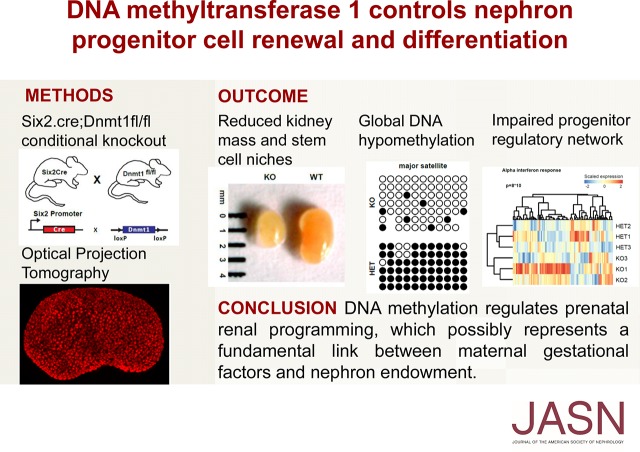
Abstract
Background
Nephron number is a major determinant of long-term renal function and cardiovascular risk. Observational studies suggest that maternal nutritional and metabolic factors during gestation contribute to the high variability of nephron endowment. However, the underlying molecular mechanisms have been unclear.
Methods
We used mouse models, including DNA methyltransferase (Dnmt1, Dnmt3a, and Dnmt3b) knockout mice, optical projection tomography, three-dimensional reconstructions of the nephrogenic niche, and transcriptome and DNA methylation analysis to characterize the role of DNA methylation for kidney development.
Results
We demonstrate that DNA hypomethylation is a key feature of nutritional kidney growth restriction in vitro and in vivo, and that DNA methyltransferases Dnmt1 and Dnmt3a are highly enriched in the nephrogenic zone of the developing kidneys. Deletion of Dnmt1 in nephron progenitor cells (in contrast to deletion of Dnmt3a or Dnm3b) mimics nutritional models of kidney growth restriction and results in a substantial reduction of nephron number as well as renal hypoplasia at birth. In Dnmt1-deficient mice, optical projection tomography and three-dimensional reconstructions uncovered a significant reduction of stem cell niches and progenitor cells. RNA sequencing analysis revealed that global DNA hypomethylation interferes in the progenitor cell regulatory network, leading to downregulation of genes crucial for initiation of nephrogenesis, Wt1 and its target Wnt4. Derepression of germline genes, protocadherins, Rhox genes, and endogenous retroviral elements resulted in the upregulation of IFN targets and inhibitors of cell cycle progression.
Conclusions
These findings establish DNA methylation as a key regulatory event of prenatal renal programming, which possibly represents a fundamental link between maternal nutritional factors during gestation and reduced nephron number.
Keywords: Dnmt1, Dnmt3a/b, nephron progenitor cells, epigenetics, kidney development, DNA methylation
Visual Abstract
In humans, the number of nephrons is highly variable, ranging from 250,000 to >2 million nephrons per kidney.1 A clear inverse correlation between nephron number and the risk of developing hypertension and secondary renal diseases has been shown.2,3 Known factors contributing to lower nephron endowment include low birth weight and in utero exposure to high-glucose, low-protein diet and in utero growth restriction.4–8 During development, reciprocal interactions between the ureteric bud (UB) tip and the surrounding cap mesenchyme (CM) lead to induction of the renal progenitor population. The CM represents a pluripotent stem cell niche which gives rise to the nephron starting from a pretubular cell aggregate, which ultimately elongates and segments to form into podocytes, parietal epithelial cells, proximal tubule, loop of Henle, and distal tubule.9–11 Previous studies have shown that the stem cell niche undergoes physiologic aging by changing its preference from self-renewal toward differentiation.12,13 In mice, the niche population exists until shortly after birth, when a last burst of differentiation into nephrons leads to its depletion.14
Nephron development is known to be influenced by prenatal exposure to environmental conditions, which in turn can induce changes in epigenetic patterns such as DNA methylation and histone modification. However, there is a paucity of information related to the role of DNA methylation for nephron development and function. During embryonic development, DNA methylation is a dynamic yet tightly controlled process, which contributes to the regulation of cell fate transitions.15,16 It is widely acknowledged that DNA methylation can lead to changes in DNA conformation, thus influencing transcription factor binding.17 In mammals, DNA methylation occurs mostly at CpG sites of which 60%–80% are methylated.18 CpG islands at promoters of constitutively active genes are usually hypomethylated, whereas long-term silencing of gene expression can be established by hypermethylated promoter CpG islands. Additionally, methylation at enhancers and insulators is crucial for the silencing of imprinted loci, which confer parent allele-specific gene expression.19 Importantly, methylation at pericentromeric regions leads to the repression of endogenous transposable elements, which make up 37.5% of the murine and 45% of the human genome.20,21 DNA methylation is catalyzed by DNA methyltransferases using S-adenosyl-l-methionine as a substrate to convert cytosine into 5-methylcytosine.22 DNA methyltransferase 1 (Dnmt1) preferentially binds to hemimethylated DNA and is crucial for restoring DNA methylation patterns at replication foci, therefore accounting for maintenance of DNA methylation during S-phase.23–25 In contrast, Dnmt3 family members Dnmt3a and Dnmt3b are not only essential for de novo DNA methylation at specific sites in early postimplantation embryos, but also for subsequent embryonic development and tissue differentiation.26
In this study, we investigate the link between environment, epigenetics, and nephron formation.
Methods
Animals
All animal experiments were conducted according to the guidelines of the American Physiologic Society, as well as the German law for the welfare of animals, and were approved by local authorities (G11/51, X13/04A, G16/85, G16/148).
Metanephric Organ Culture
Timed matings were set up with hNPHS2Cre;mT/mG (Gt(ROSA)26Sortm4(ACTB-tdTomato,−EGFP)Luo/J) mice.27 Metanephric kidneys were microdissected from the embryos at embryonic day 12.5 (E12.5) and cultured in MEM containing 10% FCS and 1% penicillin and streptomycin with 55 mM α-D-glucose or 5.5 mM α-D-glucose and 55 mM mannitol, at 37°C and 5% CO2 on 0.4-µm transwell inserts. The medium was replaced every 48 hours. Kidney cultures were harvested after 7 days in culture.
Intrauterine Growth Restriction
Wistar Kyoto rats were obtained from the Australian Resource Centre (Murdoch, Western Australia, Australia) and fed standard food pellets and tap water ad libitum. Rats were housed in a 12-hour light/dark cycle and a temperature-controlled room. All experiments were approved by the University of Melbourne Animal Ethics Committee and the La Trobe animal ethics committee before commencement following the National Health and Medical Research Council Australian code for the care and use of animals for scientific purposes. Bilateral uterine vessel ligation to induce intrauterine growth restriction or sham surgery was performed on day 18 of gestation according to Wlodek et al.7,8,28 Kidneys were collected on postnatal day 1 from male pups.
Urinary and Serum Measurements
Urinary albumin and creatinine were measured at E19.5/postnatal day 1 and then once per week using mouse albumin-specific (Mikrofluoral Mikroalbumin Test; Progen) and creatinine kits (Creatinine PAP LT-SYS; LABOR+TECHNIK; Eberhard Lehmann GmbH) according to the manufacturer’s instructions. Albumin-to-creatinine ratio was calculated and expressed as milligrams of albumin per milligram of creatinine. Serum urea was measured on E19.5/postnatal day 1 using LABOR+TECHNIK (LT-SYS; Eberhard Lehmann GmbH, Berlin) Urea Kit according to the manufacturer’s instruction using a photometer.
