Abstract
The Dishevelled gene was first identified in Drosophila mutants with disoriented hair and bristle polarity [1–3]. The Dsh gene (Dsh/Dvl, in Drosophila and vertebrates respectively) gained popularity when it was discovered that it plays a key role in segment polarity during early embryonic development in Drosophila [4]. Subsequently, the vertebrate homolog of Dishevelled genes were identified in Xenopus (Xdsh), mice (Dvl1, Dvl2, Dvl3), and in humans (DVL1, DVL2, DVL3) [5–10]. Dishevelled functions as a principal component of Wnt signaling pathway and governs several cellular processes including cell proliferation, survival, migration, differentiation, polarity and stem cell renewal. This review will revisit seminal discoveries and also summarize recent advances in characterizing the role of Dishevelled in both normal and pathophysiological settings.
Keywords: Wnt, Dishevelled, β-catenin, cancer, development, disease
1. Introduction
It is fascinating to consider how the identity of DVL is largely linked with its ability to integrate and relay complex Wnt signals in tissues and cells yet how it conducts this symphony of activity still remains poorly understood. While we know that the final outcome of Wnt signaling will depend on the abundance of and ratios of Wnt ligands, antagonists and receptors, DVL plays a key role in integrating and transmitting these instructions regardless of whether they are correct or aberrant in nature. This propagation of information may initiate a signaling cascade that ultimately leads to β-catenin stabilization or a β-catenin-independent effect. While this review will recap some of the landmark discoveries and recent advances in characterizing the role of Dishevelled, we point the reader to previous reviews that have highlighted its role in signal transduction [11, 12], developmental biology [3], and nuclear shuttling [13]. Additionally, the concept of the mystery that still surrounds DVL and what it means for it to be “activated” after more than two decades investigation was nicely addressed previously [14]. This review will discuss the function of DVL in the Wnt signaling pathway in normal and aberrant cellular contexts. It will also highlight novel roles within the nucleus and highlight new binding partners that have been discovered and novel mechanisms of post-translational regulation.
2. Function of DVL in Wnt signaling pathway
Early genetic studies demonstrated that Dishevelled proteins are involved in canonical (β-catenin-dependent) and non-canonical (β-catenin-independent) Wnt signaling pathways that govern the segment polarity in the fly. However, it was not clearly understood how DVL participates simultaneously in both pathways. It was later demonstrated that DVL acts as a branch-point and is an essential component of both arms of Wnt signaling [3]. In addition to the canonical and non-canonical pathways, DVL has been associated with other Wnt-related signaling pathways like the Wnt-GSKβ-microtubule, Wnt-calcium, Wnt-RYK (Related to tyrosine kinase), Wnt-aPKC (atypical Protein Kinase C), and Wnt-mTOR (mammalian target of rapamycin) signaling pathway [11].
2.1. Role of DVL in Canonical Wnt pathway
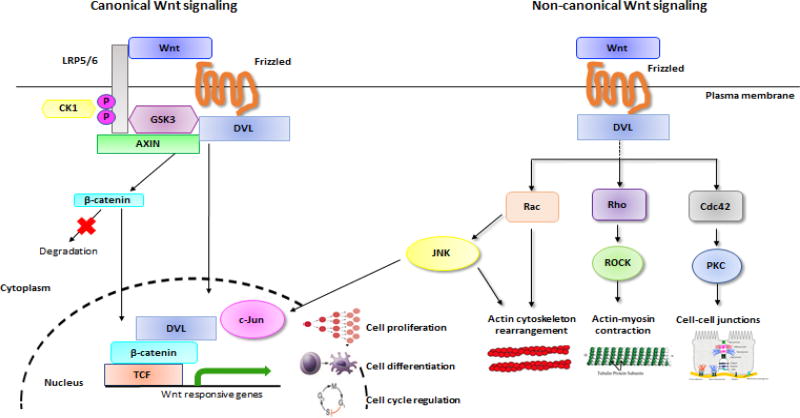
In the canonical Wnt signaling pathway (also called Wnt/β-catenin pathway), one model suggests that binding of Wnt to a seven-pass transmembrane Frizzled receptors helps recruit DVL to the plasma membrane [15]. Recruitment of DVL to the membrane provides a platform for Axin and GSK3β to bind and phosphorylate LRP5/6 thereby preventing constitutive degradation of β-catenin. Inhibition of Axin-mediated degradation by DVL allows β-catenin to accumulate in the nucleus where it serves as a coactivator for TCF to activate Wnt responsive genes (Figure 1). It is also important to note that DVL proteins have been shown to shuttle between the cytoplasm and nucleus [16–18]. A recent study reported that DVL proteins contain a conserved nuclear export sequence (NES) and a nuclear localization sequence (NLS) which is critical for its proper functioning in the canonical Wnt signaling pathway [18]. Another study demonstrated that interaction of DVL-2 with c-Jun and β-catenin, which was followed by formation of stable DVL-2/c-Jun/β-catenin/TCF complex leading to transcriptional activation of Wnt target genes in the nucleus [16] (Figure 1). In 2014, DVL-1 was shown to bind to a 2kb region upstream of the transcription start site of the Frizzled 7 gene in T-47D breast cancer cells. Interestingly, this binding was shown to be diminished with SIRT1/2 pharmacological inhibition, suggesting that DVL-1 protein levels and promoter occupancy could be regulated by at least one type of lysine deacetylase [19]. In addition, DVL was shown to modulate Wnt signaling by interacting with nuclear proteins such as HIPK1(Homeodomain-interacting protein kinase 1), xNET1 (Xenpous nucleotide exchange factor), and FOXKs (Forkhead box transcription factor) [20–22]. These studies elucidate the transcriptional function of DVL in the nucleus. A study in 2015 shed light on a possible mechanism of regulating DVL localization to the nucleus. Interestingly, they demonstrated that Forkhead box (FOX) transcription factors positively regulate Wnt/β-catenin pathway by translocating DVL into the nucleus [22]. Therefore, there seems to be two pools of DVL in a cell – one in the nucleus and another in the cytoplasm regulating the canonical Wnt pathway in a cell.
Figure 1. The role of DVL in canonical and non-canonical Wnt signaling pathway.
In canonical Wnt pathway, DVL promotes clustering of Wnt-LRP5/6-Frizzled to form signalosomes, which results in phosphorylation of LRP5/6 and recruitment of Axin to the plasma membrane, further stabilizing the β–catenin levels in the cytoplasm. DVL has also been found to shuttle between the cytoplasm and the nucleus, acting as a transcriptional activator of Wnt target genes. In non-canonical pathway, Wnt-Frizzled complex interacts with DVL which relays signal to downstream effectors. Multiple pathways downstream of DVL regulate gene transcription, polarity and actin cytoskeleton remodeling of a cell.
2.2. Role of DVL in Non-canonical Wnt pathway
In the non-canonical Wnt signaling pathway (also called Wnt/PCP pathway), DVL plays a key role in governing polarity and cytoskeletal rearrangements of a cell. The Wnt signal is received by the Frizzled receptor which relays signal to DVL-Diego (Diversin in vertebrate) located distally, and Strabismus (Vangl in vertrebrates)-Prickle (Pk) located proximally in a cell, which helps define the polarity of an epithelial cell [11, 23–25].
In the non-canonical pathway, Dishevelled acts as a branchpoint for two independent pathways which leads to activation of small GTPase Rho and Rac (Figure 1). For the activation of the Rho branch of signaling, Wnt signal induces DVL to form a complex with Daam1 (Dishevelled associated activator of morphogenesis 1). The DVL-Daam1 complex further interacts with Rho guanine nucleotide exchange factor WGEF (weak-similarity GEF) which activates downstream effectors like Rho GTPase and Rho-associated kinase (ROCK) [26, 27]. Once activated, this pathway modifies actin and cytoskeleton architecture of a cell. In addition to activating Rho/ROCK, DVL activates the Rac GTPase independent of Daam1. Studies have reported that an activated Rac stimulates the downstream effector c-Jun N-terminal kinase (JNK) which regulates the polarity and movement of a cell during Xenopus gastrulation [28].
3. Involvement of DVL in other signaling pathways
It has been shown that DVL associates with proteins of other signaling pathways to regulate key cell processes. For instance, DVL regulates the cytoskeleton remodeling of a cell by inhibiting GSK3β and stabilizing microtubule-associated proteins MAP1B and MAP2 [29, 30]. Furthermore, DVL interacts with a Ring finger protein called XRNF185 to mediate cell migration during gastrulation. In the Wnt/ Ca2+ pathway, DVL proteins have also shown to cause an increase in intracellular calcium flux which leads to activation of various calcium-sensitive enzymes such as protein kinase C (PKC) (Figure 1), and calcium-calmodulin-dependent kinase (CamKII) [31]. Accumulation of calcium in a cell regulates tissue separation and convergent extension movements during early embryonic development [32]. DVL also participates in axonal repulsion and cell migration by physically interacting with Related to tyrosine kinase (RYK) receptor [33]. Additionally, DVL binds to and stabilizes atypical PKC (aPKC) which leads to microtubule assembly and axon formation. This study in hippocampal neurons demonstrated that downregulation of DVL reduced axon differentiation, whereas overexpression of DVL induced formation of multiple axons [34].
It is evident that DVL plays a central role in propagating Wnt signaling pathway and can affect various development processes of a cell. Given its critical involvement in different pathways, DVL can be called a master integrator of diverse and complex signals.
4. DVL protein structure
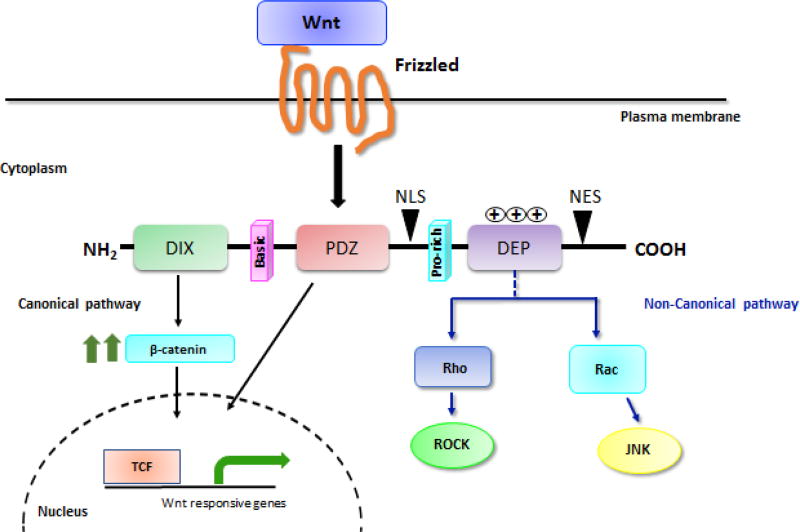
All the DVL proteins ranging from drosophila to humans possess three conserved domains: an amino-terminal DIX domain, a central PDZ, and a carboxyl-terminal DEP domain [35]. In addition to these three domains, DVL harbors two regions with positively charged amino acid residues. The first one is a ‘basic region’ comprising of conserved serine and threonine residues, situated between the DIX and PDZ domains. The second is a ‘proline-rich region’ which is present downstream of the PDZ domain. These conserved domains mediate protein-protein interactions [36] and help DVL channel signals into either canonical or non-canonical Wnt pathway (Figure 2).
Figure 2. The structure of DVL proteins.
DVL is made up of three conserved motifs namely an amino-terminal DIX domain, a central PDZ, and a carboxyl-terminal DEP domain. In addition to these three domains, DVL harbors two regions with positively charged amino acid residues (basic and proline-rich domain) plus a nuclear import (NLS) and a nuclear export signal (NES). The DIX and PDZ domain relay signal to canonical pathway (marked in black arrows), whereas DEP domain mainly regulates membrane localization of DVL and propagates non-canonical pathway (marked in blue arrows).
4.1. The DIX domain
Present on the N-terminal region of the DVL protein, it encodes 82–85 amino acids for human DVL proteins. The DIX domain is also found in proteins like Axin and coiled-coil protein DIX-domain-containing 1 (called DIXdc1 or Ccd1) [37]. The DVL proteins (both at endogenous and at overexpressed levels) have a striking ability to form cytoplasmic puncta (For review, see Gao and Chen 2010) [11]. The DIX domain mediates dynamic polymerization of DVL puncta which enables the DVL proteins to activate the Wnt/β-catenin pathway [35]. The DIX domain not only aids in assembly of signalosomes near the plasma membrane but also mediates protein-protein interactions. DVL protein can interact with Axin via DIX domain which inhibits Axin-promoted β-catenin destruction, thereby stabilizing β-catenin and inducing transcriptional activation of Wnt responsive genes [38]. The crystal structure of the DVL DIX domain has not been identified, however the DIX domain of rat Axin has been solved. Since the DIX domain of DVL and Axin share similarities, the structural properties of DVL DIX domain can be predicted. The DIX domain has five β–strands, one α–helix [39] with highly conserved amino acid residues which are critical for structural and functional roles of DVL. Mutations in some of the key residues (Y27D, F43S, V67A, K68A, E69A) have been shown to inhibit the canonical Wnt pathway [35, 40–42].
