Abstract
Ribonucleotide reductase (RNR) catalyzes the only known de-novo pathway for production of all four deoxyribonucleotides required for DNA synthesis1,2. It is essential for all organisms with DNA as genetic material and a current drug target3,4. Since the discovery that iron is required for function in the aerobic, class I RNR found in all eukaryotes and many bacteria, a di-nuclear metal site has been viewed as a requirement for generating and stabilizing a catalytic radical, essential for RNR activity5,6,7. Here, we describe a new group of RNR proteins in Mollicutes, including Mycoplasma pathogens, which possesses a metal-independent stable radical residing on a modified tyrosyl residue. Structural, biochemical and spectroscopic characterization reveal an unprecedented stable DOPA radical species that directly supports ribonucleotide reduction in vitro and in vivo. This observation overturns the presumed requirement of a dinuclear metal site in aerobic RNR. The metal-independent radical compels completely novel mechanisms for radical generation and stabilization, processes that are targeted by RNR inhibitors. Conceivably, this RNR variant provides an advantage under metal starvation induced by the immune system. Organisms encoding this type of RNR are involved in diseases of the respiratory, urinary and genital tracts, some with developing resistance to antibiotics. Further characterization of this novel RNR family and its mechanism for cofactor generation will provide insight into new enzymatic chemistry and be of value to devise strategies to combat the pathogens that utilize it. We propose that the new RNR subclass is denoted class Ie.
Three RNR classes have been discovered, all requiring transition metals for function2. Class III is strictly anaerobic and uses a 4Fe-4S cluster for radical generation. Class II is indifferent to oxygen and utilizes an adenosyl cobalamin cofactor. In all hitherto studied class I RNRs, the catalytic radical is generated and stabilized by a dinuclear metal site in protein R2 in an oxygen dependent reaction, and then reversibly shuttled to protein R1 where ribonucleotide reduction occurs8–10. The dinuclear metal site is coordinated by four carboxylate residues and two histidines. Depending on subclass, the metal site is di-iron (class Ia), di-manganese (class Ib), or heterodinuclear Mn/Fe (class Ic)11,12. Classes Ia and Ib generate a stable tyrosyl radical while class Ic forms a radical-equivalent MnIV/FeIII high-valent oxidation state of the metal site13,14. A class Id, containing a MnIV/MnIII cofactor, was also recently proposed15–17. The metal sites in classes Ia and Ic perform direct oxygen activation while class Ib requires a flavoprotein, NrdI, to generate superoxide that oxidizes the di-manganese site18–20.
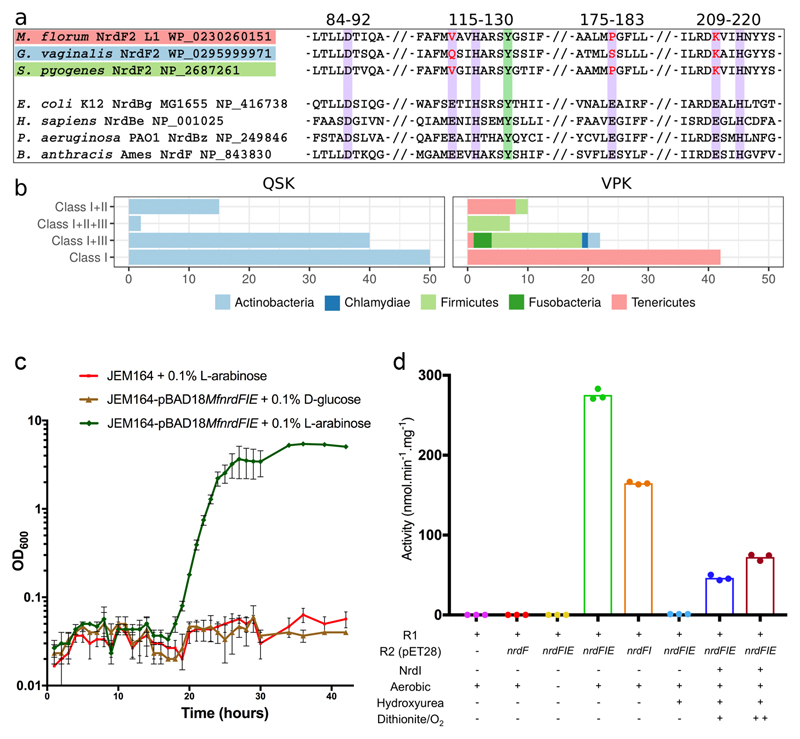
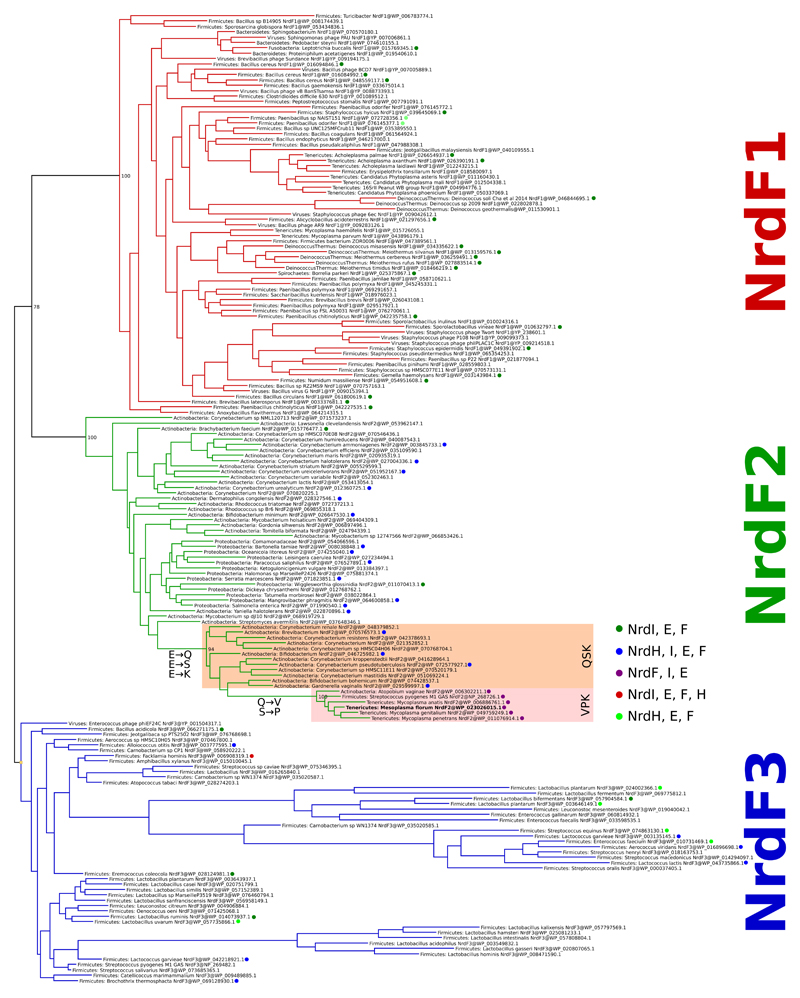
Sequence analysis revealed a group of class I RNR operons, present in common human pathogens e.g. Mycoplasma genitalium, Mycoplasma pneumoniae, and Streptococcus pyogenes. Analogous to standard class Ib RNRs, the operons contain the genes nrdE, nrdF and nrdI, coding for proteins R1, R2 and NrdI, respectively. Phylogenetically, the group forms a clade derived from class Ib proteins (Extended Data Fig. 1). Strikingly, the R2 proteins in this group retain only 3 of the 6 metal binding residues that are otherwise completely conserved, and individually essential, regardless of R2 subclass (Fig. 1a). These substitutions appear to exclude a metal site and a radical generation mechanism even remotely similar to any thus far studied ribonucleotide reductase. In many cases the QSK/VPK variants represent the only aerotolerant RNR found in the genome, for example in Mycoplasma genitalium and Mycoplasma pneumoniae (VPK) and Gardnerella vaginalis (QSK) (Fig. 1b).
Fig 1. A new RNR subclass able to rescue an Escherichia coli strain lacking aerobic RNR.
a) Sequence alignment of the new R2 protein groups to a number of standard, di-metal containing, R2 proteins. Purple background indicates the 6 normally essential metal-binding residues, only 3 of which are conserved. Two variants are observed in which 3 carboxylate metal ligands are either substituted for valine, proline and lysine (VPK variant) or for glutamine, serine and lysine (QSK variant). The normally radical harboring tyrosine residue is shown with a green background.
b) Taxonomic distribution of NrdF2. QSK/VPK encoding organisms and their collected RNR class repertoire. As common for class I RNRs, several genomes encoding the QSK or VPK variant also harbor other RNRs. The QSK clusters are found only in Actinobacteria, whereas the VPK clusters are also found in Firmicutes, Tenericutes, Chlamydiae and Fusobacteria.
c) Expression of the MfnrdFIE operon induced by addition of 0.1% v/v L-arabinose (green) rescued the JEM164 double knockout (ΔnrdAB, ΔnrdEF) strain while when gene expression was suppressed with 0.1% v/v D-glucose (brown) the strain failed to recover, as did the strain lacking the vector (red). Growth curves (average of 3 experiments with SD) are shown.
d) MfNrdI activates MfR2 in an O2 dependent reaction. HPLC based in vitro RNR activity assays show no activity for R2 protein expressed separately in E. coli (red), while aerobic co-expression with MfnrdI and MfnrdE (green) or MfnrdI (orange) produced an active R2 protein. Anaerobic co-expression (yellow) or incubation of the active R2 with hydroxyurea (light blue) abolishes the activity. Partial activity could be regenerated by the addition of MfNrdI and redox cycling with dithionite and oxygen, blue and maroon for one and two reduction-oxidation cycles respectively. Data points are shown for triplicate experiments.
We investigated if a VPK variant operon could rescue an E. coli strain lacking aerobic RNR (ΔnrdAB, ΔnrdEF)21, thus otherwise unable to grow in the presence of oxygen. A tunable arabinose-induced pBAD plasmid containing the nrdFIE operon from Mesoplasma florum was constructed and transformed into the (ΔnrdAB, ΔnrdEF) strain. Cultures grown under anaerobic conditions were subsequently exposed to oxygen. The MfnrdFIE plasmid rescued the knockout strain and provided competence to grow under aerobic conditions (Fig. 1c). This result is consistent with our previous observations of the Streptococcus pyogenes nrdFIE operon22.
We proceeded to quantify the in vitro activity of the enzyme. We were unable to obtain in vitro ribonucleotide reductase activity using the MfR2 protein expressed separately in E. coli. However, purification of MfR2 after co-expression of the entire MfnrdFIE operon under aerobic conditions rendered a deep-blue protein that exhibited RNR activity together with MfR1 (Fig. 1d). The color and activity were also observed when MfR2 was co-expressed with only MfnrdI under aerobic conditions, while co-expression under anaerobic conditions rendered an inactive and colorless MfR2. Under the current assay conditions, the turnover number is 0.18 s-1 or > 300 for the duration of the assay. Once the MfR2 protein is activated, MfNrdI is thus not required for multi turnover activity in vitro. The specific activity was determined to 275 ± 7 nmol×min-1×mg-1, on par with typical class I RNRs16,20. Incubation of the active MfR2 protein with hydroxyurea, a radical-quenching RNR inhibitor, rendered a colorless and inactive protein. The activity of the quenched protein could be partially restored if MfNrdI was added to the inactivated MfR2 and subjected to reduction-oxidation cycles using dithionite and oxygen-containing buffer (Fig. 1d).