Histologic Analysis
Kidneys were dissected, fixed in 4% paraformaldehyde, and embedded in paraffin. For periodic acid–Schiff (PAS) and acid fuchsin–Orange G stainings, 3-µm sections were cut on a Leica microtome and analyzed and photographed with an Axioplan 2 microscope (Zeiss) and an AxioCam camera (Zeiss).
Transmission Electron Microscopy
For ultrastructural analysis, kidneys were fixed in 4% PFA/0.01% glutaraldehyde for 24 hours (Serva, Heidelberg, Germany). Samples were postfixed in 1% osmium tetroxide in the same buffer for 1 hour and stained en bloc in 1% uranyl acetate in 70% ethanol for 1 hour, dehydrated in ethanol, and embedded in Durcopan (Plano, Wetzlar, Germany). Thin sections were stained with lead citrate and examined in a Zeiss Leo-906 transmission electron microscope.
Nephron Count
Sections of 8 µm were cut from paraffin-embedded kidneys. Because the glomerular diameter equals approximately 60 µm, every fourth section was stained with PAS (E19.5) or anti-Nephrin immunofluorescence (E14.5, E15.5). The glomeruli on all sections were counted.
Immunofluorescence Staining of Kidney Sections
Kidneys were fixed in 4% paraformaldehyde overnight, dehydrated, and embedded in paraffin. The embedded tissue was sectioned at 6 μm with a Leica Microtome. The sections were deparaffinized in xylol/histoclear and rehydrated. Heat-mediated antigen retrieval was performed using citrate buffer pH 6 or Tris buffer pH 9 (caspase-3) in a steamer. The sections were blocked with PBS containing 5% BSA, and incubated for 1 hour with primary antibodies. After three PBS rinses, fluorophore-conjugated Alexa secondary antibodies (Invitrogen) were applied for 30 minutes. Microscopy and acquisition of images were performed using a Zeiss microscope.
Antibodies
The following primary antibodies were used: anti-5-methylcytosine (5mC) (1:100, ab10805; Abcam), rabbit anti-Six2 (1:200, 11562–1-AP; Proteintech), rabbit anti-Pax2 (1:100, ab37129; Abcam), guinea pig anti-Nephrin (1:200, GP-N2; Progen), rabbit anti-Jag1 (1:100, #2620; Cell Signaling Technology), rabbit anti-WT1 (1:300, ab15249; Abcam), mouse anti-WT1 (1:300, clone 6F-H2, Millipore), rabbit anti-Podocin (1:600, P0372; Sigma), mouse anti-Pancytokeratin (1:100, ab11213; Abcam), mouse anti-pHH3 (1:100, Ser10 6G3; Cell Signaling Techology), anti-BrdU (1:100; AbdSerotec), and rabbit anti-active caspase 3 (1:1000, AF835; R&D). Secondary antibodies and nuclear staining reagents were obtained from Invitrogen.
Whole Mount Staining and Optical Projection Tomography
Kidneys were dissected from E15.5 embryos and E19.5 pups and fixed and stained as described previously.29 Optical projection tomography (OPT) and niche counts were performed as described29 with the following variations: Some whole kidney samples scanned for niche counts by OPT contained regions where the niches were damaged and unable to be counted. The surface area of these regions was estimated and niche number adjusted accordingly (two samples, approximately 5% damage to each). Confocal imaging and cell counts per niche were performed as described.29
DNA Methylation
DNA from E15.5 FACS-sorted Six2Cre, Tomato/EGFP heterozygous, or homozygous Dnmt1 KO animals was bisulfite converted using EpiTect Bisulfite Kit from Qiagen according to the manufacturer’s instructions. Converted DNA (20 ng) was used as template in a PCR reaction using AmpliTaq Gold (Invitrogen). PCR products were ligated into a pCR4-TOPO-Vector using the TOPO TA Cloning Kit for sequencing (Invitrogen) and transformed into DH10 bacteria. Clones were sent for sequencing (GATC, Konstanz). Inspection, alignment, visualization, and statistics were performed with QUMA: quantification tool for methylation analysis.30 DNA from growth-restricted rats was used for global DNA methylation analysis using a 5mC DNA ELISA Kit (Zymo Research) according to the manufacturer’s instructions.
RNA-Seq and Data Analysis
RNA from E18.5 sorted CM (n=3) was extracted using the chloroform/phenol method. Then, 50 ng was sent to the DKFZ (Heidelberg) for ultra-low RNA sequencing. Fastq reads were pseudoaligned to the mm10 genome assembly using Kallisto31 and transcript read counts were aggregated to Ensembl Gene IDs for further analysis. Differential gene expression was analyzed using the DESeq2 R library.32 P values were adjusted (q-values) for the false-discovery rate according to Benjamini and Hochberg. GEO accession number: GSE94089.
Preliminary link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94089.
Statistical Analyses
Data are presented as mean±SEM throughout the text unless otherwise specified. Statistical comparisons were performed using unpaired two-tailed t test where applicable and unless otherwise specified. A value of P<0.05 was considered to represent statistically significant differences. Analyses were done using GraphPad Prism.
Results
DNA Methylation Is Reduced in Models of Environmental and Intrauterine Growth Restriction
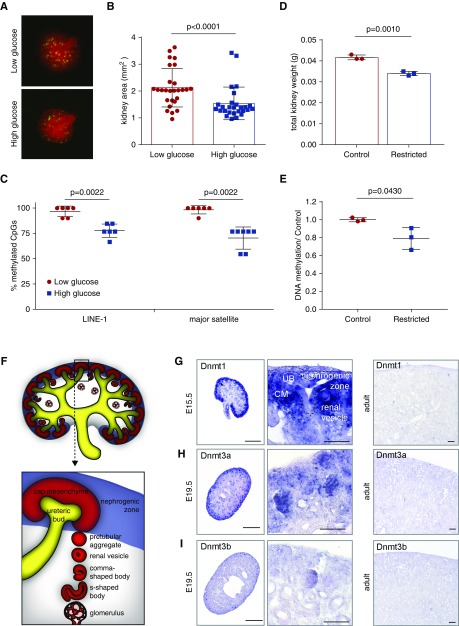
In utero exposure to environmental stressors such as gestational diabetes and dietary restriction has been shown to influence renal growth.33 To gain understanding of the effect of the environment on DNA methylation, E12.5 metanephric organs were cultured under low- and high-glucose conditions (Figure 1A). After 7 days, the kidney size and nephron number showed a significant reduction under high-glucose conditions (Figure 1B, Supplemental Figure 1). At the same time, DNA methylation at two repetitive sequences, long interspersed nuclear element-1 (LINE-1) and major satellite elements, showed partial DNA demethylation of 20% and 28%, respectively, suggesting global DNA methylation changes in the renal tissue (Figure 1C).
Figure 1.
