Abstract
Ring rot, one of the most destructive diseases of apple worldwide, is caused primarily by Botryosphaeria dothidea and B. kuwatsukai. Here, we sequenced the genomes of B. dothidea strain PG45 (44.3 Mb with 5.12 % repeat rate) and B. kuwatsukai epitype strain PG2 (48.0 Mb with 13.02 % repeat rate), and conducted a comparative analysis of these two genomes, as well as other sequenced fungal genomes, in order to understand speciation and distinctive patterns of evolution of pathogenicity-related genes. Pair-wise genome alignments revealed that the two species are highly syntenic (96.74 % average sequence identity). Both species encode a significant number of pathogenicity-related genes, e.g. carbohydrate active enzymes (CAZYs), plant cell wall degrading enzymes (PCWDEs), secondary metabolites (SMs) biosynthetic enzymes, cytochrome P450 enzymes (CYPs), and secreted peptidases, in comparison to all additional sequenced fungal species involved in various life-styles. The number of pathogenicity-related genes in B. dothidea and B. kuwatsukai is higher than other genomes of Botryosphaeriaceae pathogens (Macrophomina phaseolina and Neofusicoccum parvum), suggesting a secondary round of Botryosphaeria-lineage expansion in the family. There were, however, also significant differences in the genomes of the two Botryosphaeria species. Botryosphaeria kuwatsukai, which infects only apple and pear, apparently lost a set of SMs genes, CAZYs and PCWDEs, possibly as a result of host specialization. Botryosphaeria kuwatsukai contained significantly more transposable elements and higher value of repeat induced point (RIP) index than B. dothidea. Our results will be instrumental in understanding how both phytopathogens interact with their plant hosts and in designing efficient strategies for disease control and molecular breeding to help ensure global apple production and food security.
Keywords: genome sequencing, virulence factors, pathogenicity, Botryosphaeriaceae, genome evolution
INTRODUCTION
Fungi in the family Botryosphaeriaceae are amongst the most widespread and important canker and dieback pathogens of trees worldwide. Botryosphaeria dothidea s. lat. is one of the most common species and occurs on a large number of hosts, including more than 24 host genera, with 312 records in the literature database (Marsberg et al. 2017; Fungal Databases, US National Fungus Collections, https://nt.ars-grin.gov/fungaldatabases/).
The interaction of B. dothidea s. lat. with host plants includes a latent or endophytic phase. A basic understanding of the ecology is particularly important because the fungus can easily pass undetected by plant quarantine systems that rely on visual inspection (Marsberg et al. 2017). B. dothidea s. lat. is considered to be a stress-associated pathogen (Ma et al. 2011). Infections typically become symptomatic only under conditions of host stress, such as drought, physical damage, waterlogging, frost or unsuitable growing environments (Bostock et al. 2014, Marsberg et al. 2017). Disease symptoms include twig, branch and stem cankers, tip and branch dieback, fruit rots, blue stain or, in extreme cases, the death of the host plant (Michailides 1991, Slippers & Wingfield 2007). Such symptoms have been observed on a variety of hosts, such as ring rot of apple, fruit rot of olive, grapevine trunk disease, leaf spots and lesions on ornamental plants, and dieback and stem cankers on acacia and other shade and fruit trees (Marsberg et al. 2017).
Ring rot is one of the most destructive apple diseases worldwide, including China, Japan, South Korea, the USA, Australia, and South Africa (Ogata et al. 2000, Park 2005, Guo et al. 2009, Tang et al. 2012, Xu et al. 2015). Symptoms of the disease appear as a soft, light-coloured rot on fruit, especially during storage, and extensive cankers and/or warts on branches and trunks (Chen 1999, Kang et al. 2009). Xu et al. (2015) reappraised the etiology of apple ring rot and considered B. kuwatsukai and B. dothidea to be the main causal agents. B. kuwatsukai was previously designated as Botryosphaeria berengeriana f. sp. pyricola (Hara 1930, Koganezawa & Sakuma 1980, 1984 Xu et al. 2015). This cryptic species demonstrated substantial genetic and biological distinctions from B. dothidea. For example, the two species possessed different number and length of group I introns in the primary structures of the 18S rDNA (Xu et al. 2013, 2015). Morphologically, B. kuwatsukai presents as an appressed mycelial mat on PDA whereas B. dothidea displays columns of aerial mycelia reaching the lids of the Petri plates, and conidia of B. kuwatsukai are longer than those of B. dothidea, whereas B. dothidea had a faster growth rate than B. kuwatsukai at 35 °C and 37 °C (Xu et al. 2015). Pathogenicity tests showed that on pear stems B. kawatsukai caused large-scale cankers along with blisters whereas B. dothidea was non-pathogenic (Xu et al. 2015), but on apple shoots the two fungi induced large and small wart-like swellings, respectively, on bark (Xu et al. 2015). B. kuwatsukai apparently has a narrow host range; until now, it has been reported only from apple and pear (Xu et al. 2015).
In this study, we sequenced the genomes of one strain (PG45) of B. dothidea and an epitype strain (PG2) of B. kuwatsukai. The objectives of this study were to: (1) understand the degree of divergence between two species; (2) compare these two species to other Botryosphaeriaceae plant pathogenic fungi and to fungi with other life-styles; and (3) understand variations of pathogenesis-related gene content (e.g. CAZYs, SMs, CYPs), secreted peptidases, and candidate effectors between B. dothidea and B. kuwatsukai by comparative genomics.
MATERIALS AND METHODS
Fungal strains and culture conditions
Strain PG45 of Botryosphaeria dothidea was originally isolated from the trunk of a symptomatic apple (Malus ×domestica) tree in Shaanxi Province, China. Strain PG2 of B. kuwatsukai was originally isolated from a symptomatic apple (Malus ×domestica) fruit in Shaanxi Province, China. The cultures were purified by single spore isolation, maintained on potato dextrose agar (PDA) at 25 oC and stored as glycerol stock (15 %) at –80 oC in the Fungal Laboratory of Northwest A&F University, Yangling, Shaanxi Province, China.
DNA isolation, genome sequencing and assembly
Highly purified total genomic DNA was isolated from the fungal mycelia collected from a 2wk-old PDA culture following the modified cetyltrimethyl ammonium bromide (CTAB) protocol (Murray & Thompson 1980). The genomes were sequenced with the Illumina HiSeq2500 platform (Novogene, Beijing). The insertion size of the sequencing library was 500 bp and the sequencing strategy was 125 bp pair-ended. Filtered clean reads were assembled into scaffolds using the SPAdes v. 3.9.0 (Anton et al. 2012). In order to detect the best assembly(s), SPAdes is run at many different kmer levels (21, 33, 55, 77, 99). The completeness of assembly was assessed using BUSCO v.1.2 (Simão et al. 2015). MUMmer v. 3.23 was used to make synteny analyses at the nucleotide level (NUCmer) (Kurtz et al. 2004). The assembled scaffolds generated by the two species were aligned and oriented using Mauve, with default settings (Darling et al. 2004).