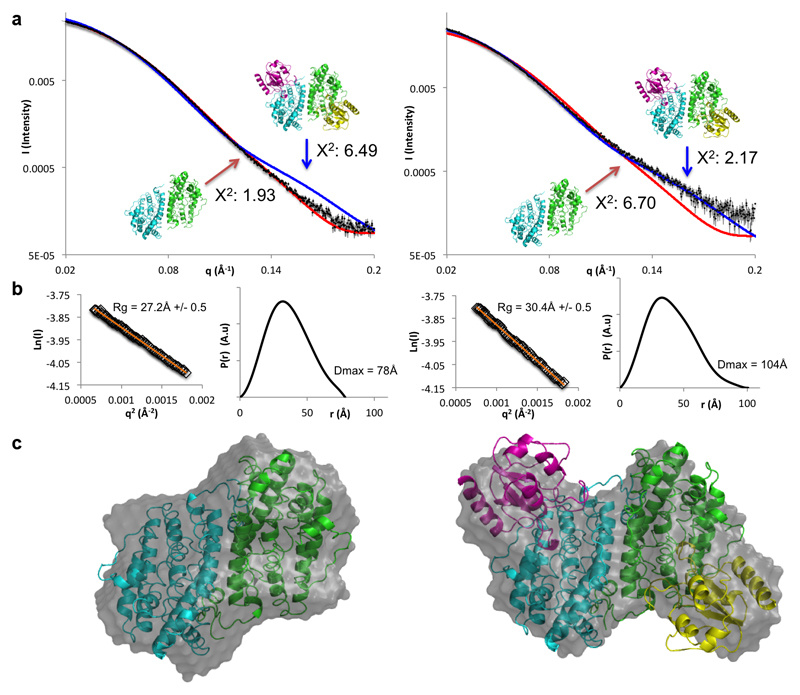
MfR2 and MfR1 thus constitute an active RNR system, but only after MfR2 has undergone an NrdI and O2 dependent activation step. This is principally similar to class Ib RNRs. Moreover, Small Angle X-ray Scattering (SAXS) measurements showed that MfR2 and MfNrdI forms a well-defined 2:2 complex with the same interaction geometry as standard class Ib RNR proteins (Extended data Fig. 2)19,23. Remarkably, however, for class Ib RNR, the role of NrdI is to provide an oxidant for the di-manganese metal site that subsequently generates the catalytic tyrosyl radical. A mechanism that appears implausible, given the substitution of the metal binding residues in MfR2.
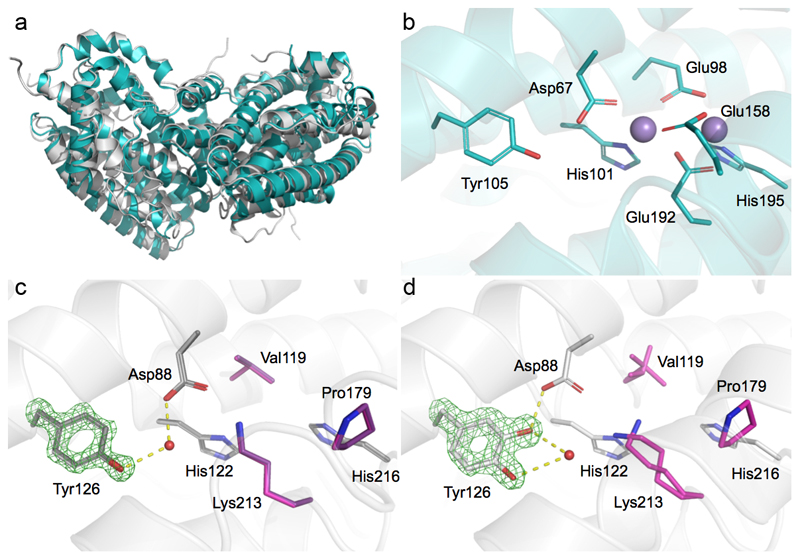
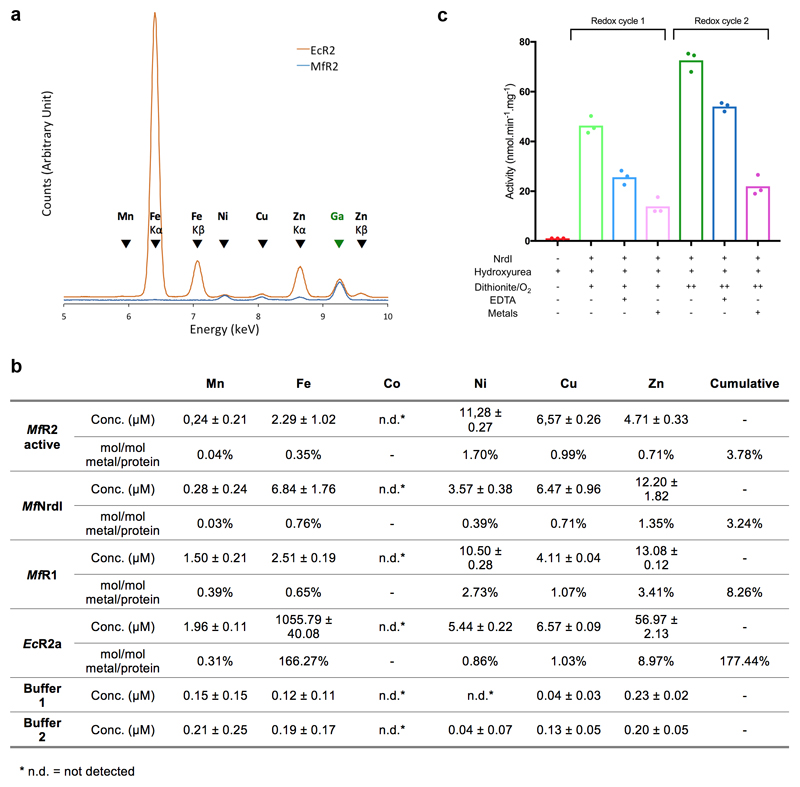
The crystal structure of MfR2 in its active form was determined to 1.5 Å resolution and two crystal structures of the inactive form, either expressed alone aerobically or co-expressed with the entire operon under anaerobic conditions, to 1.2 Å (Extended data Table 1). MfR2 shows the same fold and dimeric arrangement as standard metal-binding R2 proteins (Fig. 2a). As expected from sequence, the site normally occupied by the di-nuclear metal cofactor is remarkably atypical (Fig. 2b,c). No electron density corresponding to a metal ion could be observed in the site in any of the structures. We also collected X-ray anomalous scattering data at a wavelength of 0.97 Å. Anomalous difference maps would reveal even low occupancy metal binding. Still, no signal above noise could be observed in the vicinity of the site. To rule out that metal was lost during crystallization, we conducted Total-reflection X-Ray Fluorescence (TXRF) analysis on the active MfR2 protein solution, quantitatively detecting all elements from aluminum to uranium (with the exception of Zr, Nb, Mo, Tc, Ru and Rh). Only trace amounts of transition metals could be detected in the active protein sample (e.g. mol/mol metal/protein: Mn 0.04%, Fe 0.35%, Co 0.00%, Ni 1.70%, Cu 0.99%, Zn 0.71%, (Extended data Fig. 3)). Cumulatively, the metal content corresponded to less than 0.04 per protein monomer. TXRF analyses were also performed for the purified MfR1 and MfNrdI proteins in solution, again with only trace amounts of metals detected.
Fig 2. The active R2 protein is metal-free but covalently modified.
a) Overall structure of the Mesoplasma florum VPK R2 protein (gray) compared to the standard class Ib R2 from Escherichia coli (PDB: 3n37) (cyan).
b) Structure of the dinuclear metal site and conserved metal coordinating residues in standard class I RNR R2.
c) Structures of inactive MfR2 after expression without MfNrdI (gray) or with MfNrdI under anaerobic conditions (dark gray), both determined to 1.2 Å resolution. These structures are identical within experimental error. The 3 residues substituting the normally conserved carboxylate metal-ligands in canonical class I R2s are shown in pink and purple respectively. In the MfR2 crystal structure, the canonical metal positions are occupied: 1) by a water molecule in a tetrahedral coordination, involving the conserved His216, with distances of 2.8 ± 0.1 Å, as expected for a hydrogen-bonded water, but very unlikely for a metal; 2) by the ε-amino group of Lys213, replacing the conserved metal-bridging glutamate present in all class I R2s. This lysine forms a hydrogen bond with Asp88, the only remaining carboxylate residue. Asp88 also interacts via a H-bonded water with Tyr126, corresponding to the tyrosine harboring the metal-coupled radical in standard class Ia and Ib R2 proteins.
d) Structure of the active MfR2 after aerobic co-expression with MfNrdI and MfR1. Here, the tyrosine is covalently modified in the meta-position. Mass spectrometry determined that the tyrosine is hydroxylated in the active protein.
Simulated annealing omit Fo-Fc electron density maps for the unmodified or modified Tyr126 is shown in green and contoured at 8σ. Carbons are in cyan, gray/pink for R2 from E. coli and M. florum, respectively. Oxygens and nitrogens are colored red and blue, respectively. Mn(II) ions are represented as purple spheres. H-bond interactions to the Tyr126 are indicated.
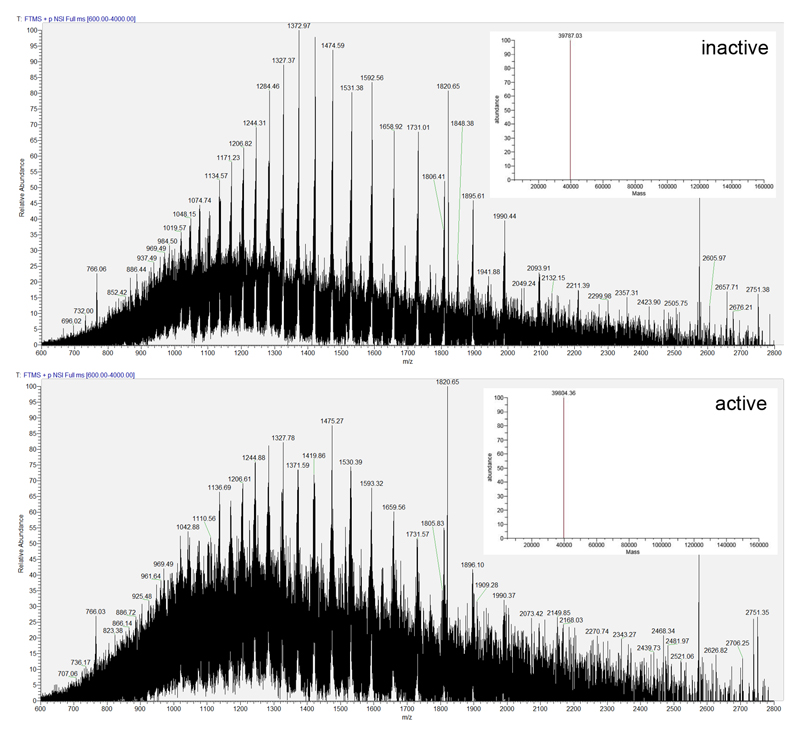
Notably, the tyrosine appears modified in the meta-position in the active MfR2 protein, but not in the inactive protein (Fig. 2c,d). Mass spectrometry analysis confirmed the covalent modification. The full-length active MfR2 protein is 17 ± 2 Da heavier than the inactive, 39804 ± 2 Da vs. 39787 ± 1.4 Da, respectively (Extended data Fig. 4). Proteolytic cleavage and peptide analysis pinpoints the modification to + 15.995 ± 0.003 Da (monoisotopic mass, corresponding to one additional oxygen atom) at Tyr126 (Extended data Fig. 5). Based on the X-ray crystallographic and mass spectrometry data, we thus conclude that Tyr126 is meta-hydroxylated in the active protein.