DNA methyltransferases are enriched in the nephrogenic zone of the developing kidney. (A) E12.5 metanephric kidneys from Nphs2Cre;mT/mG (Gt(ROSA)26Sortm4(ACTB-tdTomato,−EGFP)Luo/J) reporter mice (mEGFP: podocytes, mTomato: all other cells) were treated with 5.5 or 55 mM glucose. (B) Kidney area after 7 days of glucose treatment. n=27. Paired t test. (C) Bisulfite-sequencing quantitation shows a reduction in DNA methylation levels of LINE-1 and major satellite elements. n≥6. Mann–Whitney U test. (D) Total kidney weight of postnatal day 1 rats with intrauterine growth restriction. (E) ELISA quantification of DNA methylation of control and growth-restricted rat kidneys. (F) Schematic of renal development. (G) In situ hybridization shows prominent expression of Dnmt1 in the nephrogenic zone of embryonic kidney sections and reduced expression in 8-week adult tissue. (H) In situ hybridization shows expression of Dnmt3a in the nephrogenic zone of embryonic kidney sections and reduced expression in adult tissue. (I) In situ hybridization of embryonic and adult kidney sections of Dnmt3b shows weak overall expression. Left column: scale bar, 500 µm. Middle column: scale bar, 50 µm. Right column: scale bar, 100 µm.
As an additional in vivo model of renal hypoplasia, the kidneys of postnatal day 1 rat offspring were analyzed. This model of intrauterine growth restriction mimicking uteroplacental insufficiency has been previously shown to lead to a decreased nephron number.7,8,34 Corresponding to a reduction in kidney weight of the same animals, analysis of global DNA methylation also showed a decreased methylation rate in the kidneys 8,34 (Figure 1, D and E).
DNA Methyltransferases Are Enriched in the Nephrogenic Zone of the Developing Kidney
During renal development, the metanephric mesenchyme undergoes dynamic changes in the nephrogenic zone, where UB branching, induction of the CM, and the births of new nephrons take place (Figure 1F). To understand the role of DNA methylation in the kidney, we determined the expression patterns of Dnmt1, Dnmt3a, and Dnmt3b during renal development. Analysis of RNA-seq data showed high expression of Dnmt1 and Dnmt3a in developing kidneys, whereas Dnmt3b showed lower expression levels (Supplemental Figure 2). In adult kidneys, expression levels of all Dnmts were low. To determine the localization of Dnmt expression, we performed in situ hybridization on embryonic and adult kidney sections. Dnmt1-, Dnmt3a-, and Dnmt3b-positive cells were mainly localized to the highly proliferative nephrogenic zone of the developing kidney (Figure 1, G–I). Dnmt1 expression could be detected in cells of the nephrogenic niche (CM), UB tips, and early stages of the developing nephrons, such as renal vesicles and comma-shaped and s-shaped bodies, but not in mature podocytes and tubules (Figure 1G). It was also evident in the stromal elements of the nephrogenic zone. Expression patterns for Dnmt3a were equally enriched in the nephrogenic zone and nephron precursors, whereas Dnmt3b was only weakly expressed (Figure 1, H and I). After cessation of nephrogenesis, expression of all Dnmts was greatly reduced, resulting in only residual expression in the adult renal tissue (Figure 1, G–I).
Deletion of Dnmt1, but Not Dnmt3a or Dnmt3b, in Nephron Progenitor Cells Results in Reduced Kidney Mass
To investigate the role of Dnmts for nephron formation, we generated conditional transgenic Six2-TGCtg(Six2Cre);Dnmt1fl/fl KO mice and Six2-TGCtg(Six2Cre);Dnmt3afl/fl;Dnmt3bfl/fl KO mice (Figure 2A, Supplemental Figure 3A).35 Utilizing the Cre-loxP system, the catalytic domains are excised resulting in loss of Dnmt1 and Dnmt3a/b function, respectively, in the CM and its derivatives starting from about E10.5. Deletion of Dnmt3a/b in the nephron progenitor cells resulted in normal renal development and function, as indicated by kidney size, long-term follow-up kidney morphology, body weight, and urinary analysis (Supplemental Figure 3, B–F).
Figure 2.
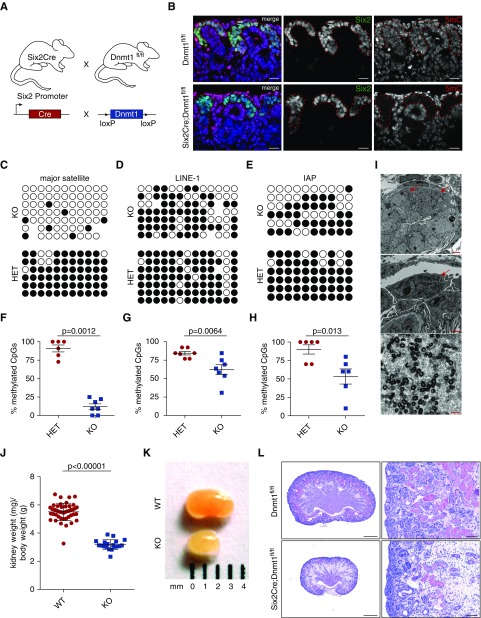
Deletion of Dnmt1, but not Dnmt3a or Dnmt3b, in nephron progenitor cells results in reduced kidney mass. (A) Six2Cre mice were bred with Dnmt1 floxed mice. (B) anti-5-methylcytosine (5mC) colocalizes with Six2 in WT cells but is absent in Dnmt1 KO cells. Scale bar, 50 µm. (C–E) Bisulfite sequencing of major satellite regions, LINE-1 elements, and intracisternal A particles (IAPs). (F–H) quantification of methylation levels of major satellite regions, LINE-1, and IAP elements. Statistical significance was evaluated using Mann–Whitney U test. (I) Production of retrotransposon particles can be seen in electron microscopy. Scale bars: top, 2.5 µm; middle, 1 µm; bottom, 250 nm. Arrows, particle-producing cells. (J) Kidney-to–body weight ratio is reduced in Dnmt1 cKO mice at E19.5 (WT: n=45 and KO: n=19). (K) Macroscopic images of E19.5 WT and Dnmt1 KO kidneys. (L) PAS staining of E19.5 kidney sections shows increased stroma and lack of epithelial structures in Dnmt1 cKO. Scale bars: left, 500 µm; right, 50 µm. HET, heterozygous; KO, knockout; WT, wildtype.
Dnmt1 is crucial for maintaining DNA methylation patterns after cell division. Therefore, we verified passive demethylation caused by Dnmt1 deletion from nephron progenitor cells in the kidney by immunofluorescence staining using an antibody against methylated cytosine (5mC) on E19.5 kidneys sections. In Dnmt1 conditional knockout (cKO) tissues, 5mC intensity was drastically reduced in cells of the CM and their descendants, whereas the surrounding stromal cells and cells from the ureter showed a specific nuclear staining (Figure 2B). Intensity of 5mC was also reduced in more mature, medullary nephrons, with very few cells retaining the 5mC signal (Supplemental Figure 4). To further quantify demethylation levels in DNA from FACS-sorted CM cells, specific loci in the mouse chromosomes were tested for methylation by bisulfite PCR. Major satellite regions around mouse centromeres showed the most severe changes in their methylation patterns (87% demethylation) (Figure 2, C and F). Likewise, transposable retroviral elements such as LINE-1 and intracisternal A particles (IAPs) were also partially demethylated in the absence of Dnmt1 (26% and 41%, respectively; Figure 2, D, E, G, and H). Electron microscopic analysis on E19.5 (postnatal day 0) confirmed a partial lack of transcriptional silencing of repetitive elements such as IAPs in cells originating from the CM such as nephron precursors, as well as tubular cells and podocytes, by showing pseudoviral vesicle production (Figure 2I).
