Abstract
Background
Several obesity-related factors have been associated with renal cell carcinoma (RCC), but it is unclear which individual factors directly influence risk. We addressed this question using genetic markers as proxies for putative risk factors and evaluated their relation to RCC risk in a mendelian randomization (MR) framework. This methodology limits bias due to confounding and is not affected by reverse causation.
Methods and findings
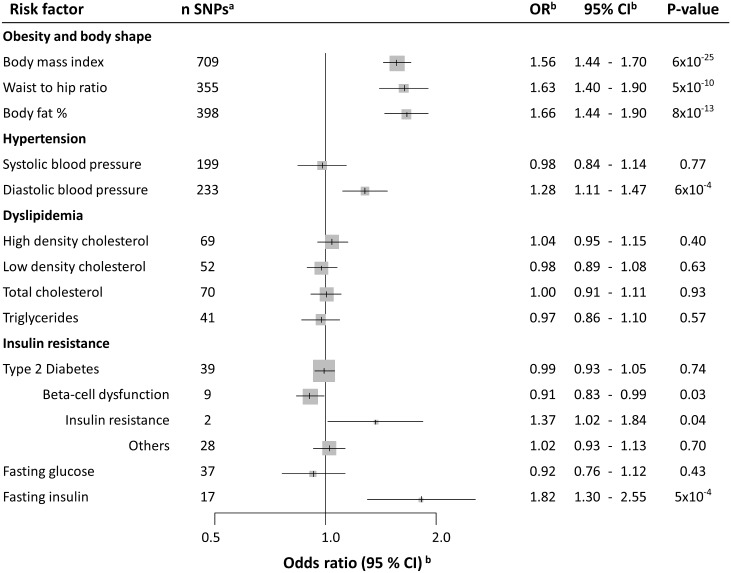
Genetic markers associated with obesity measures, blood pressure, lipids, type 2 diabetes, insulin, and glucose were initially identified as instrumental variables, and their association with RCC risk was subsequently evaluated in a genome-wide association study (GWAS) of 10,784 RCC patients and 20,406 control participants in a 2-sample MR framework. The effect on RCC risk was estimated by calculating odds ratios (ORSD) for a standard deviation (SD) increment in each risk factor. The MR analysis indicated that higher body mass index increases the risk of RCC (ORSD: 1.56, 95% confidence interval [CI] 1.44–1.70), with comparable results for waist-to-hip ratio (ORSD: 1.63, 95% CI 1.40–1.90) and body fat percentage (ORSD: 1.66, 95% CI 1.44–1.90). This analysis further indicated that higher fasting insulin (ORSD: 1.82, 95% CI 1.30–2.55) and diastolic blood pressure (DBP; ORSD: 1.28, 95% CI 1.11–1.47), but not systolic blood pressure (ORSD: 0.98, 95% CI 0.84–1.14), increase the risk for RCC. No association with RCC risk was seen for lipids, overall type 2 diabetes, or fasting glucose.
Conclusions
This study provides novel evidence for an etiological role of insulin in RCC, as well as confirmatory evidence that obesity and DBP influence RCC risk.
Using mendelian randomization approaches, Paul Brennan and colleagues reveal an association between 12 obesity-related factors, including insulin and the development of renal cell carcinoma.
Author summary
Why was this study done?
Traditional observational studies wherein putative risk factors are measured directly have found several obesity-related factors associated with increased risk of renal cell carcinoma (RCC).
Traditional observational studies are subject to confounding and reverse causation and have not been able to disentangle which obesity-related risk factors directly influence RCC risk.
This study used an alternative methodology commonly referred to as mendelian randomization (MR). MR circumvents many of the inherent limitations of traditional observational study by use of genetic proxies of putative risk factors when evaluating their associations with disease risk, as they are not subject to reverse causation and are less likely to be confounded by other risk factors.
What did the researchers do and find?
First, we used large-scale genome-wide association studies (GWAS) to identify genetic variants associated with obesity measures, blood pressure, lipids, type 2 diabetes, insulin, and glucose. Second, these genetic variants were used as proxies for the above-mentioned risk factors and evaluated in relation RCC risk using GWAS data from 10,000 RCC patients and 20,000 control participants.
Based on these genetic data, we found that multiple measures of obesity, as well as diastolic blood pressure (DBP) and fasting insulin, are associated with RCC risk. In contrast, we found little evidence for an association with RCC risk for systolic blood pressure (SBP), circulating lipids, overall diabetes, or fasting glucose.
What do these findings mean?
This study provided robust and confirmatory evidence of an important role of obesity as an important risk factor of RCC.
Further confirmatory evidence was found for elevated DBP as a risk factor of RCC, but it is not clear why DBP rather than SBP is important in RCC.
The study, to our knowledge, provided novel evidence of an important role of circulating insulin in RCC etiology.
This study provided some novel insights into the pathways involved in mediating the risk increase in RCC that is caused by obesity, most notably through insulin and DBP, but further research is needed to fully elucidate the important relationship between obesity and RCC.
