced KLF9 in mouse primary hepatocytes in a dose-dependent fashion (Fig. 1C) and that T3 treatment of human primary hepatocytes (adult and neonatal) resulted in an increase in KLF9 expression (Fig. 1D). Thus, the TR/KLF9 axis is active in transformed and nontransformed hepatocytes.
KLF9 Regulates Multiple Genes in HepG2 and Influences T3 Response
To define roles of KLF9 and the TR/KLF9 network in HepG2, we used siRNA to silence KLF9 expression in HepG2-TRβ cells. We verified that KLF9 levels were reduced in specific siRNA treated cells versus cells that were treated with control siRNA (97% reduction of mRNA and protein) (Supporting Information Fig. S3A). Although KLF9 levels were almost completely suppressed, some T3 induction of KLF9 persisted (Supporting Information Fig. S3B).
Microarray analysis revealed 368 genes that displayed more than twofold change after KLF9 knockdown, with 226 downregulated and 142 upregulated (Fig. 2A). Effects were confirmed by qRT-PCR analysis of representatives of both classes of gene (downregulated Lama1 and Aldoc, upregulated Snord 3A and 3D, Fig. 2B). Thus, KLF9 regulates large numbers of genes in HepG2 and can act as a transactivator and transrepressor.
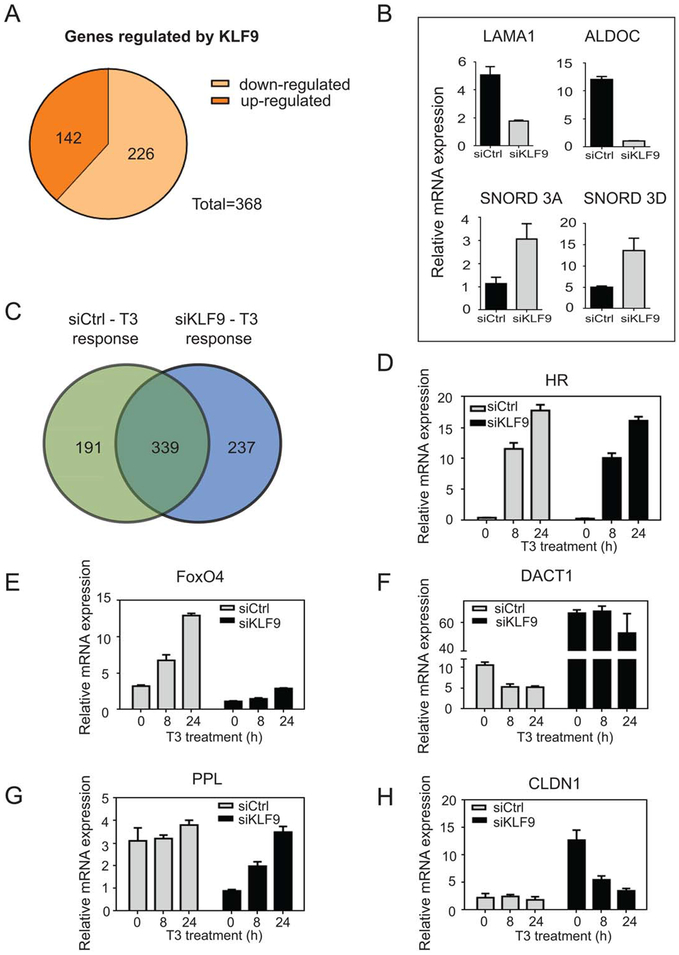
Figure 2.
TR/KLF9 axis regulates multiple genes in HepG2 cells. (A): Differential gene regulation in HepG2-TRb cells after KLF9 silencing revealed by microarray analysis. Microarray data obtained from human Illumina HT-12_v4 gene chips from control versus KLF9 knockdown were analyzed using Limma package within R. Effects determined to be significant when more than or equal to twofold with an adjusted p-value ≤ .05. (B): Effects of KLF9 knockdown confirmed at representatives of both classes of gene by quantitative real-time PCR. (C): Differential gene regulation by T3 in control and siKLF9 cells revealed by microarray analysis. (D–H): Cells were treated with 100 nM T3 and qRT-PCR was performed to verify patterns of KLF9-dependency of T3 response. Data are represented as mean ± SD. Microarray data are deposited in the Gene Expression Omnibus; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=sxkvckgwhxmfdgx&acc=GSE54699; accession number GSE54699.
To understand whether KLF9 knockdown might influence T3 response, we treated siControl and siKLF9 HepG2-TRβ cells with T3 for 8 and 24 hours and performed microarray analysis. In accordance with previous results, 530 genes displayed T3 response (more than twofold) in siControl HepG2 cells, with more genes displaying T3 response at 24 hours (Supporting Information Table S1). Comparison of genes that responded to KLF9 knockdown or T3 in siControl cells revealed significant overlap between datasets, with 68 genes regulated by both factors (Supporting Information Fig. S4). More surprisingly, KLF9 knockdown also altered the pattern of T3 response. Numbers of T3 responsive genes were similar in control and KLF9 knockdown cells (530 and 576 genes, respectively, Supporting Information Table S1). While most T3 responsive genes were unaffected by KLF9 knockdown (339 genes displayed similar responses in control and KLF9 knockdown cells), 191 T3 target genes displayed reduced T3 response after KLF9 knockdown and 237 genes gained T3 response (Fig. 2C). We confirmed that genes that responded to T3 in the presence and absence of KLF9 were truly KLF9 independent (HR, Fig. 2D). We also confirmed positive and negative T3 responses that were dependent upon KLF9 (FoxO4 and DACT1; Fig. 2E, 2F) and T3 responses that emerged after KLF9 knockdown (PPL and CLDN1, Fig. 2G, 2H). We also detected genes that showed changes in overall expression after KLF9 knockdown, but retained similar T3 response (alkaline phosphatase, intestinal (ALPI), Supporting Information Fig. S5). Thus, T3 and KLF9 display overlapping effects upon gene expression in HepG2 and many T3 responses are modulated by KLF9.