Gene prediction and genome annotation
GeneMark-ES (Alexandre et al. 2014) was first used to predict the gene structures. The gene models obtained were used to train Augustus v. 3.1 (Mario & Burkhard 2005). The predicted gene models from GeneMark-ES and Augustus and the homology proteins of the B. dothidea genome (download from Department of Energy's Joint Genome Institute) were combined in MAKER2 (Cantarel et al. 2008). Repeat sequences were identified by RepeatMasker v. 4.0.5 (http://www.repeatmasker.org) and RepeatModeler v. 1.0.7 (Saha et al. 2008) and transfer RNA was predicted by tRNAscan-SE v. 1.3.1 (Lowe & Eddy 1997) and Rfam (http://rfam.xfam.org). For calculation of RIP indices, dinucleotide frequencies were determined using the RIPCAL program (Hane & Oliver 2008).
Functional annotation of predicted genes
Carbohydrate active enzymes (CAZYs) were classified using the dbCAN Hmmer-based classification system with 1 × 10–5 as the cutoff E-value (Yin et al. 2012). Putative secondary metabolite biosynthesis genes and clusters were identified with SMURF (Khaldi et al. 2010). The ketoacyl synthase (KS) domains of PKS and condensation (C) domains of NPRS were retrieved by NAPDOS (Ziemert et al. 2012). Candidate cytochrome P450s were identified by Hmmscan with PFAM domain PF00067. Candidate secreted proteins have a secretion signal as determined by SignalP v. 4.1 (Petersen et al. 2011) and have no transmembrane domain as determined by TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM). Eventually, WoLF-PSort v. 0.2 software was used to estimate the located sites and only those proteins that were credibly positioned in the extracellular space (i.e., extracellular score >15) were included into in the final secretome (Horton et al. 2007). Small secreted proteins (SSPs) are defined here as proteins that are smaller than 200 amino acids and labeled as ‘cysteine rich’ when the percentage of cysteine residues in the protein was at least twice as high as the average percentage of cysteine residues in all predicted proteins of that organism (Ohm et al. 2012). Putative proteases were identified and classified by BLASTp querying against the MEROPS database v. 12.0 (Rawlings et al. 2018) with a cut-off E-value of 1 × 10–5.
Phylogenomic analysis
OrthoMCL v. 2.0.9 (Li et al. 2003) was used to identify ortholog pairs among compared genomes. The cutoff E-value was set as 1 × 10–5. To construct a genome-based phylogenetic tree, single-copy ortholog pairs were aligned with MAFFT v. 7 (http://mafft.cbrc.jp/alignment/server), conserved sites in the alignments were further extracted with Gblocks v. 0.91b using the default parameters (Castresama 2000), and the dataset was used for maximum likelihood tree construction in RAxML (Stamatakis 2006) with the LG+I+G+F amino acid substitution model selected by ProtTest v. 3.4 (Darriba et al. 2011). The divergence times between species were estimated using the PL method with r8s (Taylor & Berbee 2006). MEGA v. 7.0 was used to generate maximum likelihood phylogeny for KS domains, type IV siderophore C domains, CE1, AA7 families with the JTT amino acid substitution model (Jones et al. 1992, Kumar et al. 2016). Statistical support for phylogenetic grouping was assessed by 1000 bootstrap re-samplings. CAFE (Computational Analysis of gene Family Evolution) v. 3 (Han et al. 2013) was used to test whether protein family sizes were compatible with a stochastic birth and death model, and the Viterbi algorithm in the CAFE program was used to assign P-values to the expansions/contractions experienced at each branch and using a cutoff of P < 0.05. Functional enrichment tests were performed with FUNRICH v. 2.1.2 (Pathan et al. 2015).
RESULTS AND DISCUSSION
Data generated in this project has been deposited at DDBJ/EMBL/GenBank under the accession no. PRJNA394804.
Genome features of Botryoshaeria dothidea and B. kuwatsukai
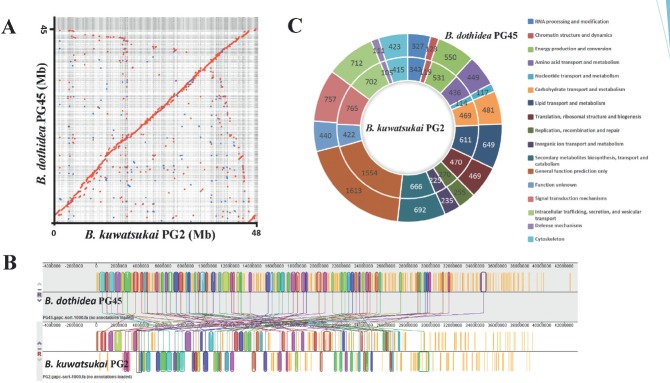
The genomes of Botryosphaeria. dothidea PG45 and B. kuwatsukai PG2 were sequenced with high coverage (163× and 156×, respectively). The B. dothidea PG45 genome was assembled into 422 scaffolds (> 1 Kb; N50, 352 Kb) with a total size of 44.3 Mb, the size is similar with the published genome of B. dothidea CBS 115476 (43.5 Mb, from Prunus sp.) (Marsberg et al. 2017). The B. kuwatsukai PG2 genome was assembled into 768 scaffolds (> 1 Kb; N50, 226 Kb) with a genome size of 48.0 Mb, the size is similar to the draft genome of B. kuwatsukai LW030101, causing apple ring rot (47.4 Mb) (Liu et al. 2016) (Table 1). The genomic GC content of B. kuwatsukai (53.01 % in strain PG2 and 53.09 % in LW030101) was lower than that of B. dothidea (54.60 % in strain PG45 and 54.69 % in CBS 115476). The completeness of the two genome assemblies in this study was assessed by BUSCO. We found1390 out of 1438 (96.7 %) and 1397 out of 1438 (97.1 %) BUSCO groups were identified in the B. dothidea PG45 genome and B. kuwatsukai PG2 genome, respectively, suggesting a high degree of completeness. The two aligned genome sequences shared 96.74 % identity at the nucleotide level and show macrosynteny (Fig. 1A). According to the genomic alignments, ~94 inverted segments were found in the two genomes (Fig. 1B). B. dothidea PG45 and B. kuwatsukai PG2 were predicted to have 15 661 and 15 306 protein coding genes, respectively. KOG analysis showed that B. dothidea PG45 had more genes involved in transport and primary and secondary metabolism than B. kuwatsukai PG2, whereas the latter taxon had more genes involved in signal transduction and DNA and RNA processing than B. dothidea PG45 (Fig. 1C). The average gene density of B. dothidea (354 in PG45 and 345 in CBS 115476, genes per Mb) was higher than that of B. kuwatsukai (321 in PG2 and 322 in LW030101, genes per Mb) (Table 1).
Table 1.