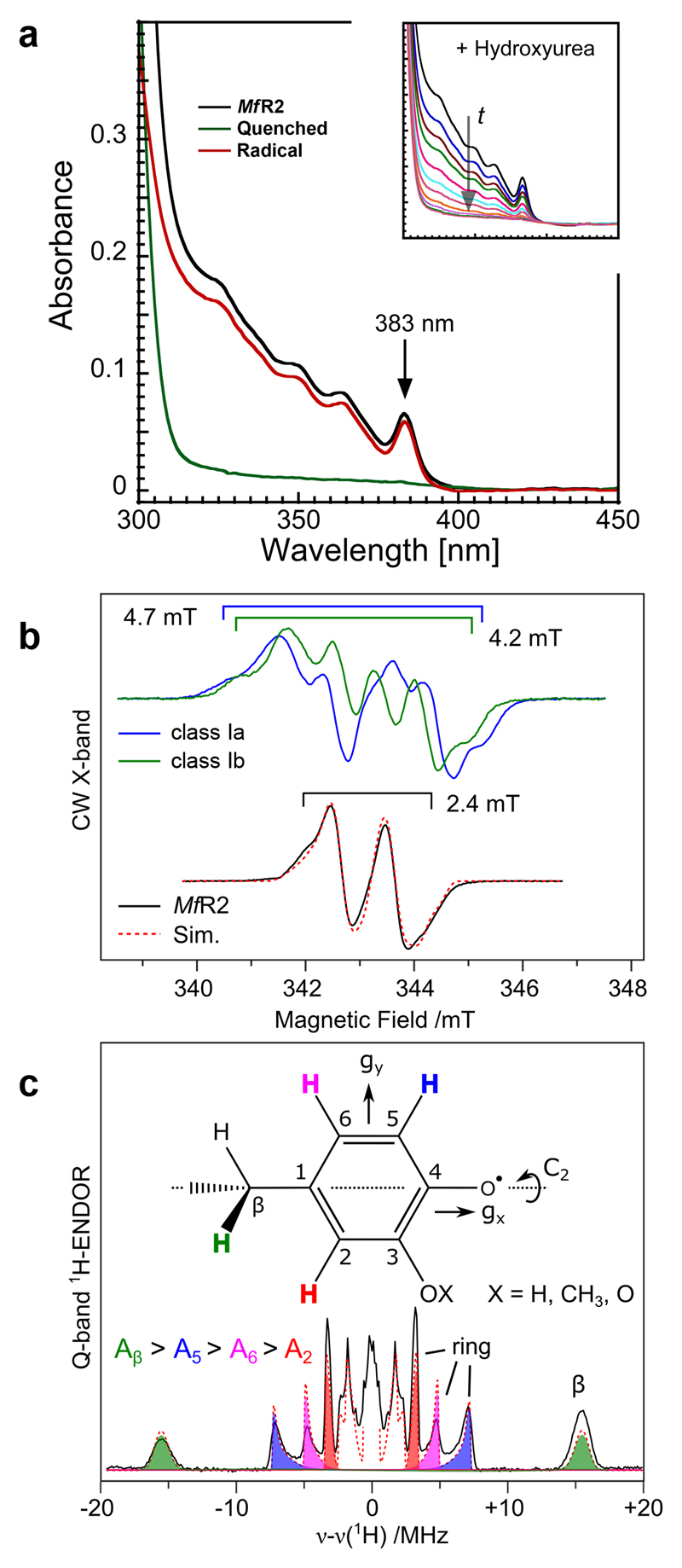
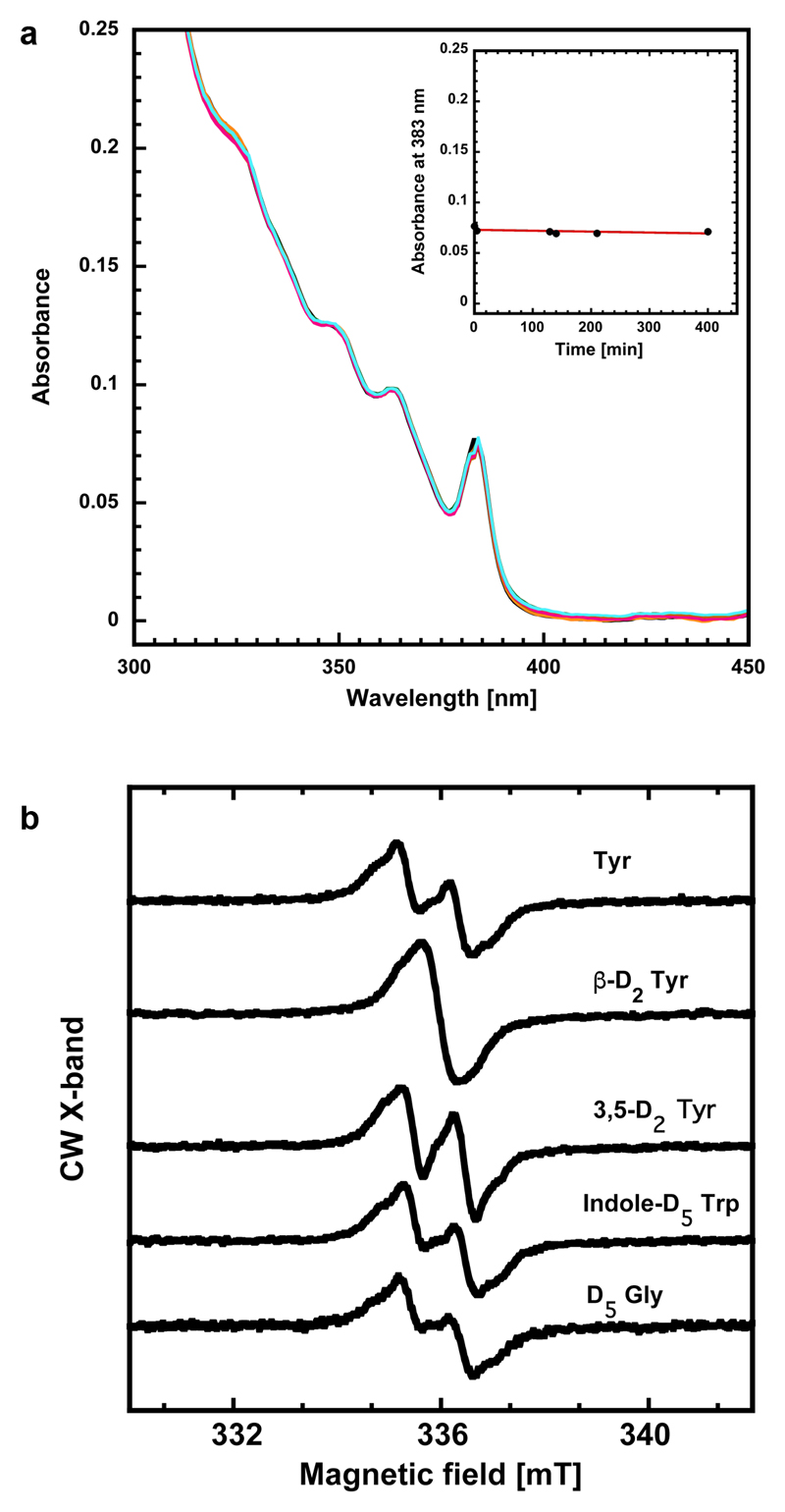
The UV/vis spectrum of active MfR2 has similar intensity and profile as the tyrosyl radical in canonical R2 proteins though significantly blue-shifted, ≈ 383 nm vs. ≈ 410 nm. Moderately intense (3000 M-1cm-1) transitions in this region are a fingerprint for an aromatic radical. It has previously been demonstrated in simpler phenoxyl radical model systems that meta OH substitution leads to a blue shift of the radical π to π* marker band24. Incubation of the protein with hydroxyurea led to the complete disappearance of the absorbance features and rendered a colorless protein (Fig. 3a). In absence of radical quenchers the features are remarkably stable with no observable decay after 400 minutes at 25°C (Extended data Fig. 6a). Incorporation of deuterium-labeled amino acids shows that the stable radical resides on a tyrosine-derived residue (Extended data Fig. 6b).
Fig 3. Characterization of a novel stable DOPA radical species in MfR2.
a) The UV/vis spectrum of the active blue protein shows a peak at 383 nm and additional structure at lower wavelengths (black). Incubation with 52 mM hydroxyurea for 20 minutes removed all features from the spectrum except the protein-related absorbance peak at 280 nm (green and inset). The red trace represents the active minus quenched spectra. The rate constant for the decay of the absorbance at 348, 364 and 383 nm was identical, consistent with all absorbance features arising from a single radical species. Experiments were repeated three times.
b) X-band EPR spectra of tyrosyl radicals observed in R2 proteins (Escherichia coli class Ia R2 and Bacillus cereus class Ib R2 reconstituted with Fe) compared to the M. florum R2 radical species reported here. X-band measurements were performed on isolated MfR2 protein (Fig. 3b) and repeated on crude cell extracts (Extended data Fig. 6b).
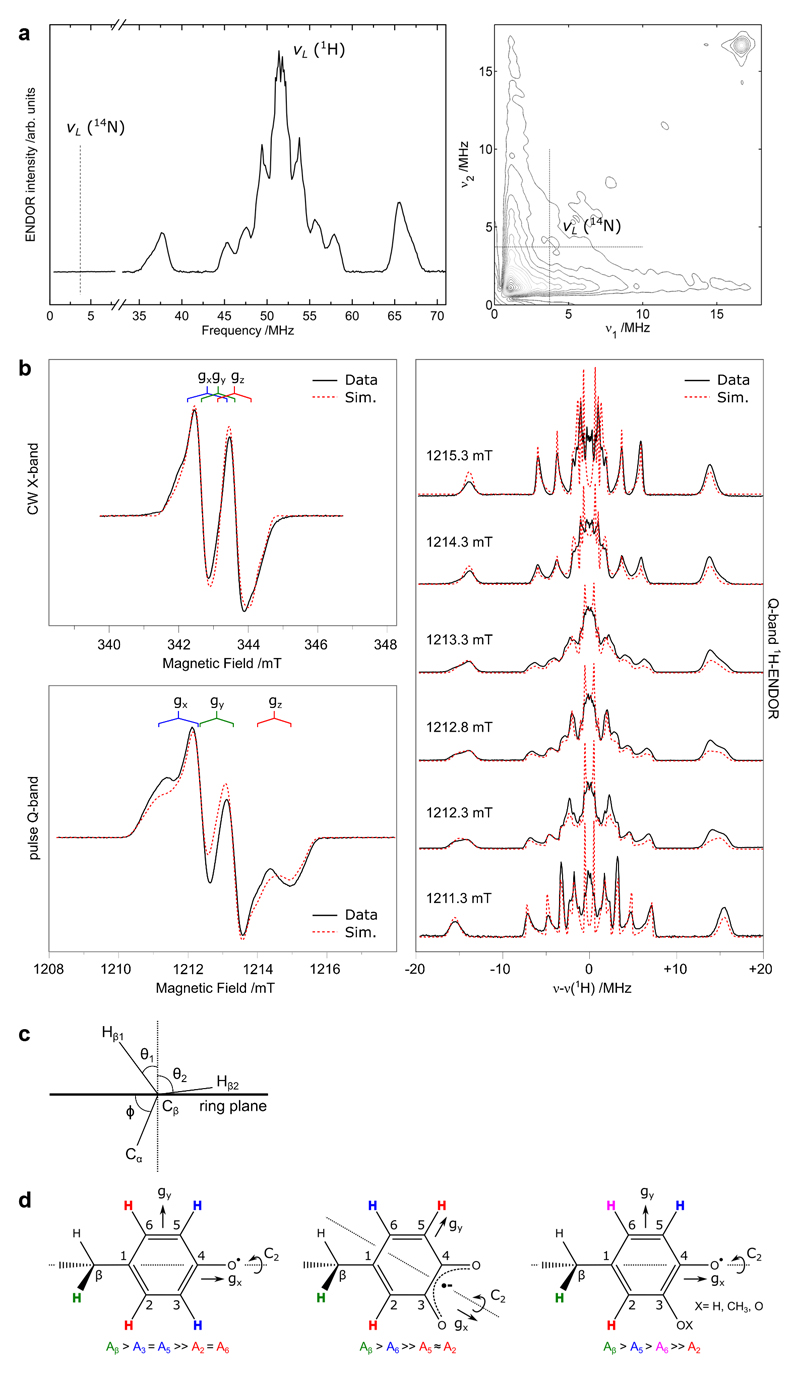
c) Q-band ENDOR spectrum recorded at the low field edge of the EPR spectrum. The red dashed lines represent a simultaneous simulation of all datasets. Spectral simulations of the multifrequency EPR and ENDOR spectra using the spin Hamiltonian formalism reveal the radical has four resolved non-equivalent proton couplings with isotropic values of 28.7, 9.8 6.5 and 4.4 MHz. The absence of a nitrogen coupling excludes the side-chains of tryptophan and histidine as the site of the stable radical. In addition, the absence of equivalent proton couplings also excludes the side-chains of tyrosine and phenylalanine. Thus, no native aromatic protein residue can explain the observed radical species. The magnitude of coupling constants are smaller than those of tyrosyl (phenoxyl) radicals, and are instead in good agreement with phenoxyl radicals with an additional oxy substituent (O-X). All ENDOR measurements were repeated at a second microwave frequency (W-band) giving similar results. Pulse EPR and ENDOR measurements represent long data accumulations/averages. EPR: 300 averages ENDOR: 600 averages.