TR/KLF9 Axis Regulates Key Signaling Pathways
To define roles of defined KLF9 target genes in HepG2 we used GeneCodis software analysis (http://genecodis.cnb.csic. es) [33–35] to examine functions of genes that displayed altered expression after KLF9 knockdown. Candidate genes were assigned to pathways and processes listed in Table 1 (upper section) and Supporting Information Table S2, respectively, with differentially expressed genes listed in Supporting Information Tables S3, S4. The main KLF9-dependent pathways were TGFβ signaling, Alzheimer’s disease-amyloid secretase pathway, FGF and Wnt signaling (Table 1). The most important processes were signal transduction, cell adhesion, aging, and cell differentiation (Supporting Information Table S2).
Table 1.
Pathway enrichment analysis of KLF9-dependent transcriptome. Canonical pathways obtained from GeneCodis using Panther pathways database. Gene co-occurrence annotation found by GeneCodis for the genes differentially expressed (FC > 2, p < .05 corrected for multiple testing) between siCtrl versus siKLF9 HepG2-TRβ samples (KLF9 pathways) and integration of KLF9 targeted and TR/KLF9 targeted pathways. p-Values have been obtained through hypergeometric analysis (Hyp) corrected by FDR method (Hyp*).
| NGR | NG | Hyp | Hyp* | Annotations (panther pathways) |
|---|---|---|---|---|
| KLF9 pathways | ||||
| 91 | 7 | 1.86E-05 | 0.00011164 | P00052: TGF-beta signaling pathway |
| 66 | 5 | 0.0003218 | 0.00096552 | P00003: Alzheimer disease-amyloid secretase pathway |
| 109 | 5 | 0.0030787 | 0.00461811 | P00021: FGF signaling pathway |
| 280 | 8 | 0.0039947 | 0.00479375 | P00057:Wnt signaling pathway |
| 107 | 5 | 0.0028423 | 0.00568463 | P00006: Apoptosis signaling pathway |
| 198 | 6 | 0.0092045 | 0.00920453 | P00031: Inflammation mediated by chemokine and cytokine signaling pathway |
| TR/KLF9 pathways | ||||
| 91 | 9 | 4.60E-05 | 0.00103529 | P00052: TGF-beta signaling pathway |
| 280 | 16 | 7.02E-05 | 0.00105367 | P00057: Wnt signaling pathway |
| 107 | 9 | 0.000162 | 0.00182779 | P00006: Apoptosis signaling pathway |
| 66 | 7 | 0.000207 | 0.00186532 | P00003: Alzheimer disease-amyloid secretase pathway |
| 70 | 8 | 4.28E-05 | 0.00192706 | P00016: Cytoskeletal regulation by Rho GTPase |
| 114 | 9 | 0.000262 | 0.0019674 | P00018: EGF receptor signaling pathway |
| 118 | 9 | 0.000339 | 0.00218076 | P00004: Alzheimer disease-presenilin pathway |
| 36 | 5 | 0.000483 | 0.00271783 | P00045: Notch signaling pathway |
| 109 | 8 | 0.000924 | 0.00415905 | P00021: FGF signaling pathway |
| 72 | 6 | 0.002094 | 0.00589054 | P00019: Endothelin signaling pathway |
| 96 | 7 | 0.001980 | 0.00594038 | P00036: Interleukin signaling pathway |
| 198 | 10 | 0.003794 | 0.00898646 | P00031: Inflammation mediated by chemokine and cytokine signaling pathway |
| 69 | 5 | 0.008789 | 0.0146485 | P04393: Ras Pathway |
| 126 | 6 | 0.029072 | 0.0408832 | P00029:Huntington disease |
| 128 | 6 | 0.031068 | 0.0411201 | P00047: PDGF signaling pathway |
NGR: Number of annotated genes in the reference list;
NG: Number of annotated genes in the input list;
Hyp: Hypergeometric pValue;
Hyp Corrected hypergeometric pValue.
We also analyzed T3 responsive pathways and processes (Supporting Information Table S5). Interestingly, only two of the top six KLF9-dependent pathways (TGFβ and FGF pathway) and three processes (transmembrane transport, cell death, and protein folding) were purely KLF9-responsive. Other KLF9 responsive pathways and processes were also flagged as T3 responsive (Table 1-green, Supporting Information Table S2-green).
Analysis of an integrated gene set that included KLF9 responsive gene targets (Fig. 2A) and T3-regulated gene targets with altered response to KLF9 knockdown (Fig. 2C) yielded more statistically enriched pathways (Table 1, lower section) and processes (Supporting Information Table S6). Together, T3 and KLF9 are active in the Notch pathway as well as the EGF, Ras, and platelet-derived growth factor (PDGF) regulated pathways (Table 1-blue). Furthermore, T3 and KLF9 influenced processes such as cell proliferation and metabolic pathways (Supporting Information Table S6-blue; differentially expressed genes in Supporting Information Tables S7, S8). Thus, there are overlaps between KLF9- and T3-dependent pathways and processes in HepG2, and TR and KLF9 cooperate to influence signaling pathways that would not be identified by consideration of actions of either T3 or KLF9 alone.
TR/KLF9 Axis Regulates Multiple Components of the Notch Pathway
Since Notch signaling emerged as a major target pathway regulated by T3 and KLF9 and because Notch signaling also plays important roles in liver development and regeneration, we explored links between T3, KLF9, and Notch in HepG2. We verified regulation patterns of the five flagged T3/KLF9 target genes with qRT-PCR (HEY1, HEY2, NOTCH3, PSEN2, and HES1) (Table 1, Supporting Information Table S7). This confirmed that HEY1, HEY2, and NOTCH3 all displayed T3 response and that expression levels of these genes were changed by KLF9 knockdown (Fig. 3A–3C). Furthermore, PSEN2 and HES1 transcript levels were, respectively, greatly reduced and elevated by KLF9 knockdown (not shown).