Genome features of Botryosphaeria dothidea and B. kuwatsukai.
| Features | B. dothidea PG45 | B. dothidea CBS 115476 | B. kuwatsukai PG2 | B. kuwatsukai LW030101 |
|---|---|---|---|---|
| Sequence coverage, fold | 163 | NA | 156 | 100 |
| Assemble Size (MB) | 44.3 | 43.5 | 48.0 | 47.4 |
| Scaffolds (size > 1000 bp) | 422 | 1711 | 768 | 932 |
| Scaffold N50 (Kb) | 352 | 86 | 226 | 288 |
| The longest length (Kb) | 1154 | 473 | 915 | 1260 |
| GC content (%) | 54.60 | 54.69 | 53.01 | 53.09 |
| Protein-coding genes | 15 661 | 14 998 | 15 306 | 15 260* |
| Gene density (per Mbp) | 354 | 345 | 321 | 322 |
| Repeat rate (%) | 5.12 | 2.09 | 13.02 | 12.41 |
| Nr (%) | 86.2 | NA | 86.1 | NA |
| KEGG (%) | 52.6 | NA | 51.7 | NA |
| KOG (%) | 48.4 | NA | 47.2 | NA |
| tRNAs | 138 | NA | 144 | NA |
| BUSCO estimates (%) | 96.7 | NA | 97.1 | NA |
* Protein-coding genes were predicted using the pipeline in this study.
Fig. 1.
Genomic alignments and synteny Botryosphaeria dothidea PG45 and B. kuwatsukai PG2. A. Dot plot showing the syntenic blocks between the scaffold sequences of B. dothidea PG45 (vertical axis) and B. kuwatsukai PG2 (horizontal axis). B. Fold differences in dinucleotide abundances and RIP indices for repeat families of four Botryosphaeria fungi compared with their nonrepetitive control sequences..
Genome sizes in both B. dothidea and B. kuwatsukai were similar to other species in the Botryosphaeriaceae (Islam et al. 2012, Blanco-Ulate et al. 2013, Yan et al. 2018) and larger than the average genome size of Ascomycota (36.9 Mb) (Mohanta & Bae 2015). The B. kuwatsukai genome size was larger than that of B. dothidea; however, the genome of B. kuwatsukai had lower gene density. Thus, the repeat content of B. kuwatsukai PG2 and LW030101 were 13.02 % and 12.41 %, respectively, compared to 5.12 % in B. dothidea PG45 and 2.09 % in CBS 115476, indicating that a larger number of repetitive elements contributed to the larger genome size and the fewer encoding genes in B. kuwatsukai.
Expansion of transposons and efficient RIP in Botryosphaeria kuwatsukai genome
Transposable elements (TEs) comprise 2.84 % and 0.38 % of Botryoshaeria dothidea PG45 and B. dothidea CBS 115476, as well as 9.13 % and 8.72 % of B. kuwatsukai PG2 and B. kuwatsukai LW030101 genomes, respectively. All classes of TEs in B. kuwatsukai were detected, and were more numerous than in B. dothidea (Fig. 2A). We observed expansion of hAT elements (~11-fold), Copia elements (~8-fold), Gypsy elements (~4-fold), LINEs elements (~4-fold), and Tourist and Tc1-IS630-Pogo elements (~2-fold) in the B. kuwatsukai PG2 genome, compared to B. dothidea PG45. B. kuwatsukai PG2 had 22 scaffolds containing more or equal 50 TEs, compared to only eight in B. dothidea PG45. As expected, TE-rich scaffolds of B. kuwatsukai PG2 encoded fewer genes (n = 1626) than those of B. kuwatsukai PG45 (n = 2093). Additionally, there were more species-specific genes in the TE-rich region than for B. kuwatsukai PG2 (151 vs. 137). The PFAM domain and GO enrichment tests revealed substantial enrichment of genes involved in secondary metabolism and several families of transcription factors were found in TE-rich scaffolds of B. dothidea PG45, whereas DNA and RNA processing were enriched in B. kuwatsukai PG2 TE-rich regions (Table S1, S4).
Fig. 2.
Analysis of repeats in Botryosphaeria dothidea and B. kuwatsukai. A. The B. kuwatsukai genome was shown to be enriched with LTR and DNA transposon-like elements compared to the B. dothidea genome. B. Fold differences of the dinucleotide frequencies of two genomes repeat elements relative to the control. It shows that both genomes have an enrichment of TpA and a depletion of CpA dinucleotides, which is characteristic of RIP.
Repeat induced point (RIP) mutation is a genome defence mechanism in fungi during which duplicated sequences are mutated from CpA to TpA (Galagan & Selker 2004). RIP in the B. kuwatsukai genome was inferred by the high value of TpA/ApT (RIP index, 1.77 in strain PG2 and 1.76 in strain LW030101), in contrast to a considerably lower ratio of TpA/ApT (RIP index, 1.27 in strain PG45 and 0.83 in strain CBS 115476) in B. dothidea genome (Fig. 2B). The higher RIP index in B. kuwatsukai suggests more active RIP defence mechanisms than in B. dothidea.
Phylogenomic analysis and evolution
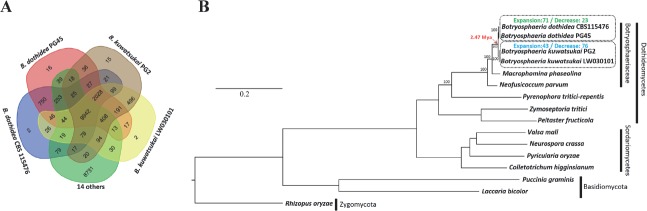
To explore gene family evolution of these two species, we clustered B. dothidea PG45 and B. kuwatsukai PG2 proteomes with those of 14 other representative fungi using OrthoMCL, which included B. dothidea CBS 115476 isolated from Prunus sp., B. kuwatsukai LW030101 isolated from apple, a biotrophic fungus (Puccinia graminis), four additional necrotrophs (Valsa mali, Pyrenophora tritici-repentis, Neofusicoccum parvum, and Macrophomina phaseolina), three hemi-biotrophic model plant pathogens (Pyricularia oryzae, Colletotrichum higginsianum, and Zymoseptoria tritici), two saprobes (Neurospora crassa and Rhizopus oryzae), a mutualistic symbiotic fungus (Laccaria bicolor), and an ectophytic fungus (Peltaster fructicola). This produced 23 584 ortholog families (groups) comprising 188 147 proteins, as well as 38 559 orphans that show no homology to any other proteins in this dataset. A total of 10 742 and 10 457 groups (B. dothidea PG45 and B. dothidea CBS 115476, respectively) and 10 602 and 10 628 groups (B. kuwatsukai PG2 and B. kuwatsukai LW030101, respectively) had homologs in the other fungi tested, of which about 9942 were conserved in all compared genomes. A total of 2028 groups involved in all four Botryposphaeria strains were Botryosphaeria lineage-specific (Fig. 3A); i.e. they were shared exclusively between the two species and had no ortholog in the other fungi we included. About 71 % these lineage-specific genes were hypothetical proteins or had no homology to sequences in GenBank/ A 81 % of them encode proteins without known PFAM domains, and the subset of genes with functional domains was enriched with genes involved in the production of secondary metabolites, transcription factors, and heterokaryon incompatibility (Table S5). The number of orphans (species-specific proteins) involved was 496 groups in B. kuwatsukai and 750 in B. dothidea (Fig. 3A). Many of these genes (79–82 %) encoded proteins without known functional domains. When we compared the Botryosphaeria genomes (2210 in PG45, 1904 in CBS 115476, 2020 in PG2 and 2005 in LW030101) to those of two other closely related members of the family Botryosphaeriaceae (2041 in M. phaseolina and 1417 in N. parvum), B. dothidea PG45 had the largest number of multi-copy genes.
