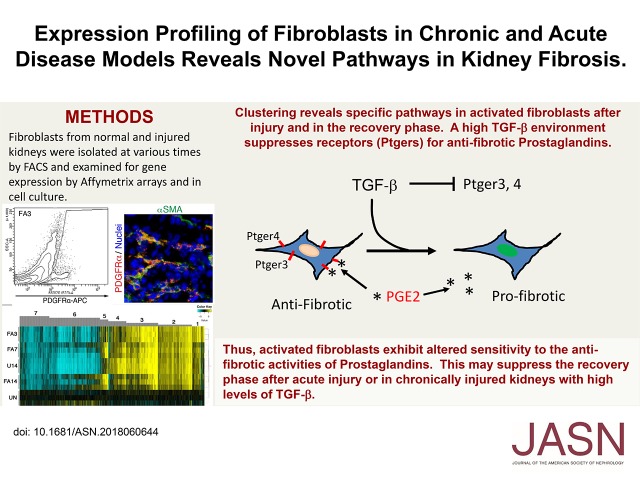
Abstract
Background
Renal interstitial fibrosis results from activation and proliferation of fibroblasts to myofibroblasts, secretion and accumulation of extracellular matrix, and displacement of normal renal tubules. In contrast to chronic renal disease, acute injury may be repaired, a process that includes a decrease in the number of myofibroblasts in the interstitium and degradation of the accumulated extracellular matrix, leaving little evidence of prior injury.
Methods
To investigate whether activated fibroblasts demonstrate changes in gene expression that correspond with regression after acute injury but are not observed in chronic models of fibrosis, we used microarrays to analyze gene expression patterns among fibroblast populations at different stages of injury or repair. We then mined the data for signaling pathways in fibroblasts corresponding to the acute proliferative, regression, and chronic phases of renal injury.
Results
We identified multiple gene clusters with changes that correlate with the three phases of renal injury, including changes in levels of receptors for the antifibrotic factor PGE2. In adult renal fibroblast cultures, PGE2 was able to upregulate many genes that are suppressed by the profibrotic cytokine TGF-β, whereas many PGE2-downregulated genes were activated by TGF-β. High levels of TGF-β suppressed expression of a subset of PG receptors in fibroblast cultures, making these cells resistant to any effects of PGE2.
Conclusions
Inherent gene expression changes in activated fibroblasts accompany the transition from AKI to repair and regeneration. In chronic models, however, activated fibroblasts are resistant to the antifibrotic effects of PGE2 due to suppression of a subset of PGE receptors.
Keywords: chronic kidney disease, acute renal failure, prostaglandins, fibrosis, myofibroblasts
Visual Abstract
Fibrosis is a common pathology due to the activation of interstitial fibroblasts to myofibroblasts, the increased secretion and accumulation of extracellular matrix (ECM), and the loss or perturbation of tissue-specific epithelial, endothelial, and other cell lineages.1,2 In CKD, this expansion and deposition of ECM compromises renal function by displacing proximal and distal tubules, by increasing glomerular mesangium to affect the filtration barrier, and by affecting the hemodynamics of the renal arterioles.3 Myofibroblast induction is also accompanied by innate immune pathways, partly through the expression and secretion of immune-mediating cytokines.4,5 Using definitive genetic cell lineage–tracing methods, the origins of renal interstitial fibroblasts and myofibroblasts have clearly been established in animal models of renal fibrosis.6 The best available data point to pericytes and perivascular fibroblastic cells, in addition to preexisting, resident interstitial fibroblasts, as the major sources of myofibroblasts in renal fibrosis models. Thus, to identify optimal therapeutic targets for CKD, it is essential to understand both the intrinsic genomic programming of pericytes and interstitial, activated fibroblasts as well as their responses to injury and genetic abnormalities.
Expansion of interstitial fibroblasts has been studied in AKI in human biopsy samples7 and in rodent models.8 AKI is the result of renal ischemia or nephrotoxic injury and results in the death of proximal tubule epithelial cells primarily from the S2 and S3 segment of the nephron. However, these segments retain the ability to regenerate and repopulate the sites of injury if the degree of injury is limited.9,10 Coincident with AKI is widespread activation and proliferation of interstitial fibroblasts. As AKI is repaired, the activation of interstitial myofibroblasts subsides and expansion regresses, often leaving little evidence of residual fibrosis. However, repeated bouts of AKI may alter the ability of activated interstitial myofibroblast to regress, leading to irreversible interstitial fibrosis and a chronic renal disease state.11,12 Nevertheless, after an initial bout of AKI the number of activated myofibroblasts decreases as injury is repaired, suggesting that these cells have the ability to reversibly deactivate or undergo apoptosis and removal by mechanisms that remain to be determined.13
In this report, we have begun to address inherent differences in renal interstitial fibroblasts after AKI and in chronic disease models. Using a folic acid (FA) model of AKI and the unilateral ureteral obstruction model (UUO) of renal fibrosis in mice, we isolated interstitial fibroblasts by fluorescence-activated cell sorting (FACS) and determined gene expression changes with Affymetrix microarrays. We compared the transcriptome of fibroblasts from uninjured kidneys and UUO-derived fibroblasts, as well as fibroblasts from different times after AKI. We hypothesized that there may be specific genes and pathways that mediate the regression of fibroblasts after AKI that are not active in chronic models of fibrosis, such as the UUO model. The data show dynamic changes in the patterns of fibroblast gene expression in the acute and recovery phases that include WNT and TGF signaling pathways, fibroblast growth factors, and genes involved in ECM deposition or turnover. Furthermore, we show that suppression of the PG receptor Ptger3 correlates with acutely injured fibroblasts but increases with recovery. Strikingly, TGF-β–cultured kidney fibroblasts suppress expression of multiple PG receptors making cells recalcitrant to PGE2 signaling. Given the proposed antifibrotic effects of PGE2,14–16 these data suggest a novel mechanism for the regression of interstitial fibroblasts that is deregulated in a high–TGF-β environment.
Methods
Animals
Mice were kept according to National Institutes of Health guidelines and all procedures were approved by the University Committee on Use and Care of Animals at the University of Michigan. Mice were housed in a specific pathogen–free facility with a 12-hour light/12-hour dark cycle and given free access to food and water.
For the UUO model, mice were anesthetized by isoflurane inhalation. Through a midline abdominal incision, the left ureter was tied off at the level of the lower pole and the midureteral level with fine suture materials (5–0 silk), then cut between the two ligated points to induce a complete obstruction. Mice were allowed to recover from anesthesia and were supplied food and water ad libitum until they were euthanized at 14 days after the surgery. For the acute tubular necrosis model, mice were injected intraperitoneally with a single dose of 250 mg/kg FA in 0.15 M NaHCO3. Mice were euthanized at 3, 7, and 14 days after injury. Mice were put under constant observation for the first 48 hours after injection.
Cell Sorting
In brief, kidneys were minced and digested with 0.5 mg/ml Liberase DH (#05401054001; Roche) and 100 U/ml DNaseI (11284932001; Roche) in PBS solution supplemented with 10% FBS at 37°C for 30 minutes with triturate by P1000 tip every 5 minutes, followed by centrifugation at 300×g for 5 minutes at 4°C. The resulting pellet was resuspended in ice-cold PBS with 2 mM EDTA and was passed through a 70-μm cell strainer (Falcon), then centrifuged at 300×g for 5 minutes at 4°C. The pellet was incubated with 0.5× red blood cell lysis buffer (0.15 M NH4Cl, 14 mM NaHCO3, and 0.1 mM Na2 EDTA) for 5 minutes on ice, to remove red blood cells, and centrifuged at 300×g for 5 minutes at 4°C. The single isolated kidney cells were stained with APC-conjugated anti-PDGFRα Antibodies (Ab) (#A18382; Life Technologies) and PE-Cyanine7–conjugated anti-CD11b Ab (#25–0112–81; eBioscience) and subjected to cell sorting using FACS Aria II cell sorter (BD Biosciences). FACS Aria II was also used for surface pattern analysis. The antibodies used for surface pattern are anti-PDGFRα, APC-Cyanine7–conjugated anti-CD45 Ab (#103115; Biolegend), PE-Cyanine7–conjugated anti-CD105 Ab (#120409; Biolegend), PE-conjugated anti-CD44 Ab (#103007; Biolegend), PE-conjugated anti-PDGFrβ Ab (#136005; Biolegend), and PE-conjugated anti-CD31 Ab (#102507; Biolegend).