4.2. The PDZ domain
The central domain of the DVL proteins, usually encodes about 73 amino acids in each of the human DVL proteins. PDZ domain is named after the proteins in which it was first discovered – Post synaptic density-95/Discs large/Zonula-occludens-1. The PDZ domain mediates crucial protein-protein interactions and regulates multiple biological processes. For instance, it directly interacts with a conserved C-terminal region of Frizzled (KTXXXW) which is necessary for activation of Wnt pathway and for membrane localization of DVL proteins [43]. Considering its central location on DVL proteins, the PDZ domain appears to play an important role in both canonical and non-canonical Wnt pathway [44]. In fact, some studies suggest that the PDZ domain of DVL is involved in distinguishing between the two Wnt pathways [45, 46]. The PDZ domain comprises of 5 or 6 β–stands and 2 or 3 α–helices [47] with a conserved motif (R/K-XXX-G-φ-G-φ motif, where X represents any amino acid residue, and φ is hydrophobic residue) which plays a critical role in ligand binding and conformational properties of the DVL protein [47]. The special role of DVL PDZ domain has attracted a lot of attention. Using NMR spectroscopy, various compounds, peptides and antagonists (3289–8625, NSC668036) have been synthesized which could selectively inhibit PDZ-domain interactions to down-regulate the downstream Wnt signaling pathway [11, 48–51]. Interestingly, an anti-tumor inhibitor called FJ9 has been developed which downregulates the canonical Wnt pathway by selectively inhibiting DVL-Fz interaction in human tumor cell lines [52].
4.3. The DEP domain
The Dishevelled, Egl-10, Pleckstrin (DEP) domain is the C-terminal domain of DVL and consists of 75 amino acids in the human DVL proteins. It has three α - helices, a β-hairpin “arm” and two short β-strands. The DEP domain enables interaction between DVL and DAAM1 (dishevelled associated activator of morphogenesis 1) thereby activating non-canonical signaling pathway. Recent studies have shown that DEP domain is responsible for targeting DVL proteins to the membrane upon Wnt signal stimulation [53]. Moreover, some evidences suggest that DEP domain is essential for the assembly of functional signalosomes and for Wnt signal transduction to the nucleus [54]. Based on structural studies, a strong electric dipole generated by K434, D445 and D448 residues on DEP domain is crucial for protein-protein interactions. Moreover, several basic residues present on the DEP domain (K408, K458, R461, R464, K465, K472 and K482) aid in membrane localization of DVL during planar epithelial polarization [55]. Mutations of these conserved residues not only disrupt membrane localization of DVL but also strongly inhibit Rho/Rac activation during convergent extension [55, 56].
In addition to these conserved regions, DVL possesses a nuclear localization signal (NLS) and a nuclear export signal (NES) (Figure 2). The NLS and NES regulate DVL cellular localization by shuttling it in and out of the nucleus. The NLS (represented by consensus sequence IxLT; where x is any amino acid) is located between the PDZ and DEP domain, whereas, the NES (represented by consensus sequence M/LxxLxL; where x is any amino acid) is present between the DEP and C-terminus of DVL protein. Recent findings suggest that nuclear localization of DVL is important for its function in the canonical Wnt pathway [16, 18].
5. DVL-associated proteins
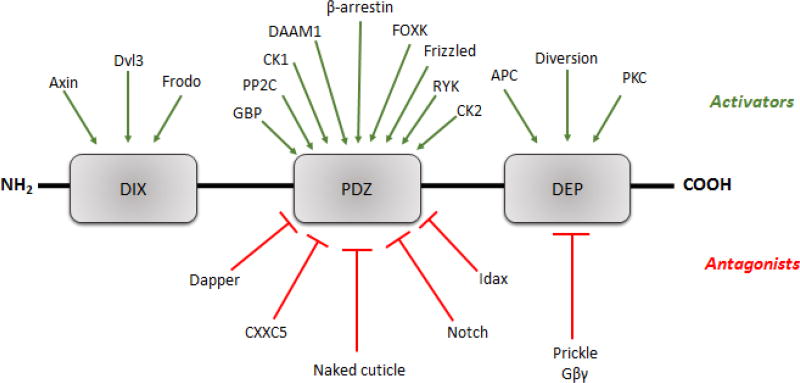
Looking at the central position of DVL proteins in the Wnt signaling pathway, it is no surprise that DVL associates with number of proteins to carry out its diverse cellular functions. Several of the DVL binding partners have been discovered over the last few decades of research (Table 1) (for review see Gao and Chen, 2010; Wallingford and Habas, 2005; Wharton 2003 and Wnt homepage) [3, 11, 12]. This leads us to an important question about how DVL generates specificity to so many proteins? The answer lies in DVL’s structural domains. The three conserved domains (DIX, PDZ and DEP) mediate protein-protein interaction and help DVL proteins to channel signals into either canonical or non-canonical Wnt pathway.
Table 1.
DVL-protein binding partners
| Binding partner | Pathway | Regulation | Function | References |
|---|---|---|---|---|
| DIX-domain binding proteins | ||||
| Axin | Canonical | Positive | β-catenin stability | [38] |
| Dishevelled | Canonical, non-canonical (PCP/Ca2+) | Positive | Enhances the ability of DVL activate Wnt pathway | [38] |
| Frodo | Canonical, Non-canonical | Positive | Secondary axis formation in Xenopus embryo | [57] |
| PDZ-domain binding proteins | ||||
| β-arrestin | Non-canonical (PCP) | Positive | Endocytosis of Frizzled receptor, β-catenin stability and activation of LEF transcription factor | [58–60] |
| Casein Kinase 1 | Canonical | Positive | Stimulate canonical pathway by phosphorylating DVL proteins | [61–63] |
| Casein Kinase 2 | Canonical | Positive | Stimulate canonical pathway by phosphorylating DVL proteins | [64] |
| CFTR | Canonical | Negative | Interacts with DVL-2, negatively regulates Wnt pathway | [81] |
| CXXC5 | Canonical | Negative | BMP4-induced inhibitor of Wnt Signaling in neural stem cell. Also plays a significant role in osteoblast differentiation and wound healing. | [36, 66–68] |
| DAAM1 | Non-canonical (PCP) | Positive | Rho A activation (Binds to both PDZ and DEP domain) | [26] |
| Dapper | Canonical, non-canonical (PCP) | Negative | Induces DVL degradation (Binds to both PDZ and DEP domain) | [82, 83] |
| Diego | Non-canonical (PCP) | Positive | Affects orientation of Drosophila wing hairs | [84] |
| FOXK1, FOXK2 (Forkhead box) | Canonical | Positive | Translocates DVL-2 to nucleus | [22] |
| Frizzled | Canonical, Non-canonical | Positive | Transduction of Wnt signal from Fz to downstream components, membrane targeting of Dishevelled | [43] |
| GSK3β-binding protein/Frat | Canonical, Non-canonical | Positive | Dissociation of Axin-GSK3β complex, activation of JNK | [85, 86] |
| IDAX/CXXC4 | Canonical | Negative | Regulate embryonic axis formation in Xenopus | [69] |
| Naked Cuticle | Non-canonical (PCP) | Negative | Repress Wnt pathway | [70, 71] |
| Notch | Non-canonical (PCP) | Negative | Regulate cell development in Drosophila eye, crosstalk between PCP and Notch pathway | [72–74] |
| PAR1 | Canonical, Non-canonical (PCP) | Positive / Negative | Phosphorylation of Dishevelled | [87–89] |
| Protein Phosphatase 2C | Canonical | Positive | Positively regulates Wnt signals by dephosphorylating Axin | [65] |
| RYK | Canonical | Positive | Essential for neurite outgrowth, activates T cell factor induced by Wnt ligands | [33] |
| Strabismus | Canonical, Non-canonical | Negative / Positive | Regulates planar cell polarity, morphogenetic movement and neural gene expression during gastrulation | [90, 91] |
| TMEM88 | Canonical | Negative | Disrupts canonical Wnt signaling | [92] |
| DEP-domain binding proteins | ||||
| APC | Non-canonical | Positive | Stabilization of microtubule, enhance cell migration | [93] |
| Diversion | Non-canonical | Positive | Regulates heart formation and gastrulation movements in zebrafish embryogenesis | [94] |
| Ephrin B1 | Non-canonical (PCP) | Positive | Regulates PCP pathway for cell movement and cell repulsion | [95–97] |
| Gpy | Canonical | Positive / Negative | Membrane localization of Dishevelled, and lysosomal degradation | [98, 99] |
| Prickle | Canonical, Non-canonical (PCP) | Negative | Degradation of DVL-3, regulate planar cell polarity | [84, 100, 101] |
| Protein Kinase C | Canonical/Non-canonical | Positive | Regulate convergent extension movements, phosphorylation and stabilization of DVL proteins | [102, 103] |
| Other associated proteins | ||||
| c-Jun | Canonical | Positive | Activates β-catenin/TCF mediated transcription of Wnt target genes in the nucleus | [16] |
| CTNNB1 | Canonical | Positive | Activate transcription of Wnt target genes | [16, 75] |
| KLHL12/Cullin-3 | Canonical | Negative | Degradation of Dishevelled 3 | [76] |
| IQGAP1 | Canonical | Positive | Promotes nuclear localization of Dishevelled which sequentially activates Wnt target genes | [104] |
| Lgl | Non-canonical | N/A | Regulate apical basal polarity | [77] |
| NFκB | NFκB signaling | Negative | DVL-1 (C-terminus) binds to p65 in nucleus and inhibits NFκB activation | [105] |
| RIPK4 | Canonical | Positive | Stimulates Wnt pathway by phosphorylating DVL proteins | [106] |
| RNF185 | Canonical | Negative | Negatively modulates osteoblast differentiation through ubiquitination and degradation of DVL-2 | [107] |
| SIRT1 | Canonical/Non-canonical | Positive | Positively regulates Wnt signaling via DVL proteins | [80] |
| TIAM1 (T-cell lymphoma invasion and metastasis 1) | Non-canonical | Positive | Mediates cell migration via Rac activation, required for neuron differentiation | [78, 79] |
Note: The table summarizes the list of proteins which bind to Dishevelled, categorized by the region of interaction with Dishevelled. The table further describes the pathway affected and the functional significance of the interaction between DVL and its associated proteins.
For instance, proteins like Axin and Frodo interact with DVL proteins via DIX domain leading to stabilization of canonical and non-canonical Wnt pathway [38, 57]. The DIX domain of DVL also interacts with itself to mediate canonical signaling pathway [38]. The central PDZ domain acts as a scaffold for many proteins to propagate signals to downstream effector molecules. It switches between the canonical and non-canonical pathway by interacting with different binding partners. For activation of canonical pathway, PDZ domain of DVL interacts with β-arrestin, Casein Kinase 1, Casein Kinase 2, Frizzled and Protein Phosphatase 2C [16, 43, 58–65]. On the other hand, proteins like IDAX, CXXC5, Notch, and Naked cuticle behave as antagonist and repress the Wnt pathway [36, 66–74] (see Table 1 and Figure 3). Similarly, the DEP domain associates with various activators (APC, diversion, protein kinase C) and antagonists (Gβγ Prickle) to regulate non-canonical pathway (see Table 1 and Figure 3). Numerous other proteins like c-Jun, CTNNB1, KLHL12/Cullin-3, Lgl, and TIAM1 interact with Dishevelled [16, 75–79]. In 2010, the SIRT1 lysine deacetylase which is known to regulate cellular responses resulting from very diverse physiological stress, was shown to serve as an important regulator of Wnt signalling [80]. SIRT1 loss of function was shown to decrease the levels of all three DVLs. Furthermore, it was demonstrated that SIRT1 and DVL proteins complex in vivo and inhibition of SIRT1 led to changes in gene expression of Wnt target genes and Wnt-stimulated cell migration. This finding was the first to link the sirtuins with DVL directly and helped to explain SIRT1-mediated regulation diverse physiological responses given its connection with a key molecular scaffold. In 2013, a subsequent report probed the SIRT1-DVL connection in greater depth and demonstrated that SIRT1 and SIRT2 positively regulate the levels of Rac1-GTP and its activator, TIAM1 [78]. This report demonstrated that SIRT1 activity was critical for the DVL-1 and TIAM1 interaction in cancer cells and positively modulates the DVL/TIAM1/Rac axis and promote sustained pathway activation. Prior to these reports, SIRT1 had only been shown to mediate the epigenetic silencing of Wnt antagonists. Collectively these reports demonstrated that SIRT1 is a novel regulator of transient and constitutive Wnt signaling. However, the specific domain(s) to which they bind is not clearly understood (see Table 1 and Figure 3).
Figure 3. DVL binding partners.
The agonists (marked in green arrows) and the antagonist (marked in red arrows) bind to specific domains of DVL to regulate Wnt pathway.
