Abstract
Biliary atresia (BA) is the most common cause of chronic cholestasis in children. The long non-coding RNA (lncRNA) Annexin A2 pseudogene 3 (ANXA2P3) and Annexin A2 (ANXA2) have been suggested to serve pivotal roles in BA; however, the clinical significance and biological roles of ANXA2P3 and ANXA2 in BA remain to be elucidated. The present study aimed to elucidate the function of ANAX2P3 and ANXA2 in BA-induced liver injury using a human liver cell line and liver tissues from patients with BA. Reverse transcription-quantitative polymerase chain reaction, western blotting and immunohistochemistry were conducted to determine the expression levels of ANXA2 and ANXA2P3 in liver tissues from patients with BA. Classification of fibrosis was analyzed by Masson staining. The functional roles of ANXA2 and ANXA2P3 in liver cells were determined by Cell Counting kit-8 assay, and flow cytometric and cell cycle analyses. Activation of the ANXA2/ANXA2P3 signaling pathway in liver cells was evaluated by western blot analysis. According to the present results, the expression levels of ANXA2 and ANXA2P3 were significantly increased in liver tissues from patients with BA. In addition, knocking down the expression of ANXA2P3 and ANXA2 may result in reduced liver cell proliferation, cell cycle arrest in G1 phase and increased apoptosis of liver cells in vitro. Furthermore, in cells in which ANXA2 and ANXA2P3 were overexpressed, cell apoptosis was reduced and cell cycle arrest in G2 phase. Taken together, these results indicated that ANXA2P3 and ANXA2 may have protective effects against liver injury progression and may be considered biomarkers in patients with BA.
Key words: Annexin A2 pseudogene 3, Annexin A2, biliary atresia, hepatic injury, long non-coding RNA
Introduction
Biliary atresia (BA) results from inflammatory and fibrotic obstruction of extrahepatic bile ducts, and is a leading cause of neonatal cholestasis. In addition, BA is the major reason for pediatric liver transplantation worldwide (1-3). The majority of children with BA succumb to liver failure within a year, and surgery is the only satisfactory treatment option, which includes Kasai surgery and liver transplantation (4). The pathogenesis of BA is currently unclear; therefore, there is an urgent requirement for more effective methods to reduce the risk of treatment, and improve diagnosis and therapeutic effects. Identification of the relevant mechanisms is conducive to the development of novel treatment strategies, and is beneficial to reduce BA-induced hepatic injury and improve clinical outcomes.
The Human Genome Project indicated that although 70% of the genome is transcribed, only ≤2% of the human genome serves as a blueprint for protein coding (5). Until recently, long non-coding RNAs (lncRNAs) were considered a by-product of the process of transcription without a biological function (6). However, recent studies have revealed that lncRNAs serve an important role in several biological events, including post-transcriptional regulation of human genes, epigenetic regulation, cell cycle regulation and regulation of cell differentiation (7,8). In previous studies that have focused on liver injury, lncRNAs have received attention, and several lncRNAs have been reported to be closely associated with the initiation and progression of liver injury (9-14).
It has been reported that the lncRNA Annexin A2 pseudo-gene 3 (ANXA2P3) encodes Annexin A2 pseudogenes. These pseudogene-expressed non-coding RNAs have a functional role in regulating their protein-coding counterparts (15). It has previously been reported that these pseudogenes can act as signaling regulators and, in most cases, pseudogenes and parental genes exhibit positive coupling and positive functional correlation (16). For example, octamer-binding transcription factor 4 (Oct4) psuedogene (Oct4P1) is a pseudogene that overexpresses Oct4, and can promote the self-renewal of mesenchymal stem cells and inhibit cell differentiation (17). A negative correlation between the expression of a psuedogene of B-Raf proto-oncogene, serine/threonine kinase (BRAF) and the degree of BRAF mutation has been detected in thyroid carcinoma; BRAF overexpression can effectively promote the transformation of NIH3T3 cells and induce tumorigenesis in nude mice (18). It has previously been reported that ANXA2 pseudogenes can be used as novel biomarkers for the diagnosis, prognosis and targeted therapy of glioma (19). However, to the best of our knowledge, the biological function of ANXA2P3 in liver injury and its expression in patients with BA have yet to be determined.
Annexin A2 (ANXA2) is a member of the connexin family, which is involved in several biological events, including cell proliferation and apoptosis; in previous studies, a close association between ANXA2 and liver damage-associated diseases has been demonstrated (20-25). Furthermore, ANXA2 is widely recognized as a promising serum marker of liver damage or liver fibrosis. In patients with chronic hepatitis B-associated hepatic fibrosis, serum ANXA2 is significantly upregulated compared with its serum levels in healthy individuals (26). Similarly, in rats with immune liver fibrosis and alcoholic liver models, ANXA2 on the membrane surface is significantly upregulated (27). In addition, ANXA2 promotes liver fibrosis by mediating von Willebrand factor secretion, which can be used to mitigate the progression of liver fibrosis (28). Hepatic overexpression of ANXA1 and ANXA2 inhibits acetaminophen-induced expansion of liver injury (29). However, to date, the biological functions of ANXA2 in liver injury and its expression in patients with BA have not been thoroughly studied.
Based on these previous findings, the present study aimed to investigate the expression profiles of ANXA2 and ANXA2P3 in liver tissue derived from patients with BA, and to elucidate its involvement in liver injury. The results may provide novel insight into the biological functions of ANXA2 and ANXA2P3, and their underlying mechanisms of action in liver injury.
Materials and methods
Patients and specimens
Liver tissue was obtained from 20 children (age 53 days-3 months old) who were diagnosed with BA and underwent Kasai surgery at the Department of General Surgery, Fudan University Children’s Hospital (Shanghai, China) between March 2016 and October 2016. Paracarcinoma liver tissue was obtained from six children (age, 2-4 years old) who were diagnosed with hepatoblastoma. Fresh tissue was immediately snap-frozen in liquid nitrogen and stored at −80°C for further analysis. The present study was approved by the Ethical Review Board of Fudan University Children’s Hospital, and written informed consent was obtained from all the parents of all participants enrolled.
Cell culture
The L0-2 normal human liver cell line was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich: Merck KGaA, Darmstadt, Germany), 50 U/ml penicillin and 50 µg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.). Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2. In the present study, cells up to passage 10 were used.
Cell transfection
All the procedures were performed at room temperature unless otherwise specified. ANXA2P3 small interfering RNA (siRNA) (si-ANXA2P3), non-targeting siRNA (si-NC) and ANXA2 siRNA (si-ANXA2) were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The siRNAs and si-NC were transfected into 2×105 L0-2 cells, after mixing with Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) for 20 min, at a final concentration of 50 nM, unless otherwise indicated. The siRNA sequences were as follows: si-NC for ANXA2P3: NR_001446.2 [National Center for Biotechnology Information (NCBI) reference sequence]; si-NC for ANXA2: NM_001002858.2 (NCBI reference sequence); scrambled negative control vector (OE-NC) for ANXA2: NM_001002858.2 (NCBI reference sequence); OE-NC for ANXA2P3: NR_001446.2 (NCBI reference sequence); si-ANXA2, 5′-GGA GTG AAG AGG AAA GGA ACT-3′; si-ANXA2P3-1, 5′-GGA TGG CTC TGT CG TTG ATT A-3′; si-ANXA2P3-2, 5′-GGT CAT CAC TCT ACA CCC TCA-3′; si-ANXA2P3-3, 5′-GGA GAG AGG ATG TTG CCT TTG-3′. In addition, pcDNA3.1 (+) expression vectors and OE-NC vectors were purchased from Shanghai GenePharma Co., Ltd. After culturing in antibiotic-free DMEM for 24 h, and once confluence reached >80%, a total of 0.5×105 cells were transfected with pcDNA vectors (multiplicity of infection, 20) and polybrene (final concentration, 5 µg/ml), according to the manufacturer’s protocol. A total of 48 h post-transfection, the transfection efficacy was assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Qualitative analysis of gene silencing was conducted by observing cells under a fluorescence microscope (Leica DMi1; Leica Microsystems GmbH, Wetzlar, Germany) after 48 h of transfection.