Introduction
The etiology of renal cell carcinoma (RCC) is only partly understood [1]. An increased risk of RCC has been observed for individuals with high body mass index (BMI), elevated blood pressure, and triglycerides [2]. However, these obesity-related exposures are inherently interrelated, and traditional epidemiological studies have not been able to untangle which individual factors directly influence RCC risk and which are merely correlated with the underlying causal factor.
Mendelian randomization (MR) is an analytical approach whereby germline genetic markers are used as proxies—or instrumental variables—for putative risk factors. These genetic markers cannot be influenced by reverse causation (i.e., the disease affecting the exposure) [3], and assuming an absence of pleiotropy (i.e., genetic variants associated with the disease through alternative pathways) can provide unconfounded estimates of disease risk [4]. MR-based studies can therefore circumvent many of the inherent limitations of traditional observational studies, and an association between genetic proxies and the disease of interest would indicate that the risk factor being proxied influences risk in a causal manner [5].
We evaluated the role of obesity-related factors in RCC etiology using a two-sample MR framework wherein genetic variants associated with 13 relevant risk factors were identified from genome-wide association studies (GWASs). Subsequently, we evaluated the association of these genetic variants with RCC risk in a large RCC GWAS comprising 10,784 RCC patients and 20,406 control participants.
Materials and methods
Analytical strategy
The goal of our analytical strategy was to clarify the role of obesity and obesity-related risk factors in RCC etiology using a two-sample MR framework. This involved reviewing the GWAS literature to identify obesity-related risk factors for which valid proxy single nucleotide polymorphisms (SNPs) could be identified or by carrying out de novo GWAS analyses (e.g., for risk factors measured in UK Biobank) (first sample). This led to assembling SNP-based instrumental variables for obesity-related factors that were evaluated in relation to risk in the largest RCC GWAS published to date (second sample) [6]. This MR-based risk analysis involved using the likelihood-based approach to estimate the RCC odds ratio (OR) associated with a standard deviation (SD) increment in each risk factor of interest, with several complementary MR methods being applied to evaluate consistency in association estimates and between-study heterogeneity. No changes to the analytical strategy were done following the initial analysis. Further details on the specific methods used are indicated in the Statistical analysis section below.
Identification of genetic markers as instrumental variables for obesity-related factors (first sample)
Genetic markers for various obesity-related risk factors comprised SNPs that were associated with the risk factor of interest (P < 5 × 10−8) based on study participants with European ancestry. Correlated SNPs were excluded based on measures of linkage disequilibrium (LD) R2 < 0.1. Instruments for BMI and waist-to-hip ratio (WHR) were identified from meta-analyses of GWASs in approximately 700,000 individuals of European ancestry. GWAS data from the Genetic Investigation of ANthropometric Traits (GIANT) consortium (approximately 325,000 participants) [7,8] were combined with GWAS data from UK Biobank (approximately 375,000 individuals). SNPs associated with body fat percentage (%) were also identified using UK Biobank GWAS data. UK Biobank released genetic data for 488,363 individuals, from which we used data on participants of European descent with valid BMI, WHR, and body fat percentage measurements (374,237, 374,722, and 368,690 individuals, respectively). Genetic instruments for systolic blood pressure (SBP) and diastolic blood pressure (DBP), as well as pulse pressure (PP), were obtained from a GWAS in 375,091 European UK Biobank samples. These GWAS analyses were performed using Plink software [9], adjusted for age, sex, genotyping array, and principal components for population stratification as well as manual or automated blood pressure measurements. Instruments for circulating high-density lipoprotein cholesterol (HDL) and low-density lipoprotein cholesterol (LDL), total cholesterol, and triglycerides were identified from the Global Lipids Genetics Consortium (GLGC) study [10]. Instruments for circulating factors related to hyperglycemia and hyperinsulinemia, including fasting glucose and fasting insulin, were identified from the Meta-Analysis of Glucose and Insulin-Related Traits Consortium (MAGIC) [11,12]. Finally, instruments for type 2 diabetes were identified from a genetic fine-mapping study by Gaulton and colleagues [13]. Because SNPs associated with risk of type 2 diabetes may act by increasing insulin resistance or affecting pancreatic beta-cell function, we also evaluated subgroups of type 2 diabetes SNPs by stratifying for these two pathophysiologic mechanisms [14].
SNPs with ambiguous strand codification (A/T or C/G) were replaced by SNPs in genetic linkage (R2 > 0.8) using the proxysnps R package (European populations) (R Project) or were removed from the analyses if the minor allele frequency was higher than 0.4. For each SNP included in the different instrumental variables, the genetic effect estimate on exposure expressed in SDs of the trait per allele (βGE) was retrieved from each respective GWAS, along with the corresponding standard errors (SEGE). Table 1 provides details on the number of SNPs that constituted the instrumental variable for each potential risk factor, the proportion of variance explained by the instrument (or cumulative SNP liability in the case of type 2 diabetes), and the mean and SD of the respective risk factors in the original discovery study. Effect estimates for each individual SNP regarding their association with risk factor (βGE) are presented in S1 Table.