6. DVL post-translational modifications
6.1. DVL Phosphorylation
DVL transmits numerous diverse signals that lead to mutually exclusive cellular processes, yet much remains unknown about the manner in which it coordinates complex signals. Some progress has been made and post-translational regulation appears to be critical for specifying how molecular signals are routed. The most well studied post-translational modification of DVL proteins is phosphorylation. Early reports first demonstrated phosphorylation of Drosophila Dsh proteins in response to Wg stimulation [108]. A couple of years later, more investigation led to the report of Casein kinase 2 (CK2) as the first Dsh kinase to be identified in Drosophila [64]. These initial discoveries were followed by a series of reports across diverse species that identified other DVL kinases including casein kinase 1 (CK1) isoforms [61, 62, 109, 110], PAR1 [111], RIPK4 [106], NEK2 [112] and other kinases [11]. Phosphorylation of DVL appears to be a dynamic process where site-specific phosphorylation elicits divergent biological responses. For example, DVL regulates both β-catenin and planar cell polarity signals and site specific phosphorylation appears to control which of the competing signals is transmitted and the strength with which they are transmitted. Par1 and CKIε-mediated phosphorylation appears to activate the arm of DVL signaling that promotes β-catenin signaling while simultaneously inhibiting the arm of DVL signaling that promotes JNK/PCP signaling [87, 113]. These studies and others demonstrated the critical role of multiple kinases such as CK1, CK2 and PAR1 in the Wnt pathway that can activate or inactivate DVL in a temporally sequential fashion [111]. Bernatik et al [111] reported that CK2 acts as a constitutive kinase whose activity is required for the further action of CK1ε to induce phosphorylation and TCF/LEF-driven transcription. This study proposed a multistep and multi-kinase model for DVL activation in the Wnt/β-catenin pathway which subsequently induces a de-activation mechanism driven by CK1δ/ε-mediated phosphorylation of DVL. The c-terminus of DVL has been implicated in the negative regulation of its own activity and of the Wnt pathway. Hyperphosphorylated DVL is also known to interact with Ror2 receptor-tyrosine kinase via its C-terminal and inactivate the canonical Wnt pathway [114]. Thus the C-terminal of DVL has been shown to be necessary and sufficient for canonical (and non-canonical) Wnt pathway inactivation.
6.2. DVL Ubiquitination
DVL ubiquitination has been linked with its degradation and activation. DVL is known to interact with proteins such as KLHL12-cullin3 complex which ubiquitinates and targets DVL for degradation [76]. The E3 ubiquitin ligase complex can polyubiquitinate and target DVL for proteasomal degradation causing an inhibition of the canonical Wnt pathway. Treatment of cells with MG132 increased the co-immunoprecipitation of KLHL12 with DVL in a Wnt3a-dependent manner [76]. ITCH is another E3-ubiquitin ligase belonging to the HECT-type E3 subfamily known to regulate DVL levels in cells by ubiquitination. ITCH negatively regulates canonical Wnt signaling by specifically targeting phosphorylated DVL for degradation [115]. Wnt-5a activation of JNK phosphorylates NEDDL4 which in turn ubiquitinates DVL for degradation via polyubiquitination at K-6, K-27 and K-29 [116]. Thus, NEDDL4 can act as a feedback regulator of Wnt pathway activation. Gao et al. [11] showed that DVL-2 can be targeted for degradation upon starvation-induced metabolic stress by Von-Hippel-Lindau protein (pVHL), an E3 ubiquitin ligase that promotes DVL-2 ubiquitination.
On the other hand, CYLD is a de-ubiquitinase and has been shown to negatively regulate β-catenin signaling. Knockdown of CYLD has been reported to stabilize β-catenin and induce β-catenin-responsive target gene activation [117]. This hyperactive Wnt signaling due to CYLD mutations was linked with human skin appendage tumors. Part of the proposed mechanism for this tumor promotion was tied to N-terminal K63-linked polyubiquitination of DVL which were proposed to potentiate the inactivation of the destruction complex and lead to β-catenin stabilization and translocation to the nucleus.
6.3. DVL methylation
Utilizing tandem mass spectrometry, Wu et al. showed that DVL-3 can be mono- or dimethylated on arginines located in DIX domain and the linker regions between DIX-PDZ and PDZ-DEP domains [118]. The precise role of DVL-3 methylation is not known but dimethylation of R698 was enhanced with Wnt3a stimulation for 15 minutes and returned to basal levels at 30 minutes.
7. Cancer associations
Wnt signaling and its association with cancer has been quite extensive in the study of intestinal and colorectal cancer, so we point the reader to previous reviews discussing this aspect of the cancer association [119–122]. In this section, we will focus primarily on the connection between Wnt signaling and breast cancer and some of the latest findings with other cancers beyond the GI tract. Somatic mutations and altered expression of Wnt ligands, antagonists or receptors can support tumorigenesis by influencing many hallmarks of cancer. Starting in the extracellular compartment, there are 19 mammalian Wnt ligands, and the genes encoding them show temporally restricted, tightly regulated, and localized expression patterns [123]. However, during tumor progression, this balance is perturbed, and altered Wnt signaling can serve as a potent stimulus for tumorigenesis. In mammals, Wnt was first identified as an oncogene in mouse mammary tumorigenesis [124]. Moreover, MMTV-Wnt-1 mice have been shown to possess a markedly expanded population of premalignant mammary tissue which is thought to arise from a discrete population of mammary stem cells [125, 126]. Wnt-1 was the first Wnt ligand demonstrated to cause mammary tumors. Early on, Wnt-1 expression was shown to cause striking changes in morphological and growth properties in mammary epithelial cell lines [127]. Wnt-1 was then shown to induce numerous mammary tumor formation in vivo [128] and was shown to transform primary human mammary epithelial cells alone without a cooperating oncogene [129]. Further study revealed that the Wnt-1 transgene preferentially induced mammary cancers from progenitor cells [130] and Wnt signaling regulated the amplification of mammary progenitor cells [131]. These Wnt-1 induced tumors contained heterogeneous cell types, expressing keratin 6 and Sca-1, and showed signs of selective targeting of mammary stem and/or progenitor cells. Further work addressing how Wnt-1 contributes to breast tumor heterogeneity demonstrated that Wnt-1 inhibits mouse mammary cell differentiation and upregulates Twist [132], a transcription factor that induces EMT to facilitate tumor metastasis [133]. However, what lies between Wnt-1 activation and the changes in programs of gene expression required for tumor formation is not known.
Interestingly, even in mice lacking ERα, Wnt-1 was shown to induce mammary gland hyperplasia and tumorigenesis, suggesting that the potency of this pathway to contribute to breast cancer may be especially relevant for the basal-like breast cancers which tend to be negative for ER, PR and do not show HER2 amplification [134]. Other Wnts such as Wnt-2, 3, 4 and 7B have now been shown to be differentially expressed in human breast tumors relative to normal tissue [135], and Wnt5a loss even correlates with loss of ERα while restoration of its expression restores tamoxifen sensitivity in ERα negative breast cancer cells [136].
The Frizzled and LRPs transmit extracellular signals and like the Wnts, members of both families of receptors contribute to tumorigenesis. For example, Wnt signaling through LRP6 was shown to be required for tumor formation and metastasis and was necessary for cancer cell self-renewal using in vivo models [137]. Moreover, inhibition of Wnt signaling resulted in re-expression of breast epithelial differentiation markers and repression of genes associated with EMT. A separate study revealed that expression of LRP6 is up-regulated in a subpopulation of human breast cancers and LRP6 inhibition in breast cancer cells reduces Wnt signaling, cell proliferation, and in vivo tumor growth. Additionally, in vivo administration of an LRP6 antagonist was shown to markedly suppress growth of Wnt1 tumors without causing appreciable side effects [138]. Aberrant splicing of the other family member, LRP5, was shown to be resistant to inhibition via the extracellular secreted antagonist, DKK1 [139]. This mutant was found to be frequently expressed in breast tumors of different cancer stage (58–100%), and an anti-LRP5 antibody was shown to inhibit cell growth, and induce apoptosis in breast cancer cells expressing the mutant suggesting its potential as a new therapeutic target. Finally, in an elegant study, expression of LRP5 in mouse mammary stem cells was shown to be required to maintain the basal lineage [140]. This is important because LRP5 may serve as a single biomarker that has been demonstrated to be functionally involved in stem cell maintenance.
Extracellular Wnt ligands can interact with several key antagonists that are also secreted. Two families of antagonists include the secreted Frizzled related proteins (SFRPs), the Dickkopf proteins (DKKs) and the Wnt inhibitor factors (WIFs) [141–143]. The final outcome of extracellular Wnt signaling depends on the relative stoichiometry of ligands:receptors:antagonists. If the local concentration of Wnts extends beyond the buffering capacity of their antagonists, Wnt ligands bind the Fzd/LRP receptors and may initiate a signaling cascade that ultimately leads to β-catenin stabilization [142]. Antagonists of Wnt signaling participate in several dimensions of tumor suppression and their deregulation has been linked with tumorigenesis. For example, in 79 of 130 (61%) primary breast tumors the SFRP1 promoter was methylated and Kaplan-Meier analyses showed SFRP1 gene hypermethylation was associated with shorter patient OS (overall survival) with invasive breast cancer [144]. Further, analysis of 168 primary breast carcinomas revealed that 73% had a methylated SFRP5 promoter and strikingly, SFRP5 methylation was associated with reduced OS and was an independent risk factor affecting OS in a multivariate Cox proportional hazard model. Thus, the SFRPs are targeted for epigenetic inactivation in human breast cancer. Based on other studies this same trend is consistent for SFRP1, 2, 5, DKK1, WIF1 [145–147] suggesting that these gene silencing events may not be strictly random and stochastic, but rather part of a coordinated program of gene expression. Epigenetic silencing of Wnt inhibitors is another mechanism employed in tumor progression [148]. Promoter hypermethylation or histone deacetylation of Wnt inhibitory factors like WIF1, sFRP1-5, DKK1 and DKK3 have been observed in breast, lung and colon cancers among others [146, 149, 150].
Inactivating or loss-of-function mutations in APC, a Wnt signaling protein classically known to cause familial adenomatous polyposis (FAP), can lead to colorectal cancer upon concurrent KRAS and p53 activation [151, 152]. Hyperactivation of the Wnt pathway in non-FAP colorectal cancer patients has also been reported due to APC and β-catenin mutations. Likewise, missense or other mutations of β-catenin, deletions and truncations of Axin1 and mutations in TCF4 are also seen in multiple cancer types including hepatocellular, medulloblastoma, colorectal, gastric, ovarian, pancreatic etc. [153].
Dishevelled alterations have now been reported for diverse cancer types. One report examined the protein levels in 67 human glioma and 3 normal brain specimens by Western blotting and immunohistochemistry. The DVL immunoreactivity score (IRS) was assessed to investigate a possible association of DVL with the malignant phenotype in glioma. The DVL IRS increased significantly with the pathologic grade of glioma and the proliferation index and tumor invasion index were significantly higher for the DVL-positive group than the DVL-negative group. This study concluded that DVL overexpression may contribute to the malignant proliferation and invasion of human glioma [154].
The Wnt signaling pathway plays a critical role in normal cell development as well as in tumorigenesis [155]. Dishevelled proteins transduce the upstream signals from Frizzled receptors via PDZ domain to downstream components. Thus, the critical role of dishevelled PDZ domain makes it an ideal pharmaceutical target for treatment of various diseases. Since fibrotic lung exhibits aberrant activation of Wnt signaling pathway, a study showed that inhibiting the pathway at DVL level could be an effective approach for the treatment of fibrotic lung cancer. Their data demonstrated that a competitive inhibitor of DVL PDZ domain, called NSC668036, suppressed β-catenin driven gene transcription in fibroblasts. Furthermore, NSC668036 abolished TGF-β1 induced migration, proliferation, and expression of collagen I and α-smooth muscle actin (α-SMA) in pulmonary fibroblasts. In vivo studies concluded that NSC668036 significantly reduced collagen I, α-SMA, and TGF-β1 levels but increased expression of epithelial markers such as E-cadherin, CK19 and occludin that could inhibit pulmonary fibrogenesis [156]. Moreover, another study reported a novel DVL PDZ domain binding peptide ligand, known as pen-N3, which inhibits the Wnt/β-catenin pathway. This suggests that interference with DVL PDZ domains may be a suitable therapeutic strategy for inhibiting Wnt signaling in diseases that are dependent on DVL function [49]. On a similar note, a recent study reported that miR-103, a short non-coding RNA, down regulates DVL-1 by binding with its 3’ UTR region which results in β-catenin degradation and transcription inhibition of c-Myc. The study further demonstrated DVL-mediated mechanism of making tumor cells more sensitive for glucocorticoid-induced apoptosis in hematopoietic malignancies [157].
