Abstract
Pancreatic ductal adenocarcinoma (PDAC) remains a particularly lethal disease that is resistant to targeted therapies. Tyrosine kinase inhibitors (TKIs), including erlotinib and gefitinib, which block the action of the human epidermal growth factor receptor type 1 receptor, provide small increases in patient survival when administered with gemcitabine. The retinoblastoma (Rb) tumor suppressor protein is an additional target in pancreatic cancer, due to its documented inactivation in PDAC. The present study, using cell number, apoptosis and immunoblotting assays, aimed to evaluate the effects of activation of the Rb tumor suppressor via dephosphorylation by small interfering RNA-mediated phosphatase activation. In the Panc1, MIAPaCa-2 and Capan-2 pancreatic cancer cell lines, and in normal H6c7 cells, the effects of phosphatase activation on Rb were revealed to be dependent on expression of the p16 tumor suppressor, which regulates Rb phosphorylation. Phosphatase activation had no effect on non-transformed pancreatic epithelial cells. When comparing kinase inhibition with phosphatase activation, it was demonstrated that kinase inhibition reduced proliferation, whereas phosphatase activation induced apoptosis. Both treatments together resulted in a greater reduction of pancreatic cancer cells than either treatment alone. In addition, the effects of combination treatment of phosphatase activation with TKIs on cell number and activation of the signal transducer and activator of transcription 3 (STAT3) resistance pathway were determined. The combination of Rb phosphatase activation with TKIs resulted in a greater reduction in cell number compared with either treatment alone, without STAT3 pathway activation. These data suggested that targeting Rb phosphorylation by activating phosphatase may be a rational strategy to inhibit pancreatic tumor cell growth, without activation of acquired resistance.
Keywords: pancreatic cancer, Rb phosphorylation, p16, erlotinib, gefitinib, STAT3
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is associated with a high mortality rate, as it is often diagnosed at an advanced stage and is resistant to current therapies (1,2). Current treatment strategies largely comprise surgical and chemotherapy regimens, which have yielded only modest improvements in survival. Notably, survival of patients with PDAC has shown little improvement in the last four decades (3). Therefore, novel targeted therapies are urgently required for the treatment of patients with these conditions. Metastatic disease is often treated with the chemotherapeutic DNA synthesis inhibitor gemcitabine, in combination with the small molecule inhibitor tyrosine kinase inhibitor (TKI) erlotinib (4,5). Erlotinib acts as an inhibitor of the human epidermal growth factor (EGF) receptor type 1 receptor (EGFR), which is overexpressed in several types of cancer, including PDAC (6). EGFR activation stimulates downstream signaling pathways that promote proliferation and metastasis (3). Clinically, erlotinib plus gemcitabine treatment provides a modest increase in patient outcome over gemcitabine alone (5). However, further preclinical and clinical studies are required to address the significant problem of resistance that develops in response to several targeted therapies, also known as acquired resistance (7). One such drug-resistance mechanism activated during erlotinib treatment is the signal transducer and activator of transcription 3 (STAT3) pathway, which promotes proliferation, as well as differentiation, survival, inflammation and angiogenesis (8). Previous studies on lung and pancreatic cancer cells combining STAT3 inhibition with EGFR-targeted therapy exhibit increased efficacy (9,10).
Activating mutations of KRAS proto-oncogene, GTPase (KRAS), and inactivating mutations of the tumor suppressor genes cyclin-dependent kinase (CDK) inhibitor 2A (CDKN2A; also known as p16INK4a or p16), tumor protein p53 and SMAD family member 4 have been reported to promote carcinogenesis in PDAC (2). In particular, CDKN2A is most commonly inactivated by a homozygous deletion that leads to p16INK4a loss of function in >90% of PDAC cases (11,12). Inactivation of CDKN2A/p16 is believed to be an early event in pancreatic cancer progression, since its inactivation is detected in 40% of precursor pancreatic intraepithelial neoplastic lesions (13,14). In addition, CDKN2A has been identified as a gatekeeper gene in PDAC, which indicates its importance in this cancer type (15). Furthermore, recent evidence has suggested that the progression of PDAC may be due to high genomic instability in the form of chromothripsis, and CDKN2A has been identified as one of the genes lost by this mechanism (16). Finally, while KRAS mutation is thought to be the first and most frequent genetic disruption in PDAC, it has been reported that oncogenic KRAS function is controlled by the tumor suppressor function of p16INK4a (17). Therefore, downregulation of p16INK4a together with oncogenic activation of KRAS may cooperate to promote pancreatic tumorigenesis (18). p16INK4a blocks cell cycle progression by interacting with and inhibiting CDK4/6, thus resulting in reduced phosphorylation of the retinoblastoma (Rb) protein. Unphosphorylated Rb associates with the E2F transcription factor to inhibit the G1 to S transition (19). Treatments that target Rb phosphorylation in cancer cells have been developed and exhibit efficacy in Rb-positive cells. For example, palbociclib is an orally active CDK4/6-specific inhibitor that causes cell cycle arrest in PDAC and other cancer cell types (20-23). Notably, palbociclib was the first CDK4/6 inhibitor approved by the United States Food and Drug Administration for the treatment of advanced breast cancer in women with estrogen receptor-positive human epidermal growth factor receptor 2-negative disease (24). Notwithstanding the development of resistance that occurs in response to palbociclib, clinical trials testing CDK4/6 inhibitors for efficacy in PDAC are underway.
A novel approach has been developed that targets the Rb hyperphosphorylation present in cancer cells. Protein phosphatase 1 (PP1) is the major Rb phosphatase (25), and specificity toward substrates is imparted onto PP1 by various interacting proteins (26,27). In proliferating cells, PP1 is associated with a regulatory protein known as phosphatase nuclear targeting subunit (PNUTS) (28,29). Our previous study demonstrated that PNUTS inhibits PP1 activity toward Rb (30). It was further revealed that PNUTS blocks Rb binding to PP1, thus identifying the PNUTS:PP1 complex as a putative cancer drug target (31,32). PNUTS dissociation from PP1, which permits Rb dephosphorylation, occurs due to alterations in protein kinase A-mediated phosphorylation of PNUTS (33). Our previous study reported that when cells are exposed to stress, including hypoxia or treatment with chemotherapeutic drugs, PNUTS dissociates from PP1 and Rb is dephosphorylated (34). Furthermore, small interfering RNA (siRNA)-mediated PNUTS depletion in breast and colon cancer cells leads to an increase in PP1 phosphatase activity towards Rb, Rb dephosphorylation on several sites, and a 3-4-fold increase in apoptosis (35). Another study revealed that apoptosis induced by PNUTS depletion involved the phosphatase and tensin homolog tumor suppressor (36). However, in our previous studies the ability of PNUTS depletion to induce apoptosis was demonstrated to be dependent on Rb expression. For example, PNUTS knockdown has no effect on Rb-null Saos2 cells; however, sensitivity is restored upon stable expression of Rb (35). That the effect of PNUTS knockdown requires phosphorylated (p)-Rb has been revealed in studies of non-transformed cells. In cells that do not express hyperphosphorylated Rb, for example in MCF10A breast and CCD-18Co colon cells, PNUTS depletion has no effect on cell number (35). Furthermore, in a study using non-transformed epithelial breast or breast cancer cells grown in 3D culture, it was demonstrated that the effect of PNUTS knockdown is dependent on the expression of hyperphosphorylated Rb. PNUTS depletion reduces the number of MCF7 and MDA-MB-231 breast cancer cells (containing hyperphosphorylated Rb) but does not affect non-transformed MCF10A cells that lack hyperphosphorylated Rb or MDA-MB-468 breast cancer cells that are Rb-null (37). Therefore, our laboratory has developed a method to activate the Rb phosphatase targeting Rb phosphorylation in cells via knockdown of the PP1-binding protein, PNUTS.