Microarray Expression Analysis
Total RNA was extracted from FACS-sorted cells using TRIzol reagent (Life Technologies) and purified by RNeasy Mini Kit (Qiagen). Microarray expression analysis was performed using three independent mice and carried out by the University of Michigan Comprehensive Cancer Center Affymetrix and Microarray Core Facility using Mouse 430 2.0 Affymetrix Gene Chip 3 expression arrays (Affymetrix), as described.17 Expression values for each gene were calculated using the robust multiarray average method and fitted to weighted linear models in R using the Affymetrix package of Bioconductor.17,18 The Affymetrix datasets have been submitted to the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), accession number GSE121190.
RNA Reverse Transcription and Quantitative Real-Time PCR
Total RNA samples of FACS-sorted cells were reverse transcribed into cDNA using SuperScript III Reverse Transcription (Life Technologies). cDNA templates were amplified with iTaq Universal SYBR Green Supermix (Bio-Rad) on an Applied Biosystems 7500 Real-Time PCR System. The primers (5′ to 3′) used: mPDGFRa: forward, cattgaccctgttccagaggag, reverse, ggtggaactactggaacctgtc; mPtger3 (EP3): forward, gaaccagatcttggatccctgg, reverse, cagggaaacaggtactgcaatg; mFGF9: forward, tcatttagagatcttccccaacg, reverse, gttcatgccgaggtagagtcc; mGja1: forward, gtgccggcttcactttcattaag, reverse, gaaaatgaagagcaccgacagc; mGapdh: forward, ccagaacatcatccctgcatc, reverse, cctgcttcaccaccttcttga. The relative standard method was used to obtain relative expression for each grouping comparison, where the amount of target was normalized to endogenous GAPDH.
Histology and Immunohistochemistry
The kidneys were fixed in Carnoy solution, embedded in paraffin, and sections (6-μm thick) were stained with Hematoxylin and Eosin for routine histologic examination. For immunofluorescent stainings, tissues from adult animals were frozen in tissue-Tek O.C.T. compound (Sakura Finetek) and sectioned using a cryostat. Frozen sections (6 µm) were treated with 100% methanol for 20 minutes at −4°C and subsequently blocked with 5% normal goat serum in PBS/0.05% Tween for 1 hour at room temperature. The following primary antibodies were used: anti-PDGFRa Ab (#14–1401–82; eBioscience), anti-PDGFRb Ab (#14–1402; eBioscience), anti-fibronectin Ab (sc-9068; Santa Cruz), anti-laminin Ab (#L9393; Sigma-Aldrich), anti-collagen I Ab (#ab34710; Abcam), anti-CD31 Ab (#ab28364; abcam), Fluorescein-labeled Lotus Tetragonolobus Lectin (#FL-1321; Vector Labs), Cyanine3-conjugated anti–α-Smooth Muscle Actin Ab (#C6198; Sigma-Aldrich), and anti-Gja1 Ab (sc-9059; Santa Cruz). All images were taken on an Olympus IX71 epifluorescent microscope. Morphometric analysis was carried out using ImageJ software.
Kidney Primary Fibroblast Culture
Tissue culture dishes were coated with 1% gelatin solution before tissue explanting. Sections of kidney cortex were diced and digested in 0.05% trypsin/EDTA (Gibco) for 10 minutes at 37°C. The kidney pieces were placed in gelatin-coated dishes and incubated in DMEM/F12 (Gibco) supplemented with 20% FBS, 1× ITS-X (Gibco), 1× Glutamax (Gibco), 1× sodium pyruvate (Gibco), and 5 µM Y-27632 (Cayman Chemical) for 3 days. After 3 days, kidney pieces were gently removed and medium was changed. Culture medium was renewed twice weekly until the monolayer had covered approximately 75% of the dish surface. For passage, cells were dissociated with 0.05% trypsin/EDTA. Y-27632 was not added to the medium after passage 5. Cells could be continuously cultured for >36 passages.
For expression analyses, cells were cultured in six-well plates to confluency then shifted to a low-serum (0.05%) media for 24 hours before addition of 10 ng/ml TGF-β or 1 μM PGE2 or both factors. RNA was harvested with TRIzol after 48 hours in culture and analyzed by Affymetrix microarray gene chips as above.
Results
Expansion of PDGFrα-Positive Cells in the Interstitium of Acute and Chronically Injured Kidneys
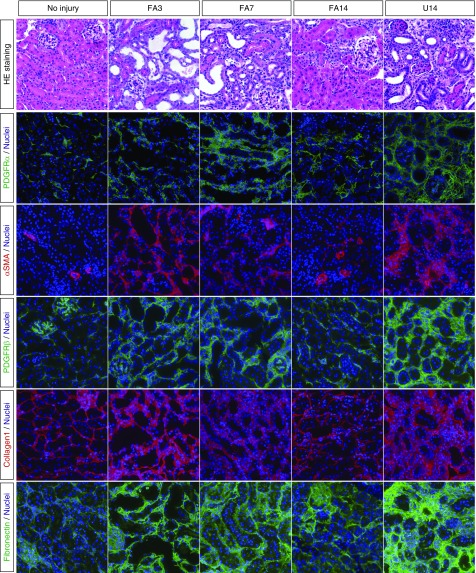
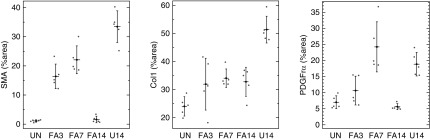
In order to examine renal interstitial fibroblasts from both acutely injured and chronically injured kidneys, we used the established FA nephrotoxicity model and the UUO model of fibrosis. To induce AKI, mice were injected with 250 mg/kg FA and kidneys were isolated at 3 days (FA3), 7 days (FA7), and 14 days (FA14) postinjection. The chronic injury was examined in UUO kidneys 14 days after ligation of a single ureter (U14). We initially confirmed kidney injury and expansion of fibroblasts and myofibroblasts by histology and immunostaining for PDGFrα, PDGFrβ, smooth muscle actin (αSMA), collagen1, and fibronectin (Figure 1). Representative images show dilated renal tubules and significant expansion of fibroblasts and ECM in FA-treated kidneys (FA3 and FA7). However, by FA14, the acute injury has resolved because the injured renal tubules have regenerated and the interstitial expansion of fibroblasts and myofibroblasts has regressed. Both PDGFrα-positive cells and αSMA-positive cells are greatly reduced in number at FA14, compared with FA3 or FA7. Staining for the ECM proteins collagen1 and fibronectin is also reduced by FA14, although it remains stronger than in uninjured controls. In the U14 kidney sections, staining for αSMA, collagen1, and PDGFrα was more widespread than even in the FA3 or FA7 kidneys. The immunostained micrographs were subject to quantitative morphometry, whereby the stained area was calculated on multiple independent images from different animals (Figure 2). These data confirm that there is a rapid expansion of interstitial fibroblasts and myofibroblasts in the acute injury phase at 3 and 7 days post–FA injection that resolves by 14 days upon recovery from injury. However, the chronic UUO model continues to exhibit interstitial expansion of PDGFrα- and αSMA-positive cells with no evidence for regression.
