Abstract
During malaria infection, Plasmodium spp. parasites are cyclically invading red blood cells (RBCs) where they can follow two different developmental pathways: replicate asexually to sustain the infection or differentiate into gametocytes - sexual stages which can be taken by a mosquito ultimately leading to disease transmission. Despite its key importance for malaria control, the process of gametocytogenesis remains poorly understood, partially due to the difficulty of generating high numbers of sexually committed parasites in laboratory conditions1. Recently an apicomplexa-specific transcription factor (AP2-G) was identified as necessary for gametocyte production in multiple Plasmodium species2,3.and suggested to be an epigenetically regulated master switch initiating gametocytogenesis4,5. Here we show that in a rodent malaria parasite Plasmodium berghei conditional overexpression of AP2-G can be used to synchronously convert the great majority of the population into fertile gametocytes. This discovery allowed us to redefine the time frame of sexual commitment, identify a number of putative AP2-G targets and chart the sequence of transcriptional changes through the gametocyte development including the observation that gender partitioned transcription within 6 h of induction. These data provide entry points for further detailed characterization of the key process required for malaria transmission.
The ability to produce multiple, specialised cell types based on one genotype is typically associated with multicellular organisms but can be found in all branches of the tree of life. The causative agent of malaria, an apicomplexan parasite from the Plasmodium genus, is characterised by a complex life cycle involving a number of morphologically different stages adapted to different niches in its mammalian host or anopheline mosquito vector. These stages undergo linear transitions with one notable exception: intracellular blood stages, which are associated with all symptoms of malaria, enter one of two developmental pathways. Upon entry into a red blood cell (RBC), parasites either replicate asexually forming a schizont (which contains multiple merozoites able to invade new RBCs leading to an increase in parasitaemia), or develop into sexual forms, male or female gametocytes, responsible for transmission to the mosquito vector1. The extent of gametocytogenesis varies in response to multiple environmental factors, implying a significant flexibility in fate determination at the level of the individual cell. The molecular mechanisms and stimuli regulating this process, however, remain poorly understood, the recent advance demonstrating the role of the human serum component, lysophosphatidylcholine in repression of gametocytogenesis notwithstanding6.
In multiple model systems, differentiation into a particular cell type is triggered by key transcription factors acting as switches between different developmental pathways. Previously AP2-G, a transcription factor from the apicomplexa-specific apiAP2 family, was shown to be necessary for gametocytogenesis in Plasmodium spp.2,3, and its absence resulted in parasites unable to commit to sexual development (Fig. 1A). Here, we tested if overexpression of this factor could increase gametocyte production and enable the investigation of the uncharacterised, earliest stages of gametocyte development.
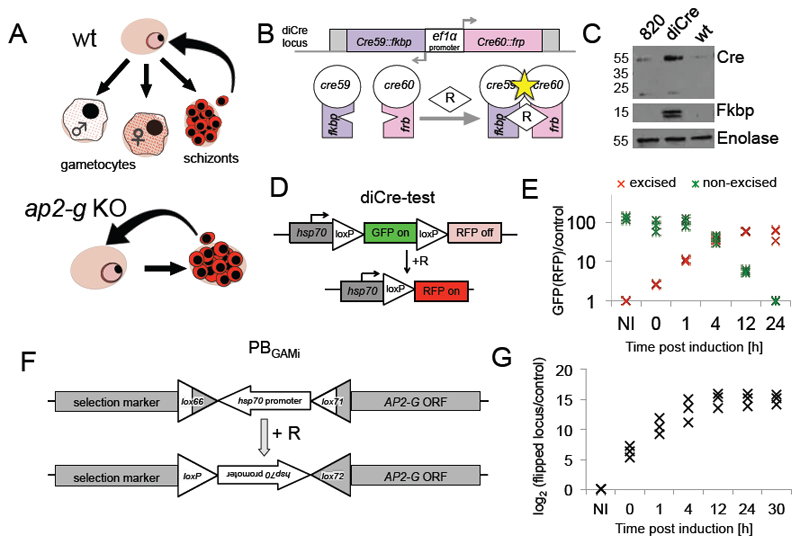
Fig. 1. Establishment of AP2-G overexpression system.
A. Top: cartoon depicting the three developmental fates of a newly invaded ring stage parasite, from left: male gametocyte, female gametocyte and schizont (comprised of daughter merozoites, capable of invading new red blood cells and forming new rings). Bottom: Parasites missing ap2-g undertake only asexual development.
B. Top: the cassette inserted in the P. berghei 820 line expressing two Cre fragments fused to FKBP and FRB respectively driven by the bidirectional ef1a promoter. Bottom: Principle of reconstitution of Cre recombinase activity upon addition of Rapamycin (R).
C. Western analysis showing expression of the two DiCre system fragments in the cloned PBGAMi line. Cre antibody shows Cre60::FRB and FKBP antibody shows Cre59::FKBP. Three independant blots were completed with similar results.
D. Design of construct to test DiCre activity.
E. QPCR quantification of edited and unedited test plasmid sequences in diCre-test parasites at different time points after rapamycin induction. Three technical replicates of the samples generated within the same time course are shown. At each time point DNA was harvested from an independently infected animal.
F. Cartoon of ap2-g locus in PBGAMi parasites before (top) and after (bottom) editing event.
G. qPCR quantification of the edited ap2-g locus in the PBGAMi parasites. Three technical replicates of the samples generated within the same time course are shown. At each time point DNA was harvested from an independently infected animal.