The present study aimed to determine the effects of activating phosphatase activity toward Rb in pancreatic cancer cells. The results demonstrated that PNUTS depletion caused apoptosis in p16-deficient pancreatic cancer cells, but had no effect on p16-positive pancreatic cancer cells or human pancreatic ductal epithelial cells. The effects of palbociclib treatment were compared with those of PNUTS depletion with regards to cell number, and an additive effect was detected for the two treatments, with palbociclib inhibiting proliferation and PNUTS depletion inducing apoptosis. Using the currently utilized clinical treatments for PDAC, TKIs erlotinib and gefitinib, cell proliferation was measured in response to these treatments in combination with PNUTS depletion, in order to evaluate the usefulness of targeting Rb phosphorylation in reducing pancreatic cancer cell proliferation. Finally, it was revealed that activation of phosphatase in combination with erlotinib does not stimulate drug resistance in pancreatic cancer cells.
Materials and methods
Cell culture
Cell culture materials were obtained from Gibco; Thermo Fisher Scientific, Inc. (Waltham, MA, USA), unless otherwise indicated. The Panc1, MIAPaCa-2 and Capan-2 cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and were used within 4 months of receipt. Panc1 and MIAPaCa-2 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 µg/ml streptomycin and 2 mM glutamine (PSG). Capan-2 cells were grown in McCoys 5A media (ATCC) supplemented with 10% FBS and PSG. The H6c7 human pancreatic duct epithelial cell line was obtained from Kerafast, Inc. (Boston, MA, USA) and was used within 4 months of purchase. H6c7 cells were grown in keratinocyte serum-free media containing EGF and bovine pituitary extract (cat. no. 17005042; Gibco; Thermo Fisher Scientific, Inc.). Cells were routinely maintained at 37°C in a humidified atmosphere containing 5% CO, and were split 2-3 times weekly to maintain sub-confluent cultures. Cultures were routinely tested for mycoplasma contamination using the MycoFluor™ Mycoplasma Detection kit (Invitrogen; Thermo Fisher Scientific, Inc.).
siRNA transfection and immunoblotting
PNUTS depletion was performed using Dharmafect II (GE Healthcare Dharmacon, Inc., Lafayette, CO, USA). The RNA oligonucleotides used were generated based on the human mRNA for PNUTS, and the sequences were as follows: PNUTS RNA interference (RNAi) sequence 5, 5'-CAGCUAAACUGGUGAAGCA-3'; PNUTS RNAi sequence 7, 5'-CCUAAUGCCACCAAAGAGA-3'; or nontargeting RNAi (siCONTROL non-targeting siRNA ≥1; GE Healthcare Dharmacon, Inc.). The nontargeting RNAi has at least four mismatches with all known human, mouse and rat genes, which is sufficient to eliminate nonspecific silencing of genes with similar sequences. Sequence 5 was used in all experiments shown. Rb depletion was performed using SignalSilence® Rb siRNA1 (cat. no. 6451) from Cell Signaling Technology, Inc. (Danvers, MA, USA). Transfection mixtures (final concentration, 100 nM RNA) were added to 40-60% confluent Panc1, MIA PaCa-2, Capan-2 or H6c7 cells. After 48 h, the transfection mixtures were removed from the cells and the cells underwent cell counting and immunoblotting. Cell extracts were prepared after washing in ice-cold Tris-buffered saline [TBS; 25 mM Tris-HCl (pH 8.0) and 150 mM NaCl] and were lysed for 15 min at 4°C in EBC buffer [50 mM Tris-HCl (pH 8.0), 120 mM NaCl and 0.5% Nonidet P-40] containing 10 µg/ml protease inhibitors aprotinin and phenylmethylsulfonylfluoride. The lysates were cleared by centrifugation at 13,000 × g for 10 min at 4°C. Protein concentration was determined using the Bradford protein assay. Electrophoresis was performed using 4-20% gradient SDS-polyacrylamide gels containing 30 µg total cell protein in each sample lane. Following electrophoresis, the proteins were transferred to nitrocellulose membranes. Residual protein binding sites on the nitrocellulose membranes were blocked by incubation with TBS-0.5% Tween-20 (TBST) containing 4% non-fat dry milk for 30-60 min at room temperature (RT). Subsequently, the nitrocellulose membranes were incubated in TBST containing 2% non-fat dry milk and 1 µg/ml primary antibody overnight at 4°C. Antibodies were all used at a concentration of 1 µg/ml. After three washes with TBST (10 min/wash), the nitrocellulose membranes were probed with horseradish peroxidase-conjugated anti-immunoglobulin G antibodies [1:2,000; cat. nos. 1031-05 (anti-mouse) and 4050-05 (anti-rabbit); SouthernBiotech, Birmingham, AL, USA] for 1.5 h at RT and developed using enhanced chemiluminescence detection (Pierce; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The following primary antibodies were used in this study: p16 (cat. no. 554079), cyclin D3 (cat. no. 610279) and PNUTS (cat. no. 611060) (all from BD Biosciences, Franklin Lakes, NJ, USA); p-Rb 807/811 (cat. no. 8516), cleaved poly(ADP-ribose) polymerase (Parp; cat. no. 9541), c-jun (cat. no. 9165), p-c-jun (cat. no. 2361), EGFR (cat. no. 4267), p-EGFR (cat. no. 3777), STAT3 (cat. no. 4904), p-STAT3 (cat. no. 9145), proliferating cell nuclear antigen (PCNA; cat. no. 13110) (all from Cell Signaling Technology, Inc.); β-actin (cat. no. A1978; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany); Rb (cat. no. sc-102) and minichromosome maintenance complex component 7 (mcm7; cat. no. sc-9966) (both from Santa Cruz Biotechnology, Inc., Dallas, TX, USA).
Cell number and apoptosis assays
Erlotinib, gefitinib and palbociclib were obtained from Selleck Chemicals (Houston, TX, USA), and were used within 3 months of receipt. Erlotinib and gefitinib were dissolved in dimethyl sulfoxide (DMSO), palbociclib was dissolved in water. Dose-response curves were generated that identified the concentration of each drug required to reduce cell numbers between 20 and 60% after a 24-h treatment: 40 µM erlotinib, 4 µM gefitinib and 20 µM palbociclib. These concentrations are within the range of half maximal inhibitory concentrations reported to inhibit proliferation of pancreatic cancer cells in previous studies (21,38-40). Panc1 or MIAPaCa-2 cells were plated (3,000/well) in 96-well plates and allowed to proliferate for 24 h. Treated cells were subjected to PNUTS depletion for 48 h and control cells were treated with nontargeting control (NTC) RNA. After 2 days, cells were treated with erlotinib, gefitinib or palbociclib for 24 h, or with a vehicle control (DMSO for erlotinib/gefitinib, water for palbociclib). Cell number was measured using the CellTiter Glo-2 assay (Promega Corporation, Madison, WI, USA). Apoptosis was measured using the Cell Death Detection ELISA (cat. no. 11544675001; Roche Diagnostics, Indianapolis, IN, USA), which detects degraded DNA released from the nucleus into the cytoplasm. Both assays were performed according to the manufacturers' protocols. Briefly, in the apoptosis assay, 104 cells from each condition were lysed and subjected to a slow-spin centrifugation (300 × g for 5 min at 4°C) to pellet nuclei. Extracts from the cytoplasmic fraction were used to detect fragmented DNA and quantified on a microplate reader at 405 nm (Promega Corporation).