EPR and ENDOR spectroscopy is consistent with a DOPA radical, but not an unmodified tyrosyl radical (Figure 3b,c, for a full description see Extended data Fig. 7 and Supplementary information). Spectral simulations suggest that the new substituent is an oxy group (O-X) and are fully consistent with a hydroxyl substituent or a strong hydrogen bond as indicated by crystallography and mass spectrometry.
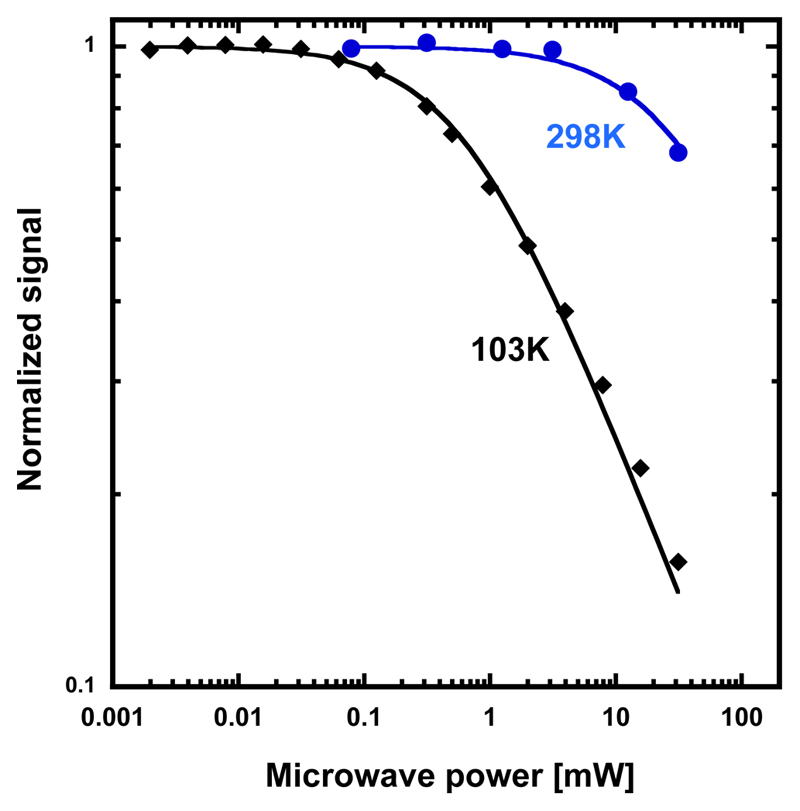
Unlike typical R2 proteins, EPR saturation data also demonstrate that the radical has no interaction with a metal (Extended data Fig. 8), consistent with the absence of a metal cofactor8,25,26. Quantification of the radical species by EPR shows that the active MfR2 sample contains ≈ 52% of radical per protein monomer. As described previously, the cumulative amount of transition metals measured by TXRF for the same sample is less than 4% per protein. It is thus not possible that a metal ion is required for stabilization of the observed radical species.
Comprehensive structural, EPR, UV/vis, TXRF and mass spectrometry data thus support that the novel radical species is metal-independent, located on a meta-hydroxylated tyrosyl residue and represents an unprecedented intrinsic DOPA radical within an RNR system.
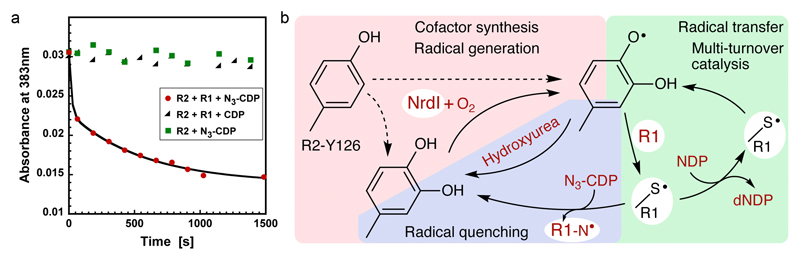
To investigate whether the observed radical species is catalytically competent, we utilized the mechanism-based inhibitor 2’-azido-2’deoxycytidine-5’-diphosphate (N3-CDP). Incubation of RNR with N3-CDP under turnover conditions leads to the loss of the R2 radical, concomitant with the formation of a nitrogen-centered radical in the active site of protein R127–29. Incubation of active MfR2 with MfR1 and N3-CDP led to the disappearance of the R2-centered radical species. Catalytic turnover with the substrate (CDP) or incubation of protein R2 with N3-CDP in the absence of protein R1 did not lead to the loss of the R2 radical (Fig. 4a). These results prove that the observed radical in protein MfR2 initiates the catalytic radical chemistry in protein R1 in this new RNR subclass.
Fig 4. Catalytic competency of the radical and proposed mechanistic scheme in class Ie RNR.
a) UV/vis absorbance at 383 nm vs time. The radical signal is quenched in the presence of protein R1 and the mechanism-based inhibitor N3-CDP (red). Protein R2 with N3-CDP alone (green) or turnover conditions with protein R1 and CDP (black) does not quench the MfR2 radical. The experiment shows that the observed radical can be reversibly transferred to the active site and support catalysis in protein R1. Experiments were repeated three times.
b) Proposed mechanistic steps in class Ie RNR. The radical-harboring cofactor is first post-translationally generated by hydroxylation of Tyr126 in an NrdI and O2 dependent process. Dashed lines indicate alternative paths for the post-translational modification of Tyr126. It is presently unknown if this reaction also directly forms the radical species. Once the DOPA radical is formed in protein R2 it supports multi-turnover ribonucleotide reduction together with protein R1, presumably analogous to other class I RNR systems. If the radical is lost, activity can be restored in the covalently modified R2 protein by NrdI, again in an O2 dependent process.
Here we identify a novel type of class I RNRs, found in human pathogens, which performs aerobic multiple-turnover ribonucleotide reduction using a DOPA initiator radical. We propose that this RNR group is denoted class Ie. Figure 4b summarizes our current understanding of the system. Remarkably, NrdI achieves two different oxygen-dependent reactions in this system, tyrosine hydroxylation and DOPA radical generation, 2- and 1- electron oxidations respectively. Detailed characterization of the covalent modification and radical generation mechanisms are exciting future prospects.
It has previously been shown that a synthetically introduced DOPA residue in the radical transfer path in E. coli RNR functions as a radical trap preventing RNR turnover, likely due to that the reduction potential is ≈ 260 mV lower than a tyrosine at pH 7.030. The EPR characteristics of the stable radical in MfR2 show that it is electronically more similar to a substituted tyrosyl radical than an o-semiquinone. It appears likely that the asymmetry is induced by the protein to tune the redox potential to allow reversible transfer to the cysteinyl radical in the R1 subunit.
Ribonucleotide reductase was the first enzyme identified to utilize a protein-derived radical for catalysis26,31. While the radical generation systems differ, all types of RNR, in all domains of life, have been considered to have an absolute metal dependence in order to generate the essential catalytic radical. Differences in metal requirement among RNRs, of which many organisms possess more than one, are believed to provide an advantage in environments where some metals are limiting. Metal starvation, including restricting access to both iron and manganese, is a central strategy in innate immunity to combat invading pathogens32. It is tempting to speculate that class Ie RNR evolved in response to such extremely metal-restricted environments. The post-translational modification of Tyr126 that generates the intrinsic DOPA cofactor is NrdI and O2 dependent, and may potentially also require other factors, including metals. Still, even if this would be the case, it would require only catalytic amounts of metal in relation to the R2 protein, drastically reducing the amount of metal required for the RNR machinery in the organisms encoding this type of RNR.
Methods
Bioinformatics
NCBI’s RefSeq database was searched with custom NrdF HMMER33 profiles on 17 February 2017 resulting in 4620 sequences. QSK/VPK variants were manually identified. To reduce the number of highly similar sequences in the phylogeny, the sequences were clustered at 0.75 identity using USEARCH34 and poor quality sequences were manually removed. The remaining 181 sequences were aligned with ProbCons35 and reliable columns in the alignment were identified with the BMGE program using the BLOSUM30 matrix36. A maximum likelihood phylogeny was estimated from the alignment with RAxML version 8.2.437, using the PROTGAMMAAUTO model with maximum likelihood estimation starting from a population of 750 rapid bootstrap trees. The number of bootstrap trees was determined determined with the MRE-based bootstopping criterium.