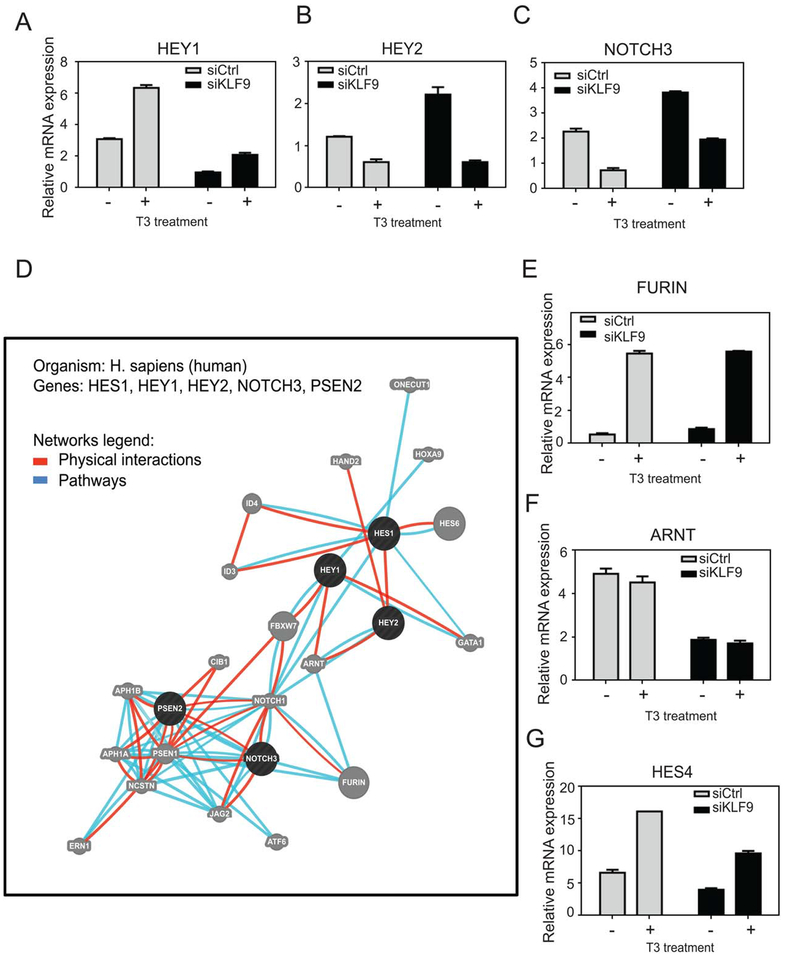
Figure 3.
TR/KLF9 axis and the Notch pathway. (A–C): Cells were treated with 100 nM T3 and qRT-PCR was performed to verify TR/KLF9-dependency of identified Notch pathway genes. (D): Network of interactions among Notch pathway KLF9 targets, as retrieved by the GeneMania. Circles represent genes and connecting lines represent interactions between genes. GeneMania retrieved known and predicted interactions between these genes and added extra genes (gray circles) that are strongly connected to query genes. (E–G): Quantitative real-time PCR verification of genes identified by GeneMania as part of TR/KLF9-Notch network. All data are represented as mean ± SD.
We next performed GeneMania analysis to identify Notch networking partners (http://www.genemania.org); this approach uncovers genes related to input genes, using a large set of functional interaction data [36–38]. We generated the network using a query of the five Notch signaling components identified as T3 and/or KLF9 regulated (Fig. 3A–3C, Supporting Information Table S7). We focused analysis only on high confidence pathway interactions (reactome pathway database; Fig. 3D, blue lines) and physical interactions (various protein interaction databases; GeneMANIA; Fig. 3D, red lines). This strategy uncovered tight networking of the 5 input genes with 20 associated genes. Of these 20 genes, three were also flagged as T3 and/or KLF9 targets in microarray analysis (ID3, Furin, ARNT), underscoring links between TR/KLF9 and Notch. We used qRT-PCR to confirm that ID3 exhibited strong regulation by T3 and KLF9 response (not shown) and that Furin and ARNT were responsive only to T3 and KLF9, respectively (Fig. 3E, 3F). A limited survey of additional genes implicated in Notch signaling revealed that KLF9 and T3 also cooperate to regulate HES4 (Fig. 3G). Thus, T3 and KLF9 regulate multiple Notch pathway genes in HepG2 and TR/KLF9 targets are involved in multiple aspects of Notch signaling (Discussion).
TR/KLF9 Axis Is Active in Embryonic Stem Cells and Definitive Endoderm
Since TR regulates KLF9 in hepatocytes, we asked whether TR might also regulate KLF9 during differentiation along the hepatocyte lineage in culture. We assessed KLF9 expression in two hiPSC lines, iKCL004 and iKCL011 [40] and the hESC line KCL034 [41] (Fig. 4A). While KLF9 is detectable in all three cell lines (iKCL004, iKCL011, and KCL034), its expression was profoundly reduced in comparison to dermal BJ fibroblasts from which the iPSCs were reprogrammed (Fig. 4A). Even though TRa was the predominant TR isoform in these cell lines (Supporting Information Fig. S6), T3 treatment for 6 and 18 hours resulted in robust increase in KLF9 mRNA in all three pluripo-tent stem cell lines, but not BJ fibroblasts (Fig. 4A).