For the phenotypic analysis, Dnmt1 cKO mice and their littermates were examined at E19.5. The pups were born at the expected Mendelian rates and no differences could be found in the body weights (Supplemental Figure 5, A and B). However, analysis of the kidneys showed a 42% reduction of kidney weight–to–body weight ratio in homozygous cKO animals compared with wildtype (WT) animals (Figure 2, J and K). Histologic analysis of sectioned E19.5 kidneys showed a reduction of epithelial structures in the Dnmt1 cKO kidneys. PAS-positive proximal tubules were decreased, whereas stromal tissue increased substantially (Figure 2L, Supplemental Figure 5D). Nevertheless, no fibrotic regions were detectable. Furthermore, renal functional parameters, such as albuminuria and serum urea, did not differ from WT littermate controls (Supplemental Figure 5, E and F).
In order to understand the role of maintenance and de novo DNA methylation in differentiated renal cells, such as podocytes, we crossed Nphs2Cre mice with Dnmt1 floxed, Dnmt3a floxed, and Dnmt3b floxed mice to generate podocyte-specific Dnmt1 single, Dnmt3a/b double, and Dnmt1/3a/3b triple knockout mice (Supplemental Figure 6, A–C). None of these mouse lines showed impaired renal development and function, as indicated by their renal morphology after 9 months and long-term urinary follow-up (Supplemental Figure 6, D–M).
Dnmt1 Is Required for Efficient Nephron Formation
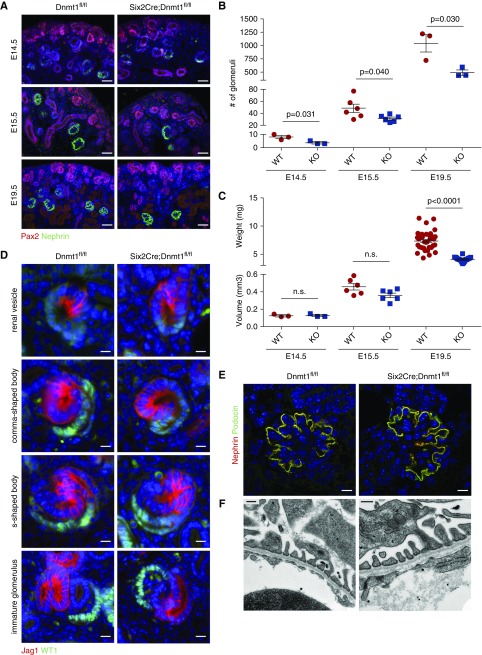
The decrease of epithelial structures in the Dnmt1 cKO kidneys indicated a reduced number of nephrons. Therefore, kidneys of WT and cKO animals were examined with podocyte marker Nephrin and the total number of glomeruli of each kidney was counted. In cKO animals, the number of glomeruli was reduced to almost half of the numbers measured in WT animals at E14.5, E15.5, and E19.5 (Figure 3, A and B). Because kidney size was still equal between cKO and WT animals at E14.5 and E15.5, the reduction of nephrogenesis seemed to precede the retardation of renal growth in cKO mice (Figure 3C). At E19.5, nephron number was reduced even in relation to kidney weight. To confirm that the process of nephron differentiation itself was not impaired but followed the normal developmental program, cKO and WT kidney sections were stained for the proximal nephron marker WT1 and the distal/intermediate marker Jag1. All precursor stages of the nephron, such as renal vesicles, comma-shaped bodies, s-shaped bodies, and immature/mature glomeruli, were found in the Dnmt1 cKO animals with correct polarization (Figure 3D). The filtration unit of the nephron, the glomerulus, from Dnmt1 cKO animals expressed mature markers of the slit diaphragm, such as Nephrin and Podocin, and mature podocyte foot processes and slit diaphragms could be seen in electron microscopy, indicating normal glomerular development (Figure 3, E and F).
Figure 3.
Dnmt1 is required for efficient nephron formation. (A) Pax2 and Nephrin immunofluorescence stainings of Six2Cre;Dnmt1 WT and KO E14.5, E15.5, and E19.5 kidney sections. Scale bar, 50 µm. (B) Total number of glomeruli determined from kidney cross-sections at E14.5 (n=3), E15.5 (n=6), and E19.5 (n=3). (C) Volume of E14.5 (n=3) and E15.5 (n=6) WT and KO kidneys determined from sections, weight of E19.5 kidneys (WT: n=45 and KO: n=19). (D) Immunofluorescence staining of distal/intermediate nephron marker Jag1 and proximal marker WT1 showing all stages of nephron precursor stadiums. Scale bar, 10 µm. (E) Immunofluorescence stainings of slit diaphragm markers Nephrin and Podocin, showing mature podocytes. Scale bar, 10 µm. (F) Electron microscopy of the glomerular filtration barrier showing mature podocyte foot processes. Scale bar, 250 nm. KO, knockout; WT, wildtype.
Dnmt1 Depletion Leads to Reduced Self-Renewal of the Progenitor Pool
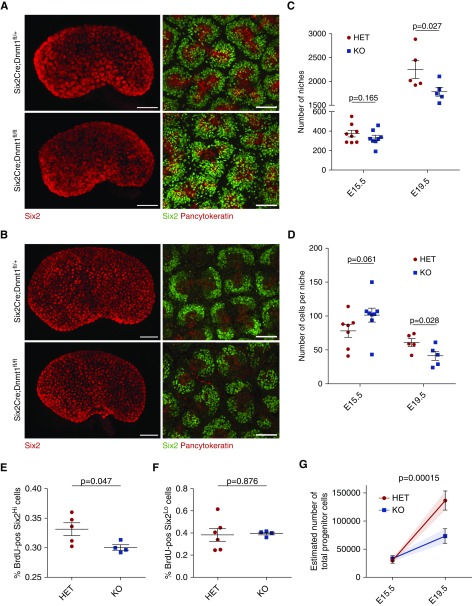
The differentiation of nephrons is initiated by condensation of cells from the CM. To analyze the nephron progenitor pool in detail, we performed whole mount stainings of Dnmt1 heterozygous and homozygous KO kidneys for the niche marker Six2 and the ureter marker Pancytokeratin and analyzed 3D reconstructions of the kidneys from OPT and confocal imaging.29,36 In E15.5 kidneys, kidney length, niche number, and cells per niche were the same in Dnmt1 cKO and heterozygous animals (Figure 4A). However, at E19.5 the morphology of the caps was changed in the Dnmt1 cKO animals (Figure 4B, Supplemental Movies 1 and 2). The total niche number of 3D-reconstructed kidneys at E19.5 indicated a reduction of 20% in the cKO animals (Figure 4C). This indicates a reduction in ureteric branching. Furthermore, at E19.5, the cap cell numbers per niche were reduced in cKO mice (Figure 4D).
Figure 4.
Dnmt1 depletion leads to reduced self-renewal of the progenitor pool. (A) OPT 3D reconstruction of the Six2-positive nephron progenitor niches and Six2 and pan-cytokeratin confocal images of Six2Cre;Dnmt1 HET and KO kidneys at E15.5. Scale bars, 300 and 50 µm, respectively. (B) OPT 3D reconstruction of the Six2-positive nephron progenitor niches and Six2 and pan-cytokeratin confocal images of HET and KO kidneys at E19.5. Scale bars, 400 and 50 µm, respectively. (C) Total number of niches at E15.5 and E19.5 using OPT analysis (n=8 and n=5, respectively). One-tailed P value. (D) Number of cells per niche, determined by confocal imaging (E15.5, HET: n=7, KO: n=8, and E19.5: n=5). One-tailed P value. (E) Quantification of BrdU-positive Six2Hi cells on E18.5 kidney sections (HET: n=5 and KO: n=4). (F) Quantification of BrdU-positive Six2Lo cells on E18.5 kidney sections (HET: n=5 and KO: n=4). (G) Estimated total progenitor cell number at E15.5 and E19.5 from niche number and cell number per niche (HET: n=7 and KO: n=5). HET, heterozygous; KO, knockout.