Fig. 3.
Homology and phylogenomic relationships of Botryosphaeria dothidea and B. kuwatsukai. A. Predicted proteins in B. dothidea and B. kuwatsukai were compared with the genome encoding proteins of other 12 species shown in phylogenetic tree. B. A maximum likelihood phylogenetic tree was constructed from concatenated alignment of 748 single-copy orthologs conserved across all species using RAxML with the best-fit model of LG+I+G+F.
Among the ortholog families, 748 single copy orthologs that were conserved across all fungi analyzed were identified and subjected to maximum likelihood phylogenomic analysis. A maximum likelihood phylogenomic tree was generated using a 173 008 amino acid position alignment and calibrated with the origin of Ascomycota clade around 500–650 Mya. The phylogenomic tree, as expected, placed B. dothidea and B. kuwatsukai in a monophyletic clade closest to M. phaseolina, a devastating necrotrophic fungal pathogen worldwide and infects more than 500 plant hosts (Islam et al. 2012) (Fig. 3B). According to the phylogenomic tree and analysis by r8s software, B. dothidea and B. kuwatsukai diverged about 2.47 Mya, suggesting a relatively recent speciation event in Botryosphaeria.
By CAFE analysis, at the most recent common ancestor (MRCA) of B. dothidea and B. kuwatsukai, the expected number of group gain (expanded groups of MRCA of two species) was 265, which exceeds the loss (76). A total of 126 groups experienced significant expansion at the Botryosphaeria MRCA (branch P < 0.05), with gains in putative glucose methanol choline (GMC) reductases and SM-related genes (Table S6). The GMC family includes extracellular alcohol oxidases and cellobiose dehydrogenase, enzymes directly involved in lignocellulose degradation (Henriksson et al. 2000, Martinez et al. 2004). In the B. kuwatsukai clade, gene family decrease exceeded expansions according to the CAFE analysis, whereas the opposite trend occurred in the B. dothidea clade (Fig. 3B). One possible explanation of this divergence in pathways is that genes required for infection of different host plants may have existed in ancestral Botryosphaeria species, and that losses from existing families occurred during the process of host specialization (Gan et al. 2016); i.e. B. kuwatsukai is specific to only apple and pear.
Genes involved in sexual and asexual development
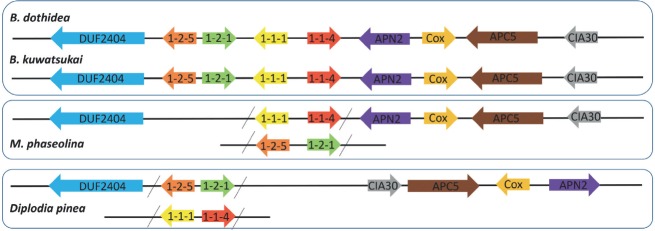
Both sexual and asexual morphs have been reported in Botyrosphaeria dothidea (Slippers et al. 2004). The first MAT gene sequence for a species in Botryosphaeriales was described for Diplodia sapinea, a well-known pathogen of Pinus species (Bihon et al. 2012, Swart & Wingfield 1991). In this study, the MAT genes of B. dothidea and B. kuwatsukai, as well as those of their closest relative, M. phaseolina, were compared with those of D. sapinea. In the B. dothidea and B. kuwatsukai genomes, all four characterized MAT genes (MAT1-1-1, MAT1-1-4, MAT 1-2-1 and MAT1-2-5) grouped together in the genome at a single locus (Fig. 4), indicating that they have a homothallic mating system. However, only MAT1-1-1 and MAT1-1-4 genes were found in M. phaseolina, and only MAT 1-2-1 and MAT1-2-5 genes were found in D. sapinea, indicating a heterothallic mating system (Fig. 4). In addition, the genes adjacent to the MAT genes on this locus were oriented in the same direction when comparing B. dothidea and B. kuwatsukai with M. phaseolina, whereas they were inverted with D. sapinea (Fig. 4). Apart from mating type, a series of other ‘sex-related’ genes have been identified as being involved in various stages of mating and ascoma production in B. dothidea and B. kuwatsukai (Table S7). In particular, the high-mobility group box (AN1962) involved in ascoma development was found only in B. dothidea PG45.
Fig. 4.
Comparison of the genomic architecture of the MAT locus and surrounding genes among Botryosphaeria dothidea, B. kuwatsukai, Macrophomina. phaseolina and Diplodia sapinea. All four characterized MAT genes group together at a single locus, suggesting a homothallic mating-type system in both B. dothidea and B. kuwatsukai. Arrows represent gene order and orientation, but genes and intergenic regions are not to scale. Abbreviations: CIA30, complex I intermediate-associated protein 30; Cox, cytochrome C oxidase subunit VIa; DUF2404, putative integral membrane protein containing DUF2404 domain; APN2, DNA lyase; APC5, anaphase-promoting complex.
Developmental features that distinguished B. dothidea from B. kuwatsukai included mycelial and conidial morphology. The latter taxon exhibited an appressed mycelial mat on PDA, whereas B. dothidea displayed columns of aerial mycelia reaching the lids of the Petri plates. Furthermore, conidia of B. dothidea were longer than those of B. dothidea (Xu et al. 2015). Conidiophore pattern and cell-identity regulators in Aspergillus nidulans include medusa (medA), stunted (stuA), abacus (abaA) and bristle (brlA) (Borkovich et al. 2004, Amselem et al. 2011). Orthologs of medA and stuA were present in B. dothidea and B. kuwatsukai, whereas an unambiguous ortholog of abaA and brlA was not present in either genome (Table S8). However, genes known to function upstream of brlA, including fadA flbC and flbD, were all present in both B. dothidea and B. kuwatsukai. FluG is present only in B. dothidea. Functional analyses will be required to determine the role of these genes in biology.
In addition to structures involved in sexual and asexual development unrelated to pathogenesis, the tetraspanin-encoding gene (pls1) required for appressorium function in P. oryzae (Xue et al. 2002) was also present in B. dothidea and B. kuwatsukai (Table S9). Some authors suggested that B. dothidea might be capable of infecting plant leaves by direct penetration via the formation of appressorium-like structures (Marsberg et al. 2017). However, the genes involved in appressorium formation (emp1), appressorium penetration (mas1) and adhesion (mpg1, MAD1 and MAD2) were absent in both B. dothidea and B. kuwatsukai.