Statistical analysis
All experiments performed in this study were conducted at least three times. Numerical data are presented as the means ± standard deviation. All data from each experiment were analyzed by Student's t-test or one-way analysis of variance with Tukey's post hoc test for multiple comparisons using SPSS 19.0 (IBM Corp., Armonk, NY USA). P<0.05 was considered to indicate a statistically significant difference.
Results
PNUTS depletion in pancreatic cancer cells reduces cell number by activation of apoptosis
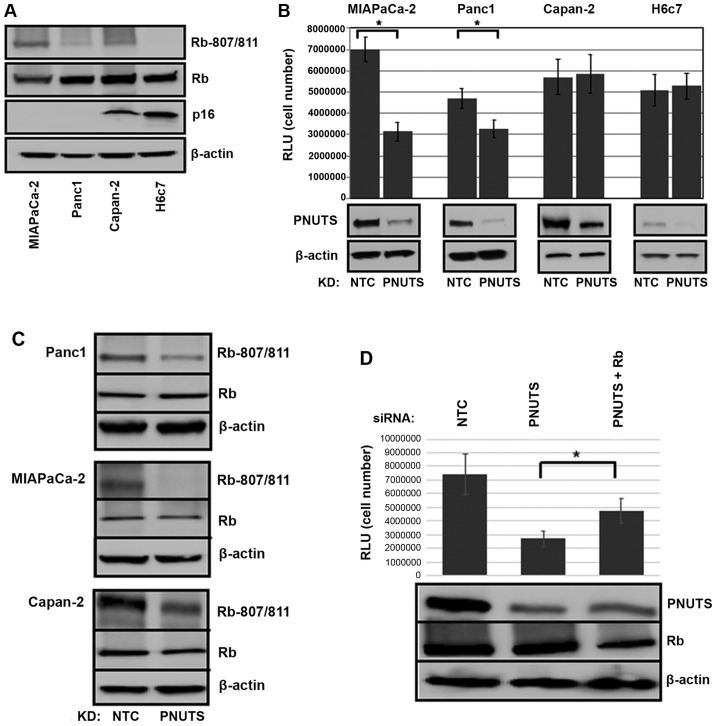
Hyperphosphorylation of Rb is a hallmark of most types of human cancer; therefore, we devised a method to target Rb phosphorylation by phosphatase activation. Our previous studies revealed that activation of phosphatase by PNUTS depletion is dependent on Rb (35,37). The function of p16 is disrupted in the majority of pancreatic carcinoma cases (11); in the present study it was demonstrated that the effects of PNUTS depletion on cell number were dependent on p16 expression in pancreatic cancer cells. Three pancreatic cancer cell types were used in this study: MIAPaCa-2 and Panc1 cells, which carry a homozygous deletion of the p16 gene; and Capan-2 cells, which are p16-positive (41-43). These cells have been extensively used to model pancreatic cancer. For comparison, experiments were also performed using the human pancreatic duct epithelial cell line H6c7 (44). As shown in Fig. 1A, Capan-2 and non-transformed H6c7 cells expressed p16, whereas MIAPaCa-2 and Panc1 cells were p16-deficient. p-Rb was also detected in the three cancer cell types, but was absent in the non-transformed cell line. As shown in Fig. 1B, Rb dephosphorylation initiated by PNUTS knockdown reduced cell number by 55% in MIAPaCa-2 and by 30% in Panc-1 cancer cells that contain hyperphosphorylated Rb; however, it had no effect on cell number in p16-positive Capan-2 cancer cells and in non-transformed H6c7 cells that lack hyperphosphorylated Rb. As shown in Fig. 1C, Rb was dephosphorylated in response to PNUTS depletion, and as shown in Fig. 1D, Rb depletion partially reversed the reduction in cell number induced by PNUTS depletion, indicating that the reduction in cell number due to PNUTS depletion may be Rb-dependent. Transfection with Rb siRNA alone was also confirmed to be successful (data not shown). The reduction in cell number observed may be due to cell proliferation arrest or an increase in apoptotic cell death. Assays using Panc1 and MIAPaCa-2 pancreatic cancer cells revealed that apoptosis was increased by ~4-fold due to PNUTS depletion in Panc1 cells and by ~7-fold in MIAPaCa-2 cells. Furthermore, the appearance of cleaved Parp in response to PNUTS depletion confirmed that apoptosis was induced (Fig. 2).
Figure 1.
(A) Immunoblotting was performed on whole cell lysates from MIAPaCa-2, Panc1, Capan-2 and H6c7 cell lines. Antibodies against p-Rb (S807/811), total Rb and p16 were used; β-actin was used as a loading control. Results shown are representative of three separate experiments. (B) MIAPaCa-2, Panc-1 and Capan-2 pancreatic cancer cells, and H6c7 normal pancreatic duct epithelial cells were subjected to PNUTS depletion for 48 h. Cell number was measured using the CellTiter Glo assay, with RLU proportional to cell number. Data shown are representative of three separate experiments. Results are expressed as the means ± standard deviation, n=8 replicates/experiment. *P<0.05, Student’s t-test. (C) Panc1, MIAPaCa-2 and Capan-2 cells were analyzed by immunoblotting with antibodies against p-Rb (Rb-807/811) or total Rb (Rb). (D) MIAPaCa-2 cells were transfected with PNUTS siRNA, or PNUTS and Rb siRNA. Cell number was measured using the CellTiter Glo assay. Data shown are representative of three separate experiments. *P<0.05, one-way analysis of variance and Tukey honestly significant difference test. KD, knockdown; NTC, nontargeting control; p, phosphorylated; PNUTs, phosphatase nuclear targeting subunit; Rb, retinoblastoma; RLU, relative light units; siRNA, small interfering RNA.
Figure 2.