Table 1. Description of SNPs used as instrumental variables for obesity-related factors.
| Risk factor (source) | Mean (SD)a | Units | n SNPb | Variance (%)c |
|---|---|---|---|---|
| BMI [7,8] (UK Biobank) | 27.2 (4.7) | kg/m2 | 709 | 9.5 |
| WHR [7,8] (UK Biobank) | 1.0 (0.1) | cm/cm | 355 | 2.9 |
| Body fat percentage (UK Biobank) | 31.8 (6.6) | (%) | 398 | 3.5 |
| SBP (UK Biobank) | 140.3 (19.6) | mmHg | 199 | 3.1 |
| DBP (UK Biobank) | 82.2 (10.9) | mmHg | 233 | 3.7 |
| PP (UK Biobank) | 59.1 (14.5) | mmHg | 260 | 4.6 |
| HDL [10] | 53.3 (15.5) | mg/dL | 69 | 13.7 |
| LDL [10] | 133.6 (38.0) | mg/dL | 52 | 14.6 |
| Total cholesterol [10] | 213.28 (42.6) | mg/dL | 70 | 15.0 |
| Triglycerides [10] | 140.85 (87.8) | mg/dL | 41 | 11.7 |
| Fasting glucose [11,12] | 5.2 (0.8) | mmol/L | 37 | 4.8 |
| Fasting insulin [11,12] | 56.9 (44.4) | pmol/L | 17 | 1.2 |
| Type 2 diabetes [13] | - | - | 39 | 5.7 |
aMean and SD for each risk factor in the original discovery study for the SNPs used as instrumental variables.
bNumber of SNPs used in the instrument variable when evaluating each risk factor of interest with risk.
cVariance explained by the instrumental variable for each risk factor as indicated in the discovery GWAS (see marker publication).
Abbreviations: BMI, body mass index; DBP, diastolic blood pressure; GWAS, genome-wide association study; HDL, high-density lipoprotein cholesterol; LDL, low-density lipoprotein cholesterol; PP, pulse pressure; SBP, systolic blood pressure; SD, standard deviation; SNP, single nucleotide polymorphism; WHR, waist-to-hip ratio.
RCC association results (second sample)
To evaluate the association of each SNP used in the respective instrumental variables with RCC risk, we used summary genetic effect estimates on RCC risk (βGD) with the corresponding standard errors (SEGD) from the most recent GWAS on RCC [15]. This study encompassed data from 31,190 study participants, including 5,586 RCC patients and 13,075 control participants from previous GWASs [16–20], as well as an additional 5,198 RCC patients and 7,331 control participants with new GWA data, resulting in a total of 10,784 RCC patients and 20,406 control participants. These samples comprised prospective and retrospective case-control studies coordinated by four institutes, including the International Agency for Research on Cancer (IARC), the United States National Cancer Institute (NCI), MD Anderson Cancer Center (MDA), and United Kingdom cancer research centers (UK) (S2 Table). Sex-stratified analyses were performed for study participants from the IARC study sample (3,227 male RCC patients and 4,915 male control participants; 1,992 female RCC patients and 3,095 female control participants). SNPs with an imputation quality score (R2 or info score) lower than 0.7 were not used in the MR analysis. Effect estimates for association with disease risk for each SNP (βGD) can be observed in S1 Table.
Statistical analysis
A priori power calculations for the MR analysis to detect an association of nominal statistical significance (P < 0.05) were performed for instrumental variables explaining a range in variance of a causal risk factor, using the method proposed by Burgess [21]. Fig A in S1 Text depicts the statistical power of instrumental variables for different levels of explained variance for the risk factor of interest.
ORSD were calculated as effect estimates on RCC risk for an SD increment for each risk factor of interest using the corresponding instrumental variable. The primary MR analysis and the instrumental SNP heterogeneity analysis were conducted using the likelihood-based approach described by Burgess and colleagues [6]. Heterogeneity of initial effect estimates between the four data sources were investigated by estimating the percentage of variance that is attributable to study heterogeneity (I2 statistic). The P value for heterogeneity (PStudy-Heterogeneity) assumed a fixed-effect model with 3 degrees of freedom. For the sex-stratified risk analysis, the P value for heterogeneity (PSex-Heterogeneity) was obtained from a fixed-effect model with 1 degree of freedom.
As a sensitivity test, the presence of pleiotropy (i.e., genetic contribution to disease risk through a separate pathway) and potential outlier SNPs among genetic instruments were assessed using a novel approach labeled “MR pleiotropy residual sum and outlier” (MR-PRESSO) [22]. SNPs behaving as outliers were excluded from the instrument, and the effect estimate for the relevant risk factor of interest was reassessed. We also provided OR estimates using the complementary weighted-median method, wherein the effect estimate is weighted toward the median of the distribution of SNPs used in the instrumental variable [23]. This approach is less sensitive to individual SNPs strongly influencing the overall effect estimate. Furthermore, to evaluate the extent to which directional pleiotropy (nonbalanced horizontal pleiotropy in the MR risk estimates) may affect the ORSD estimates, we used an Egger regression approach (SIMEX version that does not assume the absence of measurement error on the βGE estimate) [24]. As a visual evaluation of pleiotropy, we also provided funnel plots depicting the weight exerted on the effect estimate along the y-axis (βGE/SEGD) and estimates of the effect on RCC along the x-axis (exp[βGD/βGE]) for each SNP used in the corresponding instrumental variable. Finally, we removed one SNP at a time from the instrument and reestimated the risk estimate to evaluate whether individual SNPs dominated the overall effect estimate. Statistical analyses were performed using R (R Project) and Plink [9].