The nuclear localization of DVL proteins can play a critical role in tumorigenesis. A study reported that co-expression of IQGAP1, a key regulator of cell adhesion and migration, and DVL correlated with the lymph nodal metastasis and poor prognosis of NSCLC. Interestingly, coexpression of IQGAP1 and DVL in the cytoplasm and nucleus was reported to be significantly higher in lymph nodal metastases than in primary tumors, correlating with poor prognosis. Moreover, co-localization in the nucleus was proposed to play a critical role in the activation of canonical Wnt pathway [158]. A recent study demonstrated that Wnt5A represses ribosomal RNA (rRNA) synthesis in breast cancer cells by promoting nucleolar accumulation of DVL-1. The nucleolar DVL-1 proteins suppress tumor growth by negatively regulating rRNA synthesis through loss of SIRT-7 from chromatin regions containing rDNA [159]. Yet another study reported nuclear localization of DVL protein and demonstrated a protective role of DVL-2 in rheumatoid arthritis patients. This study explored the impact of DVL-2 on proliferation and inflammatory cytokine secretion in rheumatoid arthritis fibroblast-like synoviocytes (RA-FLS). The authors demonstrated that over-expression of DVL-2 increased apoptosis and inhibited inflammatory cytokine secretion by RA-FLS, both in vivo and in vitro, possibly by downregulating NFκB pathway [160]. On other hand, a group proposed that loss of histone methyltransferase, SETD2, leads to upregulation of DVL-2 expression, thereby augmenting Wnt signaling to facilitate tumor malignancies. Their data demonstrated that SETD2 depletion enhanced mRNA expression and nuclear accumulation of DVL-2 which leads to hyper activation of Wnt signaling pathway resulting in colorectal cancer [161]. Therefore, the critical role of DVL in different cancer types makes it a suitable target for cancer therapy. Additional investigation of DVL expression in NSCLC revealed expression of all DVLs in primary tumors was 36.3% (41/113) for DVL-1, 36.3% (41/113) for DVL-2 and 41.6% (47/113) for DVL-3, while normal adult bronchial and alveolar epithelia showed negative expression of all these proteins. Moreover, the expression levels of all three DVL was significantly higher in adenocarcinomas than in squamous carcinomas, and were associated with poor tumor differentiation, and DVL-1 & DVL-3 were significantly higher in nodal metastases than in primary growths, with the DVL-1 expression correlating to β-catenin expression in the metastases [162]. Additionally, in a study of brain metastases that originated from primary lung carcinomas, the expression of DVL-1 and DVL-3 were analyzed by IHC. DVL-1 and DVL-3 showed over expression in brain metastasis in 87.1% and 90.3% of samples respectively. Nuclear staining was observed in 54.8% of cases for DVL-1 and 53.3% for DVL-3, and when DVL-1 and DVL-3 were up-regulated there was a significant increase in the number of cases with nuclear beta-catenin [163].
Yet another tumor type where DVL levels are altered is chronic lymphocytic leukemia (CLL). Khan et al reported that DVL-1, −2 & −3 were exclusively expressed in CLL cells as compared to normal peripheral blood mononuclear cells (PBMCs) [164]. The expression of DVL-1 and DVL-3 proteins was significantly more pronounced in progressive than in non-progressive disease (p < 0.01), whereas the level of DVL-2 was significantly higher in non-progressive as compared to progressive disease (p < 0.001). This alteration in DVL expression was also extended to breast cancers. Nagahata et al demonstrated amplification of DVL-1 in 13 of 24 primary breast cancers examined, and increased expression of this gene in 11 of those tumors in comparison to corresponding non-cancerous breast tissues. These data suggest that amplification and increased expression of the DVL-1 gene may play some role in human breast carcinogenesis through derangement of the Wnt signaling pathway [165]. A few years later in 2007, another report demonstrated an association between nuclear localization of DVL and β-catenin. Their analysis revealed that of the 98 IDCs analyzed, 30% of tumors displayed both nuclear and cytoplasmic staining of DVL, while 52% showed nuclear localization and demonstrated a significant association between nuclear localization of DVL and β-catenin [166]. Finally, in a study involving hepatocellular carcinoma (HCC), Western blotting and immunohistochemistry were used to measure DVL-2 protein expression in HCC and adjacent normal tissues of 101 patients. In this study, DVL-2 expression was found upregulated in HCC tissues compared to the adjacent normal tissues, and its expression level was significantly correlated with histological grade, metastasis, and vein invasion (P = 0.009) [167].
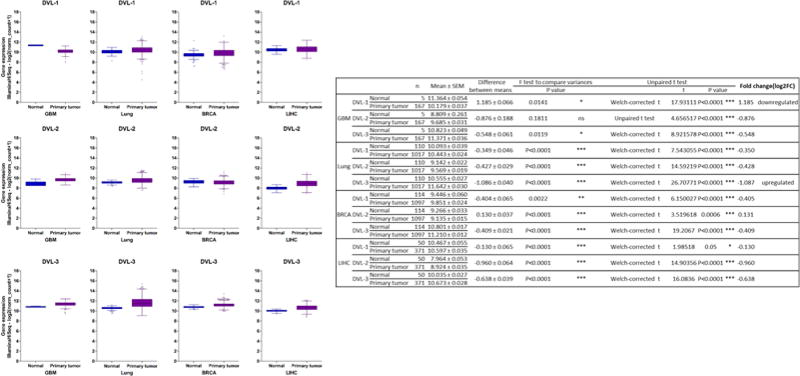
Interestingly, we performed a TCGA analysis of DVL-1, DVL-2 and DVL-3 expression across of 4 different types of cancer (Glioblastoma or GBM, Lung, Breast or BRCA and Liver or LIHC) that shows no dysregulation of DVL RNA expression in majority of those cancers compared to adjacent normal tissues except DVL-1 that appears to be downregulated in GBM and DVL-3 upregulated in lung cancer (Figure 4). Even though this data appears to be contradictory to the previous reports where DVL proteins are upregulated [154, 162, 165, 168], this suggests the important role of the post-transcriptional regulation of DVL proteins. For instance, few mechanisms like RNA transport and storage, RNA degradation and stability, or translational and protein stability may be altered in the tumor cells.
Figure 4. DVL mRNA expression in GBM, Lung, BRCA and LIHC.
DVL-1, DVL-2 and DVL-3 RNA-seq data were analyzed using TCGA downloaded from Xena tool of UCSC Cancer Genomics Browser (http://xena.ucsc.edu/). Normalized RNA expression is plotted as log2 (norm_count+1). Dysregulated DVL expression between tumor and normal tissue were identified using t-test (p value < 0.001, log2FC > 1 or < −1).
After post-transcriptional processing, the mature mRNA must be transported from the nucleus to the cytosol so that it can be translated into a protein, this step is a key point for regulation of gene expression. The different RNA species that are produced in the nucleus are exported through the nuclear pore complexes (NPCs) by nuclear transport proteins known as importins and exportins, which belong to the karyopherin-beta family proteins. The expression of karyopherin-beta proteins are dysregulated in multiple tumors such as in melanoma, pancreatic, breast, colon, gastric, prostate, esophageal, lymphoma and lung cancer which may have consequences in the differential expression in the tumor tissues with respect to the normal tissue [169]. On the other hand, RNA stability determines its half-life and therefore the time that would be available for translation. In cancer, among other pathological conditions, the dysregulation of RNA stability has been already reported to affect genes like growth factors, oncogenes, cell cycle regulators and inflammatory cytokines that can contribute to cancer development and/or progression [170, 171].
Post-translational modifications not only play a role in regulating the folding of proteins, their transport or function but also the stability of the protein. In this review, we show that DVL proteins can be phosphorylated, ubiquitinated, or methylated. Of these post-translational modifications, ubiquitination has a key role modulating DVL protein degradation [11, 76, 115]. The KLHL12-Cullin-3 ubiquitin ligase complex negatively regulates Dishevelled [76]. Interestingly, Cullin-3 act as tumor suppressor and is downregulated in lung, liver, and breast cancer [172–175] which may correlate with the elevated levels of DVL proteins in those cancers.
8. DVL and other human diseases
Since DVL is the central mediator of the Wnt signaling pathway that coordinate cell development processes and adult tissue homeostasis, it is certain that its deregulation can be linked with development disorders and syndromes. DVL was originally identified based on the phenotype of disorientation in wing hair of Drosophila. Later it was discovered that mutation in DVL signaling could perturb the segment polarity in Xenopus embryo [3]. All three DVL genes are broadly expressed in various tissues of the body. It is suggested that DVL genes work in network and there may be redundancy to some extent [176]. Several knockdown studies have been employed to elucidate the specific role of each DVL.
As a result of high similarity, DVL (DVL-1, −2 and −3) in mice and human have been proposed to have functional redundancy. Several mice models have been extensively used to understand the phenotype of DVL knockout mice. DVL-1 KO mice display normal skeletal phenotype but exhibit abnormal social interaction in nest building, and home cage huddling [177]. Additionally, lack of DVL-1 genes can induce myocardial infarction in mice [178]. Furthermore, recent studies consider DVL-1 as a candidate gene for cardiovascular malformations associated with 1p36 deletions [179]. While DVL-1 null mice showed unique feature in social interaction abnormalities, DVL-2 null mice display defects in cardiac outflow tract formation. Almost half of the DVL-2 null die in perinatal period due to cardiac anomalies. In addition, DVL-2 null mice display defects in somite segmentation and neural tube closure [180].
DVL-3 null mice do not display any skeletal defects, however these mice die perinatally likely due to cardiac tract abnormalities. Knockdown studies of DVL-3 suggest that this gene is essential for cochlea and neural tube development [181]. To elucidate the specific roles of DVL genes, rescue studies were conducted. These studies indicate the double knockout mice have severe phenotype. For instance, double knockout of either DVL-1, DVL-2, DVL-3 lead to defect in cardiovascular outflow tract, neural crest development, cochlear defect and skeletal defects suggesting redundant roles between the DVL homologs. (Refer Gao et al., 2010 for review) [11, 181]. Interestingly, double knockout of DVL-1 and DVL-3 does not display neural tube defects which indicates that each DVL has a unique role to play. Aberrant expression of DVL genes have been linked to human disorders. In human development disorders, DVL-1 has been reported to be a candidate gene in Schwartz-Jampel syndrome, Charcot-Marie-Tooth disease type 2A and DiGeorge Syndrome [6, 182]. Additionally, over-expression of DVL-1 and DVL-3 have been linked to Hirschsprung’s disease, a congenital disorder characterized by absence of ganglion cells in terminal regions of the gut during development [183].
Several studies have helped establish the fact that mutation in DVL genes can cause Robinow syndrome. Robinow Syndrome is a genetic disease which is characterized by skeletal abnormalities which can be caused by mutation in genes encoding components of Wnt signaling pathway [184]. White et al. uncovered that frameshift mutation in penultimate and final exons of DVL-1 and DVL-3 can be a common cause of autosomal-dominant Robinow syndrome [185, 186]. The data suggests that there are six frameshift mutations, out of which five are de-novo, leading to truncation of the C-terminal domain of DVL-1 disrupting the downstream canonical pathway [185]. Another study indicates that frameshift mutation on exon 15 replaces the C-terminal tail of DVL-1 with 142 highly basic amino acid. And these de-novo mutations on DVL-1 result in osteosclerotic form of Robinow syndrome [187]. To summarize, DVL plays an important role in development processes and mutations in DVL gene can lead to severe phenotypic defects.
9. Concluding remarks
Significant advances have been made regarding domain specific functions of DVL, the binding partners involved in molecular interactions, and the role it plays during development of diverse organisms. However, many significant gaps in knowledge remain regarding the mechanism by which Dishevelled relays information to different intracellular compartments, the role of post-translational regulation beyond phosphorylation and the functional significance of DVL nuclear localization. Given the central importance of this family of proteins to normal physiology and pathophysiology, many of these critical unknowns will likely be addressed in the years to come.
Highlights.
The Dishevelled gene was first identified in Drosophila mutants with disoriented hair and bristle polarity and its study gained popularity when it was discovered to play a key role in segment polarity during early embryonic development. Further research revealed that Dishevelled (DVL) proteins function as principal components of Wnt signaling pathway in many organisms and govern several cellular processes including proliferation, survival, migration, differentiation and stem cell renewal. It is fascinating to consider how the identity of DVL is largely linked with its ability to integrate and relay complex Wnt signals in tissues and cells yet how it conducts this symphony of activity still remains poorly understood. Additionally, the concept of the mystery that still surrounds DVL and what it means for it to be “activated” after more than two decades investigation is addressed. This review discusses the function of DVL in the Wnt signaling pathway in normal and aberrant cellular contexts. It also highlights novel roles within the nucleus and discusses new binding partners that have been discovered and novel mechanisms of post-translational regulation.