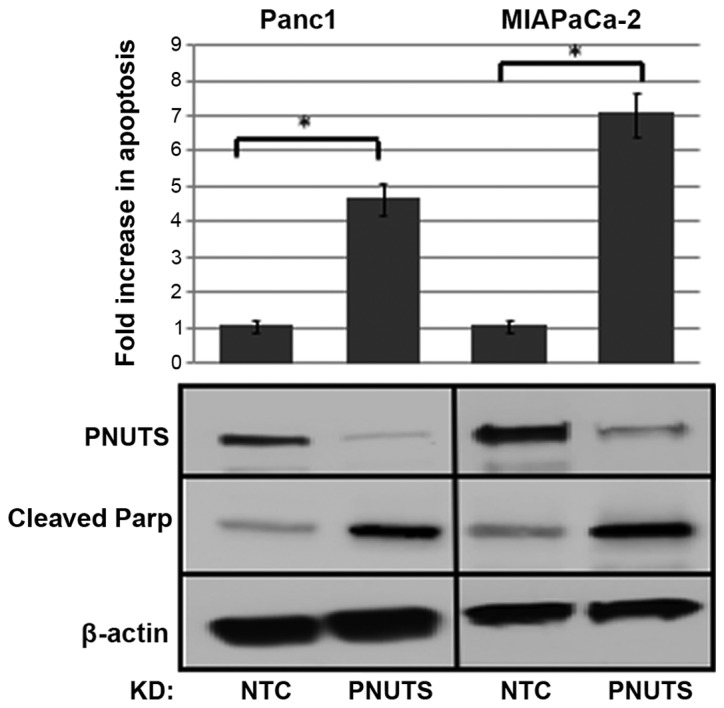
Panc1 and MIAPaCa-2 cells were subjected to PNUTS knockdown; after 48 h, apoptosis was measured using the Cell Death ELISA assay. Fold increase in apoptosis in treated cells relative to controls was shown. Data shown are representative of three separate experiments. Results are expressed as the means ± standard deviation, n=6 replicates/experiment. *P<0.05, Student’s t-test. Immunoblotting of PNUTS confirmed PNUTS depletion, whereas immunoblotting of cleaved Parp confirmed induction of apoptosis. KD, knockdown; NTC, nontargeting control; Parp, poly(ADP-ribose) polymerase; PNUTs, phosphatase nuclear targeting subunit.
Comparison of CDK inhibition with PNUTS depletion in pancreatic cancer cells
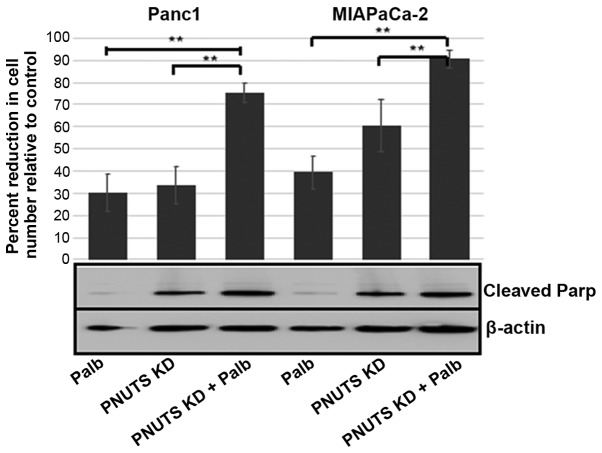
Since Rb is phosphorylated on 16 amino acid sites in vivo, it is difficult to elucidate the effects of phosphatases and kinases on the phosphorylation of Rb. To the best of our knowledge, the specific CDK responsible for each modification has only been partially elucidated to date (45). Furthermore, there appear to be overlapping functions of CDKs with regards to specific Rb sites. Our previous study demonstrated that activation of phosphatase toward Rb causes dephosphorylation of Rb at certain sites, and that these are distinct from those affected by the CDK2/5 inhibitor roscovitine (46). To compare the effects of CDK4/6 inhibition with that of PNUTS depletion on cell number in pancreatic cancer cells, it was first determined that 20 µM (24-h treatment) of palbociclib was an appropriate concentration to cause a 30-50% drop in cell number, and immunoblotting confirmed that Rb was dephosphorylated using this concentration (data not shown). The efficacy of palbociclib to reduce Panc1 cell number was comparable to the efficacy of PNUTS depletion (Fig. 3). To examine the combination of palbociclib and PNUTS knockdown, 48 h following PNUTS depletion (or NTC transfection), the medium was replaced with either 20 µM palbociclib or fresh medium (control). Finally, after 24 h, cells were counted using the CellTiter Glo assay. PNUTS depletion or palbociclib treatment each reduced cell number relative to controls by 30% in Panc1 cells; a higher efficiency was observed in response to PNUTS depletion in MIAPaCa-2 cells. However, when combining the two treatments, cell number was reduced by 75-90% in the two cell lines. In addition, while PNUTS depletion induced Parp cleavage, palbociclib treatment did not. These results suggested that the inhibition of CDK activity towards Rb and activation of phosphatase activity towards Rb are not functionally equivalent, and may influence Rb phosphorylation at distinct sites.
Figure 3.
Panc1 or MIAPaCa-2 cells were subjected to either NTC or PNUTS knockdown for 48 h and/or treatment with Palb (20 µM) for 24 h. Combination treatments were conducted by performing KD first, followed by Palb or control treatment. Cells were counted using the CellTiter Glo assay. The percentage reduction in cell number is relative to controls (NTC-transfected cells not treated with Palb). Data shown are representative of three separate experiments. Results are expressed as the means ± standard deviation, n=8 replicates/experiment. **P<0.01, one-way analysis of variance and Tukey honestly significant difference test. Immunoblotting of cleaved Parp was also conducted. KD, knockdown; NTC, nontargeting control; Palb, palbociclib; Parp, poly(ADP-ribose) polymerase; PNUTs, phosphatase nuclear targeting subunit.
Differential phosphorylation of Rb may impart distinctive consequences in cells. Using Panc1 cells, the effects of 20 µM palbociclib and PNUTS depletion on proteins that control the processes of proliferation and apoptosis were determined. As shown in Fig. 4, palbociclib treatment induced a decrease in proteins involved in proliferation (cyclin D3, mcm7 and PCNA), but did not increase the expression of proteins involved in apoptosis (cleaved Parp, c-jun and p-c-jun), thus suggesting that palbociclib inhibited proliferation but had no effect on cell death. Conversely, PNUTS depletion led to an increase in apoptosis-associated proteins but did not affect proliferation-associated proteins.
Figure 4.
Panc1 cells were treated with Palb (20 µM) for 24 h, or were transfected with NTC or PNUTS siRNA for 48 h. Subsequently, immunoblotting was conducted to analyze the expression of markers of proliferation and apoptosis. Rb phosphorylation (S807/S811), total Rb, β-actin and PNUTS expression was also detected. Data shown are representative of three independent experiments. CT, control; KD, knockdown; mcm7, minichromosome maintenance complex component 7; NTC, nontargeting control; Palb, palbociclib; p, phosphorylated; Parp, poly(ADP-ribose) polymerase; PCNA, proliferating cell nuclear antigen; PNUTs, phosphatase nuclear targeting subunit; Rb, retinoblastoma.