Total RNA extraction and RT-qPCR
Total RNA was isolated from liver samples from patients with BA and L0-2 liver cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, cDNA was synthesized from 200 ng extracted total RNA using the PrimeScript RT reagent kit (Qiagen China Co., Ltd., Shanghai, China), according to the manufacturer’s protocol. Briefly, 1 µg total RNA mixed with nuclease-free water was used for cDNA synthesis with the PrimeScript RT reagent kit (Takara Bio, Inc., Otsu, Japan), according to the manufacturer’s protocol. All RT procedures and no-template controls were run at the same time. Following RT, qPCR was conducted to determine the expression of ANXA2 and ANXA2P3 using SYBR Premix Ex Taq (Takara Bio, Inc.) on an ABI-7300 instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.). According to the manufacturer’s protocol, a final 20 µl reaction mixture was amplified using the following reaction conditions: Predenaturation at 94°C for 5 min; amplification for 35 cycles (denaturation at 94°C for 30 sec, annealing at 58°C for 30 sec and extension at 72°C for 30 sec); final extension at 72°C for 5 min. The 20 µl PCR mixture included 2 µl cDNA, 10 µl 2X QuantiTect SYBR-Green PCR Mix, 2 µl 10X upstream primer (forward), 2 µl 10X downstream primer (reverse) and 4 µl RNase-free water. GAPDH served as the control gene. The 2−ΔΔCq method was used to determine gene expression levels (30). Primer sequences were as follows: ANXA2P3 forward, 5′-GAG AGG ATG TTG CCT TTG-3′ reverse, 5′-TAC TGA GCA GGT GTC TTC-3′; GAPDH forward, 5′-AAT CCC ATC ACC ATC TTC-3′ and reverse, 5′-AGG CTG TTG TCA TAC TTC-3′; and ANXA2 forward, 5′-ATG GTC TCC CGC AGT GAA GTG-3′ and reverse, 5′-TCG CCC TTA GTG TCT TGC TGG-3′.
Immunohistochemical analysis
All procedures were performed at room temperature unless otherwise specified. Liver tissue (~20 mm) was dissected, post-fixed in 0.1 M PBS containing 4% paraformaldehyde for 24 h at 4°C and embedded in paraffin. Paraffin-embedded liver samples were cut longitudinally with a microtome, to obtain 4-µm sections. Sections were rehydrated in a microwave oven at 37°C for 30 min, and sections on microscope slides were immersed in xylene and a diluted alcohol series (95, 90, 80 and 70%) for 15 min, in order to dewax and rehydrate paraffin-embedded sections. Subsequently sections were immersed in 3% H2O2 for 10 min to quench endogenous peroxidase activity. Sections were then rinsed two times in distilled water (5 min/wash), and antigen retrieval was performed using 0.01 mol/l citrate buffer solution (pH 6.0) in a microwave oven for 20 min. Once sections were cooled to room temperature, they were rinsed a further three times in 0.1 M PBS (5 min/wash), and were incubated with 5% bovine serum albumin blocking solution for 20 min to block non-specific binding. Subsequently, sections were incubated overnight with an ANXA2 antibody (1:100; cat. no. ab41803; Abcam, Cambridge, MA, USA) at 4°C. Sections were then rinsed a further three times in 0.1 M PBS (5 min/wash), and were incubated with IRDye 800-conjugated immunoglobulin G secondary antibodies (1:1,000; cat. no. ab150077; Abcam) in Tris-buffered saline containing 0.1% Tween-20 (TBST) for 30 min at room temperature. The slides were stained with DAB and counterstained with hematoxylin, prior to visualization under a Leica microscope (DFC490; Leica Microsystems GmbH). LAS AF Lite V3.6 software (Leica Microsystems GmbH) was used to analyze the images.
Histopathological examination
Collagen deposition in the liver was evaluated by Masson staining. All procedures were performed at room temperature unless otherwise specified. Liver tissues were fixed with 10% neutral formalin for 48-72 h, embedded in paraffin, continuously sectioned at a thickness of 5 µm, and stained with Masson stains. Briefly, slides were placed in Bouin solution (Richard Allen Scientific; Thermo Fisher Scientific, Inc.) for 1 h at 56°C, after which, the slides were stained with Weigert hematoxylin (Sigma-Aldrich; Merck KGaA) for 10 min, followed by Biebrich scarlet-acid fuchsin for 5 min, phosphomolybdic/phosphotungstic acid solution for 10 min and aniline blue (all Sigma-Aldrich; Merck KGaA) for 5 min. Hepatic fibrosis was graded based on the internationally used Metavir scoring system (31).
Western blot analysis
All the procedures were performed at room temperature unless otherwise specified. Total protein was extracted using mammalian protein extraction reagent (Pierce; Thermo Fisher Scientific, Inc.) supplemented with a protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA). Total protein concentration was determined using a bicinchoninic acid assay kit. Protein samples (50 µg) were resolved by 10% SDS-PAGE and were then transferred onto polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). Membranes were blocked in TBST containing 5% w/v non-fat milk at room temperature for 1 h, and were then probed with the following antibodies overnight: Anti-ANXA2 (1:1,000; rabbit; cat. no. ab41803) and anti-GAPDH (1:2,500; rabbit; cat. no. ab9485; both Abcam). Subsequently, membranes were incubated for 2 h with specific horseradish peroxidase-conjugated secondary antibody (1:1,000; anti-rabbit immunoglobulin G; cat. no. ab150077; Abcam) in TBST. Proteins were visualized using an enhanced chemiluminescence kit (Pierce; Thermo Fisher Scientific, Inc.) and densitometric analysis was conducted using ImageJ software (V1.51; National Institutes of Health, Bethesda, MD, USA).
Cell viability assay
All the procedures were performed at room temperature unless otherwise specified. Transfected L0-2 cells were seeded into a 96-well plate at a density of 2,000 cells/well and were incubated at 37°C. Proliferation was determined using a Cell Counting kit (CCK)-8 kit (Nanjing Keygen Biotech Co., Ltd., Nanjing, China) at 24, 48 (optional), 72 and 96 h post-transfection, according to the manufacturer’s protocol. Optical density was measured at a wavelength of 450 nm using a microtiter plate reader.
Cell cycle analysis
All the procedures were performed at room temperature unless otherwise specified. Transfected L0-2 cells were washed in PBS and fixed in 70% ethanol for 2 h at 4°C. DNA staining was conducted with propidium iodide for 15 min in the dark using a Cellular DNA Flow Cytometric Analysis kit (Roche Diagnostics). Cell cycle profiles were generated using a FACSCalibur flow cytometer with ModFit 3.0 software (both BD Biosciences, San Jose, CA, USA).
Cell apoptosis assay
Transfected L0-2 cells were stained using an Annexin V-fluorescein isothiocyanate Apoptosis Detection kit I (BD Biosciences), according to the manufacturer’s protocol. Subsequently, cells were analyzed using a FACSCalibur flow cytometer equipped with CellQuest software (version 5.1; BD Biosciences). Cells were divided into viable cells, necrotic cells, early apoptotic cells and late apoptotic cells.
Statistical analysis
Statistical analyses were performed using SPSS version 18.0 software (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism (version 6.01) software (GraphPad Software, Inc., La Jolla, CA, USA). ImageJ software (National Institutes of Health) was used for immunohistochemical analysis and densitometric analysis of western blotting, which was normalized to the respective loading controls. Data are presented as the means ± standard deviation of at least three independent experiments. The statistical significance between groups was determined using the Student’s t-test and one-way analysis of variance followed by a post hoc Tukey’s test for multiple comparisons. All P-values are two-sided and P<0.05 was considered to indicate a statistically significant difference.