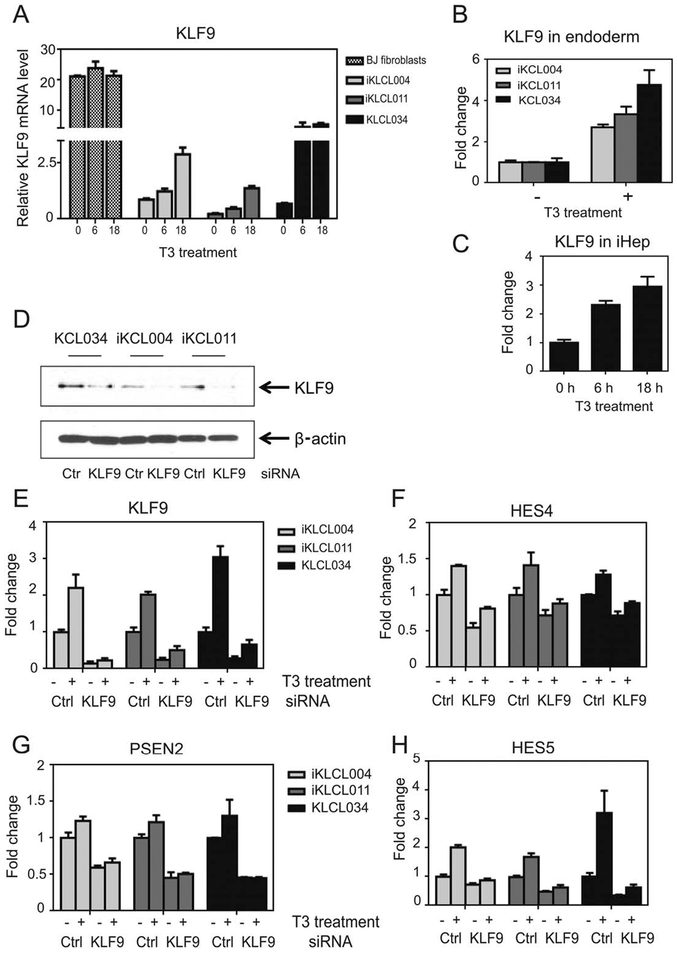
Figure 4.
TR/KLF9 axis is active in embryonic stem cells, definitive endoderm, and human induced pluripotent stem cell (hiPSC)-derived hepatocytes. (A–C): Quantitative real-time PCR (qPCR) analysis of KLF9 expression levels 2/1T3 in BJ fibroblasts, iKCL004, iKCL011, and KCL034 cells (A), endoderm differentiated from iKCL004, iKCL011, and KCL034 (B), and terminally differentiated hiPSC-derived hepatocytes (C). KLF9 mRNA levels were expressed as fold change. All data are represented as mean ± SD. (D–H): TR/KLF9 axis is involved in regulation of Notch signaling in hiPSC and human embryonic stem cell. (D): Western blot for KLF9 from cell lysates from iKCL004, iKCL011, and KCL034 transfected with Ctrl or KLF9 siRNA. (E–H): iKCL004, iKCL011, and KCL034 cells transfected with Ctrl or KLF9 siRNA were treated with 100 nM T3 for 18 hours. KLF9, HES4, PSEN2, and HES5 mRNA levels were determined by qPCR. The data are presented as fold change of mRNA levels in Ctrl nontreated samples. All data are represented as mean ± SD.
We differentiated lines into definitive endoderm [28]. As expected, expression of pluripotency markers Nanog and Oct4 decreased during definitive endoderm differentiation and endoderm markers MIXL1 and SOX17 emerged (Supporting Information Fig. S7A). We also confirmed that the majority of cells turned into endoderm by GATA4+ immunostaining (Supporting Information Fig. S7B). TRα remained the prevalent iso-form during this process (Supporting Information Fig. S6) and T3 treatment again induced KLF9 in definitive endoderm from all stem cell lines (Fig. 4B).
Finally, we asked whether TR/KLF9 axis is active in highly purified terminally differentiated hiPSC-derived hepatocytes. Like mature hepatocytes, these cells predominantly expressed TRβ (Supporting Information Fig. S6) and, here, T3 also increased KLF9 expression (Fig. 4C). Thus, T3 induces KLF9 in human ESCs and at early and late stages of hepatocyte differentiation in vitro in a manner that is independent of TR subtype.
T3 Exhibits Cell-Specific Effects on Notch Genes in ESCs
To explore TR function in ESCs, we performed microarray analysis on KCL034, iKCL004, and BJ lines treated with T3 for 6 hours. The analysis revealed 820 and 826 genes changed in KCL034 and iKCL004, respectively, and no change in BJ cells (Supporting Information Table S9).
Analysis of T3 target genes using GeneCodis analysis confirmed that T3 regulates many key pathways previously recognized as targets of TR or the TR/KLF9 axis in HepG2, including Notch, Wnt, and FGF (Supporting Information Table S10) along with others such as angiogenesis, cadherin, and endothelin pathways. However, closer investigation of T3 effects upon individual Notch pathway genes revealed differences from HepG2. HEY2 and HES4 appeared as T3 targets in ESCs, as seen in HepG2 (Supporting Information Table S11), but Notch3, HEY1, and FURIN did not respond to T3 in any of pluripotent stem lines tested (not shown). Conversely, several Notch genes responded to T3 in stem cells and not in HepG2 (HES5, LFNG, and DLK1) (Fig. 4H, Supporting Information Fig. S8). Thus, T3 displays cell-specific effects upon key Notch pathway genes.
KLF9-Dependency of Notch Genes in ESCs
To determine whether KLF9 might regulate Notch signaling in human pluripotent stem cells, we silenced KLF9 in iKCL004, iKCL011 and KCL034 cells (Fig. 4D, 4E). RNA and Western analysis showed an average of 85%, 75%, and 72% knockdown of KLF9 in iKCL004, iKCL011, and KCL034, respectively, relative to scrambled siRNA control (Fig. 4D) with T3-dependent induction of KLF9 also significantly reduced (Fig. 4E). In these cells, KLF9 knockdown reduced basal levels and T3 response of T3/KLF9 targets HES4 and HES5 (Fig. 4F, 4H). Additionally, PSEN2 and ARNT, pure KLF9-targets in HepG2 cells (Fig. 3), also exhibited similar pure KLF9-dependency in stem cells (Fig. 4G and not shown). There was no change in the expression of T3 target genes LFNG and DLK1 after KLF9 knockdown (not shown). Thus, TR and KLF9 cooperate to regulate key Notch pathway genes in stem cell lines, and work independently to regulate others, but precise effects are cell type specific (Discussion).
T3-Dependent Changes in Differentiation-Related Gene Expression in Stem Cells
To gain insight into T3 regulated biological processes in ESCs, we performed IPA (version Fall 2013) to determine functional pathways of identified genes. IPA scans input genes to identify networks using the IPKB for interactions between “target genes” and known and hypothetical interacting genes stored in the IPA software (in our study, input genes were differently expressed after T3 treatment). This revealed that T3 target genes clustered into categories related to embryonic development and differentiation (Table 2).