Figure 1.
Interstitial fibroblasts expand and regress after acute kidney injury (FA3–FA14) but not in the chronic obstruction model (U14). Representative sections of kidneys taken at the indicated times post-FA or -UUO or from uninjured controls are shown. Sections were stained with hematoxylin/eosin (HE) or with the indicated antibodies in color. Note the increase in PDGFrα/β- and αSMA-positive cells, and the matrix proteins Col1 and fibronectin at FA3, FA7, and U14. Note the decreases in matrix and fibroblasts by FA14.
Figure 2.
Quantitative morphometry of interstitial area shows regression after AKI. Multiple random images were analyzed by calculating the total positive area of micrographs stained with the indicated antibodies. Scatter plots show data points with the mean and 1 SD indicated. Note the significant decrease in interstitial fibroblast area by FA14 treatment, whereas U14 remains high. SMA, Smooth Muscle Actin; UN, Uninjured.
Enrichment of PDGFrα-Positive Cells by FACS
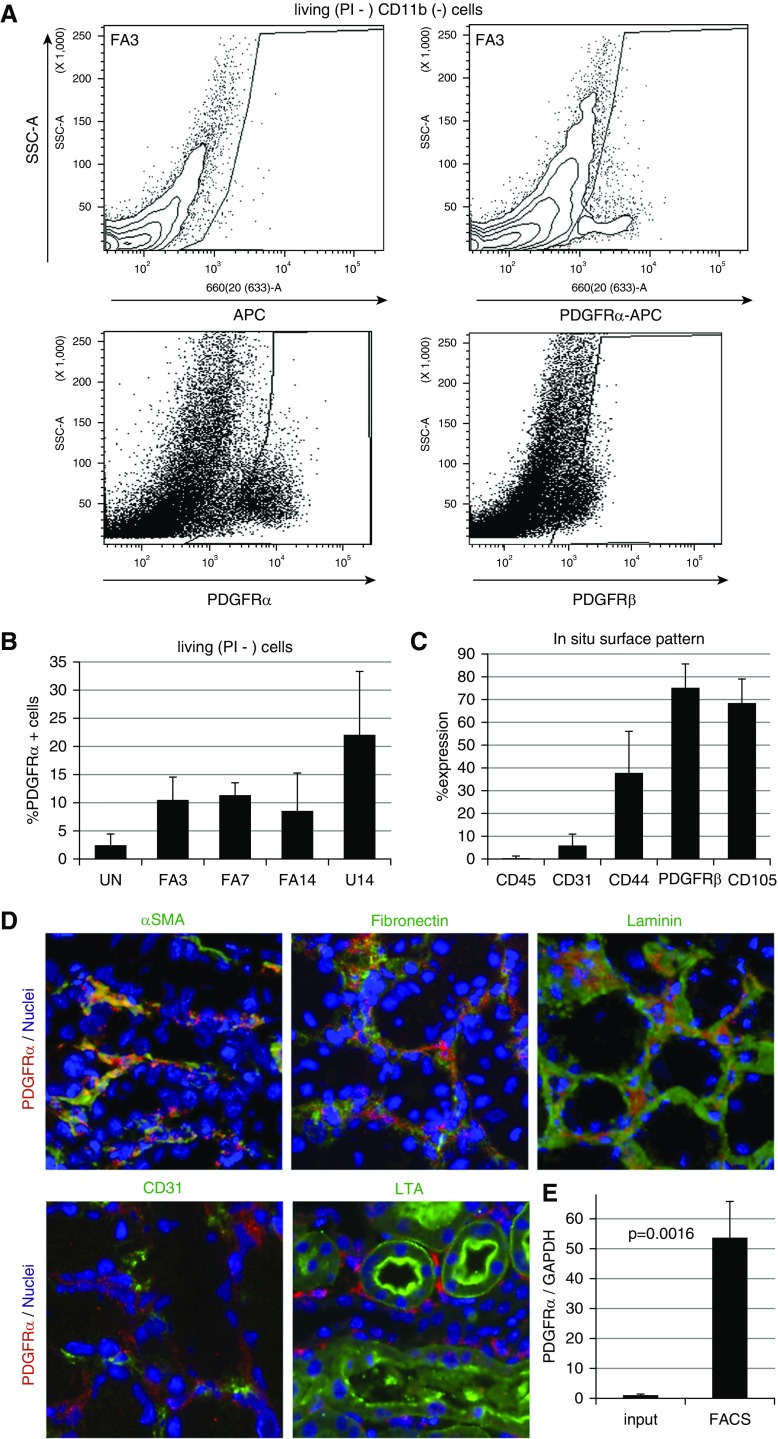
In order to determine what inherent differences among the fibroblast populations could account for the regression of cells after acute injury, we isolated populations of PDGFrα-positive cells from the kidney cortex of both acute and chronic models using FACS (Figure 3A). Uninjured control kidneys were particularly challenging because very few PDGFrα cells are observed in tissue sections. However, sufficient numbers of cells could be isolated by FACS by pooling multiple uninjured adult kidney preparations. We also tried PDGFrβ antibodies for separating cells, but the affinity was lower and gating could not separate the positive cells as cleanly as anti-PDGFrα. The PDGFrα-positive cells separated by FACS represent a population ranging from approximately 2% in uninjured to >20% in U14 kidneys (Figure 3B). The PDGFrα-sorted population was also costained for endothelial cell markers (CD31), hematopoietic cells markers (CD45), and pericyte/mesenchymal stem cell markers (CD44, PDGFrβ, and CD105) to ensure a specific population (Figure 3C). These data indicate that our PDGFrα+ population contains few endothelial and hematopoietic cells but does overlap with pericytes. Immunostaining of kidney sections, with the same antibody as used for FACS, also demonstrates that the vast majority of PDGFrα+ cells were localized within the interstitium and associated with ECM, as marked by αSMA, fibronectin, and laminin (Figure 3D). However, PDGFrα+ cells were distinct from CD31+ endothelial cells or LTA+ proximal tubule epithelium (Figure 3D). Total RNA was isolated from sorted cells and assayed for expression of the PDGFrα mRNA and compared with whole-kidney RNA to assess the degree of enrichment for PDGFrα fibroblasts (Figure 3E). These data show a 50–100-fold increase for PDGFrα expression, again indicating that a significantly enriched population was obtained.
Figure 3.
PDGFrα-positive cells can be sorted with purity and specificity from adult kidneys. (A) Representative flow cytometry plot for sorting PDGFRα-positive cells from whole adult kidney 3 days post-FA, using either a control (left panel) or anti-PDGFRα mAb (APA5, right panel). Cells in gated area were selected. (B) The percentage of PDGFRα-positive cells from whole kidneys sorted from each disease model and uninjured control kidneys (no injury, n=7; FA3, n=9, FA7, n=5; FA14, n=5; U14, n=4). (C) Flow cytometry analysis shows that PDGFRα-sorted cells include pericyte/MSC marker–positive cells (CD44, PDGFRβ, and CD105), but not endothelial (CD31) and hematopoietic (CD45) cell populations (n=3). (D) Immunostaining demonstrates that anti-PDGFRα mAb (APA5) recognizes cells specifically confined to the tubulointerstitial compartment. In FA3 kidney, PDGFRα-positive (red) cells stain positive for αSMA, overlap with ECM (fibronectin and laminin), but do not costain with endothelial marker (CD31) and proximal tubular marker (LTL-lectin). (E) Quantitative RT-PCR analyses of PDGFRα mRNA from whole FA3-treated kidneys and PDGFRα-sorted cells show approximately 50-fold enrichment (n=3). SSC-A, Side scatter; UN, Uninjured.