Effects of PNUTS depletion combined with erlotinib or gefitinib
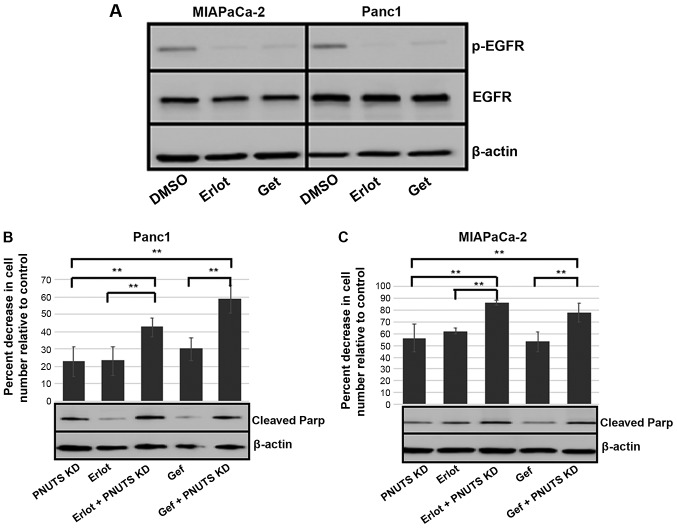
It was speculated that erlotinib, acting as an inhibitor of EGFR, blocked proliferation of cancer cells; therefore, the combination of erlotinib with PNUTS depletion, which induced apoptosis, may be an efficacious method to reduce pancreatic cancer cell numbers. For these experiments, gefitinib was also employed, which is an additional clinical treatment that blocks proliferation by inhibiting EGFR (47). Although MIAPaCa-2 and Panc1 cells are considered erlotinib-insensitive compared with other cancer cell lines, with the concentrations of erlotinib and gefitinib used, activation of EGFR was inhibited, as demonstrated by reduced phosphorylation (Fig. 5A). Subsequently, MIAPaCa-2 and Panc1 cells were subjected to 48 h of PNUTS depletion, followed by 24-h treatment with either DMSO, erlotinib or gefitinib. In both cell lines, when compared to each treatment alone, the combination of PNUTS depletion plus EGFR inhibition resulted in a significant decrease in cell number relative to the control (Fig. 5B and C). Concomitantly, increased apoptosis was indicated by Parp cleavage, thus suggesting that targeting the Rb tumor suppressor protein in combination with EGFR inhibition may be a rational strategy to reduce pancreatic cancer cell growth.
Figure 5.
(A) MIAPACa-2 and Panc1 cells were treated with Erlot (40 µM), Gef (4 µM) or DMSO (control). After 24 h, immunoblotting was performed on cell lysates obtained from cells. EGFR, p-EGFR and β-actin expression is shown. Results are representative of two separate experiments. (B) Panc1 and (C) MIAPaCa-2 cells were subjected to PNUTS depletion for 48 h and/or Erlot or Gef treatment, and cell counting assays were performed using CellTiter Glo. The percentage reduction in cell number is shown relative to nontargeting control/DMSO controls. Results are expressed as the means ± standard deviation, n=8 replicates/experiment. **P<0.01, one-way analysis of variance and Tukey honestly significant difference test. Data shown are representative of three independent experiments. Cleaved Parp and β-actin expression was determined by immunoblotting. DMSO, dimethyl sulfoxide; EGFR, human epidermal growth factor receptor type 1 receptor; Erlot, erlotinib; Gef, gefitinib; p, phosphorylated; Parp, poly(ADP-ribose) polymerase; PNUTs, phosphatase nuclear targeting subunit.
Effects of PNUTS depletion and erlotinib or gefitinib on the STAT3 pathway
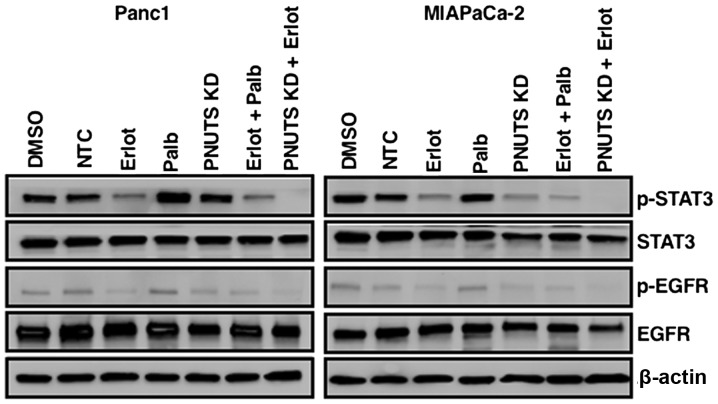
Because STAT3 pathway activation is often present in cancer cells, and since erlotinib has been reported to induce resistance in lung and pancreatic cancer via STAT3 activation, which leads to increased cell survival (9,10), the present study determined the effects of a combination of erlotinib and PNUTS depletion on STAT3 activation by immunoblotting. As shown in Fig. 6, erlotinib in Panc1 and MIAPaCa-2 cells reduced phosphorylation of EGFR, as expected. STAT3 appeared to be constitutively activated in Panc1 and MIAPaCa-2 cells (DMSO and NTC groups), and stimulated to various extents by erlotinib, palbociclib and PNUTS depletion, as evidenced by phosphorylation of STAT3. Although the various treatments affected STAT3 phosphorylation to different degrees, the combination of erlotinib with palbociclib resulted in STAT3 pathway activation, whereas the combination of erlotinib with PNUTS depletion did not activate STAT3, thus suggesting that this strategy may be an effective method to target EGFR and Rb phosphorylation without the development of resistance in pancreatic cancer cells.
Figure 6.
Panc1 or MIAPaCa-2 cells were treated with DMSO, erlotinib, NTC, PNUTS KD, palbociclib, or combinations of erlotinib + palbociclib or erlotinib + PNUTS knockdown. STAT3, p-STAT3, EGFR, p-EGFR and β-actin expression was determined by immunoblotting. Data shown are representative of three separate experiments. DMSO, dimethyl sulfoxide; EGFR, human epidermal growth factor receptor type 1 receptor; Erlot, erlotinib; KD, knockdown; NTC, nontargeting control; Palb, palbociclib; p, phosphorylated; PNUTs, phosphatase nuclear targeting subunit; STAT3, signal transducer and activator of transcription 3.
Discussion
Resistance to targeted therapy is a major issue in cancer, and resistance to the small molecule EGFR TKIs erlotinib and gefitinib has been investigated thoroughly in non-small cell lung cancer (NSCLC) (48). In NSCLC, marked initial responses to TKIs are attenuated by 6-12 months when patients experience tumor progression due to acquired resistance (49,50). Approximately 50% of the acquired resistance to erlotinib and gefitinib is due to mutations of the TKI-binding site of EGFR; however, resistance may also be due to compensatory growth promoting pathways that are activated by treatment. For example, activation of STAT3 in NSCLC occurs in response to erlotinib treatment and promotes resistance (9). In human pancreatic cancer tumors, STAT3 activation is exhibited in the majority of cases (51,52). STAT3 is activated by phosphorylation by Janus-activated kinases, interleukin-6, EGFRs and Src kinases. Upon phosphorylation, STAT3 dimerization stimulates gene transcription. STAT3 activation consequently promotes pancreatic tumorigenesis, cell invasion and metastatic potential (53,54). Notably, high expression of active STAT3 is correlated with poor prognosis (55,56). The importance of the STAT3 pathway in treatment resistance has been confirmed by the fact that acquired resistance of pancreatic cancer cells to EGFR TKIs, including erlotinib, can be abrogated by STAT3 inhibition (10).
In p16-negative cancer, hyperphosphorylated Rb is the target of CDK4/6 inhibitors; however, acquired resistance often develops in response to CDK4/6 inhibition. Several mechanisms have been proposed to be involved in CDK inhibition resistance; for example, biomarkers of treatment resistance include loss of Rb function, hyperactivity of the cyclin E-CDK2 axis and increased CDK6 activity (57-59). In addition, two recent studies demonstrated a role for the mechanistic target of rapamycin (mTOR) pathway in acquired resistance to CDK4/6 inhibition. Dysregulation of mTOR is a hallmark of adaptive resistance to several targeted therapies (60). In experiments using pancreatic cancer cells, CDK4/6 inhibition leads to mTOR complex 1 activation and metabolic alterations, including increased mitochondrial mass, and upregulation of glycolysis and oxidative metabolism (61). In other studies, it has been reported that CDK4/6 inhibition leads to activation of the pro-survival kinase protein kinase B (62). Finally, promotion of epithelial to mesenchymal transition by CDK4/6 inhibition has been demonstrated to contribute to drug resistance in pancreatic cancer (22,63).