To create a tightly controlled conditional overexpression system, two different gene expression modules were introduced into the genome of the rodent malaria parasite Plasmodium berghei. First, a constitutively expressed split Cre recombinase (diCre) able to catalyse recombination between loxP sequences when activated with rapamycin was introduced into the p230p neutral locus of the parasite (Fig. 1B,C and Supplementary Figure 1)7. The resulting parasite line provided efficient and rapid control over diCre activity, as shown by the ability to completely recombine a test plasmid both in vitro and in vivo, switching the expression from green to red fluorescent protein (Fig. 1D, E and Supplementary Figure 2). The second module included modified loxP sites (lox66 and lox71)8 in a head-to-head orientation flanking a strong constitutive promoter (hsp70) used to express a selection marker (Fig. 1F). This module was inserted in front of the ap2-g ORF, interrupting and uncoupling its native promoter (Supplementary Figure 3A). In this position the unidirectional induced lox recombination event would invert the hsp70 promoter and overexpress ap2-g.
Both modules were successfully inserted into a P. berghei reporter line containing a third module that expressed green and red fluorescent reporter markers in male and female gametocytes, respectively9, generating PBGAMi. PBGAMi schizonts were synchronised in vitro and grown in vivo with (PBGAMiR+) or without (PBGAMiR-) rapamycin. Both populations were analysed at different time points using qPCR, light microscopy and flow cytometry (FACS). Recombination was detected in the PBGAMiR+ population immediately after induction and reached its maximum at ~12 h, eliminating the unedited locus (Fig. 1G and Supplementary Figure 3B,C). In the PBGAMiR- population, within 24 h all parasites transformed into asexual schizonts, replicating the ap2-g ko phenotype and resulting in an increase of parasitaemia (Fig. 2 A-D). In the PBGAMiR+ population, in contrast, PBGAMiR+ parasites did not produce schizonts (Fig. 2A) or increase in parasitaemia (Fig. 2D). Instead, many cells with gametocyte-like morphology appeared (Fig. 2A,B). Flow cytometry confirmed expression of male and female reporter proteins in PBGAMiR+ from 12 h post induction (hpi; Fig. 2C and Supplementary Figure 4). The combined percentage of male and female gametocytes in the population reached 70% in some experiments, and the total gametocyte proportion was always significantly (P-value = 0.0021, Student’s unpaired t-test) higher than in wt (Fig. 2B). Importantly, GFP and RFP-positive cells emerged upon activation and formed male and female gametes, respectively, confirming they are functional gametocytes (Fig. 2E). The relative ability of induced male gametocytes to replicate their genomes upon activation was similar to wild type, although their ability to go on to complete egress was reduced if compared to wild type (Fig. 2E). This most likely reflects the fact that a reporter gene is necessarily an imperfect proxy for functional maturity. In our system expression of the male marker from 12 h after induction (Supplementary Figure 4) precedes functional maturity, and as a consequence, differences in the age distribution between synchronous induced and asynchronous wildtype marker-positive male gametocytes must result in differences in apparent functional maturity.
Fig. 2. AP2-G overexpression results in gametocyte conversion.
A. Parasite morphology in Giemsa-stained thin blood smears in PBGAMiR- (top) and PBGAMiR+ (bottom) population demonstrating evident production of gametocytes in the induced parasites. Representative pictures from 3 independent biological replicates for each time point are shown. Scale bar = 10μm
B. Gametocyte conversion rates in PBGAMiR- and PBGAMiR+ parasites compared to the parental line. Gametocytaemia shown as percentage of infected cells expressing male (blue)/female (red) markers in FACS analysis 30 h post induction. Means, standard deviations and individual data points from 5 independent biological replicates are shown. Statistical significance in total gametocyte numbers determined with a two tailed, unpaired Student’s t-test.
C. FACS profiles of PBGAMiR- and PBGAMiR+ parasites 30h after induction, representative for 5 biologically independent experiments. Male and female parasite populations gating is shown.
D. Parasitaemia in PBGAMiR-/R+ population compared to wt parasites defined as percentage of DNA positive red blood cells in FACS analysis. Median and individual measurements from 3 independent biological replicates are shown.
E. Percentage of unactivated, activated and partially activated cells in wild-type (WT) and PBGAMiR+ gametocytes. Full activation is defined as successful production of male gametes and their emergence out of the blood cells (exflagellation). Partial activation of male gametocytes is identified when DNA replication has occurred (increase in Hoechst 33342 intensity) but exflagellation from the red blood cell has not occurred. Means, standard deviations and individual data points from five independent biological replicates are shown. Statistical significance assessed using unpaired two-tailed Student’s t-test.
F. Gametocyte conversion rates in PBGAMi parasites induced in vivo at the different time points post-invasion. Total proportion of gametocytes generated within the same cycle (maximum measurement until 40hpi) and after subsequent reinvasion (64 hpi) is shown for the same infection. NI =non-induced. Means, standard deviations and individual data points from 3 independent biological replicates are shown.
Experimental evidence from P. falciparum has suggested that commitment to gametocytogenesis occurs in the blood stage cycle prior to the appearance of gametocytes resulting in the production of a committed schizont/merozoite population destined to develop into gametocytes following erythrocyte invasion10. To determine the period during which P. berghei asexual parasites are sensitive to reprogramming, we induced ap2-g overexpression at multiple time points during the 24 h cycle of asexual development (at 2, 4, 8, 12, 18, 22 h post merozoite invasion). Parasites induced before 12 hpi could be transformed with decreasing efficiency into gametocytes within the same developmental cycle (Fig. 2F and Supplementary Figure 5A). In marked contrast, parasites induced after 12 hpi all developed into asexual schizonts and gametocyte conversion was observed only following invasion, in the next cycle. The timing of induction did not appear to have a marked effect on the cumulative number of gametocytes formed (Supplementary Figure 5A,B). The male to female ratio in induced gametocytes was within the range observed in the parental population (Fig. 2B), and any apparent shifts in sex ratio with time of induction (Fig. 2F) may simply reflect the earlier upregulation of the male marker, as discussed above.