Table 2.
Functional categorization of T3 target genes in KCL034 and iKCL004 cells; pathway enrichment determination using ingenuity pathway analysis identify enriched development-related functional themes The number of genes and statistical values are shown for each cell line. Microarray data have been deposited in NCBI’s gene expression omnibus; http://www.ncbi.nlm.nih.gov/ geo/query/acc.cgi?tacc=GSE58273; accession number GSE58273
| Function | KCL034 | IKCL004 | ||
|---|---|---|---|---|
| NG | p-Value | NG | p-Value | |
| Gene expression | 126 | 1.39E-10 | 103 | 7.08E-06 |
| Transcription | 146 | 7.90E-10 | 124 | 4.56E-06 |
| Cellular development | ||||
| Cell differentiation | 178 | 1.09E-17 | 160 | 3.69E-14 |
| Cell proliferation | 271 | 1.02E-14 | 248 | 4.34E-12 |
| Cell commitment | 16 | 9.05E-05 | 16 | 4.39E-05 |
| Cell cycle | 73 | 2.87E-04 | 71 | 1.28E-04 |
| Cell morphology | 155 | 4.40E-10 | 154 | 6.62E-12 |
| Cell death | 242 | 6.23E-11 | 213 | 3.48E-07 |
| Cell viability | 104 | 5.55E-08 | 84 | 2.96E-04 |
| Embryonic development | ||||
| Development of body axis | 86 | 1.96E-09 | 81 | 5.51E-09 |
| Development of head | 80 | 1.38E-09 | 76 | 2.31E-09 |
| Lung development | 33 | 8.85E-07 | 25 | 2.20E-08 |
| Development of sensory organ | 51 | 5.03E-07 | 51 | 7.57E-08 |
| Cardiogenesis | 39 | 1.56E-07 | 27 | 4.14E-04 |
| Development of abdomen | 28 | 1.15E-07 | 46 | 3.39E-06 |
| Movement of neural crest cells | 11 | 6.95E-06 | 29 | 5.22E-07 |
| Eye development | 42 | 1.88E-06 | 30 | 2.46E-06 |
| Mesoderm development | 31 | 1.59E-05 | 31 | 5.25E-04 |
| Cell movement of embryonic cells | 32 | 7.17E-04 | 32 | 4.03E-04 |
| Development of epidermis | 17 | 6.96E-04 | 22 | 7.73E-07 |
| Skin development | 34 | 6.35E-04 | 34 | 4.35E-08 |
| Development of atrium | 6 | 2.46E-04 | 6 | 1.76E-04 |
| Development of lymphatic system component | 32 | 1.68E-04 | 30 | 2.83E-04 |
| Development of brain | 39 | 1.30E-04 | 45 | 1.85E-07 |
| Development of forebrain | 23 | 1.11E-04 | 21 | 3.30E-04 |
NG, Number of annotated genes in the input list.
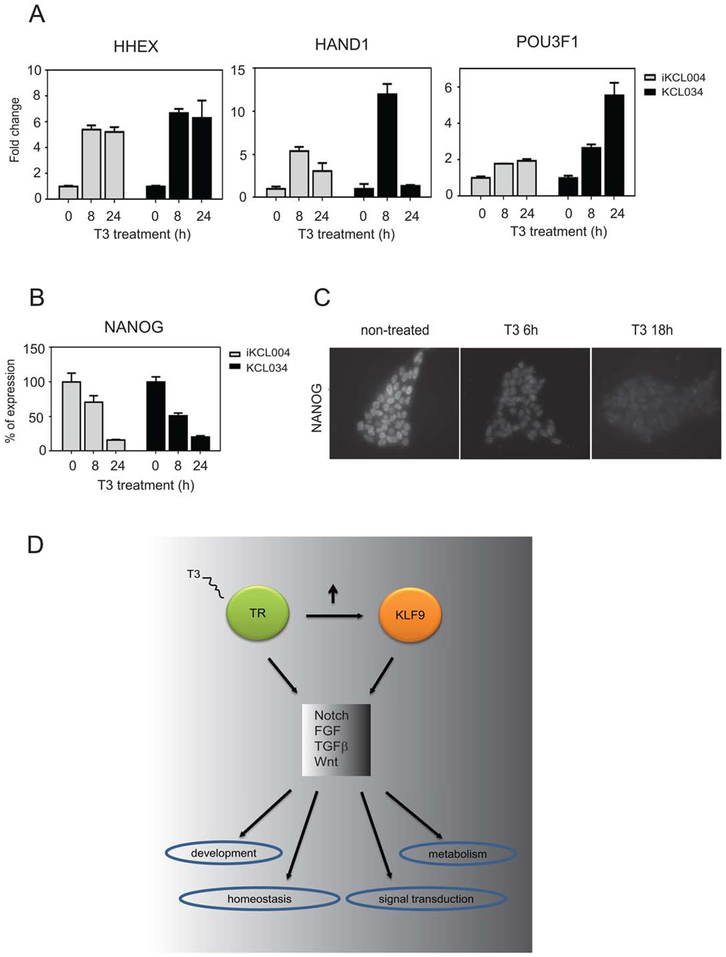
We used qRT-PCR to confirm selected T3 responses. T3 induces important differentiation-related genes such as HHEX, one of the earliest markers of anterior endoderm, which gives rise to foregut organs such as the liver, ventral pancreas, thyroid, and lungs [42], HAND1, a TF that is critical for the development of three embryologically distinct lineages: trophoblast of the placenta, extraembryonic mesoderm derivatives and cardiomyocytes [43], and the octamer TF POU3F1, which participates in cell fate determination [44] (Fig. 5A). Conversely, T3 represses the crucial ES pluripotency regulator Nanog (Fig. 5B, 5C). Thus, T3 triggers changes in genes involved in ESC differentiation.
Figure 5.