To investigate whether changes in the proliferation rate were responsible for the reduced nephron progenitor cell number in Dnmt1 cKO, Dnmt1 heterozygous and cKO kidneys were tested for Bromodeoxyuridine (BrdU) incorporation. For this, the number of Six2Hi cells representing the nephron progenitor cell population was counted and the percentage of cells that had incorporated BrdU quantified at E18.5 (Figure 4E, Supplemental Figure 7A). A significant difference was found between heterozygous and homozygous Dnmt1 cKO niche populations, indicating that loss of Dnmt1 led to a reduction in CM proliferation. Furthermore, the number of Six2Lo cells, representing cells undergoing differentiation into renal vesicles, was quantified for BrdU incorporation. However, no differences in the proliferation rate could be detected between heterozygous and cKO animals (Figure 4F).
Estimation of the total progenitor numbers at E15.5 and E19.5 from niche number and cell counts shows that the lower niche number and reduced cell count add up to an overall progenitor deficit of 46% in the organ as a whole (Figure 4G).
To exclude enhanced apoptosis as a cause for the reduced CM population and nephrogenesis upon differentiation of the nephron progenitors, E19.5 renal sections were stained for the apoptosis marker active caspase-3 and the number of positive cells in the nephrogenic zone was quantified. Here, no differences were detectable between WT and cKO (Supplemental Figure 7, B–D).
Absence of Dnmt1 Disrupts Transcriptional Programs Regulating Progenitor Identity
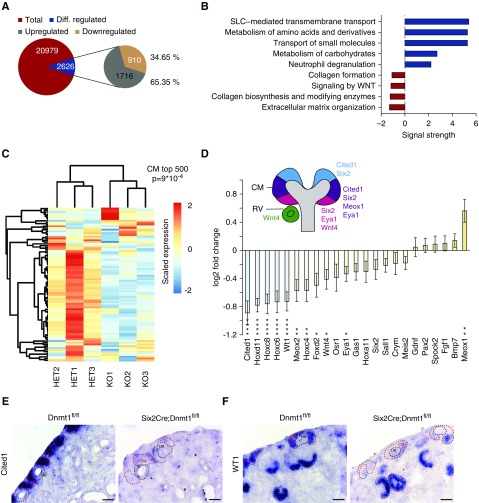
The consequences of DNA hypomethylation on transcriptional activity were examined by next generation RNA sequencing of FACS-sorted cells from the nephrogenic niche. Of all genes, 2626 genes (12.52%) were differentially regulated (q-value <0, 1), 1716 genes (65.35%) were upregulated, and only 910 genes (34.65%) were downregulated (Figure 5A). Pathway analysis on the basis of the REACTOME database was performed with differentially regulated genes to explore the functional implications of the alterations observed (Figure 5B). Collagen formation and extracellular matrix organization as well as Wnt signaling pathways were downregulated, suggesting a crucial role of stromal formation and Wnt communication in renal development. Upregulated genes were involved in carbohydrate and amino acid metabolism as well as inflammatory response (neutrophil degranulation) (Figure 5B). Strikingly, a gene set of the top 500 enriched genes in the P0 CM showed a strong negative correlation (P=9×10−4) and downregulation in KO cells (Figure 5C). Whereas CM marker genes such as Cited1, Hoxd11, Hoxc8, Wt1, and Meox2 were significantly downregulated, other markers, such as Six2, Eya1, or Osr1, showed only slight declines in expression levels (Figure 5D). Validation of CM markers Cited1 and Wt1 mRNA showed a lack of expression of both genes in the CM, whereas Wt1 seemed to be re-expressed in the developing nephron (Figure 5, E and F, Supplemental Figure 7B). Overall, these data suggest that key developmental programs are dysregulated in the nephron progenitor cells.
Figure 5.
DNA methylation controls transcriptional programs regulating progenitor identity and differentiation. (A) Pie chart of differentially regulated genes (q-value<0.1) (B) Top up- and downregulated REACTOME pathways. (C) Heatmap of P0 CM-specific genes ranked top 500 and corresponding gene set enrichment analysis score. (D) Log2 fold-change of CM marker genes. Asterisks, differentially regulated genes. (E) In situ hybridization of Cited1 and (F) Wt1 in WT and KO kidney sections. Scale bar, 50 µm. Diff., differentially; RV, renal vesicle.
DNA Hypomethylation Leads to Transcriptional Derepression of Germline Genes and ERVs
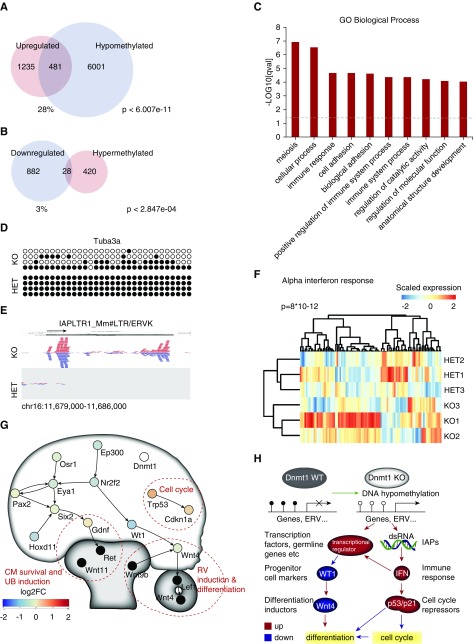
To understand the direct effect of Dnmt1 cKO–dependent hypomethylation, we compared the differentially expressed genes with DNA methylation data from Dnmt1 cKO versus WT (external data36). From the 1716 upregulated genes, the promoter regions of 481 genes were found to be significantly hypomethylated in the external data set (28%, P<6.01e−11; Figure 6A). From the downregulated genes, only 28 were found to be hypermethylated (P<2.85e−04; Figure 6B). In comparison, transcriptome data from whole kidney Six2Cre;Dnmt1 cKO animals showed little overlap (upregulated and hypomethylated genes, P<0.06; downregulated and hypermethylated genes, P<0.002). In the nephron progenitor cells, the upregulated and hypermethylated genes were involved in meiosis, immune response, adhesion, and metabolism (Figure 6C). Among the upregulated and hypermethylated genes were many genes normally not expressed in the kidney, such as the germline genes Dazl, Sohlh2, Mov10l1, Tex19.1, and Tuba3a,38 which have been described to be upregulated during DNA hypomethylation in cancer. Furthermore, protocadherins, Rhox homeobox genes, and heat shock proteins were upregulated and hypomethylated. The upregulation of several genes was validated by RT-qPCR analysis (Supplemental Figure 8) and demethylation of the promoter region of Tuba3a was validated by bisulfite sequencing, showing global loss of methylation in an otherwise fully methylated locus (Figure 6D). Alignment of the RNA-seq reads also showed expression of endogenous retroviral transcripts, specifically IAPs, whereas LINE elements were not derepressed (Figure 6E, Supplemental Figure 9). Transcription of endogenous retroviral DNA was accompanied by the enrichment of gene upregulation in response to interferon (IFN) type I in gene set enrichment analysis (P=8×10−12) (Figure 6F), including genes transmitting an antiviral response, such as Irf1, Il15, Ifit1, and Oas1a. Cell cycle regulators p53 (Trp53) and p21 (Cdkn1a) were upregulated, but not hypomethylated. Altogether, the data indicate that Dnmt1 deficiency leads to global DNA hypomethylation and upregulation of ectopic gene and retroviral elements, interfering with the progenitor cell regulatory network (Figure 6, G and H).