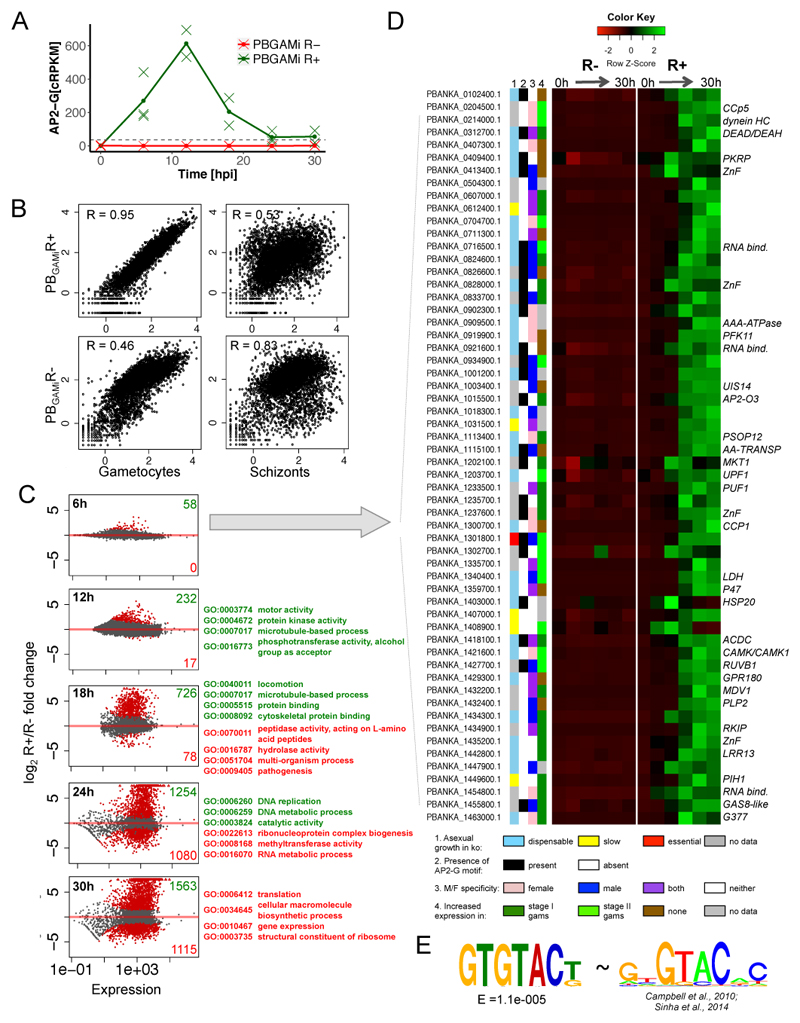
The inducible and synchronised sexual commitment provided an opportunity to assess transcriptional changes in developing gametocytes. Synchronous schizonts (22hpi) were induced in vivo with rapamycin to obtain a single wave of commitment after reinvasion and RNA-seq libraries were prepared from PBGAMiR- and PBGAMiR+ parasites harvested at 6h intervals between 0 and 30h post induction. Initially, data was used to examine the level of expression of functional ap2-g transcript which was absent in PBGAMiR- population but detected in the PBGAMiR+ population reaching its maximum at 12 h and exceeding native expression within the population11 within first 6 h (Fig. 3A and Supplementary Figure 6). Analysis of the remainder of the transcriptome confirmed that at 30 hpi the PBGAMiR- population was similar to the transcriptome of asexual schizonts, while PBGAMiR+ was indistinguishable from purified gametocytes indicating that the cells that did not develop into gametocytes either produced the gametocyte transcriptome or were transcriptionally silent (Fig. 3B). The PBGAMiR +/- populations diverged gradually starting from 6 h (58 genes) to 30 h, when 2676 genes showed some level of differential regulation (Fig. 3C and Supplementary Table 2). The dynamics of gene expression in PBGAMiR+ parasites mirrored the expected patterns of developing wild type gametocytes closely as indicated by gene ontology (GO) terms associated with different time points (Fig. 3C and Supplementary Table 3), which shifted from protein kinases and motor proteins of the male gametocyte at 12 h, to cytoskeleton organisation (18 h) and the DNA replication machinery (24 h). As expected, translationally repressed transcripts which are known to be stored by the mature female gametocyte12 were induced at later time points (Supplementary Figure 7).
Fig. 3. Transcriptome changes in PBGAMiR- and PBGAMR+ parasites reveal gametocyte-specific transcriptional program.
A. Expression of ap2-g transcript in PBGAMiR- and PBGAMiR+ populations. Corrected rpkm calculated as seen in Supplementary Figure 6. Means and individual data points from 3 (0, 6 and 24h) or 2 (12,18 and 30h) independent biological replicates are shown. Horizontal, black dashed line indicates maximal native ap2-g RNA expression in WT population11.
B. Comparison of transcriptomes (rpkm’s) from PBGAMiR- and PBGAMiR+ populations 30 hpi to purified gametocytes and schizonts transcriptomes. R = Spearman’s rank correlation coefficient. Correlation values representative for n=3 experiments.
C. Differential expression analysis between PBGAMiR- and PBGAMiR+ populations at different time points. Plots show log2 fold change of gene expression vs. expression levels, with differentially expressed genes marked in red. Numbers of genes overexpressed in PBGAMiR- (red) and PBGAMiR+ (green) and example GO terms enriched within each group next to each graph. All samples for transcriptome analysis were generated from three independent time-course experiments.