Secondary metabolism
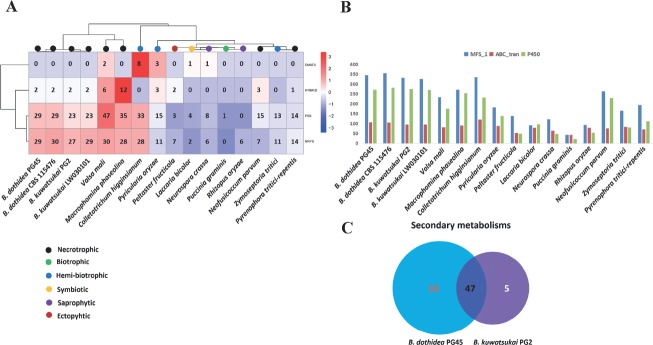
One of the crucial weapons that necrotrophic, polyphagous pathogens possess is the production of phytotoxic compounds to kill cells of a range of plant species (Amselem et al. 2011). To identify the pathways involved in the production of secondary metabolites in B. dothidea and B. kuwatsukai, we searched the genomes for genes encoding key enzymes such as NRPS (non-ribosomal peptide synthetase), PKS (polyketide synthase), HYBRID (PKS-NRPS) and DMATS (dimethylallyl tryptophane synthase). Botryosphaeria strains were found to contain a significant number of genes encoding key secondary metabolism (SMs) biosynthesis enzymes except 52 in B. kuwatsukai PG2 (P = 0.056, t-test), 60 in B. dothidea PG45 (**, P = 0.005, t-test), 61 in B. dothidea CBS 115476 (**, P = 0.004, t-test), and 54 in B. kuwatsukai LW030101 (*, P = 0.032, t-test). Compared to fungi with different life-styles, these numbers are lower than for the hemi-biotrophic model plant pathogen Colletotrichum higginsianum (69) and the necrotrophic fungi Valsa mali (85) and Macrophomina phaseolina (75), significantly higher than for all biotrophic, ectophytic, saprobic, and mutualistic symbiotic fungi that have been sequenced (Fig. 5A).
Fig. 5.
Comparison secondary metabolism backbone genes and related genes in Botryosphaeria dothidea and B. kuwatsukai to other 12 fungi species involved in different lifestyles. (A) For each secondary metabolism backbone gene family and predicted total gene numbers are shown in cells. Over-represented (+3 to 0) and under-represented (0 to -3) numbers are depicted as Z-scores for each line in heatmap. (B) The number of MFS_1 (PF07690) family, ABC_tran (PF00005) family and CYP family were compared between B. dothidea or B. kuwatsukai and all the genomes. (B) Comparative analyses of orthologous of secondary metabolism backbone genes in B. dothidea PG45 and B. kuwatsukai PG2. Orthologs were determined by BDBHs.
Generally in fungi, most key SM genes belong to clusters that encode biosynthesis enzymes, CYPs, regulators and/or transporters (Keller et al. 2005, Fox & Howlett 2008). CYP enzymes catalyze the conversion of hydrophobic intermediates of primary and secondary metabolic pathways and play essential roles in fungi. A total of 273, 283, 276 and 270 CYPs were found in B. dothidea PG45, B. dothidea CBS 115476, B. kuwatsukai PG2 and B. kuwatsukai LW030101, respectively. These numbers are higher than for the other fungi with which we compared them (Fig. 5B). The SM clusters contain a gene encoding the ATP-binding cassette (ABC) superfamily or the major facilitator superfamily (MFS) transporter that could export the metabolites produced by the enzymes encoded by the gene cluster. Both B. dothidea and B. kuwatsukai have larger sizes of MFS_1 (PF07690, 327–355) and ABC_tran (PF00005, 95-107) families than all the other fungi studied except for Colletotrichum higginsianum (335 of MFS_1 and 120 of ABC_tran) (Fig. 5B).
All key SM genes present in both B. dothidea and B. kuwatsukai and their orthologs were found in other fungal genomes, indicating the apparent absence of species-specific genes involved in the production of secondary metabolites. Botryosphaeria dothidea had more genes encoding key SM enzymes than B. kuwatsukai. This difference is even more striking when considering orthologs and paralogs; only 47 key SM genes corresponded to orthologous pairs in both genomes, whereas 13 genes were found only in B. dothidea PG45 and 5 only in B. kuwatsukai PG2 (Fig. 5C). Based on phylogenetic analysis of well-characterized PKS protein sequences from other species (Noar & Daub 2016), we hypothesize that B. dothidea PG45 and B. kuwatsukai PG2 produce ochratoxin, alternapyrone, zearalenones, cercosporin, citrinin, 6-methylsalicylic acid and melanin, among which, zearalenones are found only in B. dothidea PG45 and cercosporin only in B. kuwatsukai PG2 (Fig. S1). Both species contained NRPS genes and are orthologous to the type IV fungal siderophore synthetase (Fig. S2), whose products are essential for fungal virulence (Bushley et al. 2008, Scharf et al. 2014). Together with the SM enzymes, CYP proteins, ABC and MFS transporters are also expanded in B. dothidea, and thus, the size and diversity of these families may have evolved concomitantly with secondary metabolism genes.
Carbohydrate active enzymes
Botryosphaeria dothidea and B. kuwatsukai are particularly well equipped with genes encoding carbohydrate-active enzymes (CAZYs) (Table S10). The CAZY content in B. dothidea PG45 (823), B. dothidea CBS 115476 (825), B. kuwatsukai LW030101 (789) and B. kuwatsukai PG2 (791) genomes is larger than for any of the other 12 fungal genomes we examined (Fig. 6A), suggesting that the evolution of these species has led to different degrees of reduction in their carbohydrate degrading capabilities. These expanded CAZY arsenals are more similar to those of other hemibiotrophic and necrotrophic pathogens than to the highly reduced set found in biotrophs (e.g. Puccinia graminis) and ectophytes (e.g. Peltaster fructicola). In particular, the CAZY repertoire of B. dothidea and B. kuwatsukai is extremely expanded relative to that of Z. tritici, although all these three pathogens have a latent infection phase (Goodwin et al. 2011), suggests that they use a different mechanism for avoidance of host defences. Analysis of the genome showed that both B. dothidea and B. kuwatsukai have a glycoside hydrolase family GH33, of which GH33 is not present in the Z. tritici genome. The GH33 hydrolase family consists of sialidases which hydrolyse the glycosidic linkages of terminal sialic residues in oligosaccharides. Sialidases can act as pathogenicity factors, which can assist in host adaptation by avoiding host recognition or by inhibiting host defence responses (Alviano et al. 2004). Experimental verification is needed to understand how these two pathogens infect hosts without resulting in symptoms and can exist as endophytes.
Fig. 6.
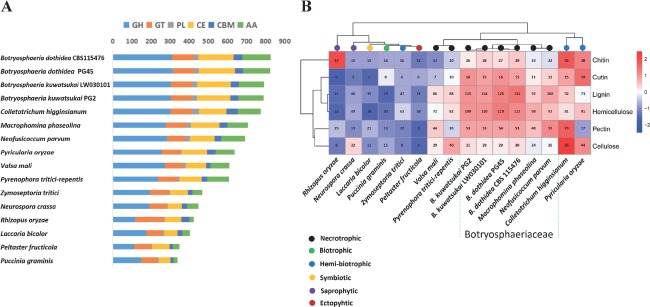
Gene expansion of CAZYs in the Botryosphaeria lineage. A. Overall comparison of glycoside hydrolases (GHs), glycosyl transferases (GTs), polysaccharide lyases (PLs), carbohydrate esterases (CEs), auxiliary activities (AAs), and carbohydrate-binding modules (CBMs) against other lifestyles fungi. B. CAZYs expanded in the Botryosphaeria lineage mainly involved in hemicellulose and lignin degradation. Over-represented (+2 to 0) and under-represented (0 to -2) numbers are depicted as Z-scores for each line in heatmap.