Figure 6.
DNA hypomethylation leads to transcriptional derepression of germline genes and ERVs. (A) Overlap of upregulated genes in the Dnmt1 KO progenitor cells and DNA hypomethylated regions. (B) Overlap of downregulated genes in the Dnmt1 KO progenitor cells and hypermethylated regions. (C) GO terms “Biological Process” of upregulated and hypomethylated genes. (D) Bisulfite sequencing of the Tuba3a promoter region shows DNA hypomethylation. (E) RNA sequencing shows transcription of reads aligning to IAP elements. ERVK, endogenous retrovirus-K; LTR, long terminal repeat; Mm, Mus musculus. (F) Heatmap of genes upregulated in response to α-IFN proteins and corresponding GSEA score. (G) Regulatory network of the CM with potentially inhibited cellular functions. RV, renal vesicle. (H) Dnmt1 KO results in reactivation of genes and endogenous retrovirus-like particles (ERVs) inducing an IFN response, influencing cell cycle progression and Wt1 gene expression and resulting in reduced differentiation of nephrons. FC, fold change; GO, gene ontology; HET, heterozygous; IAP, intracisternal A particle; KO, knockout; WT, wildtype.
Discussion
Environmental influences, such as low-protein diet and dietary restriction, have been shown to lead to renal hypoplasia and lower nephron endowment in utero.7,8,39 Because DNA methyltransferases rely on substrates derived from folate and methionine, a direct link between environmental conditions and epigenetic regulation has been established.40 Several studies show the effect of nutrition, such as dietary or protein restriction, on the expression of Dnmts and global DNA methylation.41,42 Here, we report the role of DNA methylation in nephron progenitor cell renewal and nephron formation. Our own data confirmed decreased methylation levels under in vitro high-glucose conditions, as well as in an intrauterine growth restriction model of placental insufficiency. Conditional depletion of DNA maintenance methyltransferase Dnmt1 but not de novo methyltransferases Dnmt3a and Dnmt3b in the murine nephron progenitor population led to hypoplastic kidneys, with reduced nephrogenesis and transcriptional changes in the CM population.
Although Dnmt1 depletion clearly affects nephron development, de novo DNA methylation seems to be dispensable for nephron development because deletion of both Dnmt3a and Dnmt3b did not perturb nephrogenesis and long-term renal function, despite the fact that Dnmt3a is highly expressed in the nephrogenic zone and both have been shown to play important roles in stem cell differentiation and stem cell proliferation.43 Furthermore, loss of Dnmt3a and Dnmt3b has previously been shown to lead to demethylation due to a minor contribution to maintenance methylation.26 Therefore, the lack of phenotype indicates that at this stage of nephrogenesis de novo DNA methylation might not be further required for cell fate decisions of the nephrogenic niche.
Likewise, our analysis of Dnmt1-deficient differentiated podocytes showed that these cells were not functionally dependent on Dnmt1 activity under physiologic conditions. Because they are terminally differentiated, there seems to be no requirement for maintenance of DNA methylation, which concords with the low Dnmt1 expression status. Furthermore, Dnmt3a/b function in podocytes was dispensable, as is the loss of function of all three Dnmts together. The stable renal function observed until 9 months of age in Dnmt cKO animals indicates no physiologic long-term adverse effects, although a change in sensitivity to renal injury cannot be excluded and is an interesting line for future investigation.44 Indeed, several studies indicate that DNA methylation plays a role in diabetic nephropathy or hypertensive nephrosclerosis, both of which are known to be podocyte dependent.45,46
In contrast to differentiated cells, we show that maintenance of DNA methylation plays a key role during nephron development. Loss of Dnmt1 in nephron progenitor cells led to a strong reduction of DNA methylation in all cells originating from the CM. This is in accordance with previous reports showing a global approximately 80% decrease of DNA methylation after deletion of Dnmt1.47 Furthermore, decreased DNA methylation greatly influenced the behavior of the nephron progenitor population. First of all, we observed a greatly diminished capacity for nephrogenesis starting as early as E14.5, and hence before any detectable slowdown in branching or niche reduction. This resulted in a decrease of nephron generation of about 50% at birth. Although the expression of only a few CM-specific genes, such as Cited1,48 Eya1, and Meox2, as well as many genes from the Hox gene cluster, was significantly downregulated, an overall shift in gene expression of marker genes such as Six2,49 Osr1, and Sall1 was detectable, indicating a broad dysregulation of progenitor cell identity. Because initiation of nephron differentiation is likely dependent on a threshold in expression levels, this might be the cause of delay in nephrogenesis under Dnmt1 cKO conditions. Of the genes responsible for nephron induction and activation of mesenchymal-to-epithelial transition, expression of Wt1 and its target Wnt4, crucial for initiation of nephrogenesis, were reduced.50,51 However, because DNA hypomethylation usually results in derepression of gene activation, downregulated gene expression is most likely a secondary effect of gene dysregulation. Besides the diminished capacity for nephrogenesis, the capacity for cap mesenchymal proliferation in Dnmt1 cKO renal progenitor cells was also decreased, leading to gradual changes in CM morphology and reduced niche numbers. Even though the effect on cell proliferation is subtle, the immense proliferative activity of the progenitor cells causes an accumulative reduction and premature exhaustion of the niche, which also affects ureteric branching. Upregulated as a response to endogenous retrovirus-like particle production, IFN signaling is known to have a strong antiproliferative/antitumoral effect via activation of p53/p21.52–56 Overall, the derepression of genes and endogenous retroviral elements seems to result in the activation of pathways dealing with changes in cell cycle, immune system, and metabolism, thereby changing the regulation of proliferation and differentiation.57 Thus, our study on the role of DNA methyltransferases in kidney development describes a possible mediating role for DNA methylation between gestational environmental stressors, such as metabolic influences, and renal hypoplasia.
To our knowledge, this study is the first comprehensive analysis of the regulatory function of DNA methylation during renal development and podocyte homeostasis showing that Dnmt1, but not Dnmt3a or Dnmt3b, is a key regulator of the renal stem cell niche during glomerular development, whereas long-term podocyte homeostasis and function do not depend on DNA methyltransferase activity. Our study indicates that DNA demethylation results in derepression and transcriptional activation of genes and endogenous retroviral particles in the CM, leading to reduced progenitor cell renewal and differentiation capacity by the nephrogenic niche.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank L. Hein for providing Dnmt3a/bfl/fl mice. We thank Betina Kiefer, Charlotte Meyer, and Barbara Joch for expert technical assistance and all members of our laboratory for helpful discussions and support. The authors greatly acknowledge the Genomics and Proteomics Core Facility, German Cancer Research Center/DKFZ, Heidelberg, Germany for their sequencing service.