D. Genes responding early to AP2-G overexpression. Shown are: knock-out growth phenotype in asexual blood stages14, sex specificity of expression in P.berghei15, gametocyte specificity in P.falciparum27, presence of AP2-G motif(s) in 2 kb upstream of the start codon, and gene expression profile through the time course. Available gene symbols are shown on the right of the expression profile.
E. The DNA motif enriched upstream of genes in panel D (left) compared with the known AP2-G binding motif2,16 (right).
Of particular interest was the group of 58 genes responding already at the 6 h time point. It included conserved early gametocyte markers like mdv113 as well as proteins potentially involved in nucleic acid binding, including four zinc finger proteins, three RNA binding proteins, two helicases and one other apiAP2 transcription factor (Fig. 3D). In comparison with the rest of the genome this subset was significantly enriched (P= 1.387 e-05, Fisher exact tests) in genes dispensable for asexual blood stages14 and many of its members are known to be specific to the male or female sex in mature gametocytes15. The promoter regions of these genes were also significantly (E = 1.1e-005) enriched in a GTGTAC(T/G) motif resembling closely the known DNA-binding motif for the AP2 domain of AP2-G (Fig. 4E)16, suggesting at least some of them may be direct downstream targets of AP2-G. Comparison with the available datasets from human malaria Plasmodium falciparum revealed that almost all of these genes (35 out of the 49 with identified syntenic orthologs) are upregulated in the early phase of gametocytogenesis (days 1 & 3) suggesting that the function of these genes is shared between different Plasmodium species (Fig. 3D and Supplementary Figure 8). In contrast the genes that are up-regulated and co-expressed with ap2-g in P. falciparum3,17 predominantly encode secreted proteins that are not present in the P. berghei genome and thought to be associated with the extensive remodelling of the erythrocyte that is uniquely undertaken by P. falciparum gametocytes18.
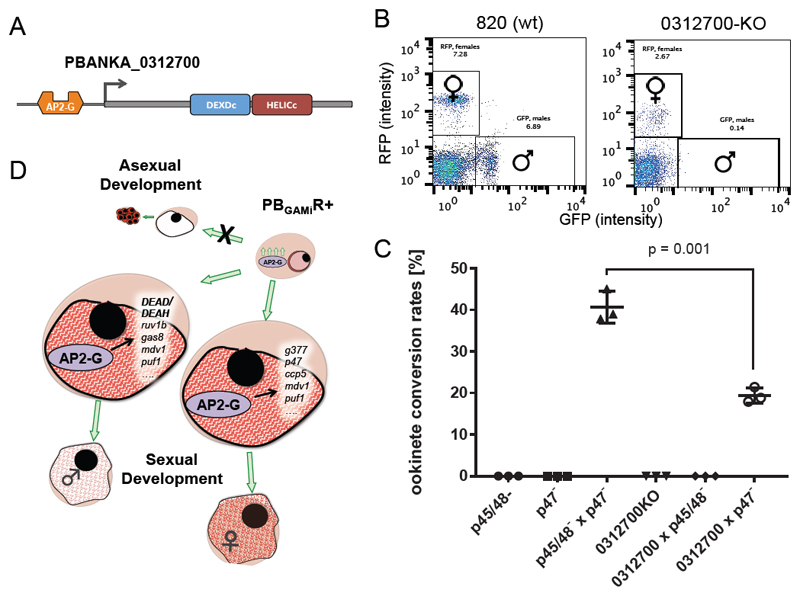
Fig. 4. PBANKA_0312700 is an AP2-G-induced gene involved in gametocyte development.
A. PBANKA_0312700 gene structure with conserved helicase domains and putative AP2-G binding motif marked.
B. Representative FACS analysis (from 4 independent experiments) showing loss of male gametocytes in the PBANKA_0312700 KO.
C. Ookinete conversion in PBANKA_0312700 KO crossed with line producing only viable females (p48/45-) or males (p47-), showing that the defect gametocyte development is male specific. Means, standard deviations and individual data points from three independent biological replicates are shown. Statistical significance was determined with a two tailed, unpaired Student’s t-test.
D. Model of AP2-G function in gametocyte commitment.
To confirm a role of one of these conserved, induced genes in gametocytogenesis, we disrupted the predicted DEAD/DEAH helicase, PBANKA_0312700 (Fig. 4A and Supplementary Figure 9), which has a putative AP2-G binding site 650 bp upstream of its start codon and shows a robust transcriptional response at 6 hpi and a high level of expression in both male and female gametocytes. The mutant completely lacked mature male gametocytes as tested by FACS (Fig. 4B) and blood smears. Female gametocytes were, however, viable and able to transmit when crossed with a strain producing viable males (Fig. 4C). These findings were consistent with a predominant function for PBANKA_0312700 in male gametocyte development, although the reduction in ookinete conversion when compared to a fully viable female only strain (Fig. 4C) could also indicate a downstream role for this helicase in female gametocyte development or fertilisation.