Gene Signatures of PDGFrα-Positive Cells from Acute and Chronically Injured Kidneys
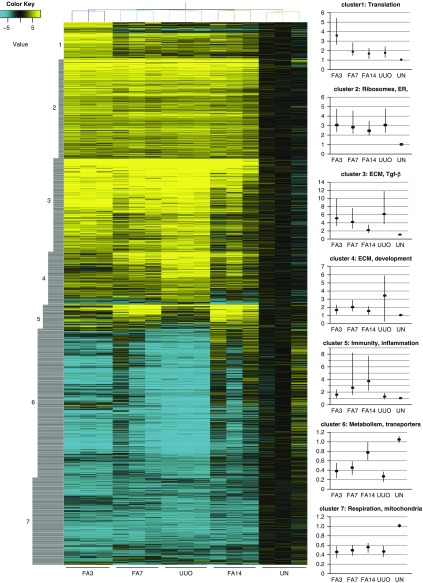
Total RNAs obtained from PDGFrα-positive cells from FA3, FA7, FA14, and U14 kidneys and from uninjured kidneys were used to assess gene expression differences between any and all of the five samples to determine what unique signatures might be expressed in the different populations. For each time point, samples were done in triplicate. The entire Affymetrix expression dataset is available as an Excel file (Supplemental Table 1) or can be accessed at https://www.ncbi.nlm.nih.gov/geo/ (accession number GSE121190). A total of 2460 probe sets, representing 1885 unique transcription units, showed statistically significant differences in expression in at least one of the samples. The differential gene expression dataset is available as a single Excel file (Supplemental Table 2). These genes were clustered to group genes depending on their level of expression under the different conditions (Figure 4). Gene expression changes showed seven distinct clusters as follows: cluster 1 genes were highest at FA3 and lowest in uninjured, cluster 2 genes were high in all injured kidneys, cluster 3 genes were high at FA3 and lower at FA7 and FA14 and high in the U14 kidneys, cluster 4 genes were highest in U14, cluster 5 genes were increased at FA7 and FA14 but low at FA3 and in U14 kidneys, cluster 6 genes were highest at FA14 and in uninjured kidneys, and cluster 7 genes were low in all injured kidneys. We utilized both ToppGene19 and Genomatix (www.genomatix.de) for gene ontology and pathway analyses to determine specific themes for each cluster (Table 1). Clusters 1 and 2 are enriched for genes involved in protein synthesis or membrane targeting. Clusters 3 and 4 are enriched in genes for matrix synthesis, deposition, and signaling. Cluster 5 highlights the inflammatory response, whereas clusters 6 and 7 are enriched in genes for metabolisms, cellular respiration, and transporters.
Figure 4.
Gene expression changes in isolated fibroblasts after renal injury exhibit distinct clustering. The normalized relative expression values for 2640 Affymetrix probe sets that show statistically significant changes in at least one of the renal disease models were clustered. The graphs on the left show mean expression values for individual clusters (dots) with the 25th and 75th percentiles represented by the vertical lines. The complete dataset with gene names is available as a supplemental Excel file (Supplemental Table 2). ER, Endoplasmic reticulum; UN, Uninjured.
Table 1.
Pathway analyses of clustered gene sets
| Cluster | Pathways | P Value | # Genes | Sourcea |
|---|---|---|---|---|
| 1 | Translation, ribosomal proteins | |||
| RNA binding | 9.6E−6 | 29 of 1632 | T | |
| Translation initiation | 2.4E−8 | 12 of 194 | T | |
| rRNA processing | 5.9E−7 | 12 of 260 | T | |
| Translation initiation | 8.1E−9 | 12 of 191 | G | |
| rRNA processing | 1.6E−6 | 11 of 257 | G | |
| Protein metabolism | 1.1E−4 | 32 of 2061 | G | |
| 2 | Ribosomes, translation, ER targeting | |||
| Structural constituent of ribosome | 6.4E−36 | 46 of 216 | T | |
| Translation | 3.5E−50 | 54 of 165 | T | |
| Translation termination | 4.8E−50 | 45 of 97 | T | |
| Translation elongation | 8.6E−50 | 45 of 98 | T | |
| Translation | 1.7E−40 | 35 of 91 | G | |
| ER targeting, membrane targeting | 1.6E−39 | 35 of 96 | G | |
| Translation initiation | 5.4E−33 | 39 of 191 | G | |
| Ribosome | 1.8E−36 | 35 of 119 | G | |
| 3 | ECM, collagens, integrins | |||
| ECM organization | 6.5E−38 | 56 of 354 | T | |
| TGF-β | 6.7E−10 | 44 of 824 | G | |
| MMPs | 2.1E−6 | 21 of 331 | G | |
| Integrin signaling | 6.6E−13 | 15 of 70 | G | |
| ECM organization | 5.5E−39 | 53 of 331 | G | |
| Smad2 | 2.9E−12 | 31 of 436 | G | |
| 4 | ECM, development | |||
| Regulation of development | 1.7E−14 | 60 of 1893 | T | |
| ECM | 2.0E−18 | 33 of 444 | T | |
| ECM | 3.4E−15 | 29 of 530 | G | |
| Smad signaling | 1.3E−7 | 20 of 491 | G | |
| TGF-β | 8.2E−7 | 25 of 824 | G | |
| 5 | Immunity, inflammatory response | |||
| Antigen processing and presentation | 2.1E−11 | 10 of 94 | T | |
| Defense response | 3.8E−11 | 30 of 1651 | T | |
| Immune, inflammatory response | 1.9E−22 | 34 of 549 | T | |
| Innate immune system | 8.8E−5 | 18 of 1459 | G | |
| Immune response | 7.1E−9 | 25 of 2042 | G | |
| 6 | Metabolism, transporters, kidney disease | |||
| Transmembrane transporters | 1.4E−19 | 81 of 1014 | T | |
| Organic acid metabolic process | 3.0E−30 | 104 of 1131 | T | |
| Acidosis | 2.5E−12 | 50 of 475 | T | |
| Metabolic pathways | 1.1E−23 | 109 of 1272 | T | |
| Kidney disease | 1.3E−15 | 53 of 569 | T | |
| Metabolism | 6.6E−20 | 126 of 2100 | G | |
| TCA cycle | 1.1E−13 | 27 of 169 | G | |
| Transmembrane transporters | 2.6E−21 | 68 of 1034 | G | |
| Renal tissue | 2.7E−49 | 122 of 1247 | G | |
| Metabolic disease | 3.6E−14 | 71 of 903 | G | |
| 7 | Respiration, mitochondria | |||
| Cellular respiration | 1.1E−7 | 16 of 178 | T | |
| Response to hypoxia | 2.5E−7 | 14 of 143 | T | |
| TCA cycle | 6.4E−6 | 14 of 171 | T | |
| Cellular respiration | 3.2E−7 | 14 of 168 | G |
ER, Endoplasmic reticulum; MMPs, Matrix metalloproteinase; rRNA, Ribosomal RNA; TCA, tricarboxylic acid.
T, ToppGene; G, Genomatix.