Results
MR results of RCC for obesity-related risk factors
BMI, WHR, and body fat percentage
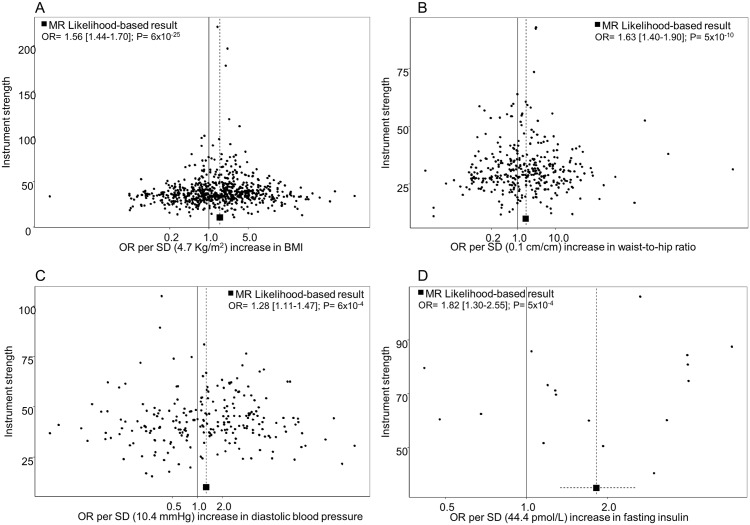
We estimated that each SD increment in BMI (1 SD: 4.7 kg/m2) increased the risk of RCC by 56% (ORSD: 1.56, 95% confidence interval [CI] 1.44–1.70) (Fig 1; Fig B in S1 Text for study stratification). A similar increase in risk was also seen for WHR (1 SD: 0.1 cm/cm) (ORSD: 1.63, 95% CI 1.40–1.90) (Fig 1; Fig C in S1 Text for study stratification) and for body fat percentage (1 SD: 6.6%) (ORSD: 1.66, 95% CI 1.44–1.90) (Fig 1; Fig D in S1 Text for study stratification). SNP heterogeneity (PSNP_heterogeneity = 9 × 10−8) and horizontal pleiotropy (PMR_PRESSO < 1 × 10−4) were observed for each obesity instrument (S3 Table), but there was little evidence of outlier SNPs or directional pleiotropy (PMR-Egger intercept > 0.12, S3 Table). The funnel plot for the BMI instruments (Fig 2A) indicated a symmetric distribution of effect estimates, and the leave-one-out histogram (Fig E in S1 Text) did not indicate that any individual SNPs were driving the overall association with risk, with similar results for WHR (Fig 2B and Fig F in S1 Text) and body fat percentage (Fig G in S1 Text). Accordingly, the complementary weighted-median MR method provided similar results (ORSD: 1.75, 95% CI 1.50–2.03, for BMI; ORSD: 1.45, 95% CI 1.14–1.86, for WHR; ORSD: 1.63, 95% CI 1.31–2.03, for body fat %; S3 Table). Sex heterogeneity was not observed in the IARC study sample (P > 0.41; Fig B–D in S1 Text).
Fig 1. Forest plot depicts OR estimates of RCC for the instrumental variables defined by genetic markers of obesity-related risk factors.
aNumber of SNPs used in each instrumental variable. bOR of RCC associated with one SD increment for each risk factor, as estimated using the instrumental variable. These ORs were estimated using the likelihood method. CI, confidence interval; OR, odds ratio; RCC, renal cell carcinoma; SD, standard deviation; SNP, single nucleotide polymorphism.
Fig 2. Funnel plots depict the weight exerted for each SNP used in the genetic instruments, along the y-axis (βGP/SEGD), into the estimated effect on RCC odds, along the x-axis (exp[βGD/βGP]), for (A) BMI, (B) waist-to-hip ratio, (C) diastolic blood pressure, and (D) fasting insulin.
Funnel plot x-axis is in logarithmic scale. BMI, body mass index; MR, mendelian randomization; OR, odds ratio; RCC, renal cell carcinoma; SD, standard deviation; SNP, single nucleotide polymorphism.