Analysis of representative gene expression in KCL034 and iKCL004 cells. (A): T3-induced expression of HHEX, HAND1, and POU3F1 in iKCL004 and KCL034. (B, C): Nanog display T3-dependent reduction in expression levels in KCL034 and iKCL004 as verified by qPCR (B) and confirmed by immunostaining in KCL034 cells (C). (D): A model for TR/KLF9 action. TR activates transcription of KLF9 and both transcription factors modulate each other’s activity in multiple pathways leading to cell-specific responses to different signals. Abbreviation: TR, thyroid hormone receptor.
DISCUSSION
It is important to understand how TR-dependent cascades of alternate TFs contribute to T3 regulation of physiologic processes. This study was initially prompted by our observation that TRs regulate KLF9 in HepG2 liver cells and nontrans-formed hepatocytes. Previous studies implicate KLF9 in regulation of the balance of cellular renewal and differentiation and TR-dependent induction of KLF9 in neural development [5, 8,9], but the role of their cooperation in hepatocytes is unknown. TR activation regulates hepatocyte proliferation and development [45, 46] and, conversely, induces regression of carcinoma-induced hepatic nodules and reduce incidence of hepatocarcinoma [47, 48]. Moreover, KLF9 regulates the balance between cell differentiation and self-renewal [6, 9]. Thus, investigation of the TR/KLF9 axis in hepatocytes could yield insights into the ways that TRs influence liver regeneration and cancer.
We used the HepG2 system to define TR/KLF9 effects in a hepatocyte background. siRNA knockdown revealed hundreds of KLF9-dependent genes in this cell type with 10%–15% of T3 regulated genes identified in this study, and previous studies [25, 49], also being KLF9 targets (Supporting Information Fig. S4). KLF9 knockdown also reduced T3 response of a large proportion of TR target genes (Fig. 2C). Thus, we suggest that some T3 responsive HepG2 target genes are regulated indirectly through induction of KLF9. In this regard, 35% of T3 responses in HepG2 cells are indirect, as judged by sensitivity to the protein synthesis inhibitor CHX [25]. Inspection of this CHX sensitive dataset reveals many genes flagged as KLF9 targets in this study (not shown).
Surprisingly, some genes displayed emergent T3 response after KLF9 knockdown. We have not assessed mechanisms of this effect, but we note that KLF9 knockdown often results in large alterations of basal expression of this gene class (Fig. 2G, 2H). Thus, we suggest that emergent T3 responses may occur at genes that display close to maximal KLF9 response in HepG2; T3-dependent increases in remaining low levels of KLF9 that persist after siRNA treatment are sufficient to partly rescue effects of KLF9 knockdown.
The fact that large numbers of genes respond to T3 and KLF9 in HepG2 allowed us to define pathways that, respectively, depend upon T3, KLF9, and T3+KLF9. KLF9 target genes are active in pathways implicated in stem cell pluripotency and differentiation, including TGFβ, FGF, and Wnt (Table 1, Fig. 5D). Analysis of an integrated dataset of KLF9- and T3-target genes flagged additional pathways, including Notch, crucial for liver development and regeneration [50–56]. Detailed analysis of T3+KLF9 targets in the Notch pathway revealed multiple genes coordinately regulated by T3 and KLF9 (HEY1, HEY2, HES4, Notch3, and ID3) and others that were purely regulated by T3 (Furin) or KLF9 (PSEN2 and ARNT).
Given connections between TR, KLF9, and Notch signaling in hepatocytes, we determined whether T3 also regulates KLF9 during hepatocyte differentiation from ESCs. Unexpectedly, T3 induced KLF9 in ESCs and hiPSC derived from BJ fibro-blasts, but not BJ fibroblasts themselves, and this effect persisted throughout differentiation to definitive endoderm and mature hepatocytes (Fig. 4). Microarray analysis revealed more than 800 T3-regulated genes in hiPSC and these genes cluster into many of the pathways recognized as TR/KLF9 responsive in HepG2, including Notch, FGF, and Wnt. Silencing of KLF9 in hESC and hiPSC confirmed that the TR/KLF9 axis regulates Notch pathway genes in ESCs, as seen in HepG2, albeit in cell-type-specific fashion. Thus, the TR/KLF9 axis is active in ESCs, remains active through early and late stages of hepatocyte differentiation, and plays important cell-specific roles in regulation of ESC Notch signaling genes.
Our studies do not yet allow us to precisely define how T3 and KLF9 affect Notch-dependent behavior of different cell types. We note, however, that T3 and/or KLF9 regulate genes involved in upstream Notch signaling events (Notch3, Furin, and PSEN2), members of the HES/HEY family of TFs that are downstream targets of the Notch pathway and a gene that is part of noncanonical Notch pathways (ARNT) (Supporting Information Fig. S9) [57–59]. Thus, TR and KLF9 regulate multiple branches of the Notch pathway and the fact that T3 and KLF9 cooperate to regulate genes of the HES/HEY family, which constitute outputs of Notch signaling, in a cell-type-specific manner suggests that T3 and KLF9 could influence Notch pathway activity in a cell type-specific manner. HEY/HES TFs are differentially and specifically expressed in different cell types [60], display an oscillatory expression pattern, and control timing of biological events [61, 62]. Thus, TR/KLF9-dependent changes in HES/HEY expression could have profound influences on cell fate, depending on cell type and context.
While we focused on TR/KLF9-dependent effects upon the Notch signaling pathway, such effects must be considered in the context of widespread effects on differentiation-related pathways (Fig. 5D). Indeed, IPA-based analysis of TR function in ESCs provides strong evidence of T3 involvement in crucial development-related processes (Table 2), suggesting that TR may orchestrate transitions from the embryonic to adult transcription programs (Fig. 5A–5C). While more experiments will be needed to dissect mechanisms and consequences of these effects, our data suggest that TR/KLF9 axis could orchestrate key biological processes during differentiation through direct transcription regulation or through signaling pathways such as Notch (Supporting Information Fig. S9). It is not clear how the TR/KLF9 axis influences dysregulated Notch signaling that occurs in pathological conditions such as congenital disorders, metabolic syndrome, and cancer [11, 12, 63]. We propose that it will be important to examine roles of KLF9 and Notch pathway genes in T3-dependent enhancement of liver proliferation and inhibition of hepatocarcinoma proliferation using gene ablation or knockdown.