Results
ANXA2 and ANXA2P3 expression is increased in liver tissues from patients with BA
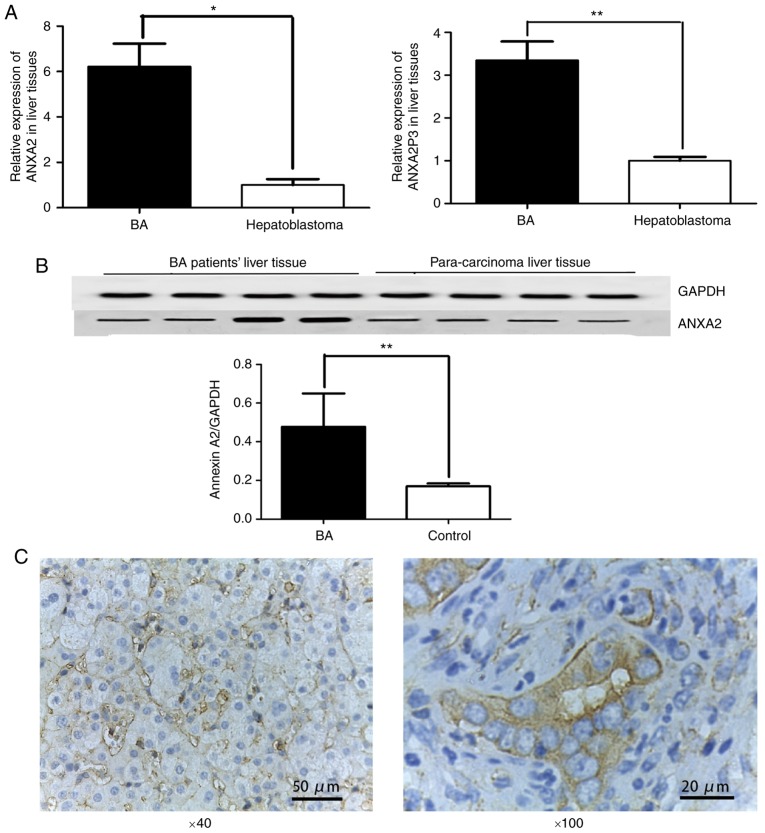
RT-qPCR was performed to determine the expression levels of ANXA2 and ANXA2P3 in clinical samples normalized to GAPDH. The results revealed that the expression levels of ANXA2 and ANXA2P3 were significantly increased in liver tissues from patients with BA compared with in paracarcinoma liver tissues from patients with hepatoblastoma (P=0.0210 and 0.0083, respectively; Fig. 1A). Western blot analysis indicated that ANXA2 protein expression was increased in liver tissues from patients with BA when compared with paracarcinoma liver tissues from patients with hepatoblastoma (Fig. 1B). Immunohistochemical staining of liver tissues further confirmed that the protein expression levels of ANXA2 were abundant in the cell membrane in patients with BA (Fig. 1C). The present study also evaluated the clinicopathological characteristics of patients with BA, and revealed that all patients had liver fibrosis, varying between stages 1 and 3. A total of four patients were at stage 1, 14 patients were at stage 2 and two patients were at stage 3 (data not shown). No significant associations were determined between the expression levels of ANXA2 and ANXA2P3 and other clinicopathological factors, including sex and age.
Figure 1.
ANXA2 and ANXA2P3 expression was detected in liver tissues from patients with BA. (A) Liver tissue from patients with BA (n=20) exhibited significantly higher ANXA2 and ANXA2P3 expression compared with in liver tissue from patients with hepatoblastoma (n=6). *P<0.05, **P<0.01. (B) Western blot analysis indicated that ANXA2 protein expression was increased in liver tissue from patients with BA compared with in liver tissue from patients with hepatoblastoma (n=4). **P<0.01. (C) Immunohistochemical staining revealed that ANXA2 protein was expressed in liver tissue from patients with BA. Data are presented as the means ± standard deviation. P-values were obtained by Student’s t-test. ANXA2, Annexin A2; ANXA2P3, ANXA2 pseudogene 3; BA, biliary atresia.
ANXA2 and ANXA2P3 knockdown inhibits liver cell proliferation, increases cell apoptosis and induces cell cycle arrest in G1 phase in vitro
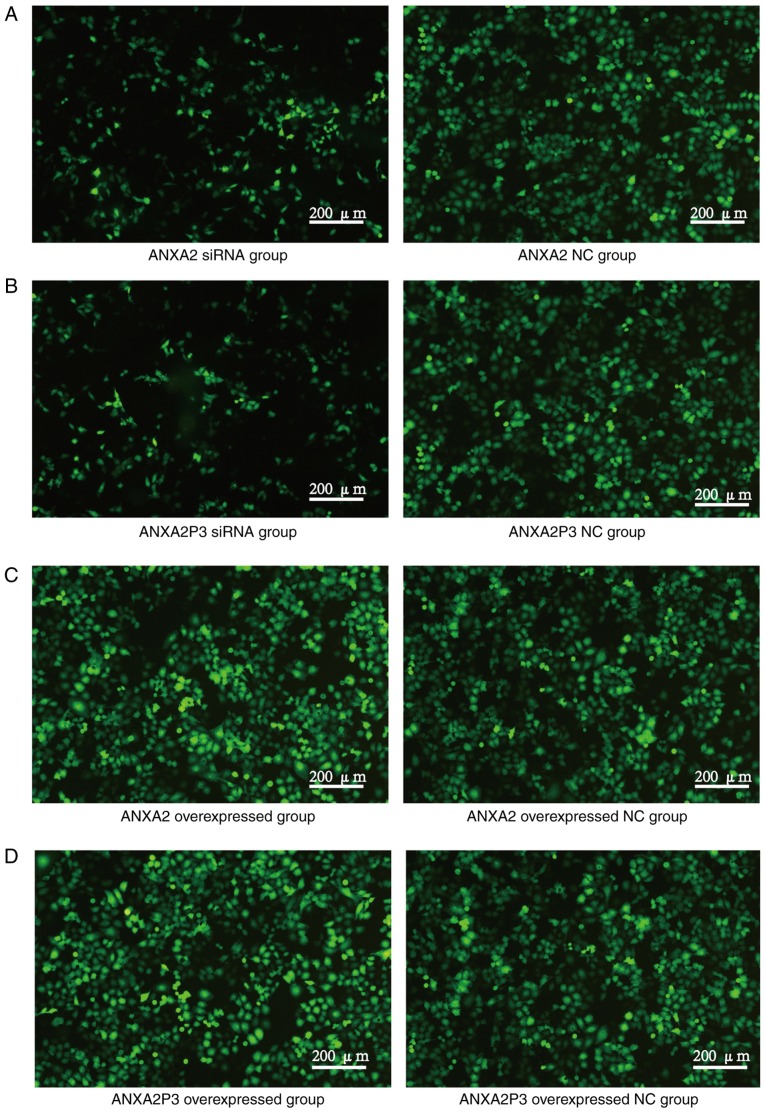
To elucidate the role of ANXA2 and ANXA2P3 in liver injury progression, the expression levels of ANXA2 and ANXA2P3 were downregulated via siRNA transfection and overexpressed via pcDNA3.1 plasmid vector transfection in normal liver cells. Cells were transfected with green fluorescent protein to detect fluorescence (Fig. 2). RT-qPCR results revealed that in siRNA-transfected liver cells the expression levels of ANXA2 and ANXA2P3 were markedly reduced compared with in si-NC-transfected cells (Fig. 3A and B). Conversely, in pcDNA3.1 plasmid vector-transfected liver cells (OE-ANXA2 and OE-ANXA2P) the expression levels of ANXA2 and ANXA2P3 were significantly increased compared with in the OE-NC group (Fig. 3A and B). Using the CCK-8 kit, it was revealed that knockdown of ANXA2P3 or ANXA2 in liver cells inhibited cell proliferation; however, the results were not significant (Fig. 4).