Blood pressure
The MR analysis indicated that DBP (1 SD of DBP: 10.9 mmHg) (ORSD: 1.28, 95% CI 1.11–1.47) (Fig 1; Fig H in S1 Text for study stratification), but not SBP (ORSD: 0.98, 95% CI 0.84–1.14) (Fig 1; Fig I in S1 Text for study stratification), influences RCC risk (P for difference in OR: 0.01). In accordance with the risk association of DBP, we also observed an inverse association for PP with RCC risk (ORSD: 0.77, 95% CI 0.68–0.88) (Fig J in S1 Text for study stratification). Sensitivity analyses did not detect any nonbalanced pleiotropic effect biasing the DBP risk estimates (S3 Table), and the funnel plot demonstrated a symmetric distribution of effect estimates from DBP SNPs (Fig 2C). The leave-one-out histogram for DBP can be seen in Fig K in S1 Text. Funnel plots and leave-one-out histograms for SBP and PP can be observed in Fig L and Fig M in S1 Text, respectively. In the IARC study samples, sex heterogeneity was not observed for DBP and SBP. However, the inverse association for PP with RCC risk was observed in men (ORSD: 0.74, 95% CI 0.58–0.94) but not in women (ORSD: 1.25, 95% CI 0.93–1.69) (PSex-heterogeneity = 7 × 10−3) (Fig J in S1 Text).
Blood lipids
For blood lipid levels—including HDL, LDL, total cholesterol, and triglycerides—the MR analysis indicated little evidence for an influence on RCC risk, with the overall ORSD estimates ranging from 0.97 to 1.04 (P≥ 0.40, Fig 1, Fig N-Q in S1 Text for stratified analyses, Fig R–U in S1 Text for sensitivity analyses).
Type 2 diabetes, insulin, and glucose
Overall type 2 diabetes was not associated with RCC risk (ORSD: 0.99, 95% CI 0.93–1.05), but when analyzing subgroups related to type 2 diabetes in a secondary analysis, SNPs related to beta-cell dysfunction were nominally inversely associated with risk (ORSD: 0.91, 95% CI 0.83–0.99), and SNPs related to insulin resistance were nominally positively associated with risk (ORSD: 1.37, 95% CI 1.02–1.84) (Fig 1). Notably, fasting insulin was positively associated with RCC risk, with each SD increment (44.4 pmol/L) increasing RCC risk by 82% (ORSD: 1.82, 95% CI 1.30–2.55) whereas little evidence was seen for a role of fasting glucose in RCC (P = 0.43) (Fig 1). No clear evidence of heterogeneity by study or sex was observed for any of the diabetes-related risk factors (Fig V–X in S1 Text), nor did the Egger regression or other sensitivity analyses indicate the presence of pleiotropy (S3 Table and Fig Y–AA in S1 Text).
Discussion
This MR analysis confirmed a role for higher BMI and DBP in RCC etiology and provided novel evidence for a role of fasting insulin. In contrast, we found little evidence for a role of other obesity-related risk factors RCC etiology, with evaluated risk factors including diagnosis of type 2 diabetes, SBP, and blood lipid levels.
There is an abundance of observational studies implicating obesity in RCC development, and several reviews have concluded that there is convincing evidence that being overweight or obese increases RCC risk [25,26]. Although a previous MR analysis on obesity and RCC did not provide unequivocal support for such a relation, the statistical power of that study was limited by a small sample size [27]. The current study leveraged results of several large-scale GWAS initiatives on both the metabolic risk factors of interest and RCC risk. We identified 709 SNPs as instrumental variables for BMI through meta-analyses of the existing GIANT GWAS and the novel UK Biobank GWAS, as well as results from our recent RCC GWAS initiative of over 10,000 RCC patients and 20,000 control participants. This enabled a well-powered two-sample MR analysis that provided strong support for a role of obesity and being overweight in RCC etiology (Fig 1 and Fig A in S1 Text). In particular, we estimated that an SD increment in BMI—approximately equivalent to 5 kg/m2—increases the risk of RCC by 56% (Fig 1). Considering that most observational studies have estimated risk increases of approximately 30% per 5 units of BMI [25,26], these results are notable, as they suggest that the impact of higher BMI and obesity in RCC etiology may be even more important than previously thought. This difference in effect estimates may reflect an inherent weakness in observational studies that use single direct measures of the risk factor of interest, whereas the genetic variants used to proxy the risk factor in an MR study would be expected to reflect the life-course exposure, thus better capturing the cumulative exposure.