Acknowledgments
This work was supported by the National Institute of Health [CA155223 to K.P.] and a Cancer Prevention and Research Institute of Texas (CPRIT) Award to K.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fahmy OG, Fahmy MJ. Complementation among the subgenic mutants in the r-locus of Drosophila melangogaster. Nature. 1959;184:1927–1929. doi: 10.1038/1841927a0. [DOI] [PubMed] [Google Scholar]
- 2.Fahmy OG, Fahmy MJ. Differential Gene Response to Mutagens in Drosophila Melanogaster. Genetics. 1959;44:1149–1171. doi: 10.1093/genetics/44.6.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallingford JB, Habas R. The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development. 2005;132:4421–4436. doi: 10.1242/dev.02068. [DOI] [PubMed] [Google Scholar]
- 4.Perrimon N, Mahowald AP. Multiple functions of segment polarity genes in Drosophila. Dev Biol. 1987;119:587–600. doi: 10.1016/0012-1606(87)90061-3. [DOI] [PubMed] [Google Scholar]
- 5.Klingensmith J, Yang Y, Axelrod JD, Beier DR, Perrimon N, Sussman DJ. Conservation of dishevelled structure and function between flies and mice: isolation and characterization of Dvl2. Mech Dev. 1996;58:15–26. doi: 10.1016/s0925-4773(96)00549-7. [DOI] [PubMed] [Google Scholar]
- 6.Pizzuti A, Amati F, Calabrese G, Mari A, Colosimo A, Silani V, Giardino L, Ratti A, Penso D, Calza L, Palka G, Scarlato G, Novelli G, Dallapiccola B. cDNA characterization and chromosomal mapping of two human homologues of the Drosophila dishevelled polarity gene. Hum Mol Genet. 1996;5:953–958. doi: 10.1093/hmg/5.7.953. [DOI] [PubMed] [Google Scholar]
- 7.Semenov MV, Snyder M. Human dishevelled genes constitute a DHR-containing multigene family. Genomics. 1997;42:302–310. doi: 10.1006/geno.1997.4713. [DOI] [PubMed] [Google Scholar]
- 8.Sokol SY, Klingensmith J, Perrimon N, Itoh K. Dorsalizing and neuralizing properties of Xdsh, a maternally expressed Xenopus homolog of dishevelled. Development. 1995;121:1637–1647. doi: 10.1242/dev.121.6.1637. [DOI] [PubMed] [Google Scholar]
- 9.Sussman DJ, Klingensmith J, Salinas P, Adams PS, Nusse R, Perrimon N. Isolation and characterization of a mouse homolog of the Drosophila segment polarity gene dishevelled. Dev Biol. 1994;166:73–86. doi: 10.1006/dbio.1994.1297. [DOI] [PubMed] [Google Scholar]
- 10.Tsang M, Lijam N, Yang Y, Beier DR, Wynshaw-Boris A, Sussman DJ. Isolation and characterization of mouse dishevelled-3. Dev Dyn. 1996;207:253–262. doi: 10.1002/(SICI)1097-0177(199611)207:3<253::AID-AJA2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 11.Gao C, Chen YG. Dishevelled: The hub of Wnt signaling. Cell Signal. 2010;22:717–727. doi: 10.1016/j.cellsig.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 12.Wharton KA., Jr Runnin’ with the Dvl: proteins that associate with Dsh/Dvl and their significance to Wnt signal transduction. Dev Biol. 2003;253:1–17. doi: 10.1006/dbio.2002.0869. [DOI] [PubMed] [Google Scholar]
- 13.Weitzman JB. Dishevelled nuclear shuttling. J Biol. 2005;4:1. doi: 10.1186/jbiol21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mlodzik M. The Dishevelled Protein Family: Still Rather a Mystery After Over 20 Years of Molecular Studies. Curr Top Dev Biol. 2016;117:75–91. doi: 10.1016/bs.ctdb.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacDonald BT, He X. Frizzled and LRP5/6 receptors for Wnt/beta-catenin signaling. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a007880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gan XQ, Wang JY, Xi Y, Wu ZL, Li YP, Li L. Nuclear Dvl, c-Jun, beta-catenin, and TCF form a complex leading to stabilization of beta-catenin-TCF interaction. J Cell Biol. 2008;180:1087–1100. doi: 10.1083/jcb.200710050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habas R, Dawid IB. Dishevelled and Wnt signaling: is the nucleus the final frontier? J Biol. 2005;4:2. doi: 10.1186/jbiol22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoh K, Brott BK, Bae GU, Ratcliffe MJ, Sokol SY. Nuclear localization is required for Dishevelled function in Wnt/beta-catenin signaling. J Biol. 2005;4:3. doi: 10.1186/jbiol20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simmons GE, Jr, Pandey S, Nedeljkovic-Kurepa A, Saxena M, Wang A, Pruitt K. Frizzled 7 expression is positively regulated by SIRT1 and beta-catenin in breast cancer cells. PLoS One. 2014;9:e98861. doi: 10.1371/journal.pone.0098861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louie SH, Yang XY, Conrad WH, Muster J, Angers S, Moon RT, Cheyette BN. Modulation of the beta-catenin signaling pathway by the dishevelled-associated protein Hipk1. PLoS One. 2009;4:e4310. doi: 10.1371/journal.pone.0004310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyakoshi A, Ueno N, Kinoshita N. Rho guanine nucleotide exchange factor xNET1 implicated in gastrulation movements during Xenopus development. Differentiation. 2004;72:48–55. doi: 10.1111/j.1432-0436.2004.07201004.x. [DOI] [PubMed] [Google Scholar]
- 22.Wang W, Li X, Lee M, Jun S, Aziz KE, Feng L, Tran MK, Li N, McCrea PD, Park JI, Chen J. FOXKs promote Wnt/beta-catenin signaling by translocating DVL into the nucleus. Dev Cell. 2015;32:707–718. doi: 10.1016/j.devcel.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein TJ, Mlodzik M. Planar cell polarization: an emerging model points in the right direction. Annu Rev Cell Dev Biol. 2005;21:155–176. doi: 10.1146/annurev.cellbio.21.012704.132806. [DOI] [PubMed] [Google Scholar]
- 24.Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell. 2003;5:367–377. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 25.Wallingford JB, Fraser SE, Harland RM. Convergent extension: the molecular control of polarized cell movement during embryonic development. Dev Cell. 2002;2:695–706. doi: 10.1016/s1534-5807(02)00197-1. [DOI] [PubMed] [Google Scholar]
- 26.Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001;107:843–854. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- 27.Tanegashima K, Zhao H, Dawid IB. WGEF activates Rho in the Wnt-PCP pathway and controls convergent extension in Xenopus gastrulation. EMBO J. 2008;27:606–617. doi: 10.1038/emboj.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Habas R, Dawid IB, He X. Coactivation of Rac and Rho by Wnt/Frizzled signaling is required for vertebrate gastrulation. Genes Dev. 2003;17:295–309. doi: 10.1101/gad.1022203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goold RG, Owen R, Gordon-Weeks PR. Glycogen synthase kinase 3beta phosphorylation of microtubule-associated protein 1B regulates the stability of microtubules in growth cones. J Cell Sci. 1999;112(Pt 19):3373–3384. doi: 10.1242/jcs.112.19.3373. [DOI] [PubMed] [Google Scholar]
- 30.Lucas FR, Goold RG, Gordon-Weeks PR, Salinas PC. Inhibition of GSK-3beta leading to the loss of phosphorylated MAP-1B is an early event in axonal remodelling induced by WNT-7a or lithium. J Cell Sci. 1998;111(Pt 10):1351–1361. doi: 10.1242/jcs.111.10.1351. [DOI] [PubMed] [Google Scholar]
- 31.Sheldahl LC, Slusarski DC, Pandur P, Miller JR, Kuhl M, Moon RT. Dishevelled activates Ca2+ flux, PKC, and CamKII in vertebrate embryos. J Cell Biol. 2003;161:769–777. doi: 10.1083/jcb.200211094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slusarski DC, Pelegri F. Calcium signaling in vertebrate embryonic patterning and morphogenesis. Dev Biol. 2007;307:1–13. doi: 10.1016/j.ydbio.2007.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu W, Yamamoto V, Ortega B, Baltimore D. Mammalian Ryk is a Wnt coreceptor required for stimulation of neurite outgrowth. Cell. 2004;119:97–108. doi: 10.1016/j.cell.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Zhu J, Yang GY, Wang QJ, Qian L, Chen YM, Chen F, Tao Y, Hu HS, Wang T, Luo ZG. Dishevelled promotes axon differentiation by regulating atypical protein kinase C. Nat Cell Biol. 2007;9:743–754. doi: 10.1038/ncb1603. [DOI] [PubMed] [Google Scholar]
- 35.Schwarz-Romond T, Fiedler M, Shibata N, Butler PJ, Kikuchi A, Higuchi Y, Bienz M. The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol. 2007;14:484–492. doi: 10.1038/nsmb1247. [DOI] [PubMed] [Google Scholar]
- 36.Lee I, Choi S, Yun JH, Seo SH, Choi S, Choi KY, Lee W. Crystal structure of the PDZ domain of mouse Dishevelled 1 and its interaction with CXXC5. Biochem Biophys Res Commun. 2017;485:584–590. doi: 10.1016/j.bbrc.2016.12.023. [DOI] [PubMed] [Google Scholar]
- 37.Shiomi K, Uchida H, Keino-Masu K, Masu M. Ccd1, a novel protein with a DIX domain, is a positive regulator in the Wnt signaling during zebrafish neural patterning. Curr Biol. 2003;13:73–77. doi: 10.1016/s0960-9822(02)01398-2. [DOI] [PubMed] [Google Scholar]
- 38.Kishida S, Yamamoto H, Hino S, Ikeda S, Kishida M, Kikuchi A. DIX domains of Dvl and axin are necessary for protein interactions and their ability to regulate beta-catenin stability. Mol Cell Biol. 1999;19:4414–4422. doi: 10.1128/mcb.19.6.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwarz-Romond T, Metcalfe C, Bienz M. Dynamic recruitment of axin by Dishevelled protein assemblies. J Cell Sci. 2007;120:2402–2412. doi: 10.1242/jcs.002956. [DOI] [PubMed] [Google Scholar]
- 40.Capelluto DG, Kutateladze TG, Habas R, Finkielstein CV, He X, Overduin M. The DIX domain targets dishevelled to actin stress fibres and vesicular membranes. Nature. 2002;419:726–729. doi: 10.1038/nature01056. [DOI] [PubMed] [Google Scholar]
- 41.Capelluto DG, Overduin M, Secondary structure 1H. 13C and 15N resonance assignments and molecular interactions of the dishevelled DIX domain. J Biochem Mol Biol. 2005;38:243–247. doi: 10.5483/bmbrep.2005.38.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ehebauer MT, Arias AM. The structural and functional determinants of the Axin and Dishevelled DIX domains. BMC Struct Biol. 2009;9:70. doi: 10.1186/1472-6807-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong HC, Bourdelas A, Krauss A, Lee HJ, Shao Y, Wu D, Mlodzik M, Shi DL, Zheng J. Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol Cell. 2003;12:1251–1260. doi: 10.1016/s1097-2765(03)00427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moon RT, Shah K. Developmental biology: signalling polarity. Nature. 2002;417:239–240. doi: 10.1038/417239a. [DOI] [PubMed] [Google Scholar]
- 45.Boutros M, Mlodzik M. Dishevelled: at the crossroads of divergent intracellular signaling pathways. Mech Dev. 1999;83:27–37. doi: 10.1016/s0925-4773(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 46.Weston CR, Davis RJ. Signal transduction: signaling specificity- a complex affair. Science. 2001;292:2439–2440. doi: 10.1126/science.1063279. [DOI] [PubMed] [Google Scholar]
- 47.Lee HJ, Zheng JJ. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal. 2010;8:8. doi: 10.1186/1478-811X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grandy D, Shan J, Zhang X, Rao S, Akunuru S, Li H, Zhang Y, Alpatov I, Zhang XA, Lang RA, Shi DL, Zheng JJ. Discovery and characterization of a small molecule inhibitor of the PDZ domain of dishevelled. J Biol Chem. 2009;284:16256–16263. doi: 10.1074/jbc.M109.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Appleton BA, Wiesmann C, Lau T, Costa M, Hannoush RN, Sidhu SS. Inhibition of Wnt signaling by Dishevelled PDZ peptides. Nat Chem Biol. 2009;5:217–219. doi: 10.1038/nchembio.152. [DOI] [PubMed] [Google Scholar]