Figure 2.
Detection of green fluorescent protein in transfected liver cells. (A) ANXA2 and (B) ANXA2P3 siRNA groups. (C) ANXA2 and (D) ANXA2P3 overexpressed groups. ANXA2, Annexin A2; ANXA2P3, ANXA2 pseudogene 3; NC, negative control; siRNA, small interfering RNA.
Figure 3.
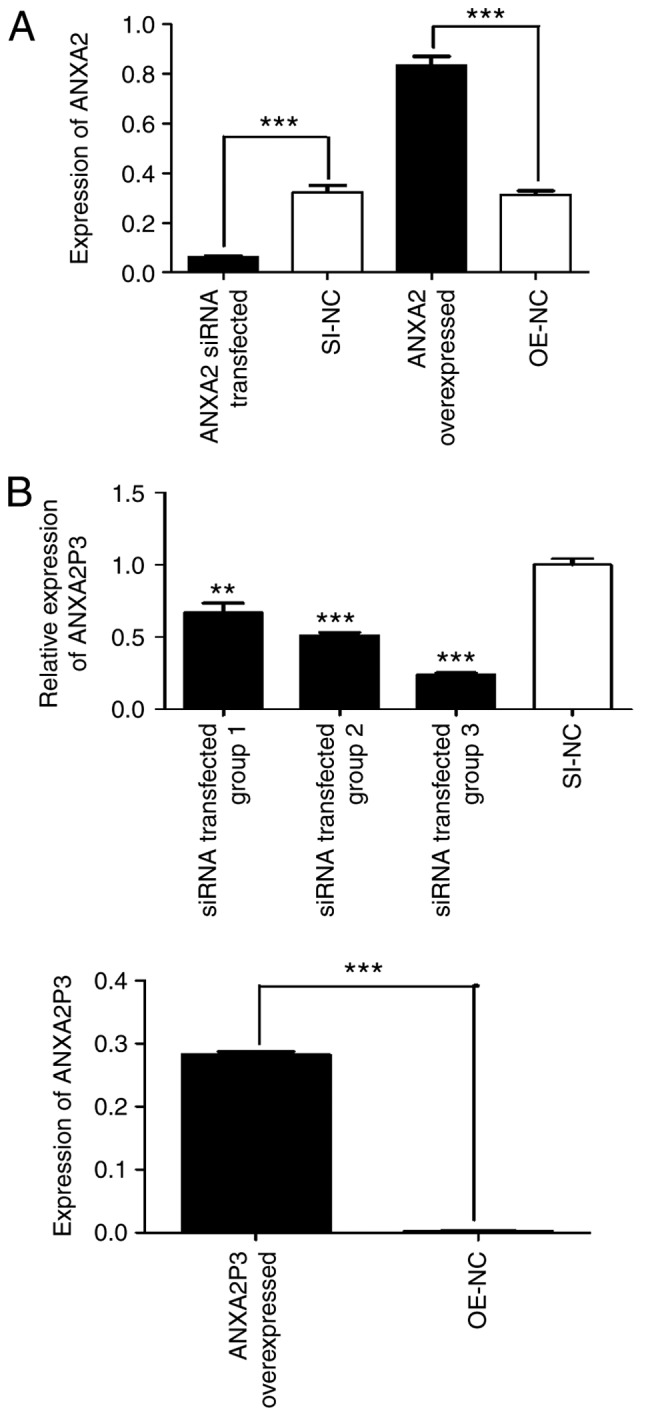
ANXA2 and ANXA2P3 expression was detected using reverse transcription-quantitative polymerase chain reaction. (A) Expression levels of ANXA2 and (B) ANXA2P3 in siRNA- and pcDNA3.1 plasmid vector-transfected liver cells. Data are presented as the means ± standard deviation. P-values were obtained by Student’s t-test. For all experiments, n=3. **P<0.01, ***P<0.001. ANXA2, Annexin A2; ANXA2P3, ANXA2 pseudogene 3; NC, negative control; OE, overexpression; SI/siRNA, small interfering RNA.
Figure 4.
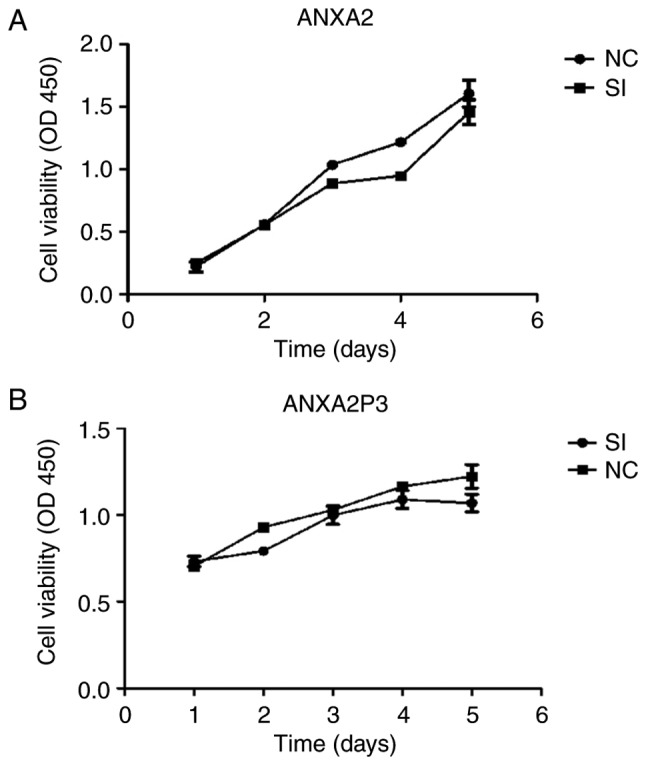
Cell Counting kit-8 assay was used to determine cell viability. Cell viability of (A) si-ANXA2- and (B) si-ANXA2P3-transfected cells was detected. ANXA2, Annexin A2; ANXA2P3, ANXA2 pseudogene 3; NC, negative control; OD, optical density; SI/siRNA, small interfering RNA.
Using flow cytometry, it was revealed that si-ANXA2P3-transfected liver cells were arrested in G1 phase compared with si-NC-transfected cells; conversely, OE-ANXA2P3-transfected liver cells were arrested in G2 phase compared with OE-NC-transfected cells (Fig. 5). When compared with the si-NC group, flow cytometric analysis of si-ANXA2-transfected liver cells exhibited similar results as in the si-ANXA2P3 group; an increased proportion of liver cells was arrested in G1 phase compared with si-NC-transfected cells. Furthermore, OE-ANXA2-transfected liver cells were arrested in G2 phase compared with OE-NC-transfected cells (Fig. 6). According to the results of flow cytometric analysis, it was revealed that siANXA2P3-transfected liver cells exhibited an increased apoptotic rate when compared with the si-NC group, whereas opposite findings were detected in the OE-ANXA2P3 group, in which the apoptotic rate was reduced compared with in the OE-NC group (Fig. 7). Similarly, siANXA2-transfected liver cells exhibited an increased apoptotic rate compared with the si-NC group, whereas the OE-ANXA2 group exhibited a reduced apoptotic rate compared with the OE-NC group (Fig. 8).
Figure 5.