Several additional obesity-related factors have been consistently implicated in RCC—in particular, factors traditionally associated with the metabolic syndrome, which is defined as a cluster of factors that increase the risk of type 2 diabetes and cardiovascular disease [28]. In addition to obesity, these risk factors typically include impaired glucose tolerance or diabetes, hypertension, and dyslipidemia [29,30]. The Metabolic Syndrome and Cancer (Me-Can) consortium pooled individual-level prospective data on over half a million study participants and estimated that men with high BMI, blood pressure, and blood triglycerides have up to a 3-fold risk increase of RCC compared to men without these conditions, and some have suggested that these risk factors exert independent effects on RCC risk [2,29]. When we used genetic markers for these risk factors, we observed a positive association between DBP and RCC, whereas no association with risk was seen for SBP nor for blood lipids, including HDL, LDL, total cholesterol, and triglycerides. Based on the fraction of variance explained by the instrumental variables of these factors (Table 1), we estimated that there was sufficient statistical power (>80%) to detect even modest log-linear relative risks of 1.2 per SD increment in a risk factor and close to complete statistical power (100%) to detect relative risks of 1.5 (Fig A in S1 Text). As such, this MR-based analysis did not support a role of SBP and blood lipids in RCC [2]. However, the positive association between the instrumental variable of DBP with risk is in line with previous epidemiological evidence. We also note that the difference in risk effect estimates between the DBP and SBP was significant, suggesting that it was not due to chance and lack of statistical power [2,31]. The mechanisms by which elevated blood pressure might influence RCC development are not established, but several plausible mechanisms have been suggested, including by influencing angiogenesis, growth factors, and renal function, thereby making the kidney more susceptible to carcinogens [31]. We are not aware of any hypothesis for why DBP rather than SBP would be important in RCC, but note that some epidemiological studies have indicated a stronger association with risk for DBP than for SBP [31].
We further observed a strong association between the instrumental variables of fasting insulin and RCC risk (Fig 1). In particular, we estimated that one SD increment in fasting insulin results in 82% increased risk of RCC. Whereas no association with RCC risk was seen for type 2 diabetes, we found an inverse association with risk for SNPs related to beta-cell dysfunction and a positive association with risk for SNPs related to insulin resistance, even though these subanalyses were conducted with limited statistical power. Insulin resistance may lead to compensatory hyperinsulinemia when pancreatic beta cells increase insulin secretion to maintain normal blood glucose. The contrasting associations with risk for the beta-cell dysfunction and insulin-resistance SNPs would therefore lend further support for a role of insulin in RCC etiology, as well as explain the lack of association with RCC risk for overall type 2 diabetes. Based on previously published data on the relationship between fasting insulin and BMI [32], we estimated that approximately one-fifth of the effect of BMI on RCC risk would be mediated by fasting insulin. Whereas the experimental data on the role of insulin in RCC tumorigenesis are still limited [33], there is ample in vivo and in vitro data describing pro-proliferative and antiapoptotic properties of insulin together with insulin-like growth factor 1 (IGF1) [33–35]. Prospective studies evaluating the association between directly measured fasting insulin (prediagnostically) and RCC risk are further warranted, and improving our understanding of insulin and IGF1 signaling in RCC development and progression may also offer therapeutic opportunities [36].
An overarching observation was that little evidence was found for sex heterogeneity in the relation between any of the risk-associated factors and RCC. This result is interesting, as it contrasts with some traditional observational studies that have reported stronger associations for obesity-related factors with RCC risk among women [37,38].
In conclusion, this study confirmed the important role of being overweight and having elevated DBP in affecting RCC risk and provided novel evidence for an etiological role of elevated insulin. The study gave little support for SBP or blood lipids and glucose being important in RCC. Taken together, these results advance our understanding of RCC etiology but highlight the need for further research focused on understanding how DBP and insulin-related pathways affect RCC risk, as well as complementary research aiming to identify additional pathways explaining the mechanisms by which obesity influences RCC development.
Supporting information
aBeta-cell dysfunction SNPs within type 2 diabetes. bInsulin resistance SNPs within type 2 diabetes. Β, beta estimate; BMI, body mass index; BP, base position; CHR, chromosome; DBP, diastolic blood pressure; EffAl, effect allele; GD, genotype-to-disease; GE, genotype-to-exposure; HDL, high-density lipoprotein cholesterol; LDL, low-density lipoprotein cholesterol; OthAl, other allele; PP, pulse pressure; SBP, systolic blood pressure; SE, standard error.
(PDF)
GWAS, genome-wide association study; RCC, renal cell carcinoma.
(PDF)
LCI, lower confidence interval; n SNPs, number of SNPs; OR, odds ratio; P, P value; PSNP-Heterogeneity, heterogeneity P value between instrumental SNP causal estimates (βGD/βGE) from genetic effects in S1 Table; UCI, upper confidence interval.
(PDF)
(PDF)
Acknowledgments
The authors thank all of the participants who took part in this research and the funders and support staff who made this study possible. We further thank Paul Pharoah for his valuable contribution to the study.
The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Abbreviations
- BMI
body mass index
- CI
confidence interval
- DBP
diastolic blood pressure
- GIANT
Genetic Investigation of ANthropometric Traits
- GLGC
Global Lipids Genetics Consortium
- GWAS
genome-wide association study
- HDL
high-density lipoprotein cholesterol
- IARC
International Agency for Research on Cancer
- IGF1
insulin-like growth factor 1
- LD
linkage disequilibrium
- LDL
low-density lipoprotein cholesterol
- MAGIC
Meta-Analysis of Glucose and Insulin-Related Traits Consortium
- MDA
MD Anderson Cancer Center
- Me-Can
Metabolic Syndrome and Cancer
- MR
mendelian randomization
- MR-PRESSO
MR pleiotropy residual sum and outlier
- NCI
National Cancer Institute
- OR
odds ratio
- PP
pulse pressure
- RCC
renal cell carcinoma
- SBP
systolic blood pressure
- SD
standard deviation
- SNP
single nucleotide polymorphism
- WHR
waist-to-hip ratio
Data Availability
All data needed to reproduce our results are included in S1 Table.