Dysregulation of the tumor suppressor protein Rb is commonly found in human cancer. Hyperphosphorylation of Rb, rather than mutation of Rb, is thought to promote tumorigenesis. The CDKs that accomplish Rb phosphorylation are regulated by cyclin binding and by the association of endogenous CDK inhibitors, such as p16INK4a. Overexpression of cyclin D1/CDK4 or the loss of p16INK4a proteins are common genetic alterations in cancer that result in hyperphosphorylation of Rb (64,65). It has been reported that phosphorylation of Rb stimulates proliferation, blocks apoptosis and promotes invasion (37,66-69). By targeting PNUTS, an inhibitor of phosphatase activity toward Rb, dephosphorylation of Rb in cancer cells can be achieved. The present study demonstrated that PNUTS depletion in p16-negative pancreatic cancer cells reduced cell numbers, due to stimulation of apoptosis. In addition, non-transformed pancreatic duct epithelial cells that express p16 remained impervious to PNUTS depletion. When PNUTS depletion was compared with palbociclib-induced CDK4/6 inhibition, it was revealed that the combination of these treatments reduced cell numbers more effectively than either treatment alone. These results suggested that activation of phosphatase compared with inhibition of kinase are not functionally equivalent, perhaps by affecting different subsets of Rb phosphorylation sites, and thus Rb activity. Specific patterns of Rb phosphorylation may lead to distinctive Rb protein-binding abilities that lead to functional consequences in the cell. The present study demonstrated that, in Panc1 pancreatic cancer cells, CDK4/6 inhibition with palbociclib reduced proliferation; however, PNUTS depletion-mediated phosphatase activation resulted in apoptosis. It may be the case that in response to these two treatments the Rb phosphorylation pattern is altered such that in one case proliferation is inhibited, whereas in the other apoptosis is stimulated. This may be due to Rb interaction with transcriptional regulators that promote either proliferation arrest or apoptosis.
Because the TKIs (erlotinib and gefitinib) that block EGFR in pancreatic cancer cells inhibit proliferation and are used in the clinic, the present study tested how a combination of these TKIs with PNUTS depletion-mediated Rb phosphatase activation would affect p16-positive pancreatic cancer cells. It was revealed that in MIAPaCa-2 and Panc1 cells, which are considered erlotinib-resistant, targeting Rb phosphorylation by this method alongside EGFR inhibition resulted in a greater reduction in cell number than either treatment alone. Furthermore, whereas activation of the STAT3 pathway was constitutive in these pancreatic cancer cells, the combination of erlotinib with PNUTS depletion abrogated activation of this pathway, thus suggesting that targeting Rb phosphatase activity may be a strategy that circumvents erlotinib-induced acquired resistance. Additional studies may further elucidate the usefulness of this strategy in the clinical setting.
Acknowledgments
Not applicable.
Funding
This work was supported by the National Cancer Institute of the National Institutes of Health (grant nos. R15CA182723 and R15CA231372) awarded to NAK.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
NAK initiated the work, performed the apoptosis experiments, analyzed data and co-wrote the manuscript with NAT. NAT compared the effects of PNUTS knockdown with those of palbociclib on cell proliferation, and performed the PNUTS depletion + gefitinib study. RGA performed and analyzed the erlotinib + PNUTS knockdown experiments. BD performed and analyzed the PNUTS knockdown study using cell proliferation and immunoblotting assays. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–1049. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 3.Philip PA, Lutz MP. Targeting Epidermal Growth factor receptor-related signaling pathways in pancreatic cancer. Pancreas. 2015;44:1046–1052. doi: 10.1097/MPA.0000000000000389. [DOI] [PubMed] [Google Scholar]
- 4.Yang ZY, Yuan JQ, Di MY, Zheng DY, Chen JZ, Ding H, Wu XY, Huang YF, Mao C, Tang JL. Gemcitabine plus erlotinib for advanced pancreatic cancer: A systematic review with meta-analysis. PLoS One. 2013;8:e57528. doi: 10.1371/journal.pone.0057528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, Au HJ, Murawa P, Walde D, Wolff RA, et al. National Cancer Institute of Canada Clinical Trials Group: Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: A phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 6.Troiani T1, Martinelli E, Capasso A, Morgillo F, Orditura M, De Vita F, Ciardiello F. Targeting EGFR in pancreatic cancer treatment. Curr Drug Targets. 2012;13:802–810. doi: 10.2174/138945012800564158. [DOI] [PubMed] [Google Scholar]
- 7.Nedaeinia R, Avan A, Manian M, Salehi R, Ghayour- Mobarhan M. EGFR as a potential target for the treatment of pancreatic cancer: Dilemma and controversies. Curr Drug Targets. 2014;15:1293–1301. doi: 10.2174/1389450115666141125123003. [DOI] [PubMed] [Google Scholar]
- 8.Zhao C, Li H, Lin HJ, Yang S, Lin J, Liang G. Feedback activation of STAT3 as a cancer drug-resistance mechanism. Trends Pharmacol Sci. 2016;37:47–61. doi: 10.1016/j.tips.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Lee H-J, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26:207–221. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 10.Ioannou N, Seddon AM, Dalgleish A, Mackintosh D, Solca F, Modjtahedi H. Acquired resistance of pancreatic cancer cells to treatment with gemcitabine and HER-inhibitors is accompanied by increased sensitivity to STAT3 inhibition. Int J Oncol. 2016;48:908–918. doi: 10.3892/ijo.2016.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schutte M, Hruban RH, Geradts J, Maynard R, Hilgers W, Rabindran SK, Moskaluk CA, Hahn SA, Schwarte-Waldhoff I, Schmiegel W, et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- 12.Hustinx SR, Leoni LM, Yeo CJ, Brown PN, Goggins M, Kern SE, Hruban RH, Maitra A. Concordant loss of MTAP and p16/CDKN2A expression in pancreatic intraepithelial neoplasia: Evidence of homozygous deletion in a noninvasive precursor lesion. Mod Pathol. 2005;18:959–963. doi: 10.1038/modpathol.3800377. [DOI] [PubMed] [Google Scholar]
- 13.Fukushima N, Sato N, Ueki T, Rosty C, Walter KM, Wilentz RE, Yeo CJ, Hruban RH, Goggins M. Aberrant methylation of preproenkephalin and p16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol. 2002;160:1573–1581. doi: 10.1016/S0002-9440(10)61104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerdes B, Ramaswamy A, Kersting M, Ernst M, Lang S, Schuermann M, Wild A, Bartsch DK. p16(INK4a) alterations in chronic pancreatitis-indicator for high-risk lesions for pancreatic cancer. Surgery. 2001;129:490–497. doi: 10.1016/S0039-6060(01)01158-8. [DOI] [PubMed] [Google Scholar]