Cloning
Genomic DNA of Mesoplasma florum was extracted from a Mesoplasma florum L1 (NCTC 11704) culture obtained from the National Collection of Type Cultures operated by Public Health England. The M. florum nrdF gene was amplified from M. florum genomic DNA by PCR using MfnrdF-fow and MfnrdF-rev primers and ligated between NheI-BamHI restriction sites of a modified pET-28a plasmid (Novagen), in which the thrombin cleavage site following the N-terminal His6 tag has been replaced by a TEV site. All codons translating to Tryptophan in the MfnrdF gene were mutated from TGA to TGG to correct for the codon usage difference between M. florum and E. coli. The QuikChange Lightning Multi Site-Directed Mutagenesis Kit from Agilent Technologies, using MfnrdF-mut1, MfnrdF-mut2, MfnrdF-mut3 and MfnrdF-mut4 primers (Extended data Fig. 9a), as per the protocol suggested by the manufactures, were used for these changes. Similarly, the pET28MfnrdI plasmid was generated by ligating the MfnrdI PCR amplicon resulting from MfnrdI-Fow and MfnrdI-Rev primers, into the modified pET-28a plasmid between NdeI-BamHI restriction sites. A synthetic gene coding for M. florum nrdE with a N-terminal His6 tag and a TEV cleavable site, into a pET-21a vector in between the restriction sites NdeI-EcoRI and codon optimized for overexpression in E. coli was ordered from GenScript (pET21MfnrdE). To generate the pET28MfnrdFIE plasmid the MfnrdF, MfnrdI and MfnrdE genes were individually amplified with primer pairs of MfnrdF-Fow-MfnrdF-Rev, MfnrdI2-Fow-MfnrdI2-Rev and MfnrdE-Fow-MfnrdE-Rev and double digested by restriction enzymes pairs of NheI-BamHI, BamHI-SacI and SacI-SalI respectively. The amplicons were then ligated into the NheI and SalI restriction site of a modified pET-28a plasmid with MfnrdF placed with a N-terminal TEV cleavable HIS-tag and ribosome binding sites placed ahead of both MfNrdI and MfNrdE genes (Extended data Fig. 9b). This entire operon assembly was double digested from pET28MfnrdFIE and ligated between KpnI and SalI restriction digestion sites of pBAD18 to generate the pBAD18MfnrdFIE plasmid. pET28MfnrdFI was prepared similarly by double digesting pET28MfnrdFIE with NheI and SacI and ligating it into a modified pET-28a plasmid. All of the above-mentioned plasmids were sequenced to check for any unintended mutations.
In vivo studies
A 4 ml LB (ForMedium) culture of E. coli double knockout JEM164 (ΔnrdAB, ΔnrdEF~srlD::Tn10) supplemented with tetracycline was grown in an anerobic glovebox (MBraun Unilab Plus SP model) with O2 < 10 ppm, washed twice with ice cold water to prepare electro competent cells, transformed with pBAD18MfnrdFIE plasmid and plated on a LB-agar media with tetracycline and carbenicillin. Primary cultures of both JEM164 and JEM164-pBAD18MfnrdFIE were grown at 37°C overnight anaerobically, supplemented with respective antibiotics. The cultures were removed from the glovebox and inoculated into aerobic LB media supplemented with tetracycline and 0.1% L-arabinose for JEM164 and tetracycline, carbenicillin and either 0.1% D-glucose or 0.1% L-arabinose for JEM164-pBAD18MfnrdFIE and grown for 48 hours at 37°C in a bench-top bioreactor system (Harbinger). Tetracycline was used at a concentration of 12.5 μg/ml and carbenicillin was used at a concentration of 50 μg/ml for all of the above-mentioned cultures.
Protein expression in aerobic conditions
E. coli BL21(DE3) (NEB) carrying the plasmid pRARE camR (Novagen) were transformed with each of the pET28MfnrdF, pET28MfnrdI, pET28MfnrdFI and pET28MfnrdFIE plasmids individually. Glycerol stocks of the transformed cells were flash-frozen and stored at -80°C. All cultures were grown in LB media at 37° C, supplemented with 50 μg/ml kanamycin and 25 μg/ml chloramphenicol, in a bench-top bioreactor system (Harbinger) until an OD600 of ~ 0.7 was reached followed by induction with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The cultures were then allowed to grow overnight at room temperature. pET21MfnrdE –BL21(DE3) was grown similarly, but with an antibiotic selection of 50 μg/ml carbenicillin instead. Post-expression the cells were harvested by centrifugation and stored at -20°C until further use.
Protein purification
The cell pellets were thawed and re-suspended in lysis buffer (25 mM HEPES-Na pH 7, 20 mM imidazole and 300 mM NaCl) and lysed using a high-pressure homogenizer (EmulsiFlex-C3). The un-lysed cells and cell debris was pelleted by centrifugation and the clear supernatant was applied to a lysis buffer-equilibrated gravity-flow Ni-NTA agarose resin (Protino) column and washed with lysis buffer of 40 mM imadizole concentration. The bound protein was eluted with elution buffer (25 mM HEPES-Na pH 7, 250 mM imidazole and 300 mM NaCl), concentrated and loaded onto a HiLoad 16/60 Superdex 200 size exclusion column (GE Healthcare) attached to an ÄktaPrime Plus (GE Healthcare) equilibrated with SEC buffer (25 mM HEPES-Na pH 7 and 50 mM NaCl). The fractions corresponding to MfR2 was pooled and treated to TEV protease cleavage overnight, at a molar ratio of one TEV protease for every 50 MfR2 monomers. TEV protease and any un-cleaved HIS-tagged MfR2 was removed by performing an additional reverse Ni-NTA step. The purity of the protein was evaluated using SDS-PAGE, the protein was then concentrated using a Vivaspin 20 centrifugal concentrators 30,000 Da molecular weight cut-off polyethersulfone membrane (Sartorius) to a desired concentration. All of the above purifications steps were performed at 4° C. The purified MfR2 protein was then aliquoted, flash-frozen in liquid nitrogen and stored at -80°C. MfNrdI and MfR1 were purified similarly but with a buffer system of 25 mM Tris-HCl pH 8 instead of 25 mM HEPES-Na pH 7.
Anaerobic MfR2 production and purification
The glycerol stock of E. coli transformed with the pET28MfnrdFIE was used to inoculate a primary culture of LB media supplemented with 50 μg/ml kanamycin and 25 μg/ml chloramphenicol and grown in aerobic conditions at 37°C overnight. All following steps of protein production and purification were carried out anaerobically in an anaerobic chamber. The primary culture was transferred into the anaerobic chamber and used to inoculate at 2% (v/v) a secondary culture of TB media (ForMedium) deoxygenated by N2-saturation and supplemented with 50 μg/ml kanamycin and 25 μg/ml chloramphenicol, and grown anaerobically at 37°C until an OD600 of 1.1 was reached. This secondary culture was then used to inoculate deoxygenated TB media supplemented with 50 μg/ml kanamycin and 25 μg/ml chloramphenicol. As the cell culture reached an OD600 of 0.7, overexpression was induced with 0.5 mM IPTG and allowed to grow overnight at 37°C, the cells were then harvested by centrifugation. In order to extract the soluble fraction, the cell pellet was resuspended at room temperature using 5 ml of deoxygenated BugBuster Protein Extraction Reagent (Novagen) per gram of wet cell paste. The cell suspension was stirred for 40 min at room temperature. The insoluble cell debris were removed by centrifugation at 13,000 × g for 20 min. From the supernatant, the MfR2 protein was purified, as described in the aerobic procedure, using deoxygenated buffers. Instead of size exclusion chromatography, a HiTrap Desalting column (GE Healthcare) was used to remove the imidazole. The protein was then subjected to TEV protease treatment at a molar ratio of one TEV protease for every 10 MfR2 monomers at room temperature for 1 h. TEV protease and any un-cleaved HIS-tagged MfR2 was removed by passing the sample over a Ni-NTA agarose (Protino) gravity flow column equilibrated in the SEC buffer (25 mM HEPES-Na pH 7.0, 50 mM NaCl). The flow-through containing pure cleaved MfR2 was concentrated to a desired concentration, aliquoted, flash-frozen in liquid nitrogen and stored at −80 °C.
In vitro activity
The in-vitro M. florum R1-R2 catalyzed reduction of CDP to deoxy-CDP was measured in a HPLC system (Agilent 1260 Infinity) using a Waters Symmetry C18 column. A 50 μl reaction mixture consists of 5 μM MfR1 protein, 5 μM MfR2 protein, 50 mM of Tris-HCl pH 8.0, 20 mM MgCl2, 50 mM DTT and 0.5 mM of dATP. The reaction was started with the addition of 2 mM CDP and allowed to run at room temperature for 30-60 minutes, then quenched with 50 μL of Methanol. Additional 200 μL of Milli-Q water was added to the reaction mixture. The concentration of the deoxy-CDP product was measured as described previously16,38. All of the in-vitro RNR enzymatic reactions were performed with three replicates. Regeneration of quenched MfR2 in presence of EDTA or metals were performed by incubating MfR2 (0.3 mM) with either EDTA pH 7.0 (0.3 mM) or a mixture of metals (Mn, Fe, Co, Ni, Cu and Zn at 0.2 mM each), before adding MfNrdI and dithionite at 0.6 mM and 2 mM, respectively.
Crystallization
M. florum R2 was crystallized by sitting drop vapor diffusion method at a protein concentration of 25 mg/ml. A Mosquito nanoliter pipetting robot (TTP Labtech) was used to set up MRC 2-well crystallization plates (Swissci) with 200:200 nl protein to reservoir volume ratios and reservoir volume of 50 μl. Initial crystallization hits were obtained in condition number C4 of the PEG/Ion HT crystallization screen (Hampton Research). The crystallization condition was further optimized using the Additive screen HT (Hampton Research) with condition number B3 producing the best hits. The crystallization condition is as follows: 100-200 mM Calcium acetate, 100 mM Ammonium sulfate and 15-20% PEG 3350, with cuboidal crystals appearing within 2-3 days of incubation at room temperature. Reservoir solution supplemented with 20% glycerol was used as cryoprotectant. Crystals were flash-frozen in liquid nitrogen before data collection
Data collection and structure determination
X-ray diffraction data were collected at beamline I24 of the Diamond Light Source (Oxfordshire, United Kingdom) at a wavelength of 0.96859 Å at 100 K. Data reduction was carried out using XDS39.
MfR2 crystal structures were solved using PHASER40 by molecular replacement using the atomic coordinates of the R2 protein from Corynebacterium ammoniagenes (PDB: 3dhz)41 as a starting model. A well-contrasted solution was obtained with 2 molecules per asymmetric unit in the space group C2 for all the datasets. Crystallographic refinement was performed using PHENIX42 applying anisotropic B-factor and TLS, and was similarly but independently conducted for the 3 models. The 3D models were examined and modified using the program COOT43 and validated using MolProbity44. The core root-mean-square deviation values between structures were calculated by the Secondary-Structure Matching (SSM) tool45. Extended data Table 1 was generated with phenix.table_one and lists the crystallographic statistics in which the test set represents 5% of the reflections. The Ramachandran statistics are: favored 98.5%, 99.2% and 99.5% for the active, inactive aerobic and inactive anaerobic MfR2 crystal structures, respectively. Ramachandran outliers were 0.0% in all cases. Figure 2 was prepared using the PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
For anomalous signal analysis, data were processed with XDS keeping the Friedel pair reflections unmerged (FRIEDEL'S_LAW=FALSE) to preserve any anomalous contributions. Anomalous difference maps were calculated with PHENIX.
EPR sample preparation
Class Ia E. coli nrdB was PCR amplified using primers pairs of EcnrdB-Fow and EcnrdB-Rev from genomic DNA of E. coli BL21 (DE3) (Novagen). The PCR amplicon was then double digested with restriction enzymes pair of NheI-BamHI and ligated into a modified pET-28a plasmid. Class Ib Bacillus cereus nrdF gene was restriction digested out of pET-22bBcnrdF using NdeI-HindIII restriction enzymes and ligated into a modified pET-28a plasmid. The resulting plasmids were sequenced for any unintended mutations. The E. coli R2a and B. cereus R2b proteins were produced and purified using protocols as described earlier. A few modifications to the protocols were made in order to obtain metal-free R2 proteins i.e., addition of 0.5 mM of ethylenediaminetetraacetic acid (EDTA) to the growth cultures before induction of overexpression with 0.5 mM IPTG and addition of 0.5 mM of EDTA to the lysis buffer. The metal-free E. coli R2 and B. cereus R2 proteins were then metal loaded with (NH4)2Fe(SO4)2 solution with a final concentration of 1.5 molar equivalent of Fe(II) per monomer and left to incubate at room temperature for 60 minutes under aerobic conditions. The R2 proteins were later diluted with 100% glycerol to a final protein concentration of 0.575 mM and 0.695 mM respectively. Active M. florum R2 CW-X band EPR sample was also diluted with 100% glycerol to a final protein concentration of 0.675 mM.