Funding Statement
This analysis was supported by the World Cancer Research Fund International—https://www.wcrf.org/ (2014/1193, to MJ) and Cancer Research UK—https://www.cancerresearchuk.org/ (C18281/A19169, to RMM and CR). Funding for the genome-wide genotyping was provided by the US National Institutes of Health (NIH), National Cancer Institute—https://www.cancer.gov (U01CA155309) for those studies coordinated by IARC, and by the intramural research program of the National Cancer Institute, US NIH, for those studies coordinated by the NCI; and the MD Anderson GWAS was supported in part by the NIH (grant R01 CA170298) and the Center for Translational and Public Health Genomics, Duncan Family Institute for Cancer Prevention and Risk Assessment, The University of Texas MD Anderson Cancer Center. RMM is supported by the National Institute for Health Research (NIHR) Bristol Nutritional Biomedical Research Unit based at University Hospitals Bristol NHS Foundation Trust and the University of Bristol. RCT, PCH, CR, RMM, GDS, MJ, and PB are investigators or researchers on a Cancer Research UK (C18281/A19169) Programme Grant (the Integrative Cancer Epidemiology Programme). CR, GDS, and NT work within the MRC Integrative Epidemiology Unit at the University of Bristol (MC_UU_12013/1, MC_UU_12013/2, MC_UU_12013/3). JBR is supported by the Canadian Institutes of Health Research and the Fonds du Recherche Québec-Santé. JL is supported by the RM/ICR NIHR Biomedical Research Centre for Cancer. The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65(2):87–108. 10.3322/caac.21262 . [DOI] [PubMed] [Google Scholar]
- 2.Haggstrom C, Rapp K, Stocks T, Manjer J, Bjorge T, Ulmer H, et al. Metabolic factors associated with risk of renal cell carcinoma. PLoS ONE. 2013;8(2):e57475 10.1371/journal.pone.0057475 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith GD, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. Epub 2003/04/12. . [DOI] [PubMed] [Google Scholar]
- 4.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–98. Epub 2014/07/30. 10.1093/hmg/ddu328 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgess S, Butterworth AS, Thompson JR. Beyond Mendelian randomization: how to interpret evidence of shared genetic predictors. J Clin Epidemiol. 2016;69:208–16. 10.1016/j.jclinepi.2015.08.001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–65. 10.1002/gepi.21758 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. 10.1038/nature14177 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–96. 10.1038/nature14132 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. 10.1086/519795 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Global Lipids Genetics C, Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274–83. 10.1038/ng.2797 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44(9):991–1005. 10.1038/ng.2385 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44(9):981–90. 10.1038/ng.2383 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaulton KJ, Ferreira T, Lee Y, Raimondo A, Magi R, Reschen ME, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47(12):1415–25. 10.1038/ng.3437 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimas AS, Lagou V, Barker A, Knowles JW, Magi R, Hivert MF, et al. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 2014;63(6):2158–71. 10.2337/db13-0949 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scelo G, Purdue MP, Brown KM, Johansson M, Wang Z, Eckel-Passow JE, et al. Genome-wide association study identifies multiple risk loci for renal cell carcinoma. Nature communications. 2017;8:15724 Epub 2017/06/10. 10.1038/ncomms15724 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han SS, Yeager M, Moore LE, Wei MH, Pfeiffer R, Toure O, et al. The chromosome 2p21 region harbors a complex genetic architecture for association with risk for renal cell carcinoma. Hum Mol Genet. 2012;21(5):1190–200. Epub 2011/11/25. 10.1093/hmg/ddr551 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henrion MY, Purdue MP, Scelo G, Broderick P, Frampton M, Ritchie A, et al. Common Variation at 1q24.1 (ALDH9A1) Is a Potential Risk Factor for Renal Cancer. PLoS ONE. 2015;10(3):e0122589 Epub 2015/04/01. 10.1371/journal.pone.0122589 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purdue MP, Johansson M, Zelenika D, Toro JR, Scelo G, Moore LE, et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat Genet. 2011;43(1):60–5. 10.1038/ng.723 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu XF, Scelo G, Purdue MP, Rothman N, Johansson M, Ye YQ, et al. A genome-wide association study identifies a novel susceptibility locus for renal cell carcinoma on 12p11.23. Hum Mol Genet. 2012;21(2):456–62. 