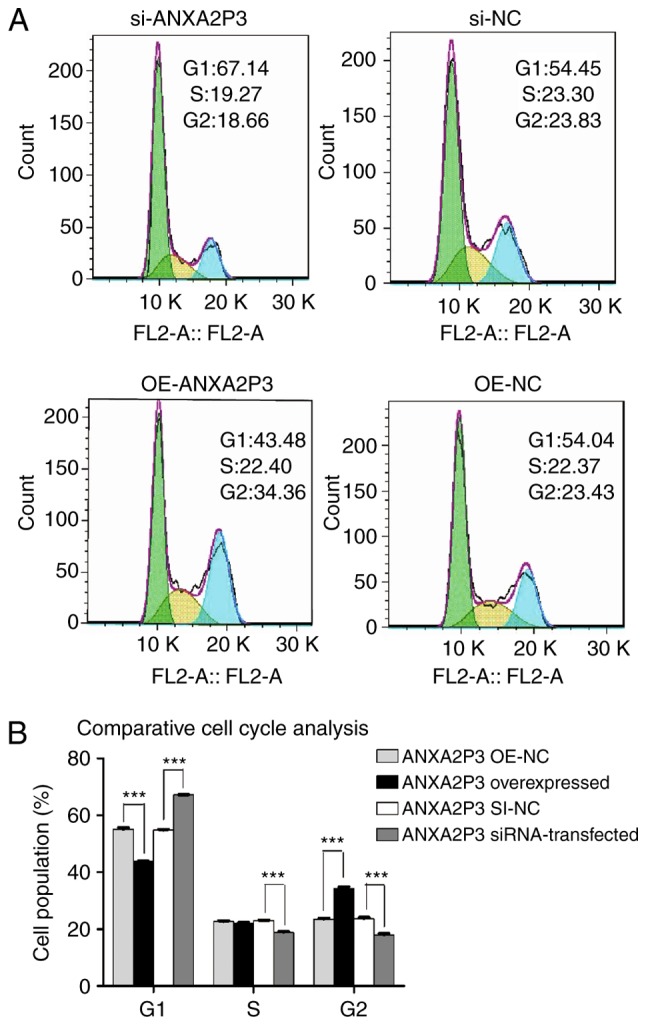
Results of flow cytometric analysis of cell cycle progression. (A) si-ANXA2P3-transfected liver cells exhibited cell cycle arrest in the G1 phase compared with si-NC cells. si-ANXA2P3 group: 67.14% in G1 phase, 19.27% in S phase and 18.66% in G2 phase; si-NC group: 54.45, 23.3 and 23.83%, respectively. In the OE-ANXA2P3 group, the percentage of cells in G1 phase, S phase and G2 phase was 43.48, 22.40 and 34.36%, respectively. Conversely, in the OE-NC group, the percentage of cells in G1 phase, S phase and G2 phase was 54.04, 22.37 and 23.43%, respectively. (B) Summary of cell cycle distribution in transfected liver cells. Data are presented as the means ± standard deviation. P-values were obtained by Student’s t-test. For all experiments, n=3. ***P<0.001. ANXA2P3, Annexin A2 pseudogene 3; NC, negative control; OE, overexpression; SI/siRNA, small interfering RNA.
Figure 6.
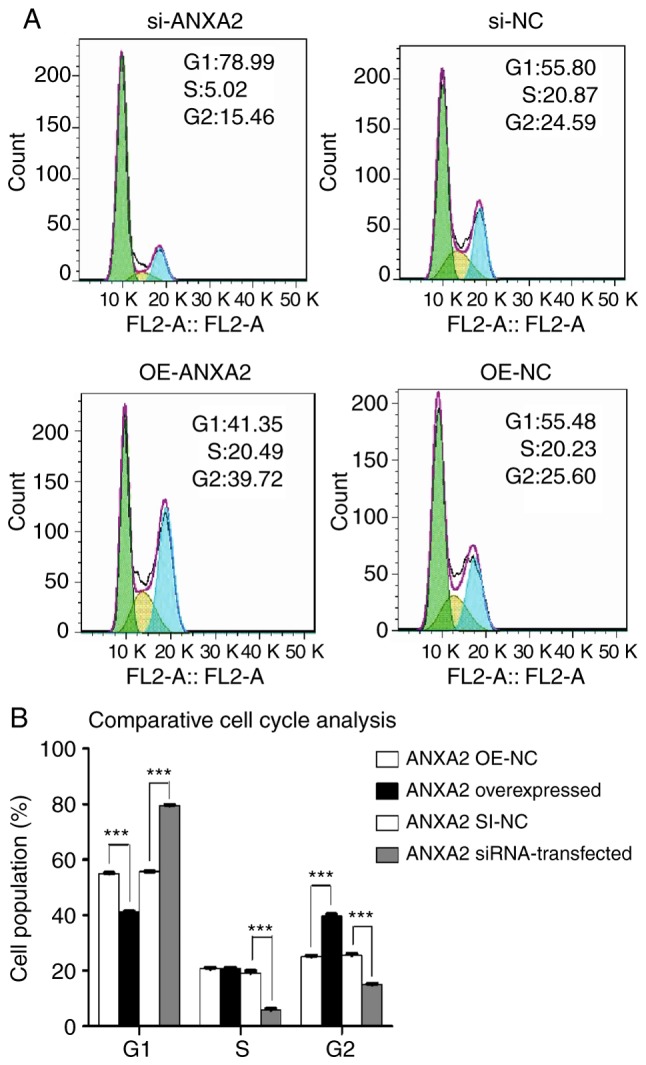
Results of flow cytometric analysis of cell cycle progression. (A) Of the si-ANXA2-transfected human liver cells, 78.99% were in G1 phase, 5.02% were in S phase and 15.46% were in G2 phase. In the si-NC group, 55.80, 20.87 and 24.59% cells were in G1, S and G2 phases, respectively. In the OE-ANXA2 group, the percentage of cells in G1 phase, S phase and G2 phase was 41.35, 20.49 and 39.72%, respectively. Conversely, in the OE-NC group, the percentage of cells in G1 phase, S phase and G2 phase was 55.48, 20.23 and 25.6%, respectively. (B) Summary of cell cycle distribution in transfected liver cells. Data are presented as the means ± standard deviation. P-values were obtained by Student’s t-test. For all experiments, n=3. ***P<0.001. ANXA2, Annexin A2; NC, negative control; OE, overexpression; SI/siRNA, small interfering RNA.
Figure 7.
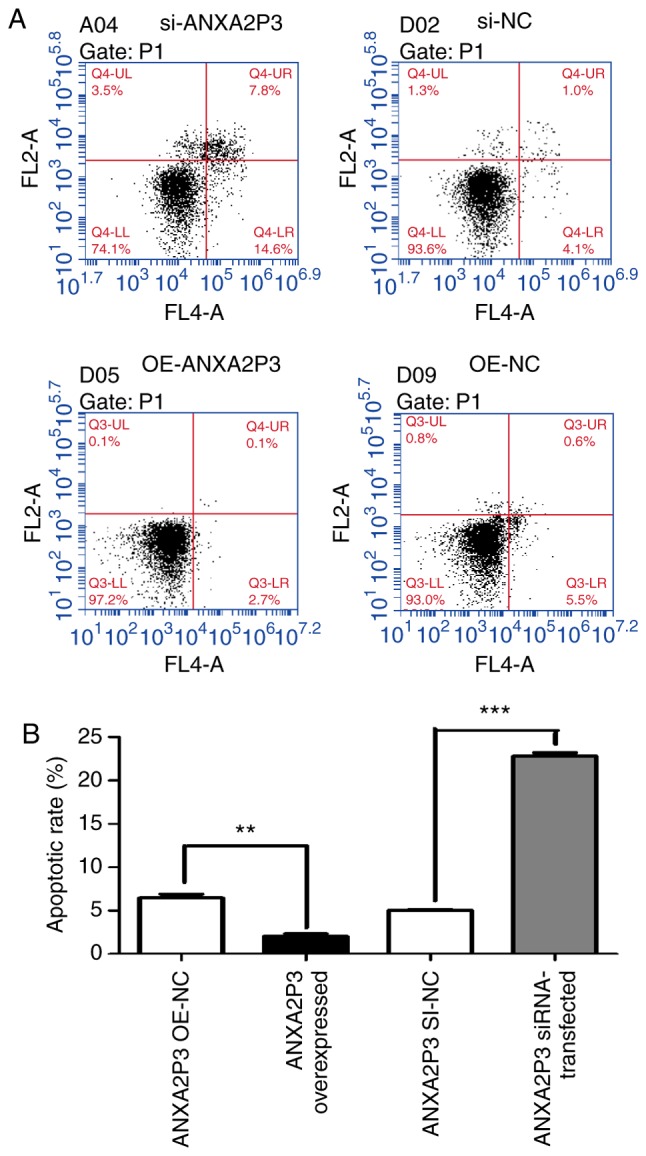
Results of flow cytometric analysis of apoptosis. (A) Apoptotic rate of si-ANXA2P3-transfected liver cells was increased compared with si-NC-transfected liver cells. In the si-ANXA2P3 group, the values were as follows: Normal cells, 74.1%; early apoptotic cells, 14.6%; late apoptotic cells, 7.8%; and necrotic cells, 3.5%. Conversely, in the si-NC group, the values were as follows: Normal cells, 93.6%; early apoptotic cells, 4.1%; late apoptotic cells, 1.0%; and necrotic cells, 1.3%. In the OE-ANXA2P3 group, the values were as follows: Normal cells, 97.2%; early apoptotic cells, 2.7%; late apoptotic cells, 0.1%; and necrotic cells, 0.1%. Conversely, in the OE-NC group, the values were as follows: Normal cells, 93.0%; early apoptotic cells, 5.5%; late apoptotic cells, 0.6%; and necrotic cells and 0.8%. (B) Summary of cell apoptotic rates. Cells in the upper right quadrant are late apoptotic cells and cells in the lower right quadrant are early apoptotic cells. The total apoptotic rate of cells is the sum of the apoptotic rates of the upper right and lower right quadrants. Data are presented as the means ± standard deviation. P-values were obtained by Student’s t-test. For all experiments, n=3. **P<0.01, ***P<0.001. ANXA2P3, Annexin A2 pseudogene 3; NC, negative control; OE, overexpression; si/siRNA, small interfering RNA.