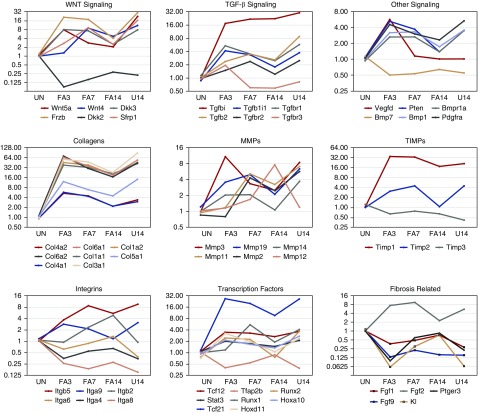
We examined candidate genes associated with signaling pathways, ECM secretion, or transcription, or previously implicated in fibrosis (Figure 5). Receptors for the profibrotic cytokine TGF-β were up slightly at FA3 and U14, but lower at FA14. Most strikingly, the TGF-β–induced gene was up 15-fold at FA3 and continued to increase despite a reduction of PDGFrα-positive cells by FA14. For WNT ligands, Wnt5a increased seven-fold by FA3 and declined by FA14, whereas Wnt4 peaked at FA7 in the acute model. WNT inhibitors Dkk3 and Frzb also peaked at FA3 in the acute model with even higher levels in the chronic model at U14, whereas the inhibitor Dkk2 was down 3–4-fold at FA3. These data suggest that Wnt4 may be more specific to the resolution phase after AKI and that dynamic changes in WNT inhibitors could modulate signaling. Other signaling effectors that increased during the acute phase include Bmp1 and Vegfd, whereas Bmp7 was reduced at FA3 and FA7. The tumor suppressor Pten, a phosphatidylinositol phosphatase, also increases four-fold at FA3 and declines by FA14, but remains high in the U14 kidneys.
Figure 5.
Expression profiles of selected genes and pathways exhibit dynamic changes. Normalized RNA expression levels from PDGFrα-selected fibroblasts isolated from uninjured (UN) kidneys; or AKI kidneys at 3, 7, or 14 days post-FA; or kidneys 4 days post-UUO. RNAs were assayed with Affymetrix microarrays in triplicate from three independent sorted samples, with data points representing the mean. Expression levels are relative to UN and graphed on a log2 scale. Signaling pathways include TGF-β–related, WNT-related, and other ligands or receptors. ECM genes include the collagen family, the matrix metalloproteinase (MMPs), and the tissue inhibitors of MMPs (TIMPs).
As expected, almost all collagen chains showed differential expression, which increased at FA3 and declined slowly thereafter, but remained high in the U14 samples. The matrix metalloproteases that are involved in matrix turnover showed different patterns, with Mmp3 increasing rapidly at FA3 and then declining, but with Mmp12 showing maximum levels at FA14. Again, this suggests that Mmp12 may be involved primarily in the degradation of matrix that accumulates as a result of injury. Integrins also show dynamic changes, with Itgb5 and Itga9 strongly induced at FA3, FA7, and U14, whereas Itgb2 peaks at FA14 and is low at U14. Again, these data suggest that Itgb2 may be associated with the regression of fibroblasts. Of transcription factors, the Tcf21 gene is the most highly upregulated in AKI and declines in the resolution phase at FA14. Similarly, kidney-specific Hox genes (HoxD11 and HoxA10) are also upregulated 2–3-fold in the acute phase and decline back to uninjured levels by FA14.
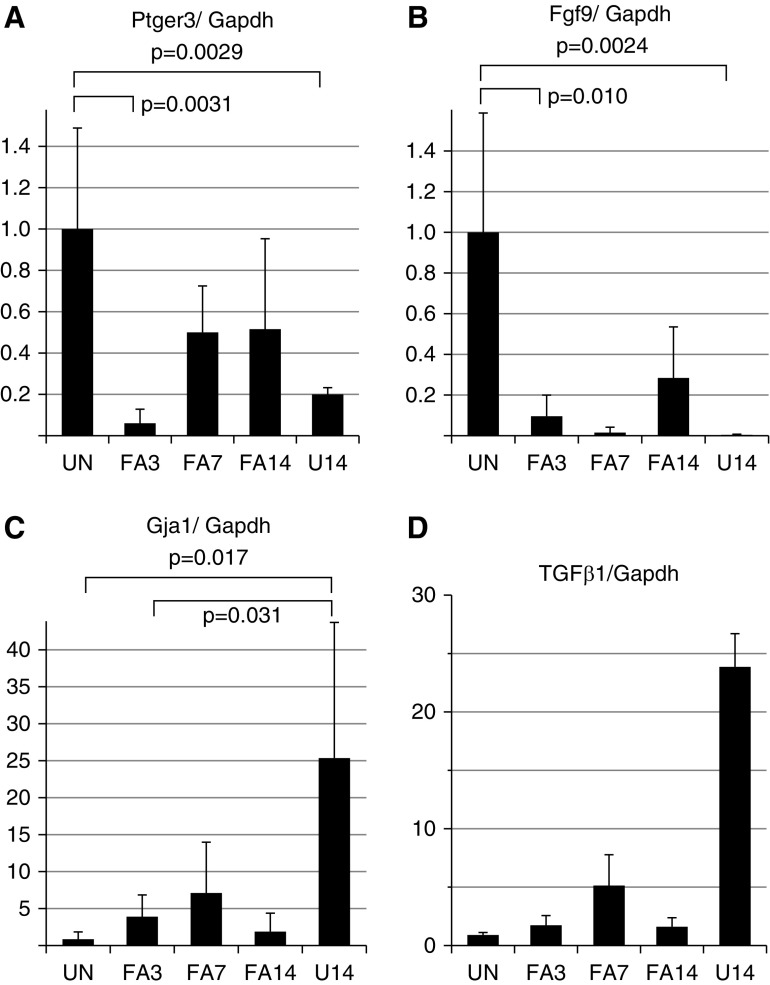
Of genes associated with fibrosis, the Fgf2 but not Fgf1 expression increased strongly at FA3, whereas Fgf1 and Fgf9 declined in the AKI model, suggesting that Fgf2 is a primary driver of fibroblast proliferation. Among the objectives of the screen were to examine the expression levels of potential antifibrotic genes during the recovery phase after acute injury. PGs are lipid compounds with hormone-like activity, of which PGE2 has been implicated in tissue repair and ECM degradation.15,16 Thus, PGE2 has been implicated as an antifibrotic factor.5,14 We noted that one of its receptors, Ptger3, was down more than three-fold at FA3 and began to increase by FA7 and FA14, whereas the U14 samples remained low. The Klotho gene has also been implicated in renal disease20,21 and its expression was also strongly reduced at FA3 and U14 but approached normal levels by FA14. We confirmed by quantitative RT-PCR that expression levels of the PGE2 receptor Ptger3 were low in acutely injured fibroblasts at FA3, but began to increase at FA7 and FA14, while remaining low in the UUO model (Figure 6). Similarly, we independently confirmed the reduction of Fgf9 and the marked increase in Gja1, a gap junction protein highly expressed in U14 kidneys (Figure 6, B and C). These data suggest that the antifibrotic activities of PGE2 may be limited in the UUO model due to a specific decrease in Ptger3 receptor expression. We also examined the levels of the profibrotic factor TGF-β1 in whole-tissue RNA from kidneys isolated before and after injury, which increased at FA3 and FA7, decreased by FA14, but was >20-fold higher in U14 compared with uninjured (Figure 6D).
Figure 6.
Quantitative RT-PCR for selected genes confirms expression changes. (A–C) Total RNA samples from PDGFRα-sorted cells were analyzed by quantitative RT-PCR for the indicated genes to confirm Affymetrix array expression differences. Note decline of Ptger3 in the early acute phase (FA3) and its increase during the recovery phase (FA7, FA14). The gap junction protein Gja1, expressed in fibroblasts, shows an expression pattern opposite that of Ptger or Fgf9, with maximum levels in the chronic UUO model (U14). (D) Total RNA was isolated from whole kidneys at the indicated times postinjury and assayed for TGF-β1 by quantitative RT-PCR. The y axis is relative RNA levels in arbitrary units.