The Galaxy server that was used for some calculations is in part funded by Collaborative Research Centre 992 Medical Epigenetics (Deutsche Forschungsgesellschaft [DFG] grant SFB 992/1 2012) and the German Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung [BMBF] grants 031 A538A/A538C RBC, 031L0101B/031L0101C de.NBI-epi, 031L0106 de.STAIR [de.NBI]). This work is supported by a Marie Curie European Union grant (CIG 293568) (to W.B.-W.), the Margarete von Wrangell Habilitationsprogramm (Ministerium für Wissenschaft Baden-Württemberg) (to W.B.-W.), and the Mathilde-Wagner-Habilitationspreis (to W.B.-W.). Furthermore, this study was supported by the German Research Foundation (DFG): CRC 1140 (to T.B.H.), CRC 1192 (to T.B.H.), CRC 992 (to T.B.H.), EXE306 (to H.B.), and CRC 850 (to M.B.); by the Heisenberg program (to T.B.H.); by the European Research Council grant 616891 (to T.B.H.); by the H2020-IMI2 consortium BEAt-DKD (Biomarker Enterprise to Attack Diabetic Kidney Disease) (115974, to T.B.H.); by STOP-FSGS (Speed Translation-Oriented Progress to Treat Focal Segmental Glomerulosclerosis) 01GM1518C (to T.B.H.); by the Else-Kröner Fresenius Stiftung (iPRIME [innovative Promotionsförderung im Bereich translationale Entzündungsforschung] and NAKSYS [Nierenfunktionsstörungen als Komplikation von Systemerkrankungen]); by the Excellence Initiative of the German Federal and State Governments (EXC294, BIOSS II to T.B.H.); and by the Freiburg Institute for Advanced Studies (to T.B.H.). M.H.L. is a Senior Principal Research Fellow and V.G.P. is an Early Career Research Fellow of the National Health and Medical Research Council of Australia. A.C. is a Discovery Early Career Researcher Award Fellow of the Australian Research Council. M.H.L., A.C., and S.W. are funded by the National Health and Medical Research Council (ID1063989). V.G.P. is also funded by the Alexander von Humboldt Foundation and the German Society of Nephrology (DGfN). Murdoch Children's Research Institute is supported by the Victorian Government’s Operational Infrastructure Support Program. M.B. is funded by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (FKZ (Forschungskennzeichen) 01ZX1409B).
Conceptualization: N.W., W.B.-W., and T.B.H. Investigation: N.W., J.V., A.C., S.W., J.P., G.R., S.L., V.G.P., O.K., M.E.W., T.R., K.M.M., and W.B.-W. Data curation: N.W., M.B., H.B., T.L., R.-U.R., and S.B. Writing—original draft: N.W. Writing—review and editing: N.W., M.H.L., V.G.P., W.B.-W., and T.B.H. Funding acquisition: M.E.W., T.R, W.B.-W., and T.B.H. Supervision: T.B.H.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2018070736/-/DCSupplemental.
supplemental material
Supplemental Figure 1. Nephrin number is decreased in kidneys grown on high glucose medium.
Supplemental Figure 2. Dnmt1 and Dnmt3a are highly expressed during kidney development.
Supplemental Figure 3. Dnmt3a/b function is dispensable for nephron development.
Supplemental Figure 4. Lineage tracing of Dnmt1 KO cells using 5mC.
Supplemental Figure 5. Renal parameters of Dnmt1 KO animals.
Supplemental Figure 6. Podocyte function does not rely on DNA methyltransferase activity.
Supplemental Figure 7. Proliferation and survival rate of renal progenitor cells.
Supplemental Figure 8. Germline genes are upregulated in the Dnmt1 KO kidney.
Supplemental Figure 9. IAPs are specifically expressed upon loss of Dnmt1.
References
- 1.Hughson M, Farris AB 3rd, Douglas-Denton R, Hoy WE, Bertram JF: Glomerular number and size in autopsy kidneys: The relationship to birth weight. Kidney Int 63: 2113–2122, 2003 [DOI] [PubMed] [Google Scholar]
- 2.Brenner BM, Garcia DL, Anderson S: Glomeruli and blood pressure. Less of one, more the other? Am J Hypertens 1: 335–347, 1988 [DOI] [PubMed] [Google Scholar]
- 3.Keller G, Zimmer G, Mall G, Ritz E, Amann K: Nephron number in patients with primary hypertension. N Engl J Med 348: 101–108, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Tran S, Chen YW, Chenier I, Chan JS, Quaggin S, Hébert MJ, et al.: Maternal diabetes modulates renal morphogenesis in offspring. J Am Soc Nephrol 19: 943–952, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luyckx VA, Brenner BM: Low birth weight, nephron number, and kidney disease. Kidney Int Suppl 68(97): S68–S77, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Langley-Evans SC, Welham SJ, Jackson AA: Fetal exposure to a maternal low protein diet impairs nephrogenesis and promotes hypertension in the rat. Life Sci 64: 965–974, 1999 [DOI] [PubMed] [Google Scholar]
- 7.Wlodek ME, Westcott K, Siebel AL, Owens JA, Moritz KM: Growth restriction before or after birth reduces nephron number and increases blood pressure in male rats. Kidney Int 74: 187–195, 2008 [DOI] [PubMed] [Google Scholar]
- 8.Moritz KM, Mazzuca MQ, Siebel AL, Mibus A, Arena D, Tare M, et al.: Uteroplacental insufficiency causes a nephron deficit, modest renal insufficiency but no hypertension with ageing in female rats. J Physiol 587: 2635–2646, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costantini F, Kopan R: Patterning a complex organ: Branching morphogenesis and nephron segmentation in kidney development. Dev Cell 18: 698–712, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Little MH, McMahon AP: Mammalian kidney development: Principles, progress, and projections. Cold Spring Harb Perspect Biol 4, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schell C, Wanner N, Huber TB: Glomerular development--shaping the multi-cellular filtration unit. Semin Cell Dev Biol 36: 39–49, 2014 [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Brunskill EW, Potter SS, Dexheimer PJ, Salomonis N, Aronow BJ, et al.: Intrinsic age-dependent changes and cell-cell contacts regulate nephron progenitor lifespan. Dev Cell 35: 49–62, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopan R, Chen S, Little M: Nephron progenitor cells: Shifting the balance of self-renewal and differentiation. Curr Top Dev Biol 107: 293–331, 2014 [DOI] [PubMed] [Google Scholar]
- 14.Rumballe BA, Georgas KM, Combes AN, Ju AL, Gilbert T, Little MH: Nephron formation adopts a novel spatial topology at cessation of nephrogenesis. Dev Biol 360: 110–122, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holliday R, Pugh JE: DNA modification mechanisms and gene activity during development. Science 187: 226–232, 1975 [PubMed] [Google Scholar]
- 16.Bechtel-Walz W, Huber TB: Chromatin dynamics in kidney development and function. Cell Tissue Res 356: 601–608, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Keshet I, Lieman-Hurwitz J, Cedar H: DNA methylation affects the formation of active chromatin. Cell 44: 535–543, 1986 [DOI] [PubMed] [Google Scholar]
- 18.Smith ZD, Meissner A: DNA methylation: Roles in mammalian development. Nat Rev Genet 14: 204–220, 2013 [DOI] [PubMed] [Google Scholar]
- 19.Li E, Beard C, Jaenisch R: Role for DNA methylation in genomic imprinting. Nature 366: 362–365, 1993 [DOI] [PubMed] [Google Scholar]
- 20.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, et al.: Mouse Genome Sequencing Consortium : Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562, 2002 [DOI] [PubMed] [Google Scholar]