Figure 8.
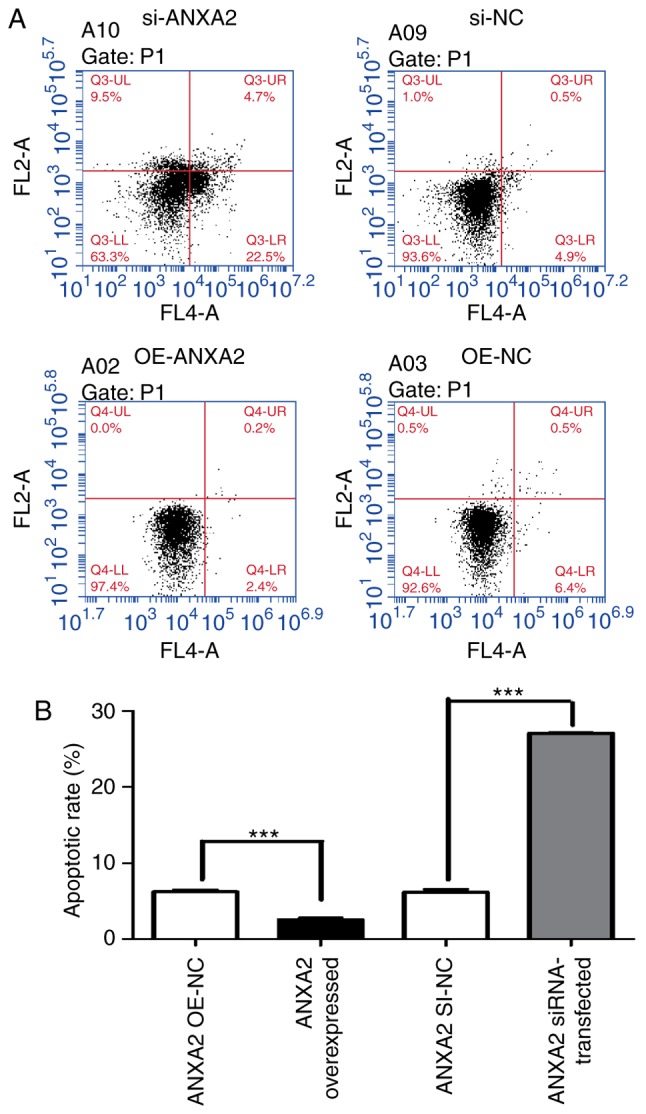
Results of flow cytometric analysis of apoptosis. (A) si-ANXA2- transfected liver cells exhibited a higher apoptotic rate compared with si-NC-transfected liver cells. The values for the si-ANXA2 group were as follows: Normal cells, 63.3%; early apoptotic cells, 22.5%; late apoptotic cells, 4.7%; and necrotic cells 9.5%. In the si-NC group, the values were as follows: Normal cells, 93.6%; early apoptotic cells, 4.9%; late apoptotic cells, 0.5%; and necrotic cells, 1.0%. In the OE-ANXA2 group, the values were as follows: Normal cells, 97.4%; early apoptotic cells, 2.4%; late apoptotic cells, 0.2%; and necrotic cells, 0.0%. Conversely, in the OE-NC group, the values were as follows: Normal cells, 92.6%; early apoptotic cells, 6.4%; late apoptotic cells, 0.5%; and necrotic cells, 0.5%. (B) Summary of cell apoptotic rates. Cells in the upper right quadrant are late apoptotic cells and cells in the lower right quadrant are early apoptotic cells. The total apoptotic rate of cells is the sum of the apoptotic rates of the upper right and lower right quadrants Data are presented as the means ± standard deviation. P-values were obtained by Student’s t-test. For all experiments, n=3. ***P<0.001. ANXA2, Annexin A2; NC, negative control; OE, overexpression; si/siRNA, small interfering RNA.
These in vitro findings suggested that overexpression of ANXA2 and ANXA2P3 may induce a more active liver cell phenotype. Conversely, inhibition of ANXA2 and ANXA2P3 may result in more negative consequences towards normal liver cells.
ANXA2P3 may activate ANXA2/ANXA2P3 signaling in liver cells
Western blot analysis revealed that knockdown of ANXA2P3 expression using si-ANXA2P3-2 and si-ANXA2P3-3 inhibited the protein expression levels of ANXA2 in vitro. Conversely, si-ANXA2P3-1 increased ANXA2 expression, which may be due to its reduced ANXA2P3 knockdown efficiency. Overexpression of ANXA2P3 markedly increased ANXA2 expression in vitro (Fig. 9). Taken together, these findings indicated that ANXA2P3 may affect the activity of the ANXA2/ANXA2P3 pathway in human liver cells in vitro.
Figure 9.
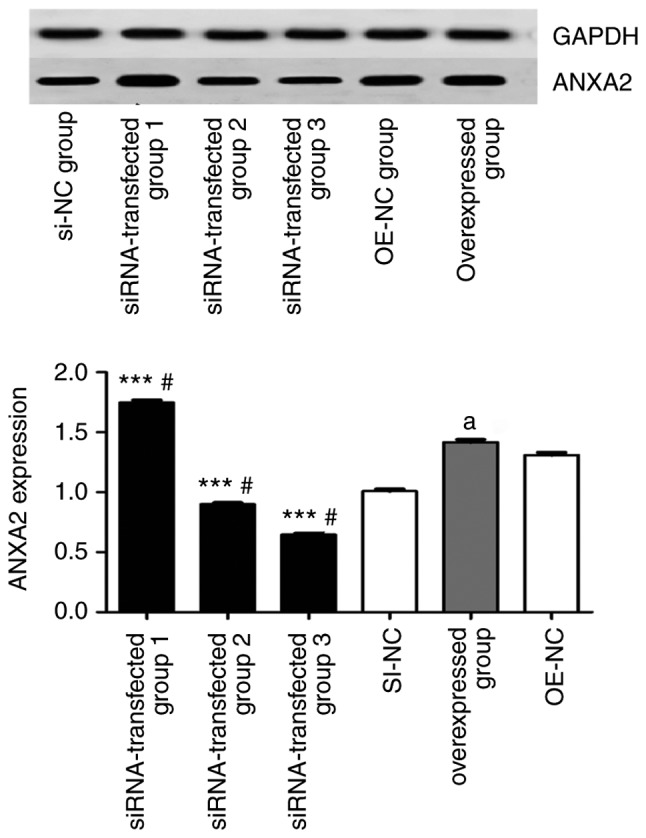
Inhibition of ANXA2P3 in siRNA groups 2 and 3 attenuates the expression of ANXA2, whereas overexpression of ANXA2P3 increases the expression of ANXA2 in vitro. Data are presented as the means ± standard deviation (n=3). P-values were obtained by one-way analysis of variance followed by a post hoc Tukey’s test for multiple comparisons. #P<0.05 compared with the SI-NC group; aP<0.05 compared with the OE-NC group; ***P<0.001 compared with the overexpressed group. ANXA2, Annexin A2; ANXA2P3, ANXA2 pseudogene 3; NC, negative control; OE, overexpression; si/siRNA, small interfering RNA.