Effects of TGF-β and PGE2 on Cultured Kidney Fibroblasts
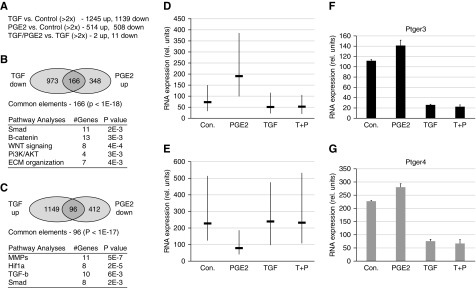
To examine and compare the effects of profibrotic and potential antifibrotic agents, we cultured immortalized adult kidney fibroblasts with PGE2, TGF-β, or both compounds for 48 hours and measured gene expression profiles with Affymetrix microarrays (Supplemental Tables 3–5). PGE2 treatment resulted in >1000 genes with statistically significant altered expression values; approximately 514 genes were upregulated and 508 genes were downregulated two-fold or more (Figure 7). TGF-β also greatly altered gene expression patterns in adult fibroblast cultures with >2500 genes affected (Figure 7). Analyses of the TGF-β–regulated gene set show great similarity (P<3.8E−43) to the response seen in mouse embryo fibroblasts (MEFs) in Gene Set Enrichment Analysis M2445 (PLASRI_TGFB_TARGETS_10HR_UP), as reported by Plasari et al.20 (Table 2). Significant overlap is also seen with genes downregulated by TGF-β in kidney fibroblasts and MEFs. Conversely, the gene set upregulated by PGE2 overlaps with genes suppressed by TGF-β in MEFs, whereas the gene set downregulated by PGE2 is most similar to genes upregulated by TGF-β. Within our datasets, of the 1139 genes downregulated with TGF-β, approximately 166 were upregulated by PGE2, a highly significant number of overlapping genes some of which are associated with TGF-β signaling (Smad), Wnt signaling (β-catenin), and extracellular matrix remodeling. Similarly, 96 genes that are upregulated by TGF-β were also downregulated by PGE2 and correlated with matrix metalloproteinase, Hif1a, as well as TGF-β pathways. The list of overlapping genes can be found in Supplemental Table 6.
Figure 7.
TGF-β suppresses the effects of PGE2 on adult renal fibroblast gene expression. (A) The number of genes up- or downregulated by TGF-β or PGE2 is compared with untreated cells after 48 hours in culture. However, TGF-β treatment alone abrogates almost all of the effects of PGE2 treatment. The complete datasets are in Supplemental Tables 3–5. (B) Pathway analyses of the 166 genes that are downregulated by TGF-β and upregulated by PGE2. (C) Pathway analyses of 96 genes that are upregulated by TGF-β and downregulated by PGE2 in adult kidney fibroblasts. (D) Median expression values of genes upregulated two-fold by PGE2 (−) with vertical lines representing the 75th and 25th percentiles. Note that TGF-β treatment suppresses any increase by PGE2 on average gene expression values (T+P). (E) Median expression values of genes downregulated two-fold by PGE2 (−) with vertical lines representing the 75th and 25th percentiles. Note that TGF-β treatment suppresses any decrease by PGE2 (T+P) on average gene expression values. (F) Relative RNA expression levels of the PGE2 receptor Ptger3 in adult kidney fibroblasts after treatment with PGE2, TGF-β, or both factors (T+P) compared with controls. (G) Similar analyses as in (F) but for the PGE2 receptor Ptger4.
Table 2.
Pathway analyses of gene expression in response to TGFβ or PGE2
| Gene Cluster | Pathways | P Value | # Genes | Sourcea |
|---|---|---|---|---|
| TGF-β: Genes upregulated | ||||
| Genes upregulated in MEFs by TGFβ | 3.8E−43 | 71 of 199 | T | |
| Hif1α transcription | 2.1E−14 | 25 of 65 | T | |
| Glucose metabolism | 2.9E−10 | 23 of 81 | T | |
| Hif1α | 1.6E−11 | 21 of 67 | G | |
| Carbohydrate metabolism | 1.9E−8 | 39 of 276 | G | |
| TGF-β: Genes downregulated | ||||
| Genes downregulated in MEFs by TGFβ | 1.4E−43 | 77 of 244 | T | |
| Cell adhesion | 2.1E−21 | 176 of 1530 | T | |
| Cell migration | 2,6E−20 | 155 of 1300 | T | |
| ECM and associated proteins | 7.2E−14 | 119 of 1028 | T | |
| ECM organization | 5.7E−7 | 37 of 298 | G | |
| Cell invasion | 6.0E−9 | 76 of 845 | G | |
| PGE2: Genes upregulated | ||||
| Genes downregulated in MEFs by TGFβ | 4.3E−37 | 51 of 244 | T | |
| Proteinaceous ECM | 1.2E−7 | 28 of 379 | T | |
| Smad-related signaling | 2.1E−4 | 25 of 491 | G | |
| ECM components | 3.4E−6 | 12 of 120 | G | |
| PGE2: Genes downregulated | ||||
| Genes upregulated in MEFs by TGFβ | 1.8E−69 | 71 of 199 | T | |
| Regulation of cell death | 4.6E−18 | 100 of 1650 | T | |
| Smad signaling | 2.9E−6 | 31 of 491 | G | |
| Matrix metalloproteases | 1.3E−5 | 23 of 331 | G | |
| Fibrosis | 6.4E−11 | 25 of 268 | G |
T, ToppGene; G, Genomatix.
Given that PGE2 and TGF-β may have opposite effects on gene expression in fibroblasts and the progression of fibrosis, we also examined whether PGE2 could counter the effects of TGF-β by comparing cells cultured with TGF-β to those cultured with TGF-β and PGE2 together. Surprisingly, there were few changes in gene expression between these two sets after 48 hours in culture (Figure 7). For both genes that were upregulated by PGE2 (Figure 7D) and genes downregulated by PGE2 (Figure 7E), TGFβ inhibited the effects of added PGE2, suggesting that PGE2 had little effect on gene expression when present in a high–TGF-β environment, as seen in the U14 kidneys. Given that TGF-β appeared to suppress almost all of the effects of PGE2, we examined the level of PG receptor expression in fibroblasts before and after treatment with factors (Figure 7, D and E). Strikingly, TGF-β suppressed the expression of PG receptors Ptger3 and Ptger4, although there was no effect on Ptger1 or Ptger2. These data suggest that TGF-β suppression of a subset of PG receptors and presumably subsequent downstream signaling contribute to drive the profibrotic phenotype of activated fibroblasts in chronically injured kidneys and inhibit the antifibrotic effects of PG.
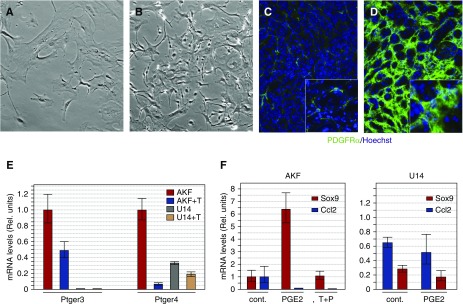
To test whether fibroblasts isolated from healthy or diseased kidneys showed differences in their morphology and ability to respond to PGE2, we examined primary fibroblasts in culture (Figure 8). Interestingly, cells isolated from U14 kidneys showed different morphology in culture, with less cytoplasm, more lamellipodia, and more compact, dense monolayers (Figure 8, A and B), suggesting that more cells can be compacted into the interstitial space. This increased density is also observed in vivo when kidney sections were stained with anti-PDGFrα (Figure 8, C and D). We next examined adult kidney fibroblasts (AKF) from the U14 kidneys and from uninjured kidneys to determine whether the ability to respond to PGE2 was a stable trait in cultured cells. When treated with TGF-β, AKF cells suppressed both Ptger3 and Ptger4, whereas the U14 fibroblasts had virtually no Ptger3 expression and significantly less Ptger4, compared with AKFs (Figure 8E). We then asked whether the U14 fibroblasts could respond to PGE2 by assaying for two responsive genes identified in the AKFs (see Supplemental Table 4). Sox9 is significantly upregulated, whereas Ccl2 is suppressed by PGE2 in AKFs (Figure 8F). In the U14 fibroblasts, baseline expression of Sox9 is approximately 60% of AKF levels and there is no response to PGE2. Ccl2 expression is downregulated by PGE2 in AKFs and is also suppressed by TGF-β alone, whereas in U14 fibroblasts, PGE2 again has no effect on Ccl2 expression. These data suggest that the inability to respond to PGE2 is a direct result of low levels of Ptger3 and/or Ptger4 and that this is an innate property of fibrotic PDGFrα+ cells.