CW X-band EPR measurements
X-Band CW-EPR measurements were performed at 70 K using a Bruker E500 spectrometer equipped with a Bruker ER 4116DM TE102/TE012 resonator, Oxford Instruments ESR 935 cryostat and ITC-503 temperature controller. Microwave power was 20 μW and magnetic field modulation amplitude was 0.5 G. The static magnetic field was corrected (for X- and Q-band measurements) using a Bruker ER 035M NMR Gaussmeter. For measurements at 103K the temperature was controlled by flowing gas of known temperature over the sample in a dewar in the cavity. For spectra at 298 K the sample was contained in a capillary. Quantifications were performed by comparing double integrals of the samples with that a standard Cu2+/EDTA (1 mM and 10 mM, respectively) sample at non-saturating conditions.
Microwave saturation measurements
The amplitude of the first derivative of an EPR signal is a function of the applied microwave power (P). Plotting the normalized intensity of the EPR signal X'=(Y'/P1/2)/(Y'0/P01/2) as a function of P in the logarithmic form allows determination of the half-saturation power (P1/2)25. Under non-saturating conditions X' equals 1. As the signal starts to saturate, X’ decreases. The half-saturation power is calculated from the equation X'= 1/(1+P/P1/2)1/2 for an inhomogeneously broadened signal. The value of P1/2 is affected by the magnetic environment of the studied radical. The temperature dependence of P1/2 can provide information on an adjacent metal center, or it may reflect the absence of metals in the vicinity of the radical. R2Tyr• with adjacent diiron centers have P1/2 values that are considerably higher than those for isolated Tyr• radicals at temperatures above 20-30 K25.
EPR radical measurements of MfR2 with specifically deuterated amino acids
E. coli BL21(DE3) (NEB) carrying the plasmid pRARE camR (Novagen) transformed with the pET28MfnrdFIE plasmid was grown at 37 °C in a bench-top bioreactor system (Harbinger) in M9 minimal media (Minimal Salts Base from ForMedium containing Na2HPO4, KH2PO4, NaCl and NH4Cl) with 4 mM MgSO4, 0.2 mM CaCl2, 1% D-glucose and 50 μg/mL kanamycin, without addition of trace metals. The media was supplemented with either 50 mg/l L-Tyrosine-(ring-3,5-D2) or 50 mg/l Glycine-D5 or 40 mg/l L-Tryptophan-(indole-D5) (Cambridge Isotope Laboratories) or 100 mg/l DL-Tyrosine-(β,β-D2)46. As the cell cultures reached OD600 of 0.6-0.9, 0.5 mM EDTA was added, followed by induction with 0.5 mM IPTG. Induction continued 36 h at 37 °C. MfR2 overexpression was checked by SDS-PAGE. The cells were harvested by centrifugation and loaded into EPR tubes before flash freezing. Spectra were recorded at 100K in a nitrogen flow system at 1 mW and 2 G modulation amplitude. The spectra have been normalized to the same double integrals, i.e same number of spins in the cavity.
Pulse-Q band EPR measurements
Q-band pulse-EPR measurements were performed in the temperature range of 50 to 70 K using a Bruker ELEXSYS E580 Q-band pulse-EPR spectrometer, equipped with a homebuilt TE011 microwave cavity47 Oxford-CF935 liquid helium cryostat and an ITC-502S temperature controller. Electron spin echo-detected (ESE) field-swept spectra were measured using the pulse sequence: tp−τ−2tp−τ−echo. The length of the π/2 microwave pulse was generally set to tp = 22 ns and the interpulse distance was τ = 260 ns. Pseudomodulated EPR spectra were generated by convoluting the original absorption spectra with a Bessel function of the 1st kind. The peak-to-peak amplitude used was 0.8 G. 1H-ENDOR spectra were collected using the Davis pulse sequence: tinv−tRF−T−tp−τ−2tp−τ−echo with an inversion microwave pulse length of tinv = 140 ns, a radio frequency pulse length of tRF = 20 μs (optimized for 1H). The length of the π/2 microwave pulse in the detection sequence was set to tp = 70 ns and the interpulse delays to T = 22 μs and τ = 400 ns. The RF frequency was swept 40 MHz around the 1H-Larmor frequency of about 52 MHz (at 1.2 T) in 100 or 50 kHz steps. The RF amplifier used for ENDOR measurements was ENI 3200L. 1H- and 14N-ENDOR spectra were collected using the Davis pulse sequence: tinv - tRF-T- tp -t- 2tp -t-echo with an inversion microwave pulse length of tinv = 140 ns, a radio frequency pulse length of tRF = 20 ms (optimized for 1H) and 30 ms (optimized for 14N). HYSCORE spectra were collected using the pulse sequence: tp -t- 2tp - t1 -2tp - t2-tp- t-echo. Two values of t where used, 208 and 220 ns. t1 and t2 were incremented in 4 ns steps.
Total-reflection X-ray fluorescence (TXRF) metal quantification
The metal contents of protein and buffer solutions were quantified using TXRF analysis on a Bruker PicoFox S2 instrument. For each solution, measurements on 3 independently prepared samples were carried out. A gallium internal standard at 2 mg/l was added to the samples (v/v 1:1) prior to the measurements. TXRF spectra were analyzed using the software provided with the spectrometer. As a control, the metal-free E. coli class Ia R2 protein was Fe-reconstituted by incubation with 2 molar equivalent of Fe(II) per monomer at room temperature for 60 min under aerobic conditions. The protein solution was then applied on a HiTrap Desalting column (GE Healthcare) in order to remove unbound iron, and was concentrated using a Vivaspin centrifugal concentrator, to a concentration similar to the MfR2 sample used for TXRF measurements.
Small Angle X-Ray Scattering
SAXS measurements were carried out on beamline B21 at the Diamond Light Source at 12.4 keV in the momentum transfer range 0.0038 < q < 0.41 Å−1 (q = 4π sin (θ)/λ, 2θ is the scattering angle) using a Pilatus 2M hybrid photon-counting detector (Dectris Ltd., Baden, Switzerland). Prior to measurements, samples of MfR2 alone or MfR2 incubated with MfNrdI, with a molar ratio R2:NrdI of 1:1.5, were run on a HiLoad 16/60 Superdex 200 prep grade size exclusion chromatography column (GE Healthcare) equilibrated in the SEC buffer 25 mM HEPES pH 7.0, 50 mM NaCl. Fractions corresponding to MfR2 or to the complex of MfR2-MfNrdI were pooled, diluted to 3 different concentrations with the SEC buffer and flash-frozen until measurements. A volume of 30 μl of either MfR2 alone or MfR2 incubated with MfNrdI was loaded onto the sample capillary using the EMBL Arinax sample handling robot. Each dataset comprised 18 exposures each of 180 seconds. Identical buffer samples were measured before and after each protein measurement and used for background subtraction. Data averaging and subtracting used the data processing tools of the EMBL-Hamburg ATSAS package48. The radius of gyration (Rg) and maximum particle size (Dmax) were determined from tools in the PRIMUS program suite49. Twenty independent ab initio models of either MfR2 or the MfR2-NrdI complex were derived from the experimental scattering curves using DAMMIF50. For each protein, the models were aligned, averaged and filtered using the DAMAVER program suite51. Theoretical scattering profiles of the crystal structure of MfR2 and the MfR2-NrdI homology model were calculated and fitted against the experimental data using CRYSOL52. The homology model of the MfR2-NrdI complex was created by superposing the MfR2 dimer crystal structure and an MfNrdI homology model prepared based on the crystal structure of the R2-NrdI complex from E. coli (PDB: 3N3A)19.
Proteolytic digestion and peptide analysis by LC-MS
The protein of interest was cleaned from contaminants in the SEC buffer following a modified version of the SP3 protocol53,54. Samples were digested and subjected to LC-MS/MS analysis. Unless noted otherwise, all reagents were purchased from Sigma Aldrich.
The SP3 beads stock suspension was prepared freshly prior to sample processing. The two SP3 bead bottles (Sera-Mag Speed beads – carboxylate modified particles, P/N 65152105050250 + P/N 45152105050250 from Thermo Scientific) were shaken gently until the suspension looked homogenous, and then 50 μl from each bottle was taken to one tube. The 100μl of beads was then washed three times with water (in each wash, tubes were taken off the magnetic rack, beads were mixed with 500 μl H2O by pipetting, then placed back in the rack, and after a 30 s delay, the supernatant was removed). Beads were finally resuspended in 500 μl H2O after washing.
To each aliquot of 30 μl of purified protein solution in SEC buffer at 70 μM protein concentration, 10 μl of SP3 stock suspension was added. MeCN (acetonitrile) was added to reach a final concentration of 50 % (v/v). The reaction was allowed to stand for 20 min. The beads were collected on the magnetic rack and the supernatant was discarded. The beads were washed two times with 70 % (v/v) EtOH and once with MeCN. The beads were then resuspended in 100 μl of digestion buffer containing either i) 1 μg of Chymotrypsin in 50 mM HEPES, pH 8, 10 mM CaCl2, or ii) 1 μg of Pepsin in 50 mM HCl, pH 1.37. Samples were digested at 37°C overnight. The beads were removed and where necessary HCl was neutralized with one equivalent of triethylammonium bicarbonate buffer (1M, pH 8.5). From each sample, 8μl was injected to LC-MS/MS analysis on an LTQ-Orbitrap Velos Pro coupled to an Agilent 1200 nano-LC system. Samples were trapped on a Zorbax 300SB-C18 column (0.3x5 mm, 5 μm particle size) and separated on a NTCC-360/100-5-153 (100 μm internal diameter, 150 mm long, 5 μm particle size, Nikkyo Technos., Ltd, Tokyo, Japan, (http://www.nikkyo-tec.co.jp) picofrit column using mobile phases A (3% MeCN, 0.1% FA) and B (95% MeCN, 0.1% FA) with a gradient ranging from 3% to 40% B in 45 min at a flowrate of 0.4 μl/min. The LTQ-Orbitrap Velos was operated in a data-dependent manner, selecting up to 3 precursors for sequential fragmentation by both CID and HCD, analyzed in the Orbitrap. The survey scan was performed in the Orbitrap at 30 000 resolution (profile mode) from 300-1700 m/z with a max injection time of 200 ms and AGC set to 1 x 106 ions. MS2 scans were acquired at 15 000 resolution in the Orbitrap. Peptides for dissociation were accumulated for a max ion injection time of 500 ms and AGC of 5 x 104 with 35% collision energy. Precursors were isolated with a width of 2 m/z and put on the exclusion list for 30 s after two repetitions. Unassigned charge states were rejected from precursor selection. The raw files were searched against a database only containing the sequence of NrdF R2 protein with Proteome Discoverer 1.4 and the Sequest algorithm55 allowing for a precursor mass tolerance of 15 ppm and fragment mass tolerance of 0.02 Da. Non-enzyme searches were conducted and methionine and tyrosine oxidations were included as variable modifications.
For the MODa56 searches, raw files were first converted to mzXML files using msconvert from ProteoWizard57 using vendor peak picking at the MS2 level and otherwise standard settings. The mzXML files were searched against the sequence of NrdF with MODa, allowing for an arbitrary number of mods with sizes within the +/- 200 Da interval, and using a fragment mass tolerance of 0.05 Da. Otherwise the same settings as for the Sequest searches were chosen. The high-resolution mode was used.