Here we have shown that inducible expression of AP2-G is sufficient to induce synchronous gametocytogenesis in a Plasmodium parasite, overriding the default asexual replication program and leading to the generation of functional gametocytes. Importantly, this conversion could be initiated within the same developmental cycle, proving that the fate of the young intraerythrocytic parasites is not irreversibly determined. This finding is seemingly at odds with previous reports from human parasites where the existence of sexually pre-committed schizonts producing next generation gametocytes has been reported4,10,17. Our data may simply reflect significant differences in life cycle regulation between the two Plasmodium parasites - gametocytogenesis in P. falciparum uniquely takes ~12 days which is significantly longer than other Plasmodium spp. and P. falciparum gametocytes also possess a distinctive morphology. Alternatively our discovery suggests a more general plasticity in commitment and an extended time window during which environmental factors6 and epigenetic pre-programming4,5 can impact the number of gametocytes. In that scenario a proportion of the schizonts would be predisposed to generate gametocytes but the final, irreversible commitment, would be a late event in asexual blood stage development in the next cycle depending solely on AP2-G acting as a switch between the two transcriptional fates of the cell. According to that theory, also P. falciparum parasites could be converted within the same cycle, but assays which would capture this possibility have not been developed or tested. Instead our ability to bypass any upstream events allowed us to reveal the early transcriptional programme downstream of AP2-G which appears to be shared between the two parasites. Intriguingly, many of these early response genes have a strongly sex specific expression profile15 or knock-out phenotype (including PBANKA_0312700 tested here), suggesting that AP2-G may regulate different subsets of genes in both male and female precursors (Fig. 4D). This raises the possibility that commitment to the male or female sex either precedes or is simultaneous with AP2-G mediated gametocytogenesis. This hypothesis has also been proposed in recent work capturing early transcription events in P. falciparum19 where certain genes with sex-specific phenotypes were shown to be regulated independently of AP2-G.
In summary, the AP2-G overexpression system allows full control of Plasmodium gametocytogenesis and generated findings related to sexual commitment for future investigation. Equally the application of the concept of the conditional inducible commitment may be extended to other apicomplexan parasites in which sexual development is less tractable, leading to useful insights regarding the transmission in these species.
Materials and Methods
Rodent malaria parasites and their maintenance
All experiments were performed using Plasmodium berghei 2.34 ANKA strain and its modifications maintained in female, outbred mice (Theiler’s Original (TO)) 8 -12 weeks of age. The infections were initiated by intraperitoneal or intravenous injection of either the frozen parasite stock or blood from an infected donor mouse and monitored daily using Giemsa stained thin blood smears. All animal procedures were conducted under project licenses issued by the UK Home Office and with local ethical approval of the Animal Welfare and Ethical Review Body of the Wellcome Trust Sanger Institute and the University of Glasgow Ethics Committee. For qualitative experiments (i.e genotyping, Western blots) one mouse per strain was used in each of the experiments. For quantitative experiments, the numbers were adjusted based on the variability of the studied trait and the effect size of interest and are included in the description of experimental results. All animals/samples were assigned a number and the data was initially analyzed without the knowledge of the infection type or conditions/treatment to ensure minimal bias.
Generation of the mutant parasite lines
The non-fluorescent strain c115cy1 HP and its derivate 820, expressing GFP and RFP markers in male and female gametocytes respectively9, were used to create diCre-expressing lines. The marker-free HP diCre and 820 diCre lines were generated using the GIMO (gene-in marker out) approach described previously20 and presented in Supplementary Figure 1. Briefly, a positive-negative selection marker was inserted into the parasite’s neutral p230p locus via homologous recombination. After positive selection and cloning by serial dilution, the second transfection followed by negative selection was used to replace the marker with the diCre expression cassette, and a second cloning step was performed. The diCre-test line was generated by transfecting HP diCre with a centromeric, single-copy plasmid containing a hsp70 promoter, a floxed GFP and RFP separated by a loxP site, as shown in Fig. 1G. The 820 diCre line was transfected with linear DNA modifying the ap2-g locus as shown in Supplementary Figure 3 to generate PBGAMi. A PBANKA_0312700 KO line was generated in 820 parasites by replacing the gene with a positive-negative selection marker as shown in Supplementary Figure 8.
All transfections were performed using an established P. berghei schizont electroporation protocol. Parasites were grown to 1-6% parasitaemia in mice treated with 1.2 mg of phenylhydrazine (Sigma) two days prior the infection, and collected by cardiac puncture before transfer to schizont media (RPMI 1640 + L-glutamine + 25 mM HEPES, supplemented with 25% of foetal bovine serum, 10 mM sodium bicarbonate and penicillin-streptomycin). After overnight culture at 37°C in an atmosphere of 5% CO2, 5% O2 and 90% N2, schizonts were isolated by 15 min of centrifugation at 500 g on a cushion of 55% Nycodenz (Lucron Bioproducts). The schizonts were harvested from the interface were then electroporated with 1-10 μg of linear DNA or plasmid using Amaxa Nucleofector device and Basic Parasite Nucleofector Kit 2 according to manufacturer’s instructions. Parasites were intravenously injected into new mice and recovered for 25 h before selection with pyrimethamine (7 μg/ml in drinking water) or 5-fluorocytosine (1.5 mg/ml in the drinking water, HP diCre and 820 diCre lines only). Selected parasites were recovered ~7 days post transfection. All lines were cloned by limiting dilution before the next modification step.
Genome editing in diCre parasites
P. berghei parasites were synchronised by overnight culture in schizont media and a Nycodenz gradient was used to harvest schizonts, as described above. Purified schizonts were intravenously injected into naive mice, where they maintained their synchronicity for 48 h. Rapamycin (Sigma) was dissolved in DMSO to 4 mg/ml to generate the stock solution. DiCre test parasites were induced either in vitro and in vivo. For in vitro induction, parasites were removed from the host by cardiac puncture 2 h post invasion (hpi) and cultured in schizont media with 200 mM rapamycin. For in vivo induction the host animals were injected intraperitoneally with 4 mg/kg of rapamycin 2 hpi. Parasites were harvested at various time points via tail bleed or cardiac puncture for DNA and flow cytometry analysis. To generate a population of invading merozoites overexpressing AP2-G, the PBGAMi line was induced 22 hpi by 4 mg/kg of rapamycin injection and its development was analysed in the next invasion cycle, unless specified otherwise. In each case, an uninduced control population of parasites was analysed in parallel.