- 50.Mahindroo N, Punchihewa C, Bail AM, Fujii N. Indole-2-amide based biochemical antagonist of Dishevelled PDZ domain interaction down-regulates Dishevelled-driven Tcf transcriptional activity. Bioorg Med Chem Lett. 2008;18:946–949. doi: 10.1016/j.bmcl.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 51.Tran FH, Zheng JJ. Modulating the wnt signaling pathway with small molecules. Protein Sci. 2017;26:650–661. doi: 10.1002/pro.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujii N, You L, Xu Z, Uematsu K, Shan J, He B, Mikami I, Edmondson LR, Neale G, Zheng J, Guy RK, Jablons DM. An antagonist of dishevelled protein-protein interaction suppresses beta-catenin-dependent tumor cell growth. Cancer Res. 2007;67:573–579. doi: 10.1158/0008-5472.CAN-06-2726. [DOI] [PubMed] [Google Scholar]
- 53.Pan WJ, Pang SZ, Huang T, Guo HY, Wu D, Li L. Characterization of function of three domains in dishevelled-1: DEP domain is responsible for membrane translocation of dishevelled-1. Cell Res. 2004;14:324–330. doi: 10.1038/sj.cr.7290232. [DOI] [PubMed] [Google Scholar]
- 54.Gammons MV, Renko M, Johnson CM, Rutherford TJ, Bienz M. Wnt Signalosome Assembly by DEP Domain Swapping of Dishevelled. Mol Cell. 2016;64:92–104. doi: 10.1016/j.molcel.2016.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong HC, Mao J, Nguyen JT, Srinivas S, Zhang W, Liu B, Li L, Wu D, Zheng J. Structural basis of the recognition of the dishevelled DEP domain in the Wnt signaling pathway. Nat Struct Biol. 2000;7:1178–1184. doi: 10.1038/82047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park TJ, Gray RS, Sato A, Habas R, Wallingford JB. Subcellular localization and signaling properties of dishevelled in developing vertebrate embryos. Curr Biol. 2005;15:1039–1044. doi: 10.1016/j.cub.2005.04.062. [DOI] [PubMed] [Google Scholar]
- 57.Gloy J, Hikasa H, Sokol SY. Frodo interacts with Dishevelled to transduce Wnt signals. Nat Cell Biol. 2002;4:351–357. doi: 10.1038/ncb784. [DOI] [PubMed] [Google Scholar]
- 58.Bryja V, Gradl D, Schambony A, Arenas E, Schulte G. Beta-arrestin is a necessary component of Wnt/beta-catenin signaling in vitro and in vivo. Proc Natl Acad Sci U S A. 2007;104:6690–6695. doi: 10.1073/pnas.0611356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen W, ten Berge D, Brown J, Ahn S, Hu LA, Miller WE, Caron MG, Barak LS, Nusse R, Lefkowitz RJ. Dishevelled 2 recruits beta-arrestin 2 to mediate Wnt5A–stimulated endocytosis of Frizzled 4. Science. 2003;301:1391–1394. doi: 10.1126/science.1082808. [DOI] [PubMed] [Google Scholar]
- 60.Chen W, Hu LA, Semenov MV, Yanagawa S, Kikuchi A, Lefkowitz RJ, Miller WE. beta-Arrestin1 modulates lymphoid enhancer factor transcriptional activity through interaction with phosphorylated dishevelled proteins. Proc Natl Acad Sci U S A. 2001;98:14889–14894. doi: 10.1073/pnas.211572798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klimowski LK, Garcia BA, Shabanowitz J, Hunt DF, Virshup DM. Site-specific casein kinase 1epsilon-dependent phosphorylation of Dishevelled modulates beta-catenin signaling. FEBS J. 2006;273:4594–4602. doi: 10.1111/j.1742-4658.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- 62.Peters JM, McKay RM, McKay JP, Graff JM. Casein kinase I transduces Wnt signals. Nature. 1999;401:345–350. doi: 10.1038/43830. [DOI] [PubMed] [Google Scholar]
- 63.Sakanaka C, Leong P, Xu L, Harrison SD, Williams LT. Casein kinase iepsilon in the wnt pathway: regulation of beta-catenin function. Proc Natl Acad Sci U S A. 1999;96:12548–12552. doi: 10.1073/pnas.96.22.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Willert K, Brink M, Wodarz A, Varmus H, Nusse R. Casein kinase 2 associates with and phosphorylates dishevelled. EMBO J. 1997;16:3089–3096. doi: 10.1093/emboj/16.11.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strovel ET, Wu D, Sussman DJ. Protein phosphatase 2Calpha dephosphorylates axin and activates LEF-1-dependent transcription. J Biol Chem. 2000;275:2399–2403. doi: 10.1074/jbc.275.4.2399. [DOI] [PubMed] [Google Scholar]
- 66.Lee SH, Kim MY, Kim HY, Lee YM, Kim H, Nam KA, Roh MR, Min do S, Chung KY, Choi KY. The Dishevelled-binding protein CXXC5 negatively regulates cutaneous wound healing. J Exp Med. 2015;212:1061–1080. doi: 10.1084/jem.20141601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim HY, Yoon JY, Yun JH, Cho KW, Lee SH, Rhee YM, Jung HS, Lim HJ, Lee H, Choi J, Heo JN, Lee W, No KT, Min D, Choi KY. CXXC5 is a negative-feedback regulator of the Wnt/beta-catenin pathway involved in osteoblast differentiation. Cell Death Differ. 2015;22:912–920. doi: 10.1038/cdd.2014.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andersson T, Sodersten E, Duckworth JK, Cascante A, Fritz N, Sacchetti P, Cervenka I, Bryja V, Hermanson O. CXXC5 is a novel BMP4-regulated modulator of Wnt signaling in neural stem cells. J Biol Chem. 2009;284:3672–3681. doi: 10.1074/jbc.M808119200. [DOI] [PubMed] [Google Scholar]
- 69.Hino S, Kishida S, Michiue T, Fukui A, Sakamoto I, Takada S, Asashima M, Kikuchi A. Inhibition of the Wnt signaling pathway by Idax, a novel Dvl-binding protein. Mol Cell Biol. 2001;21:330–342. doi: 10.1128/MCB.21.1.330-342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rousset R, Mack JA, Wharton KA, Jr, Axelrod JD, Cadigan KM, Fish MP, Nusse R, Scott MP. Naked cuticle targets dishevelled to antagonize Wnt signal transduction. Genes Dev. 2001;15:658–671. doi: 10.1101/gad.869201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wharton KA, Jr, Zimmermann G, Rousset R, Scott MP. Vertebrate proteins related to Drosophila Naked Cuticle bind Dishevelled and antagonize Wnt signaling. Dev Biol. 2001;234:93–106. doi: 10.1006/dbio.2001.0238. [DOI] [PubMed] [Google Scholar]
- 72.Axelrod JD, Matsuno K, Artavanis-Tsakonas S, Perrimon N. Interaction between Wingless and Notch signaling pathways mediated by dishevelled. Science. 1996;271:1826–1832. doi: 10.1126/science.271.5257.1826. [DOI] [PubMed] [Google Scholar]
- 73.Strutt D, Johnson R, Cooper K, Bray S. Asymmetric localization of frizzled and the determination of notch-dependent cell fate in the Drosophila eye. Curr Biol. 2002;12:813–824. doi: 10.1016/s0960-9822(02)00841-2. [DOI] [PubMed] [Google Scholar]
- 74.Le Garrec JF, Kerszberg M. Modeling polarity buildup and cell fate decision in the fly eye: insight into the connection between the PCP and Notch pathways. Dev Genes Evol. 2008;218:413–426. doi: 10.1007/s00427-008-0235-y. [DOI] [PubMed] [Google Scholar]
- 75.Song DH, Sussman DJ, Seldin DC. Endogenous protein kinase CK2 participates in Wnt signaling in mammary epithelial cells. J Biol Chem. 2000;275:23790–23797. doi: 10.1074/jbc.M909107199. [DOI] [PubMed] [Google Scholar]
- 76.Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, MacCoss MJ, Moon RT. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol. 2006;8:348–357. doi: 10.1038/ncb1381. [DOI] [PubMed] [Google Scholar]
- 77.Dollar GL, Weber U, Mlodzik M, Sokol SY. Regulation of Lethal giant larvae by Dishevelled. Nature. 2005;437:1376–1380. doi: 10.1038/nature04116. [DOI] [PubMed] [Google Scholar]
- 78.Saxena M, Dykes SS, Malyarchuk S, Wang AE, Cardelli JA, Pruitt K. The sirtuins promote Dishevelled-1 scaffolding of TIAM1, Rac activation and cell migration. Oncogene. 2015;34:188–198. doi: 10.1038/onc.2013.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cajanek L, Ganji RS, Henriques-Oliveira C, Theofilopoulos S, Konik P, Bryja V, Arenas E. Tiam1 regulates the Wnt/Dvl/Rac1 signaling pathway and the differentiation of midbrain dopaminergic neurons. Mol Cell Biol. 2013;33:59–70. doi: 10.1128/MCB.00745-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holloway KR, Calhoun TN, Saxena M, Metoyer CF, Kandler EF, Rivera CA, Pruitt K. SIRT1 regulates Dishevelled proteins and promotes transient and constitutive Wnt signaling. Proc Natl Acad Sci U S A. 2010;107:9216–9221. doi: 10.1073/pnas.0911325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang JT, Wang Y, Chen JJ, Zhang XH, Dong JD, Tsang LL, Huang XR, Cai Z, Lan HY, Jiang XH, Chan HC. Defective CFTR leads to aberrant beta-catenin activation and kidney fibrosis. Sci Rep. 2017;7:5233. doi: 10.1038/s41598-017-05435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang L, Gao X, Wen J, Ning Y, Chen YG. Dapper 1 antagonizes Wnt signaling by promoting dishevelled degradation. J Biol Chem. 2006;281:8607–8612. doi: 10.1074/jbc.M600274200. [DOI] [PubMed] [Google Scholar]
- 83.Cheyette BN, Waxman JS, Miller JR, Takemaru K, Sheldahl LC, Khlebtsova N, Fox EP, Earnest T, Moon RT. Dapper, a Dishevelled-associated antagonist of beta-catenin and JNK signaling, is required for notochord formation. Dev Cell. 2002;2:449–461. doi: 10.1016/s1534-5807(02)00140-5. [DOI] [PubMed] [Google Scholar]
- 84.Jenny A, Reynolds-Kenneally J, Das G, Burnett M, Mlodzik M. Diego and Prickle regulate Frizzled planar cell polarity signalling by competing for Dishevelled binding. Nat Cell Biol. 2005;7:691–697. doi: 10.1038/ncb1271. [DOI] [PubMed] [Google Scholar]
- 85.Li L, Yuan H, Weaver CD, Mao J, Farr GH, 3rd, Sussman DJ, Jonkers J, Kimelman D, Wu D. Axin and Frat1 interact with dvl and GSK, bridging Dvl to GSK in Wnt-mediated regulation of LEF-1. EMBO J. 1999;18:4233–4240. doi: 10.1093/emboj/18.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Amerongen R, Nawijn MC, Lambooij JP, Proost N, Jonkers J, Berns A. Frat oncoproteins act at the crossroad of canonical and noncanonical Wnt-signaling pathways. Oncogene. 2010;29:93–104. doi: 10.1038/onc.2009.310. [DOI] [PubMed] [Google Scholar]
- 87.Sun TQ, Lu B, Feng JJ, Reinhard C, Jan YN, Fantl WJ, Williams LT. PAR-1 is a Dishevelled-associated kinase and a positive regulator of Wnt signalling. Nat Cell Biol. 2001;3:628–636. doi: 10.1038/35083016. [DOI] [PubMed] [Google Scholar]
- 88.Ossipova O, Dhawan S, Sokol S, Green JB. Distinct PAR-1 proteins function in different branches of Wnt signaling during vertebrate development. Dev Cell. 2005;8:829–841. doi: 10.1016/j.devcel.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 89.Elbert M, Cohen D, Musch A. PAR1b promotes cell-cell adhesion and inhibits dishevelled-mediated transformation of Madin-Darby canine kidney cells. Mol Biol Cell. 2006;17:3345–3355. doi: 10.1091/mbc.E06-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park M, Moon RT. The planar cell-polarity gene stbm regulates cell behaviour and cell fate in vertebrate embryos. Nat Cell Biol. 2002;4:20–25. doi: 10.1038/ncb716. [DOI] [PubMed] [Google Scholar]
- 91.Momose T, Kraus Y, Houliston E. A conserved function for Strabismus in establishing planar cell polarity in the ciliated ectoderm during cnidarian larval development. Development. 2012;139:4374–4382. doi: 10.1242/dev.084251. [DOI] [PubMed] [Google Scholar]
- 92.Lee HJ, Finkelstein D, Li X, Wu D, Shi DL, Zheng JJ. Identification of transmembrane protein 88 (TMEM88) as a dishevelled-binding protein. J Biol Chem. 2010;285:41549–41556. doi: 10.1074/jbc.M110.193383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Matsumoto S, Fumoto K, Okamoto T, Kaibuchi K, Kikuchi A. Binding of APC and dishevelled mediates Wnt5a–regulated focal adhesion dynamics in migrating cells. EMBO J. 2010;29:1192–1204. doi: 10.1038/emboj.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moeller H, Jenny A, Schaeffer HJ, Schwarz-Romond T, Mlodzik M, Hammerschmidt M, Birchmeier W. Diversin regulates heart formation and gastrulation movements in development. Proc Natl Acad Sci U S A. 2006;103:15900–15905. doi: 10.1073/pnas.0603808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee HS, Bong YS, Moore KB, Soria K, Moody SA, Daar IO. Dishevelled mediates ephrinB1 signalling in the eye field through the planar cell polarity pathway. Nat Cell Biol. 2006;8:55–63. doi: 10.1038/ncb1344. [DOI] [PubMed] [Google Scholar]