Intact protein analysis by LC-MS
A volume of 2 μl of inactive MfR2 and active R2, at a concentration of 70 μM, were diluted in 200μl of LC-MS mobile phase A (3 % MeCN, 0.1% FA). From this, 3 μl was injected in each LC-MS run, which was set up as described in the previous section, but with the following differences: the trapping column was a Zorbax 300SB-C8 column (0.3x5mm, 5μm particle size); the gradient went from 3% B to 99% B in 10 min at a flow rate of 0.6 μl/min; the MS only acquired full scans of the 600-4000 m/z high mass range at 100 000 resolution. The raw MS files were processed with Protein Deconvolution 4.0 (Thermo Scientific), using the ReSpect algorithm therein. Default parameters were used except: the relative abundance threshold was set to 40%, the m/z range set to 800-2400 m/z and the charge state range set to 6-100.
Extended Data
Extended data Figure 1. Unrooted maximum likelihood phylogeny of representative NrdF (RNR subclass Ib radical-generating subunit) sequences.
All RefSeq NrdF sequences were clustered at 75% identity to reduce redundancy and a maximum likelihood phylogeny was estimated. Sequences with non-canonical amino acids in the positions involved in coordinating the metal center of the enzyme formed a well-supported clan in the NrdF2 group of sequences. We identified two variants, one in which three of the glutamates were replaced by Gln, Ser, and Lys (NrdF2.QSK) and the other in which they were replaced by Val, Pro and Lys (NrdF2.VPK). Both variants thus have a substitution of a Lys for the normally metal-bridging Glu (residue 213 in M. florum NrdF2.VPK). Together, the two variants form a well-supported (96% bootstrap support) clan in the phylogeny inside the NrdF2 diversity. The NrdF2.VPK clan appears to be derived from the NrdF2.QSK clan. Behind the sequences in the tree are a set of sequences more than 75% identical to each represented sequence. The VPK and QSK sequences in the phylogeny represent 138 and 182 sequences in RefSeq respectively.
Extended data Figure 2. Small angle X-ray scattering characterization of the MfR2-NrdI complex.
Solution scattering data for MfR2 (left) and MfR2 incubated with MfNrdI (right).
a) Experimental solution scattering profiles (black spheres) for MfR2 alone and incubated with MfNrdI superposed with the theoretical scattering profile of the MfR2 crystal structure (red line) and the theoretical scattering profile from the homology model based on the E. coli R2-NrdI complex structure (blue line). Theoretical scattering curves and goodness of fit values were calculated by CRYSOL.
b) Guinier fit and p(r) function of MfR2 alone and incubated with MfNrdI. The fit to the data is shown as an orange line. The shift in invariant parameters Rg and Dmax indicate that an increase in dimensions occurred as MfR2 was incubated with MfNrdI. Radius of gyration statistics were derived from 60 data points within the Guinier region for MfR2 and 55 for MfNrdI-MfR2.
c) ab initio models as calculated by DAMMIN of MfR2 alone and with NrdI (grey surface) overlaid with the crystal structure of MfR2 (left) and the homology model based on the E. coli R2-NrdI complex structure model (right).
Extended data Figure 3. Metal analysis and radical generation in the presence of chelator.
a) Representative TXRF spectra measured for MfR2 (blue, at 664 μM) and Fe-reconstituted class Ia EcR2 (orange, at 635 μM), on the 5 to 10 keV energy range. The spectra have been scaled using the peak size of the Ga internal standard and offset slightly in the Y-direction for clarity. K-level X-ray emission lines are indicated with arrows. For elements, where both Kα and Kβ lines are present, they are specified. Otherwise, arrows indicate Kα lines. Experiments were repeated three times.
b) Concentrations of Mn, Fe, Co, Ni, Cu and Zn were measured in the active MfR2, MfNrdI and MfR1 protein solutions and in their respective buffers, as well as in a solution of E. coli class Ia R2 protein reconstituted with Fe in vitro. Mean concentrations and SD of measurements on 3 independently prepared samples for each sample are reported. The concentrations were converted to metal to protein molar ratio. The measurements show that none of the MfRNR proteins contain a significant amount of metal as opposed to EcR2a which as expected contains on the order of 2 metal ions per monomer also after a desalting step. Buffer 1 is the buffer system used for MfR2, i.e. 25 mM HEPES-Na pH 7, 50 mM NaCl. Buffer 2 is the buffer system used for MfR1 and MfNrdI, i.e. 25 mM Tris-HCl pH 8, 50 mM NaCl. The protein purification involves a Ni-affinity step which is likely the reason for nickel being the dominating metal species in the sample.
c) HPLC based in vitro assays show that RNR activity can be restored after MfR2 is quenched by hydroxyurea. MfR2 is regenerated by the addition of MfNrdI followed by redox cycling with dithionite and oxygen containing buffer (green) (see main Fig. 2d). Reactivation and activity are observed also in the presence of a metal chelator (EDTA 0.3 mM, blue). Addition of extra metals (0.2 mM of each Mn, Fe, Co, Ni, Cu, Zn) does not improve the activity recovery (pink).
Extended data Figure 4. Mass spectrometric characterization of intact proteins.
Intact protein mass spectra obtained from purified MfR2 proteins. Inactive protein (above) and active protein (below). Insets represent the decharged and deisotoped mass as calculated by the program Protein Deconvolution (version 4.0) using the ReSpect algorithm therein. The result from the deconvolution of n = 30 consecutive scans in one LC/MS run per protein form is shown. Each protein form was analyzed in duplicate LC/MS runs. Protein intact masses are given as mean +/- standard deviation (SD). SD was 1.4 Da for the “inactive” protein and 2 Da for the “active” form. The results show that the active protein is 17 ± 2 Da heavier than the inactive MfR2.
Extended data Figure 5. MS2 fragmentation spectra of peptides with oxidized tyrosine.
a) Annotated MS2 fragmentation spectra and respective theoretical fragment ion tables of the doubly charged precursor ion 661.8458 m/z corresponding to peptide VAVHARSY(+15.995)GSIF, and c) the doubly charged precursor ion 458.7279 m/z corresponding to peptide ARSY(+15.995)GSIF, both with the oxidized (+16) Y126 residue. The peptides shown in a) and c) were obtained by proteolytic digestion of the active form of the MfR2 protein with chymotrypsin and pepsin, respectively. The mass error is typically less than 0.01 m/z, in accordance with the high resolution used (15 000). Errors in ppm are indicated for the corresponding fragment ions when detected. Among the fragment ions observed, the most relevant are the b7 and b8 ions for peptide a). The experimental m/z values, the annotation, theoretical m/z values and ppm errors are shown in b), including the peaks for the corresponding isotope envelope. In d) the Y(+O) immonium ion for c) is shown, which demonstrate that Tyr126 is modified by a mass of +15.995 and the absence of the corresponding immonium ion for the unmodified Y. Four independent experiments per peptidase treatment were performed, confirming the modified peptide sequences shown. Figures are taken from one representative experiment.
Extended data Figure 6. Radical stability and Isotope labeling.
a) Superimposed UV/vis spectra at timepoints between 0 and 400 minutes.
Inset, absorbance at 383 nm at 0, 4, 129, 140, 210 and 400 minutes. Experiments were repeated three times.
b) X-band spectra of the radical observed in harvested cells grown in minimal media supplemented with deuterated amino acids. EDTA (0.5 mM) was added before induction.
From top: non-labeled tyrosine, β,β-D2 tyrosine, 3,5-D2 tyrosine, indole-D5 tryptophan and D5 glycine. The doublet signal collapses to a singlet when β,β-deuterated tyrosine is incorporated in the protein. Furthermore, the additional coupling to the remaining 3/5 proton in the 3,5-D2 tyrosine grown cells disappears, which is also in line with the radical being tyrosine-derived. Finally, the exclusion of tryptophan and glycine as source for the observed radical is evident from the two lower traces in the figure, which are identical to the top spectrum. Five independent cultures were grown each including the indicated deuterated amino acid. Spectra were recorded at 100K in a nitrogen flow system. The spectra have been normalized to the same double integrals, i.e. same number of spins in the cavity.
Extended data Figure 7. EPR/ENDOR characterization, see Supplementary information.
a) Q-band ENDOR and HYSCORE spectra of the spectral region where an 14N hyperfine coupling should be observed. Experimental parameters are listed in the materials and methods section.
b) Full multifrequency (X-/Q-band) EPR dataset and corresponding field dependent Q-band ENDOR spectra. Experimental parameters are listed in the materials and methods section. The red dashed lines represent a simultaneous simulation of all datasets using the spin Hamiltonian formalism. Simulation parameters are listed in Supplementary information Table 1.
c) Inferred orientation (θ1, θ2) of the Cβ protons relative to the phenoxyl radical ring plane as determined by the dihedral angle (ϕ) between the ring plane (C1) and Cα.
d) Candidates discussed for the MfR2 radical species, see Supplementary information.
All ENDOR measurements were repeated at a second microwave frequency (W-band) giving similar results. Pulse EPR and ENDOR measurements represent long data accumulations/averages. EPR: 300 averages (6 scans/50 shots). ENDOR: 600 averages (600 scans/1 shot).
Extended data Figure 8. EPR saturation.
EPR saturation behavior of the MfR2 radical at 103 and 298 K. Saturation curves at different temperatures determine the microwave power at half saturation P1/2. The temperature dependence of P1/2 gives information about possible relaxing transition metals in the vicinity of the radical. A fast relaxing metal site will give a higher P1/2 than for an isolated radical. The microwave saturation behavior of the MfR2 is similar to that for an hv-irradiated tyrosine solution. Here we evaluate P1/2 ≈ 0.6 mW at 103 K and P1/2 ≈ 30 mW at 298 K for MfR2. This can be compared to hν Tyr• with P1/2 ≈ 0.4mW at 93 K and E. coli Tyr• with P1/2 ≈ 150 mW at 106 K and not possible to saturate at 298K.
Extended data Figure 9. Primers and operon construct.
a) Primers used in this study.
b) Mesoplasma florum class Ie RNR operon construction.
Extended data Table 1. Data collection and refinement statistics.
*Values in parentheses are for highest-resolution shell.
A single crystal was used for each dataset.
| Active MfR2 | Inactive MfR2 (aerobic) | Inactive MfR2 (anaerobic) | |
|---|---|---|---|
| Data collection | |||
| Space group | C2 | C2 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 176.16, 53.74, 79.28 | 176.19, 53.43, 79.13 | 176.30 53.51 79.10 |
| α, β, γ (°) | 90, 108.58, 90 | 90, 108.53, 90 | 90 108.49 90 |
| Resolution (Å) | 42.73-1.48 (1.53-1.48)* | 42.71-1.23 (1.27-1.23) | 44. 19-1.24 (1.28-1.24) |
| Rmerge | 0.066 (1.403) | 0.054 (1.092) | 0.052 (0.951) |
| I / σI | 12.67 (1.17) | 15.13 (1.35) | 15.15 (1.50) |
| Completeness (%) | 99.8 (99.7) | 98.9 (97.3) | 97.4 (94.4) |
| Redundancy | 6.6 (6.4) | 6.4 (6.0) | 6.5 (6.2) |
| Refinement | |||
| Resolution (Å) | 42.73-1.43 (1.53-1.43) | 42.71-1.23 (1.27-1.23) | 44. 19-1.24 (1.28-1.24) |
| No. reflections | 117311 (11659) | 200307 (19622) | 192936 (18564) |
| Rwork / Rfree | 0.1543 (0.3738) / 0.1845 (0.4083) | 0.1466 (0.3296) / 0.1688 (0.3434) | 0.1472 (0.3008) / 0.1630 (0.3105) |
| No. atoms | 6137 | 6848 | 6743 |
| Protein | 5610 | 6128 | 6097 |
| Ligand/ion | 2 | 2 | 2 |
| Water | 525 | 718 | 644 |
| B-factors | 39.00 | 27.28 | 28.61 |
| Protein | 38.07 | 25.71 | 27.45 |
| Ligand/ion | 29.44 | 20.44 | 22.58 |
| Water | 48.89 | 40.65 | 39.67 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.007 | 0.006 | 0.006 |
| Bond angles (°) | 1.13 | 1.12 | 1.13 |
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
We would like to acknowledge the foundational contributions of P Reichard to the RNR field. We wish to thank Prof. James A Imlay for the kind gift of the E. coli (ΔnrdAB, ΔnrdEF) strain, Prof. K Kristoffer Andersson and Dr. Marta Hammerstad for the pET-22bBcnrdF plasmid and Prof. Eduard Torrents for the pBAD18 plasmid. Financial support to M.H. was provided by the Swedish Research Council (2017-04018), the European Research Council (HIGH-GEAR 724394), and the Knut and Alice Wallenberg Foundation (Wallenberg Academy Fellows (2012.0233 and 2017.0275)), to B.M.S. by the Swedish Research Council (2016-01920), the Swedish Cancer Foundation (CAN 2016/670), and the Wenner-Gren Foundations, to N.C. by the Australian Research Council (FT140100834) and the Max Planck Society. We would like to thank Diamond Light Source for beamtime (proposals mx11265 and mx15806) and particularly the staff from beamlines I24 and B21.