The ability to degrade complex plant carbohydrates is an important aspect of the life-styles of plant-associated fungi. Plant cell wall carbohydrates form a complex network of different polysaccharides that includes cellulose, hemicellulose, pectin, and lignin. This network is the target of carbohydrate-active enzymes and auxiliary proteins (jointly referred to as CAZY) needed to access internal plant tissues and to degrade plant cell wall components to simple monomers serving as carbon sources. Both species contains a much more extensive set of glycoside hydrolases (GHs), polysaccharide lyases (PL), carbohydrate esterases (CE), auxiliary activities (AAs), and carbohydrate binding modules (CBMs) than the other species (Fig. 6A). The strong expansion of GHs, PLs, CEs, AAs, and CBMs suggests an expanded capacity to degrade plant cell walls. The genomes of B. dothidea (PG45 and CBS 115476) and B. kuwatsukai (PG2 and LW030101) contain, respectively, (370 and 372) and (350 and 349) genes associated with plant cell wall degradation (Table S11); these numbers are larger than the average of all plant pathogens analysed (n = 290) as well as the average of all non-phytopathogens (n = 118). Their potential pectin-degrading capacity is comparable to that of N. parvum and M. phaseolina, and exceeds the other fungi except for C. higginsianum (Fig. 6B). Botryosphaeria dothidea and B. kuwatsukai have an intermediate number of enzymes putatively involved in degradation of cellulose; however, they possess more hemicellulose-degrading enzymes (119 in B. dothidea PG45, 113 in B. dothidea CBS 115476, 109 in B. kuwatsukai PG2 and 110 in B. kuwatsukai LW030101) than all of the other fungi (average = 68) (Fig. 6B). As expected, both species, which are woody-tissue colonizing phytopathogens, encoded more genes involved in lignin degradation than all of the other fungi (Fig. 6B). Interestingly, the number of cutinases (15–16) is higher than for all the other fungi except for P. oryzae, which suggests that these Botryosphaeria species possess a relatively high degree of adaptability to fruit cuticles. Hierarchical clustering analysis showed that the PCWDEs profile of two species most closely related to that of M. phaseolina and N. parvum in Botryosphaeriaceae (Fig. 7). A cluster involved in 25 different modules is remarkably expanded in the Botryosphaeria lineage, including 11 hemicellucose, 6 lignin, 5 pectin, 1 cutin, 1 chitin, and 1 cellulose (Fig. 7). Moderate expansion also occurring in M. phaseolina and N. parvum suggests that the ancient ancestor of Botryosphaeriaceae possessed many genes involved in hemicellulose, lignin, and pectin degradation, whereas the Botryosphaeria lineage underwent a second round of expansion during evolution.
Fig. 7.
Distribution of selected corresponding plant cell wall degrading enzymes among different fungi. Red dashed box shows a cluster involved in different modules expanded in the Botryosphaeria lineage, which mainly include hemicellulose (11), lignin (6), pectin (5), cellulose (1), cutin (1), and chitin (1). Over-represented (+3 to 0) and under-represented (0 to -3) numbers are depicted as Z-scores for each line in heatmap.
Comparing the two Botryospheria species, B. dothidea (823 and 825 in two strains) had a higher number of genes encoding CAZYs compared to B. kuwatsukai (789 and 791 in two strains). When compared B. kuwatsukai PG2 and B. dothidea PG45, only 771 CAZYs corresponded orthologous pairs in both genomes by Bidirectional Best BLAST Hits (BDBHs) analysis, whereas 18 were found only in B. kuwatsukai PG2, and 52 only in B. dothidea PG45. In particular, B. dothidea PG45 expanded CE1 and AA7 modules involved in hemicellulose and lignin degradation compared to those of B. kuwatsukai PG2 (8 vs. 1 and 11 vs. 2). Phylogenetic analysis shows that these modules occurred mainly in lineage-specific expansions (Fig. S3).
Secretome
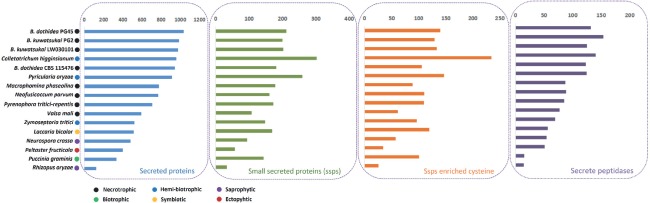
Pathogens can secrete a series of proteins that are deployed to the host-pathogen interface during infection, and secretome proteins play an important role in pathogenicity (O’Connell et al. 2012). In the current study, a remarkable number of 986 and 975 secreted proteins in the B. kuwatsukai PG2 and B. kuwatsukai LW030101 genome, as well as, 1032 and 940 in the B. dothidea PG45 and B. dothidea CBS 115476 genome were predicted (*, P < 0.05, t-test) (Fig. 8). The number of secreted proteins in both genomes exceeded that of M. phaseolina (776) and N. parvum (768), indicating a genus-lineage expansion in Botryosphaeria.
Fig. 8.
Comparison a series of secreted proteins in Botryosphaeria dothidea, B. kuwatsukai and to other 12 fungi species involved in different lifestyles.
Secreted effector proteins that are transferred into plant host cells are essential for pathogenesis by many plant pathogenic microorganisms. The larger size of the B. kuwatsukai secretome was also evident for small secreted proteins (SSPs): 199 and 202 in B. kuwatsukai (PG2 and LW030101) and 211 and 181 in B. dothidea (PG45 and CBS 115476) were smaller in size than 200 amino acids. The number of SSPs in B. dothidea PG45 was the highest among the Botryosphaeriaceae in this study (Fig. 8). In addition, 129 and 133 in B. kuwatsukai (PG2 and LW030101) and 139 and 105 in B. dothidea (PG45 and CBS 115476) were cysteine-enriched SSPs, which were considered as candidate secreted effectors (Fig. 8). Except B. dothidea CBS 115476 candidate secreted effectors were fewer than in N. parvum; these were more numerous in four Botryosphaeria strains than N. parvum (110) and M. phaseolina (88) PFAM domain analysis reveals that 7, 9, 10, and 11 known domains were identified in the predicted secreted effectors (B. dothidea PG45, B. dothidea CBS 115476, B. kuwatsukai PG2, and B. kuwatsukai LW030101) respectively (Table S12 and S13). Ribonuclease and the cerato-platanin domain were found only in B. kuwatsukai; the WSC domain may be involved in carbohydrate binding (Ponting & Hofmann 1999) and occurred only in B. dothidea PG45; the CAP domain related to pathogenesis proteins occurred only in B. kuwatsukai LW030101 (Fig. S4). The cerato-platanin family of proteins includes the phytotoxin which causes severe plant disease accompanied by canker stain symptoms (Sbrana et al. 2007). One cerato-platanin family effector gene was found only in B. kuwatsukai, which may explain why it can cause large cankers and blistering on pear stems, whereas as B. dothidea induces only localized necrotic spots (Xu et al. 2015). Pathogenic as well as saprotrophic fungi secrete peptidases to degrade a variety of proteases in their environment. This degradation is potentially beneficial in eliminating the activity of antifungal host proteins and in providing nutrients. A total of 131, 123, 153, and 125 secreted protease-encoding sequences were present in B. dothidea PG45, B. dothidea CBS 115476, B. kuwatsukai PG2 and B. kuwatsukai LW030101 respectively (Fig. 8), more than all of the other fungi in the study and similar to the hemi-biotrophic plant pathogens P. oryzae (127) and C higginsianum (143).