- 96.Lee HS, Mood K, Battu G, Ji YJ, Singh A, Daar IO. Fibroblast growth factor receptor-induced phosphorylation of ephrinB1 modulates its interaction with Dishevelled. Mol Biol Cell. 2009;20:124–133. doi: 10.1091/mbc.E08-06-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tanaka M, Kamo T, Ota S, Sugimura H. Association of Dishevelled with Eph tyrosine kinase receptor and ephrin mediates cell repulsion. EMBO J. 2003;22:847–858. doi: 10.1093/emboj/cdg088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Egger-Adam D, Katanaev VL. The trimeric G protein Go inflicts a double impact on axin in the Wnt/frizzled signaling pathway. Dev Dyn. 2010;239:168–183. doi: 10.1002/dvdy.22060. [DOI] [PubMed] [Google Scholar]
- 99.Jung H, Kim HJ, Lee SK, Kim R, Kopachik W, Han JK, Jho EH. Negative feedback regulation of Wnt signaling by Gbetagamma-mediated reduction of Dishevelled. Exp Mol Med. 2009;41:695–706. doi: 10.3858/emm.2009.41.10.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tree DR, Shulman JM, Rousset R, Scott MP, Gubb D, Axelrod JD. Prickle mediates feedback amplification to generate asymmetric planar cell polarity signaling. Cell. 2002;109:371–381. doi: 10.1016/s0092-8674(02)00715-8. [DOI] [PubMed] [Google Scholar]
- 101.Chan DW, Chan CY, Yam JW, Ching YP, Ng IO. Prickle-1 negatively regulates Wnt/beta-catenin pathway by promoting Dishevelled ubiquitination/degradation in liver cancer. Gastroenterology. 2006;131:1218–1227. doi: 10.1053/j.gastro.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 102.Kinoshita N, Iioka H, Miyakoshi A, Ueno N. PKC delta is essential for Dishevelled function in a noncanonical Wnt pathway that regulates Xenopus convergent extension movements. Genes Dev. 2003;17:1663–1676. doi: 10.1101/gad.1101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Velazquez DM, Castaneda-Patlan MC, Robles-Flores M. Dishevelled stability is positively regulated by PKCzeta-mediated phosphorylation induced by Wnt agonists. Cell Signal. 2017;35:107–117. doi: 10.1016/j.cellsig.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 104.Goto T, Sato A, Shimizu M, Adachi S, Satoh K, Iemura S, Natsume T, Shibuya H. IQGAP1 functions as a modulator of dishevelled nuclear localization in Wnt signaling. PLoS One. 2013;8:e60865. doi: 10.1371/journal.pone.0060865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Deng N, Ye Y, Wang W, Li L. Dishevelled interacts with p65 and acts as a repressor of NF-kappaB-mediated transcription. Cell Res. 2010;20:1117–1127. doi: 10.1038/cr.2010.108. [DOI] [PubMed] [Google Scholar]
- 106.Huang X, McGann JC, Liu BY, Hannoush RN, Lill JR, Pham V, Newton K, Kakunda M, Liu J, Yu C, Hymowitz SG, Hongo JA, Wynshaw-Boris A, Polakis P, Harland RM, Dixit VM. Phosphorylation of Dishevelled by protein kinase RIPK4 regulates Wnt signaling. Science. 2013;339:1441–1445. doi: 10.1126/science.1232253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou Y, Shang H, Zhang C, Liu Y, Zhao Y, Shuang F, Zhong H, Tang J, Hou S. The E3 ligase RNF185 negatively regulates osteogenic differentiation by targeting Dvl2 for degradation. Biochem Biophys Res Commun. 2014;447:431–436. doi: 10.1016/j.bbrc.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 108.Yanagawa S, van Leeuwen F, Wodarz A, Klingensmith J, Nusse R. The dishevelled protein is modified by wingless signaling in Drosophila. Genes Dev. 1995;9:1087–1097. doi: 10.1101/gad.9.9.1087. [DOI] [PubMed] [Google Scholar]
- 109.Bernatik O, Sedova K, Schille C, Ganji RS, Cervenka I, Trantirek L, Schambony A, Zdrahal Z, Bryja V. Functional analysis of dishevelled-3 phosphorylation identifies distinct mechanisms driven by casein kinase 1 and frizzled5. J Biol Chem. 2014;289:23520–23533. doi: 10.1074/jbc.M114.590638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sakanaka C, Weiss JB, Williams LT. Bridging of beta-catenin and glycogen synthase kinase-3beta by axin and inhibition of beta-catenin-mediated transcription. Proc Natl Acad Sci U S A. 1998;95:3020–3023. doi: 10.1073/pnas.95.6.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bernatik O, Ganji RS, Dijksterhuis JP, Konik P, Cervenka I, Polonio T, Krejci P, Schulte G, Bryja V. Sequential activation and inactivation of Dishevelled in the Wnt/beta-catenin pathway by casein kinases. J Biol Chem. 2011;286:10396–10410. doi: 10.1074/jbc.M110.169870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cervenka I, Valnohova J, Bernatik O, Harnos J, Radsetoulal M, Sedova K, Hanakova K, Potesil D, Sedlackova M, Salasova A, Steinhart Z, Angers S, Schulte G, Hampl A, Zdrahal Z, Bryja V. Dishevelled is a NEK2 kinase substrate controlling dynamics of centrosomal linker proteins. Proc Natl Acad Sci U S A. 2016;113:9304–9309. doi: 10.1073/pnas.1608783113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cong F, Schweizer L, Varmus H. Casein kinase Iepsilon modulates the signaling specificities of dishevelled. Mol Cell Biol. 2004;24:2000–2011. doi: 10.1128/MCB.24.5.2000-2011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Witte F, Bernatik O, Kirchner K, Masek J, Mahl A, Krejci P, Mundlos S, Schambony A, Bryja V, Stricker S. Negative regulation of Wnt signaling mediated by CK1-phosphorylated Dishevelled via Ror2. FASEB J. 2010;24:2417–2426. doi: 10.1096/fj.09-150615. [DOI] [PubMed] [Google Scholar]
- 115.Wei W, Li M, Wang J, Nie F, Li L. The E3 ubiquitin ligase ITCH negatively regulates canonical Wnt signaling by targeting dishevelled protein. Mol Cell Biol. 2012;32:3903–3912. doi: 10.1128/MCB.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ding Y, Zhang Y, Xu C, Tao QH, Chen YG. HECT domain-containing E3 ubiquitin ligase NEDD4L negatively regulates Wnt signaling by targeting dishevelled for proteasomal degradation. J Biol Chem. 2013;288:8289–8298. doi: 10.1074/jbc.M112.433185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tauriello DV, Haegebarth A, Kuper I, Edelmann MJ, Henraat M, Canninga-van Dijk MR, Kessler BM, Clevers H, Maurice MM. Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol Cell. 2010;37:607–619. doi: 10.1016/j.molcel.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 118.Wu C, Wei W, Li C, Li Q, Sheng Q, Zeng R. Delicate analysis of post-translational modifications on Dishevelled 3. J Proteome Res. 2012;11:3829–3837. doi: 10.1021/pr300314d. [DOI] [PubMed] [Google Scholar]
- 119.Deitrick J, Pruitt WM. Wnt/beta Catenin-Mediated Signaling Commonly Altered in Colorectal Cancer. Prog Mol Biol Transl Sci. 2016;144:49–68. doi: 10.1016/bs.pmbts.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 120.Krausova M, Korinek V. Wnt signaling in adult intestinal stem cells and cancer. Cell Signal. 2014;26:570–579. doi: 10.1016/j.cellsig.2013.11.032. [DOI] [PubMed] [Google Scholar]
- 121.Stubbins RE, Hakeem A, Nunez NP. Using components of the vitamin D pathway to prevent and treat colon cancer. Nutr Rev. 2012;70:721–729. doi: 10.1111/j.1753-4887.2012.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 123.Gavin BJ, McMahon AP. Differential regulation of the Wnt gene family during pregnancy and lactation suggests a role in postnatal development of the mammary gland. Mol Cell Biol. 1992;12:2418–2423. doi: 10.1128/mcb.12.5.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nusse R, van Ooyen A, Cox D, Fung YK, Varmus H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature. 1984;307:131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- 125.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 126.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 127.Brown AM, Wildin RS, Prendergast TJ, Varmus HE. A retrovirus vector expres sing the putative mammary oncogene int-1 causes partial transformation of a mammary epithelial cell line. Cell. 1986;46:1001–1009. doi: 10.1016/0092-8674(86)90699-9. [DOI] [PubMed] [Google Scholar]
- 128.Nusse R, Varmus HE. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 129.Ayyanan A, Civenni G, Ciarloni L, Morel C, Mueller N, Lefort K, Mandinova A, Raffoul W, Fiche M, Dotto GP, Brisken C. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc Natl Acad Sci U S A. 2006;103:3799–3804. doi: 10.1073/pnas.0600065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, Rowlands T, Egeblad M, Cowin P, Werb Z, Tan LK, Rosen JM, Varmus HE. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci U S A. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu BY, McDermott SP, Khwaja SS, Alexander CM. The transforming activity of Wnt effectors correlates with their ability to induce the accumulation of mammary progenitor cells. Proc Natl Acad Sci U S A. 2004;101:4158–4163. doi: 10.1073/pnas.0400699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Howe LR, Watanabe O, Leonard J, Brown AM. Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res. 2003;63:1906–1913. [PubMed] [Google Scholar]
- 133.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenes is, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 134.Bocchinfuso WP, Hively WP, Couse JF, Varmus HE, Korach KS. A mouse mammary tumor virus-Wnt-1 transgene induces mammary gland hyperplasia and tumorigenesis in mice lacking estrogen receptor-alpha. Cancer Res. 1999;59:1869–1876. [PubMed] [Google Scholar]
- 135.Huguet EL, McMahon JA, McMahon AP, Bicknell R, Harris AL. Differential expression of human Wnt genes 2, 3, 4, and 7B in human breast cell lines and normal and disease states of human breast tissue. Cancer Res. 1994;54:2615–2621. [PubMed] [Google Scholar]
- 136.Ford CE, Ekstrom EJ, Andersson T. Wnt-5a signaling restores tamoxifen sensitivity in estrogen receptor-negative breast cancer cells. Proc Natl Acad Sci U S A. 2009;106:3919–3924. doi: 10.1073/pnas.0809516106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 137.DiMeo TA, Anderson K, Phadke P, Fan C, Perou CM, Naber S, Kuperwasser C. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009;69:5364–5373. doi: 10.1158/0008-5472.CAN-08-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu CC, Prior J, Piwnica-Worms D, Bu G. LRP6 overexpression defines a class of breast cancer subtype and is a target for therapy. Proc Natl Acad Sci U S A. 2010;107:5136–5141. doi: 10.1073/pnas.0911220107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bjorklund P, Svedlund J, Olsson AK, Akerstrom G, Westin G. The internally truncated LRP5 receptor presents a therapeutic target in breast cancer. PLoS One. 2009;4:e4243. doi: 10.1371/journal.pone.0004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Badders NM, Goel S, Clark RJ, Klos KS, Kim S, Bafico A, Lindvall C, Williams BO, Alexander CM. The Wnt receptor, Lrp5, is expressed by mouse mammary stem cells and is required to maintain the basal lineage. PLoS One. 2009;4:e6594. doi: 10.1371/journal.pone.0006594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hsieh JC, Kodjabachian L, Rebbert ML, Rattner A, Smallwood PM, Samos CH, Nusse R, Dawid IB, Nathans J. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature. 1999;398:431–436. doi: 10.1038/18899. [DOI] [PubMed] [Google Scholar]
- 142.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 143.Hall CL, Kang S, MacDougald OA, Keller ET. Role of Wnts in prostate cancer bone metastases. J Cell Biochem. 2006;97:661–672. doi: 10.1002/jcb.20735. [DOI] [PubMed] [Google Scholar]
- 144.Veeck J, Niederacher D, An H, Klopocki E, Wiesmann F, Betz B, Galm O, Camara O, Durst M, Kristiansen G, Huszka C, Knuchel R, Dahl E. Aberrant methylation of the Wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene. 2006;25:3479–3488. doi: 10.1038/sj.onc.1209386. [DOI] [PubMed] [Google Scholar]