DNA extraction for PCR or qPCR genotyping
To extract parasites from host blood, ~1 ml of heparinised whole blood was collected by cardiac puncture. Leucocyte depletion was achieved by filtration through two sequential Plasmodipur filters (Europroxima) according to the manufacturer’s instructions. Remaining cells were resuspended in 10 volumes of pre-chilled erythrocyte lysis buffer (150 mM NH4Cl; 10 mM KHCO3; 1 mM EDTA) and incubated on ice for 15 min. After lysis, parasites were pelleted by centrifugation for 8 min at 450 g and washed with 1x phosphate buffered saline until complete removal of the coloration of supernatant and either stored at -20°C for later DNA isolation or processed immediately.
In the second part of the protocol, the fresh or frozen parasite pellet was resuspended in 700 μl TNE buffer (50 mm Tris–HCl, pH 7.4, 100 mm NaCl, 0.1 mm EDTA), supplemented with 200 μg RNase and 1% SDS. This mixture was incubated for 10 – 15 min at 37°C after which 200 μg proteinase K was added and the solution incubated for a further 1 hour. Then a standard phenol and chloroform extraction protocol followed by ethanol precipitation21 was used to extract the DNA from the lysate. Quality and concentration of the nucleic acids were measured using a UV-Vis spectrophotometer (NanoDrop, Thermo Scientific).
PCR genotyping
Genotyping strategies for genetically modified parasites are presented in Supplementary Figures 1, 2, 3 and 9, and the primer sequences are given in the Supplementary Table 1. All PCR reactions were performed using Taq DNA Polymerase (NEB) and the following PCR program: 94°C 30 s//94°C 30 s/Tm°C 30 s/72°C 1min // x 30/72°C 10 s/4°C, where Tm°C was a annealing temperature specific for each primer pair. PCR products were resolved on a 1% agarose gel supplemented with 1:10,000 SYBR Safe reagent and visualised using Gel Doc XR+ Gel Documentation System.
QPCR genotyping
QCR primers were designed as shown in Supplementary Figures 2 and 3 and are shown in Supplementary Table 1. The amplification reaction was set up using 50 ng of DNA and QuantiTect Sybr Green PCR Kit (Qiagen) according to the manufacturer’s instructions. The reaction was incubated in StepOne Real-Time PCR System (Applied Biosystems) using the following program: 95°C 10min s//95°C 15 s/60°C 1 min//x 40/95°C 15 s/60°C 1 min/gradient +0.3°C/95°C 15 s.
At least three technical replicates of each measurement were taken. A neutral sequence present in both induced and uninduced sample was amplified as a reference and DNA from uninduced population was used as control. The fold change calculations were based on the ΔΔCt method (docs.appliedbiosystems.com/pebiodocs/04371095.pdf).
Western blot analysis of diCre production
Protein pellets (isolated as described for the DNA extraction) were suspended in 5x pellet volume of RIPA lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5 % sodium deoxycholate, 0.1 % SDS, 1 % triton X-100) and incubated on ice for ~30 min. The lysate was spun for 10 min at 4oC, at 14,000 rpm and the supernatant was combined with 4x SDS gel-loading buffer (62.5 mM Tris-H3PO4, pH 7.5, 1 mM EDTA, 2 % SDS, 10 mM DTT, 1 mM NaN3 and 33% glycerol) and fresh 15% β-mercaptoethanol. Samples were boiled at 100oC for 5 min and loaded on SDS-PAGE gradient gel (Biorad). Electrophoresis was performed using Mini-PROTEAN Tetra cell electrophoresis chamber (Qiagen) for 2 h at 120 V. The resolved proteins were transferred on Whatman nitrocellulose membrane using a trans-blot electrophoretic transfer system according to the manufacturer’s protocol, blocked with 5% milk PBST and probed with the antibodies against FKBP-12 (1:500), Cre (1:500) and enolase (1:1000) (all Abcam), followed by a compatible HRP-labelled secondary antibody. Enhanced chemiluminescence system (ECL) was used to visualise the proteins on X-ray film.
Flow cytometry analysis diCre activity and gametocyte production
Red blood cells for analysis were collected from tail drops, cardiac puncture or parasite cultures. Cells were suspended in pre-warmed rich PBS with Hoechst 33342 dye and stained for 30 min at 37°C. Stained samples were washed and resuspended in flow cytometry buffer (10% rich PBS, 1 mM EDTA in PBS) and analysed using LSR-II flow cytometer (Becton Dickinson) with following emission/excitation settings: Hoechst (DAPI, yellow laser, 350 nm) 450/50, GFP (FITC 488 nm) 488/10 and RFP (PE 561 nm) 585/15. At least 500,000 events were acquired for each sample. Initial gating was performed using forward and side scatter to exclude the events below the size and granularity thresholds of red blood cells. Then FCS-H FCS-W gating served to isolate single red blood cells and followed by Hoechst staining selecting parasite-infected cells as shown in Supplementary Figure 4A. For the 820 line and its derivatives, GFP and RFP gates were used to select the male and female gametocytes respectively. For the diCre-test line the GFP, RFP and bifluorescent populations were selected to show different stages of excision as shown in Supplementary Figure 2D. In each experiment involving 820 modifications uninfected blood and wt 820 line were used as controls. In fluorescence switching experiments both non-fluorescent parasites and lines constitutively expressing GFP or RFP only (with appropriate strength promoters) only were used as controls (Supplementary Figure 2E).