- 15.Makohon-Moore A, Iacobuzio-Donahue CA. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat Rev Cancer. 2016;16:553–565. doi: 10.1038/nrc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Notta F, Chan-Seng-Yue M, Lemire M, Li Y, Wilson GW, Connor AA, Denroche RE, Liang SB, Brown AM, Kim JC, et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538:378–382. doi: 10.1038/nature19823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabien A, Sanchez-Ruderisch H, Schulz P, Otto N, Wimmel A, Wiedenmann B, Detjen KM. Tumor suppressor p16INK4a controls oncogenic K-Ras function in human pancreatic cancer cells. Cancer Sci. 2012;103:169–175. doi: 10.1111/j.1349-7006.2011.02140.x. [DOI] [PubMed] [Google Scholar]
- 18.Chang Z, Ju H, Ling J, Zhuang Z, Li Z, Wang H, Fleming JB, Freeman JW, Yu D, Huang P, et al. Cooperativity of oncogenic K-ras and downregulated p16/INK4A in human pancreatic tumorigenesis. PLoS One. 2014;9:e101452. doi: 10.1371/journal.pone.0101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Poi MJ, Tsai MD. Regulatory mechanisms of tumor suppressor P16(INK4A) and their relevance to cancer. Biochemistry. 2011;50:5566–5582. doi: 10.1021/bi200642e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franco J, Witkiewicz AK, Knudsen ES. CDK4/6 inhibitors have potent activity in combination with pathway selective therapeutic agents in models of pancreatic cancer. Oncotarget. 2014;5:6512–6525. doi: 10.18632/oncotarget.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heilmann AM, Perera RM, Ecker V, Nicolay BN, Bardeesy N, Benes CH, Dyson NJ. CDK4/6 and IGF1 receptor inhibitors synergize to suppress the growth of p16INK4A-deficient pancreatic cancers. Cancer Res. 2014;74:3947–3958. doi: 10.1158/0008-5472.CAN-13-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu F, Korc M. Cdk4/6 inhibition induces epithelial-mesenchymal transition and enhances invasiveness in pancreatic cancer cells. Mol Cancer Ther. 2012;11:2138–2148. doi: 10.1158/1535-7163.MCT-12-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witkiewicz AK, Borja NA, Franco J, Brody JR, Yeo CJ, Mansour J, Choti MA, McCue P, Knudsen ES. Selective impact of CDK4/6 suppression on patient-derived models of pancreatic cancer. Oncotarget. 2015;6:15788–15801. doi: 10.18632/oncotarget.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finn RS, Martin M, Rugo HS, Jones S, Im SA, Gelmon K, Harbeck N, Lipatov ON, Walshe JM, Moulder S, et al. Palbociclib and Letrozole in advanced breast cancer. N Engl J Med. 2016;375:1925–1936. doi: 10.1056/NEJMoa1607303. [DOI] [PubMed] [Google Scholar]
- 25.Nelson DA, Krucher NA, Ludlow JW. High molecular weight protein phosphatase type 1 dephosphorylates the retinoblastoma protein. J Biol Chem. 1997;272:4528–4535. doi: 10.1074/jbc.272.7.4528. [DOI] [PubMed] [Google Scholar]
- 26.Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: Designed to create specificity. Trends Biochem Sci. 2003;33:113–121. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolupaeva V, Janssens V. PP1 and PP2A phosphatases - cooperating partners in modulating retinoblastoma protein activation. FEBS J. 2013;280:627–643. doi: 10.1111/j.1742-4658.2012.08511.x. [DOI] [PubMed] [Google Scholar]
- 28.Allen PB, Kwon YG, Nairn AC, Greengard P. Isolation and characterization of PNUTS, a putative protein phosphatase 1 nuclear targeting subunit. J Biol Chem. 1998;273:4089–4095. doi: 10.1074/jbc.273.7.4089. [DOI] [PubMed] [Google Scholar]
- 29.Kreivi JP, Trinkle-Mulcahy L, Lyon CE, Morrice NA, Cohen P, Lamond AI. Purification and characterisation of p99, a nuclear modulator of protein phosphatase 1 activity. FEBS Lett. 1997;420:57–62. doi: 10.1016/S0014-5793(97)01485-3. [DOI] [PubMed] [Google Scholar]
- 30.Udho E, Tedesco VC, Zygmunt A, Krucher NA. PNUTS (phosphatase nuclear targeting subunit) inhibits retinoblastoma-directed PP1 activity. Biochem Biophys Res Commun. 2002;297:463–467. doi: 10.1016/S0006-291X(02)02236-2. [DOI] [PubMed] [Google Scholar]
- 31.Choy MS, Hieke M, Kumar GS, Lewis GR, Gonzalez-DeWhitt KR, Kessler RP, Stein BJ, Hessenberger M, Nairn AC, Peti W, et al. Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code. Proc Natl Acad Sci USA. 2014;111:4097–4102. doi: 10.1073/pnas.1317395111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoekstra E, Peppelenbosch MP, Fuhler GM. Meeting report Europhosphatase 2015: Phosphatases as drug targets in cancer. Cancer Res. 2016;76:193–196. doi: 10.1158/0008-5472.CAN-15-2091. [DOI] [PubMed] [Google Scholar]
- 33.Kim YM, Watanabe T, Allen PB, Kim YM, Lee SJ, Greengard P, Nairn AC, Kwon YG. PNUTS, a protein phosphatase 1 (PP1) nuclear targeting subunit. Characterization of its PP1- and RNA-binding domains and regulation by phosphorylation. J Biol Chem. 2003;278:13819–13828. doi: 10.1074/jbc.M209621200. [DOI] [PubMed] [Google Scholar]
- 34.Krucher NA, Rubin E, Tedesco VC, Roberts MH, Sherry TC, De Leon G. Dephosphorylation of Rb (Thr-821) in response to cell stress. Exp Cell Res. 2006;312:2757–2763. doi: 10.1016/j.yexcr.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 35.De Leon G, Sherry TC, Krucher NA. Reduced expression of PNUTS leads to activation of Rb-phosphatase and caspase- mediated apoptosis. Cancer Biol Ther. 2008;7:833–841. doi: 10.4161/cbt.7.6.5839. [DOI] [PubMed] [Google Scholar]
- 36.Kavela S, Shinde SR, Ratheesh R, Viswakalyan K, Bashyam MD, Gowrishankar S, Vamsy M, Pattnaik S, Rao S, Sastry RA, et al. PNUTS functions as a proto-oncogene by sequestering PTEN. Cancer Res. 2013;73:205–214. doi: 10.1158/0008-5472.CAN-12-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egger JV, Lane MV, Antonucci LA, Dedi B, Krucher NA. Dephosphorylation of the Retinoblastoma protein (Rb) inhibits cancer cell EMT via Zeb. Cancer Biol Ther. 2016;17:1197–1205. doi: 10.1080/15384047.2016.1235668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pino MS, Shrader M, Baker CH, Cognetti F, Xiong HQ, Abbruzzese JL, McConkey DJ. Transforming growth factor alpha expression drives constitutive epidermal growth factor receptor pathway activation and sensitivity to gefitinib (Iressa) in human pancreatic cancer cell lines. Cancer Res. 2006;66:3802–3812. doi: 10.1158/0008-5472.CAN-05-3753. [DOI] [PubMed] [Google Scholar]
- 39.Buck E, Eyzaguirre A, Haley JD, Gibson NW, Cagnoni P, Iwata KK. Inactivation of Akt by the epidermal growth factor receptor inhibitor erlotinib is mediated by HER-3 in pancreatic and colorectal tumor cell lines and contributes to erlotinib sensitivity. Mol Cancer Ther. 2006;5:2051–2059. doi: 10.1158/1535-7163.MCT-06-0007. [DOI] [PubMed] [Google Scholar]