- 145.Veeck J, Geisler C, Noetzel E, Alkaya S, Hartmann A, Knuchel R, Dahl E. Epigenetic inactivation of the secreted frizzled-related protein-5 (SFRP5) gene in human breast cancer is associated with unfavorable prognosis. Carcinogenesis. 2008;29:991–998. doi: 10.1093/carcin/bgn076. [DOI] [PubMed] [Google Scholar]
- 146.Suzuki H, Toyota M, Carraway H, Gabrielson E, Ohmura T, Fujikane T, Nishikawa N, Sogabe Y, Nojima M, Sonoda T, Mori M, Hirata K, Imai K, Shinomura Y, Baylin SB, Tokino T. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 2008;98:1147–1156. doi: 10.1038/sj.bjc.6604259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lo PK, Mehrotra J, D’Costa A, Fackler MJ, Garrett-Mayer E, Argani P, Sukumar S. Epigenetic suppression of secreted frizzled related protein 1 (SFRP1) expression in human breast cancer. Cancer Biol Ther. 2006;5:281–286. doi: 10.4161/cbt.5.3.2384. [DOI] [PubMed] [Google Scholar]
- 148.Klarmann GJ, Decker A, Farrar WL. Epigenetic gene silencing in the Wnt pathway in breast cancer. Epigenetics. 2008;3:59–63. doi: 10.4161/epi.3.2.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hussain M, Rao M, Humphries AE, Hong JA, Liu F, Yang M, Caragacianu D, Schrump DS. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 2009;69:3570–3578. doi: 10.1158/0008-5472.CAN-08-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 152.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 153.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 154.Li J, Guo G, Li J, Hao J, Zhang J, Guo Y, Yu H. The expression and significance of dishevelled in human glioma. J Surg Res. 2014;192:509–514. doi: 10.1016/j.jss.2014.06.034. [DOI] [PubMed] [Google Scholar]
- 155.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 156.Wang C, Dai J, Sun Z, Shi C, Cao H, Chen X, Gu S, Li Z, Qian W, Han X. Targeted inhibition of disheveled PDZ domain via NSC668036 depresses fibrotic process. Exp Cell Res. 2015;331:115–122. doi: 10.1016/j.yexcr.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 157.Kfir-Erenfeld S, Haggiag N, Biton M, Stepensky P, Assayag-Asherie N, Yefenof E. miR-103 inhibits proliferation and sensitizes hemopoietic tumor cells for glucocorticoid-induced apoptosis. Oncotarget. 2017;8:472–489. doi: 10.18632/oncotarget.13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhao H, Xie C, Lin X, Zhao Y, Han Y, Fan C, Zhang X, Du J, Han Y, Han Q, Wu G, Wang E. Coexpression of IQ-domain GTPase-activating protein 1 (IQGAP1) and Dishevelled (Dvl) is correlated with poor prognosis in non-small cell lung cancer. PLoS One. 2014;9:e113713. doi: 10.1371/journal.pone.0113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dass RA, Sarshad AA, Carson BB, Feenstra JM, Kaur A, Obrdlik A, Parks MM, Prakash V, Love DK, Pietras K, Serra R, Blanchard SC, Percipalle P, Brown AM, Vincent CT. Wnt5a Signals through DVL1 to Repress Ribosomal DNA Transcription by RNA Polymerase I. PLoS Genet. 2016;12:e1006217. doi: 10.1371/journal.pgen.1006217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liu XZ, Fan J, Qi K, Liu SP, Xu WD, Gao Y, Gu XD, Li J, Bai CG, Shi YQ, Zhang LL, Zhao DB. Dishevelled2 promotes apoptosis and inhibits inflammatory cytokine secretion in rheumatoid arthritis fibroblast-like synoviocytes through crosstalk with the NF-kappaB pathway. Oncotarget. 2017;8:12649–12663. doi: 10.18632/oncotarget.15172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yuan H, Li N, Fu D, Ren J, Hui J, Peng J, Liu Y, Qiu T, Jiang M, Pan Q, Han Y, Wang X, Li Q, Qin J. Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis. J Clin Invest. 2017;127:3375–3391. doi: 10.1172/JCI94292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wei Q, Zhao Y, Yang ZQ, Dong QZ, Dong XJ, Han Y, Zhao C, Wang EH. Dishevelled family proteins are expressed in non-small cell lung cancer and function differentially on tumor progression. Lung Cancer. 2008;62:181–192. doi: 10.1016/j.lungcan.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 163.Kafka A, Tomas D, Beros V, Pecina HI, Zeljko M, Pecina-Slaus N. Brain metastases from lung cancer show increased expression of DVL1, DVL3 and beta-catenin and down-regulation of E-cadherin. Int J Mol Sci. 2014;15:10635–10651. doi: 10.3390/ijms150610635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Khan AS, Hojjat-Farsangi M, Daneshmanesh AH, Hansson L, Kokhaei P, Osterborg A, Mellstedt H, Moshfegh A. Dishevelled proteins are significantly upregulated in chronic lymphocytic leukaemia. Tumour Biol. 2016;37:11947–11957. doi: 10.1007/s13277-016-5039-5. [DOI] [PubMed] [Google Scholar]
- 165.Nagahata T, Shimada T, Harada A, Nagai H, Onda M, Yokoyama S, Shiba T, Jin E, Kawanami O, Emi M. Amplification, up-regulation and over-expression of DVL-1, the human counterpart of the Drosophila disheveled gene, in primary breast cancers. Cancer Sci. 2003;94:515–518. doi: 10.1111/j.1349-7006.2003.tb01475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Prasad CP, Gupta SD, Rath G, Ralhan R. Wnt signaling pathway in invasive ductal carcinoma of the breast: relationship between beta-catenin, dishevelled and cyclin D1 expression. Oncology. 2007;73:112–117. doi: 10.1159/000120999. [DOI] [PubMed] [Google Scholar]
- 167.Zhang C, Li C, Chen X, Zhou Y, Yin B, Ni R, Zhang Y, Liu J. Overexpression of dishevelled 2 is involved in tumor metastasis and is associated with poor prognosis in hepatocellular carcinoma. Clin Transl Oncol. 2017 doi: 10.1007/s12094-017-1697-z. [DOI] [PubMed] [Google Scholar]
- 168.Zhang C, Li C, Chen X, Zhou Y, Yin B, Ni R, Zhang Y, Liu J. Overexpression of dishevelled 2 is involved in tumor metastasis and is associated with poor prognosis in hepatocellular carcinoma. Clin Transl Oncol. 2017;19:1507–1517. doi: 10.1007/s12094-017-1697-z. [DOI] [PubMed] [Google Scholar]
- 169.Mahipal A, Malafa M. Importins and exportins as therapeutic targets in cancer. Pharmacol Ther. 2016;164:135–143. doi: 10.1016/j.pharmthera.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 170.Benjamin D, Moroni C. mRNA stability and cancer: an emerging link? Expert Opin Biol Ther. 2007;7:1515–1529. doi: 10.1517/14712598.7.10.1515. [DOI] [PubMed] [Google Scholar]
- 171.Griseri P, Pages G. Regulation of the mRNA half-life in breast cancer. World J Clin Oncol. 2014;5:323–334. doi: 10.5306/wjco.v5.i3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang C, Liu J, Huang G, Zhao Y, Yue X, Wu H, Li J, Zhu J, Shen Z, Haffty BG, Hu W, Feng Z. Cullin3-KLHL25 ubiquitin ligase targets ACLY for degradation to inhibit lipid synthesis and tumor progression. Genes Dev. 2016;30:1956–1970. doi: 10.1101/gad.283283.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Berthold J, Schenkova K, Ramos S, Miura Y, Furukawa M, Aspenstrom P, Rivero F. Characterization of RhoBTB-dependent Cul3 ubiquitin ligase complexes--evidence for an autoregulatory mechanism. Exp Cell Res. 2008;314:3453–3465. doi: 10.1016/j.yexcr.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kossatz U, Breuhahn K, Wolf B, Hardtke-Wolenski M, Wilkens L, Steinemann D, Singer S, Brass F, Kubicka S, Schlegelberger B, Schirmacher P, Manns MP, Singer JD, Malek NP. The cyclin E regulator cullin 3 prevents mouse hepatic progenitor cells from becoming tumor-initiating cells. J Clin Invest. 2010;120:3820–3833. doi: 10.1172/JCI41959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lee J, Zhou P. Cullins and cancer. Genes Cancer. 2010;1:690–699. doi: 10.1177/1947601910382899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lee YN, Gao Y, Wang HY. Differential mediation of the Wnt canonical pathway by mammalian Dishevelleds-1, -2, and −3. Cell Signal. 2008;20:443–452. doi: 10.1016/j.cellsig.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lijam N, Paylor R, McDonald MP, Crawley JN, Deng CX, Herrup K, Stevens KE, Maccaferri G, McBain CJ, Sussman DJ, Wynshaw-Boris A. Social interaction and sensorimotor gating abnormalities in mice lacking Dvl1. Cell. 1997;90:895–905. doi: 10.1016/s0092-8674(00)80354-2. [DOI] [PubMed] [Google Scholar]
- 178.van Gijn ME, Daemen MJ, Smits JF, Blankesteijn WM. The wnt-frizzled cascade in cardiovascular disease. Cardiovasc Res. 2002;55:16–24. doi: 10.1016/s0008-6363(02)00221-3. [DOI] [PubMed] [Google Scholar]
- 179.Zaveri HP, Beck TF, Hernandez-Garcia A, Shelly KE, Montgomery T, van Haeringen A, Anderlid BM, Patel C, Goel H, Houge G, Morrow BE, Cheung SW, Lalani SR, Scott DA. Identification of critical regions and candidate genes for cardiovascular malformations and cardiomyopathy associated with deletions of chromosome 1p36. PLoS One. 2014;9:e85600. doi: 10.1371/journal.pone.0085600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hamblet NS, Lijam N, Ruiz-Lozano P, Wang J, Yang Y, Luo Z, Mei L, Chien KR, Sussman DJ, Wynshaw-Boris A. Dishevelled 2 is essential for cardiac outflow tract development, somite segmentation and neural tube closure. Development. 2002;129:5827–5838. doi: 10.1242/dev.00164. [DOI] [PubMed] [Google Scholar]
- 181.Etheridge SL, Ray S, Li S, Hamblet NS, Lijam N, Tsang M, Greer J, Kardos N, Wang J, Sussman DJ, Chen P, Wynshaw-Boris A. Murine dishevelled 3 functions in redundant pathways with dishevelled 1 and 2 in normal cardiac outflow tract, cochlea, and neural tube development. PLoS Genet. 2008;4:e1000259. doi: 10.1371/journal.pgen.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pizzuti A, Novelli G, Mari A, Ratti A, Colosimo A, Amati F, Penso D, Sangiuolo F, Calabrese G, Palka G, Silani V, Gennarelli M, Mingarelli R, Scarlato G, Scambler P, Dallapiccola B. Human homologue sequences to the Drosophila dishevelled segment-polarity gene are deleted in the DiGeorge syndrome. Am J Hum Genet. 1996;58:722–729. [PMC free article] [PubMed] [Google Scholar]
- 183.Chen D, Mi J, Wu M, Wang W, Gao H. Expression of dishevelled gene in Hirschsprung’s disease. Int J Clin Exp Pathol. 2013;6:1791–1798. [PMC free article] [PubMed] [Google Scholar]
- 184.Person AD, Beiraghi S, Sieben CM, Hermanson S, Neumann AN, Robu ME, Schleiffarth JR, Billington CJ, Jr, van Bokhoven H, Hoogeboom JM, Mazzeu JF, Petryk A, Schimmenti LA, Brunner HG, Ekker SC, Lohr JL. WNT5A mutations in patients with autosomal dominant Robinow syndrome. Dev Dyn. 2010;239:327–337. doi: 10.1002/dvdy.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.White J, Mazzeu JF, Hoischen A, Jhangiani SN, Gambin T, Alcino MC, Penney S, Saraiva JM, Hove H, Skovby F, Kayserili H, Estrella E, Vulto-van Silfhout AT, Steehouwer M, Muzny DM, Sutton VR, Gibbs RA, Baylor-Hopkins G, Center for Mendelian. Lupski JR, Brunner HG, van Bon BW, Carvalho CM. DVL1 frameshift mutations clustering in the penultimate exon cause autosomal-dominant Robinow syndrome. Am J Hum Genet. 2015;96:612–622. doi: 10.1016/j.ajhg.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.White JJ, Mazzeu JF, Hoischen A, Bayram Y, Withers M, Gezdirici A, Kimonis V, Steehouwer M, Jhangiani SN, Muzny DM, Gibbs RA, Baylor-Hopkins G, Center for Mendelian. van Bon BWM, Sutton VR, Lupski JR, Brunner HG, Carvalho CMB. DVL3 Alleles Resulting in a −1 Frameshift of the Last Exon Mediate Autosomal-Dominant Robinow Syndrome. Am J Hum Genet. 2016;98:553–561. doi: 10.1016/j.ajhg.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bunn KJ, Daniel P, Rosken HS, O’Neill AC, Cameron-Christie SR, Morgan T, Brunner HG, Lai A, Kunst HP, Markie DM, Robertson SP. Mutations in DVL1 cause an osteosclerotic form of Robinow syndrome. Am J Hum Genet. 2015;96:623–630. doi: 10.1016/j.ajhg.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]