Exflagellation/emergence
Tail drops of blood were collected in 500 μl 37°C schizont media (as above, unactivated) or 21°C ookinete media (RPMI1640, 10% FCS, xanthurenic acid, activated) and incubated at the appropriate temperature for 30 min. Ter119 PEcy7 (https://www.thermofisher.com/antibody/product/TER-119-Antibody-clone-TER-119-Monoclonal/25-5921-82) and Hoechst were added to a final concentration of 1:200 and a total volume of 700 μl and intensively vortexed. Samples were incubated at their appropriate temperature for an additional 20 min, vortexing every 5 min. Stained samples were washed in 500 μl of rPBS and resuspended in 500 μl of FACS buffer for analysis. Male and female gametocytes were identified by their GFP and RFP signal respectively using the gating presented in Supplementary Figure 4A. Gametocytes were considered unactivated if positive for PEcy7 Ter119 staining and activated if this staining is negative. Only gametocytes were included in quantification of activation.
Time course of variable gametocyte induction and flow cytometry quantification
Mice were injected with synchronised PBGAMi parasites and induced in vivo with rapamycin at multiple time points post invasion (2, 6, 8, 12, 18, 22 h) as described previously. Samples were obtained from tail drops and the parasitaemia (Hoechst 33342-positive cells) and percentage of male (GFP) and female (RFP) gametocytes were quantified every 4 h from 8 h post invasion using flow cytometry. Since sex specific fluorescence makers became detectable only from 16 h post invasion (Supplementary Figure 4B), any gametocytes originating from the second cycle after induction would become detectable only after 40 hpi post invasion (24 h cycle + 16 h maturation;). Therefore we considered the highest measurement taken before 40 hpi as the number of gametocytes produced within the first cycle. The last measurement taken 64 hpi was considered as the total number of gametocytes produced within both cycles.
Time course of gametocyte induction, RNA extraction and RNA-seq library preparation
Mice were injected with synchronised PBGAMi parasites and induced with rapamycin at 22 h as described previously. Parasites were harvested at different time points post induction via cardiac puncture, filtered, extracted from RBC and washed in 1xPBS as for DNA isolation. Parasite pallets were resuspended in 1ml of Trizol reagent (Ambion) lysed for 10 min at room temperature and stored at -80°C for later RNA extraction.
Complete RNA was isolated from the samples using Trizol/chloroform extraction followed by isopropanol precipitation21 and its concentration and integrity was verified using Agilent Bioanalyzer (RNA 6000 Nano kit) and NanoDrop 1000 spectrophotometer. 1-2 μg of total RNA from each sample (or complete sample if the yield was lower) was used for mRNA isolation (Magnetic mRNA Isolation Kit, NEB). First strand cDNA synthesis was performed using the SuperScript III First-Strand Synthesis System and a 1:1 mix of Oligo(dT) and random primers (Invitrogen). The DNA-RNA hybrids were purified using Agencourt RNACleanXP beads (Beckman Coulter) and the second cDNA strand was synthetized using a 10 mM dUTP nucleotide mix, DNA Polymerase I (Invitrogen) and RNAseH (NEB) for 2.5 h at 16°C. The long cDNA fragments were purified and fragmented using a Covaris S220 system (duty cycles = 20, intensity = 5, cycles/burst = 200, time = 30s). The ~200 bp long fragments were end-repaired, dA-tailed and ligated to “PCR-free” adapters (Kozarewa et al., 2009) with index tags using NEBNext Modules according to the manufacturer’s instructions. Excess adapters were removed by two rounds of clean-up with 1 volume of Agencourt AMPure XP beads. Final libraries were eluted in 30 μl water, quality-controlled using Agilent Bioanalyzer (High Sensitivity DNA chip) digested with USER enzyme (NEB) and quantified by qPCR. For some libraries additional 5 cycles of PCR amplification were performed, using KAPA HiFi HotStart PCR mix and Illumina tag-specific primers to obtain enough material for sequencing. Pools of indexed libraries were sequenced using an Illumina HiSeq2500 system (100 bp paired-end reads) according to manufacturer’s manual. All samples were generated in duplicates or triplicates and uninduced controls were always generated and processed in parallel. Raw data is available through GEO database repository (study GSE110201).
RNAseq data analysis
The generation of raw data in the form of *.cram files quality control and adapter trimming was performed using the default analysis pipelines of the Sanger Institute. The raw data was transformed into paired *. fastq files using Samtools software (ver. 1.3.1). The generated reads were re-aligned to Plasmodium berghei genome (PlasmoDB-30 release) in a splice aware manner with HISAT222 using --known-splicesite-infile option within the splicing sites file generated based on the current genome annotation. Resulting *.bam files were sorted and indexed using Samtools and inspected visually using Integrated Genome Viewer (ver. 2.3.91). HT-seq python library23 was used to generate reads counts for all genes for further processing. Differential expression calculation and correlation analysis was performed and visualised using R studio software (v. 1.0.136) with DESeq2, ggplot2 and GMD packages24.
The reference schizont and gametocytes transcriptome datasets were downloaded from25 and compared to the generated samples using Spearman's rank correlation coefficient. The enrichment for the translationally repressed genes in different differentially expressed datasets was performed using Fisher exact test. For the analysis of male and female specificity and growth rates the data from15 and14 respectively was used. Growth rate above 0.8 was considered normal, between 0.8 and 0.2 – decreased and below 0. 2 the gene was considered essential. Gene was considered male/female specific if its expression in the gametocytes of given sex was at least 10 fold greater than in the opposite and sex as well as in the asexual parasites. DOZI translationally repressed genes were defined as the ones enriched in both DOZI and CITH RIP-ChIP datasets 26. Comparison with P.falciparum data was performed using the available PF datasets3,27 and synteny information11. Genes were considered as overexpressed in early gametocytes stages in their expression was at least 2-fold greater than in matched asexual population27.
De novo regulatory motif discovery was performed using DREME software28 and sequences 2 kb upstream of the translation sites of the genes upregulated 6 h post rapamycin induction as the input set and the promoters of the remaining genes in the genome as the reference set.