Discussion
LncRNAs are mRNA-like transcripts >200 nucleotides in length that lack protein-coding functions (32). In recent decades, overwhelming evidence has indicated that lncRNAs are implicated in a wide range of biological functions, including cell proliferation, apoptosis and metastasis (33,34). The association between lncRNA dysregulation and the development of fibrosis is considered a major focus of hepatology studies. The present study aimed to detect ANXA2 and ANXA2P3 expression in clinical samples and to investigate their effects on liver cells in vitro.
Aberrant activation of the canonical ANXA2P3/ANXA2 signaling pathway is often observed during the initiation and progression of liver fibrosis. In previous liver fibrosis studies, the expression of ANXA2 was revealed to be positively correlated with ANXA2P3 (26,27). Therefore, it was hypothesized that an ANXA2P3/ANXA2 signaling pathway exists, which may serve an important role in the process of BA-induced liver injury, which is characterized by liver fibrosis. According to a previous study regarding the functions of pseudogenes (35), it was suggested that abnormal expression of ANXA2P3 may influence the ANXA2P3/ANXA2 signaling pathway. In addition, the present study revealed that downregulation of ANXA2P3 in liver cells, using si-ANXA2P3-2 and si-ANXA2P3-3, resulted in suppression of the ANXA2P3/ANXA2 signaling pathway. Since there is a close association between these genes, functional analyses of both ANXA2P3 and ANXA2 were conducted.
The present study identified the role of the ANXA2 and ANXA2P3 in BA-induced liver injury, which is characterized by liver fibrosis. The results revealed that the expression levels of ANXA2 and ANXA2P3 were significantly elevated in tissues derived from patients with BA when compared with in paracarcinoma liver tissues from patients with hepatoblastoma. These findings indicated that upregulation of ANXA2P3 and ANXA2 may be positively associated with the characteristics of liver injury. Since aberrant cell proliferation and dysregulated cell cycle progression are two main features of fibrosis-associated liver injury, further in vitro mechanistic experiments were conducted. Previous studies reported that ANXA2 is closely associated with cell proliferation and cell cycle progression, although the cells used in these studies differ (23,24,36). In the present in vitro mechanistic studies, it was revealed that knockdown of ANXA2 and ANXA2P3 reduced cell proliferation and promoted cell apoptosis. Furthermore, overexpression of ANXA2 and ANXA2P3 accelerated cell cycle progression and slightly inhibited cell apoptosis. Therefore, ANXA2 and ANXA2P3 expression may have promising effects on liver cell cycle progression and cell proliferation. To the best of our knowledge, the present study is the first to report the biological function of ANXA2 and ANXA2P3 in liver cells, which contributes toward improving the understanding of the mechanisms underlying liver injury progression and development. The present study indicated that, according to flow cytometry and cell cycle progression analysis, overexpression of ANXA2 and ANXA2P3 reduced cell apoptosis rates and induced cell cycle arrest in G2 phase; therefore, ANXA2 and ANXA2P3 may serve a protective role in the process of liver injury. In addition, the expression levels of ANXA2 and ANXA2P3 were significantly increased in patients with BA; all of these patients suffered from liver injury caused by liver fibrosis, as demonstrated by Masson staining. Therefore, it may be hypothesized that these genes are upregulated in order to exert protective effects and to hinder the process of liver injury, thus reversing the negative consequences of injury. However, the specific details and underlying mechanism of action require further elucidation.
Although the results of this study are promising, there are two potential limitations. Firstly, the clinical study involved retrospective validation, and the cohort of patients was with BA relatively small. Secondly, although the possible association between ANXA2P3 and ANXA2P3/ANXA2 signaling was revealed in liver cell lines, the underlying mechanisms by which ANXA2P3 exerts protective effects against liver injury remains to be elucidated.
In conclusion, to the best of our knowledge, the present study is the first to reveal that the expression levels of ANXA2P3 and ANXA2 are increased in the liver tissue of patients with BA, and their association with liver injury. ANXA2P3 expression may influence the biological behavior of liver cells through activation of ANXA2. In addition, the expression of both genes had a positive effect on cell proliferation and inhibited cell apoptosis. These findings suggested that ANXA2P3 and ANXA2 may be considered novel molecular targets for the prognosis and treatment of liver injury. However, a further investigation with a larger sample size is required to support these results.
Acknowledgments
Not applicable.
Funding
The present study received financial support from the Shanghai Key Disciplines (grant no. 2017ZZ02022), the National Natural Science Foundation of China (grant nos. 81770519 and 81771633) and the Shanghai Natural Science Foundation (grant no. 17ZR1403000).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors’ contributions
YN carried out the experiments and wrote the manuscript, with support from ZS and SZ. RD contributed to sample preparation, performed the calculations and designed the figures. ZS and SZ helped supervise the project and conceived the original idea. All authors discussed the results and contributed to the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethical Review Board of Fudan University Children’s Hospital, and written informed consent was obtained from the parents of all participants enrolled.