Figure 8.
Activated fibroblasts from fibrotic kidneys exhibit alterations in PGE2 responses. Fibroblast from control adult kidneys (AKF) or from fibrotic kidneys were isolated by FACS and cultured as primary cell lines. Morphologies of PDGRa+ fibroblasts in primary cultures from (A) control AKFs or (B) U14 fibroblasts show higher densities of cells with less cytoplasm and more lamellipodia in U14 cell cultures. (C) Densities of PDGFRa+ cells from (C) control and (D) FA14 kidney sections are shown. Insets in (C and D) are higher magnifications. (E) Quantitative RT-PCR for PG receptors Ptger3 and Ptger4 in AKFs and U14-isolated fibroblasts cultured with or without TGF-β (T). (F) Sensitivity to PGE2 (1 μM) in control AKFs and U14 fibroblasts as measured by two PGE2-responsive genes, Sox9 and Ccl2. Cont., control media; Rel., relative; T+P, TGF-β and PGE2.
Discussion
Wound healing is a natural response after injury and inflammation such that damaged cells and tissues can be repaired. Essential for repair is the activation of fibroblasts to myofibroblasts, driven in part by profibrotic cytokines such as TGF-β. This results in increased secretion of ECM proteins, which promote rigidity and form a scaffold for tissue repair.5 In the kidney, much of the current literature addressing the tissue repair and regeneration after AKI has focused on the renal epithelium. However, dramatic changes in the numbers of interstitial activated fibroblasts, myofibroblasts, and immune cells are observed after AKI, even at sites far removed from localized injury. Yet, by 2–4 weeks post-AKI in mouse models, this expansion of interstitial cells is no longer observed because the numbers of αSMA- and PDGFrα-positive fibroblasts have regressed nearly to uninjured levels. We proposed that there are inherent gene expression differences in fibroblasts that correlate with active proliferation during the acute injury phase when compared with fibroblasts that are regressing during the repair process. In this report, we compared populations of interstitial fibroblasts from uninjured kidneys with fibroblasts isolated from acutely injured and chronically injured kidneys. Given that expression values were derived from relatively pure populations of PDFGrα-positive cells and were normalized, these differences in gene expression do not reflect differences in cell number; rather, they must represent inherent differences in gene expression levels per cell.
These data allow clustering of gene sets that correlate with critical steps in the progression or resolution of fibrosis. Among these are genes in cluster 3, which are high in the early acute phase of AKI, but decline to near normal levels after 14 days, and yet remain high in the UUO model of fibrosis. Many cluster 3 genes are associated with ECM organization and synthesis or signaling from the ECM to cells through integrins. Thus, reduced ECM expression and integrin signaling within fibroblasts correlate with the resolution phase of the fibrotic response after AKI. Similarly, genes associated with the inflammatory response in cluster 5 are upregulated in the resolution phase, suggesting that PDGFrα-positive fibroblasts can regulate the immune response that may be necessary for clearing activated fibroblasts and help to degrade the associated ECM.
Among critical signaling pathways, dynamic changes in WNT components can be seen in the acute injury phase and during recovery. WNT5a and Wnt4 are increased at 3 and 7 days post-AKI and decline by 14 days, whereas the WNT inhibitor Dkk2 is down more than five-fold 3 days post-AKI. Consistent with our data, genetic fibroblast-specific ablation of β-catenin, the canonical WNT signaling effector, results in less injury and inflammation after renal ischemia, suggesting that increased WNT signaling in activated fibroblasts inhibits tubular repair and promotes immune cell infiltration.23 Many of these pathways have previously been identified in animal models of renal injury and fibrosis using other methods, such as sophisticated genetic labeling of different cell populations,24,25 demonstrating that FACS for PDGFrα+ cells is a viable alternative to genetic lineage markers for fibroblasts.
A novel finding among fibroblasts isolated from the acute phase 3 days after AKI was the reduction of PG receptor Ptger3. During the recovery phase, Ptger3 levels increased but remained low in the chronic UUO model at 14 days. In lung fibrosis models, PGE2 is thought to promote repair in part by inhibition of fibroblast proliferation,26 migration,27 and/or differentiation into myofibroblasts.28 The suppression of fibroblast activation by PGE2 is thought to occur primarily through the cAMP and protein kinase A pathways.26,28–30 In samples isolated from patients with idiopathic pulmonary fibrosis, low levels of PGE2 correlate with enhanced fibrosis.31,32 However, in our adult fibroblast cultures, these antifibrotic effects of PGE2 are diminished, likely because the receptors Ptger3 and Ptger4 are significantly downregulated by TGF-β. In the presence of TGF-β, PGE2 has little effect on the transcriptional program of adult kidney fibroblasts in cell culture. In the absence of TGF-b, PGE2 activates many genes that are suppressed by TGF-β alone, whereas many genes downregulated by PGE2 are activated by TGF-β. These reciprocal effects on potential target genes could explain in part the antifibrotic effects of PGE2. However, those effects are likely diminished, because a high concentration of TGF-β suppresses expression of PGE2 receptors in fibroblasts. In cultured fibroblasts isolated from UUO kidneys after 14 days, the expression of Ptger3 and 4 remains low, which strongly attenuates any response to PGE2. This would suggest that there may be stable silencing mechanisms in activated fibroblasts derived from high–TGF-β environments.
In summary, our data point to inherent differences in gene expression patterns among PDGFrα-positive fibroblasts isolated from different phases of renal injury. These changes result in activation of profibrotic pathways and suppression of antifibrotic pathways early after injury. Recovery from the acute phase correlates with regression of activated fibroblasts and a shift in gene expression toward a resolution phase that includes proteins involved in ECM remodeling and promotion of an inflammatory response, processes not observed in the chronic fibrosis model. With respect to potential antifibrotic factors such as PG, our data also demonstrate that it is important to consider the context and the level of receptor expression before pursuing any therapeutic strategies on the basis of secreted factors or hormone-like small molecules. The data also point to the potential for combining anti–TGF-b strategies with PG and/or their downstream targets.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank the University of Michigan Flow Cytometry Core Facility for help with FACS analyses, J. Woolforth for help with the unilateral ureteral obstruction models, and A. Soofi for helpful advice and mouse colony management.
This work was supported in part by National Institutes of Health grants DK073722 and DK054740 to G.R.D. and by the Young Scientist Fellowship of the Japan Society for the Promotion of Science to A.Y.H.
A.Y.H. designed experiments and did the FACS analyses, RNA isolation, immunostaining, and quantitative RT-PCR; B.J.A. did the bioinformatics analyses, gene clustering, and gene ontology; G.R.D. designed experiments, did the fibroblast cell culture and quantitative RT-PCR, and wrote the text.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2018060644/-/DCSupplemental.
Supplemental Materials
Supplemental Table 1. AKF FACS raw data from the Affymetrix Arrays.
Supplemental Table 2. Clustering of gene expression changes in AKFs FACS sorted from kidneys.