CONCLUSIONS
TRs and KLF9 work together to regulate multiple genes and important developmental processes. The TR/KLF9 axis is active in ESCs and cell types that are representative of the hepatocyte lineage and influences Notch signaling. Better understanding of synergy between TR and KLF9 is crucial for understanding decisions between stemness and differentiation and to uncover regulatory patterns of cell homeostasis. Given the likely importance of KLF9 and Notch signaling in regulation of the balance between proliferation and differentiation, we propose that it will be important to further explore how the TR/KLF9 axis regulates hepatocellular cancer development and important physiological processes such as liver regeneration.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by UO1 GM094614 subcomponent and RC4 DK090849 to P.W. L.N. has studentship from Saudi Arabia government.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
REFERENCES
- 1.Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev 2001;81:1097–1142. [DOI] [PubMed] [Google Scholar]
- 2.Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev 2010;31:139–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lomberk G, Urrutia R. The family feud: Turning off Sp1 by Sp1-like KLF proteins. Biochem J 2005;392:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol Rev 2010;90:1337–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoopfer ED, Huang L, Denver RJ. Basic transcription element binding protein is a thyroid hormone-regulated transcription factor expressed during metamorphosis in Xenopus laevis. Dev Growth Differ 2002;44: 365–381. [DOI] [PubMed] [Google Scholar]
- 6.Ying M, Sang Y, Li Y et al. Krüppel-like family of transcription factor 9, a differentiation-associated transcription factor, suppresses Notch1 signaling and inhibits glioblastoma-initiating stem cells. Stem Cells. 2011;29:20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura H, Fujimori K. Activation of early phase of adipogenesis through Krüppel-like factor KLF9-mediated, enhanced expression of CCAAT/enhancer-binding protein b in 3T3-L1 cells. Gene 2014;534:169–176. [DOI] [PubMed] [Google Scholar]
- 8.Denver RJ, Ouellet L, Furling D et al. Basic transcription element binding protein is a thyroid hormone-regulated gene in the developing central nervous system: Evidence for a role in neurite outgrowth. J Biol Chem 1999;274:23128–23134. [DOI] [PubMed] [Google Scholar]
- 9.Dugas JC, Ibrahim A, Barres BA. The T3-induced gene KLF9 regulates oligodendrocyte differentiation and myelin regeneration. Mol Cell Neurosci 2012;50:45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang J, Chan YS, Loh YH et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol 2008;10:353–360. [DOI] [PubMed] [Google Scholar]
- 11.Katoh M Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during carcinogenesis. Stem Cell Rev 2007;3:30–38. [DOI] [PubMed] [Google Scholar]
- 12.Katoh M Network of WNT and other regulatory signaling cascades in pluripotent stem cells and cancer stem cells. Curr Pharm Biotechnol 2011;12:160–170. [DOI] [PubMed] [Google Scholar]
- 13.Lum L, Beachy PA. The Hedgehog response network: Sensors, switches, and routers. Science 2004;304:1755–1759. [DOI] [PubMed] [Google Scholar]
- 14.Varga AC, Wrana JL. The disparate role of BMP in stem cell biology. Oncogene 2005; 24:5713–5721. [DOI] [PubMed] [Google Scholar]
- 15.Dailey L, Ambrosetti D, Mansukhani A et al. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev 2005;16:233–247. [DOI] [PubMed] [Google Scholar]
- 16.Schnerch A, Cerdan C, Bhatia M. Distinguishing between mouse and human pluripo-tent stem cell regulation: The best laid plans of mice and men. Stem Cells 2010;28:419–430. [DOI] [PubMed] [Google Scholar]
- 17.Martin KM, Metcalfe JC, Kemp PR. Expression of Klf9 and Klf13 in mouse development. Mech Dev 2001;103:149–151. [DOI] [PubMed] [Google Scholar]
- 18.Bookout AL, Jeong Y, Downes M et al. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 2006;126:789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallis K, Dudazy S, van Hogerlinden M et al. The thyroid hormone receptor α1 protein is expressed in embryonic postmitotic neurons and persists in most adult neurons. Mol Endocrinol 2010;24:1904–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohguchi H, Tanaka T, Uchida A et al. Hepatocyte nuclear factor 4alpha contributes to thyroid hormone homeostasis by cooperatively regulating the type 1 iodothyronine deiodinase gene with GATA4 and Kruppel-like transcription factor 9. Mol Cell Biol 2008;28: 3917–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gamper I, Koh KR, Ruau D. GAR22: A novel target gene of thyroid hormone receptor causes growth inhibition in human erythroid cells. Exp Hematol 2009;37:539–548. [DOI] [PubMed] [Google Scholar]
- 22.Simmen FA, Xiao R, Velarde MC et al. Dysregulation of intestinal crypt cell proliferation and villus cell migration in mice lacking Krüppel-like factor 9. Am J Physiol Gastrointest Liver Physiol 2007;292:G1757–1769. [DOI] [PubMed] [Google Scholar]
- 23.Kress E, Samarut J, Plateroti M. Thyroid hormones and the control of cell prolifera tion or cell differentiation: Paradox or duality? Mol Cell Endocrinol 2009;313:36–49. [DOI] [PubMed] [Google Scholar]
- 24.Kress E, Skah S, Sirakov M et al. Cooperation between the thyroid hormone receptor TRalpha1 and the WNT pathway in the induction of intestinal tumorigenesis. Gastroenterology 2010;138:1863–1874. [DOI] [PubMed] [Google Scholar]
- 25.Lin JZ, Sieglaff DH, Yuan C et al. Gene specific actions of thyroid hormone receptor subtypes. PLoS One 2013;8:e52407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berry MN, Friend DS. High-yield preparation of isolated rat liver parenchymal cells: A biochemical and fine structural study. J Cell Biol 1969;43:506–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seglen PO. Preparation of isolated rat liver cells In: Prescott DM, ed. Methods in Cell Biology. Vol 19 New York: Academic Press, 1976:29–83. [DOI] [PubMed] [Google Scholar]
- 28.Cheng X, Ying L, Lu L et al. Self-renewing endodermal progenitor lines generated from human pluripotent stem cells. Cell Stem Cell 2012;10:371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du P, Kibbe WA, Lin SM. Lumi: A pipeline for processing Illumina microarray. Bioinformatics 2008;24:1547–1548. [DOI] [PubMed] [Google Scholar]