Data availibility
RNA-seq data generated in this study is available through GEO database repository (study accession number GSE110201).
Supplementary Material
Acknowledgments
We thank R. Menard and Daniel Bargieri for the diCre-test plasmid utilised in this study and to Mandy Sanders and WTSI sequencing service for assistance with RNA-seq samples processing. KKM is supported by Wellcome Trust and the Royal Society (Ref 202600/Z/16/Z). RSK is supported by BBSRC (Ref BB/J013854/1). APW is supported by the Wellcome Trust (Refs: 083811, & 107046). OB is supported by Wellcome Trust Sanger Institute (Ref WT098051).
Footnotes
Author’s contributions:
RSK generated and phenotyped HP diCre, diCre test and PBANKA_0312700 lines, performed phenotyping experiments on PBGAMi line and generated parasites for transcriptome sequencing. KKM generated the AP2-G overexpression construct and PBGAMi line, performed phenotyping experiments on PBGAMi line, generated RNA-seq libraries and performed RNA-seq data analysis. RC generated 820 diCre and GIMO lines and parasites for transcriptome sequencing. NP performed the ookinete conversion assay for PBANKA_0312700 line. OB and APW led and supervised the study. KKM and APW wrote the manuscript with contributions from other authors.
Competing financial interests:
Authors declare no competing financial interests.
References
- 1.Meibalan E, Marti M. Biology of Malaria Transmission. Cold Spring Harb Perspect Med. 2017;7:a025452. doi: 10.1101/cshperspect.a025452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sinha A, et al. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature. 2014;507:253–7. doi: 10.1038/nature12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kafsack BFC, et al. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature. 2014;507:248–52. doi: 10.1038/nature12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brancucci NMB, et al. Article Heterochromatin Protein 1 Secures Survival and Transmission of Malaria Parasites. Cell Host Microbe. 2014;16:165–176. doi: 10.1016/j.chom.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Filarsky M, et al. GDV1 induces sexual commitment of malaria parasites by antagonizing HP1-dependent gene silencing. Science (80-. ) 2018 doi: 10.1126/science.aan6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brancucci NMB, et al. Lysophosphatidylcholine Regulates Sexual Stage Differentiation in the Human Malaria Parasite Plasmodium falciparum. Cell. 2017;171:1532–1544.e15. doi: 10.1016/j.cell.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jullien N, Sampieri F, Enjalbert A, Herman J-P. Regulation of Cre recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Res. 2003;31:e131. doi: 10.1093/nar/gng131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albert H, Dale EC, Lee E, Ow DW. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 1995;7:649–59. doi: 10.1046/j.1365-313x.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- 9.Mair GR, et al. Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog. 2010;6:e1000767. doi: 10.1371/journal.ppat.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruce MC, Alano P, Duthie S, Carter R. Commitment of the malaria parasite Plasmodium falciparum to sexual and asexual development. Parasitology. 1990;100 Pt 2:191–200. doi: 10.1017/s0031182000061199. [DOI] [PubMed] [Google Scholar]
- 11.Otto TD, et al. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 2014;12:86. doi: 10.1186/s12915-014-0086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mair GR, et al. Regulation of sexual development of Plasmodium by translational repression. Science. 2006;313:667–9. doi: 10.1126/science.1125129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lal K, et al. Plasmodium male development gene-1 (mdv-1) is important for female sexual development and identifies a polarised plasma membrane during zygote development. Int J Parasitol. 2009;39:755–61. doi: 10.1016/j.ijpara.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Bushell E, et al. Functional Profiling of a Plasmodium Genome Reveals an Abundance of Essential Genes. Cell. 2017;170:260–272.e8. doi: 10.1016/j.cell.2017.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeoh LM, Goodman CD, Mollard V, McFadden GI, Ralph SA. Comparative transcriptomics of female and male gametocytes in Plasmodium berghei and the evolution of sex in alveolates. BMC Genomics. 2017;18:734. doi: 10.1186/s12864-017-4100-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell TL, De Silva EK, Olszewski KL, Elemento O, Llinás M. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathog. 2010;6:e1001165. doi: 10.1371/journal.ppat.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poran A, et al. Single-cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature. 2017;551:95–99. doi: 10.1038/nature24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tibúrcio M, Sauerwein R, Lavazec C, Alano P. Erythrocyte remodeling by Plasmodium falciparum gametocytes in the human host interplay. Trends Parasitol. 2015;31:270–278. doi: 10.1016/j.pt.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Painter HJ, Carrasquilla M, Llinás M. Capturing in vivo RNA transcriptional dynamics from the malaria parasite Plasmodium falciparum. Genome Res. 2017;27:1074–1086. doi: 10.1101/gr.217356.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J, et al. A Novel ‘Gene Insertion/Marker Out’ (GIMO) Method for Transgene Expression and Gene Complementation in Rodent Malaria Parasites. PLoS One. 2011;6:e29289. doi: 10.1371/journal.pone.0029289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15:532–4. 536–7. [PubMed] [Google Scholar]
- 22.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Pyl PT, Huber W. HTSeq - A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Modrzynska K, et al. A Knockout Screen of ApiAP2 Genes Reveals Networks of Interacting Transcriptional Regulators Controlling the Plasmodium Life Cycle. Cell Host Microbe. 2017;21:11–22. doi: 10.1016/j.chom.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerreiro A, et al. Genome-wide RIP-Chip analysis of translational repressor-bound mRNAs in the Plasmodium gametocyte. Genome Biol. 2014;15:493. doi: 10.1186/s13059-014-0493-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young Ja, et al. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol Biochem Parasitol. 2005;143:67–79. doi: 10.1016/j.molbiopara.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–9. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.