Footnotes
Author contributions:
B-M.S. and M.H. conceived and led the study. D.L. performed bioinformatics. V.S. did cloning and operon constructs. V.S. and H.L performed protein production, in vivo and in vitro activity assays, TXRF analysis, crystallography and sample preparation for all other methods. Y.K. M.S. and N.C. performed spectroscopy. M.L. performed SAXS experiments. J.E. and R.M.M.B. performed mass spectrometry. All authors were involved in experiment design and data analysis. M.H. wrote the manuscript with significant input from all authors.
Author Information:
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing interests.
Data availability:
Structures and crystallographic data have been deposited in the protein data bank with PDB id: 6GP2, DOPA-active form and 6GP3, inactive form. All other data can be obtained from the corresponding author upon reasonable request.
References
- 1.Hofer A, Crona M, Logan DT, Sjöberg B-M. DNA building blocks: keeping control of manufacture. Crit Rev Biochem Mol Biol. 2012;47:50–63. doi: 10.3109/10409238.2011.630372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 3.Mannargudi MB, Deb S. Clinical pharmacology and clinical trials of ribonucleotide reductase inhibitors: is it a viable cancer therapy? J Cancer Res Clin Oncol. 2017;143:1499–1529. doi: 10.1007/s00432-017-2457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aye Y, Li M, Long MJC, Weiss RS. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene. 2015;34:2011–2021. doi: 10.1038/onc.2014.155. [DOI] [PubMed] [Google Scholar]
- 5.Brown NC, Eliasson R, Reichard P, Thelander L. Nonheme iron as a cofactor in ribonucleotide reductase from E. coli. Biochem Biophys Res Commun. 1968;30:522–527. doi: 10.1016/0006-291x(68)90083-1. [DOI] [PubMed] [Google Scholar]
- 6.Lundin D, Berggren G, Logan DT, Sjöberg B-M. The origin and evolution of ribonucleotide reduction. Life (Basel) 2015;5:604–636. doi: 10.3390/life5010604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang M, Parker MJ, Stubbe J. Choosing the right metal: case studies of class I ribonucleotide reductases. J Biol Chem. 2014;289:28104–28111. doi: 10.1074/jbc.R114.596684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nordlund P, Sjöberg BM, Eklund H. Three-dimensional structure of the free radical protein of ribonucleotide reductase. Nature. 1990;345:593–598. doi: 10.1038/345593a0. [DOI] [PubMed] [Google Scholar]
- 9.Uhlin U, Eklund H. Structure of ribonucleotide reductase protein R1. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 10.Stubbe J, Nocera DG, Yee CS, Chang MCY. Radical initiation in the class I ribonucleotide reductase: Long-range proton-coupled electron transfer? Chem Rev. 2003;103:2167–2201. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 11.Cotruvo JA, Stubbe J. Class I ribonucleotide reductases: metallocofactor assembly and repair in vitro and in vivo. Annu Rev Biochem. 2011;80:733–767. doi: 10.1146/annurev-biochem-061408-095817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Högbom M. Metal use in ribonucleotide reductase R2, di-iron, di-manganese and heterodinuclear--an intricate bioinorganic workaround to use different metals for the same reaction. Metallomics. 2011;3:110–120. doi: 10.1039/c0mt00095g. [DOI] [PubMed] [Google Scholar]
- 13.Högbom M, et al. The radical site in chlamydial ribonucleotide reductase defines a new R2 subclass. Science. 2004;305:245–248. doi: 10.1126/science.1098419. [DOI] [PubMed] [Google Scholar]
- 14.Jiang W, et al. A manganese(IV)/iron(III) cofactor in Chlamydia trachomatis ribonucleotide reductase. Science. 2007;316:1188–1191. doi: 10.1126/science.1141179. [DOI] [PubMed] [Google Scholar]
- 15.Berggren G, Lundin D, Sjöberg B-M. Encyclopedia of Inorganic and Bioinorganic Chemistry. American Cancer Society; 2017. pp. 1–17. [DOI] [Google Scholar]
- 16.Rozman Grinberg I, et al. Novel ATP-cone-driven allosteric regulation of ribonucleotide reductase via the radical-generating subunit. Elife. 2018;7:389. doi: 10.7554/eLife.31529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose H, et al. Structural Basis for Superoxide Activation of Flavobacterium johnsoniae Class I Ribonucleotide Reductase and for Radical Initiation by its Dimanganese Cofactor. Biochemistry. 2018 doi: 10.1021/acs.biochem.8b00247. acs.biochem.8b00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotruvo JA, Stich TA, Britt RD, Stubbe J. Mechanism of assembly of the dimanganese-tyrosyl radical cofactor of class Ib ribonucleotide reductase: enzymatic generation of superoxide is required for tyrosine oxidation via a Mn(III)Mn(IV) intermediate. J Am Chem Soc. 2013;135:4027–4039. doi: 10.1021/ja312457t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boal AK, Cotruvo JAJ, Stubbe J, Rosenzweig AC. Structural basis for activation of class Ib ribonucleotide reductase. Science. 2010;329:1526–1530. doi: 10.1126/science.1190187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berggren G, Duraffourg N, Sahlin M, Sjöberg B-M. Semiquinone-induced maturation of Bacillus anthracis ribonucleotide reductase by a superoxide intermediate. J Biol Chem. 2014;289:31940–31949. doi: 10.1074/jbc.M114.592535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin JE, Imlay JA. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol Microbiol. 2011;80:319–334. doi: 10.1111/j.1365-2958.2011.07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roca I, Torrents E, Sahlin M, Gibert I, Sjöberg B-M. NrdI essentiality for class Ib ribonucleotide reduction in Streptococcus pyogenes. J Bacteriol. 2008;190:4849–4858. doi: 10.1128/JB.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammerstad M, Hersleth H-P, Tomter AB, Røhr AK, Andersson KK. Crystal structure of Bacillus cereus class Ib ribonucleotide reductase di-iron NrdF in complex with NrdI. ACS Chem Biol. 2014;9:526–537. doi: 10.1021/cb400757h. [DOI] [PubMed] [Google Scholar]
- 24.Land EJ, Porter G. Primary photochemical processes in aromatic molecules. Part 7.—Spectra and kinetics of some phenoxyl derivatives. Trans Faraday Soc. 1963;59:2016–2026. [Google Scholar]
- 25.Sahlin M, et al. Magnetic interaction between the tyrosyl free radical and the antiferromagnetically coupled iron center in ribonucleotide reductase. Biochemistry. 1987;26:5541–5548. doi: 10.1021/bi00391a049. [DOI] [PubMed] [Google Scholar]
- 26.Ehrenberg A, Reichard P. Electron spin resonance of the iron-containing protein B2 from ribonucleotide reductase. J Biol Chem. 1972;247:3485–3488. [PubMed] [Google Scholar]
- 27.Thelander L, Larsson B, Hobbs J, Eckstein F. Active site of ribonucleoside diphosphate reductase from Escherichia coli. Inactivation of the enzyme by 2'-substituted ribonucleoside diphosphates. J Biol Chem. 1976;251:1398–1405. [PubMed] [Google Scholar]
- 28.Eliasson R, Pontis E, Eckstein F, Reichard P. Interactions of 2‘-modified azido- and haloanalogs of deoxycytidine 5’-triphosphate with the anaerobic ribonucleotide reductase of Escherichia coli. J Biol Chem. 1994;269:26116–26120. [PubMed] [Google Scholar]
- 29.Sjöberg BM, Gräslund A, Eckstein F. A substrate radical intermediate in the reaction between ribonucleotide reductase from Escherichia coli and 2‘-azido-2’-deoxynucleoside diphosphates. J Biol Chem. 1983;258:8060–8067. [PubMed] [Google Scholar]
- 30.Seyedsayamdost MR, Stubbe J. Site-specific replacement of Y356 with 3,4-dihydroxyphenylalanine in the beta2 subunit of E. coli ribonucleotide reductase. J Am Chem Soc. 2006;128:2522–2523. doi: 10.1021/ja057776q. [DOI] [PubMed] [Google Scholar]
- 31.Sjöberg BM, Reichard P, Gräslund A, Ehrenberg A. The tyrosine free radical in ribonucleotide reductase fromEscherichia coli. J Biol Chem. 1978;253:6863–6865. [PubMed] [Google Scholar]
- 32.Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7 doi: 10.1371/journal.pcbi.1002195. e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 35.Do CB, Mahabhashyam MSP, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loderer C, et al. A unique cysteine-rich zinc finger domain present in a majority of class II ribonucleotide reductases mediates catalytic turnover. J Biol Chem. 2017;292:19044–19054. doi: 10.1074/jbc.M117.806331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kabsch W. XDS. Acta Crystallogr D-Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andersson ME, et al. Structural and mutational studies of the carboxylate cluster in iron-free ribonucleotide reductase R2. Biochemistry. 2004;43:7966–7972. doi: 10.1021/bi036088l. [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D-Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D-Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 46.Sjöberg BM, Reichard P, Gräslund A, Ehrenberg A. Nature of the free radical in ribonucleotide reductase from Escherichia coli. J Biol Chem. 1977;252:536–541. [PubMed] [Google Scholar]
- 47.Reijerse E, Lendzian F, Isaacson R, Lubitz W. A tunable general purpose Q-band resonator for CW and pulse EPR/ENDOR experiments with large sample access and optical excitation. Journal of Magnetic Resonance. 2012;214:237–243. doi: 10.1016/j.jmr.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 48.Franke D, et al. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J Appl Crystallogr. 2017;50:1212–1225. doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 50.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svergun D, Barberato C, Koch MH. CRYSOL - A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- 53.Sielaff M, et al. Evaluation of FASP, SP3, and iST Protocols for Proteomic Sample Preparation in the Low Microgram Range. J Proteome Res. 2017;16:4060–4072. doi: 10.1021/acs.jproteome.7b00433. [DOI] [PubMed] [Google Scholar]
- 54.Hughes CS, et al. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol Syst Biol. 2014;10:757–757. doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 56.Na S, Bandeira N, Paek E. Fast multi-blind modification search through tandem mass spectrometry. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.010199. M111.010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chambers MC, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.