CONCLUSIONS
Comparative genomics of Botryosphaeria dothidea and B. kuwatsukai revealed an expectedly high degree of sequence identity and synteny. We observed several similarities in gene structure and content between these closely related plant pathogens. Organization of the MAT loci and processing of a glycoside hydrolase family GH33 are the same in B. dothidea and B. kuwatsukai. Both species encodes a significant number of virulence factors, i.e. CAZYs, PCWDEs, SMs and secreted proteases, in comparison to other plant pathogenic fungi we studied. In particular, the number of CAZY in both species surpass that of all other fungi included in the comparison. They possess a large number of plant cell wall breakdown genes in cutin, hemicellulose, lignin and pectin degradation, indicating that their MCRA expanded these pathogenic genes to adapt to a wider host range. In Botryosphaeriaceae, M. phaseolina encodes a significant number of CYPs, MFS type membrane transporters, glycosidases and secondary metabolites (Islam et al. 2012). In this study, we documented that the number of pathogenicity-related genes in B. dothidea and B. kuwatsukai is higher than M. phaseolina, suggesting a secondary round of lineage expansion in Botryosphaeriaceae.
Our data also highlighted several striking differences in gene content and high genetic diversity between these plant pathogens. The first difference was in the content of TEs. The larger number of TEs in B. kuwatsukai genome resulted in larger genome size and fewer encoding genes relative to B. dothidea. Previous studies showed that B. dothidea is able to infect a wide range of hosts, whereas B. kuwatsukai is specific to apple and pear (Inderbitzin et al. 2010, Marques et al. 2013, Xu et al. 2013, 2015). The comparative genomics analysis further revealed a striking difference between these two species in the amount. B. kuwatsukai, which infects only apple and pear, apparently lost a set of SM genes, CAZYs and PCWDEs, possibly as a result of host specialization. Generating and analyzing additional genomes of location-diversity-based strains will be necessary for discerning these common genome features between B. dothidea and B. kuwatsukai. These data shed light on the evolutionary and mechanistic bases of the genetically complex traits of two main plant pathogens causing apple ring rot. With increased understanding of the differences between two main apple ring rot pathogens at a genomic level, we can begin to develop targeted disease control strategies based on molecular breeding for resistance.
Acknowledgments
We would like to thank the anonymous reviewers for their kind and helpful comments on the original manuscript. This work was supported by National Natural Science Foundation of China (31371887), the 111 Project from Education Ministry of China (B07049), and the earmarked fund for China Agriculture Research System (CARS-27).
Supplementary Material
REFERENCES
- Alexandre L, Burns PD, Mark B. (2014) Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Research 42: 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alviano DS, Rodrigues ML, Almeida CA, Santos AL, Couceiroet JN, et al. (2004) Differential expression of sialylglycoconjugates and sialidase activity in distinct morphological stages of Fonsecaea pedrosoi. Archives of Microbiology 181: 278–286. [DOI] [PubMed] [Google Scholar]
- Amselem J, Cuomo CA, van Kan JA, Viaud M, Benito EP, et al (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genetics 7: e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton B, Nurk S, Antipov D, Gurevich AA, Dvorkin M.et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihon W, Burgess T, Slippers B, Wingfield MJ, Wingfield BD. (2012) High level of genetic diversity and cryptic recombination is widespread in introduced Diplodia pinea populations. Australasian Plant Pathology 41: 41–46. [Google Scholar]
- Blanco-Ulate B, Rolshausen P, Cantu D. (2013) Draft genome sequence of Neofusicoccum parvum isolate UCR-NP2, a fungal vascular pathogen associated with grapevine cankers. Genome Announcements 1: e00339-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE. et al (2004) Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiology and Molecular Biology Reviews 68: 1–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock RM, Pye MF, Roubtsova TV. (2014) Predisposition in plant disease: exploiting the nexus in abiotic and biotic stress perception and response. Annual Review of Phytopathology 52: 517. [DOI] [PubMed] [Google Scholar]
- Bushley KE, Ripoll DR, Turgeon BG. (2008) Module evolution and substrate specificity of fungal nonribosomal peptide synthetases involved in siderophore biosynthesis. BMC Evolutionary Biology 8: 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SM, Parra G, Ross E. et al (2008) MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Research 18: 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresama J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Chen C. (1999) Advances in the research of apple ring rot. Acta Phytopathologica Sinica 29: 1–7 [in Chinese]. [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT. (2004) MAUVE: multiple alignment of conserved genomic sequence with rearrangements. Genome Research 14: 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox EM, Howlett BJ. (2008) Secondary metabolism: regulation and role in fungal biology. Current Opinion in Microbiology 11: 481–487. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Selker EU. (2004) Rip: the evolutionary cost of genome defense. Trends in Genetics 20: 417–423. [DOI] [PubMed] [Google Scholar]
- Gan P, Narusaka M, Kumakura N, Tsushima A, Takano Y. et al (2016) Genus-wide comparative genome analyses of Colletotrichum species reveal specific gene family losses and gains during adaptation to specific infection lifestyles. Genome Biology and Evolution 8: 1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin SB, M’Barek SB, Dhillon B, Wittenberg AHJ, Crane CF, et al (2011) Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genetics 7: e1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Li J, Li B, Zhang X, Zhou Z. et al (2009) Investigations on the occurrence and chemical control of Botryosphaeria canker of apple in China. Plant Protection 35: 120–123. [Google Scholar]
- Han MV, Thomas GWC, Lugo-Martinez J, Hahn MW. (2013) Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Molecular Biology and Evolution 30: 1987–1997. [DOI] [PubMed] [Google Scholar]
- Hane J, Oliver R. (2008) RIPCAL: a tool for alignment-based analysis of repeat-induced point mutations in fungal genomic sequences. BMC Bioinformatics 9: 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K. (1930) Pathologia Agriculturalis Plantarum. Tokyo: Yokendo. [in Japanese.] [Google Scholar]
- Henriksson G, Johansson G, Pettersson G. (2000) A critical review of cellobiose dehydrogenases. Journal of Biotechnology 78: 93–113. [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H. et al (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Research 35: 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inderbitzin P, Bostock RM, Trouillas FP, Michailides TJ. (2010) A six locus phylogeny reveals high species diversity in Botryosphaeriaceae from California almond. Mycologia 102: 1350–1368. [DOI] [PubMed] [Google Scholar]