- 21.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al.: International Human Genome Sequencing Consortium : Initial sequencing and analysis of the human genome. Nature 409: 860–921, 2001 [DOI] [PubMed] [Google Scholar]
- 22.Bestor TH: The DNA methyltransferases of mammals. Hum Mol Genet 9: 2395–2402, 2000 [DOI] [PubMed] [Google Scholar]
- 23.Bestor T, Laudano A, Mattaliano R, Ingram V: Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol 203: 971–983, 1988 [DOI] [PubMed] [Google Scholar]
- 24.Yoder JA, Soman NS, Verdine GL, Bestor TH: DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J Mol Biol 270: 385–395, 1997 [DOI] [PubMed] [Google Scholar]
- 25.Leonhardt H, Page AW, Weier HU, Bestor TH: A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71: 865–873, 1992 [DOI] [PubMed] [Google Scholar]
- 26.Okano M, Bell DW, Haber DA, Li E: DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257, 1999 [DOI] [PubMed] [Google Scholar]
- 27.Moeller MJ, Sanden SK, Soofi A, Wiggins RC, Holzman LB: Podocyte-specific expression of cre recombinase in transgenic mice. Genesis 35: 39–42, 2003 [DOI] [PubMed] [Google Scholar]
- 28.Wlodek ME, Westcott KT, O’Dowd R, Serruto A, Wassef L, Moritz KM, et al.: Uteroplacental restriction in the rat impairs fetal growth in association with alterations in placental growth factors including PTHrP. Am J Physiol Regul Integr Comp Physiol 288: R1620–R1627, 2005 [DOI] [PubMed] [Google Scholar]
- 29.Combes AN, Short KM, Lefevre J, Hamilton NA, Little MH, Smyth IM: An integrated pipeline for the multidimensional analysis of branching morphogenesis. Nat Protoc 9: 2859–2879, 2014 [DOI] [PubMed] [Google Scholar]
- 30.Kumaki Y, Oda M, Okano M: QUMA: Quantification tool for methylation analysis. Nucleic Acids Res 36: W170–W175, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bray NL, Pimentel H, Melsted P, Pachter L: Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527, 2016 [DOI] [PubMed] [Google Scholar]
- 32.Love MI, Huber W, Anders S: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amri K, Freund N, Vilar J, Merlet-Bénichou C, Lelièvre-Pégorier M: Adverse effects of hyperglycemia on kidney development in rats: In vivo and in vitro studies. Diabetes 48: 2240–2245, 1999 [DOI] [PubMed] [Google Scholar]
- 34.Romano T, Wark JD, Owens JA, Wlodek ME: Prenatal growth restriction and postnatal growth restriction followed by accelerated growth independently program reduced bone growth and strength. Bone 45: 132–141, 2009 [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, et al.: Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3: 169–181, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Short KM, Combes AN, Lefevre J, Ju AL, Georgas KM, Lamberton T, et al.: Global quantification of tissue dynamics in the developing mouse kidney. Dev Cell 29: 188–202, 2014 [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen BG, Berent RM, Ha SE, Horiguchi K, Sasse KC, Becker LS, et al.: DNA methylation, through DNMT1, has an essential role in the development of gastrointestinal smooth muscle cells and disease. Cell Death Dis 9: 474, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Smet C, Loriot A: DNA hypomethylation and activation of germline-specific genes in cancer. Adv Exp Med Biol 754: 149–166, 2013 [DOI] [PubMed] [Google Scholar]
- 39.Zeman FJ: Effects of maternal protein restriction on the kidney of the newborn young of rats. J Nutr 94: 111–116, 1968 [DOI] [PubMed] [Google Scholar]
- 40.Duthie SJ, Narayanan S, Sharp L, Little J, Basten G, Powers H: Folate, DNA stability and colo-rectal neoplasia. Proc Nutr Soc 63: 571–578, 2004 [DOI] [PubMed] [Google Scholar]
- 41.Unterberger A, Szyf M, Nathanielsz PW, Cox LA: Organ and gestational age effects of maternal nutrient restriction on global methylation in fetal baboons. J Med Primatol 38: 219–227, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altmann S, Murani E, Schwerin M, Metges CC, Wimmers K, Ponsuksili S: Maternal dietary protein restriction and excess affects offspring gene expression and methylation of non-SMC subunits of condensin I in liver and skeletal muscle. Epigenetics 7: 239–252, 2012 [DOI] [PubMed] [Google Scholar]
- 43.Umehara Y, Hanaoka K, Watanabe D: Distinct functions of Dnmt3a and Dnmt3b de novo DNA methyltransferases in ES cell proliferation and differentiation. Stem Cell Discovery 3: 127–132, 2013 [Google Scholar]
- 44.Bechtel W, McGoohan S, Zeisberg EM, Müller GA, Kalbacher H, Salant DJ, et al.: Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med 16: 544–550, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Majumder S, Advani A: The epigenetic regulation of podocyte function in diabetes. J Diabetes Complications 29: 1337–1344, 2015 [DOI] [PubMed] [Google Scholar]
- 46.Hayashi K, Sasamura H, Nakamura M, Sakamaki Y, Azegami T, Oguchi H, et al.: Renin-angiotensin blockade resets podocyte epigenome through Kruppel-like Factor 4 and attenuates proteinuria. Kidney Int 88: 745–753, 2015 [DOI] [PubMed] [Google Scholar]
- 47.Lei H, Oh SP, Okano M, Jüttermann R, Goss KA, Jaenisch R, et al.: De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122: 3195–3205, 1996 [DOI] [PubMed] [Google Scholar]
- 48.Boyle S, Shioda T, Perantoni AO, de Caestecker M: Cited1 and Cited2 are differentially expressed in the developing kidney but are not required for nephrogenesis. Dev Dyn 236: 2321–2330, 2007 [DOI] [PubMed] [Google Scholar]
- 49.Combes AN, Wilson S, Phipson B, Binnie BB, Ju A, Lawlor KT, et al.: Haploinsufficiency for the Six2 gene increases nephron progenitor proliferation promoting branching and nephron number. Kidney Int 93: 589–598, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sim EU, Smith A, Szilagi E, Rae F, Ioannou P, Lindsay MH, et al.: Wnt-4 regulation by the Wilms’ tumour suppressor gene, WT1. Oncogene 21: 2948–2960, 2002 [DOI] [PubMed] [Google Scholar]
- 51.Essafi A, Webb A, Berry RL, Slight J, Burn SF, Spraggon L, et al.: A wt1-controlled chromatin switching mechanism underpins tissue-specific wnt4 activation and repression. Dev Cell 21: 559–574, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sangfelt O, Erickson S, Grander D: Mechanisms of interferon-induced cell cycle arrest. Front Biosci 5: D479–D487, 2000 [DOI] [PubMed] [Google Scholar]
- 53.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al.: Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162: 974–986, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giandomenico V, Vaccari G, Fiorucci G, Percario Z, Vannuchi S, Matarrese P, et al.: Apoptosis and growth inhibition of squamous carcinoma cells treated with interferon-alpha, IFN-beta and retinoic acid are associated with induction of the cyclin-dependent kinase inhibitor p21. Eur Cytokine Netw 9: 619–631, 1998 [PubMed] [Google Scholar]
- 55.Schroeder M, Mass MJ: CpG methylation inactivates the transcriptional activity of the promoter of the human p53 tumor suppressor gene. Biochem Biophys Res Commun 235: 403–406, 1997 [DOI] [PubMed] [Google Scholar]
- 56.Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, et al.: Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet 27: 31–39, 2001 [DOI] [PubMed] [Google Scholar]
- 57.Buck MD, O’Sullivan D, Klein Geltink RI, Curtis JD, Chang CH, Sanin DE, et al.: Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166: 63–76, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.