10.1093/hmg/ddr479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henrion M, Frampton M, Scelo G, Purdue M, Ye Y, Broderick P, et al. Common variation at 2q22.3 (ZEB2) influences the risk of renal cancer. Hum Mol Genet. 2013;22(4):825–31. Epub 2012/11/28. 10.1093/hmg/dds489 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol. 2014;43(3):922–9. 10.1093/ije/dyu005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8. 10.1038/s41588-018-0099-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40(4):304–14. 10.1002/gepi.21965 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25. 10.1093/ije/dyv080 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauby-Secretan B, Scoccianti C, Loomis D, Grosse Y, Bianchini F, Straif K, et al. Body Fatness and Cancer—Viewpoint of the IARC Working Group. N Engl J Med. 2016;375(8):794–8. 10.1056/NEJMsr1606602 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Cancer Research Fund International/American Institute for Cancer Research. Continuous Update Project Report: Diet, Nutrition, Physical Activity and Kidney Cancer. 2015. https://www.wcrf.org/sites/default/files/Kidney-cancer-report.pdf
- 27.Brennan P, McKay J, Moore L, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, et al. Obesity and cancer: Mendelian randomization approach utilizing the FTO genotype. Int J Epidemiol. 2009;38(4):971–5. 10.1093/ije/dyp162 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aguilar-Salinas CA, Rojas R, Gomez-Perez FJ, Mehta R, Franco A, Olaiz G, et al. The metabolic syndrome: a concept hard to define. Arch Med Res. 2005;36(3):223–31. 10.1016/j.arcmed.2004.12.003 . [DOI] [PubMed] [Google Scholar]
- 29.Chow WH, Dong LM, Devesa SS. Epidemiology and risk factors for kidney cancer. Nat Rev Urol. 2010;7(5):245–57. 10.1038/nrurol.2010.46 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckel RH, Alberti KG, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet. 2010;375(9710):181–3. 10.1016/S0140-6736(09)61794-3 . [DOI] [PubMed] [Google Scholar]
- 31.Chow WH, Gridley G, Fraumeni JF Jr., Jarvholm B. Obesity, hypertension, and the risk of kidney cancer in men. N Engl J Med. 2000;343(18):1305–11. 10.1056/NEJM200011023431804 . [DOI] [PubMed] [Google Scholar]
- 32.Richmond RC, Wade KH, Corbin L, Bowden J, Hemani G, Timpson NJ, et al. Investigating the role of insulin in increased adiposity: Bi-directional Mendelian randomization stuy. bioRxiv. 2017. Epub June 28 2017. 10.1101/155739 [DOI] [Google Scholar]
- 33.Solarek W, Czarnecka AM, Escudier B, Bielecka ZF, Lian F, Szczylik C. Insulin and IGFs in renal cancer risk and progression. Endocr Relat Cancer. 2015;22(5):R253–64. 10.1530/ERC-15-0135 . [DOI] [PubMed] [Google Scholar]
- 34.Frasca F, Pandini G, Sciacca L, Pezzino V, Squatrito S, Belfiore A, et al. The role of insulin receptors and IGF-I receptors in cancer and other diseases. Arch Physiol Biochem. 2008;114(1):23–37. 10.1080/13813450801969715 . [DOI] [PubMed] [Google Scholar]
- 35.Bowers LW, Rossi EL, O'Flanagan CH, deGraffenried LA, Hursting SD. The Role of the Insulin/IGF System in Cancer: Lessons Learned from Clinical Trials and the Energy Balance-Cancer Link. Front Endocrinol (Lausanne). 2015;6:77 Epub 2015/06/02. 10.3389/fendo.2015.00077 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallagher EJ, Fierz Y, Ferguson RD, LeRoith D. The pathway from diabetes and obesity to cancer, on the route to targeted therapy. Endocr Pract. 2010;16(5):864–73. 10.4158/EP10098.RA . [DOI] [PubMed] [Google Scholar]
- 37.Wilson KM, Cho E. Obesity and Kidney Cancer. Recent Results Cancer Res. 2016;208:81–93. Epub 2016/12/03. 10.1007/978-3-319-42542-9_5 . [DOI] [PubMed] [Google Scholar]
- 38.Graff RE, Sanchez A, Tobias DK, Rodriguez D, Barrisford GW, Blute ML, et al. Type 2 Diabetes in Relation to the Risk of Renal Cell Carcinoma Among Men and Women in Two Large Prospective Cohort Studies. Diabetes Care. 2018;41(7):1432–7. Epub 2018/04/22. 10.2337/dc17-2518 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
aBeta-cell dysfunction SNPs within type 2 diabetes. bInsulin resistance SNPs within type 2 diabetes. Β, beta estimate; BMI, body mass index; BP, base position; CHR, chromosome; DBP, diastolic blood pressure; EffAl, effect allele; GD, genotype-to-disease; GE, genotype-to-exposure; HDL, high-density lipoprotein cholesterol; LDL, low-density lipoprotein cholesterol; OthAl, other allele; PP, pulse pressure; SBP, systolic blood pressure; SE, standard error.
(PDF)
GWAS, genome-wide association study; RCC, renal cell carcinoma.
(PDF)
LCI, lower confidence interval; n SNPs, number of SNPs; OR, odds ratio; P, P value; PSNP-Heterogeneity, heterogeneity P value between instrumental SNP causal estimates (βGD/βGE) from genetic effects in S1 Table; UCI, upper confidence interval.
(PDF)
(PDF)
Data Availability Statement
All data needed to reproduce our results are included in S1 Table.