- 30.Smyth GK. Limma: Linear models for microarray data In: Gentleman R, Carey V, Dudoit S et al. , eds. Bioinformatics and Computational Biology Solutions using R and Bio-conductor. New York: Springer, 2005:397–420. [Google Scholar]
- 31.Team RDC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2009. [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300. [Google Scholar]
- 33.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: A non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res 2012;40:W478–W483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nogales-Cadenas R, Carmona-Saez P, Vazquez M et al. GeneCodis: Interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res 2009;37:W317–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carmona-Saez P, Chagoyen M, Tirado F et al. GENECODIS: A web-based tool for finding significant concurrent annotations in gene lists. Genome Biol 2007;8:R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mostafavi S, Ray D, Warde-Farley D et al. GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol 2008;9(suppl 1):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warde-Farley D, Donaldson SL, Comes O et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010; 38:W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuberi K, Franz M, Rodriguez H et al. GeneMANIA prediction server 2013 update. Nucleic Acids Res 2013;41:W115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan C, Lin JZ, Sieglaff DH et al. Identical gene regulation patterns of T3 and selective thyroid hormone receptor modulator GC-1. Endocrinology 2012;153:501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrova A, Celli A, Jacquet L et al. 3D in vitro model of a functional epidermal permeability barrier from human embryonic stem cells and induced pluripotent stem cells. Stem Cell Rep 2014;2:675–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacquet L, Stephenson E, Collins R et al. Strategy for the creation of clinical grade hESC line banks that HLA-match a target population. EMBO Mol Med 2013;5:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rankin SA, Kormish J, Kofron M et al. A gene regulatory network controlling hhex transcription in the anterior endoderm of the organizer. Dev Biol 2011;351:297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smart N, Dube KN, Riley PR. Identification of Thymosin b 4 as an effector of Hand1-mediated vascular development. Nat Commun 2010;1:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katoh Y, Katoh M. Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA. Int J Mol Med 2008;22:271–275. [PubMed] [Google Scholar]
- 45.Pibiri M, Ledda-Columbano GM, Cossu C et al. Cyclin D1 is an early target in hepato cyte proliferation induced by thyroid hormone (T3). FASEB J 2001;15:1006–1013. [DOI] [PubMed] [Google Scholar]
- 46.López-Fontal R, Zeini M, Través PG et al. Mice lacking thyroid hormone receptor Beta show enhanced apoptosis and delayed liver commitment for proliferation after partial hepatectomy. PLoS One 2010;14:e8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ledda-Columbano GM, Perra A, Loi R et al. Cell proliferation induced by triiodothyronine in rat liver is associated with nodule regression and reduction of hepatocellular carcinomas. Cancer Res 2000; 60:603–609. [PubMed] [Google Scholar]
- 48.Perra A, Kowalik MA, Pibiri M et al. Thyroid hormone receptor ligands induce regression of rat preneoplastic liver lesions causing their reversion to a differentiated phenotype. Hepatology 2009;49:1287–1296. [DOI] [PubMed] [Google Scholar]
- 49.Chan IH, Privalsky ML. Isoform-specific transcriptional activity of overlapping target genes that respond to thyroid hormone receptors alpha1 and beta1. Mol Endocrinol 2009;23:1758–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanimizu N, Miyajima A. Notch signaling controls hepatoblast differentiation by altering the expression of liver-enriched transcription factors. J Cell Sci 2004;117:3165–3174. [DOI] [PubMed] [Google Scholar]
- 51.Wang T, You T, Tao K et al. Notch is the key factor in the process of fetal liver stem/progenitor cells differentiation into hepatocytes. Dev Growth Differ 2012;54:605–617. [DOI] [PubMed] [Google Scholar]
- 52.Ortica S, Tarantino N, Aulner N et al. The 4 Notch receptors play distinct and antagonistic roles in the proliferation and hepatocytic differentiation of liver progenitors. FASEB J 2014;28:603–614. [DOI] [PubMed] [Google Scholar]
- 53.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science 1999; 284:770–776. [DOI] [PubMed] [Google Scholar]
- 54.Liu J, Sato C, Cerletti M et al. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr Top Dev Biol 2010;92:367–409. [DOI] [PubMed] [Google Scholar]
- 55.Bigas A, Espinosa L. Hematopoietic stem cells: To be or Notch to be. Blood 2012;119: 3226–3235. [DOI] [PubMed] [Google Scholar]
- 56.Koch U, Lehal R, Radtke F. Stem cells living with a Notch. Development 2013;140: 689–704. [DOI] [PubMed] [Google Scholar]
- 57.Chin MT, Maemura K, Fukumoto S et al. Cardiovascular basic helix loop helix factor 1, a novel transcriptional repressor expressed preferentially in the developing and adult cardiovascular system. J Biol Chem 2000;275: 6381–6387. [DOI] [PubMed] [Google Scholar]
- 58.Fischer A, Gessler M. Delta-Notch—And then?. Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res 2007;35:4583–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Medina M, Dotti CG. RIPped out by presenilin-dependent gamma-secretase. Cell Signal 2003;15:829–841. [DOI] [PubMed] [Google Scholar]
- 60.Katoh M, Katoh M. Integrative genomic analyses on HES/HEY family: Notch-independent HES1, HES3 transcription in undifferentiated ES cells, and Notch-dependent HES1, HES5, HEY1, HEY2, HEYL transcription in fetal tissues, adult tissues, or cancer. Int J Oncol 2007;31:461–466. [PubMed] [Google Scholar]
- 61.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: Repressors and oscillators that orchestrate embryogenesis. Development 2007;134:1243–1251. [DOI] [PubMed] [Google Scholar]
- 62.Kobayashi T, Mizuno H, Imayoshi I et al. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev 2009;23:1870–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boulter L, Govaere O, Bird TG et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med 2012;18: 572–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.