Patient consent for publication
Written informed consent for publication of the data was obtained from the participant and the patient’s family, according to federal and institutional guidelines.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Asai A, Miethke A, Bezerra JA. Pathogenesis of biliary atresia: Defining biology to understand clinical phenotypes. Nat Rev Gastroenterol Hepatol. 2015;12:342–352. doi: 10.1038/nrgastro.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dehghani SM, Efazati N, Shahramian I, Haghighat M, Imanieh MH. Evaluation of cholestasis in Iranian infants less than three months of age. Gastroenterol Hepatol Bed Bench. 2015;8:42–48. [PMC free article] [PubMed] [Google Scholar]
- 3.Hoerning A, Raub S, Dechêne A, Brosch MN, Kathemann S, Hoyer PF, Gerner P. Diversity of disorders causing neonatal cholestasis-the experience of a tertiary pediatric center in Germany. Front Pediatr. 2014;2:65. doi: 10.3389/fped.2014.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lishuang M, Zhen C, Guoliang Q, Zhen Z, Chen W, Long L, Shuli L. Laparoscopic portoenterostomy versus open portoenterostomy for the treatment of biliary atresia: A systematic review and meta-analysis of comparative studies. Pediatr Surg Int. 2015;31:261–269. doi: 10.1007/s00383-015-3662-7. [DOI] [PubMed] [Google Scholar]
- 5.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y, Wen L, Zhu H. Unveiling the hidden function of long non-coding RNA by identifying its major partner-protein. Cell Biosci. 2015;5:59. doi: 10.1186/s13578-015-0050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilczynska A, Bushell M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015;22:22–33. doi: 10.1038/cdd.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang XY, Pan HF, Leng RX, Ye DQ. Long noncoding RNAs: Novel insights into gastric cancer. Cancer Lett. 2015;356:357–366. doi: 10.1016/j.canlet.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Luo Y, Yang W, Ding L, Wang J, Tu J, Geng B, Cui Q, Yang J. Comparison analysis of dysregulated LncRNA profile in mouse plasma and liver after hepatic ischemia/reperfusion injury. PLoS One. 2015;10:e0133462. doi: 10.1371/journal.pone.0133462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Y, Wu YT, Huang C, Meng XM, Ma TT, Wu BM, Xu FY, Zhang L, Lv XW, Li J. Inhibitory effects of long noncoding RNA MEG3 on hepatic stellate cells activation and liver fibro-genesis. Biochim Biophys Acta. 2014;1842:2204–2215. doi: 10.1016/j.bbadis.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Qiao J, Yao H, Xia Y, Chu P, Li M, Wu Y, Li W, Ding L, Qi K, Li D, et al. Long non-coding RNAs expression profiles in hepatocytes of mice after hematopoietic stem cell transplantation. IUBMB Life. 2016;68:232–241. doi: 10.1002/iub.1479. [DOI] [PubMed] [Google Scholar]
- 12.Su S, Liu J, He K, Zhang M, Feng C, Peng F, Li B, Xia X. Overexpression of the long noncoding RNA TUG1 protects against cold-induced injury of mouse livers by inhibiting apoptosis and inflammation. FEBS J. 2016;283:1261–1274. doi: 10.1111/febs.13660. [DOI] [PubMed] [Google Scholar]
- 13.Liu H, Song G, Zhou L, Hu X, Liu M, Nie J, Lu S, Wu X, Cao Y, Tao L, et al. Compared analysis of LncRNA expression profiling in pdk1 gene knockout mice at two time points. Cell Physiol Biochem. 2013;32:1497–1508. doi: 10.1159/000356586. [DOI] [PubMed] [Google Scholar]
- 14.Xu D, Yang F, Yuan JH, Zhang L, Bi HS, Zhou CC, Liu F, Wang F, Sun SH. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/beta-catenin signaling. Hepatology. 2013;58:739–751. doi: 10.1002/hep.26361. [DOI] [PubMed] [Google Scholar]
- 15.Groen JN, Capraro D, Morris KV. The emerging role of pseudogene expressed non-coding RNAs in cellular functions. Int J Biochem Cell Biol. 2014;54:350–355. doi: 10.1016/j.biocel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piehler AP, Hellum M, Wenzel JJ, Kaminski E, Haug KB, Kierulf P, Kaminski WE. The human ABC transporter pseudogene family: Evidence for transcription and gene-pseudogene interference. BMC Genomics. 2008;9:165. doi: 10.1186/1471-2164-9-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin H, Shabbir A, Molnar M, Lee T. Stem cell regulatory function mediated by expression of a novel mouse Oct4 pseudogene. Biochem Biophys Res Commun. 2007;355:111–116. doi: 10.1016/j.bbrc.2007.01.106. [DOI] [PubMed] [Google Scholar]
- 18.Karreth FA, Reschke M, Ruocco A, Ng C, Chapuy B, Léopold V, Sjoberg M, Keane TM, Verma A, Ala U, et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161:319–332. doi: 10.1016/j.cell.2015.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Zou H, Shao YY, Mei Y, Cheng Y, Hu DL, Tan ZR, Zhou HH. Pseudogenes of annexin A2, novel prognosis biomarkers for diffuse gliomas. Oncotarget. 2017;8:106962–106975. doi: 10.18632/oncotarget.22197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan BL, Tong ZW, Wu L, Pan L, Li JE, Huang YG, Li SD, Du SX, Li XD. Effects of MicroRNA-206 on osteosarcoma cell proliferation, apoptosis, migration and invasion by targeting ANXA2 through the AKT signaling pathway. Cell Physiol Biochem. 2018;45:1410–1422. doi: 10.1159/000487567. [DOI] [PubMed] [Google Scholar]
- 21.Pérez-Sánchez G, Jiménez A, Quezada-Ramírez MA, Estudillo E, Ayala-Sarmiento AE, Mendoza-Hernández G, Hernández-Soto J, Hernández-Hernández FC, Cázares-Raga FE, Segovia J. Annexin A1, Annexin A2, and Dyrk 1B are upregulated during GAS1-induced cell cycle arrest. J Cell Physiol. 2018;233:4166–4182. doi: 10.1002/jcp.26226. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Cui Z, Yang S, Wu C, Li W, Bao G, Xu G, Sun Y, Wang L, Zhang J. The upregulation of annexin A2 after spinal cord injury in rats may have implication for astrocyte proliferation. Neuropeptides. 2017;61:67–76. doi: 10.1016/j.npep.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Stewart AG, Xia YC, Harris T, Royce S, Hamilton JA, Schuliga M. Plasminogen-stimulated airway smooth muscle cell proliferation is mediated by urokinase and annexin A2, involving plasmin-activated cell signalling. Br J Pharmacol. 2013;170:1421–1435. doi: 10.1111/bph.12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong Z, Yao M, Zhang H, Wang L, Huang H, Yan M, Wu W, Yao D. Inhibition of Annexin A2 gene transcription is a promising molecular target for hepatoma cell proliferation and metastasis. Oncol Lett. 2014;7:28–34. doi: 10.3892/ol.2013.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang SL, Pan DY, Gu C, Qin HF, Zhao SH. Annexin A2 silencing enhances apoptosis of human umbilical vein endothelial cells in vitro. Asian Pac J Trop Med. 2015;8:952–957. doi: 10.1016/j.apjtm.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Kolgelier S, Demir NA, Inkaya AC, Sumer S, Ozcimen S, Demir LS, Pehlivan FS, Arslan M, Arpaci A. Serum levels of Annexin A2 as a candidate biomarker for hepatic fibrosis in patients with chronic hepatitis B. Hepat Mon. 2015;15:e30655. doi: 10.5812/hepatmon.30655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Peng X, Zhang Z, Feng Y, Jia X, Shi Y, Yang H, Zhang Z, Zhang X, Liu L, et al. Subcellular proteome analysis unraveled annexin A2 related to immune liver fibrosis. J Cell Biochem. 2010;110:219–228. doi: 10.1002/jcb.22529. [DOI] [PubMed] [Google Scholar]
- 28.Yang M, Wang C, Li S, Xv X, She S, Ran X, Li S, Hu H, Hu P, Zhang D, et al. Annexin A2 promotes liver fibrosis by mediating von Willebrand factor secretion. Dig Liver Dis. 2017;49:780–788. doi: 10.1016/j.dld.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Dadhania VP, Muskhelishvili L, Latendresse JR, Mehendale HM. Hepatic overexpression of Annexin A1 and A2 in thioacetamide-primed mice protects them against acetaminophen-induced liver failure and death. Int J Toxicol. 2016;35:654–665. doi: 10.1177/1091581816659067. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Weerasooriya VS, White FV, Shepherd RW. Hepatic fibrosis and survival in biliary atresia. J Pediatr. 2004;144:123–125. doi: 10.1016/j.jpeds.2003.09.042. [DOI] [PubMed] [Google Scholar]
- 32.Blythe AJ, Fox AH, Bond CS. The ins and outs of lncRNA structure: How, why and what comes next. Biochim Biophys Acta. 2016;1859:46–58. doi: 10.1016/j.bbagrm.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 33.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non-coding RNA: A new player in cancer. J Hematol Oncol. 2013;6:37. doi: 10.1186/1756-8722-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pink RC, Carter DR. Pseudogenes as regulators of biological function. Essays Biochem. 2013;54:103–112. doi: 10.1042/bse0540103. [DOI] [PubMed] [Google Scholar]
- 36.Zhou X, Deng S, Liu H, Liu Y, Yang Z, Xing T, Jing B, Zhang X. Knockdown of ubiquitin protein ligase E3A affects proliferation and invasion, and induces apoptosis of breast cancer cells through regulation of annexin A2. Mol Med Rep. 2015;12:1107–1113. doi: 10.3892/mmr.2015.3549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.