- 40.Ali S, Banerjee S, Ahmad A, El-Rayes BF, Philip PA, Sarkar FH. Apoptosis-inducing effect of erlotinib is potentiated by 3,3'-diindolylmethane in vitro and in vivo using an orthotopic model of pancreatic cancer. Mol Cancer Ther. 2008;7:1708–1719. doi: 10.1158/1535-7163.MCT-08-0354. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Deer EL, Gonzalez-Hernandez J, Coursen JD, Shea JE, Ngatia J, Scaife CL, Firpo MA, Mulvihill SJ. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gradiz R, Silva HC, Carvalho L, Botelho MF, Mota-Pinto A. MIA PaCa-2 and PANC-1 - pancreas ductal adenocarcinoma cell lines with neuroendocrine differentiation and somatostatin receptors. Sci Rep. 2016;6:21648. doi: 10.1038/srep21648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loukopoulos P, Kanetaka K, Takamura M, Shibata T, Sakamoto M, Hirohashi S. Orthotopic transplantation models of pancreatic adenocarcinoma derived from cell lines and primary tumors and displaying varying metastatic activity. Pancreas. 2004;29:193–203. doi: 10.1097/00006676-200410000-00004. [DOI] [PubMed] [Google Scholar]
- 44.Furukawa T, Duguid WP, Rosenberg L, Viallet J, Galloway DA, Tsao MS. Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am J Pathol. 1996;148:1763–1770. [PMC free article] [PubMed] [Google Scholar]
- 45.Macdonald JI, Dick FA. Posttranslational modifications of the retinoblastoma tumor suppressor protein as determinants of function. Genes Cancer. 2012;3:619–633. doi: 10.1177/1947601912473305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Leon G, Cavino M, D'Angelo M, Krucher NA. PNUTS knockdown potentiates the apoptotic effect of Roscovitine in breast and colon cancer cells. Int J Oncol. 2010;36:1269–1275. doi: 10.3892/ijo_00000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohammed A, Janakiram NB, Li Q, Madka V, Ely M, Lightfoot S, Crawford H, Steele VE, Rao CV. EGFR inhibitor gefitinib prevents progression of pancreatic lesions to carcinoma in a conditional LSL-KrasG12D/+ transgenic mice model. Cancer Prev Res (Phila) 2010;3:1417–1426. doi: 10.1158/1940-6207.CAPR-10-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin Y, Wang X, Jin H. EGFR-TKI resistance in NSCLC patients: Mechanisms and strategies. Am J Cancer Res. 2014;4:411–435. [PMC free article] [PubMed] [Google Scholar]
- 49.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 50.Jackman D, Pao W, Riely GJ, Engelman JA, Kris MG, Jänne PA, Lynch T, Johnson BE, Miller VA. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol. 2010;28:357–360. doi: 10.1200/JCO.2009.24.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scholz A, Heinze S, Detjen KM, Peters M, Welzel M, Hauff P, Schirner M, Wiedenmann B, Rosewicz S. Activated signal transducer and activator of transcription 3 (STAT3) supports the malignant phenotype of human pancreatic cancer. Gastroenterology. 2003;125:891–905. doi: 10.1016/S0016-5085(03)01064-3. [DOI] [PubMed] [Google Scholar]
- 52.Toyonaga T, Nakano K, Nagano M, Zhao G, Yamaguchi K, Kuroki S, Eguchi T, Chijiiwa K, Tsuneyoshi M, Tanaka M. Blockade of constitutively activated Janus kinase/signal transducer and activator of transcription-3 pathway inhibits growth of human pancreatic cancer. Cancer Lett. 2003;201:107–116. doi: 10.1016/S0304-3835(03)00482-8. [DOI] [PubMed] [Google Scholar]
- 53.Corcoran RB, Contino G, Deshpande V, Tzatsos A, Conrad C, Benes CH, Levy DE, Settleman J, Engelman JA, Bardeesy N. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res. 2011;71:5020–5029. doi: 10.1158/0008-5472.CAN-11-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fofaria NM, Srivastava SK. STAT3 induces anoikis resistance, promotes cell invasion and metastatic potential in pancreatic cancer cells. Carcinogenesis. 2015;36:142–150. doi: 10.1093/carcin/bgu233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu X, Tang W, Marquez RT, Li K, Highfill CA, He F, Lian J, Lin J, Fuchs JR, Ji M, et al. Overcoming chemo/radio-resistance of pancreatic cancer by inhibiting STAT3 signaling. Oncotarget. 2016;7:11708–11723. doi: 10.18632/oncotarget.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang X, Ren D, Wu X, Lin X, Ye L, Lin C, Wu S, Zhu J, Peng X, Song L. miR-1266 contributes to pancreatic cancer progression and chemoresistance by the STAT3 and NF-kB signaling pathways. Mol Ther Nucleic Acids. 2018;11:142–158. doi: 10.1016/j.omtn.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dean JL, Thangavel C, McClendon AK, Reed CA, Knudsen ES. Therapeutic CDK4/6 inhibition in breast cancer: Key mechanisms of response and failure. Oncogene. 2010;29:4018–4032. doi: 10.1038/onc.2010.154. [DOI] [PubMed] [Google Scholar]
- 59.Yang C, Li Z, Bhatt T, Dickler M, Giri D, Scaltriti M, Baselga J, Rosen N, Chandarlapaty S. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene. 2017;36:2255–2264. doi: 10.1038/onc.2016.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guri Y, Hall MN. mTOR signaling confers resistance to targeted cancer drugs. Trends Cancer. 2016;2:688–697. doi: 10.1016/j.trecan.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 61.Franco J, Balaji U, Freinkman E, Witkiewicz AK, Knudsen ES. Metabolic reprogramming of pancreatic cancer mediated by CDK4/6 inhibition elicits unique vulnerabilities. Cell Rep. 2016;14:979–990. doi: 10.1016/j.celrep.2015.12.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J, Xu K, Liu P, Geng Y, Wang B, Gan W, Guo J, Wu F, Chin YR, Berrios C, et al. Inhibition of Rb phosphorylation leads to mTORC2 mediated activation of AKT. Mol Cell. 2016;62:929–942. doi: 10.1016/j.molcel.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arumugam T, Ramachandran V, Fournier KF, Wang H, Marquis L, Abbruzzese JL, Gallick GE, Logsdon CD, McConkey DJ, Choi W. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 2009;69:5820–5828. doi: 10.1158/0008-5472.CAN-08-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 65.Mittnacht S. The retinoblastoma protein--from bench to bedside. Eur J Cell Biol. 2005;84:97–107. doi: 10.1016/j.ejcb.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 66.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/S1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 67.Harbour JW, Dean DC. The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 68.Thangavel C, Boopathi E, Liu Y, Haber A, Ertel A, Bhardwaj A, Addya S, Williams N, Ciment SJ, Cotzia P, et al. RB loss promotes prostate cancer metastasis. Cancer Res. 2017;77:982–995. doi: 10.1158/0008-5472.CAN-16-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arima Y, Hayashi H, Sasaki M, Hosonaga M, Goto TM, Chiyoda T, Kuninaka S, Shibata T, Ohata H, Nakagama H, et al. Induction of ZEB proteins by inactivation of RB protein is key determinant of mesenchymal phenotype of breast cancer. J Biol Chem. 2012;287:7896–7906. doi: 10.1074/jbc.M111.313759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.