- Islam MS, Samiul HM, Moinul IM, Mannan EE, Abdul H. et al (2012) Tools to kill: genome of one of the most destructive plant pathogenic fungi Macrophomina phaseolina. BMC Genomics 13: 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AL, Aldwinckle HS. (1990) Compendium of Apple and Pear Diseases. St Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. (1992) The rapid generation of mutation data matrices from protein sequences. CABIOS 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kang L, Hao H, Yang Z, Li X, Kang G. (2009) The advances in the research of apple ring rot. Chinese Agricultural Science Bulletin 25: 188–191 [in Chinese]. [Google Scholar]
- Keller NP, Turner G, Bennett JW. (2005) Fungal secondary metabolism - from biochemistry to genomics. Nature Reviews Microbiology 3: 937–947. [DOI] [PubMed] [Google Scholar]
- Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC. et al (2010) SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genetics and Biology 7: 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koganezawa H, Sakuma T. (1980) Fungi associated with blister canker and internal bark necrosis of apple trees. Bulletin of the Fruit Tree Research Station C 7: 83–99. [Google Scholar]
- Koganezawa H, Sakuma T. (1984) Causal fungi of apple fruit rot. Bulletin of the Fruit Tree Research Station C 11: 49–62. [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M. et al (2004) Versatile and open software for comparing large genomes. Genome Biology 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Research 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lian S, Li B, Lu H, Dong X. et al (2016) Draft genome sequence of Botryosphaeria dothidea, the pathogen of apple ring rot. Genome Announcements 4: e01142-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Morgan DP, Michailides TJ. (2001) Effects of water stress on Botryosphaeria blight of Pistachio caused by Botryosphaeria dothidea. Plant Disease 85: 745–749. [DOI] [PubMed] [Google Scholar]
- Mario S, Burkhard M. (2005) AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Research 33: W465–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques MW, Lima NB, de Morais MA, Michereff SJ, Phillips AJL, et al (2013) Botryosphaeria, Neofusicoccum, Neoscytalidium and Pseudofusicoccum species associated with mango in Brazil. Fungal Diversity 61: 195–208. [Google Scholar]
- Marsberg A, Kemler M, Jami F, Nagel JH, Postma-Smidt A, et al (2017) Botryosphaeria dothidea: a latent pathogen of global importance to woody plant health. Molecular Plant Pathology 18: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D, Larrondo LF, Putnam N, Gelpke MD, Huang K. et al (2004) Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain rp78. Nature Biotechnology 22: 695–700. [DOI] [PubMed] [Google Scholar]
- Michailides TJ. (1991) Pathogenicity, distribution, sources of inoculum, and infection courts of Botryosphaeria dothidea on Pistachio. Phytopathology 81: 566–573. [Google Scholar]
- Mohanta TK, Bae H. (2015) The diversity of fungal genome. Biological Procedures Online 17: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noar RD, Daub ME. (2016) Bioinformatics prediction of polyketide synthase gene clusters from Mycosphaerella fijiensis. PLoS One 11: e0158471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose T. (1933) On the ring rot of pears and the causal organism, especially on its perfect generation Physalospora piricola. Money Marketing 7: 156–163 [in Japanese]. [Google Scholar]
- O’Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J, et al (2012) Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nature Genetics 44: 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata T, Sano T, Harada Y. (2000) Botryosphaeria spp. isolated from apple and several deciduous fruit trees are divided into three groups based on the production of warts on twigs, size of conidia, and nucleotide sequences of nuclear ribosomal DNA ITS regions. Mycoscience 41: 331–337. [Google Scholar]
- Ohm RA, Feau N, Henrissat B, Schoch CL, Horwitz BA. et al (2012) Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathogens 8: e1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EW. (2005) An infection model of apple white rot based on conidial germination and appressorium formation of Botryosphaeria dothidea. The Plant Pathology Journal 21: 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CYJ. et al (2015) FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics 15: 2597–2601. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. (2011) SignalP 4.0: discriminating signal protease from transmembrane regions. Nature Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Hofmann K, Bork P. (1999) A latrophilin/CL-1-like GPS domain in polycystin-1. Current biology 9: R585–R588. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A. et al (2018) The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Research 46: D624–D632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Bridges S, Magbanua ZV, Peterson DG. (2008) Empirical comparison of ab initio repeat finding programs. Nucleic Acids Research 36: 2284–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrana F, Bongini L, Cappugi G, Fanelli D, Guarino A. et al (2007) Atomic force microscopy images suggest aggregation mechanism in cerato-platanin. European Biophysics Journal 36: 727–732. [DOI] [PubMed] [Google Scholar]
- Scharf DH, Heinekamp T, Brakhage AA. (2014) Human and plant fungal pathogens: the role of secondary metabolites. PLoS Pathogens 10: e1003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. [DOI] [PubMed] [Google Scholar]
- Slippers B, Crous PW, Denman S, Coutinho TA, Wingfield BD. et al (2004) Combined multiple gene genealogies and phenotypic characters differentiate several species previously identified as Botryosphaeria dothidea. Mycologia 96: 83–101. [PubMed] [Google Scholar]
- Slippers B, Wingfield MJ. (2007) Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity, ecology and impact. Fungal Biology Reviews 21: 90–106. [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Swart WJ, Wingfield MJ. (1991) Biology and control of Sphaeropsis sapinea on Pinus species in South Africa. Plant Disease 75: 761–766. [Google Scholar]
- Tang W, Ding Z, Zhou Z, Wang Y, Guo L. (2012) Phylogenetic and pathogenic analyses show that the causal agent of apple ring rot in China is Botryosphaeria dothidea. Plant Disease 96: 486–496. [DOI] [PubMed] [Google Scholar]
- Taylor JW, Berbee ML. (2006) Dating divergences in the Fungal Tree of Life: review and new analyses. Mycologia 98: 838–849. [DOI] [PubMed] [Google Scholar]
- Xu C, Wang C, Sun X, Zhang R, Gleason ML. et al (2013) Multiple group I introns in the small-subunit rDNA of Botryosphaeria dothidea: implication for intraspecific genetic diversity. PLoS One 8: e67808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang C, Ju L, Zhang R, Biggs AR. et al (2015) Multiple locus genealogies and phenotypic characters reappraise the causal agents of apple ring rot in China. Fungal Diversity 71: 215–231. [Google Scholar]
- Xue C, Park G, Choi W, Zheng L, Dean RA. et al (2002) Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell 14: 2107–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan JY, Zhao WS, Chen Z, Xing QK, Zhang W. et al (2018) Comparative genome and transcriptome analyses reveal adaptations to opportunistic infections in woody plant degrading pathogens of Botryosphaeriaceae. DNA Research 25: 87–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Mao X, Yang J, Chen X, Mao F. et al (2012) dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Research 40: W445–W451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, Podell S, Penn K, Badger JH, Allen E. et al (2012) The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One 7: e34064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.