Supplemental Table 3. AKF cell culture, Affymetrix raw data with and without PGE2 or Tgf-β.
Supplemental Table 4. AKF controls versus PGE2, expression changes.
Supplemental Table 5. AKF controls versus Tgf-β, expression changes.
Supplemental Table 6. Overlap between Tgf-β and PGE2 gene expression changes.
References
- 1.Kaissling B, Le Hir M: The renal cortical interstitium: Morphological and functional aspects. Histochem Cell Biol 130: 247–262, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leaf IA, Duffield JS: What can target kidney fibrosis? Nephrol Dial Transplant 32[Suppl 1]: i89–i97, 2017 [DOI] [PubMed] [Google Scholar]
- 3.Rockey DC, Bell PD, Hill JA: Fibrosis--a common pathway to organ injury and failure. N Engl J Med 372: 1138–1149, 2015 [DOI] [PubMed] [Google Scholar]
- 4.Leaf IA, Nakagawa S, Johnson BG, Cha JJ, Mittelsteadt K, Guckian KM, et al.: Pericyte MyD88 and IRAK4 control inflammatory and fibrotic responses to tissue injury. J Clin Invest 127: 321–334, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerarduzzi C, Di Battista JA: Myofibroblast repair mechanisms post-inflammatory response: A fibrotic perspective. Inflamm Res 66: 451–465, 2017 [DOI] [PubMed] [Google Scholar]
- 6.Humphreys BD, Lin SL, Kobayashi A, Hudson TE, Nowlin BT, Bonventre JV, et al.: Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol 176: 85–97, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moyses Neto M, Costa RS, Volpini RA, Garcia TM, Rodrigues FF, Coimbra TM: Interstitial alterations in renal cortex in acute tubular necrosis (ATN) post-renal transplantation and in patients with ATN not related to renal transplant. Clin Transplant 18: 156–165, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Vincent IS, Okusa MD: Biology of renal recovery: Molecules, mechanisms, and pathways. Nephron Clin Pract 127: 10–14, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humphreys BD, Czerniak S, DiRocco DP, Hasnain W, Cheema R, Bonventre JV: Repair of injured proximal tubule does not involve specialized progenitors. Proc Natl Acad Sci U S A 108: 9226–9231, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Humphreys BD, Valerius MT, Kobayashi A, Mugford JW, Soeung S, Duffield JS, et al.: Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell 2: 284–291, 2008 [DOI] [PubMed] [Google Scholar]
- 11.Yang L, Humphreys BD, Bonventre JV: Pathophysiology of acute kidney injury to chronic kidney disease: Maladaptive repair. Contrib Nephrol 174: 149–155, 2011 [DOI] [PubMed] [Google Scholar]
- 12.Zuk A, Bonventre JV: Acute kidney injury. Annu Rev Med 67: 293–307, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jun JI, Lau LF: Resolution of organ fibrosis. J Clin Invest 128: 97–107, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bozyk PD, Moore BB: Prostaglandin E2 and the pathogenesis of pulmonary fibrosis. Am J Respir Cell Mol Biol 45: 445–452, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasrallah R, Hassouneh R, Hébert RL: Chronic kidney disease: Targeting prostaglandin E2 receptors. Am J Physiol Renal Physiol 307: F243–F250, 2014 [DOI] [PubMed] [Google Scholar]
- 16.Nasrallah R, Hassouneh R, Hébert RL: PGE2, kidney disease, and cardiovascular risk: Beyond hypertension and diabetes. J Am Soc Nephrol 27: 666–676, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang P, Cai Y, Soofi A, et al.: Activation of Wnt11 by Transforming Growth Factor-beta Drives Mesenchymal Gene Expression through Non-canonical Wnt Protein Signaling in Renal Epithelial Cells. J Biol Chem 287: 21290–21302, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irizarry RA, Wu Z, Jaffee HA. Comparison of Affymetrix GeneChip expression measures. Bioinformatics 22: 789–794, 2006 [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Bardes EE, Aronow BJ, Jegga AG: ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37: W305–W311, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doi S, Masaki T: Klotho as a therapeutic target during the development of renal fibrosis. Contrib Nephrol 189: 178–183, 2017 [DOI] [PubMed] [Google Scholar]
- 21.Mencke R, Olauson H, Hillebrands JL: Effects of Klotho on fibrosis and cancer: A renal focus on mechanisms and therapeutic strategies. Adv Drug Deliv Rev 121: 85–100, 2017 [DOI] [PubMed] [Google Scholar]
- 22.Plasari G, Calabrese A, Dusserre Y, Gronostajski RM, McNair A, Michalik L, et al.: Nuclear factor I-C links platelet-derived growth factor and transforming growth factor beta1 signaling to skin wound healing progression. Mol Cell Biol 29: 6006–6017, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou D, Fu H, Xiao L, Mo H, Zhuo H, Tian X, et al.: Fibroblast-specific β-catenin signaling dictates the outcome of AKI. J Am Soc Nephrol 29: 1257–1271, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Krautzberger AM, Sui SH, Hofmann OM, Chen Y, Baetscher M, et al.: Cell-specific translational profiling in acute kidney injury. J Clin Invest 124: 1242–1254, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Kumar S, Dolzhenko E, Alvarado GF, Guo J, Lu C, et al.: Molecular characterization of the transition from acute to chronic kidney injury following ischemia/reperfusion. JCI Insight 2(18): e94716, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang SK, Wettlaufer SH, Chung J, Peters-Golden M: Prostaglandin E2 inhibits specific lung fibroblast functions via selective actions of PKA and Epac-1. Am J Respir Cell Mol Biol 39: 482–489, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohyama T, Ertl RF, Valenti V, Spurzem J, Kawamoto M, Nakamura Y, et al.: Prostaglandin E(2) inhibits fibroblast chemotaxis. Am J Physiol Lung Cell Mol Physiol 281: L1257–L1263, 2001 [DOI] [PubMed] [Google Scholar]
- 28.Kolodsick JE, Peters-Golden M, Larios J, Toews GB, Thannickal VJ, Moore BB: Prostaglandin E2 inhibits fibroblast to myofibroblast transition via E. prostanoid receptor 2 signaling and cyclic adenosine monophosphate elevation. Am J Respir Cell Mol Biol 29: 537–544, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Gerarduzzi C, He Q, Antoniou J, Di Battista JA: Quantitative phosphoproteomic analysis of signaling downstream of the prostaglandin e2/g-protein coupled receptor in human synovial fibroblasts: Potential antifibrotic networks. J Proteome Res 13: 5262–5280, 2014 [DOI] [PubMed] [Google Scholar]
- 30.Huang S, Wettlaufer SH, Hogaboam C, Aronoff DM, Peters-Golden M: Prostaglandin E(2) inhibits collagen expression and proliferation in patient-derived normal lung fibroblasts via E prostanoid 2 receptor and cAMP signaling. Am J Physiol Lung Cell Mol Physiol 292: L405–L413, 2007 [DOI] [PubMed] [Google Scholar]
- 31.Borok Z, Gillissen A, Buhl R, Hoyt RF, Hubbard RC, Ozaki T, et al.: Augmentation of functional prostaglandin E levels on the respiratory epithelial surface by aerosol administration of prostaglandin E. Am Rev Respir Dis 144: 1080–1084, 1991 [DOI] [PubMed] [Google Scholar]
- 32.Wilborn J, Crofford LJ, Burdick MD, Kunkel SL, Strieter RM, Peters-Golden M: Cultured lung fibroblasts isolated from patients with idiopathic pulmonary fibrosis have a diminished capacity to synthesize prostaglandin E2 and to express cyclooxygenase-2. J Clin Invest 95: 1861–1868, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.